PURPOSE
Discordant responses between brain metastases and extracranial tumors can arise from branched tumor evolution, underscoring the importance of profiling mutations to optimize therapy. However, the morbidity of brain biopsies limits their use. We investigated whether cell-free DNA (cfDNA) in CSF could serve as an effective surrogate marker for genomic profiling of intraparenchymal (IP) brain metastases.
METHODS
CSF and blood were collected simultaneously from patients with progressive brain metastases undergoing a craniotomy or lumbar puncture. Mutations in both biofluids were measured using an error-suppressed deep sequencing method previously published by our group. Forty-three regions of 24 cancer-associated genes were assayed.
RESULTS
This study enrolled 14 patients with either IP brain metastases (n = 12) or cytology-positive leptomeningeal disease (LMD, n = 2) and two controls with normal pressure hydrocephalus. Primary cancer types were lung, melanoma, renal cell, and colorectal. cfDNA was measurable in all sixteen samples of CSF. Cancer-associated mutations were found in the CSF of ten patients (eight with IP [67%] and two with LMD [100%]) and plasma of five patients (five with IP [42%] and none with LMD). All patients with plasma cfDNA had extracranial tumors. Among the five patients in the cohort who also had mutation data from time-matched brain metastasis tissue, four patients (80%) had matching mutations detected in CSF and brain, whereas only one patient (20%) had matching mutations detected in plasma and brain.
CONCLUSION
The detection of mutational DNA in CSF is not restricted to LMD and was found in two thirds of patients with IP brain metastases in our cohort. Analysis of CSF can be a viable alternative to biopsy for detection of somatic mutations in brain metastases.
INTRODUCTION
Brain metastases are diagnosed in approximately 9%-17% of patients with cancer and represent a significant and devastating cause of morbidity and mortality in patients with metastatic cancer.1 With the advent of targeted therapy and immunotherapy, overall survival in some patient populations can now extend beyond two years and the incidence of brain metastasis increases in patients with longer survival.2-5 Although some of these newer drugs have CNS penetrating ability, CNS response is still often less robust and/or less durable than systemic response.6,7 It remains unclear why CNS resistance occurs particularly in the case of targeted therapies. Currently proposed mechanisms are either (1) that drug levels are lower in the CNS allowing tolerance to develop or (2) that molecular drivers in the CNS lesions may be different than those in the body.8,9 Recent studies indicate that late-stage brain metastases from solid cancers may follow a branched evolution model, where the primary tumor and matched metastases evolve separately despite sharing a common clonal origin. Progression of brain metastases may thus be mediated by acquisition of unique driver mutations.10 Accordingly, several therapeutically actionable mutations have been detected in human brain metastases and not in matched extra-cranial tumors.11 These findings indicate that optimal treatment decisions for patients with CNS metastasis will require specific molecular profiling of brain metastasis tissue.11,12 However, unlike primary brain cancers, for which tumors are often resected as part of treatment, tissue is less frequently obtained from brain metastases because patients with disseminated disease are less likely to undergo craniotomies or even biopsies.
CONTEXT
Key Objective
Can cell-free DNA in CSF be used to detect mutations that are derived from intraparenchymal brain metastases?
Knowledge Generated
This study found that tumor-specific mutations were detectable in CSF of 67% (8 of 12) of patients with intraparenchymal brain metastases and that mutations were highly concordant between CSF and brain metastases tissue.
Relevance
Our findings suggest that CSF can be used as a complement to or surrogate for brain tissue biopsy to detect actionable mutations in patients with brain metastases.
Cell-free DNA (cfDNA) from various biofluids has been shown to contain tumor-derived DNA and has emerged as a less invasive alternative to tissue biopsy for profiling of somatic mutations to guide therapy. Previous studies have shown that tumor-specific mutations can frequently be detected in the CSF of patients with primary brain tumors such as glioblastoma.13-15 Importantly, when compared with plasma, the CSF of patients with glioblastoma more accurately reflected the mutations detected in the brain tumor, potentially because cfDNA may not easily traverse the blood-brain barrier.13,15 Other studies have shown that tumor-specific mutations can also be reliably detected in the CSF of patients with metastatic leptomeningeal disease (LMD).16,17 However, the evidence is less robust for the detection of tumor-derived DNA in the CSF of patients with intraparenchymal (IP) brain metastases, which are the most common intracranial tumors. Such studies have been challenging because CSF is not routinely sampled for clinical management of these patients. We undertook this current study to determine whether tumor-derived DNA can be detected in the CSF of patients with IP brain metastases and to compare this with the detectability of tumor-derived DNA in plasma.
METHODS
Patient Recruitment
Patients with known solid organ metastatic cancers either (1) with IP brain metastases undergoing a craniotomy or (2) with suspected LMD undergoing lumbar puncture at a single institution were approached for inclusion into our study between July 2018 and April 2019. Informed consent was obtained from all patients or legally authorized representatives under the existing institutional review board–approved clinical and tissue collection protocols. Two patients who had lumbar drain trials for the evaluation of normal pressure hydrocephalus (NPH) served as controls. These control subjects did not have any history of cancer, and magnetic resonance imaging of the brain obtained within 3 months of collection showed no evidence of CNS mass lesions.
Sample Collection
Time-matched CSF and plasma were collected from each patient. Tissues from brain metastases were also obtained from patients who underwent craniotomy. Time-matched was defined as samples from the same patient collected within 12 hours of each other to minimize temporal variability or treatment-related changes. For patients undergoing a craniotomy, CSF was collected via a ventricular catheter when indicated for decompression or by meticulous subarachnoid dissection and aspiration at a site distant to the tumor before surgical resection of the brain metastasis. Lumbar punctures were performed by licensed physicians as per standard of care. Every effort was made to minimize blood contamination of the CSF. CSF appearance at collection was described as clear or contaminated with blood, and CSF samples with visible blood contamination were excluded. Blood was collected through a standard peripheral venipuncture or from an arterial line. CSF was placed on ice, and blood was kept at room temperature during immediate transport to the research laboratory for processing. Up to 10 mL of blood and CSF were collected at each time point in EDTA-containing tubes. Plasma was isolated by centrifugation at 1,000 × g for 10 minutes within 4 hours of collection and was stored at −80°C. CSF samples were also centrifuged at 1,000 × g for 10 minutes to remove any cellular debris and stored at −80°C until further use.
Brain Metastasis Tissue Processing
Brain metastasis tissue was sent directly from the operating room to the Yale Department of Pathology. A clinical neuropathologist performed histological diagnosis of brain metastasis tissue, which is standard practice for all brain tumor surgeries. In non–small-cell lung cancer (NSCLC) and melanoma cases, the tissue was also sent to the CLIA-certified Yale Tumor Profiling Laboratory for genotyping using either a 50-gene Ion AmpliSeq Cancer Hotspot Panel v2 or a 161-gene Oncomine Comprehensive Assay v3 (Thermo Fisher Scientific).
cfDNA Isolation and Quantification of Mutations
cfDNA was extracted from 1 mL of plasma as described by Goldberg et al.18 cfDNA from CSF was extracted using a QIAamp Circulating Nucleic Acid kit (QIAGEN, Santa Clarita, CA) and processed according to the manufacturer's protocol for 5-mL input volume. The DNA extracted from plasma and CSF was eluted in 25 and 50 μL, respectively. Tumor-derived somatic mutations within cfDNA were identified and quantified using the error-suppressed deep sequencing method previously published by our group.18,19 The assay simultaneously queries thousands of possible point mutations and insertions or deletions within 43 mutation-prone regions of 24 cancer-associated genes (Appendix Table 1). Targeted deep sequencing was performed in 75 base-pair, paired-end mode on an Illumina HiSeq2500 instrument. Computational analysis of resulting raw sequence data, which includes suppression of background polymerase chain reaction-amplification errors and sequencer errors to enhance the sensitivity and accuracy of mutation calling, has been previously described.18-19
Clinical Information and Radiographic Assessment of Cranial and Systemic Disease
Demographics, imaging, and clinical information were retrospectively reviewed for each patient from the electronic medical record as approved by the study protocols.
IP tumor volumes were estimated using the formula20:
Volume in cm3 = (length × width × depth)/2.
Total tumor volume was the sum of all untreated parenchymal lesion volumes.
The presence and progression of systemic disease burden were determined by evaluating two most recent sequential computed tomography scans of the chest, abdomen, and pelvis obtained before sample collection. Patients suspected to have LMD on imaging were confirmed by CSF cytology.
Tumor volumes of patients with and without CSF mutational DNA detected were compared using the Mann-Whitney test.
RESULTS
Patient and Tumor Characteristics
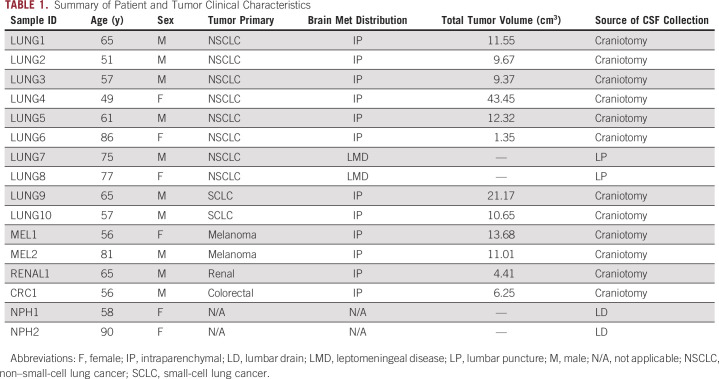
A total of 16 patients were enrolled in this study (Table 1)—12 patients with IP brain metastases, two patients with CSF cytology-positive LMD, and two noncancer control patients with NPH. Ten of 14 patients with cancer were male, and two NPH patients were female. The median age of the patients with cancer was 63 years (range 49-86 years), and the age of the two NPH patients was 58 and 90 years, respectively.
TABLE 1.
Summary of Patient and Tumor Clinical Characteristics
Of the 12 patients with IP disease, six patients had NSCLC, two patients had small-cell lung cancer, two patients had melanoma, one patient had renal cell carcinoma, and one patient had colorectal carcinoma. Both patients with LMD had NSCLC as a primary diagnosis. Total volumes of IP brain metastasis per patient ranged from 1.35 to 43.45 cm3 (median 10.83 cm3) (Table 1).
cfDNA Results
Time-matched CSF and plasma samples from all sixteen patients were subjected to cfDNA isolation and mutation quantification using the error-suppressed deep sequencing assay.18-19 cfDNA was measurable in all samples. The concentration of CSF cfDNA ranged from 0.05 to 352.91 ng/mL. Of the 12 patients with IP brain metastases, mutant DNA was found in CSF of eight patients (67%), whereas mutant DNA was found in plasma of only five patients (42%). Of the two patients with LMD, mutant DNA was found in the CSF, but not in the plasma of both patients. As expected, no mutations were identified in the CSF or blood from NPH patients (Fig. 1).
FIG 1.
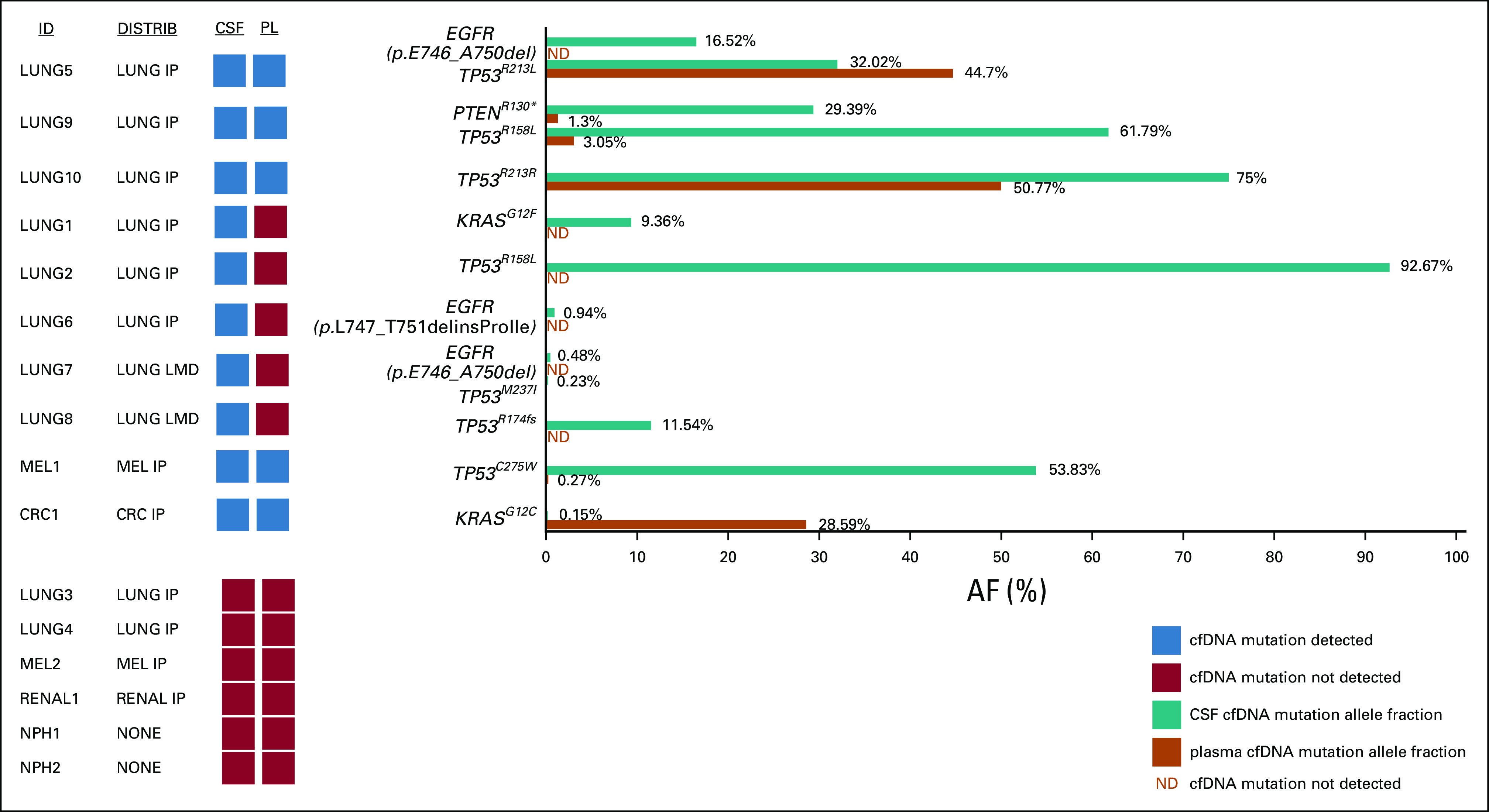
Quantification of cfDNA mutations in time-matched CSF and plasma of patients with brain metastasis. The amount of mutated DNA within the total cell-free DNA is shown as the mutant allele fraction (% AF). Blue and red squares denote the presence or absence, respectively, of detectable mutations in cfDNA in CSF or PL. AF, allele fraction; cfDNA, cell-free DNA; IP, intraparenchymal; LMD, leptomeningeal disease; ND, not detected; PL, plasma.
Among the ten patients with one or more mutations detected in CSF, four patients (40%) had the same mutation(s) identified in the plasma, whereas five patients (50%) had no mutations detectable in the plasma. Interestingly, one patient (10%; patient LUNG5) had a TP53 mutation that was identified in both plasma and CSF but had an additional EGFR exon 19 deletion mutation that was identified in CSF only. This patient did not have an EGFR mutation in previous tumor profiling of his lung mass.
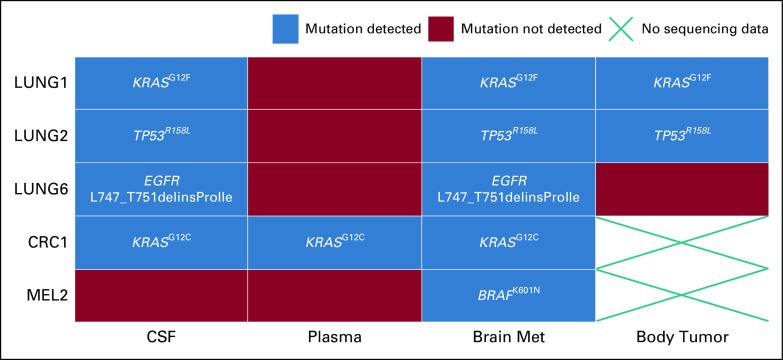
We also collected time-matched brain metastasis tissue from five patients who had undergone craniotomies. These tumor specimens were profiled using a clinical-grade assay (either 50- or 161-gene panel) in the CLIA-certified Yale Tumor Profiling Laboratory. Extracranial biopsy mutations were also included when available from archival tissue (analysis was done either at Yale or elsewhere if the patient was referred from an outside institution) (Fig. 2). We evaluated mutations covered by both our 24-gene and clinical sequencing panels. Among the five patients who had mutation data from brain metastasis tissue, matching mutations were found in the CSF of 4 of 5 patients and in the plasma of 1 of 5 patients. In patient LUNG6, an EGFR exon 19 deletion mutation was detected in the brain metastasis tissue and CSF but was missed in the profiling of her previous lung tumor biopsy (the specific deletion was not queried by the TaqMan-based PCR assay used at that time). She was started on an EGFR tyrosine kinase inhibitor shortly after tumor resection.
FIG 2.
Comparison of mutations among time-matched CSF, plasma, brain metastasis tissue, and previously sampled extracranial tumor tissue (when available). Shown are the five patients for whom brain metastasis tissue mutation results were available.
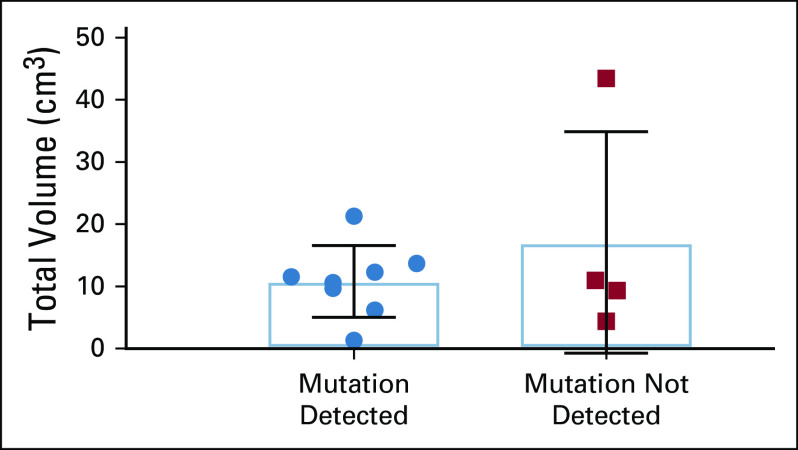
The allele fraction of mutations in CSF and plasma covered a broad range, spanning from 0.15% (CRC1) to 92.67% (LUNG2) in the CSF and from 0.27% (MEL1) to 50.77% (LUNG10) in the plasma. Although one might have expected a higher allele fraction of mutations in the CSF of patients with LMD (LUNG7 and LUNG8) compared with patients with IP brain metastases, this did not appear to be the case (Fig. 1). Furthermore, there was no significant association between total IP tumor volume and ability to detect mutant cfDNA in CSF (P = .93) (Fig. 3).
FIG 3.
Total tumor volume of IP brain metastasis in patients with versus without detectable mutations in cell-free DNA in CSF (P = .93). IP, intraparenchymal.
cfDNA Detectability in CSF and Plasma Relative to Intracranial and Extracranial Disease
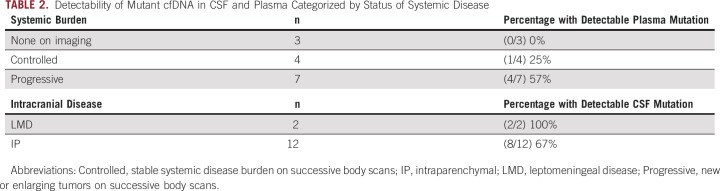
The presence of detectable mutant DNA in the plasma and CSF was compared with the presence of systemic disease and the distribution of CNS disease (Table 2). Of the 14 patients with cancer, seven had progressive systemic disease, four patients had stable systemic disease, and three patients had no evidence of extracranial disease. Among patients with progressive systemic disease, four of seven patients (57%) had detectable mutant DNA in plasma. By contrast, only one of four patients (25%) with stable systemic disease, and none of the patients without evidence of systemic disease, had detectable mutant DNA in plasma. It is noteworthy that the three patients with no evidence of systemic disease had large brain metastases with tumor volumes of 9.67 and 11 cm3 (LUNG2 and MEL2, respectively) or cytology-proven LMD (LUNG7).
TABLE 2.
Detectability of Mutant cfDNA in CSF and Plasma Categorized by Status of Systemic Disease
DISCUSSION
Despite significant improvements in efficacy of some anticancer agents in the CNS, the development of resistant brain metastases continues to be a major cause of morbidity and mortality for many types of cancers. As survival of patients with cancer increases in duration, the likelihood of developing brain metastases that progress discordant to the systemic disease in the body also increases.21 Thus, profiling of brain-specific somatic mutations can be important in guiding the optimal therapy for resistant CNS lesions. Because brain biopsies are inherently high risk, there has been substantial interest in using cfDNA fragments in CSF as an alternative source of tumor-derived genetic material. Although previous studies have demonstrated the feasibility of detecting somatic mutations in CSF of patients with primary brain gliomas or with LMD, data for IP brain metastasis have been limited.22 Here, we show that CSF can be used as a surrogate for brain biopsy to determine somatic mutation status of CNS lesions in patients with purely IP brain metastases.
In our cohort, CSF mutations were identified in two thirds of patients with purely IP brain metastases and in both patients with LMD who served as positive controls. As expected, no mutations were found in the CSF of both negative control NPH patients. The rate of mutation detection was higher in the CSF compared with time-matched plasma (67% v 42%). Importantly, two patients (LUNG5 and LUNG6) had new and targetable EGFR mutations detectable in the CSF, but not in the plasma or previous tissue biopsies. In patient LUNG6, the EGFR exon 19 deletion in CSF was concordant with the mutation found in brain metastasis tissue but was either missed or not present in the analysis of the original primary tumor biopsy because the specific mutation was not covered by the PCR-based assay that was used. This finding resulted in a change in treatment to targeted therapy. In both patients with LMD, driver mutations were identified in CSF that were not present in plasma. There did not appear to be a correlation between volume of IP tumors and ability to detect mutations in either CSF or plasma.
Our study had several limitations. First, blood contamination of CSF at the time of its collection could artificially increase the detection of mutational DNA. Although all CSF samples looked clear, cell counts were not obtained, and therefore, the degree of contamination was not known. Second, the small sample size of only 14 patients with brain metastases stemming from a variety of cancer types did not allow for the comparisons of mutation detectability and allelic fractions across different primary tumor subtypes or any meaningful statistical analysis of our results. In addition, our panel included mutation hotspot regions in 24 genes but did not include coverage of many commonly mutated tumor-suppressor genes in which the mutations could be more broadly distributed. Thus, we may have missed detecting mutations in CSF in some cases, not only because the tumor-derived DNA was not sufficiently abundant but also because some mutations may not have been covered by our panel. However, our assay was designed to balance sequencing cost and breadth of coverage, and it thus focuses on detection of more clinically relevant, actionable oncogene alterations that can guide therapeutic decisions.
Study of a larger number of patients with a more comprehensive panel of mutations is clearly needed to statistically validate our findings and confirm the relevance of this line of investigation. Finally, although we were able to collect time-matched CSF and plasma, brain metastasis tissue, comparisons of primary tumor mutation data were not available for all samples. With different assays (with only partially overlapping gene panels) being used to analyze brain metastasis tissue and primary tissue, this inherently limits the comparisons that could be made. A more comprehensive analysis using broader mutation coverage of matched CSF, tissue, and plasma may reveal additional differences in their mutational profiles.
Although not all patients with IP brain metastases may have tumor-derived mutations detectable in their CSF, given the significantly lower risks and costs of performing lumbar punctures compared with brain biopsies, CSF could be considered a first-line alternative for identifying driver mutations in IP brain metastases. This approach would be most pertinent in patients with discordant CNS and systemic responses to therapy. For instance, NSCLC patients with mutations in EGFR and ALK are more prone to relapses in the CNS than patients with other driver mutations.23 Detection of resistance mutations in the CSF would be particularly beneficial as new therapies are increasingly being designed to penetrate the brain and historical studies have shown that drug-resistant mutations in the CNS may differ from those detected in extracranial tissues.24,25
In conclusion, the findings of this study indicate that mutant DNA from IP brain metastases is more likely to be detectable in cfDNA in CSF than in plasma. If validated, these findings could change the paradigm for the management of brain metastases, not only for those resistant to first-line therapies, but perhaps also for personalized molecular diagnosis at the time of first-line therapy.
APPENDIX
TABLE A1.
Twenty-Four Gene Panel and Mutation-Prone Regions Profiled for CSF and Plasma Sequencing
EQUAL CONTRIBUTIONS
S.K.C. and A.N. contributed equally to this work and are co-first authors. A.P. and V.C. are co-lead authors.
PRIOR PRESENTATION
Presented at the Congress of Neurological Surgery annual meeting from October 19 to 23, 2019; presented at the Society of NeuroOncology annual meeting from August 13 to 15, 2020.
SUPPORT
Supported by the following grants: NCI grant P50-CA196530 to V.C., D.N., and A.A.P., R01-CA197486 to A.A.P, and P50-CA121974, R01-CA227432, and R01-CA216846 to H.K. The funding sources of this article did not have any role in the decision making, study design, data collection, data interpretation, and writing of this text. Dr. Stephanie Cheok and Dr. Veronica Chiang had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
AUTHOR CONTRIBUTIONS
Conception and design: Stephanie Kim Cheok, Azeet Narayan, Scott Gettinger, Sarah B. Goldberg, Harriet M. Kluger, Don Nguyen, Abhijit Patel, Veronica Chiang
Financial support: Don Nguyen, Abhijit Patel
Administrative support: Don Nguyen
Provision of study materials or patients: Sarah B. Goldberg
Collection and assembly of data: Stephanie Kim Cheok, Azeet Narayan, Anna Arnal-Estape, Scott Gettinger, Sarah B. Goldberg, Harriet M. Kluger, Abhijit Patel, Veronica Chiang
Data analysis and interpretation: Stephanie Kim Cheok, Azeet Narayan, Scott Gettinger, Sarah B. Goldberg, Harriet M. Kluger, Don Nguyen, Abhijit Patel, Veronica Chiang
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Azeet Narayan
Employment: Vanessa Research, Inc
Stock and Other Ownership Interests: Guardant health, crispr therapeutics
Consulting or Advisory Role: www.arch-road.com
Scott Gettinger
Consulting or Advisory Role: Bristol-Myers Squibb, NEKTAR
Research Funding: Bristol-Myers Squibb, Genentech, ARIAD/Takeda, Iovance Biotherapeutics
Sarah B. Goldberg
Consulting or Advisory Role: AstraZeneca/MedImmune, Bristol-Myers Squibb, Boehringer Ingelheim, Genentech/Roche, Blueprint Medicines, Genzyme, Daiichi Sankyo, Regeneron
Research Funding: AstraZeneca, Merck, Genentech, Pfizer, Bristol-Myers Squibb, Spectrum Pharmaceuticals, Boehringer Ingelheim
Harriet M. Kluger
Consulting or Advisory Role: Nektar, Celldex, Iovance Biotherapeutics, Merck, Immunocore, Array BioPharma, Elevate Bio, Instil Bio, Clinigen Group, Shionogi, Bristol-Myers Squibb, Bristol-Myers Squibb
Research Funding: Merck, Bristol-Myers Squibb, Apexigen
Travel, Accommodations, Expenses: Apexigen
Don Nguyen
Research Funding: AstraZeneca
Abhijit Patel
Stock and Other Ownership Interests: Binary Genomics
Honoraria: NuGEN
Consulting or Advisory Role: NuGEN, NuProbe, Kohlberg Kravis Roberts & Co Inc
Research Funding: AstraZeneca
Patents, Royalties, Other Intellectual Property: Inventor and assignee of patents and patent applications covering ultra sensitive nucleic acid analysis technologies.
Travel, Accommodations, Expenses: NuGEN, Statum Fund
Veronica Chiang
Consulting or Advisory Role: Monteris Medical, MRI Interventions
Speakers' Bureau: Monteris Medical
No other potential conflicts of interest were reported.
REFERENCES
- 1.Nayak L, Lee EQ, Wen PY: Epidemiology of brain metastases. Curr Oncol Rep 14:48-54, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Choong ES, Lo S, Drummond M, et al. : Survival of patients with melanoma brain metastasis treated with stereotactic radiosurgery and active systemic drug therapies. Eur J Cancer 75:169-178, 2017 [DOI] [PubMed] [Google Scholar]
- 3.Johung KL, Yeh N, Desai NB, et al. : Extended survival and prognostic factors for patients with ALK-rearranged non-small-cell lung cancer and brain metastasis. J Clin Oncol 34:123-129, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian SK, Sharma M, Venur VA, et al. : Impact of EGFR mutation and ALK rearrangement on the outcomes of non-small cell lung cancer patients with brain metastasis. Neuro Oncol 22:267-277, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox BD, Cheung VJ, Patel AJ, et al. : Epidemiology of metastatic brain tumors. Neurosurg Clin North Am 22:1-6, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Peuvrel L, Saint-Jean M, Quereux G, et al. : Incidence and characteristics of melanoma brain metastases developing during treatment with vemurafenib. J Neurooncol 120:147-154, 2014 [DOI] [PubMed] [Google Scholar]
- 7.Di Lorenzo R, Ahluwalia MS: Targeted therapy of brain metastases: Latest evidence and clinical implications. Ther Adv Med Oncol 9:781-796, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benedettini E, Sholl LM, Peyton M, et al. : Met activation in non-small cell lung cancer is associated with de novo resistance to EGFR inhibitors and the development of brain metastasis. Am J Pathol 177:415-423, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen PB, Kumler I, Nielsen DL: A systematic review of trastuzumab and lapatinib in the treatment of women with brain metastases from HER2-positive breast cancer. Cancer Treat Rev 39:720-727, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Shih DJH, Nayyar N, Bihun I, et al. : Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet 52:371-377, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brastianos PK, Carter SL, Santagata S, et al. : Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov 5:1164-1177, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmer M, Werner JM, Rohn G, et al. : Discordance and conversion rates of progesterone-, estrogen-, and HER2/neu-receptor status in primary breast cancer and brain metastasis mainly triggered by hormone therapy. Anticancer Res 37:4859-4865, 2017 [DOI] [PubMed] [Google Scholar]
- 13.Miller AM, Shah RH, Pentsova EI, et al. : Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565:654-658, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piccioni DE, Achrol AS, Kiedrowski LA, et al. : Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol 8:CNS34, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Mattos-Arruda L, Mayor R, Ng CKY, et al. : Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 6:8839, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ballester LY, Glitza Oliva IC, Douse DY, et al. : Evaluating circulating tumor DNA from the cerebrospinal fluid of patients with melanoma and leptomeningeal disease. J Neuropathol Exp Neurol 77:628-635, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, He JY, Zou YL, et al. : Evaluating the cerebrospinal fluid ctDNA detection by next-generation sequencing in the diagnosis of meningeal Carcinomatosis. BMC Neurol 19:331, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg SB, Narayan A, Kole AJ, et al. : Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res 24:1872-1880, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narayan A, Carriero NJ, Gettinger SN, et al. : Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res 72:3492-3498, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel TR, McHugh BJ, Bi WL, et al. : A comprehensive review of MR imaging changes following radiosurgery to 500 brain metastases. AJNR Am J Neuroradiol 32:1885-1892, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnholtz-Sloan JS, Sloan AE, Davis FG, et al. : Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the metropolitan detroit cancer surveillance system. J Clin Oncol 22:2865-2872, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Ma C, Yang X, Xing W, et al. : Detection of circulating tumor DNA from non-small cell lung cancer brain metastasis in cerebrospinal fluid samples. Thorac Cancer 11:588-593, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singhi EK, Horn L, Sequist LV, et al. : Advanced non-small cell lung cancer: Sequencing agents in the EGFR-mutated/ALK-rearranged populations. Am Soc Clin Oncol Educ Book 39:e187-e197, 2019 [DOI] [PubMed] [Google Scholar]
- 24.Hata A, Katakami N, Yoshioka H, et al. : Rebiopsy of non-small cell lung cancer patients with acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitor: Comparison between T790M mutation-positive and mutation-negative populations. Cancer 119:4325-4332, 2013 [DOI] [PubMed] [Google Scholar]
- 25.Page S, Milner-Watts C, Perna M, et al. : Systemic treatment of brain metastases in non-small cell lung cancer. Eur J Cancer 132:187-198, 2020 [DOI] [PubMed] [Google Scholar]