PURPOSE
It has recently been described that alternative oncogenic drivers may be found in KRAS wild-type (KRASWT) pancreatic cancers. This study aimed to determine the incidence of targetable gene fusions present in KRASWT pancreatic adenocarcinoma and response to targeted therapy.
METHODS
One hundred consecutive patients with pancreatic adenocarcinoma who underwent targeted next-generation sequencing using DNA sequencing with RNA sequencing (n = 47) or without RNA sequencing (n = 53) at a single institution were included in the study. The frequency and landscape of targetable fusions in KRASWT pancreatic adenocarcinoma was characterized and compared with the frequency of fusions in KRAS-mutated (KRASMUT) pancreatic adenocarcinoma. Results were validated in two independent cohorts using data from AACR GENIE (n = 1,252) and TCGA (n = 150). The clinical history of fusion-positive patients who received targeted treatment is described.
RESULTS
Pancreatic cancers from 13 of 100 patients (13%) were found to be KRASWT. Targetable fusions were identified in 4/13 (31%) KRASWT tumors compared with 0/87 (0%) KRAS MUT pancreatic adenocarcinomas (P = .0002). One patient with a novel MET fusion had a complete response to targeted therapy with crizotinib that is ongoing at 12+ months of treatment. In the validation cohorts, gene fusions were identified in 18/97 (19%) and 2/10 (20%) KRASWT tumors reported in the AACR GENIE and TCGA cohorts, respectively.
CONCLUSION
Oncogene fusions are present in KRASWT pancreatic adenocarcinomas at an increased frequency when compared with KRASMUT pancreatic adenocarcinomas. As these fusions may be susceptible to targeted therapy, molecular analyses for the detection of fusions in KRASWT pancreatic adenocarcinomas may warrant increased consideration.
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-related death in the United States with 57,600 new cases and 47,050 deaths projected in 2020.1 Clinical studies have established the antitumor activity of polychemotherapy approaches including FOLFIRINOX (folinic acid, fluorouracil, irinotecan, and oxaliplatin) and gemcitabine combined with nab-paclitaxel for advanced or metastatic pancreatic cancer.2,3 Despite the activity of polychemotherapy, the majority of patients with advanced or metastatic pancreatic cancer develop disease progression within 6 months, and thus more effective therapies are needed.2,3
CONTEXT
Key Objective
To describe the landscape targetable gene fusions in KRAS wild-type (KRASWT) pancreatic cancer including a cohort of patients treated with matched targeted therapy.
Knowledge Generated
Targetable gene fusions are detected in KRASWT pancreatic cancers at an increased rate (approximately one out of five) compared with KRAS-mutated pancreatic adenocarcinomas. Some patients treated with targeted therapy directed at these fusions can achieve meaningful clinical benefit.
Relevance
The increased frequency of oncogene fusions in KRASWT pancreatic adenocarcinoma suggests that KRAS status may be used as a method to identify patients who are more likely to harbor targetable fusions. The identification of MET fusions in the setting of KRASWT pancreatic cancer suggests a novel target in KRASWT cases that should be interrogated with assays capable of detecting MET fusions with both known and novel partners.
In recent years, enthusiasm for targeted therapy in pancreatic cancer has grown with the approval of the PARP-inhibitor olaparib for the treatment of patients with BRCA-mutated disease and the identification of alternative oncogenic drivers in KRAS wild-type (KRASWT) tumors.4-6 Further advances have included the identification of NRG1 fusions in KRASWT pancreatic adenocarcinoma and case reports describing exceptional responders to targeted therapy in pancreatic cancers harboring a variety of oncogene fusions, all identified in KRASWT tumors.7-10
Although KRASWT tumors represent a minority of pancreatic cancer cases, they may possess potentially targetable alterations, making their identification a therapeutic opportunity.5,6 This study sought to characterize the landscape of targetable oncogene fusions detected in KRASWT pancreatic adenocarcinoma through a retrospective analysis of 100 pancreatic adenocarcinoma cases sequenced at a single institution. Results were validated through two other pancreatic adenocarcinoma studies. Finally, we report the clinical course of a series of fusion-positive cases treated with matched targeted therapy.
METHODS
Patient Selection and Data Collection
This study was conducted in accordance with an institutional review board–approved protocol. The protocol was approved by Moffitt Cancer Center (MCC) in accordance with the Declaration of Helsinki and the 21st Century Cures Act. A cohort comprising 100 consecutive patients (MCC 100) was identified from a Moffitt database that discretely annotates all genomic results from targeted next-generation sequencing (NGS) panels.11 Cases were required to have a pathologically confirmed diagnosis of pancreatic adenocarcinoma and had targeted NGS performed using FoundationOne, FoundationOne CDx, or Moffitt STAR assays between March 1, 2013, and August 30, 2019, as part of clinical care. Targetable gene fusions were defined as those with clinical evidence supporting the use of targeted therapy as follows: ALK, BRAF, FGFR2, FGFR3, MET, NRG1, NTRK1, NTRK2, NTRK3, RAF1, RET, and ROS1 (Data Supplement, online only).
Assays Used
Moffitt STAR.
The Illumina TruSight Tumor 170 gene (TST170) platform is a NGS platform designed to detect genetic alterations in 170 genes. The assay uses an enrichment-based hybrid capture targeted panel that simultaneously analyzes DNA for single-nucleotide variants, multi-nucleotide variants, and indels, and RNA for fusions and splice variants (55 genes).12
FoundationOne.
This assay has been described in depth previously.13 The assay uses hybridization-based capture of 4,557 exons of 287 cancer-related genes and 47 introns of 19 genes frequently rearranged in solid tumors.
FoundationOne CDx.
This assay has been described in depth previously.14 The assay uses hybridization-based capture of all coding exons from 309 cancer-related genes, one promoter region, one noncoding (ncRNA), and select intronic regions from 34 commonly rearranged genes.14 A list of genes included in each of these assays is available in the Data Supplement.
Validation Using Two Independent Cohorts
As a validation cohort, the AACR GENIE and TCGA Pancreatic Adenocarcinoma data sets were analyzed. For the AACR Genie data set, only pancreatic adenocarcinoma cases were included from sites within the AACR GENIE consortium that contributed data on both structural rearrangements (ie, fusions) and mutation data (Memorial Sloan Kettering Cancer Center [MSK] and Vanderbilt-Ingram Cancer Center [VICC]).15 Sites reporting structural and mutation data included 1,252 of 2,048 unique patients within the AACR GENIE data set. Molecular profiling assays and associated bioinformatics pipeline used by AACR GENIE consortium sites has been previously described.15 Samples identified in the AACR GENIE database without any mutations or copy number alterations identified were excluded to avoid failed genotyping samples. A complete list of included cases is available in the Data Supplement.
As a second validation cohort, the TCGA Pancreatic Adenocarcinoma data set was obtained through cBioportal.16,17 The 150 cases from TCGA were included as these cases had a pathologic confirmation of a diagnosis of pancreatic adenocarcinoma along with sufficient tumor cellularity for sequencing.18 Detailed methods for the sequencing and bioinformatics pipeline has been described elsewhere.18 A complete list of cases included is available in the Data Supplement.
Statistical Analysis
Fisher's exact test (two-tailed) was performed to test for differences in the incidence of fusions in KRASWT versus KRAS-mutated (KRASMUT) patients in each of the three cohorts (MCC100, AACR GENIE, and TCGA).
RESULTS
Patient Cohort and Molecular Characteristics
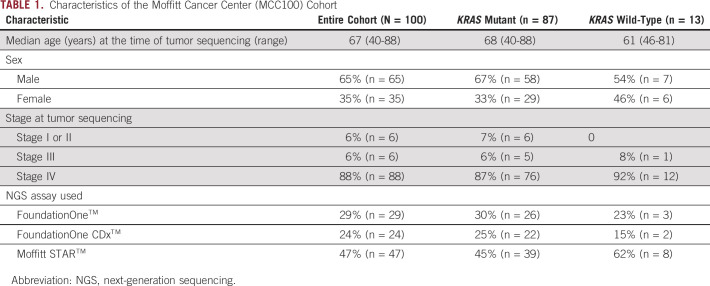
A cohort comprising 100 patients (MCC100) diagnosed with pancreatic adenocarcinoma who underwent somatic molecular sequencing with one of the three clinical NGS assays (FoundationOne, FoundationOne CDx, or Moffitt STAR) is included (Table 1). The median age for MCC100 at the time of NGS was 67 years (range, 40-88 years) with a male predominance (65%). The majority of patients (88%) had stage IV disease. Thirteen (13%) patients had KRASWT pancreatic adenocarcinoma and 87 (87%) had KRASMUT pancreatic adenocarcinoma, with KRAS (p.Gly12Asp) as the most commonly observed KRAS alteration (n = 42). Targetable oncogene fusions were identified in 31% (4 of 13) of KRASWT and 0% (0 of 87) of KRASMUT pancreatic adenocarcinomas (P = .0002) (Fig 1).
TABLE 1.
Characteristics of the Moffitt Cancer Center (MCC100) Cohort
FIG 1.
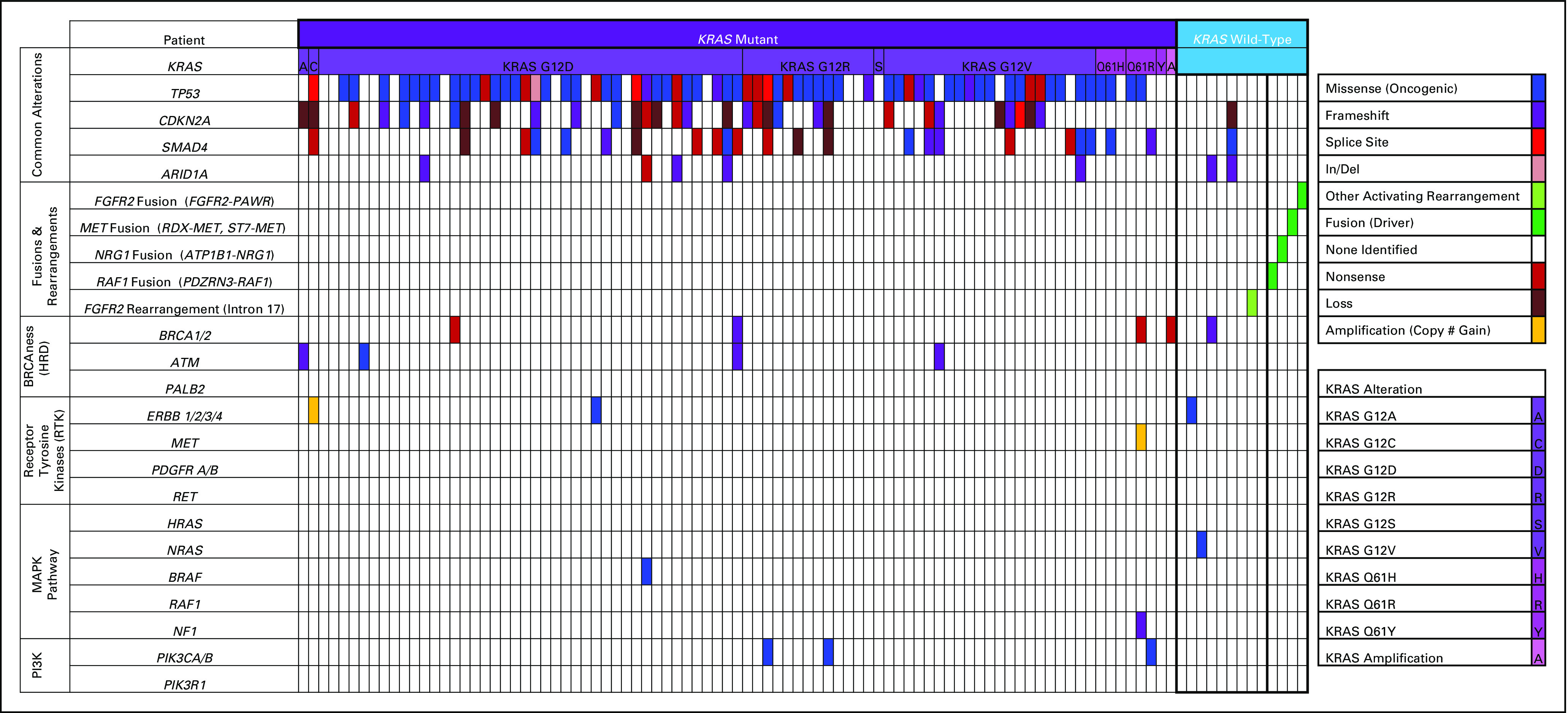
Enrichment for oncogene fusions in KRASWT pancreatic adenocarcinoma. Targetable fusions were identified in 4 of 13 KRASWT patients with no alternative driver oncogenes identified. Additionally, a FGFR2 rearrangement was identified that was predicted to be activating.
Targetable Oncogene Fusions Identified in KRASWT Pancreatic Adenocarcinoma
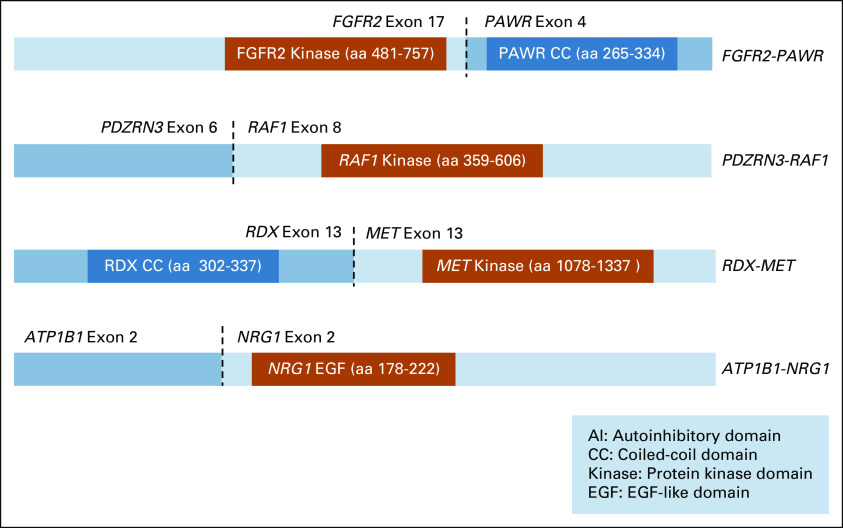
The fusion events in MCC100 involved four targetable genes. The first was an FGFR2-PAWR rearrangement (number of supporting reads not reported) in which exons 1-17 of FGFR2 are fused with exons 4-7 of PAWR (Fig 2). This fusion was consistent with other known activating FGFR2 fusions, which frequently occur with a breakpoint after exon 17 at the 3′ end of FGFR2 with a 3′ partner that typically contributes a coiled-coil domain or other domain capable of oligomerization.19-23
FIG 2.
Targetable fusions identified in the MCC100 cohort. Schematic representation of the predicted products of the four fusions identified among 13 KRASWT patients. Fusions were predicted to be activating based upon the known mechanism of activation, observed breakpoints, and relevant functional domains retained or lost in the chimeric fusion transcript.
The second fusion was a PDZRN3-RAF1 rearrangement (543 supporting reads) with the chimeric transcript involving exons 1-2 of PDZRN3 fused with the 5′ end of exon 8 of RAF1 (Fig 2). This fusion was consistent with previously observed RAF1 fusions, which frequently have an exon 8 breakpoint leading to loss of an N-terminal autoinhibitory region (amino acids, 1-147) of RAF1 and constitutive activation.24-27
A third fusion was an ATP1B1-NRG1 rearrangement (652 supporting reads) with the chimeric transcript involving exons 1-2 of ATP1B1 fused with the 5′ end of exon 2 of NRG1 (Fig 2). Across studies, the 5′ partner of NRG1 has been variable. All activating NRG1 fusions retain the EGF-like domain (exons 6 and 7) of NRG1,6,8 enabling ligand binding of the EGF-like domain of NRG1 to ERBB3, which activates ERBB2/ERBB3 heterodimerization and downstream proliferative signaling.7,8,28 The identified fusion was consistent with this known mechanism, with the EGF-like domain predicted to be retained in the chimeric protein.
The fourth identified fusion was a novel RDX-MET fusion (142 supporting reads) that has not been previously reported. The fusion involves exons 1-13 of the RDX gene fused with the 5′ end of MET exon 13 (Fig 2). The fusion was found to be consistent with previously described activating MET gene fusions, involving the intracellular domain of MET (with an intact kinase domain) fused at its amino terminus with a dimerization motif such as a coiled-coil domain.29-31 In the described case, the chimeric transcript retains an intact MET kinase domain and RDX contributes a coiled-coil domain capable of oligomerization. A second MET fusion was also detected in the same patient. This fusion involved exons 1-11 of MET as the 5′ partner of the chimeric transcript fused to the 5′ end of exon 4 of the ST7 gene. This second fusion does not retain the MET kinase domain, and thus, may be a reciprocal fusion with the first fusion, RDX-MET, serving as the oncogenic driver.
Among the KRASWT subset of MCC100, there was also a rearrangement of FGFR2 at intron 17 that was not categorized as a fusion but was predicted to be activating as a result of loss of the 3′ untranslated region (UTR) of FGFR2.21,32,33
Validation in Independent Data Sets
Among the AACR GENIE cohort of 1,252 patients with unique pancreatic adenocarcinoma, 92% (n = 1,155) were KRASMUT. Targetable oncogene fusions were identified in 19% (n = 18) of KRASWT cancers (Fig 3) compared with < 1% (n = 4) of KRASMUT cancers (P < .0001). One of the four KRASMUT cancers with fusions identified harbored an atypical KRAS alteration (KRAS [p. Ile118Leu]) that has not been characterized as activating. Among KRASWT cancers, the identified fusion was the lone mitogenic driver in 89% (16/18) of fusion-positive samples, supporting consideration of these fusions as oncogenic drivers of cellular proliferation. One notable exception was a sample harboring both a BRAF fusion and a BRAF V600E mutation.
FIG 3.
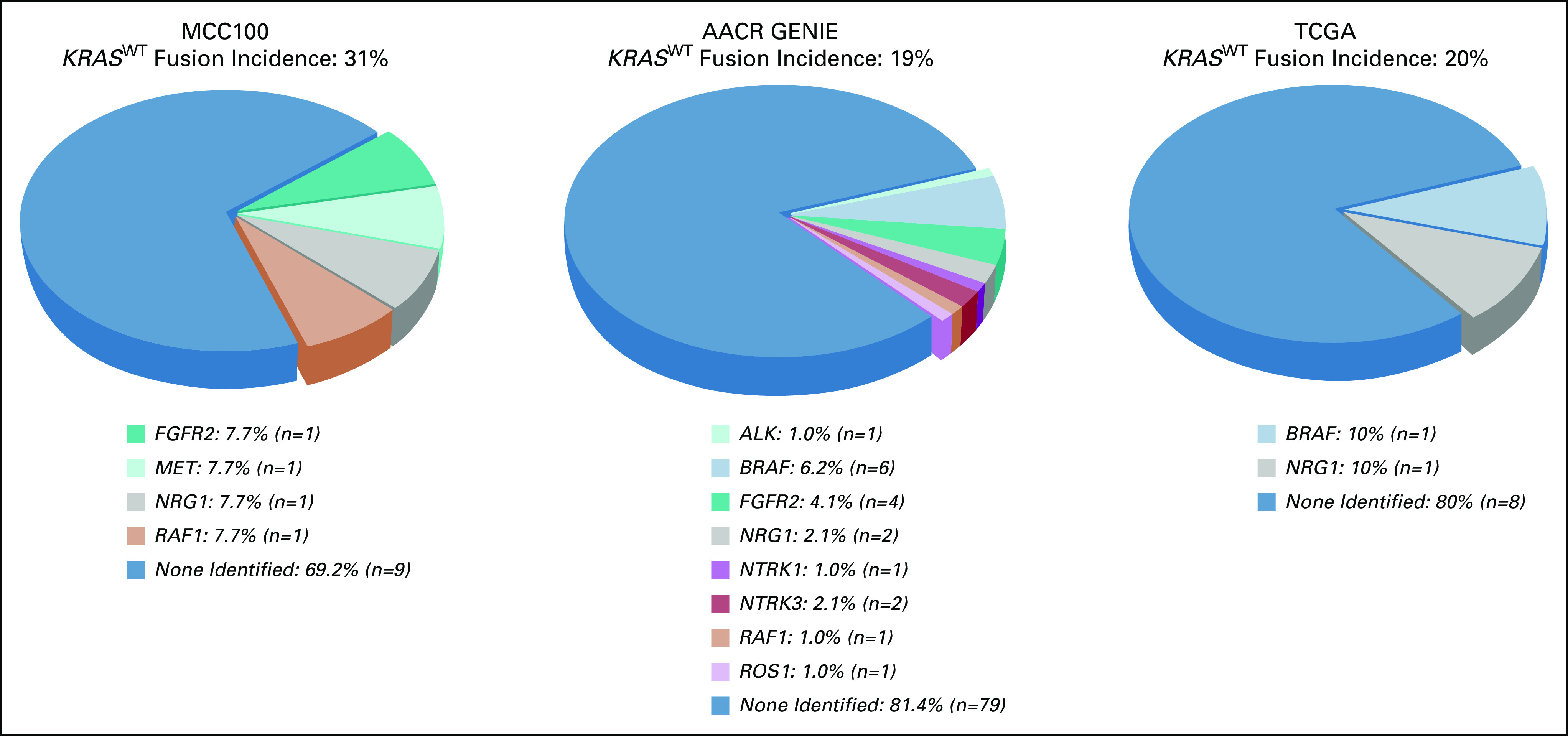
Targetable oncogene fusions identified in KRASWT pancreatic adenocarcinoma in the Moffitt Cancer Center (MCC100) and independent cohorts (AACR GENIE and TCGA). Targetable fusions were reported across all three cohorts at an incidence of at least 19% among KRASWT pancreatic adenocarcinoma patients.
In the TCGA cohort, KRAS mutations were identified in 93% (140/150) of pancreatic adenocarcinoma samples. Targetable oncogene fusions were identified in 20% (n = 2) of KRASWT cancers (Fig 3) compared with < 1% (n = 1) of KRASMUT cancers (P = .0116). Among KRASWT cancers, the identified fusion was the lone mitogenic driver in both of the fusion-positive samples.
Targeted Treatment for Oncogene Fusions in KRASWT Pancreatic Adenocarcinoma
Four cases of KRASWT pancreatic adenocarcinomas with targetable gene fusions (n = 3) or structural rearrangements (n = 1) in MCC100 received targeted treatment on the basis of the recommendations of the molecular tumor board at MCC.11
Patient Case #34: PDZRN3-RAF1
An 81-year-old male with pancreatic adenocarcinoma presented with a pancreatic head mass and multiple liver metastases. He was treated with gemcitabine plus nab-paclitaxel for 11 months. At progression, targeted NGS (Moffitt STAR) was performed, which identified a PDZRN3-RAF1 fusion. Treatment with fluorouracil and leucovorin was initiated as second-line therapy, but after 5 months cancer antigen (CA) 19-9 continued to rise and chemotherapy was stopped. On the basis of prior case reports describing responses to MEK inhibition (MEKi) in RAF1 fusion-positive patients,26,34,35 targeted therapy with trametinib (MEKi) was initiated at a dose of 2 mg daily. Treatment with trametinib was discontinued after 3 weeks as a result of the development of a left gluteal hematoma after a fall. Comparison of pre- and post-trametinib computed tomography (CT) scans revealed a decrease in the size of the primary mass in the pancreatic head (decreasing from 2.9 × 5.2 cm to 2.4 × 3.9 cm). However, an increase in the size of upper abdominal nodes and liver lesions were observed, including a right hepatic lobe lesion that increased from 3.6 cm to 4.2 cm. The patient's condition continued to decline, and 2 weeks later he was transitioned to hospice care.
Patient Case #82: ATP1B1-NRG1
A 56-year-old female, who was initially diagnosed at 50 years of age, presented with recurrent metastatic pancreatic adenocarcinoma. She previously underwent four resections and five lines of chemotherapy, including most recently HIPEC with mitomycin-C. Following debulking and HIPEC, CA 19-9 continued to rise with imaging suspicious for recurrence. Targeted NGS (Moffitt STAR) was performed, with an ATP1B1-NRG1 fusion identified. On the basis of the described clinical benefit of afatinib in prior case series of NRG1 fusion-positive cases,7,8 the patient was started on treatment with afatinib 40 mg daily. She received treatment for approximately 2 weeks before the medication was temporarily held as a result of the development of an acneiform rash that resolved with clindamycin gel and a break from treatment. Her subsequent CT scans showed stable disease and the decision was made to restart afatinib at a reduced dose of 30 mg daily, which she tolerated well. Unfortunately, at her follow-up scans 4 months after initiating treatment with afatinib, she had progressed with new liver lesions.
Patient Case #73: RDX-MET
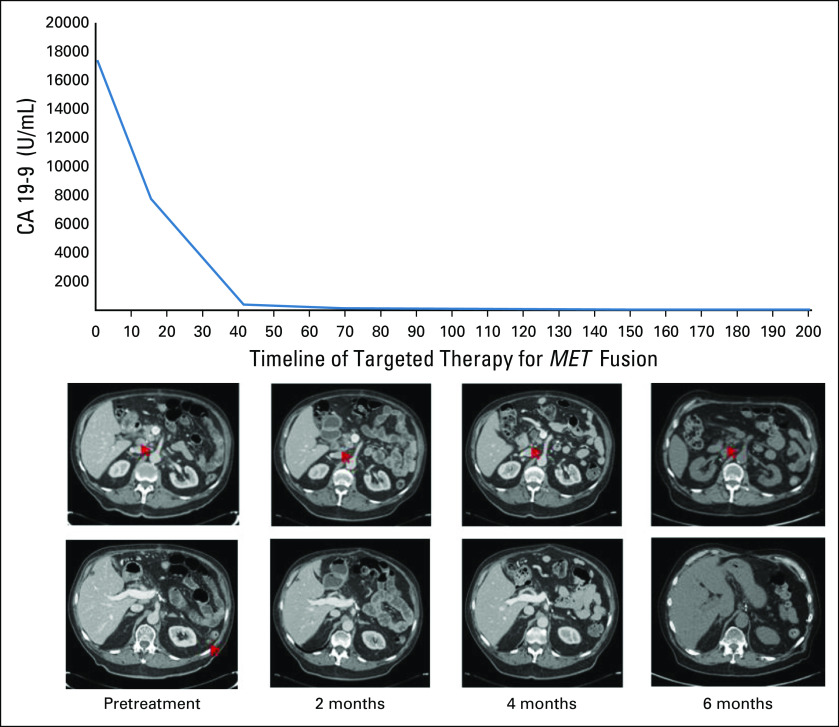
An 80-year-old male with pancreatic adenocarcinoma was diagnosed at 77 years of age. He underwent a partial pancreatectomy and completed 6 months of adjuvant gemcitabine plus capecitabine. Four months later, a peritoneal metastasis was found and treated with gemcitabine plus nab-paclitaxel. CT scans 2 months later showed progressive disease and rising CA 19-9. At this time, NGS (Moffitt STAR) identified a novel RDX-MET fusion. Subsequent CT scans showed further progression with CA 19-9 continuing to rise to a zenith of 17,406 U/mL, and treatment was initiated with the c-MET inhibitor, crizotinib, on the basis of the identified MET fusion. After 2 weeks of the targeted therapy, CA 19-9 had decreased to 7,752 U/mL. CT scans after 2 months of treatment with crizotinib demonstrated resolution of peritoneal disease in parallel to continued decline in CA 19-9 down to 123 U/mL. At approximately 7 months of treatment, a complete response was demonstrated on positron emission tomography and CT with normalization of CA 19-9 down to 32.7 U/mL. The response is ongoing now at > 12 months of treatment with crizotinib (Fig 4).
FIG 4.
Response to MET-targeted treatment with crizotinib in KRASWT pancreatic adenocarcinoma. Top: scans demonstrating the resolution of peritoneal disease in the RDX-MET fusion patient treated with crizotinib. Bottom: schematic of the patient’s CA 19-9 decline from a zenith of 17,406 U/mL down to 33 U/mL.
Patient Case #53: FGFR2 Rearrangement (Intron 17)
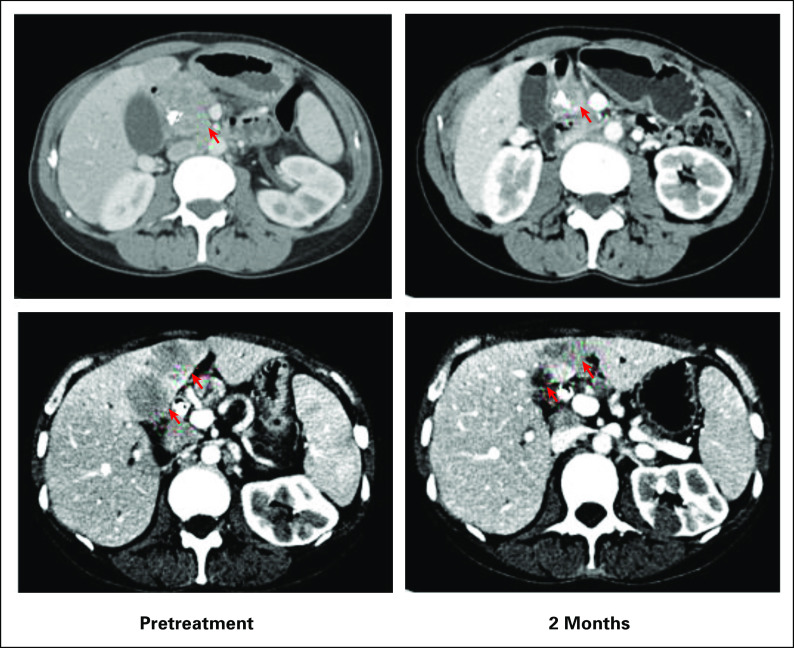
A 48-year-old female with metastatic pancreatic adenocarcinoma was diagnosed after work-up for severe abdominal pain and new-onset jaundice identified a mass in the pancreatic head. She was initially treated with FOLFIRINOX (5FU, leucovorin, irinotecan, and oxaliplatin), but treatment was switched to maintenance FOLFIRI (5FU, leucovorin, and irinotecan) as a result of toxicity after 6 months. Subsequent CT scans showed disease progression with enlargement of liver lesions and pancreatic lesion. On the basis of her NGS results (FoundationOne CDx), which identified an FGFR2 rearrangement, treatment with erdafitinib was initiated. Follow-up CT scans at 2 months demonstrated a partial response and she has continued on treatment (Fig 5).
FIG 5.
Partial response observed at 2 months in an FGFR2-rearranged case treated with FGFR-targeted treatment with erdafitinib.
DISCUSSION
It has been previously reported that alternative oncogenic drivers may be present in pancreatic cancers lacking a KRAS oncogene mutation; data from the three cohorts described here (MCC100, AACR GENIE, and TCGA) corroborate this finding. Among MCC100, targetable oncogene fusions were identified in 31% of KRASWT pancreatic adenocarcinomas and this enrichment for targetable fusions among KRASWT cases was validated through the independent AACR GENIE and TCGA cohorts. The lowest incidence of targetable fusions identified was 19% among any of the three analyzed cohorts. Across the three cohorts, fusions were identified in ALK, BRAF, FGFR2, MET, NRG1, NTRK1, NTRK3, RAF1, and ROS1 in the setting of KRASWT pancreatic adenocarcinoma. Other studies have also described fusions in EGFR, ERBB4, FGFR3, and RET.36 Recent studies incorporating RNA sequencing (RNA-seq) provide additional confirmation of our finding of targetable gene fusions in approximately one of five KRASWT pancreatic cancers.37 This enrichment for targetable fusions in KRASWT cases suggests that the identification of KRASWT status should trigger a dedicated search for targetable fusions or rearrangements in patients who would be candidates for such targeted therapies.
In this study, clinical correlation of response to targeted therapy is described. Notable among these is a novel RDX-MET fusion in which treatment with crizotinib resulted in an ongoing complete response. MET oncogene fusions responsive to targeted therapy have been identified in NSCLC and primary brain tumors.29,30,38,39 Durable responses to targeted treatment with crizotinib have been described previously in the setting of MET fusion-positive NSCLC, with responses ranging from 8 months to ongoing at over a year of targeted treatment.29,30,38 To the best of our knowledge, this is the first report of a MET fusion-positive pancreatic adenocarcinoma that has been treated with MET-targeted therapy. Additionally, a second MET fusion-positive pancreatic cancer was identified recently in another cohort, suggesting that this may be a rare, but recurrent, feature of KRASWT pancreatic cancers.37 The incidence of MET fusions in pancreatic adenocarcinoma remains unknown, but improved fusion detection with RNA-seq may reveal additional cases with targetable MET fusions.40-42
To provide meaningful treatment advances, prospective studies are needed to identify the optimal method of targeted treatment for each of oncogene fusions in KRASWT pancreatic adenocarcinoma. With the growing knowledge of the gene fusions present, it may become more feasible to open umbrella trials with targeted therapy specific for each of the oncogene fusions known to be present in KRASWT pancreatic adenocarcinoma. This would allow for more patients to receive treatment and a more robust assessment of response to targeted treatment.
In the MCC cohort, two fusions were identified with RNA-seq (NRG1 and MET) that would not have been detected by many DNA sequencing (DNA-seq) assays as a result of technical limitations. DNA-seq is capable of identifying fusions that occur at recurrent breakpoints with high sensitivity,41 but is limited in interrogating fusions in genes with large introns (eg, NRG1) or those occur outside a common intronic breakpoint (eg, RDX-MET).40,41,43 These inherent limitations of DNA-seq can be overcome with RNA-seq, as only the coding exons are targeted with the introns already spliced out.40-42
This study has limitations. Multiple assays were used, including some that used only DNA for fusion detection. Consequently, we are able to confidently report that targetable fusions are present in ≥ 19% of KRASWT cases, but additional studies using RNA-seq for all KRASWT cases would be warranted to confirm the respective incidence of each of the targetable fusions identified. A second limitation is that FGFR2 fusions are well described in other pancreaticobiliary malignancies such as intrahepatic cholangiocarcinoma, suggesting that patients with altered FGFR2 in our cohort may instead be cholangiocarcinoma.32,44 However, FGFR2 fusions have been reported in other large pancreatic cancer studies,36,37 and an additional pathology and radiology review validated the diagnosis.36,37
The clinical utility of NGS for pancreatic cancer has advanced over the last 5 years, and the National Comprehensive Cancer Network guidelines now recommend tumor or somatic gene profiling for patients with locally advanced or metastatic disease who are candidates for anticancer therapy.45 Platinum-based chemotherapy or PARP inhibitors can provide meaningful clinical benefit for patients with germline or somatic alterations in homologous recombination repair genes such as BRCA1, BRCA2, and PALB2.46,47 Microsatellite instability is relatively rare in pancreatic cancer (< 1%), but these rare cases can experience durable responses with immune checkpoint blockade.48 ERBB2 (HER2) amplification has been identified in approximately 3% of pancreatic cancers and has been identified in both KRASWT and KRASMUT tumors (approximately 3% and approximately 2%, respectively).36 HER2-targeted approaches have not been successful thus far in pancreatic cancer,49 but assessment of KRAS status may be important for future clinical trials (as has been seen in colon cancer).50 The development of novel HER2-targeted agents may also assist with the realization of clinical benefit in this population.51,52 In the setting of KRASWT tumors, there is an enrichment for BRAF mutations (V600E: approximately 3% and in-frame insertions or deletions: approximately 3%) with responses to targeted treatment described6,36,37,53 and a diverse group of oncogenic fusions including ALK, BRAF, FGFR2, FGFR3, MET, NRG1, NTRK1, NTRK3, RAF1, RET, and ROS1 (approximately 20%). Expanded access to somatic testing in appropriate patients will help optimize the treatment of each of these molecular subsets in pancreatic cancer and will accelerate the development of new treatments for patients with advanced or metastatic disease.
This report and others have shown that some patients with pancreatic adenocarcinoma treated on the basis of identified fusions can derive substantial clinical benefit.7-10,54-56 This is an important finding in a disease where clinical outcomes in the advanced stages remain poor. Additionally, the incidence of MET fusions in pancreatic adenocarcinoma remains unknown as a result of limitations of many assays and future studies may consider incorporating RNA-based testing for KRASWT cancers with inclusion of coverage of the MET oncogene. Future clinical investigations may reveal the optimal method for targeting each of the fusions reported among KRASWT pancreatic adenocarcinoma to offer more effective therapeutic options for patients in need.
ACKNOWLEDGMENT
This work has been supported in part by the Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292). T.C. Knepper is supported by the NCI of the NIH under an LRP award.
EQUAL CONTRIBUTION
T.C.K. and D.W.K. contributed equally to this work.
SUPPORT
Supported in part by the Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292); a State of Florida Endowed Chair in Cancer Research; and by funds provided by Moffitt Cancer Center in pursuit of its mission “To contribute to the prevention and cure of cancer.” T.C. Knepper is supported by the NCI of the NIH under an LRP award.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Theresa A. Boyle
Stock and Other Ownership Interests: Ions Pharmaceuticals
Honoraria: Illumina, Excel CME
Research Funding: Bristol Myers Squibb
Travel, Accommodations, Expenses: Bristol Myers Squibb, Excel CME, Illumina
Jamie K. Teer
Patents, Royalties, Other Intellectual Property: Patent application: Large Data Set Negative Information Storage Model.
Christine M. Walko
Employment: Mission Healthcare
Consulting or Advisory Role: Jackson Laboratory for Genomic Medicine, Intermountain Precision Genomics
Other Relationship: Mission Hospital
Howard L. McLeod
Leadership: Cancer Genetics
Stock and Other Ownership Interests: Cancer Genetics, Interpares Biomedicine, Clariifi, Pharmazam
Honoraria: Genentech/Roche, Illumina
Consulting or Advisory Role: Gentris, Cancer Genetics, Saladax Biomedical, NIH/NCI, Admera Health, eviCore healthcare, Pharmazam, LLC, VieCure
Speakers' Bureau: Genentech
Other Relationship: Northwestern University, Xiangya Hospital
J. Kevin Hicks
Consulting or Advisory Role: Quest Diagnostics, 23andMe
Research Funding: OneOme
Martine Extermann
Consulting or Advisory Role: Alnylam
Jason B. Fleming
Leadership: Bio-Path Holdings, Inc
Consulting or Advisory Role: Johnson & Johnson, Glycobio, Moleculin Biotech, Inc, Perthera
Patents, Royalties, Other Intellectual Property: U.S. Application No. 15/780,799, based on International Application No. PCT/US2016/065763, titled “Polymeric Drug Delivery Systems For Treatment of Disease,” by Chun Li et al.; in the Name of Board of Regents, The University of Texas System
No other potential conflicts of interest were reported.
AUTHOR CONTRIBUTIONS
Conception and design: Michael J. Fusco, Mokenge Malafa, Howard L. McLeod, J. Kevin Hicks, Jason B. Fleming, Todd C. Knepper
Financial support: Howard L. McLeod
Administrative support: Mokenge Malafa, Howard L. McLeod
Provision of study materials or patients: Mokenge Malafa
Collection and assembly of data: Michael J. Fusco, Estrella M. Carballido, Theresa A. Boyle, Mokenge Malafa, Kirsten L. Blue, Howard L. McLeod, Dae Won Kim
Data analysis and interpretation: Michael J. Fusco, Daryoush Saeed-Vafa, Theresa A. Boyle, Mokenge Malafa, Jamie K. Teer, Christine M. Walko, Howard L. McLeod, J. Kevin Hicks, Martine Extermann, Jason B. Fleming, Todd C. Knepper, Dae Won Kim
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2020. CA Cancer J Clin 70:7-30, 2020 [DOI] [PubMed] [Google Scholar]
- 2.Conroy T, Desseigne F, Ychou M, et al. : FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 364:1817-25, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Von Hoff DD, Ramanathan RK, Borad MJ, et al. : Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: A phase I/II trial. J Clin Oncol 29:4548-54, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Reilly EM, Lee JW, Zalupski M, et al. : Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol 38:1378-1388, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowery MA, Jordan EJ, Basturk O, et al. : Real-time genomic profiling of pancreatic ductal adenocarcinoma: Potential actionability and correlation with clinical phenotype. Clin Cancer Res 23:6094-6100, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Aguirre AJ, Nowak JA, Camarda ND, et al. : Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov 8:1096, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heining C, Horak P, Uhrig S, et al. : NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov 8:1087-1095, 2018 [DOI] [PubMed] [Google Scholar]
- 8.Jones MR, Williamson LM, Topham JT, et al. : NRG1 gene fusions are recurrent, clinically actionable gene rearrangements in KRAS wild-type pancreatic ductal adenocarcinoma. Clin Cancer Res 25:4674-4681, 2019 [DOI] [PubMed] [Google Scholar]
- 9.O’Reilly EM, Hechtman JF: Tumour response to TRK inhibition in a patient with pancreatic adenocarcinoma harbouring an NTRK gene fusion. Ann Oncol 30:viii36-viii40, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pishvaian MJ, Garrido-Laguna I, Liu SV, et al. : Entrectinib in TRK and ROS1 fusion-positive metastatic pancreatic cancer. JCO Precision Oncol 10.1200/PO.18.00039, 2018 [DOI] [PubMed] [Google Scholar]
- 11.Knepper TC, Bell GC, Kevin Hicks J, et al. : Key lessons learned from Moffitt's Molecular Tumor Board: The Clinical Genomics Action Committee Experience. Oncologist 22:144-151, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illumina : Trusight Tumor 170 Data Sheet. www.illumina.com/illumina/trusight-tumor-170-data-sheet-1170-2016-01, 2017 [Google Scholar]
- 13.Frampton GM, Fichtenholtz A, Otto GA, et al. : Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023-31, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foundation Medicine, Inc.: FoundationOne CDx™ Technical Information. www.accessdata.fda.gov/cdrh_docs/pdf17/P170019C.pdf [Google Scholar]
- 15.AACR Project GENIE : Powering precision medicine through an international consortium. Cancer Discov 7:818; Data Release Version 6.1., 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raphael BJ, Hruban RH, Aguirre AJ, et al. : Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32:185-203.e13, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YM, Su F, Kalyana-Sundaram S, et al. : Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 3:636-47, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arai Y, Totoki Y, Hosoda F, et al. : Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 59:1427-1434, 2014 [DOI] [PubMed] [Google Scholar]
- 21.Tuna M, Amos CI, Mills GB: Molecular mechanisms and pathobiology of oncogenic fusion transcripts in epithelial tumors. Oncotarget 10:2095-2111, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dienstmann R, Rodon J, Prat A, et al. : Genomic aberrations in the FGFR pathway: Opportunities for targeted therapies in solid tumors. Ann Oncol 25:552-563, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voss MH, Hierro C, Heist RS, et al. : A phase I, open-label, multicenter, dose-escalation study of the oral selective FGFR inhibitor Debio 1347 in patients with advanced solid tumors harboring FGFR gene alterations. Clin Cancer Res 25:2699-2707, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran NH, Wu X, Frost JA: B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. J Biol Chem 280:16244-16253, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Hartmaier RJ, Albacker LA, Chmielecki J, et al. : High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res 77:2464-2475, 2017 [DOI] [PubMed] [Google Scholar]
- 26.Kim KB, Semrad T, Schrock AB, et al. : Significant clinical response to a MEK inhibitor therapy in a patient with metastatic melanoma harboring an RAF1 fusion. JCO Precision Oncol, 10.1200/PO.17.00138, 2018 [DOI] [PubMed] [Google Scholar]
- 27.Jain P, Fierst TM, Han HJ, et al. : CRAF gene fusions in pediatric low-grade gliomas define a distinct drug response based on dimerization profiles. Oncogene 36:6348, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gay ND, Wang Y, Beadling C, et al. : Durable response to Afatinib in lung adenocarcinoma harboring NRG1 gene fusions. J Thorac Oncol 12:e107-e110, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Davies KD, Ng TL, Estrada-Bernal A, et al. : Dramatic response to Crizotinib in a patient with lung cancer positive for an HLA-DRB1-MET gene fusion. JCO Precision Oncol 10.1200/PO.17.00117, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plenker D, Bertrand M, de Langen AJ, et al. : Structural alterations of MET trigger response to MET kinase inhibition in lung adenocarcinoma patients. Clin Cancer Res 24:1337-1343, 2018 [DOI] [PubMed] [Google Scholar]
- 31.Comoglio PM, Trusolino L, Boccaccio C: Known and novel roles of the MET oncogene in cancer: A coherent approach to targeted therapy. Nat Rev Cancer 18:341-358, 2018 [DOI] [PubMed] [Google Scholar]
- 32.Abou-Alfa GK, Sahai V, Hollebecque A, et al. : Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study. Lancet Oncol 21:671-684, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jusakul A, Cutcutache I, Yong CH, et al. : Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 7:1116, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEvoy CR, Xu H, Smith K, et al. : Profound MEK inhibitor response in a cutaneous melanoma harboring a GOLGA4-RAF1 fusion. J Clin Invest 129:1940-1945, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Touat M, Younan N, Euskirchen P, et al. : Successful targeting of an ATG7-RAF1 gene fusion in anaplastic pleomorphic xanthoastrocytoma with leptomeningeal dissemination. JCO Precision Oncol 10.1200/PO.18.00298, 2019 [DOI] [PubMed] [Google Scholar]
- 36.Singhi AD, George B, Greenbowe JR, et al. : Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology 156:2242-2253.e4, 2019 [DOI] [PubMed] [Google Scholar]
- 37.Philip PA, Xiu J, Hall MJ, et al. : Enrichment of alterations in targetable molecular pathways in KRAS wild-type (WT) pancreatic cancer (PC). J Clin Oncol 38:4629, 2020 [Google Scholar]
- 38.Cho JH, Ku BM, Sun JM, et al. : KIF5B-MET gene rearrangement with robust antitumor activity in response to crizotinib in lung adenocarcinoma. J Thorac Oncol 13:e29-e31, 2018 [DOI] [PubMed] [Google Scholar]
- 39.International Cancer Genome Consortium PedBrain Tumor P, Bender S, Gronych J, et al. : Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med 22:1314, 2016 [DOI] [PubMed] [Google Scholar]
- 40.Benayed R, Offin M, Mullaney K, et al. : High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res 25:4712-4722, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon JP, Linkov I, Rosado A, et al. : NTRK fusion detection across multiple assays and 33,997 cases: Diagnostic implications and pitfalls. Mod Pathol 33:38-46, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solomon JP, Hechtman JF: Detection of NTRK fusions: Merits and limitations of current diagnostic platforms. Cancer Res 79:3163-3168, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treangen TJ, Salzberg SL: Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat Rev Genet 13:36-46, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farshidfar F, Zheng S, Gingras MC, et al. : Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep 18:2780-2794, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Comprehensive Cancer Network : NCCN guidelines, Pancreatic Adenocarcinoma (Version 1.2020). https://www.nccn.org
- 46.Golan T, Hammel P, Reni M, et al. : Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 381:317-327, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Binder KAR, Mick R, Hara M, et al. : Abstract CT234: A phase II, single arm study of maintenance rucaparib in patients with platinum-sensitive advanced pancreatic cancer and a pathogenic germline or somatic mutation in BRCA1, BRCA2 or PALB2. Cancer Res 79:CT234, 2019 [DOI] [PubMed] [Google Scholar]
- 48.Hu ZI, Shia J, Stadler ZK, et al. : Evaluating mismatch repair deficiency in pancreatic adenocarcinoma: Challenges and recommendations. Clin Cancer Res 24:1326-1336, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harder J, Ihorst G, Heinemann V, et al. : Multicentre phase II trial of trastuzumab and capecitabine in patients with HER2 overexpressing metastatic pancreatic cancer. Br J Cancer 106:1033-1038, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meric-Bernstam F, Hurwitz H, Raghav KPS, et al. : Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): An updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol 20:518-530, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh DY, Hamilton E, Hanna Dm et al. : Safety, anti-tumour activity, and biomarker results of the HER2-targeted bispecific antibody ZW25 in HER2-expressing solid tumours. Ann Oncol 30:ix22-ix29, 2019(suppl 9) [Google Scholar]
- 52.Tsurutani J, Iwata H, Krop I, et al. : Targeting HER2 with trastuzumab deruxtecan: A dose-expansion, phase I study in multiple advanced solid tumors. Cancer Discovery 10:688, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seghers AK, Cuyle PJ, Van Cutsem E: Molecular targeting of a BRAF mutation in pancreatic ductal adenocarcinoma: Case report and literature review. Target Oncol 15:407-410, 2020 [DOI] [PubMed] [Google Scholar]
- 54.Singhi AD, Ali SM, Lacy J, et al. : Identification of targetable ALK rearrangements in pancreatic ductal adenocarcinoma. J Natl Compr Canc Netw 15:555-562, 2017 [DOI] [PubMed] [Google Scholar]
- 55.Pishvaian MJ, Blais EM, Brody JR, et al. : Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: A retrospective analysis of the know your tumor registry trial. Lancet Oncol 21:508-518, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subbiah V, Hu MIN, Gainor JF, et al. : Clinical Activity of the RET Inhibitor Pralsetinib (BLU-667) in Patients With RET Fusion+ Solid Tumors, ASCO 2020 Virtual Scientific Meeting, 2020 [Google Scholar]