Abstract
PURPOSE
This study was designed to assess the ability of perioperative circulating tumor DNA (ctDNA) to predict surgical outcome and recurrence following neoadjuvant chemoradiation for locally advanced rectal cancer (LARC).
MATERIALS AND METHODS
Twenty-nine patients with newly diagnosed LARC treated between January 2014 and February 2018 were enrolled. Patients received long-course neoadjuvant chemoradiation prior to surgery. Plasma ctDNA was collected at baseline, preoperatively, and postoperatively. Next-generation sequencing was used to identify mutations in the primary tumor, and mutation-specific droplet digital polymerase chain reaction was used to assess mutation fraction in ctDNA.
RESULTS
The median age was 54 years. The overall margin-negative, node-negative resection rate was 73% and was significantly higher among patients with undetectable preoperative ctDNA (n = 17, 88%) versus patients with detectable preoperative ctDNA (n = 9, 44%; P = .028). Undetectable ctDNA was also associated with more favorable neoadjuvant rectal scores (univariate linear regression, P = .029). Recurrence-free survival (RFS) was calculated for the subset (n = 19) who both underwent surgery and had postoperative ctDNA available. At a median follow-up of 20 months, patients with detectable postoperative ctDNA experienced poorer RFS (hazard ratio, 11.56; P = .007). All patients (4 of 4) with detectable postoperative ctDNA recurred (positive predictive value = 100%), whereas only 2 of 15 patients with undetectable ctDNA recurred (negative predictive value = 87%).
CONCLUSION
Among patients treated with neoadjuvant chemoradiation for LARC, patients with undetectable preoperative ctDNA were more likely to have a favorable surgical outcome as measured by the rate of margin-negative, node-negative resections and neoadjuvant rectal score. Furthermore, we have confirmed prior reports indicating that detectable postoperative ctDNA is associated with worse RFS. Future prospective study is needed to assess the potential for ctDNA to assist with personalizing treatment for LARC.
INTRODUCTION
Standard treatment for locally advanced rectal cancer (LARC) involves trimodality therapy consisting of neoadjuvant chemoradiation, surgery, and adjuvant chemotherapy. In recent years, total neoadjuvant therapy (TNT) has emerged as a novel paradigm for LARC that aims to address micrometastatic disease earlier in the treatment course.1,2 However, not all patients require adjuvant chemotherapy,3 and adopting a uniform TNT approach for LARC carries with it significant risk of overtreatment.4 Furthermore, after neoadjuvant chemoradiation, 15%-27% of patients are found to have a pathologic complete response (pCR)5,6 and interest in organ preservation (ie, omission of surgery) in these patients has gained considerable interest in recent years.7,8 Current practice uses the clinical complete response as the benchmark for identifying patients for whom preservation may be a viable option. However, the clinical response poorly correlates with the pathologic response. In the future, a reliable biomarker of therapeutic efficacy available in real time could be beneficial for assessing the response to treatment and likelihood of benefit from completion of TNT and guiding perioperative management. An National Cancer Institute panel was recently convened and highlighted the need for more studies in this area.9
CONTEXT
Key Objective
Patients with locally advanced rectal cancer (LARC) who have minimal residual disease as measured using detectable postoperative circulating tumor DNA (ctDNA) have been shown to have higher risk for recurrence compared with those with undetectable postoperative ctDNA; however, the relationship between preoperative ctDNA and surgical outcome has been less well-studied.
Knowledge Generated
In this study, we analyzed preoperative and postoperative ctDNA in a cohort of patients who received neoadjuvant chemoradiation for LARC. We confirm prior work showing that those who harbor minimal residual disease as measured by postoperative ctDNA following resection are at increased risk for recurrence. Additionally, we show that undetectable preoperative ctDNA was associated with improved surgical outcomes, including lower and more favorable neoadjuvant rectal scores and better margin-negative, node-negative resection rates.
Relevance
Further prospective study is necessary to determine the utility of incorporating ctDNA for clinical decision making for patients with LARC.
Novel high-sensitivity assays have been developed in recent years to detect fragments of circulating tumor DNA (ctDNA) among a background of cell-free DNA in the plasma of individuals with both localized and metastatic cancers.10-15 Detectable postoperative ctDNA has been shown to accurately identify patients with resected stage II colorectal cancer at risk for recurrence with improved sensitivity compared with standard clinical risk factors or tumor markers.15 Similarly, detectable postchemoradiotherapy and postoperative ctDNA identify patients with LARC who are likely to subsequently recur.16 However, studies investigating preoperative ctDNA assessments measured after neoadjuvant therapy are fewer in number and conflicting with respect to whether preoperative ctDNA can be useful for predicting the outcome of surgery.16,17
Therefore, in this study, we sought to perform a proof-of-concept analysis evaluating whether ctDNA can identify patients who are likely to have a favorable surgical outcome using composite end points of neoadjuvant rectal (NAR) score18 and margin-negative, node-negative (R0-NN) resection rates after neoadjuvant chemoradiation. The NAR is a validated surrogate composite end point for early-phase clinical trials involving neoadjuvant therapy for rectal cancer and has been shown to predict overall survival (OS) better than pCR in a previously published independent validation cohort.18-20 Additionally, we sought to validate prior work16 showing that those who harbor minimal residual disease (MRD), as defined by ctDNA detection after curative-intent surgery for rectal cancer, are at increased risk for recurrence.
MATERIALS AND METHODS
Patient Population and Treatment
Between January 2014 and February 2018, 29 patients with newly diagnosed locally advanced rectal adenocarcinoma who were being treated at either the Massachusetts General Hospital Center for GI Cancers or the University of Michigan Multidisciplinary Colorectal Cancer Clinic were enrolled onto an institutional-specific institutional review board–approved protocol for biospecimen collection. The present study focused on ctDNA analysis was retrospective. Patients were treated per the recommendations of the treating physician with long-course chemoradiation (conformal external beam radiation therapy [CRT]) consisting of 45 Gy to the pelvis followed by a boost to the mesorectum to 50.4 Gy in 1.8 Gy fractions with concurrent capecitabine or infusional fluorouracil (5FU). Three of the patients who received infusional 5FU also received midostaurin on a separate interventional protocol.
Per the recommendation of the treating physician, some patients received neoadjuvant chemotherapy prior to planned surgical resection and some received adjuvant chemotherapy. Reductions in the dose of chemotherapy prescribed, timing of chemotherapy delivery, and number of cycles received were decided by the treating physician as needed to manage acute chemotherapy–related toxicities.
Tumor Sequencing and ctDNA Analysis
At the time of initial biopsy or surgical resection, patients underwent next-generation sequencing (NGS) of their primary tumor to identify tumor-specific mutations to personalize the ctDNA assay, as described previously.21,22 Whole blood for ctDNA analysis was collected in two 10 mL Streck tubes generally at baseline prior to neoadjuvant CRT, preoperatively, and postoperatively. Plasma was isolated through two centrifugation steps: the first at room temperature for 10 minutes at 1,600×g and the second at room temperature for 10 minutes at 3,000×g. Plasma was then stored at −80°C until ctDNA isolation. At the time of analysis, ctDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit according to the manufacturer's instructions, increasing the protein lysis step from 30 minutes to 60 minutes according to recommendations from the Streck tube manufacturer. ctDNA was analyzed using a highly sensitive and specific droplet digital polymerase chain reaction (ddPCR) method, which allows for detection of one mutant DNA molecule among a background of 10,000 wild-type molecules.23-25 Methods for ctDNA isolation and analysis have been previously described in detail.26 Each assay was constructed to detect one or more mutations in peripheral blood specific to individual patients. Specific mutations for ctDNA analysis were chosen after NGS review on the basis of determining that a given mutation was somatic (not germline) and on the basis of probe availability. Probe information is available on request.
Assessment of preoperative clinical response and determination of surgical outcome.
As directed by the treating physician, patients also had standard tumor markers (carcinoembryonic antigen [CEA]) drawn before and after neoadjuvant chemotherapy and CRT. The following clinical and treatment factors were noted for each patient: age, sex, tumor location, clinical American Joint Committee on Cancer stage, types and number of cycles of neoadjuvant chemotherapy, radiation technique (three-dimensional conformal, intensity-modulated radiation therapy, etc), total dose received, dose per fraction, and concurrent and adjuvant chemotherapy type.
After neoadjuvant treatment, most patients proceeded to curative-intent surgical resection. However, a subset of patients developed metastatic disease while undergoing neoadjuvant therapy. For patients who underwent surgical resection, the following pathologic outcomes were noted: clinical and pathologic American Joint Committee on Cancer T and N stage, tumor grade, tumor regression grade,27 R-resection status on the basis of margin status,28 lymphovascular invasion, and perineural invasion. Surgical specimens from Massachusetts General Hospital were centrally reviewed by a dedicated GI pathologist (J.K.L.) to confirm the tumor regression grade assessment.
Follow-up and determination of recurrence and death.
After neoadjuvant treatment and surgery, patients were followed for disease recurrence with serial history and physical examinations, tumor marker assessment (CEA), and computed tomography of the chest, abdomen, and pelvis, or magnetic resonance imaging per treating clinician.29 Patients were followed from the time of enrollment until the time of death or last follow-up.
Statistical Methods
Prediction of surgical outcomes using ctDNA.
Patients were grouped according to whether they had a detectable (≥ 2 mutant ctDNA alleles among a minimum of 2,000 wild-type alleles) versus undetectable (0 mutant ctDNA alleles among a minimum of 2,000 wild-type alleles) preoperative ctDNA. Conservatively, if only one mutant ctDNA allele was detected, the patient's ctDNA status was classified as negative.
Composite surgical outcome end points.
R0-NN resection.
Patients were classified as achieving an R0-node negative (R0-NN) resection if surgical margins were negative, and there were no positive lymph nodes on surgical pathology. In addition to pathologic node–positive disease, non-R0-NN resection included patients who had an R1 or R2 resection or those patients who were not resected because of either poor response to neoadjuvant therapy or development of metastatic disease during neoadjuvant therapy.
NAR score.
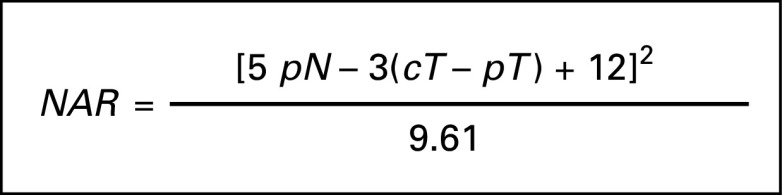
The NAR score18 was computed for patients who were able to undergo resection of the primary tumor. The NAR score is a composite end point, which reflects the response to neoadjuvant therapy, taking into account downstaging of the primary tumor and pathologic nodal status (Fig 1).18 As in the work of George et al,18 patients were grouped into NAR-low or NAR-intermediate and NAR-high categories if their NAR score fell 8-16 and > 16, respectively.
FIG 1.
Formula used for calculating NAR score as developed by George et al.18 cT, clinical tumor stage; NAR, neoadjuvant rectal; pN, pathologic nodal stage; pT, pathologic tumor stage.
Fisher's exact tests were used to compare the rate of R0-NN resection and the rate of NAR-low or NAR-intermediate score among patients with detectable versus undetectable preoperative ctDNA. Univariate logistic regression with R0-NN status as the dependent variable was conducted separately for the following independent variables: preoperative ctDNA status and preoperative CEA. Univariate linear regression with NAR score as the dependent variable was conducted separately for the following independent variables: preoperative ctDNA status and preoperative CEA.
Predictors of progression-free survival.
Patients were grouped according to whether they had a detectable (≥ 2 mutant ctDNA alleles among a minimum of 2,000 wild-type alleles) versus undetectable (0 mutant ctDNA alleles among a minimum of 2,000 wild-type alleles) postoperative ctDNA. Conservatively, if only one mutant ctDNA allele was detected, the patient's ctDNA status was classified as negative. Progression-free survival (PFS) was estimated using the Kaplan-Meier method from the date of surgery until the time of radiographic progression (as determined by the interpreting radiologist and confirmed by the treating physician) or date of last follow-up.30 Patients without radiographic progression at the time of last follow-up were then censored from the PFS estimate. Estimates of PFS were calculated separately for patients with detectable versus undetectable postoperative ctDNA, and the Cox proportional hazard model was used to compare the PFS between groups. Positive predictive value (PPV) and negative predictive value (NPV) of postoperative ctDNA for the prediction of recurrence were calculated. PPV was defined as the probability that postoperative ctDNA predicts recurrence (ie, the number of patients with subsequent recurrence and detectable postoperative ctDNA divided by all patients with detectable postoperative ctDNA). NPV was defined as the probability that undetectable postoperative ctDNA predicts subsequent freedom from recurrence (ie, the number of patients without subsequent recurrence and with negative postoperative ctDNA divided by all patients with negative postoperative ctDNA). Statistical analyses were performed using STATA (StataCorp, College Station, TX) and SAS version 9.4 (SAS Institute, Cary, NC).
RESULTS
Patient Characteristics
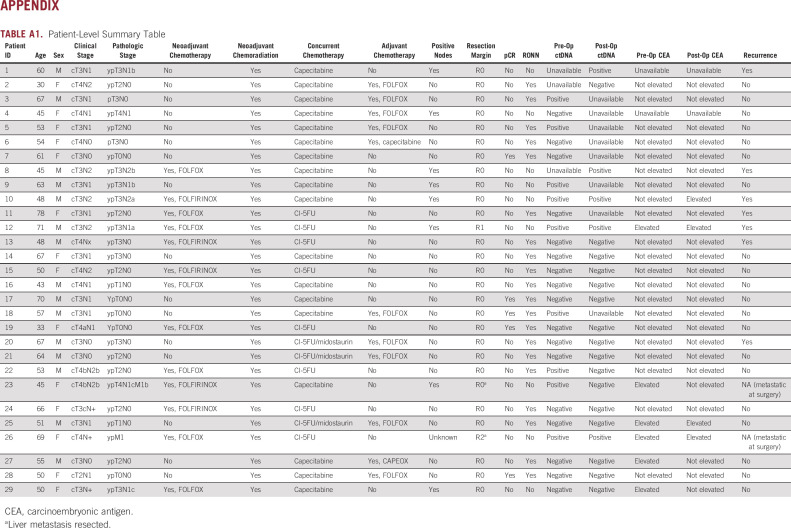
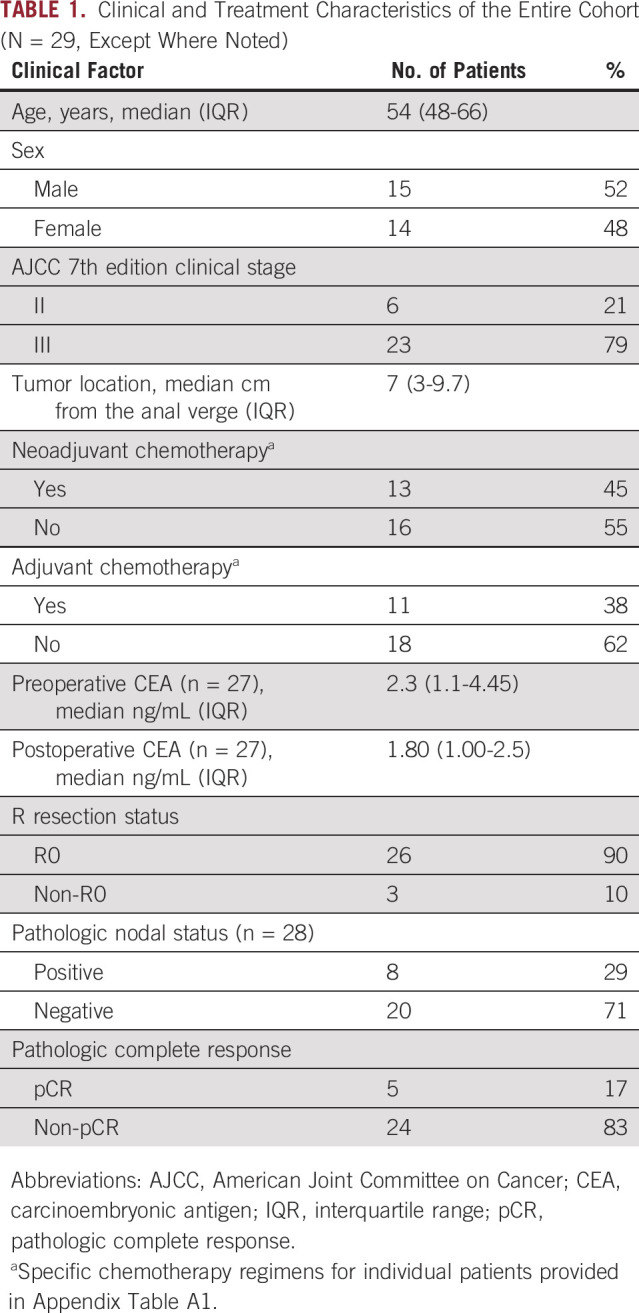
Table 1 illustrates clinical characteristics for the entire cohort. The median age of the cohort was 54 years (range, 45-78 years). All patients received neoadjuvant long-course chemoradiation. Preoperative ctDNA assessments were performed between 0 and 17 weeks preoperatively. Postoperative ctDNA assessments were performed between 1 and 5 months postoperatively. Notably, some patients received neoadjuvant chemotherapy prior to chemoradiation followed by surgery, one patient received neoadjuvant chemoradiation followed by chemotherapy prior to surgery, and some patients received adjuvant chemotherapy after surgery. A patient-level summary table detailing individual treatment course per patient is included in Appendix Table A1.
TABLE 1.
Clinical and Treatment Characteristics of the Entire Cohort (N = 29, Except Where Noted)
Analysis of ctDNA Trajectory over Time and Treatment
As described above, ddPCR was used to assess one or more tumor-specific mutations (identified by tumor tissue sequencing) in ctDNA at available timepoints for each patient. Figure 2 illustrates examples of ctDNA trajectories for five representative patients, illustrating examples of ctDNA trajectories for favorable and unfavorable surgical outcomes.
FIG 2.
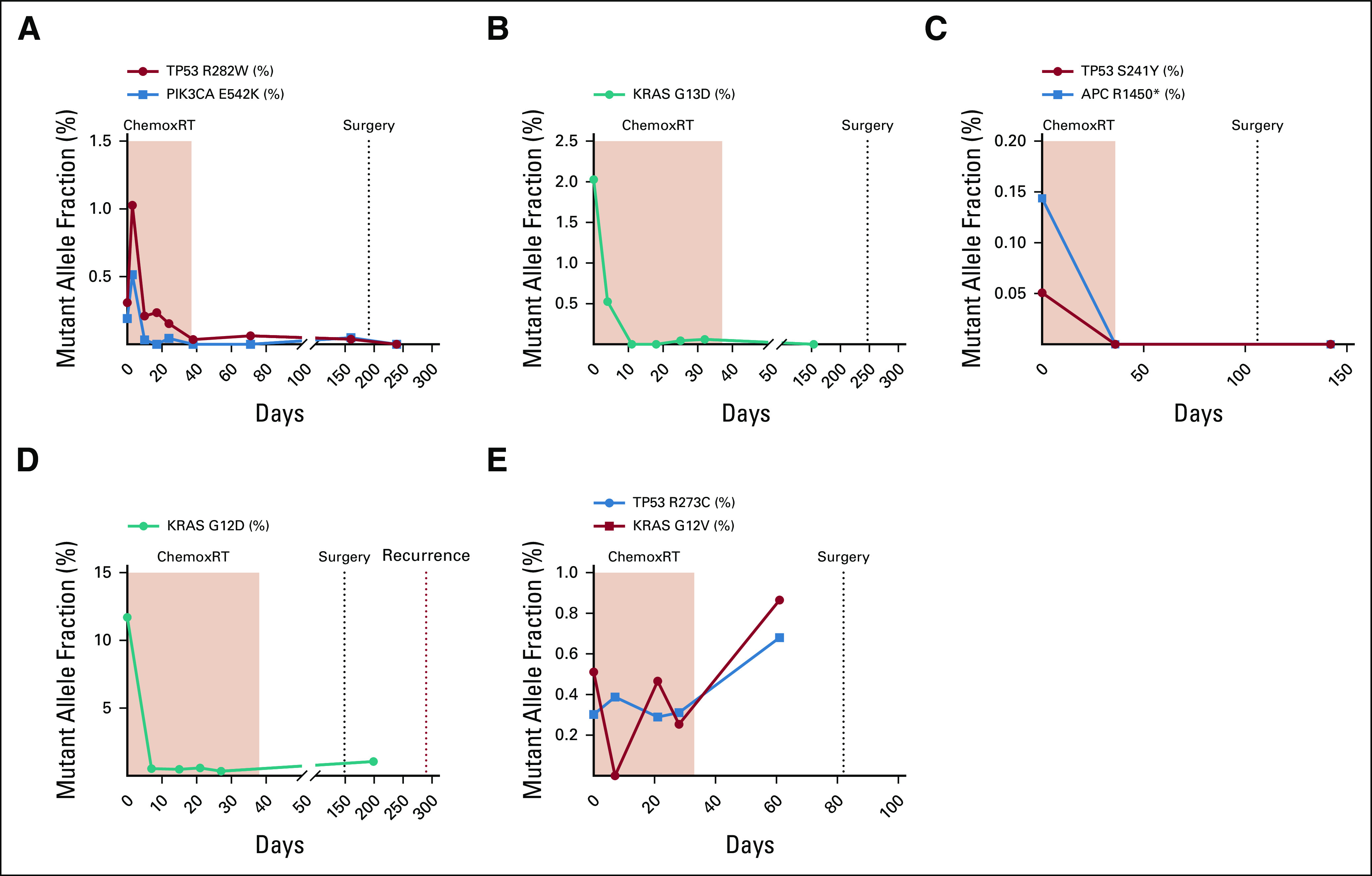
Five representative patient examples. Panel A displays the circulating tumor DNA (ctDNA) trajectory for two mutations (TP53 R282W and PIK3CA E542K) over time for a 53-year-old who received eight cycles of FOLFOX followed by chemoradiotherapy for a TP53 and PIK3CA cT4bN2b locally advanced rectal cancer (LARC). As shown in the panel, his ctDNA decreased during chemoradiation; however, his preoperative computed tomography scan showed a mixed response where the tumor appeared tethered to presacral soft tissue. Surgical pathology revealed significant tumor regression, negative lymph nodes, and negative margins. His postoperative ctDNA assay was negative, and the patient is currently NED 20 months postoperatively. Panel B displays the ctDNA trajectory for a 78-year-old who received neoadjuvant chemoradiation for a T3N1 LARC. Her KRAS G13D ctDNA dropped preoperatively, and she was found to have significant treatment effect with 1-2 mm of residual tumor foci in a 1.5-cm tumor bed. However, her carcinoembryonic antigen (CEA) remained elevated pre- and postoperatively. Panel C displays the ctDNA trajectory for a 50-year-old with a cT2N1 TP53– and APC-mutant LARC, treated with neoadjuvant FOLFOX and conformal external beam radiation therapy (CRT). Her ctDNA decreased during CRT, and she was found to have a pathologic complete response at the time of surgery. Her postoperative ctDNA was negative, and she is currently NED 14 months postoperatively. Panel D displays the ctDNA trajectory for a 48-year-old who received eight cycles of FOLFIRINOX and CRT for KRAS G12D–mutant cT3N2 LARC. His CEA decreased during CRT, but his preoperative ctDNA was positive. Surgical pathology revealed a poor tumor response and six of 14 involved lymph nodes. His postoperative ctDNA was positive, which preceded a radiographic recurrence by 3 months. Panel E displays the ctDNA trajectory for a 45-year-old with TP53- and KRAS-mutant cT4bN2b LARC who received neoadjuvant FOLFIRINOX and chemoradiation prior to surgical resection. Her ctDNA increased during CRT. She underwent resection of rectal primary and partial hepatectomy, which revealed liver metastases (despite a negative liver biopsy prior to chemoradiation).
ctDNA as a Predictor of Surgical Outcome
Twenty-six patients had preoperative plasma available for analysis. The overall R0-NN resection rate was 73% for the entire cohort. The rate of R0-NN resection was significantly higher among patients with an undetectable preoperative ctDNA (n = 17) compared with those with a detectable (n = 9) preoperative ctDNA (88% R0-NN v 44% R0-NN, respectively, P = .028, Table 2). On univariate logistic regression, both ctDNA status and CEA were significantly associated with R0-NN resection (P = .027 and .034, respectively).
TABLE 2.
Results Table Summarizing Relationship Between Preoperative ctDNA, CEA and Surgical Outcomes
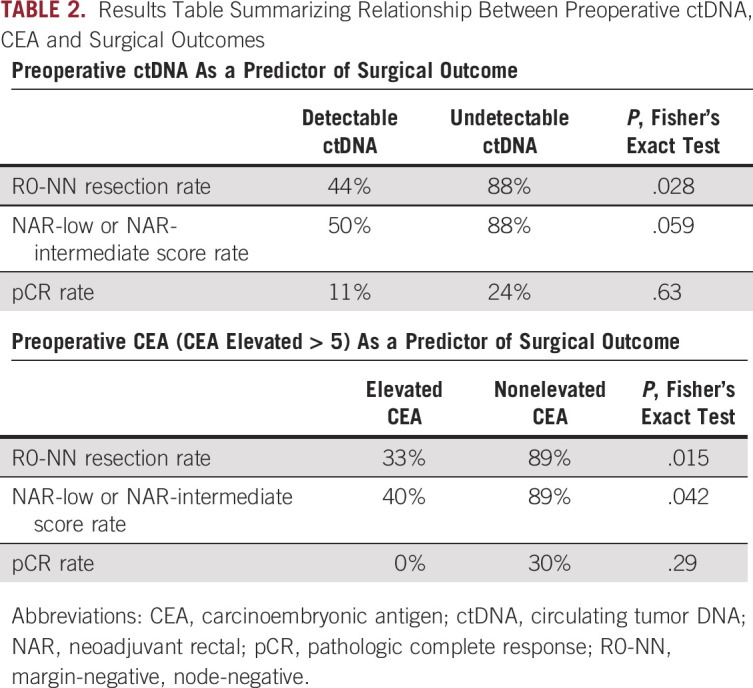
The NAR score was able to be computed for 25 patients, and 19 were in the NAR-low or NAR-intermediate category, whereas six were in the NAR-high category. Patients with preoperative undetectable ctDNA had a higher rate of more favorable NAR-low and NAR-intermediate scores (88%) as compared with those with detectable ctDNA (50%, P = .059, Table 2). On univariate linear regression, both ctDNA status and CEA were significantly associated with NAR score (P = .029 and .044, respectively). The pCR rate among patients with detectable versus undetectable preoperative ctDNA was not significantly different, 11% versus 24% (P = .63), respectively (Table 2).
Predictors of PFS
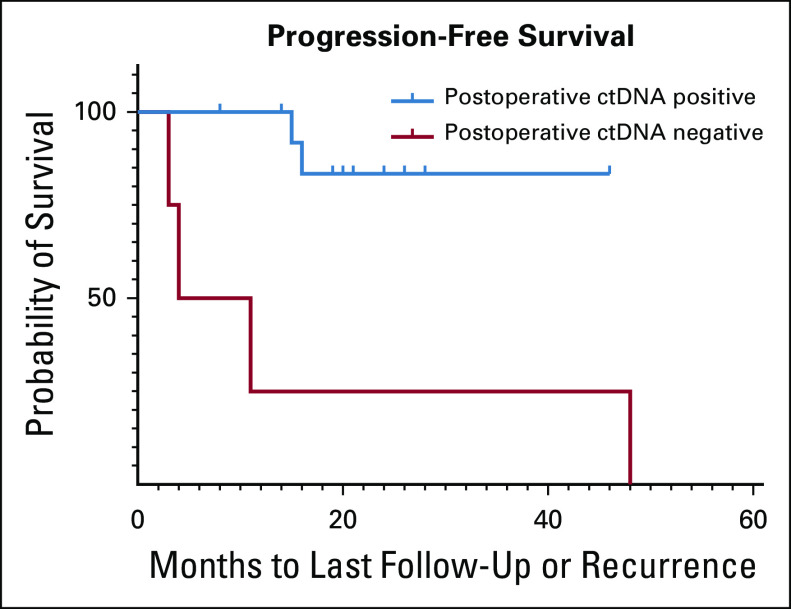
For the subset of patients who underwent curative-intent surgery with postoperative ctDNA available (n = 19), recurrence-free survival was calculated and patients were stratified by detectable versus undetectable postoperative ctDNA. After a median follow-up of 20 months (interquartile range, 14.0-43 months), 4 of 17 (24%) patients had died and 6 of 19 (32%) had experienced local or distant recurrence. Cumulative incidence estimates of PFS for patients with detectable (n = 4) versus undetectable (n = 15) postoperative ctDNA are shown in Figure 3. Notably, four of four patients with detectable postoperative ctDNA recurred (PPV = 100%), whereas only two of 15 (13%) patients with undetectable postoperative ctDNA recurred (NPV = 87%). Patients with detectable postoperative ctDNA had significantly worse PFS than patients with undetectable postoperative ctDNA (hazard ratio, 11.56; P = .007). None of the patients with positive postoperative ctDNA MRD received adjuvant chemotherapy in this cohort.
FIG 3.
Progression-free survival stratified by postoperative detectable versus undetectable ctDNA status (hazard ratio, 11.56; P = .007). ctDNA, circulating tumor DNA.
DISCUSSION
In this proof-of-concept, confirmatory study, undetectable preoperative ctDNA was associated with improved surgical outcomes, including a lower and more favorable NAR score and better R0-NN rates in a cohort of patients treated with neoadjuvant CRT for LARC. We chose to evaluate the relationship between ctDNA and NAR score as this score was developed as a composite short-term end point and predicts OS better than pCR.18 Additionally, consistent with many studies,16 we also found that postoperative analysis of ctDNA strongly predicted risk for recurrence following curative-intent surgery for LARC as the recurrence rate among patients with detectable postoperative ctDNA was 100%, but it was only 13% among patients with undetectable ctDNA.
Although further prospective studies are needed, these findings illustrate the potential for ctDNA to predict the treatment response and thus could affect real-time clinical decision making in three main ways. First, as TNT emerges as a paradigm for treatment of LARC,1,2 widespread adoption of a standard TNT approach may result in overtreatment as some patients may be adequately treated with neoadjuvant chemoradiation and surgical resection alone. This was shown by the ADORE trial, which randomly assigned patients after CRT and surgical resection to adjuvant chemotherapy with FOLFOX versus fluorouracil plus leucovorin.3,4 Although there was an overall significant improvement in progression-free survival among patients receiving FOLFOX, subgroup analysis revealed that patients with pathologic stage III benefitted from FOLFOX, whereas those with pathologic stage II disease did not,3 highlighting the need to determine which patients will benefit from intensified adjuvant chemotherapy upfront. The use of ctDNA could potentially help guide which patients are at the highest risk of recurrence and help determine who should receive chemotherapy. Multiple studies have shown that consolidation chemotherapy (after chemoradiation) is safe and effective and the ongoing Organ Preservation of Rectal Adenocarcinoma (OPRA) trial is currently assessing the optimal order of preoperative treatment.31-35 If TNT is planned with chemoradiation before chemotherapy and ctDNA is predictive of outcome, ctDNA could potentially be used to identify patients who would not necessarily benefit from further chemotherapy with the goal of preventing overtreatment.
Second, a subset of patients with a clinical complete response to neoadjuvant therapy may not require surgery to achieve a cure.7,8 However, it is challenging to identify good candidates for a watch-and-wait approach as standard clinical response assessment does not often correlate with the outcome of surgery. We demonstrated that patients with undetectable preoperative ctDNA following neoadjuvant therapy were more likely to have an R0-NN surgery and favorable NAR score than patients with detectable preoperative ctDNA. Thus, combined with clinical response assessment, ctDNA may represent a promising additional tool to guide future studies for tailoring the TNT approach for individual patients and improving selection for organ preservation.
Third, elevated postoperative ctDNA could possibly help identify a high-risk subset of patients who would potentially benefit from increased surveillance or escalation of adjuvant therapy. The recently published COLOFOL36 and GILDA37 randomized trials, which found that intensive surveillance does not prolong survival in patients with nonselected colorectal cancer, have called into question the need for intensive surveillance. Based on prior studies of intensive surveillance, it seems likely that there are high-risk subgroups who will still benefit from more intensive surveillance, which could potentially be selected using postoperative ctDNA. Additionally, it is unknown whether patients with MRD identified with postoperative ctDNA assays would benefit from escalation of adjuvant therapy. In this cohort, none of the patients with positive ctDNA MRD received adjuvant chemotherapy, and it is unknown whether adjuvant chemotherapy would have changed their ultimate disease trajectory. Prospective randomized clinical trials are needed in the MRD space, incorporating ctDNA to determine the utility of ctDNA to monitor the treatment response and whether escalation of adjuvant therapy for patients with positive ctDNA is beneficial.
Several points require further consideration. First, our findings corroborate and complement ctDNA analysis from a larger cohort of patients with LARC treated with neoadjuvant therapy.16 Tie et al16 analyzed plasma for ctDNA from 159 patients before and after neoadjuvant therapy and following surgical resection. Although they observed that detectable postoperative ctDNA predicted subsequent recurrence, they did not observe a correlation between post-CRT ctDNA and pCR surgical outcome and concluded that ctDNA measured after CRT cannot differentiate between minimal and no residual disease or be used to select patients for a nonoperative approach.16 Murahashi et al17 also investigated ctDNA following neoadjuvant treatment for LARC. They also did not observe a significant association between preoperative ctDNA and pCR; however, they did observe that change in ctDNA was associated with patients who achieved a pCR or those who were managed by the watch-and-wait approach for more than 12 months after achieving a clinical complete response.17 Although our study was limited by small numbers, we also did not observe a significant relationship between preoperative ctDNA and pCR. However, we did observe a significant relationship between preoperative ctDNA and composite end point markers of surgical outcome, namely, favorable NAR score and R0-NN resection, which have not been previously assessed. We chose to evaluate composite end points (NAR and R0-NN resection) in our analysis instead of individual surgical outcomes to capture more aspects of downstaging. The NAR score is a validated surrogate end point, which has been shown to predict OS better than pCR by considering both tumor and nodal downstaging and was chosen as the primary end point for the ongoing NRG GI002 trial (ClinicalTrials.gov identifier: NCT02921256) evaluating veliparib and pembrolizumab in the neoadjuvant setting for LARC. Further prospective study of multiple assays will be necessary to further understand these differences and further elucidate the utility of incorporating such assays into clinical decision making.
Our study has several limitations. First, the sample size of this cohort is small. As a result, although we were able to show that both ctDNA and CEA are correlated with surgical outcome, the small sample size of our cohort precluded the ability to ascertain the extent to which ctDNA adds over CEA and other clinical factors to predict surgical and oncologic outcomes. This relationship will be important for further study in larger prospective cohorts. Second, although many patients in our cohort received neoadjuvant chemotherapy prior to chemoradiation, not all these individuals had plasma available for ctDNA analysis prior to initiation of any systemic therapy. Thus, for the small subset of patients who had a negative ctDNA assay at all timepoints collected, it is unclear whether the negative assay reflected the treatment response to initial systemic therapy or whether these patients had undetectable ctDNA because of low rates of ctDNA shedding. To mitigate this concern, we assayed ctDNA to a limit of detection of 0.05% in each case, although some patients may have harbored ctDNA levels below this limit. Although our study used custom ddPCR to assess ctDNA in each patient, newer technologies assessing multiple mutations by NGS and/or methylation and epigenomic markers could represent more optimal approaches for assessment of residual disease through ctDNA.38 A third limitation is that there was variability in the timing of collection of preoperative and postoperative timepoints, but our small sample size precluded assessment of whether this altered our result. Future prospective studies should minimize this heterogeneity with prespecified collection times.
Despite these limitations, our study suggests that ctDNA may be useful for identifying patients who will have a favorable surgical outcome following neoadjuvant chemoradiation for LARC and confirms prior studies indicating that patients with postoperative ctDNA MRD are at higher risk for recurrence. Further study is necessary to examine the utility of incorporating a ctDNA assay for tailoring the TNT approach for individual patients or using ctDNA along with other clinical factors to aid in identification of patients who may be suitable for an organ preservation approach following neoadjuvant treatment. Additionally, identifying patients early following surgery who are at high risk of recurrence could provide an opportunity for alteration of surveillance, potential early intervention, and thus offer a window for a second chance of cure for these individuals, particularly in patients where adjuvant therapy may not otherwise be used.39 Further prospective study is necessary to determine the utility of ctDNA in personalized therapy for patients with LARC.
APPENDIX
TABLE A1.
Patient-Level Summary Table
EQUAL CONTRIBUTION
S.G.R.M. and K.M.H. contributed equally to this work as first authors. R.B.C. and T.S.H. contributed equally to this work as last authors.
PRIOR PRESENTATION
Presented at the 2019 Annual Meeting of the ASCO Gastrointestinal Cancers Symposium, San Francisco, CA, January 17-19.
AUTHOR CONTRIBUTIONS
Conception and design: Susan G. R. McDuff, Karin M. Hardiman, Aparna R. Parikh, Emily E. Van Seventer, David P. Ryan, Jeffrey W. Clark, James C. Cusack, Ryan B. Corcoran, Theodore S. Hong
Financial support: Karin M. Hardiman, Ryan B. Corcoran, Theodore S. Hong
Administrative support: Ryan B. Corcoran, Theodore S. Hong
Provision of study materials or patients: Aparna R. Parikh, Jochen K. Lennerz, Colin D. Weekes, Jeffrey W. Clark, James C. Cusack, Andrew X. Zhu, Ryan B. Corcoran, Theodore S. Hong
Collection and assembly of data: Susan G. R. McDuff, Karin M. Hardiman, Peter J. Ulintz, Jochen K. Lennerz, Mehlika Hazar-Rethinam, Emily E. Van Seventer, Isobel J. Fetter, Brandon Nadres, Christine E. Eyler, David P. Ryan, Colin D. Weekes, Jeffrey W. Clark, Lipika Goyal, Andrew X. Zhu, Jennifer Y. Wo, Lawrence S. Blaszkowsky, Jill Allen, Ryan B. Corcoran, Theodore S. Hong
Data analysis and interpretation: Susan G. R. McDuff, Karin M. Hardiman, Aparna R. Parikh, Hui Zheng, Daniel W. Kim, Jochen K. Lennerz, Mehlika Hazar-Rethinam, Emily E. Van Seventer, Jeffrey W. Clark, James C. Cusack, Lawrence S. Blaszkowsky, Theodore S. Hong
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Aparna R. Parikh
Consulting or Advisory Role: PureTech, Foundation Medicine, Lilly, Natera
Research Funding: Plexxikon, Bristol-Myers Squibb, Genentech, Guardant Health, Array BioPharma, Lilly, Novartis Pharmaceuticals UK Ltd., Tesaro
Mehlika Hazar-Rethinam
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Travel, Accommodations, Expenses: Foundation Medicine, Roche
David P. Ryan
Stock and Other Ownership Interests: MPM Capital, Acworth Pharmaceuticals Inc., Thrive
Honoraria: UpToDate, Research to Practice
Consulting or Advisory Role: MPM Capital, Oncorus, Gritstone Oncology, Maverick Therapeutics, TCR2 Therapeutics, 28/7 Therapeutics
Research Funding: SU2C
Patents, Royalties, Other Intellectual Property: McGraw Hill Chapter Royalties, Johns Hopkins University Press
Expert Testimony: Boehringer Ingelheim
Other Relationship: TCR2 Therapeutics
Colin D. Weekes
Honoraria: Celgene, Merrimack, Lilly, Bayer
Consulting or Advisory Role: Celgene, Merrimack, Ipsen
Research Funding: Millennium, Celgene, Bayer, Genentech/Roche, Abbvie, Lilly, AstraZeneca, Gilead Sciences, Ipsen, Halozyme
Travel, Accommodations, Expenses: Lilly, Celgene, Bayer
Jeffrey W. Clark
Research Funding: Pfizer
James C. Cusack
Consulting or Advisory Role: Verthermia
Research Funding: Lumicell
Lipika Goyal
Consulting or Advisory Role: Debiopharm Group, Alentis Therapeutics AG, QED, H3 Biomedicine, AstraZeneca, Klus Pharmaceuticals, Agios, Taiho Pharmaceutical, Incyte, Sirtex Medical
Uncompensated Relationships: Agios, Debiopharm Group, Taiho Pharmaceutical
Andrew X. Zhu
Consulting or Advisory Role: Eisai, Merck, AstraZeneca, Bayer, Exelixis, Lilly, Roche/Genentech, Sanofi/Aventis, Gilead Sciences
Research Funding: Lilly, Bayer, Bristol-Myers Squibb, Novartis, Merck
Lawrence S. Blaszkowsky
Stock and Other Ownership Interests: Pfizer
Ryan B. Corcoran
Stock and Other Ownership Interests: Avidity Biosciences, nRichDX, Fount Therapeutics, C4 Therapeutics, Revolution Medicines, Kinnate Biopharma
Consulting or Advisory Role: Avidity Nanomedicines, Taiho Pharmaceutical, Merrimack, Genentech, N-of-One, Astex Pharmaceuticals, Amgen, Bristol-Myers Squibb, Roche, Shire, FOG Pharma, Loxo, Roivant, Warp Drive Bio, Spectrum Pharmaceuticals, Symphogen, Array BioPharma, Chugai Pharma, nRichDX, Fount Therapeutics, Novartis, Shionogi, Elicio, C4 Therapeutics, Revolution Medicines, Zikani Therapeutics, Natera, Kinnate Biopharma, Ipsen, Abbvie, Asana Biosciences
Research Funding: AstraZeneca, Sanofi, Asana Biosciences, Lilly
Theodore S. Hong
Consulting or Advisory Role: Merck, Synthetic Biologics, Novocure
Research Funding: Novartis, Taiho Pharmaceutical, AstraZeneca, Intrao, Tesaro, Bristol-Myers Squibb, Ipsen
No other potential conflicts of interest were reported.
REFERENCES
- 1.Fokas E, Allgäuer M, Polat B, et al. Randomized phase II trial of chemoradiotherapy plus induction or consolidation chemotherapy as total neoadjuvant therapy for locally advanced rectal cancer: CAO/ARO/AIO-12 J Clin Oncol 373212–32222019 [DOI] [PubMed] [Google Scholar]
- 2.Hong TS, Ryan DP. Total neoadjuvant therapy for locally advanced rectal cancer—The new standard of care? JAMA Oncol. 2018;4:e180070. doi: 10.1001/jamaoncol.2018.0070. [DOI] [PubMed] [Google Scholar]
- 3.Hong YS, Nam BH, Kim KP, et al. Oxaliplatin, fluorouracil, and leucovorin versus fluorouracil and leucovorin as adjuvant chemotherapy for locally advanced rectal cancer after preoperative chemoradiotherapy (ADORE): An open-label, multicentre, phase 2, randomised controlled trial Lancet Oncol 151245–12532014 [DOI] [PubMed] [Google Scholar]
- 4.Hong TS, Ryan DP.Adjuvant chemotherapy for locally advanced rectal cancer: Is it a given? J Clin Oncol 331878–18802015 [DOI] [PubMed] [Google Scholar]
- 5.Ferrari L, Fichera A.Neoadjuvant chemoradiation therapy and pathological complete response in rectal cancer Gastroenterol Rep 3277–2882015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan R, Gibbons D, Hyland JMP, et al. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer Histopathology 47141–1462005 [DOI] [PubMed] [Google Scholar]
- 7.Habr-Gama A, Perez RO, Nadalin W, et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: Long-term results Ann Surg 240711–7172004discussion 717-718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maas M, Beets-Tan RGH, Lambregts DMJ, et al. Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer J Clin Oncol 294633–46402011 [DOI] [PubMed] [Google Scholar]
- 9.Dasari A, Morris VK, Allegra CJ, et al. ctDNA applications and integration in colorectal cancer: An NCI colon and rectal–anal task forces whitepaper Nat Rev Clin Oncol 17757–7702020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz LA, Bardelli A.Liquid biopsies: Genotyping circulating tumor DNA J Clin Oncol 32579–5862014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl F, Schmidt K, Choti Ma, et al. Circulating mutant DNA to assess tumor dynamics Nat Med 14985–9902008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito A, Bardelli A, Criscitiello C, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: Clinical perspectives and research opportunities Cancer Treat Rev 40648–6552014 [DOI] [PubMed] [Google Scholar]
- 14.Siravegna G, Bardelli A.Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients Mol Oncol 10475–4802016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer : A prospective biomarker study Gut 681–92019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murahashi S, Akiyoshi T, Sano T, et al. Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: Prediction of pathological response and postoperative recurrence Br J Cancer 123803–8102020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George TJ, Allegra CJ, Yothers G.Neoadjuvant rectal (NAR) score: A new surrogate endpoint in rectal cancer clinical trials Curr Colorectal Cancer Rep 11275–2802015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Zhang Y, Wu X, et al. Prognostic significance of neoadjuvant rectal score in locally advanced rectal cancer after neoadjuvant chemoradiotherapy and construction of a prediction model J Surg Oncol 117737–7442018 [DOI] [PubMed] [Google Scholar]
- 20.Yothers G, George TJ, Petrelli NJ, et al. Neoadjuvant rectal cancer (RC) score to predict survival: Potential surrogate endpoint for early-phase trials. J Clin Oncol. 2014;32:3533. [Google Scholar]
- 21.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine EMBO Mol Med 2146–1582010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frydrych LM, Ulintz P, Bankhead A, et al. Rectal cancer sub-clones respond differentially to neoadjuvant therapy Neoplasia 211051–10622019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients Nat Med 21795–8012015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number Anal Chem 838604–86102011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinert T, Schøler LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery Gut 65625–6342016 [DOI] [PubMed] [Google Scholar]
- 26.Hazar-Rethinam M, Kleyman M, Han GC, et al. Convergent therapeutic strategies to overcome the heterogeneity of acquired resistance in BRAF V600E colorectal cancer Cancer Discov 8417–4272018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thies S, Langer R. Tumor regression grading of gastrointestinal carcinomas after neoadjuvant treatment. Front Oncol. 2013;3:262. doi: 10.3389/fonc.2013.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermanek P, Wittekind C.Residual tumor (R) classification and prognosis Semin Surg Oncol 1012–201994 [DOI] [PubMed] [Google Scholar]
- 29.Tempero MA, Malafa MP, Al-Hawary M, et al. Pancreatic adenocarcinoma, version 2.2017, NCCN clinical practice guidelines in oncology J Natl Compr Canc Netw 151028–10612017 [DOI] [PubMed] [Google Scholar]
- 30.Kaplan EL, Meier P.Nonparametric estimation from incomplete observations J Am Stat Assoc 53457–4811958 [Google Scholar]
- 31.Garcia-Aguilar J, Smith DD, Avila K, et al. Optimal timing of surgery after chemoradiation for advanced rectal cancer Ann Surg 25497–1022011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Aguilar J, Chow OS, Smith DD, et al. Effect of adding mFOLFOX6 after neoadjuvant chemoradiation in locally advanced rectal cancer: A multicentre, phase 2 trial Lancet Oncol 16957–9662015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Aguilar J, Patil S, Kim JK, et al. Preliminary results of the organ preservation of rectal adenocarcinoma (OPRA) trial. J Clin Oncol. 2020;38:4008. [Google Scholar]
- 34.Conroy T, Lamfichekh N, Etienne P-L, et al. Total neoadjuvant therapy with mFOLFIRINOX versus preoperative chemoradiation in patients with locally advanced rectal cancer: Final results of PRODIGE 23 phase III trial, a UNICANCER GI trial. J Clin Oncol. 2020;38:4007. [Google Scholar]
- 35.Hospers G, Bahadoer RR, Dijkstra EA, et al. Short-course radiotherapy followed by chemotherapy before TME in locally advanced rectal cancer: The randomized RAPIDO trial. J Clin Oncol. 2020;38:4006. [Google Scholar]
- 36.Wille-Jørgensen P, Syk I, Smedh K, et al. Effect of more vs less frequent follow-up testing on overall and colorectal cancer–Specific mortality in patients with stage II or III colorectal cancer the COLOFOL randomized clinical trial JAMA 3192095–21032018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosati G, Ambrosini G, Barni S, et al. A randomized trial of intensive versus minimal surveillance of patients with resected Dukes B2-C colorectal carcinoma Ann Oncol 27274–2802016 [DOI] [PubMed] [Google Scholar]
- 38.Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer JAMA Oncol 51124–11312019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pita-Fernández S, Alhayek-Aí M, González-Martín C, et al. Intensive follow-up strategies improve outcomes in nonmetastatic colorectal cancer patients after curative surgery: A systematic review and meta-analysis Ann Oncol 26644–6562015 [DOI] [PubMed] [Google Scholar]