PURPOSE
Hereditary cancer syndromes infer high cancer risks and require intensive surveillance. Identification of high-risk individuals among patients with colorectal cancer (CRC) needs improvement.
METHODS
Three thousand three hundred ten unselected adults who underwent surgical resection for primary invasive CRC were prospectively accrued from 51 hospitals across Ohio between January 1, 2013, and December 31, 2016. Universal Tumor screening (UTS) for mismatch repair (MMR) deficiency was performed for all, and pathogenic germline variants (PGVs) were identified using multigene panel testing (MGPT) in those who met at least one inclusion criterion: MMR deficiency, diagnosed < 50 years, multiple primary tumors (CRC or endometrial cancer), or with a first-degree relative with CRC or endometrial cancer.
RESULTS
Five hundred twenty-five patients (15.9%) had MMR deficiency. Two hundred thirty-four of 3,310 (7.1%; 16% of the 1,462 who received MGPT) had 248 PGVs in cancer susceptibility genes. One hundred forty-two (4.3%) had a PGV in an MMR gene, and 101 (3.1%) had a PGV in a non-MMR gene. Ten with Lynch syndrome (LS) also had a non-MMR PGV and were included in both groups. Two (0.06%) had constitutional MLH1 hypermethylation. Of unexplained MMR-deficient patients, 88.4% (76 of 86) had double somatic MMR mutations. Testing for only MMR genes in MMR-deficient patients would have missed 18 non-MMR gene PGVs (7.3% of total PGVs identified). Had UTS been the only method used to screen for hereditary cancer syndromes, 38.6% (91 of 236) would have been missed, including 6.3% (9 of 144) of those with LS. These results have treatment implications as 5.3% (175 of 3,310) had PGVs in genes with therapeutic targets.
CONCLUSION
UTS alone is insufficient for identifying a large proportion of CRC patients with hereditary syndromes, including some with LS. At a minimum, 7.1% of individuals with CRC have a PGV and pan-cancer MGPT should be considered for all patients with CRC.
INTRODUCTION
Lynch syndrome (LS) is the most common form of hereditary colorectal cancer (CRC). Prevalence of LS among patients with CRC has been estimated to be 2.8%-3.9%.1-4 Previous Ohio studies proved that Universal Tumor Screening (UTS) for LS was feasible,1,2 and professional societies now recommend this practice.5,6 However, UTS is underutilized and only 15%-30% of patients are tested.7-9 Implementation of UTS is important for LS identification and assessment for treatment options as the use of immunotherapy is US Food and Drug Administration–approved for microsatellite-unstable tumors.10
CONTEXT
Key Objective
To improve identification of hereditary cancer syndromes in individuals with colorectal cancer (CRC), a statewide initiative was created to provide universal tumor screening for Lynch syndrome (LS) for 3,310 unselected patients with CRC (the largest cohort to date) and to provide germline pan-cancer multigene panel testing to selected patients with CRC to assess frequency and spectrum of cancer susceptibility gene mutations.
Knowledge Generated
About 16% of patients with CRC had mismatch repair deficiency, and 7.1% had a germline pathogenic variant. Had universal tumor screening for LS been the only method used to screen for hereditary cancer syndromes, 38.6% of patients who tested positive would have been missed, including 6.3% of those found to have LS.
Relevance
Pan-cancer multigene panel testing should be considered for all patients with CRC.
Germline testing for cancer predisposition was traditionally performed in high-risk families with striking cancer histories. The use of germline multigene panel testing (MGPT) among unselected patients with cancer has led to the identification of pathogenic germline variants (PGVs) causing highly penetrant cancer syndromes in seemingly low- or moderate-risk families. Additionally, PGVs are being found in families that do not fit the traditional phenotype of the associated syndrome.4,11,12 Emerging data indicate that the prevalence of PGV among unselected patients with CRC is 9.9%-15.3%.4,13
We formed a statewide initiative in Ohio to increase access to UTS and germline genetic testing for patients with CRC using MGPT.
METHODS
Methods have previously been described in detail.11 Fifty-one hospitals (Data Supplement) across Ohio participated in the Ohio Colorectal Cancer Prevention Initiative (ClinicalTrials.gov identifier: NCT01850654). This large-scale collaboration was led by The Ohio State University (OSU) Comprehensive Cancer Center. Participating hospitals were selected to include clinical centers with a high volume of patients with CRC, an affiliation with a high-volume hospital, or an interest in participation. Written informed consent was obtained from all participants. Institutional review board approval was obtained by the individual hospitals, National Cancer Institute Community Oncology Research Programs, or ceding review to the OSU Institutional Review Board (2012C0123). All study-related services were provided locally at no cost to participants.
Participants
Three thousand four hundred seventy-one adults who underwent surgical resection for primary invasive colorectal adenocarcinoma in Ohio between January 1, 2013, and December 31, 2016, were prospectively enrolled. One hundred twenty-five patients were excluded (118 ineligible and seven withdrew). Ineligibility reasons included insufficient tumor material, ineligible pathology type, diagnosis made outside of the qualifying study period, and diagnosis made outside of Ohio. Tumor screening and germline testing were completed for 3,310 patients, representing 12.4% of CRC diagnosed in Ohio during the study period per the Ohio Incidence and Surveillance System (N = 26,692).
Samples and Clinical Data
Blood and paraffin-embedded tumor block or unstained slides were obtained for each patient. Study pathologists confirmed tumor histology and marked areas with ≥ 30% tumor and normal adjacent tissue. DNA was prepared using standard methods.14 Pathology reports were reviewed for all patients, and cancer history and first-degree relative cancer histories were provided. Three-generation pedigrees were obtained for patients with PGVs. Specific subsets of data from 963 patients have been previously described11,15-21; however, none of these reports included the complete UTS and germline results of the entire cohort.
Tumor Screening for Mismatch Repair Deficiency
All tumors were assessed for mismatch repair (MMR) deficiency by microsatellite instability (MSI) testing and/or immunohistochemical (IHC) analysis at OSU, if not already completed in a Clinical Laboratory Improvement Amendments (CLIA)–approved laboratory for routine care. MSI testing was performed in a CLIA–approved lab at OSU using the Promega MSI Analysis System (version 1.2), which includes five mononucleotide repeat markers (BAT-25, BAT-26, NR-21, NR-24, and MONO-27), with ≥ 2 of 5 unstable markers classified as MSI-high (MSI-H), 1 of 5 unstable markers classified as MSI-low (MSI-L), and 0 of 5 unstable markers classified as microsatellite stable. Immunohistochemistry of the MMR proteins was performed in a research lab at OSU using the two-stain method as previously described.22 Staining for all four MMR proteins was done as routine care for some patients and attempted for all study participants if MSI could not be performed or if the MSI and two-stain IHC results were discordant. Antibodies included MLH-1 Clone: Leica ES05 (Mouse: NCL-L-MLH1), MSH-2 Clone: Calbiochem FE11 (Mouse: NA27), MSH-6 Clone: Epitomics EP49 (Rabbit: AC-0047), and PMS-2 Clone: BD Pharmingen A16-4 (Mouse: 556415). Proteins with convincing stain in > 1% of cells were considered present. Equivocal and weak stains were treated as present. Hypermethylation of the MLH1 promoter was assessed using pyrosequencing23 at four CpG sites when tumors were MSI-H and/or absent MLH1 and PMS2 proteins on IHC if not already done for routine care, with ≥ 15% methylation classified as hypermethylated.
Genetic Testing
Patients meeting selection criteria underwent germline MGPT for 25-66 cancer genes (Data Supplement). Those with MMR deficiency without MLH1 hypermethylation had either ColoSeq (January 1, 2013-July 31, 2016) or BROCA (August 1, 2016-December 31, 2016) MGPT through the University of Washington's Genetics and Solid Tumor Laboratory using methods previously described.24,25 Those with unexplained MMR deficiency underwent tumor sequencing of the MMR genes with ColoSeqTumor including loss of heterozygosity (LOH) analysis as previously described.26 Those with MMR-proficient tumors or MLH1-hypermethylated tumors meeting clinical inclusion criteria underwent myRisk MGPT through Myriad Genetics Inc using methods previously described.27 Clinical inclusion criteria were CRC diagnosed under age 50 years, a personal history of synchronous or metachronous CRC and/or endometrial cancer (EC), or a family history of a first-degree relative with CRC or EC. Patients with MLH1 hypermethylation in their tumor were assessed for constitutional MLH1 hypermethylation at OSU using extracted DNA from blood by pyrosequencing if they were diagnosed under age 50 and/or reported multiple LS-associated tumors.
Genetic counseling was provided to all patients with a PGV, and genetic counseling or testing was offered to all at-risk relatives of those with LS. Counseling was provided in-person or via telehealth by a study genetic counselor or Informed DNA. Relatives were enrolled for an additional year (through December 31, 2017) to ensure that all families had sufficient opportunity for cascade testing.
Classification of Mutations
The variant interpretation system for Myriad Genetics Inc and the University of Washington has been previously described.11,15,17,18,28-32 For tumor sequencing, cases were considered double somatic if two pathogenic or likely pathogenic somatic variants were identified or if one pathogenic or likely pathogenic somatic variant was identified with LOH. For patients with MMR-deficient tumors and a germline MMR variant of uncertain significance (VUS), tumors were assessed for additional MMR mutations or LOH to attempt to clarify the pathogenicity of the variant. Variants were reclassified as likely pathogenic when the tumor screening results supported pathogenicity and one additional pathogenic mutation was identified in the tumor using methods previously described.15
Statistics
Descriptive statistics were provided. Sensitivity and specificity for MSI and IHC were calculated using standard methods from all completed tests. Positive predictive values and negative predictive values were calculated using population prevalence via MEDCALC statistical software. Wilson score intervals with continuity correction were used to compute CIs.
RESULTS
Patients
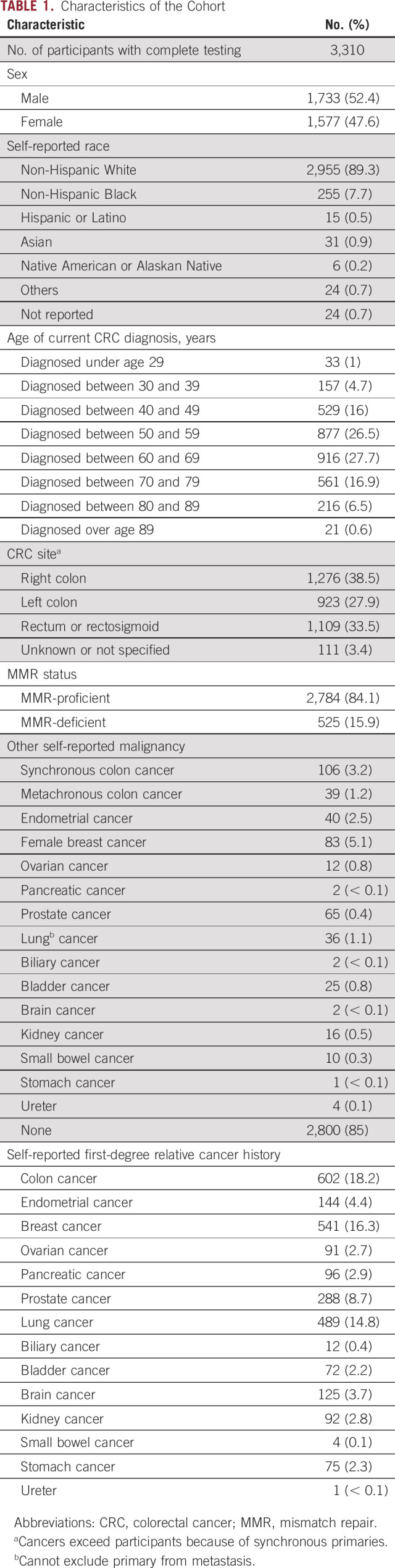
Patient characteristics are presented in Table 1. The mean age of diagnosis was 60 years (range, 17-89 years), and participants were 52% male. Self-reported race was consistent with that of the state of Ohio: 89% White, 8% Black, 2% Other, and 1% did not report. Clinical stage of the CRC was not ascertained. Fifteen percent had an additional malignancy. The most common cancers reported in first-degree relatives were colon (18.2%), breast (16.3%), and lung (14.8%).
TABLE 1.
Characteristics of the Cohort
Overall Results
See Figure 1 for study schema and overall results. Patient characteristics and family history are provided in the Data Supplement for those with PGVs.
FIG 1.
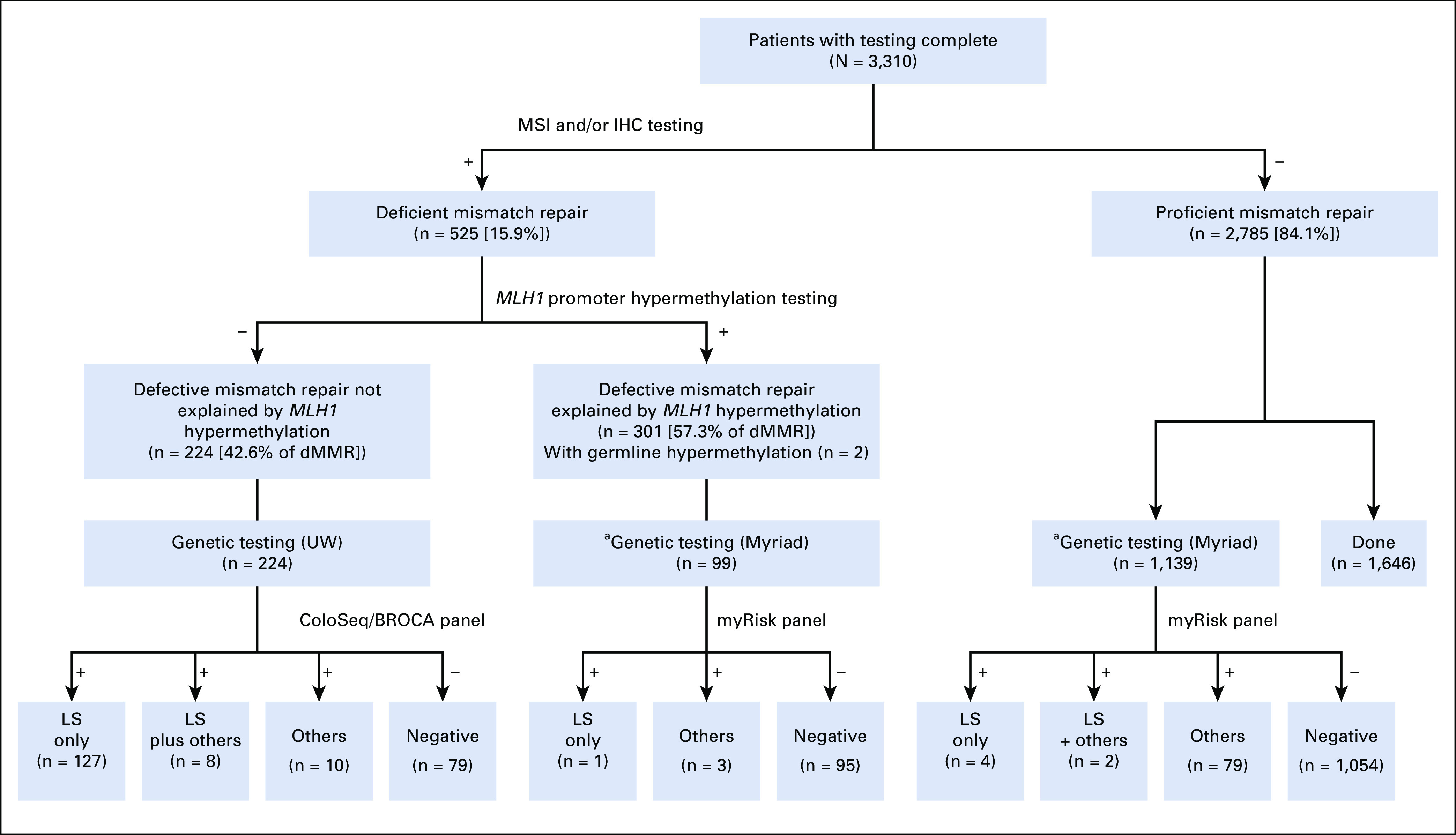
Study schema. aOnly patients who met the following clinical inclusion criteria: (1) diagnosed with CRC under age 50 or (2) personal history of synchronous or metachronous CRC or EC or (3) reported first-degree relative with CRC or EC. CRC, colorectal cancer; dMMR, deficient mismatch repair; EC, endometrial cancer; IHC, immunohistochemistry; LS, Lynch syndrome; MSI, microsatellite instability; UW, University of Washington.
Tumor Screening for MMR Deficiency
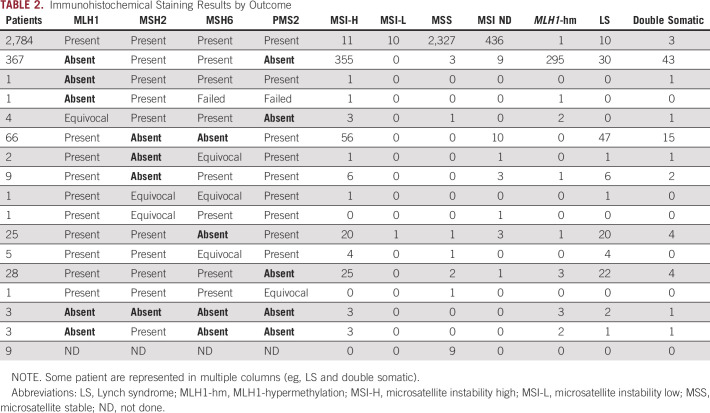
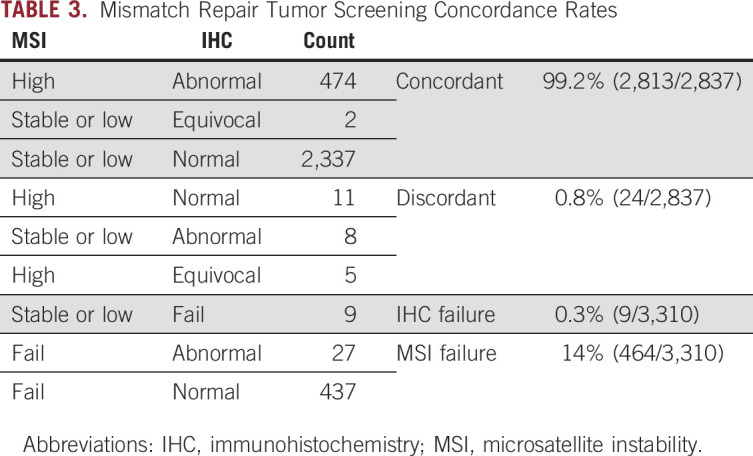
See Table 2 for tumor screening results. Overall, 15.9% (525 of 3,310) had an MMR-deficient tumor(s). Of those who received MSI testing, 17.2% (490 of 2,846) were MSI-H, 0.4% (11 of 2,846) were MSI-L, and 82.4% (2,345 of 2,846) were microsatellite stable. Fourteen percent (464 of 3,310) did not have sufficient tumor or normal DNA for MSI testing. Of those who received IHC testing, 15.4% (509 of 3,301) had abnormal staining. Nine patients (0.3%) did not receive IHC testing. Twenty-four cases (0.8%) had discordant tumor screening (Table 3). Of cases with absent MLH1/PMS2, 80.3% (301 of 375) had MLH1 hypermethylation. Eight of those had insufficient tumor for hypermethylation but had a BRAF mutation so MLH1 hypermethylation was inferred.
TABLE 2.
Immunohistochemical Staining Results by Outcome
TABLE 3.
Mismatch Repair Tumor Screening Concordance Rates
Constitutional MLH1 Hypermethylation
Two of 58 patients (3.5%) meeting criteria for germline hypermethylation analysis were found to have constitutional MLH1 hypermethylation of normal tissue (one diagnosed under age 50 and one with two LS-associated tumors). They are included among the patients with LS (n = 144), but not those with a PGV in an MMR gene (n = 142).
Germline Genetic Testing
Of the entire cohort, 1,498 patients met criteria for germline testing. Testing was completed for 1,462 patients. Two hundred forty-eight PGVs or likely PGVs in cancer susceptibility genes were identified in 234 patients (16% of those tested [95% CI, 14.2 to 18.0] and 7.1% of the overall cohort [95% CI, 6.2 to 8.0]). The spectrum of PGVs is shown in Figures 2 and 3. One hundred forty-two patients (60.7% of those with a PGV [95% CI, 54.1 to 66.9], 9.7% of those tested [95% CI, 8.3 to 11.4], and 4.3% of the overall cohort [95% CI, 3.6 to 5.1]) had a PGV in an MMR gene (54 MSH2, 30 MSH6, 29 MLH1, 28 PMS2, and one EPCAM). One hundred one patients (43.2% of those with a PGV [95% CI, 36.7 to 49.8], 6.9% of those tested [95% CI, 5.7 to 8.4], and 3.1% of the overall cohort [95% CI, 2.5 to 3.7]) had a PGV in a non-MMR gene (31 MUTYH [monoallelic], 11 APC, 10 ATM, 10 CHEK2, eight MUTYH [biallelic], six APC p.I1307K, five BRCA2, four BRIP1, four PALB2, three BRCA1, three CDKN2A, two NBN, two NTHL1 [monoallelic], one BMPR1A, one GALNT12, one FANCM, one POT1, one RAD51D, one RPS20, and one SMAD4), including four with two PGVs (one ATM/BRIP1, one ATM/NBN, one ATM/CHEK2, and one RAD51D/MUTYH [monoallelic]). Ten with LS had two PGVs (three MSH2/CHEK2, three MSH2/MUTYH [monoallelic], one MSH2/FANCM, one MSH2/NTHL1 [monoallelic], one MSH6/PALB2, and one PMS2/APC) and were included among those with MMR PGVs and non-MMR PGVs. In the overall cohort, 15.9% (114 of 719 [95% CI, 13.3 to 18.8]) of patients diagnosed under age 50 had a PGV, consistent with our previously published data, which included a subset of these cases.11
FIG 2.
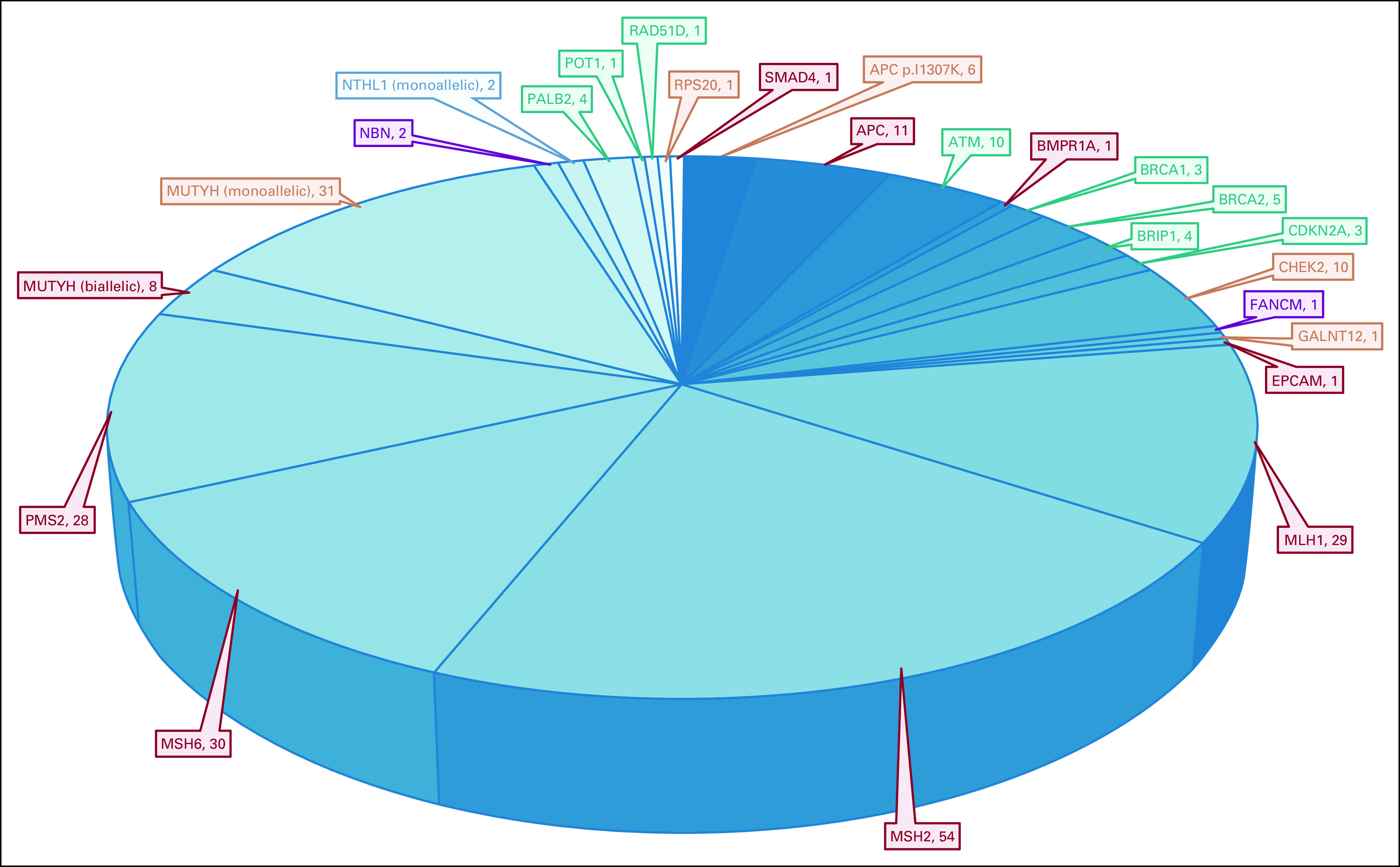
Overall spectrum and penetrance of pathogenic variants detected. Penetrance key: Red: high-risk cancer gene associated with colon cancer with or without other cancers. Orange: moderate- or low-risk cancer gene associated with colon cancer with or without other cancers. Green: high- or moderate-risk cancer gene not typically associated with colon cancer. Purple: low-risk cancer gene not typically associated with colon cancer. Blue: no increased cancer risk for monoallelic carriers.
FIG 3.
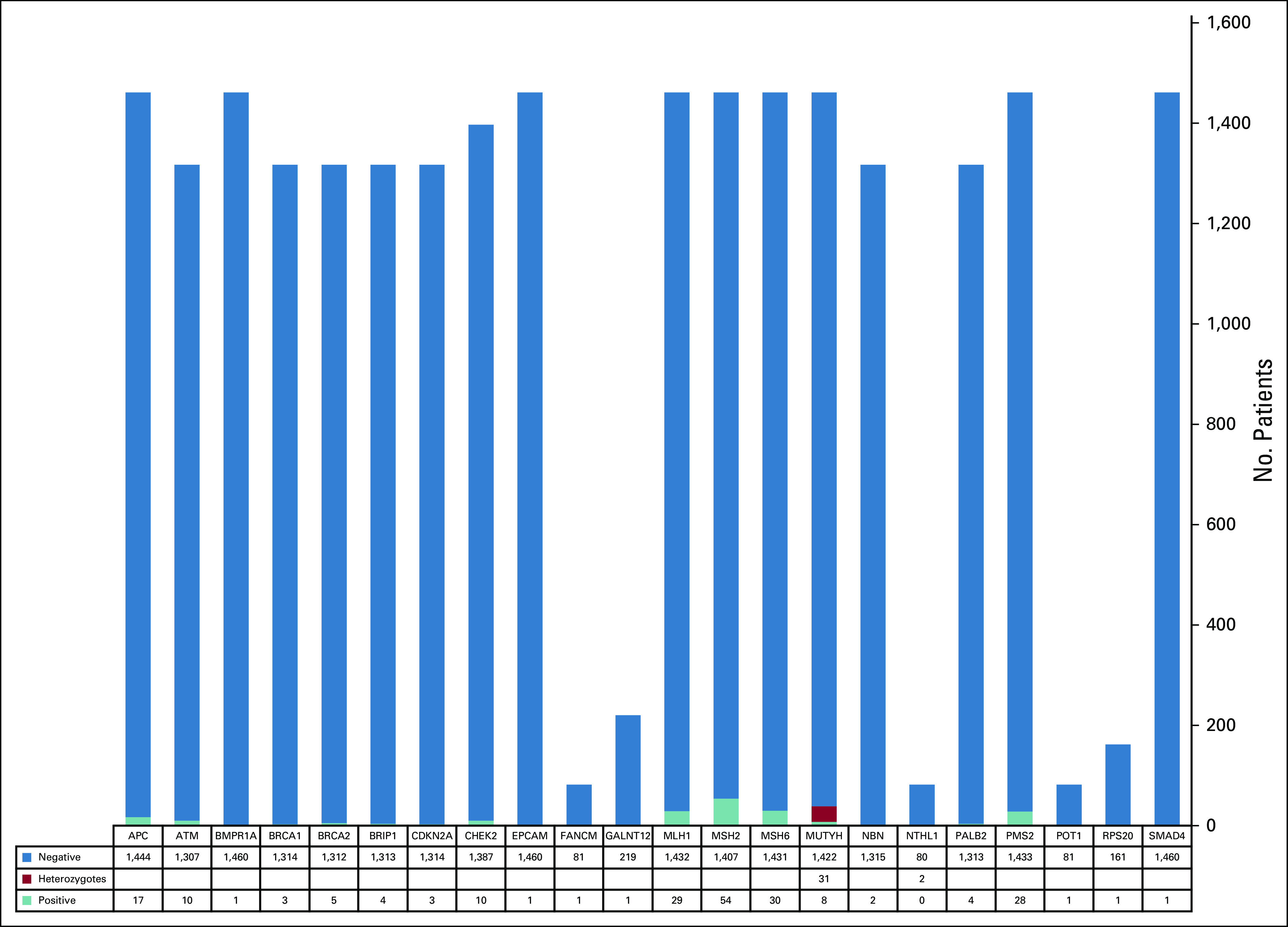
Pathogenic germline variants detected per gene based on the number of patients tested for that gene.
Of patients with MMR-deficient tumors not explained by MLH1 hypermethylation, 64.7% (145 of 224) had at least one PGV (93.1% [135 of 145] in the MMR genes). Of those with MMR-proficient tumors who underwent germline testing, 7.5% (85 of 1,139) had at least one PGV (95.3% [81 of 85] in non-MMR genes). Ninety-nine MLH1-hypermethylated cases underwent germline testing, and four (4%) had a PGV (one PMS2, one PALB2, and two MUTYH [monoallelic]).
Sensitivity and Specificity of MSI/IHC for LS Detection
Of those who found to have LS, 95.8% (138 of 144) had an MMR-deficient tumor. For MSI alone, the sensitivity was 92.9% (95% CI, 86.8 to 96.7), the specificity was 86.3% (95% CI, 84.9 to 87.6), the positive predictive value was 23.9% (95% CI, 22 to 25.9), and the negative predictive value was 99.6% (95% CI, 99.3 to 99.8). When equivocal stains were interpreted as intact, the sensitivity of IHC alone was 88.9% (95% CI, 82.6.8 to 93.5), the specificity was 87.9% (95% CI, 86.8 to 89.1), the positive predictive value was 25.2% (95% CI, 23.1 to 27.3), and the negative predictive value was 99.4% (95% CI, 99.1 to 99.6). When equivocal stains were interpreted as abnormal, the sensitivity of IHC alone was 92.4% (95% CI, 86.7 to 96.1), the specificity was 87.8% (95% CI, 86.7 to 89), the positive predictive value was 25.8% (95% CI, 23.8 to 27.8), and the negative predictive value was 99.6% (95% CI, 99.3 to 99.8).
MMR Tumor Sequencing
Ninety-one patients had unexplained MMR deficiency (including two with LS who also had the absence of another MMR protein unrelated to their MMR PGV). Eighty-six patients had sufficient tumor for sequencing, and 88.4% (76 of 86) were found to have double somatic MMR mutations. Fifteen patients' MMR-deficient tumor(s) remain unexplained including the five with insufficient tumor for sequencing. Clinical characteristics and tumor sequencing results for the patients with double somatic mutations and unexplained tumors were previously published.17
Variants of Uncertain Significance
Five hundred twenty-eight (433 unique) VUSs were found in 28.9% (422 of 1,462) of patients who underwent germline testing (Data Supplement). The most common genes with VUS were ATM and APC. One hundred forty-four patients with 102 unique variants (23.6%; 144 of 422) had their VUS reclassified between 2013 and 2020. Fifteen patients had 13 unique variants that were upgraded to likely pathogenic or pathogenic (3% [13 of 433] of all variants and 10.4% [15 of 144] of all patients with reclassified variants), and 129 patients had 87 unique variants that were downgraded to likely benign or benign (20.1% [87 of 433] of all variants and 89.6% [129 of 144] of all patients with reclassified variants). Three patients with MMR-deficient tumors have a VUS in an MMR gene that could be pathogenic (Data Supplement).
Cascade Genetic Testing
Of the 144 LS probands from 142 families, 92 (64.7%) had at least one relative participate in cascade testing. In total, 596 relatives underwent genetic counseling and testing and 223 had LS as follows: 105 of 242 (43.4%) first-degree relatives, 46 of 120 (38.3%) second-degree relatives, and 72 of 234 (30.7%) third-degree relatives and beyond. Lack of participation in cascade testing was accounted for by the following: proband unable (deceased or adopted) or unwilling to disclose the results to relatives, proband refused genetic counseling or genetic test result, and family declined testing.
Of the 144 patients with LS, 142 (98.6%) did not know that they had LS before they were diagnosed with cancer. However, 19 (13.3%) of these patients had a family member with a known diagnosis of LS before they were diagnosed with cancer, and their cancers were potentially preventable. It is unclear how many of them were aware of the LS diagnosis in their family and had chosen not to be tested or were unaware of the prior diagnosis in their family.
DISCUSSION
To our knowledge, this is the largest study of UTS to date, including 3,310 patients with CRC from 51 hospitals throughout Ohio. Although the sensitivity of MSI (92.9%) was slightly higher than that of IHC (88.9%-92.4%) for identifying LS, MSI failed more often (14%, 464 of 3,310) than IHC (0.3%, 9 of 3,310) and concordance between the two tests was high (99.1%). Tumors with equivocal IHC stains were more likely to be MSI-H (9 of 14), and many were in patients with LS (6 of 14). Therefore, we recommend that equivocal stains should be treated as abnormal or at least prompt the addition of MSI to help clarify the meaning. Overall, we prefer IHC to MSI since it can be performed in any pathology department, does not require a molecular laboratory, uses less tumor, and indicates which gene is nonfunctioning.
This study indicates that minimally, 1 of 14 (7.1%, 234 of 3,310) patients with CRC has at least one PGV in a cancer predisposition gene. Identification of a PGV provides the opportunity to prevent future cancers and sometimes allows for targeted treatment for their current cancer (PARP inhibitor or immunotherapy).10,32 Importantly, 5.3% (175 of 3,310) of patients had PGVs in genes with therapeutic targets: 4.1% (136 of 3,310) MMR PGVs and 1.2% (39 of 3,310) homologous recombination–deficient PGVs. It also provides information for family members by facilitating cascade genetic testing that can lead to life-saving surveillance and risk-reducing surgeries.
Although UTS is still important for identifying immunotherapy eligibility, it is insufficient for identifying the majority of CRC patients with hereditary syndromes, including some with LS. Had UTS been the only method used to screen patients with CRC for germline assessment, 38.6% (91 of 236) with a PGV in a cancer susceptibility gene or constitutional hypermethylation would have been missed, including 6.3% (9 of 144) of patients with LS; six had an MMR-proficient tumor (four PMS2 and two MSH6), two had constitutional MLH1 hypermethylation, and one was absent MLH1/PMS2 with MLH1 hypermethylation (PMS2). We recommend consideration of germline hypermethylation on all MLH1-hypermethylated patients under the age of 50 and those with a history of more than one LS-associated tumor given that 3.5% were found to have constitutional MLH1 hypermethylation. Additionally, the use of broad pan-cancer MGPT instead of testing for only MMR genes is important even among the patients with MMR deficiency as 8% (18/224) with a nonhypermethylated MMR-deficient tumor were found to have a PGV in a non-MMR gene (10 in addition to LS).
LS was more common (4.4%, 144 of 3,310) than previously reported.1-3 This difference may be the result of ascertainment bias as some participating hospitals provided clinical UTS and might have made extra efforts to enroll patients with abnormal results. Constitutional MLH1 hypermethylation is not a standard part of UTS so the two patients who were identified with this might have been missed in a clinical setting. Additionally, we had an excess of patients diagnosed under age 50. Since hereditary cancer syndromes are more common in younger patients with CRC, selection for younger patients might have also elevated the number of individuals with any hereditary cancer syndrome. As enrollment primarily occurred in oncology clinics and clinical stage was not collected, it is possible that there was enrichment for metastatic cases.
This study confirms that tumor testing for MMR genes is beneficial for patients with unexplained MMR deficiency. Eighty-eight percent of unexplained MMR-deficient tumors were found to have double somatic MMR mutations, the highest frequency reported to date. This is likely due to the tumor screening being centralized for quality control, limiting incorrect tumor screening results. Additionally, germline testing for MMR genes has improved, making it less likely to miss a germline mutation. Identifying double somatic MMR mutations that explain the MMR deficiency provides evidence that the individual's CRC was sporadic and they can be managed based on their personal and family history.33
Our findings of 7.1% prevalence of PGV are an underestimate of the true prevalence of PGV among all patients with CRC since 1,848 individuals in this cohort did not undergo germline testing. In the selected patients with CRC tested, 16% had a PGV. Additionally, if CHEK2 p.I157T was included as a PGV, this would add 13 cases to the mutation frequency (increasing our total to 7.5%). However, it was not included as a PGV because Myriad classifies it as a VUS, whereas most other laboratories classify it as a low-penetrance PGV. Our protocol of testing selected patients and number of genes tested likely accounts for the difference in our overall prevalence of mutations compared with two other similar studies (7.1% identified in our study versus 9.9%4 identified by testing all CRC patients with a similar-sized MGPT versus 15.3%13 identified by testing advanced-stage patients with a larger MGPT).
Minimally, patients with CRC should be referred for genetic counseling when they are diagnosed under age 50, have a personal history of multiple primaries, a family history of CRC or EC, or an MMR-deficient tumor. However, we believe that germline MGPT should be offered to all patients with CRC since 7.1%-15.3% have a PGV in a cancer susceptibility gene. This is similar to the 8.2%34 PGV rate found in patients with pancreatic cancer and 11.8%35 PGV rate found in patients with metastatic prostate cancer for whom the National Comprehensive Cancer Network recommends germline MGPT.36
ACKNOWLEDGMENT
This work is dedicated to the late Dr Albert de la Chapelle, whose passion and scholarly mind contributed to the identification of microsatellite instability and some of the MMR genes, among many other novel discoveries. His work undoubtedly saved the lives of countless individuals with Lynch syndrome. He will be greatly missed and fondly remembered. The authors are grateful to the patients and families included in this manuscript and the OCCPI study group (https://cancer.osu.edu/research-and-education/pelotonia-funded-research/statewide-colon-cancer-initiative/occpi-work-group). The authors also thank OSU past and present undergraduate student interns Angela Onorato, James Miner, Jessica Purnell, Emily McDowell, Hannah Datz, Chloe Kent, and Meghan Bartos, who assisted with database entry, as well as past OSU graduate student Abigail Schaber who assisted with database entry and pedigree analysis. We would like to acknowledge the Biospecimen Services Shared Resource, Informed DNA, the Columbus and Dayton NCORPs for assistance with accrual, the Cancer Center Support Grant at the Ohio State University Comprehensive Cancer Center (P30 CA016058), Myriad Genetics, and all the staff in the University of Washington Genetics and Solid Tumors Laboratory who assisted with sequencing studies.
APPENDIX 1. Group Information
A full list of the OCCPI study group members can be found at https://cancer.osu.edu/research-and-education/pelotonia-funded-research/statewide-colon-cancer-initiative/occpi-work-group
Below are members of the Ohio Colorectal Cancer Prevention Initiative study group.
Adena Regional Medical Center (Jeffrey VanDeusen, Linda Kight, Jeyanthi Ramanaratanan, Waheed Gul, Ganapathy Krishnan, Jaswant Madhavan, John Miller, Zion Oshikanlu, Herbert Sinning)
Akron General Medical Center (Esther Rehmus, Cathy Farmer, Debbie Thomas, Michael McNeal, Scott Awender, Walter Chlysta, Jason Fried, Mark Horattas, Kathryn Leininger, Kevin Lu, Robert Marley, Laurie Matt-Amaral, Scott McGee, Osei-Tutu Owusu, William Papouras, John Petrus, Sonia Sandhu)
Atrium Medical Center (Albert Malcolm, Sandy Fletcher, Caitlin Conaway, Ronald Hale, Nkeiruka Okoye, Radhika Rajsheker, Mridula Reddy, Cheryl Skinner, Ryan Steinmetz, Nandagopal Vrindavanam)
Aultman Hospital (Kisa Weeman, Kristin Shine, Janet Moore, Kathleen Collins, Donna Cobedesh, Juli Grove, Jen McCutchan, Steven Albertson, Nicholas Bisconti, Ferdinando Cortese, Michael Gurney, Amir Iqbal, John Jakob, Steven Kelly, Raza Khan, Sawjiv Khetarpal, Brandis Lewis, Warren Kofol, Khalil Korkor, Paul Manuszak, Alan Meshekow, Norman Rarique, Joseph Saddey, James Schmotzer, Mona Shay, Sabrina Shilad, Richard Sternjholm, Shruti Trehan, Michael Van Ness, Sunitha Vemulapalli)
Blanchard Valley Health System (Sharon Cole, Shannon Kohls, Brianne Hottinger, Christine Montgomery, Eric Browning, Ihsan Haq, Geetika Kumar, Chaoyang Li, Mohammad Mabayed, Britt Olmsted, Kevin Shannon, Thomas Stringle, Douglas Yoder)
Children's Hospital Medical Center of Akron (Catherine Ward-Melver, Julie D'attoma, Marcie Parker, Jennifer Stein, Lance Grau)
Cincinnati OHC—Mercy West, Mercy Fairfield, Mercy Anderson, Mercy Clermont, The Jewish Hospital (Lynette Hart, Brittany Hagen, Nicole Given, Sheena Chandler, Kim Brockman, Lindsay Vogel, Kimmy Miller, Alessa Hubbell, Susan Hendricks, Laura Kokenge, Brittany Gardner, Michelle Nguyen, Lisa Schmid, Dawn Abshire, Kyla Scott, Ashley Vollmer, Randy Drosick, Mark Johns, Peter Ruehlman, John Bismayer, Cynthia Chua, Edward Crane, Colleen Darnell, Patrick Ward, David Waterhouse, Paula Weisenberger, Karyn Dyehouse, Kurt Leuenberger, Suzanne Partridge)
Cleveland Clinic—Main Campus, Hillcrest, Fairview, South Pointe, Independence/Strongsville (Matthew Kalady, Timothy Spiro, Herman Kessler, Brandie Leach, Jessica Marquard, Carla Greenwood, Traci Stafford, Cathy Hugney, Samantha Kopack, Michelle Parson, Patti O-Reilly, Kathy Smolenski, Cathy Schilero, Thomas Plesec, Kimberly McDonald, Mary Oldenburgh, Donna McPeek, Cathy Schilero, Drew Abramovich, Mir Ali, Jean Ashburn, Amit Bhatt, Ravi Chari, Ronald Charles, Aneel Chowdhary, Byron Coffman, James Church, Megan Costedio, Saurabh Das, Robert Debernardo, Conner Delaney, David Dietz, Donald Eicher, Charis Eng, Bassan Estfan, Emre Gorgun, Michael Grillis, Abdo Haddah, Emina Huang, Tracy Hull, David Hykes, Dinesh Khera, Alok Khorana, Mark Kyei, Paul Laffay, Richard Latuska, Ian Lavery, Jeremy Lipman, David Kiska, Vinit Makkar, James Malgieri, Paul Masci, Neha Mitra, Michael Menunaitis, Ila Tamaskar, Brian Murphy, Gokhan Ozuner, Robert Pelley, Vitaliy Pishchik, Feza Remzi, Warren Rose, Marc Shapiro, Davendra Sohal, Luca Stocchi, Scott Strong, Albert Tsang, Michael Valente, Albert Vargas, Ravi Verma, Rich Wieseck, Hamed Daw, Bachar Dergham, Rick Gemma, David Gottesman, Kevin Kerwin, Michael Springer, Jason Collweiler, Ryan Williams, Samir Abrasksia)
Columbus NCORP (J. Philip Kuebler, Sheree Oxley, Julie O'Brian, Ed Perigo)
Dayton Clinical Oncology Program (Howard Gross, Mary Ontko, Karen Dickerson, Michele Hamann)
Doctors Hospital (Sanjay Yadav, Janet Harvey, Michele Dillow)
Fairfield Medical Center (Lisa Stevens, David Hasl, Scott Johnson, Srinvas Kolli, David Robertson, Margaret Sawyer, Kanwalijit Singh, Jeffrey Yenchar)
Genesis Cancer Care Center (Shyamal Bastola, Karen Wickham, Annette Barr, Carrie Lee, Kristen Miller, Alicia Hunt, Terence Campbell, Robert Chess, Jan Elston, Adam Rothermel, Scott Wegner)
Good Samaritan Hospital—Dayton (Charles Bane, John Haluschak, Katherine Peyton, Eileen Flynn, Jennifer Neikamp, Minia Hellan, Shamim Jilani, Jhansi Koduri, Mark Marinella, James Sabiers, Laszlo Toth)
Grady Memorial Hospital (Raymond Fuller, Kelly Reynolds, Bonnie Baker, Vickie Maggard, Patti Dunn, Eileen Troutman, Michelle McKinney-Harris, Chaoyang Li, Christopher George, Kenneth Graffeo, Arun Kumas, Erin Macrae, Jerry Mitchell, Timothy Moore, William Schirmer, Michelle Wood)
Grant Medical Center (Shakir Sarwar, Laura Reebel, Juliana Hite, Angel Spencer, Angie Mullholland, Patti Dunn, Pedro Aguilar, Joshua Braverman, Sudhathi Chennuru, Mark Jump, Munit Kapoor, Brandon Machicao, Timothy Moore, Anitha Nallari, Kristin Ryan, Dale Thomas, John Tzagournis, Michael Tzagournis)
Kettering Medical Center (Manisha Nanda, Judy Bair, Krista Bensman, Gary Anderson, Alejandro Calvo, Gregory Gordon, Minia Hellan, Priya Jain, Amanda Laubemthal, Joseph Lavelle, Stuart Merl, Malek Safa, Carol Sawmiller, Emily Vannorsdall, Stephanie Hartke)
Knox Community Hospital (Husain Rasheed, Katy Breeze, Katy Breeze, Bethany Lee, Diana Endsley, Michael Heuman, Hypi Laidlaw-Smith, Husain Rasheed, Paul Taiganides)
Licking Memorial Hospital (Arunda Gowda, Kenita Keck, Suzanne Posey, Michelle Majoy, Jacqueline Jones, D'Anna Mullins)
Marion General Hospital (Arvinder Bhinder, Eileen Troutman, Arun Kumar, Prasad Maturu, John Mcdonough, Prasad Ravi, Jose Vale)
Marietta Memorial Hospital (Kelli Cawley, Joanna Lupardus, Sheryl Corp, Utpal Bhanja, Rajendra Bhati, David Blevins, Bradley Carmen, Anirban Gupta, Matthew Matacol, Allen Mcelroy, Tuan Nguyen, M. Nill, Shinoj Pattali, Devaki Siva)
MedCentral Mansfield Hospital (Katie Stafford, Srividya Viswanathan, Robert Exten)
Mercy Medical Center—Canton (Mitchell Haut, Joan Edwards, Linda Sims, Jane Westfall, Janet Muckley, Tara Barker, Kimberly Mayle, Brooks Bolyard, Ferdinando Cortese, Khatib Jafri, Hanspreet Kaur, Scott McGee, Nagaprasad Nagajothi, Francisco Paras, Noman Rafique, Russell Ramey, Aida Safar, Mazin Shackour, Kirby Sweitzer, Steven Albertson, Steven Kelly, David Linz, Nicholas Roberts, Carol Baker, Munir Ahmad, Farouq Ahmed, Mohamad Ayoubi, Gregory Boone, Greg Manson)
MetroHealth (Joanna M. Brell, Deborah Strater, Amy Shaper, Taryn Ferber, Edward Crum, Natalie Joseph, Jeremy Lipman, Timothy O'Brien, Diane Wolf, Larisa Schwartzman, Melissa Times, Michelle Treasure, Joan Trey)
Miami Valley Hospital (Mark Marinella, M. Jane Boerger, Darinda Reis, Jenny Dillon, Ellen Cato, Minia Hallan, John Haluschak, Satheesh Kathula, Jhansi Koduri, James Ouellette, Mridula Reddy, Steve Rudich Manish Sheth, Laszlo Toth)
Mount Carmel Health System—East, West, St Ann's (Karamjit Khanduja, Mark Knapp, Mark Segal, Mark Thompson, Jenna Knoble, Taral Patel, Rosemary Zacks, Sue DeVictor, Grace Glace, Peggy Lyons, Jeffrey Zangmeister, Ellen Bailey, Glen Borchers, Tarek Chidiac, Mark Devanzo, Richard Edgin, Patrick Elwood, William Emlich, Kim Hammelberg, Jodi Lee, Seth Hoffman, Bruce Kerner, Mark Lindsey, Jaswant Madhavan, Neil Makadia, Marcus Miller, Jerry Mitchell, Timothy Moore, Ananthan Padmanabhan, Phillip Price, Jeffrey Sams, P. Kothai Sundaram, Charles Taylor, Joshua Braveman, Mark Stechschult, Mary Dillhoff, Samer Eldika, Adewale Fawole, Melinda Jack, Tasos Madhaven)
Myriad Genetics Inc (Brian Allen, Shelly Cummings)
OSUMC—Colorectal (Heather Hampel, Albert de la Chapelle, Richard Goldberg, Electra Paskett, Peter Shields, Rachel Pearlman, Wendy Frankel, Wei Chen, Benjamin Swanson, Weiqiang Zhao, Ahmet Yilmaz, Kristin Miller, Jason Bacher, Christopher Bigley, Lori Nelsen, Michael Bigley, Thomas Prior, Daniel Jones, Debbie Knight. Cheryl Reeder, Amy Glaze, Peter Stanich, Ilene Lattimer, Thelma Asare, Mark Arnold, Sandya Liyanarachchi, Christina Wu, Anne Noonan, Tony Saab, Sigurdis Haraldsdottir, Sherif Abdel-Misih, Mark Bloomston, Kristen Ciombor, William Cirocco, Cassandra Grenade, Alan Harzman, John Hays, John Howard, Syed Husain, Sameh Mikhail, Kate Shane-Carson, Anterpreet Neki, Timothy Pawlik, Sameek Roychowdhury, Robert Rupert, Carl Schmidt, Jennifer Sipos, Judith Westman)
OSUMC—Endometrial (Paul Goodfellow, Elizabeth Solinger, Molly Myers, David Cohn, David O'Malley, Adrian Suarez, Ritu Salani, Floor Backes, Larry Copeland, Jeff Fowler, Casey Cosgrove)
ProMedica Health System—Flower and Toledo Hospital (Terry Gibbs, Carissa Jock, Kelly Morse Joyce Vernier, Julie Moon, Sarah Adelsperger, Sue Mayer, Abed Alo, Sanjiv Bais, Paul Bosio, Stephanie Cole, Mark Gretsinger, Douglas Hess, Peter Klein, Michael Mcphee, Jose Parodi, Brad Sachs, Joseph Sferra, Eilynn Sipe, John Stengle, Truman Wigand)
St Luke's Hospital (participation ended December 31, 2015) (Ginger Schwyn, Abed Alo, Peter Klein, Douglas Lindsey, Asish Mukherjee, Raju Shah, Eilynn Sipe, Beth White)
Riverside Methodist Hospital (J. Philip Kuebler, Kelly Reynolds, Megan Sisson, Kristen Johnson, Amanda Adams, Portia Schiming, Patti Dunn, Sonia Abuzakhm, Jeffrey Archer, Brent Behrens, Scott Blair, Stephanie Dunkle-Blatter, Christopher George, Andrew Grainger, Joseph Hofmeister, Peter Kourlas, Peter Lee, John Leff, Erin Macrae, Nse Ntukidem, Kwang Suh, Thomas Sweeney, Robert Toscano, William Wise)
Southern Ohio Medical Center (Thomas Summers, Yinong Liu, Jamie Arnett, Murielle Brohez, Bruce Kerner, Thomas Khourg)
Springfield Regional Medical Center (Ravi Khanna, Daljeet Singh, Linda Blosser, Chaundra Foss, Zaw Bo, Filix Kencana, Sandra Victor)
St Rita's Medical Center (Chris Rhoades, Clarissa Alford, Lois Gerding, Charles Brunelle, Richard Capone, Henry Gerad, Todd Hixenbaugh, Paul Kalogerou, Chethana Kanapathi, Ewa Mrozek, Robert Neidich, David Powell, Tariq Sheikh, Abdulla Taja)
Summa Health System—Akron City and St Thomas, Barberton, Robinson Memorial, Western Reserve (Sameer Mahesh, Nicole Buie, Annie Papik, Debbie Neal, Ally Kovach, Sarah Stanaszek, John Gusz, Lynn Wojtasik, Mark Arrendondo, Elizabeth Bender, Leo Claecilla, Bradley Clifford, Michael Cullado, Arthur Dalton, Adrian Dan, Anad Desai, Edward Esber, Rachel Jenkins, Ann Stone, John Fondran, Susan Hong, John Jakob, Joseph Koenig, Erica Laipply, Melanie Lynch, Fredrick Seefeldlt, Dean Mayors, Leon Miller, Mehool Matel, Jennifer Payne, Joel Porter, Mark Pozsgay, Warren Rose, Frederick Slezak P. Torowski, Douglas Trochelman, Arjun Venkat, Darrell Widmer, Gary Williams, John Zografakis, Bradley Clifford, Andrew Haas, Dean Mayors)
The Christ Hospital (Ian Paquette, Janice Rafferty, Bradley Davis, Marla Prues, Pam Manfresca, Robert Cody, William Crafton, Randy Drokick, Jon Snider, Slobodon Stanisic)
Toledo Clinic Cancer Center (Pam Shoup, Jennifer Martinez, Jessica Ciesler, Sameh Almadani, Michael Bielefeld, Timothy Kasunic, Richard Phinney, Todd Tamlyn)
TriHealth—Bethesda North and Good Samaritan Cincinnati (David Draper, Scott Kelley, Carolyn Lindeman, Kathleen Bieniek, Antoinette Dean, Courtney Rice, Karen Huelsman, Megan Shearouse, Meghan Caldwell, Alex Sympson, Marc Alexander, Susan Alcasid, Mark Andolina, Kevin Bundy, Robert Bennett, Arcot Bhaskar, Ranga Brahmamdam, Catherine Brown, Kevin Budke, Elena Caoili, Jessica Cassady, Michael Caudy, Suhail Chaudhry, Cynthia Chua, Stephen Cleves, Francis Collins, Richard Dammel, Erik Dunki-Jacobs, James Fidelholtz, Steven Grendel, Michael Handleton, Lynn Gronbach, Douglas Hawley, Jason Hoke, Jacon Jones, David Kirkpatrick, Robert Kindle, Gennaro Labella, Steven Langdon, Jay Logeman, Michelle louis, Steven Lunz, Anjuli Mahajan, James Maher, Arturo Maldonado, Jennfier Marklay, Gina Matacia, Apurua Mehta, John Merling, Huxley Miller, Thomas Mueller, Gerald Palermo, Andrew Parchman, Alan Putrus, Janice Rafferty, Robert Rechtin, Raymond Reuss, Antonio Rojas, Mark Rudemiller, Paul Rupp, Alex Saba, Ronald Smith, Melanie Spaedy, Alan Tarshis, Kirubel Tefera, Christine Wallace, Cheryl Skinner, Christopher South, Fredrick Weber)
University at Buffalo (Jo Freudenheim)
University of Washington (Colin Pritchard, Brian Shirts, Angela Jacobson)
Upper Valley Medical Center (Rajeev Kulkarni, Heather Penwell, Minia Hellan, L. Lowry, Mohan Nuthakki)
Wayne Healthcare (Manish Sheth, Catherine Jill Brown, John Haluschak, Daniel Mckellar)
Wright-Patterson Medical Center (Roger Wood, Joyce Russo, Susannah Cooper, Brandon Cutler, Alyssa Mcmanamon, Gary Peitzmeier)
Wendy L. Frankel
Patents, Royalties, Other Intellectual Property: Patent title: Automated Identification of Tumor Buds. Application no.: 16/230,118. Filing date: December 21, 2018. Publication date: July 4, 2019. Applicant(s): Ohio State Innovation Foundation, Columbus, OH. Inventor(s): Metin Gurcan, Winston-Salem, NC; Wendy Frankel, Columbus, OH; Wei Chen, Columbus, OH; Ahmad Fauzi, Mohammad Faizal, Selangor, MY
Benjamin J. Swanson
Stock and Other Ownership Interests: Cogen Bioscience Inc
Consulting or Advisory Role: Cogen Bioscience Inc
Dan Jones
Research Funding: AbbVie, MingSight, Acerta Pharma, Pharmacyclics, Sunesis Pharmaceuticals, Sarah Cannon Research Institute
Patents, Royalties, Other Intellectual Property: SARS-CoV-2 Diagnostics, BTK Inhibitor Diagnostics Assays
Christopher Bigley
Employment: Vikor Scientific
Paul J. Goodfellow
Research Funding: Promega
Richard M. Goldberg
Stock and Other Ownership Interests: Advanced Chemotherapy Technologies
Honoraria: Amgen
Consulting or Advisory Role: Merck, Taiho Pharmaceutical, Novartis, AstraZeneca, Bayer
Expert Testimony: Taiho Pharmaceutical, Genentech/Roche
Travel, Accommodations, Expenses: Merck KGaA, Merck, Amgen
Electra Paskett
Stock and Other Ownership Interests: Pfizer, Meridian Bioscience Inc
Research Funding: Merck, Pfizer
Peter P. Stanich
Research Funding: Janssen, Pfizer, Emtora Biosciences, Freenome
Patents, Royalties, Other Intellectual Property: I am an author for UpToDate and receive royalties
Mark Arnold
Stock and Other Ownership Interests: Pfizer
Matthew F. Kalady
Consulting or Advisory Role: ACTIVARTIS Biotech
Research Funding: Freenome
Brandie Heald
Consulting or Advisory Role: InVitae
Sameer Mahesh
Stock and Other Ownership Interests: Pfizer, Merck, Moderna Therapeutics, Immunomedics, Exelixis, GTHX, Gilead Sciences
Colin C. Pritchard
Consulting or Advisory Role: Promega, AstraZeneca, Sana Biotechnology
Shelly A. Cummings
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Heather Hampel
Stock and Other Ownership Interests: Genome Medical
Consulting or Advisory Role: InVitae, Genome Medical, Promega, 23andMe
No other potential conflicts of interest were reported.
SUPPORT
The data reported here were derived from the Ohio Colorectal Cancer Prevention Initiative, supported by a grant from Pelotonia, an annual cycling event in Columbus OH, that supports cancer research at The Ohio State University Comprehensive Cancer Center (OSUCCC)—James Cancer Hospital and Solove Research Institute. This study was also supported in part by Grant No. P30 CA016058, National Cancer Institute, Bethesda, MD, and utilized the Biospecimen Services Shared Resource Biorepository of the OSUCCC. Myriad Genetics Laboratories donated germline next-generation sequencing testing for selected mismatch repair-proficient patients.
CLINICAL TRIAL INFORMATION
AUTHOR CONTRIBUTIONS
Conception and design: Rachel Pearlman, Dan Jones, Weiqiang Zhao, Jason Bacher, Richard M. Goldberg, Electra Paskett, Peter G. Shields, Jo L. Freudenheim, Ian Paquette, Albert de la Chapelle, Heather Hampel
Financial support: Peter G. Shields, Colin C. Pritchard
Administrative support: Rachel Pearlman, Colin C. Pritchard, Angela Jacobson
Provision of study materials or patients: Rachel Pearlman, Ahmet Yilmaz, Lori Nelsen, Ilene Lattimer, Mark Arnold, Mitchell Haut, David J. Draper, Kisa Weeman, Shyamal Bastola, Jeffrey Zangmeister, Aruna Gowda, Filix Kencana, Yinong Liu, Charles Bane, Esther Rehmus
Collection and assembly of data: Rachel Pearlman, Benjamin J. Swanson, Weiqiang Zhao, Ahmet Yilmaz, Kristin Miller, Jason Bacher, Christopher Bigley, Lori Nelsen, Richard M. Goldberg, Ilene Lattimer, Mark Arnold, Mitchell Haut, Brandie Heald, David J. Draper, Joanna M. Brell, Kisa Weeman, Shyamal Bastola, Jeffrey Zangmeister, Aruna Gowda, Albert Malcolm, Yinong Liu, Charles Bane, Chaoyang Li, Esther Rehmus, Colin C. Pritchard, Angela Jacobson, Heather Hampel
Data analysis and interpretation: Rachel Pearlman, Wendy L. Frankel, Dan Jones, Weiqiang Zhao, Ahmet Yilmaz, Paul J. Goodfellow, Richard M. Goldberg, Electra Paskett, Peter G. Shields, Jo L. Freudenheim, Peter P. Stanich, Ilene Lattimer, Thomas W. Prior, Matthew F. Kalady, Sameer Mahesh, Filix Kencana, Sharon Cole, Colin C. Pritchard, Brian H. Shirts, Angela Jacobson, Shelly A. Cummings, Albert de la Chapelle
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Wendy L. Frankel
Patents, Royalties, Other Intellectual Property: Patent title: Automated Identification of Tumor Buds. Application no.: 16/230,118. Filing date: December 21, 2018. Publication date: July 4, 2019. Applicant(s): Ohio State Innovation Foundation, Columbus, OH. Inventor(s): Metin Gurcan, Winston-Salem, NC; Wendy Frankel, Columbus, OH; Wei Chen, Columbus, OH; Ahmad Fauzi, Mohammad Faizal, Selangor, MY
Benjamin J. Swanson
Stock and Other Ownership Interests: Cogen Bioscience Inc
Consulting or Advisory Role: Cogen Bioscience Inc
Dan Jones
Research Funding: AbbVie, MingSight, Acerta Pharma, Pharmacyclics, Sunesis Pharmaceuticals, Sarah Cannon Research Institute
Patents, Royalties, Other Intellectual Property: SARS-CoV-2 Diagnostics, BTK Inhibitor Diagnostics Assays
Christopher Bigley
Employment: Vikor Scientific
Paul J. Goodfellow
Research Funding: Promega
Richard M. Goldberg
Stock and Other Ownership Interests: Advanced Chemotherapy Technologies
Honoraria: Amgen
Consulting or Advisory Role: Merck, Taiho Pharmaceutical, Novartis, AstraZeneca, Bayer
Expert Testimony: Taiho Pharmaceutical, Genentech/Roche
Travel, Accommodations, Expenses: Merck KGaA, Merck, Amgen
Electra Paskett
Stock and Other Ownership Interests: Pfizer, Meridian Bioscience Inc
Research Funding: Merck, Pfizer
Peter P. Stanich
Research Funding: Janssen, Pfizer, Emtora Biosciences, Freenome
Patents, Royalties, Other Intellectual Property: I am an author for UpToDate and receive royalties
Mark Arnold
Stock and Other Ownership Interests: Pfizer
Matthew F. Kalady
Consulting or Advisory Role: ACTIVARTIS Biotech
Research Funding: Freenome
Brandie Heald
Consulting or Advisory Role: InVitae
Sameer Mahesh
Stock and Other Ownership Interests: Pfizer, Merck, Moderna Therapeutics, Immunomedics, Exelixis, GTHX, Gilead Sciences
Colin C. Pritchard
Consulting or Advisory Role: Promega, AstraZeneca, Sana Biotechnology
Shelly A. Cummings
Employment: Myriad Genetics
Stock and Other Ownership Interests: Myriad Genetics
Travel, Accommodations, Expenses: Myriad Genetics
Heather Hampel
Stock and Other Ownership Interests: Genome Medical
Consulting or Advisory Role: InVitae, Genome Medical, Promega, 23andMe
No other potential conflicts of interest were reported.
REFERENCES
- 1.Hampel H, Frankel WL, Martin E, et al. : Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352:1851-1860, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, Frankel WL, Martin E, et al. : Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 26:5783-5788, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cragun D, DeBate RD, Vadaparampil ST, et al. : Comparing universal Lynch syndrome tumor-screening programs to evaluate associations between implementation strategies and patient follow-through. Genet Med 16:773-782, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yurgelun MB, Kulke MH, Fuchs CS, et al. : Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol 35:1086-1095, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palomaki GE, McClain MR, Melillo S, et al. : EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med 11:42-65, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giardiello FM, Allen JI, Axilbund JE, et al. : Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol 109:1159-1179, 2014 [DOI] [PubMed] [Google Scholar]
- 7.Beamer LC, Grant ML, Espenschied CR, et al. : Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol 30:1058-1063, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaikh T, Handorf EA, Meyer JE, et al. : Mismatch repair deficiency testing in patients with colorectal cancer and nonadherence to testing guidelines in young adults. JAMA Oncol 4:e173580, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noll A, Parekh PJ, Zhou M, et al. : Barriers to Lynch syndrome testing and preoperative result availability in early-onset colorectal cancer: A National Physician Survey Study. Clin Transl Gastroenterol 9:185, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, Uram JN, Wang H, et al. : PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372:2509-2520, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearlman R, Frankel WL, Swanson B, et al. : Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol 3:464-471, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espenschied CR, LaDuca H, Li S, et al. : Multigene panel testing provides a new perspective on Lynch syndrome. J Clin Oncol 35:2568-2575, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samadder NJ, Riegert-Johnson D, Boardman L, et al. : Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol 7:230-237, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckingham L, Flaws ML: Molecular Diagnostics Fundamentals, Methods, & Clinical Applications. Philadelphia, PA, F. A. Davis Company, 2007, pp 65-79 [Google Scholar]
- 15.Haraldsdottir S, Hampel H, Tomsic J, et al. : Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology 147:1308-1316.e1, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearlman R, Markow M, Knight D, et al. : Two-stain immunohistochemical screening for Lynch syndrome in colorectal cancer may fail to detect mismatch repair deficiency. Mod Pathol 31:1891-1900, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearlman R, Haraldsdottir S, de la Chapelle A, et al. : Clinical characteristics of patients with colorectal cancer with double somatic mismatch repair mutations compared with Lynch syndrome. J Med Genet 56:462-470, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampel H, Pearlman R, Beightol M, et al. : Assessment of tumor sequencing as a replacement for Lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol 4:806-813, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranola JMO, Pearlman R, Hampel H, et al. : Modified capture-recapture estimates of the number of families with Lynch syndrome in Central Ohio. Fam Cancer 18:67-73, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Pearlman R, Hampel H, et al. : MSH6 immunohistochemical heterogeneity in colorectal cancer: Comparative sequencing from different tumor areas. Hum Pathol 96:104-111, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W, Hampel H, Pearlman R, et al. : Unexpected expression of mismatch repair protein is more commonly seen with pathogenic missense than with other mutations in Lynch syndrome. Hum Pathol 103:34-41, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shia J, Tang LH, Vakiani E, et al. : Immunohistochemistry as first-line screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: A 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol 33:1639-1645, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Newton K, Jorgensen NM, Wallace AJ, et al. : Tumour MLH1 promoter region methylation testing is an effective prescreen for Lynch Syndrome (HNPCC). J Med Genet 51:789-796, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard CC, Smith C, Salipante SJ, et al. : ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn 14:357-366, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nord AS, Lee M, King MC, et al. : Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics 12:184, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirts BH, Konnick EQ, Upham S, et al. : Using somatic mutations from tumors to classify variants in mismatch repair genes. Am J Hum Genet 103:19-29, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Judkins T, Leclair B, Bowles K, et al. : Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC Cancer 15:215, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eggington JM, Bowles KR, Moyes K, et al. : A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet 86:229-237, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Esterling L, Wijayatunge R, Brown K, et al. : Impact of a cancer gene variant reclassification program over a 20-year period. JCO Precis Oncol doi: 10.1200/PO.20.00020 [DOI] [PMC free article] [PubMed]
- 30.Richards S, Aziz N, Bale S, et al. : Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405-424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirts BH, Casadei S, Jacobson AL, et al. : Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet Med 18:974-981, 2016 [DOI] [PubMed] [Google Scholar]
- 32.Goyal G, Fan T, Silberstein PT: Hereditary cancer syndromes: Utilizing DNA repair deficiency as therapeutic target. Fam Cancer 15:359-366, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines™). Genetic/Familial High-Risk Assessment: Colorectal (Version 1.2020). National Comprehensive Cancer Network Inc, 2020 [Google Scholar]
- 34.Hu C, Hart SN, Polley EC, et al. : Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 319:2401-2409, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritchard CC, Mateo J, Walsh MF, et al. : Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375:443-453, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines™). Genetic/Familial High-Risk Assessment: Breast, Ovarian and Pancreatic (Version 1.2021). National Comprehensive Cancer Network Inc, 2020 [DOI] [PubMed] [Google Scholar]