A point-of-care, microfluidic platform detects antibodies against three SARS-CoV-2 antigens from blood in less than an hour.
Abstract
Highly sensitive, specific, and point-of-care (POC) serological assays are an essential tool to manage coronavirus disease 2019 (COVID-19). Here, we report on a microfluidic POC test that can profile the antibody response against multiple severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens—spike S1 (S1), nucleocapsid (N), and the receptor binding domain (RBD)—simultaneously from 60 μl of blood, plasma, or serum. We assessed the levels of antibodies in plasma samples from 31 individuals (with longitudinal sampling) with severe COVID-19, 41 healthy individuals, and 18 individuals with seasonal coronavirus infections. This POC assay achieved high sensitivity and specificity, tracked seroconversion, and showed good concordance with a live virus microneutralization assay. We can also detect a prognostic biomarker of severity, IP-10 (interferon-γ–induced protein 10), on the same chip. Because our test requires minimal user intervention and is read by a handheld detector, it can be globally deployed to combat COVID-19.
INTRODUCTION
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic poses an enormous challenge to the world. SARS-CoV-2 has resulted in more than 100 million cases of coronavirus disease 2019 (COVID-19) worldwide, resulting in more than 2.3 million deaths as of 12 February 2020 (1). Unlike many other viruses, SARS-CoV-2 displays high infectivity, a large proportion of asymptomatic carriers, and a long incubation time of up to 12 days, during which carriers are infectious (2–4). As a result, transmission has been widespread, resulting in overwhelmed health care capacities across the globe (5, 6). Timely, reliable, and accurate diagnostic and surveillance tests are necessary to control the current outbreak and to prevent future spikes in transmission.
Reverse transcription polymerase chain reaction (RT-PCR), which detects viral nucleic acids, is the current gold standard for COVID-19 diagnosis (7, 8). Although RT-PCR is highly sensitive and specific (9, 10), it does not detect past infections—RNA is typically only present at high quantities during acute infection—and it does not provide insight into the host’s response to infection (11). Serological assays, which detect antibodies induced by SARS-CoV-2, are a crucial supplement to nucleic acid testing for COVID-19 management (12, 13). Specifically, serological assays are important to track the body’s immune response (14) and to potentially inform prognosis (15) or immunity status (12). Serological assays are also essential for use in epidemiological studies (16) and are a critical enabling tool for vaccine development (17).
SARS-CoV-2 is an enveloped RNA virus with four structural proteins: spike (S) protein, membrane (M) protein, enveloped (E) protein, and nucleocapsid (N) protein (18). As the pandemic unfolded, several serological binding assays were developed including enzyme-linked immunosorbent assays (ELISAs) and lateral flow assays (LFAs). These assays measure either the level of total antibody or that of specific antibody isotypes that bind to viral proteins—normally S or N. Several studies have demonstrated promising clinical sensitivity and specificity for ELISA and some LFAs (19, 20). Furthermore, several ELISAs have been shown to correlate well with neutralizing antibody titers (21, 22) and thus may be useful clinically and in vaccine development (23). However, both ELISA and LFAs have major disadvantages that limit their applicability for COVID-19 management. ELISA requires technical expertise, laboratory infrastructure, and multiple incubation and wash steps, limiting its applicability to settings outside of a centralized laboratory (24). On the other hand, LFAs are portable, but they have lower sensitivity and provide qualitative results (25), whereas a quantitative readout is preferred for clinical use, research studies, and surveillance applications. Collectively, these shortcomings of ELISAs and LFAs motivate the need for an easily deployable, point-of-care test (POCT) that can be manufactured in large volumes, has quantitative figures of merit equal to laboratory-based tests, and is as easy to use as an LFA.
To address the challenge of creating a user-friendly and widely deployable assay that can detect prior exposure to and immunological response against SARS-CoV-2, we developed a new multiplexed portable COVID-19 serological assay that is described herein. Our passive microfluidic platform provides sensitive and quantitative detection of antibodies against multiple SARS-CoV-2 viral antigens in 60 min with a single test from a single 60-μl drop of blood, plasma, or serum. We chose to quantify the antibody response against three different SARS-CoV-2 antigens because emerging studies have demonstrated that the primary antigenic target of the humoral immune response may inform disease progression and prognosis (14). Thus, being able to differentiate the viral targets of antibodies—as we can with our platform—may be especially valuable. Furthermore, our portable test is completely automated and can function at the POC independent of a centralized laboratory using only an inexpensive handheld detector. We also show that our test can be easily modified to detect additional protein biomarkers, such as cytokines/chemokines, without compromising the performance of the serological assay, which may provide further clinical insight into disease severity and/or patient outcomes (2, 26, 27). Collectively, these attributes suggest that our platform is a valuable tool for COVID-19 management both at the individual patient level (i.e., monitoring patients who may progress to severe disease) and for large-scale epidemiological studies at the population level. Furthermore, this platform is modular and can be easily modified to detect other pathogens or diagnostic markers simply by using a different set of biological reagents.
RESULTS
The DA-D4 POCT for COVID-19 serology
Our strategy to evaluate the antibody response to SARS-CoV-2 is based on the D4 assay platform, developed recently and reported elsewhere (28). The D4 platform is a completely self-contained immunoassay platform fabricated upon a “nonfouling” poly(oligoethylene glycol methyl ether methacrylate) (POEGMA) brush, where all reagents needed to complete the assay are inkjet-printed directly onto the surface. In previous work, we have used this platform for the detection of several protein biomarkers using a fluorescent sandwich immunoassay format (28). Here, we modified the design of the assay to detect antibodies against SARS-CoV-2 using a double-antigen (DA) bridging immunoassay format, which detects total antibody (all isotypes and subclasses). The DA-D4 is fabricated by inkjet printing viral antigens as stable and spatially discrete capture spots. In addition, viral antigens are labeled with a fluorescent tag and are printed nearby on an excipient pad as dissolvable spots. When a sample is added to the assay (Fig. 1A, i), the excipient pad dissolves and liberates the fluorescently labeled antigen (Fig. 1A, ii), which then diffuses across the polymer brush to the capture spots and labels any antibody that has been captured from solution by the stable capture spots of unlabeled antigen (Fig. 1A, iii). The fluorescence intensity of the capture spots is then imaged using a fluorescent detector and scales with antibody concentration in a sample (Fig. 1A, iv). Because capture spots of each antigen are printed at spatially discrete locations, this design enables multiplexed quantification of multiple target antibodies using a single fluorescent tag, which greatly simplifies the detector design and assay readout.
Fig. 1. DA-D4 POCT schematic and analytical validation.
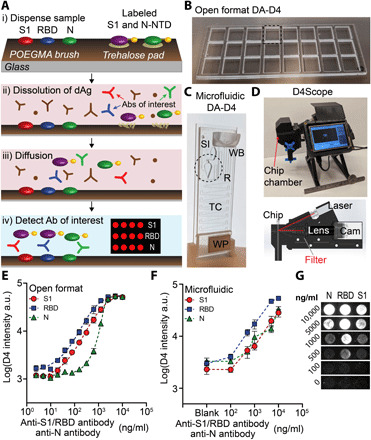
(A) DA-D4 assay chip schematic. S1, RBD, and N capture antigens and fluorescently labeled S1 and N-NTD detection antigens (dAgs) are inkjet-printed onto a POEGMA substrate. When a sample is added, dAgs are liberated from the surface owing to the dissolution of the underlying trehalose pad. Antibodies targeting each viral antigen then bridge the capture antigens to the dAgs, resulting in a fluorescence signal that scales with antibody concentration. (B) Open-format DA-D4 with 24 individual assays. (C) Microfluidic DA-D4. Sample is added to the sample inlet (SI), filling the reaction chamber (R) that contains the assay reagents. Wash buffer (WB) is added to the WB reservoir, which chases the sample through the microfluidic cassette. The timing channel (TC) sets the incubation time. All liquid is eventually soaked up by the wicking pad (WP) after the incubation process. The size is that of a standard microscope slide. (D) D4Scope and cut-away view of the optical path. The microfluidic flow cell is inserted on the left and pressing a button automates laser excitation, camera exposure, and data output. (E) Analytical validation of the open-format DA-D4. Antibodies targeting each antigen were spiked into undiluted human serum and incubated for 30 min. Each data point represents the average of three independent runs, and the error bars represent the SEM. a.u., arbitrary units. (F) Analytical validation of microfluidic DA-D4. Each data point for an antigen represents the average of four independent microfluidic flow cells and error bars represent the SEM. (G) Representative D4 spots shown for each dose. Photo credit for (B) to (D): David S. Kinnamon, Duke University.
To fabricate a serological assay for SARS-CoV-2, nucleocapsid (N), spike S1 domain (S1), and the receptor binding domain (RBD) of S1 were inkjet-printed as the “stable” capture reagents onto POEGMA-coated slides. Our rationale for simultaneously assaying the antibody response toward N, S1, and RBD antigens is that it is not fully understood which epitopes elicit an immune response in all individuals, although they are all believed to be immunogenic (29, 30) and because studies have shown that the primary target of the immune response may inform disease prognosis (14). N is expressed abundantly by SARS-CoV-2 during infection and is highly immunogenic in other coronaviruses (31, 32). The S protein—composed of the S1 and S2 domains—is exposed on the viral coat of SARS-CoV-2 and plays an essential role in viral attachment, fusion, entry, and transmission (33). Because S2 is highly conserved across many coronaviruses and is thus potentially cross-reactive, S1 was chosen for antibody detection (34). RBD—the portion of S1 that binds cells expressing viral receptor—is the target for many neutralizing antibodies and is thus a promising antigenic target for serological assays (34). Figure S1 shows the layout and dimensions of an open-format DA-D4 chip. Each chip contains 24 individual assays with S1, RBD, and N antigens arrayed as separate rows of five identical ~170-μm-diameter spots. Next, fluorescent conjugates of S1—which contains the amino acid sequence for RBD—and the N-terminal domain (NTD) of N (produced in-house, see fig. S2A for SDS–polyacrylamide gel electrophoresis of expression and purification) were mixed 1:1 and inkjet-printed as 12 identical 1-mm-diameter spots on an identically sized trehalose pad (fig. S1A). N-NTD—instead of full-length N—was chosen as the detection reagent because full-length N can dimerize in solution, potentially leading to a false-positive result in the DA format (fig. S2B) (35). Because of this choice of reagents, our assay only detects antibodies directed against the N-terminal region of N.
Analytical validation of the DA-D4 POCT using simulated samples
We first sought to demonstrate that the DA-D4 assay can detect antibodies against recombinantly expressed SARS-CoV-2 antigens. Initially, the analytical performance was characterized using the open-format DA-D4 (Fig. 1B). This is because the open-format DA-D4 assay has been extensively optimized and characterized by our group and has extremely high analytical sensitivity, which enables us to determine the figures of merit that are theoretically possible for a particular D4 assay. A disadvantage of the open-format DA-D4 assay, however, is that it requires a rinse step by the user after incubation of the sample (28).
For POC deployment and an improved user experience, we developed—in the course of this study—a new, gravity- and capillary-driven “passive” microfluidic flow cell that fully automates the assay (Fig. 1C). The microfluidic flow cell is fabricated by adhering complementary layers of precision laser-cut acrylic and adhesive sheets onto a functionalized POEGMA substrate (figs. S1B and S3 for the print layout). The resulting microfluidic flow cell features a reaction chamber, timing channel, sample inlet, wash buffer reservoir, and wicking pad that automates the sample incubation, sample removal, wash, and drying steps. This simplifies the user experience and limits the possibility of a user incorrectly carrying out the test, as it only requires the user to add the sample and a drop of wash buffer to the cassette. After ~60 min, the cassette is ready for imaging with a custom-built fluorescent detector—the D4Scope (Fig. 1D).
The D4Scope is a low-cost (<$1000), portable fluorescence detector [with dimensions of 7 inches (width) by 6 inches (height) by 5 inches (depth) and a weight of ~5 pounds; see fig. S4] built from off-the-shelf components and assembled using three dimensional (3D)–printed parts that can image microarray spots with high sensitivity. It uses coherent 638-nm red laser light set at an oblique angle (30°) relative to the surface to excite the fluorescently labeled antigens. The fluorescence wavelength emission from the labeled reagents then passes through a band-pass filter and imaged with a high-efficiency Sony IMX CMOS sensor in a Basler Ace camera (Fig. 1D). This setup provides a large field of view of 7.4 mm by 5 mm and a fine (raw) lateral resolution of ~2.4 μm. A user-friendly interface was developed in Python that runs on a 3.5″ Raspberry Pi touchscreen to control laser excitation, camera exposure, and image file output (see the Supplementary Materials for more details, fig. S4).
To mimic seropositive samples, we spiked commercially available antibodies (with known binding affinity toward SARS-CoV-2 antigens) into undiluted pooled human serum that was collected before the COVID-19 outbreak. A dilution series spanning four logs was evaluated on open-format DA-D4 chips and yielded dose-response curves with fluorescence intensities that scaled with antibody concentration and approximated a sigmoidal curve, demonstrating that the assay was responsive to the antibodies of interest (Fig. 1E). Within the microfluidic flow cell, the chamber geometry, reagent spacing/alignment, and amount of printed reagent were iteratively optimized to match the performance metrics of the open-format DA-D4. Six doses (including a blank) with varying amounts of anti-S1/RBD and anti-N antibodies were prepared and tested in quadruplicate on 24 separate microfluidic flow cells to demonstrate equivalence between the open format (Fig. 1B) and microfluidic flow cell (Fig. 1C). In the microfluidic flow cell, the fluorescence intensity of the capture antigens—imaged with the D4Scope—also scaled with antibody concentration, suggesting that the test is responsive to anti–SARS-CoV-2 antibodies (Fig. 1, F and G). Anti-S1/RBD antibodies did not cross-react with N antigen, and anti-N antibodies did not react with S1 or RBD antigens (fig. S5).
Clinical validation of the DA-D4 POCT
Next, we sought to validate the clinical performance of the DA-D4 POCT in a retrospective study using banked plasma samples from patients with PCR-confirmed COVID-19 who had been admitted to the intensive care unit (ICU) at Duke University Medical Center (Fig. 2A). A total of 46 COVID-19–positive plasma samples (heat-inactivated) from 31 patients—some of which had longitudinal samples available—and 41 negative samples (collected before the COVID-19 pandemic) were tested on the microfluidic DA-D4 and imaged with the D4Scope. We also evaluated the specificity of the DA-D4 using plasma samples from patients infected with human coronaviruses 229E (n = 2), HKU1 (n = 4), NL63 (n = 2), and OC43 (n = 10). The median age of the patients with COVID-19 was 55. Of the 31 patients, 10 were female and 21 were male. For most patients, the date of symptom onset was known (41 of 46 samples), where the average was ~20 days with a range of 6 to 48 days. The complete patient profile is provided in table S1.
Fig. 2. Clinical validation study.
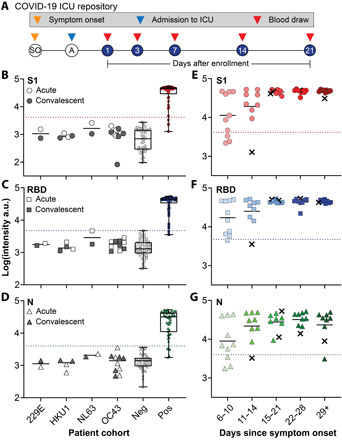
(A) Study design for COVID-19 ICU biorepository samples. Patients at Duke University Medical Center were enrolled into the study after admission to the ICU. Blood draws were taken at days 1, 3, 7, 14, and 21 after enrollment until discharge or death occurred. (B to D) Aggregated data for 46 positive samples, 41 negative controls, and 18 acute/convalescent coronavirus 229E (n = 2), HKU1 (n = 4), NL63 (n = 2), and OC43 (n = 10) samples tested for antibodies against (B) S1, (C) RBD, and (D) N. Dotted lines represent 2 SDs above the mean of the negative controls and the solid line represents the mean of each group. The box extends from the 25th to 75th percentiles and the line in the middle of the box is plotted at the median. The whiskers extend to the minimum and maximum values. (E to G) Data from (B) to (D) partitioned by days since symptom onset. For five samples, date since symptom onset was unknown, so days since first positive COVID-19 test were used (marked with an x).
Antibody reactivity toward all three viral antigens was measured on a single microfluidic flow cell for each patient sample. For validation, we assigned the threshold for a positive test result as 2 SDs above the mean of the 41 prepandemic negative samples, which we calculated individually for S1, RBD, and N. There was a statistically significant difference between the mean intensity for COVID-19–positive and –negative samples (P < 0.0001) for all three markers, as determined by a two-tailed unpaired t test (Fig. 2, B to D). Furthermore, all 41 healthy negative control samples tested below the threshold for each marker (specificity of 100%) and all samples within our specificity panel of similar coronavirus infections—both acute and convalescent—also tested below the threshold (specificity of 100%), indicating that our test is highly specific to SARS-CoV-2 (Table 1). Representative images for a high positive and negative sample are included in fig. S6.
Table 1. Specificity and sensitivity of the DA-D4.
| Specificity | |||
| Cohort | S1 | RBD | N |
| Healthy | 41/41 | 41/41 | 41/41 |
| Coronavirus 229E | 2/2 | 2/2 | 2/2 |
| Coronavirus HKU1 | 4/4 | 4/4 | 4/4 |
| Coronavirus NL63 | 2/2 | 2/2 | 2/2 |
| Coronavirus OC43 | 10/10 | 10/10 | 10/10 |
| Sensitivity | |||
| Days since symptom onset | S1 | RBD | N |
| 6–14 | 15/19 = 78.9% | 17/19 = 89.5% | 15/19 = 78.9% |
| 15+ | 27/27 = 100% | 27/27 = 100% | 26/27 = 96.3% |
Next, we partitioned the data into five groups based on days since symptom onset: 6 to 10, 11 to 14, 15 to 21, 22 to 28, and >29 days (Fig. 2, E to G). For two patients (five total samples), the date of symptom onset was unknown; hence, the days since first positive RT-PCR test result were used instead (these data points are marked with an x). The sensitivities obtained for S1, RBD, and N at various time frames are summarized in Table 1. For antibodies targeting S1 and RBD, the sensitivity reaches 100% 2 weeks after symptom onset. These results suggest that our assay spans a useful temporal range to detect the dynamic production of antibodies that typically occurs within 2 weeks of symptom onset (15). In addition, all tested patients developed a robust and sustained antibody response against S1 and RBD. For antibodies targeting N, the sensitivity reaches 96.3% 2 weeks after symptom onset. The concentration of N-targeting antibodies also appears to be more variable across all patients, especially at later time points, with some samples testing close to the threshold value (Fig. 2G). This could be due to the fact that some patients may develop a stronger response against other viral antigens/epitopes (RBD or S1) (36, 37) or against an epitope of N not within the NTD, highlighting the importance of testing for antibodies against several antigens simultaneously to maximize test sensitivity and specificity.
We also conducted a proof-of-concept study using whole human blood as the sample source for the microfluidic flow cell to demonstrate that the DA-D4 assay can be used at the POC or the point-of-sample collection without the need for any sample processing. To do so, we made minor modifications (see fig. S7) to the microfluidic timing channel and reaction chamber to account for the non-Newtonian fluid mechanics of whole blood (Fig. 3A). Briefly, a gradual slope was added to the reaction chamber to prevent accumulation of red blood cells during washing, and the incubation channel was shortened to account for a reduced flow rate. Fresh blood was collected in EDTA-coated tubes from four patients with negative COVID-19 antibody status (as determined by ELISA performed by the supplier) and from five patients with confirmed COVID-19 (from enrollments to the ICU study) (table S2). Each 60-μl blood sample was tested on the microfluidic DA-D4 assay. No complications were observed, such as coagulation of blood that can occur when testing whole blood in microfluidic systems. Figure 3B shows representative images of the reaction chamber, with the time since sample addition noted in the lower right-hand corner of each subpanel, demonstrating the ability of the microfluidic chip to process whole blood. The antibody response toward S1, RBD, and N from whole blood is shown in Fig. 3C. We set the threshold to determine a positive test as 2 SDs above the mean of the four negative samples, calculated individually for S1, RBD, and N. All negative samples tested as negative, and all positive samples tested above the threshold. These preliminary results suggest that the microfluidic DA-D4 assay is capable of detecting anti–SARS-CoV-2 antibodies in whole blood, so that the assay can be carried out immediately at the point of sample collection without the need for transport to a centralized laboratory for sample processing into serum or plasma and subsequent testing.
Fig. 3. Testing whole blood.
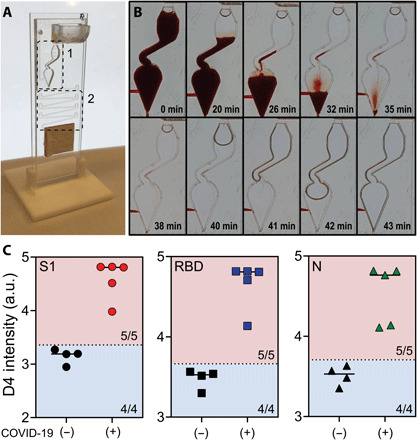
(A) Modified microfluidic flow cell for testing whole blood. Zone 1: The reaction chamber was modified to prevent red blood cells from collecting in the chamber. Zone 2: The incubation timing channel was shortened to compensate for the slower flow rate of blood and to ensure blood did not clot or clog the channels. (B) Time lapse of blood and wash buffer in the reaction chamber. (C) Aggregated data for five positive samples and four negative controls tested for antibodies against S1, RBD, and N. Dotted lines represent 2 SDs above the mean of the negative controls; 100% sensitivity (5/5) and 100% specificity (4/4) were achieved for S1, RBD, and N. Photo credit for (A) and (B): David S. Kinnamon, Duke University.
Monitoring antibody levels longitudinally
Having demonstrated the high clinical sensitivity and specificity of the microfluidic DA-D4 assay for detection of antibodies against SARS-CoV-2 antigens, as well as the ability to detect changes in antibody levels with time, we next sought to track seroconversion in individual patients. To accomplish this, we tested longitudinal plasma samples from six patients (Fig. 4, A to F). Across all six patients, the antibody response was initially low for the first time point tested and then increased and plateaued at later time points, consistent with the antibody dynamics reported in other studies (15, 38, 39). The DA-D4 readout for antibodies targeting S1 and RBD appeared to saturate by the second time point—typically 2 to 3 weeks after symptom onset—suggesting that each patient mounted a strong and robust immune response that was sustained over time. For N, the dynamics were slower in one patient (#1) and did not fully saturate in another (#3), providing insight into the primary target of the antibody response in those patients. In general, patients with severe COVID-19 often develop very high antibody titers (38), which is reflected in this ICU patient sample set by saturated signals at later time points. However, we were still able to measure seroconversion and antibody kinetics in each patient, suggesting that the DA-D4 is a useful tool for monitoring the immune response. For patients later in disease progression with high antibody titers, dilutions could be performed to adjust the concentration into the linear range of the assay. Testing a sample at various dilutions would also allow us to calculate specific antibody titers, which we are not able to do from a single undiluted sample.
Fig. 4. Longitudinal antibody tracking.
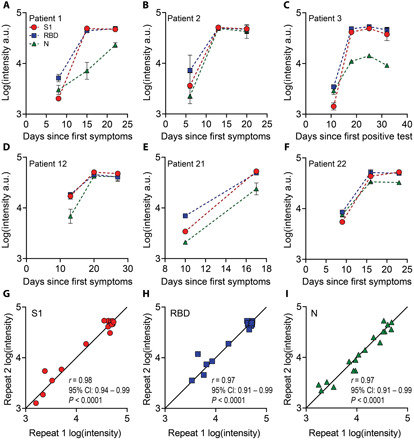
(A to F) Six patients were tracked across multiple time points for antibodies targeting S1, RBD, and N. For patient 3 (C), date since symptom onset was unknown, so days since first positive test was used. Each data point represents the average of two independent chips (with SD) run by separate users. (G to I) Data from parts (A) to (F) for each repeat. The solid line is drawn to have a slope of 1. There is a strong correlation between each repeat for (G) S1, (H) RBD, and (I) N, with a Pearson’s r of 0.98, 0.97, and 0.97, respectively (P < 0.0001).
Each sample in the longitudinal study was tested in duplicate by a different user to characterize the reproducibility and robustness of our platform (Fig. 4, G to I). We found a strong correlation for each marker, with a Pearson’s r correlation of 0.98, 0.97, and 0.97 for S1, RBD, and N, respectively. The high correlation between replicates further emphasizes the quantitative nature and reproducibility of our platform for profiling the immune response to SARS-CoV-2.
Concordance with neutralizing antibody titers
We next compared the performance of the DA-D4 with a microneutralization (MN) assay that monitors functional neutralization of SARS-CoV-2 via neutralizing antibodies binding to the RBD. All six patients that we tracked longitudinally developed robust neutralizing antibodies, and the MN titer was strongly concordant with DA-D4 assay readout for antibodies targeting S1 and the RBD of S1 (Fig. 5, A to F). Furthermore, a concordance analysis of the DA-D4 assay with the MN assay for antibodies targeting S1 and RBD showed a strong correlation across 34 plasma samples tested (fig. S8, A and B), as determined by a Pearson’s r > 0.70 (P < 0.0001). For antibodies targeting N, the concordance between the two assays was not as strong, with only a moderate correlation between the DA-D4 results and MN data (fig. S8C). This is expected, as N resides inside the capsid of SARS-CoV-2 and is not relevant for functional neutralization (40). This is also reflected in the longitudinal sample set. For example, patient 1 at day 15 after symptom onset has strong neutralizing antibodies, as seen by the MN assay, despite a weak overall antibody level for N. Although future studies are required to validate the ability of neutralizing antibodies to confer protection, these results suggest that the DA-D4 assay could be used as a supplement to live virus neutralization assays, which typically require >48 hours and biosafety level 3 containment.
Fig. 5. Correlation to MN assay.
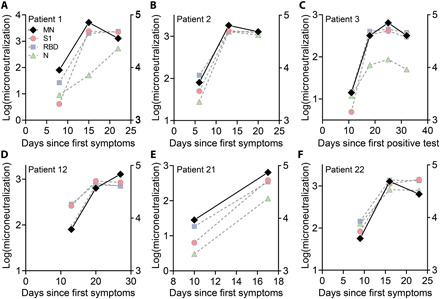
(A to F) MN assays were performed on each longitudinal sample (black diamonds) as described in Materials and Methods. The log-transformed MN titer is plotted on the left axis superimposed against the antibody data from Fig. 4 (plotted on the right axis).
Profiling prognostic biomarkers concurrent with serological testing
Last, we investigated the feasibility of detecting a prognostic protein biomarker concurrent with serological profiling. This is motivated by the fact that others have identified potentially prognostic biomarkers that correlate well with disease severity and patient outcomes (41, 42). Therefore, tracking antibody levels alongside prognostic biomarkers may provide clinically relevant information to inform interventions in the ICU for patients with a high probability of a poor outcome. As proof of concept, detection of interferon-γ (IFN-γ)–induced protein 10 (IP-10; CXCL10)—a chemokine that recruits inflammatory cells to the site of inflammation and which has been shown to be elevated in severe disease and correlates with patient prognosis (27, 42)—was integrated into the DA-D4 assay using a traditional sandwich immunoassay, as described previously (28).
Before testing patient samples, we sought to confirm that the multiplexed serological assay is compatible (not cross-reactive) with the IP-10 sandwich assay. To do so, we fabricated open-format chips containing all necessary reagents for both COVID-19 serology and human IP-10 detection. First, we prepared a 15-point dilution series of recombinant human IP-10 spiked into fetal bovine serum (FBS)—spanning the relevant physiological range for patients with COVID-19 identified elsewhere (42)—and added samples to chips in triplicate in the absence of antibodies targeting SARS-CoV-2 antigens. We observed a dose-dependent behavior for IP-10 response with a low limit of detection (LOD) of 0.12 ng/ml (43) and minimal reactivity for SARS-CoV-2 capture antigens, confirming that the IP-10 assay components do not cross-react with the serology components (Fig. 6A). Next, we prepared a dilution series of simulated seropositive samples and added them to the open-format chips. Across all concentrations of anti–SARS-CoV-2 antibodies, IP-10 capture antibody intensity was close to baseline, thus confirming that the serology components do not interfere with the IP-10 detection assay (Fig. 6B).
Fig. 6. Combined prognostic biomarker and serology detection.
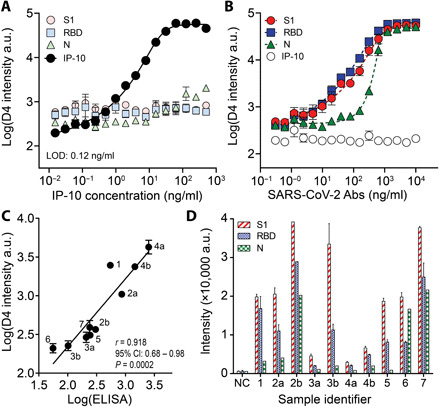
(A) Dose-response curve for recombinant IP-10 spiked into FBS. Each data point represents the average (n = 3), and error bars represent the SEM. The limit of detection (LOD) for IP-10 is 0.12 ng/ml. (B) Dose-response curve for anti–SARS-CoV-2 antibodies spiked into FBS. The highest concentration is 10 μg/ml of anti-S1/RBD and 10 μg/ml of anti-N antibodies. Each data point represents the average (n = 3) with SEM. (C) Correlation between DA-D4 readout for IP-10 with an ELISA performed separately. Samples with a letter designate samples from one individual at different time points, where b occurs later in disease than a. All samples were tested in duplicate on the DA-D4 (with SD shown) except 2b (due to insufficient volume). The solid line shows linear regression. (D) Antibody reactivity against S1, RBD, and N for sample tested in (C) (with SD shown). NC, negative control pooled healthy plasma.
Having confirmed the compatibility of the IP-10 assay with multiplexed serology in the open D4 format, we next sought to test the performance of our assay in patient samples. Ten COVID-19–positive plasma samples (from seven patients) were procured from the ICU biorepository and were added undiluted to open-format chips and then quantitatively assessed by the DA-D4. We measured IP-10 and antibodies against S1, RBD, and N simultaneously on the same device from a single sample. Separately, serum samples from the same patients were evaluated in parallel via LEGENDplex ELISA kits that report IP-10 concentration in picograms per milliliter. We observed a strong positive correlation between the DA-D4 assay for IP-10 with ELISA across all 10 pairs of measurements, with a Pearson’s r of 0.918 [P = 0.0002, 95% confidence interval (CI): 0.68 to 0.98] (Fig. 6C). We also tested for antibody reactivity toward S1, RBD, and N from the same samples and an additional sample of healthy pooled plasma (pre–COVID-19 negative control) (Fig. 6D). Although we did not observe a strong relationship between antibody and IP-10 levels, we did observe that, in the patients for which we tested multiple samples, IP-10 decreased over time, while the levels of antibodies increased.
Overall, these results clearly show that the D4 assay format can simultaneously detect antibody response to foreign SARS-CoV-2 antigens and a native protein biomarker from undiluted patient plasma. One of the benefits of detecting anti–SARS-CoV-2 antibodies from undiluted samples is that the sensitivity of the protein detection assay is not reduced because of dilution, allowing us to detect chemokines and cytokines—which are present at very low concentrations even during disease state—directly from complex biological milieu. Detection of additional prognostic biomarkers could also be implemented on the same chip, as long as there is no cross-reactivity between the assay reagents for serology and prognosis. For example, we recently developed a multiplexed assay to detect interleukin-6 (IL-6), N-terminal (NT)–pro-B-type natriuretic peptide (NT-proBNP), and D-dimer, which have all been implicated in disease progression and severity and could be added to our existing chip (fig. S9). A recent study found that the ratio of IL-6 to IL-10 can be used to guide clinical decision-making (44), which we plan to measure in the next generation of this assay.
DISCUSSION
As the COVID-19 pandemic unfolded, countries around the globe grappled with developing streamlined systems for diagnosis of acute infection using nucleic acid detection methods. Although there remains an urgent need for rapid and sensitive POCTs for acute diagnosis, developing accurate and reliable serological assays has been deemed an equally important endeavor to complement existing diagnostic strategies (12, 45). The challenge with developing an easy-to-use serology assay that can be broadly disseminated but that performs as well as centralized laboratory-based methods is highlighted by the large number of ELISA and LFA tests that have been developed. While LFAs are portable and easy to use and ELISAs are quantitative and highly sensitive, there remains a need for a technology that can merge the best attributes of each format.
The DA-D4 POCT is a promising platform to supplement existing diagnostic technologies to manage the COVID-19 pandemic because it marries the best attributes of LFAs and ELISAs—it is quantitative, easy to use, widely deployable, requires only a single 60-μl drop of blood, and can be performed with minimal user intervention. The SARS-CoV-2 DA-D4 assay can be used to measure antibody kinetics and seroconversion at the individual patient level directly from unprocessed blood or plasma. This test is highly sensitive and specific and is potentially suited for epidemiological surveillance at the population level using low-cost microfluidic cassettes that can be transported and stored for an extended period of time without a cold chain. Furthermore, it requires minimal user intervention to carry out the assay and provides a quantitative readout using a low-cost, handheld detector.
We show a strong correlation between the DA-D4 assay readout (for S1 and the RBD of S1) and neutralizing antibody titers, suggesting that this test may be useful in understanding efficacy and durability of natural or vaccine-induced humoral immunity and to potentially inform disease prognosis and population-level immunity. We also demonstrate that an additional prognostic biomarker can be easily incorporated into the test, which may be useful for monitoring disease severity and predict clinical outcomes. Combined, these attributes suggest that this platform may also be useful on the individual patient level to aid in clinical decision-making. While the results presented here mainly highlight the performance of the microfluidic chip, the open-format architecture with up to 24 individual assays per glass slide may be useful for scenarios where higher-throughput testing is demanded. The open format still has advantages compared to traditional ELISA because the open format only requires a single incubation step and one wash step, which reduces the hands-on time and equipment complexity required to complete the assay.
The DA-D4 has additional features that synergize to deliver a highly desirable serological assay. First, the DA sandwich format has advantages over other serological assay formats. Because total antibody is detected rather than a single antibody isotype or subclass, seroconversion in patients can be detected earlier, which reduces the chances of a false-negative result due to a test being administered too early in disease (39). Furthermore, because the labeled reagent does not have species specificity, the single assay kit could be used in preclinical vaccine development studies to measure antibody responses in experimental animals (23). The lack of species-specific detection antibodies also reduces the risk of high background signal caused by nonspecific antibodies binding to the surface and subsequently being labeled (46). Last, the DA-D4 can be conducted in a single step to accomplish multiplex detection without the need for an intermediate wash step, which other assay formats require.
Second, all reagents needed to complete the assay are incorporated onto the nonfouling POEGMA brush that eliminates virtually all nonspecific protein adsorption and cellular adhesion, thereby enabling an extremely low LOD directly from undiluted samples (47, 48). Although many serological assays often dilute samples, the ability to test undiluted samples is advantageous, especially when combined with prognostic biomarker testing where dilution of low-concentration analytes can lead to an undetectable signal. Testing multiple dilutions can still be performed using our test when antibody levels become high, which could be used to calculate specific titers. POEGMA also acts as a stabilizing substrate for printed reagents, enabling long-term storage of chips without a cold chain (28). In this study, results were generated over the course of 3 months from the same batch of tests stored in silica desiccated pouches at room temperature and ambient humidity.
Third, this platform can be easily multiplexed, which can be used to capture a more detailed picture of the host immune response to SARS-CoV-2 infection by quantifying the antibody level induced to multiple viral antigens—in this case, N, S1, and RBD—from a single sample without sacrificing ease of use. This is because each viral antigen is deposited at a spatially discrete location, which allows for a single fluorescent tag to be used during fluorescence imaging of the chip, thereby simplifying assay readout compared to other multiplexing technologies such as Simoa or Luminex assays, which rely on multiple different reporter molecules and a more complex readout (14, 49). This method also allows us to simultaneously measure the concentration of potential prognostic biomarkers directly from plasma (26, 27) without compromising the performance of the multiplexed serological assay. To the best of our knowledge, there are currently no tests on the market that can probe for antibodies against multiple viral antigens and prognostic protein biomarkers simultaneously.
Fourth, this platform is designed for POC deployment because it requires a single drop of blood that is readily obtained from a finger stick. This droplet is injected into the sample port of a gravity-driven microfluidic chip that requires no further user intervention beyond the concurrent addition of a few drops of wash buffer into a separate port. The assay runs by itself under the action of gravity and capillary action until all the fluid is drained from the microfluidic path by the absorbent pad at the bottom of the cassette, which fully absorbs and contains all liquid. This design eliminates the need for pumps, valves, or actuators and reduces the complexity and cost of the assay. Furthermore, it can be read out at the point of sample collection using the D4Scope, a highly sensitive and inexpensive handheld detector developed to work with the microfluidic chip. The D4Scope images a chip and provides a quantitative readout in less than 5 s, does not require an external power source or laboratory infrastructure, and can wirelessly transmit the results to a remote server over Wi-Fi. While smartphone-based diagnostics are becoming more popular, a benefit of this platform is that it does not rely on smartphone hardware and software, which change rapidly.
Where might this POC assay for COVID-19 serology and prognosis be useful? Serial quantification of antibody response and prognostic biomarkers would be most useful to monitor symptomatic and severe cases where use of available therapeutics, such as antiviral or monoclonal therapies, are indicated. Furthermore, it could be used to screen for patients with poor antibody responses who may benefit from convalescent plasma or monoclonal antibody therapy. We believe that this platform has potential utility in POC settings such as ICUs, urgent care clinics, and at the point of use—at locations where periodic surveillance of health care workers and other essential workers in close proximity to others for extended periods of time such as assembly-line manufacturing or food processing plants is desirable to assist in tracking clusters of disease and epidemiological studies. This platform could also be used as an inexpensive tool to study the longitudinal dynamics of antibody levels to inform reinfection potential, as coronavirus immunity often lasts only ~6 months (50). Similarly, it could be used to monitor vaccine-induced humoral immunity, which could help determine if boosters are needed in certain vaccinated individuals. This technology is suitable for low-resource settings across the globe, where eliminating the need for sample storage and transport to a centralized testing facility, and the attendant cold chain, is desirable and where access to expensive, high-throughput clinical analyzers that process large volumes of serology and other sandwich immunoassays is limited. Similarly, remote and austere settings—such as the field-forward position of the military or other remote locations where pandemics often emerge—can also benefit from this platform, as the testing is carried out with a disposable cassette and a low-cost, lightweight, and handheld detector whose production can easily be scaled up to enable widespread and dispersed deployment.
While the results presented here are promising, there are several issues identified during this study that require further investigation before its deployment. First, our cohort of individuals with SARS-CoV-2 infection consisted of adults with clinically severe disease, which is not representative of the entire spectrum of COVID-19 disease severity. These samples were chosen to demonstrate proof of concept of the DA-D4 assay and because these samples were locally available through an existing biobank. We recognize that a larger sample size that spans the disease severity spectrum is required to develop a more robust measure of sensitivity and specificity of the DA-D4 serology test for SARS-CoV-2. Similarly, we were not able to match demographics in our negative control group, which may have introduced confounding variables in our analyses. Because of limitations in the volume available from archived samples, we were not able to directly compare the performance of our test to ELISA or LFAs. These studies will be conducted on additional samples in future studies that are designed to address this precise issue and will allow us to assess the concordance in the clinical performance metrics between the different analytical methods. Furthermore, several of the samples we tested saturated the readout of our assay, which limits the dynamics we can measure once high antibody titers are achieved. This limitation could be addressed by testing individual samples on separate microfluidic chips at various dilutions, which would effectively increase the dynamic range of our assay and yield more precise quantitative titer. In addition, because of the DA design of our assay, we are also not able to discriminate between specific antibody subclasses or isotypes, which has been shown to be important for other diseases. This assay format also required that we use a truncated form of the N protein—expressed in Escherichia coli—as the detection reagent to avoid high signal at low antibody concentrations due to dimerization of full-length N. This may have caused our assay to underestimate the titer of anti-N antibodies for two reasons: (i) the bacterial expression system we used does not perform glycosylation, which could negatively impact antibody recognition, and (ii) the truncated form does not allow us to detect antibodies that are developed against the C-terminal domain, which also contains immunogenic epitopes (51). This limitation is compensated for by the fact that we can easily multiplex using the DA-D4 format and thus detect antibodies directed against different antigens to maintain high sensitivity and specificity. Despite these limitations, we believe that our assay is poised well to complement existing diagnostic solutions once additional validation studies encompassing larger patient cohorts are completed. We are actively developing an improved version of the test that requires less sample volume and has a shorter run time to better match the time to results and volume requirements of existing LFAs.
In summary, we have developed a COVID-19 serological assay that merges the benefits of LFAs and ELISAs. We used this test to simultaneously measure the antibody levels for multiple viral antigens and a potential prognostic biomarker directly from plasma and whole blood. For COVID-19 management, our platform may be useful to better understand patient antibody responses, provide actionable intelligence to physicians to guide interventions for hospitalized patients at the POC, to assess vaccine efficacy, and to perform epidemiological studies. Furthermore, our platform is broadly applicable to other diseases where sensitive and quantitative antibody and/or protein detection is desirable in settings without access to a centralized laboratory. Overall, we believe that our platform is a promising approach to democratize access to laboratory quality tests, by enabling rapid and decentralized testing with minimal user intervention to locations outside the hospital.
MATERIALS AND METHODS
DA-D4 assay
The DA-D4 assay is based on the design of the D4 immunoassay, reported elsewhere (28). Briefly, a polymer brush composed of POEGMA was “grafted from” a glass slide by surface-initiated atom transfer radical polymerization (48). Recombinant SARS-CoV-2 proteins were then printed onto POEGMA-coated slides as capture and detection spots. Capture spots of the following proteins were printed as ~170-μm-diameter spots using a Scienion S11 sciFLEXARRAYER (Scienion AG) inkjet printer: spike S1 (Sino Biological, catalog #40591-V05H1), spike RBD (Sino Biological, catalog #40592-V02H), and nucleocapsid protein (Leinco, catalog #S854). Each protein was printed as a row/column of five identical spots. Next, 12 excipient pads of trehalose with 1.6-mm spacing were printed from a 10% (w/v) trehalose solution in deionized water around the periphery of the capture antigen array using a BioDot AD1520 printer (BioDot Inc.). To print the detection reagents, S1 (Sino Biological, catalog #40591-V08H) and N-NTD (produced in-house) were first conjugated to Alexa Fluor 647 (per the manufacturer’s instructions) and then detection spots of the fluorescent protein conjugates of these proteins were printed on top of the excipient pads as 12 1-mm-diameter spots. A schematic of the chip that shows the spatial address and dimensions of the capture spots, trehalose pad, and detection spots is shown in fig. S1. After printing and final assembly, D4 chips were stored with desiccant until use. The amount of reagent deposited for the open format and microfluidic format was identical, with the only difference being the relative spot placement (fig. S1, A and B). For DA-D4 assays that also detected IP-10, an additional column of five spots of capture antibody (R&D Systems, catalog #MAB266) was included and anti–IP-10 detection antibody (R&D Systems, catalog #AF-266) was included in the detection cocktail for the open-format chips.
Fabrication and analytical testing of open-format DA-D4
Open-format slides were prepared by adhering acrylic wells to each slide, which splits one slide into 24 independent arrays (see fig. S1A for a schematic and Fig. 1B for an image). To validate the analytical performance of the test, dose-response curves were generated using antibodies targeting SARS-CoV-2 antigens (Sino Biological, catalog #40143-MM05, 40150-D001, and 40150-D004) spiked into undiluted pooled human serum. Open-format chips were incubated with a 13-point dilution series (run in triplicate) for 30 min, briefly rinsed in a 0.1% Tween 20/phosphate-buffered saline (PBS) wash buffer and then dried. Arrays were imaged on an Axon Genepix 4400 tabletop scanner (Molecular Devices LLC).
Fabrication and analytical testing of microfluidic DA-D4
The microfluidic chip fabrication process is described in detail in the Supplementary Materials. Briefly, the microfluidic chip was fabricated by adhering complementary layers of precision laser-cut acrylic and adhesive sheets onto a POEGMA substrate that had been functionalized with the relevant capture and detection reagents. The resulting assembly features a reaction chamber, timing channel, sample inlet, wash buffer reservoir, and wicking pad that automates the sample incubation, sample removal, wash, and drying steps. Simulated doses were prepared using antibodies targeting SARS-CoV-2 antigens (Sino Biological, catalog #40143-MM05, 40150-D001, and 40150-D004) spiked into undiluted pooled human serum. Six doses (including a blank) were tested on the microfluidic DA-D4 in the following way: (i) The user dispenses 60 μl of sample into the sample inlet using a pipette. (ii) The user dispenses 135 μl of wash buffer into the wash reservoir of the cassette using a pipette. (iii) The user waits 60 min for the cassette to run to completion. During this time, (a) fluorescently labeled antigens dissolve and form sandwiches with the antibodies of interest and the immobilized capture antigen in the reaction chamber. (b) A small volume of sample traverses the timing channel, which governs the incubation time. (c) The sample reaches an absorbent pad situated at the end of the timing channel that rapidly wicks away all sample from the reaction chamber, ending incubation. (d) As the sample clears, wash buffer enters the reaction chamber, removing residual sample and unbound reagent before it is also wicked away, leaving a cleaned and dry imaging surface. We observed a less than ±10% variation in the designed 23-min incubation time for the data presented in Fig. 1F. The remaining difference in time accounts for washing and drying time. (iv) The cassette is ready for analysis on the D4Scope. The vertical orientation of the cassette works in conjunction with the POEGMA brush to maintain low background fluorescence. Cellular and other sample debris can collect on the brush surface owing to gravitational forces, even if no binding is occurring. The vertical orientation ensures that these debris fall harmlessly toward the timing channel during the wash step. This proved especially important when testing with undiluted human whole blood samples.
Patient samples
De-identified heat-inactivated EDTA plasma samples (57°C for 30 min) were accessed from the Duke COVID-19 ICU biorepository (Pro00101196, PI Bryan Kraft) approved by the Duke Health Institutional Review Board (IRB) via an exempted protocol approved by the Duke Health IRB (Pro00105331, PI Ashutosh Chilkoti). Briefly, eligible patients included in the repository were men and women ages 18 years and above who were admitted to an adult ICU at Duke University Hospital with SARS-CoV-2 infection confirmed by PCR testing and who gave informed consent. Samples were collected on study days 1, 3, 7, 14, and 21. In addition to biological samples, clinical data on these patients were also collected including demographics, laboratory data, and clinical course. This study was performed in collaboration with the biorepository team and we have complied with all relevant ethical regulations.
Ten negative control plasma samples were collected under a normal blood donor protocol (Pro00009459, PI Tony Moody) and were collected from 2014 to 2019 (before the COVID-19 outbreak). All patient information, including demographics, is unknown to the investigator team. An additional 11 negative control samples were purchased commercially (Lee Biosolutions Inc.). Last, 20 negative control samples and 18 samples from patients infected with coronavirus 229E (n = 2), HKU1 (n = 4), NL63 (n = 2), and OC43 (n = 10) were collected under Pro00001698. All samples were accessed via an exempted protocol approved by the Duke Health IRB (Pro00105331, PI Ashutosh Chilkoti). Blood was either purchased commercially (Innovative Research Inc.) or accessed from the ICU biorepository (Pro00101196, PI Bryan Kraft) in EDTA-collection tubes and was tested within 48 hours of sample collection.
Testing of prepandemic healthy controls, specificity panel, and ICU samples on the microfluidic DA-D4
The plasma samples (prepandemic healthy controls, specificity panel, and ICU biorepository) were thawed from −80°C storage and allowed to reach room temperature before testing. Blood samples were tested at room temperature. The same procedure used to test the simulated samples as described in “Fabrication and analytical testing of microfluidic DA-D4” was used for testing of all clinical samples. The only exception was that a modified microfluidic flow cell described in the Supplementary Materials that required the use of 200 μl of wash buffer was used for testing whole blood.
D4Scope fabrication and operation
The D4Scope design, fabrication, and assembly are described in detail in the Supplementary Materials. Briefly, the D4Scope’s optical elements—the laser, band-pass filter, lens, and camera—and processing elements—the Raspberry Pi 4, touchscreen, and cabling—are mounted in a custom 3D-printed chassis. Fully assembled, it weighs ~5 pounds. The D4Scope can be powered through either a portable battery pack or wall power. Once connected to the power source, the D4Scope automatically runs our custom imaging Python program. The user removes the light protection cover from the cassette loading port and slides the microfluidic cassette with the glass side toward the detector. The light protection cover is then replaced enclosing the cassette. The user is then prompted to enter the sample ID # and chip ID # using either the touchscreen or optional attached keyboard and mouse.
The D4Scope has two fine adjustment knobs on the cassette loading port that allow for precise vertically and horizontally movement of the cassette relative to the laser source to ensure that the DA-D4 array is perfectly centered with the excitation source. Each array has coprinted two control spots that will always be uniformly bright across all tested samples and align with two superimposed alignment cross hairs on the live video feed of the D4Scope. Using the “toggle video” function on the user interface activates the laser and camera to provide a live view of the imaging area for this alignment. Once aligned, the toggle video function can be pressed again to end the live view, and the “capture image” function can be used to collect and save the resulting image onto the on-board hard drive and, optionally, to a cloud-based server defined by the end user. The live-view feature should be used sparingly to prevent photobleaching of the sample. For this study, we manually analyzed the resulting fluorescence intensity using Genepix Analysis software. However, we have developed an algorithm for automatic analysis of spot intensity and instantaneous results on our open-format platform, which will be reported elsewhere.
Live SARS-CoV-2 MN assay
The SARS-CoV-2 virus (Isolate USA-WA1/2020, NR-52281) was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH). SARS-CoV-2 MN assays were adapted from a previous study (52). In short, plasma samples are diluted twofold and incubated with 100 TCID50 (median tissue culture infectious dose) virus for 1 hour. These dilutions are transferred to a 96-well plate containing 2 × 104 Vero E6 cells per well. Following a 96-hour incubation, cells were fixed with 10% formalin and cytopathic effect (CPE) was determined after staining with 0.1% crystal violet. Each batch of MN includes a known neutralizing control antibody (clone D001; SINO, catalog #40150-D001). Data are reported as the inverse of the last dilution of plasma that protected from CPE, log10-transformed.
IP-10 experiments
Open-format DA-D4 slides were fabricated as described above using all reagents needed for antibody detection and IP-10 detection. Citrated plasma samples from 10 patients were procured from the ICU biorepository. Sixty microliters of each sample was added to two separate DA-D4 chips and incubated for 30 min, and the chips were then rinsed using 0.1% Tween 20 in 1× PBS. All slides were scanned with the Genepix tabletop scanner.
IP-10 levels were measured using the LEGENDplex Human Proinflammatory Chemokine Panel (13-plex) and LEGENDplex Human Anti-Virus Response Panel (13-plex) obtained from BioLegend. Assays were performed with patient serum per the manufacturer’s instructions. The assay was performed using a Beckman Coulter CytoFLEX flow cytometer, and data processing was performed using BioLegend’s Bio-Bits cloud-based software platform. Each sample was tested in triplicate, and the results are reported as the mean of these triplicates.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 8.4.1 (GraphPad Software Inc.). All data were log-transformed for analysis. To establish statistical significance between negative and positive cohorts (Fig. 2, B to D), unpaired t tests were used. Pearson’s r correlation was used to assess the degree of correlation between measurements and was calculated using GraphPad Prism.
Acknowledgments
We thank D. Montefiori for providing laboratory space to complete the clinical validation studies. We also thank R. Sahm for completing live SARS-CoV-2 microneutralization assays, which were performed in the Virology Unit of the Duke Regional Biocontainment Laboratory, which received partial support for construction from the NIH/NIAD (UC6AI058607; GDS). We thank the nurses in the ICUs of Duke University Hospital for collecting the blood samples used for this study and thank P. Lee for supporting the ICU Biorepository. We thank T. Moody for access to the prepandemic negative control samples used in this study, T. Denny for providing access to BSL2+ laboratory facilities to run the prepandemic negative samples, and H. Register for assistance with testing the negative samples on the DA-D4 POCT. Funding: A.C. acknowledges the support of the National Science Foundation (grant no. CBET2029361); the National Cancer Institute through grants P30-CA014236, R01-CA248491, and UH3-CA211232; Department of Defense United States Special Operations Command (grant no. W81XWH-16-C-0219); Defence Academy of the United Kingdom (grant no. ACC6010469); and the Combat Casualty Care Research Program (JPC-6) (grant no. W81XWH-17-2-0045). B.D.K. receives funding from NHLBI (K08HL130557). Author contributions: J.T.H. and D.S.K. are co–lead authors, who equally participated in experimental design, data collection, data analysis, manuscript drafting, figure creation, and manuscript revision. L.B.O. participated in experimental design and collection and analysis of data for clinical validation. J.L. developed the D4Scope used throughout the study and drafted text and figures related to the D4Scope. D.S.K., C.M.F., and A.M.H. developed the microfluidic cassette. G.K. cloned, expressed, and purified the N-NTD. S.A.W., S.O., and Z.Q. participated in data collection and analysis. C.M.F., D.Y.J., and A.M.H. were responsible for conceptualization, investigation, and manuscript revision. C.P. oversaw statistical analysis and assisted in study design. J.G.A. and T.W.B. assisted in procurement and testing of negative control samples. T.O. designed and ran live virus microneutralization assays, analyzed data, and participated in writing the manuscript. G.D.S. participated in writing the manuscript. B.D.K., C.W.W., L.C., L.G.Q., S.K.N., B.A.S., I.A.N., and L.B.O. contributed to the development of the Duke COVID-19 biorepositories and oversaw clinical data acquisition. T.W.B., B.D.K., and C.W.W. participated in manuscript revision. A.C. is the principal investigator who directed the studies, helped plan experiments, analyzed data, and participated in writing and editing the manuscript. All authors read and approved the manuscript. Competing interests: A.C., D.S.K., C.P., A.M.H., J.L., and J.T.H. are inventors on two provisional patents related to this work filed by Duke University [no. 63/068,432, filed (21 August 2020), not yet published, and no. 63/116,511, filed (20 November 2020), not yet published]. Both are entitled “Microfluidic assay device” and both describe innovations used for the D4 microfluidic cassette described in this work. A.C. and J.L. are inventors on a patent related to this work filed by Duke University [no. WO/2020/223713, filed (2 May 2020), published (5 May 2020)]. The patent is entitled “Devices and methods for imaging microarray chips” and describes innovations used for the D4Scope described in this work. Immucor Inc. has acquired the rights to the D4 assay on POEGMA brushes for in vitro diagnostics from Sentilus Inc. (cofounded by A.C. and A.M.H.). All other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/26/eabg4901/DC1
REFERENCES AND NOTES
- 1.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M., Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiersinga W. J., Rhodes A., Cheng A. C., Peacock S. J., Prescott H. C., Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA 324, 782–793 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Miller I. F., Becker A. D., Grenfell B. T., Metcalf C. J. E., Disease and healthcare burden of COVID-19 in the United States. Nat. Med. 26, 1212–1217 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Liu Q., Luo D., Haase J. E., Guo Q., Wang X. Q., Liu S., Xia L., Liu Z., Yang J., Yang B. X., The experiences of health-care providers during the COVID-19 crisis in China: A qualitative study. Lancet Glob. Health 8, e790–e798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y. W., Schmitz J. E., Persing D. H., Stratton C. W., Laboratory diagnosis of COVID-19: Current issues and challenges. J. Clin. Microbiol. 58, e00512-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu D. K. W., Pan Y., Cheng S. M. S., Hui K. P. Y., Krishnan P., Liu Y., Ng D. Y. M., Wan C. K. C., Yang P., Wang Q., Peiris M., Poon L. L. M., Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 66, 549–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman J. A., Pepper G., Naccache S. N., Huang M.-L., Jerome K. R., Greninger A. L., Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J. Clin. Microbiol. 58, e00821-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nalla A. K., Casto A. M., Huang M. L. W., Perchetti G. A., Sampoleo R., Shrestha L., Wei Y., Zhu H., Jerome K. R., Greninger A. L., Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 58, e00557-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wölfel R., Corman V. M., Guggemos W., Seilmaier M., Zange S., Müller M. A., Niemeyer D., Jones T. C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C., Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Krammer F., Simon V., Serology assays to manage COVID-19. Science 368, 1060–1061 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Bryant J. E., Azman A. S., Ferrari M. J., Arnold B. F., Boni M. F., Boum Y., Hayford K., Luquero F. J., Mina M. J., Rodriguez-Barraquer I., Wu J. T., Wade D., Vernet G., Leung D. T., Serology for SARS-CoV-2: Apprehensions, opportunities, and the path forward. Sci. Immunol. 5, eabc6347 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Atyeo C., Fischinger S., Zohar T., Slein M. D., Burke J., Loos C., McCulloch D. J., Newman K. L., Wolf C., Yu J., Shuey K., Feldman J., Hauser B. M., Caradonna T., Schmidt A. G., Suscovich T. J., Linde C., Cai Y., Barouch D., Ryan E. T., Charles R. C., Lauffenburger D., Chu H., Alter G., Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 53, 524–532.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L., Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 26, 845–848 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Yong S. E. F., Anderson D. E., Wei W. E., Pang J., Chia W. N., Tan C. W., Teoh Y. L., Rajendram P., Toh M. P. H. S., Poh C., Koh V. T. J., Lum J., Suhaimi N.-A. M., Chia P. Y., Chen M. I.-C., Vasoo S., Ong B., Leo Y. S., Wang L., Lee V. J. M., Connecting clusters of COVID-19: An epidemiological and serological investigation. Lancet Infect. Dis. 20, 809–815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipsitch M., Kahn R., Mina M. J., Antibody testing will enhance the power and accuracy of COVID-19-prevention trials. Nat. Med. 26, 818–819 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Khailany R. A., Safdar M., Ozaslan M., Genomic characterization of a novel SARS-CoV-2. Gene Rep. 19, 100682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisboa Bastos M., Tavaziva G., Abidi S. K., Campbell J. R., Haraoui L.-P., Johnston J. C., Lan Z., Law S., Lean E. M., Trajman A., Menzies D., Benedetti A., Khan F. A., Diagnostic accuracy of serological tests for covid-19: Systematic review and meta-analysis. BMJ 370, m2516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitman J. D., Hiatt J., Mowery C. T., Shy B. R., Yu R., Yamamoto T. N., Rathore U., Goldgof G. M., Whitty C., Woo J. M., Gallman A. E., Miller T. E., Levine A. G., Nguyen D. N., Bapat S. P., Balcerek J., Bylsma S. A., Lyons A. M., Li S., Wong A. W.-Y., Gillis-Buck E. M., Steinhart Z. B., Lee Y., Apathy R., Lipke M. J., Smith J. A., Zheng T., Boothby I. C., Isaza E., Chan J., Acenas D. D. II, Lee J., Macrae T. A., Kyaw T. S., Wu D., Ng D. L., Gu W., York V. A., Eskandarian H. A., Callaway P. C., Warrier L., Moreno M. E., Levan J., Torres L., Farrington L. A., Loudermilk R. P., Koshal K., Zorn K. C., Garcia-Beltran W. F., Yang D., Astudillo M. G., Bernstein B. E., Gelfand J. A., Ryan E. T., Charles R. C., Iafrate A. J., Lennerz J. K., Miller S., Chiu C. Y., Stramer S. L., Wilson M. R., Manglik A., Ye C. J., Krogan N. J., Anderson M. S., Cyster J. G., Ernst J. D., Wu A. H. B., Lynch K. L., Bern C., Hsu P. D., Marson A., Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 38, 1174–1183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T. H. O., Chromikova V., McMahon M., Jiang K., Arunkumar G. A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D. S., Lugo L. A., Kojic E. M., Stoever J., Liu S. T. H., Cunningham-Rundles C., Felgner P. L., Moran T., García-Sastre A., Caplivski D., Cheng A. C., Kedzierska K., Vapalahti O., Hepojoki J. M., Simon V., Krammer F., A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okba N. M. A., Müller M. A., Li W., Wang C., GeurtsvanKessel C. H., Corman V. M., Lamers M. M., Sikkema R. S., de Bruin E., Chandler F. D., Yazdanpanah Y., le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C. B. E. M., Bosch B.-J., Drosten C., Koopmans M. P. G., Haagmans B. L., Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 26, 1478–1488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowther J. R., The ELISA guidebook. Methods Mol. Biol. 149, III-IV, 1–III-IV413 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Posthuma-Trumpie G. A., Korf J., van Amerongen A., Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 393, 569–582 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T., Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 57, 389–399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laing A. G., Lorenc A., del Molino del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D. R., Hayday T. S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J. D., Gee S., Chan J. N. E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmägi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M. A. A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K. J., di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A. C., Author correction: A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 26, 1951 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Joh D. Y., Hucknall A. M., Wei Q., Mason K. A., Lund M. L., Fontes C. M., Hill R. T., Blair R., Zimmers Z., Achar R. K., Tseng D., Gordan R., Freemark M., Ozcan A., Chilkoti A., Inkjet-printed point-of-care immunoassay on a nanoscale polymer brush enables subpicomolar detection of analytes in blood. Proc. Natl. Acad. Sci. U.S.A. 114, E7054–E7062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravichandran S., Coyle E. M., Klenow L., Tang J., Grubbs G., Liu S., Wang T., Golding H., Khurana S., Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 12, eabc3539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed S. F., Quadeer A. A., McKay M. R., Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12, 254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutta N. K., Mazumdar K., Gordy J. T., The nucleocapsid protein of SARS-CoV-2: A target for vaccine development. J. Virol. 94, e00647-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., Thiel V., de Haan C. A. M., Reggiori F., Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 94, e01925-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang S., Hillyer C., Du L., Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses: (Trends in Immunology 41, 355-359; 2020). Trends Immunol. 41, 545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W., Tan W., Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395, 565–574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q., Yan Y., Zhang C., Shan H., Chen S., Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 10, 1228–1238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosadas C., Randell P., Khan M., McClure M. O., Tedder R. S., Testing for responses to the wrong SARS-CoV-2 antigen? Lancet 396, e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Röltgen K., Powell A. E., Wirz O. F., Stevens B. A., Hogan C. A., Najeeb J., Hunter M., Wang H., Sahoo M. K., Huang C. H., Yamamoto F., Manohar M., Manalac J., Otrelo-Cardoso A. R., Pham T. D., Rustagi A., Rogers A. J., Shah N. H., Blish C. A., Cochran J. R., Jardetzky T. S., Zehnder J. L., Wang T. T., Narasimhan B., Gombar S., Tibshirani R., Nadeau K. C., Kim P. S., Pinsky B. A., Boyd S. D., Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 5, eabe0240 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L., To K. K.-W., Chan K.-H., Wong Y.-C., Zhou R., Kwan K.-Y., Fong C. H.-Y., Chen L.-L., Choi C. Y.-K., Lu L., Tsang O. T.-Y., Leung W.-S., To W.-K., Hung I. F.-N., Yuen K.-Y., Chen Z., High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg. Microbes Infect. 9, 1664–1670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z., Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin. Infect. Dis. 71, 2027–2034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McAndrews K. M., Dowlatshahi D. P., Dai J., Becker L. M., Hensel J., Snowden L. M., Leveille J. M., Brunner M. R., Holden K. W., Hopkins N. S., Harris A. M., Kumpati J., Whitt M. A., Lee J. J., Ostrosky-Zeichner L. L., Papanna R., LeBleu V. S., Allison J. P., Kalluri R., Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity. JCI Insight 5, e142386 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaninov N., In the eye of the COVID-19 cytokine storm. Nat. Rev. Immunol. 20, 277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., Peng L., Yang M., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Gao G. F., Jiang C., Liu L., Liu Y., Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 146, 119–127.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armbruster D. A., Pry T., Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 29(suppl. 1), S49–S52 (2008). [PMC free article] [PubMed] [Google Scholar]
- 44.McElvaney O. J., Hobbs B. D., Qiao D., McElvaney O. F., Moll M., McEvoy N. L., Clarke J., O’Connor E., Walsh S., Cho M. H., Curley G. F., McElvaney N. G., A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. EBioMedicine 61, 103026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter A. K., Hegde S. T., The important role of serology for COVID-19 control. Lancet Infect. Dis. 20, 758–759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterboer T., Sehr P., Pawlita M., Suppression of non-specific binding in serological Luminex assays. J. Immunol. Methods 309, 200–204 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Heggestad J. T., Fontes C. M., Joh D. Y., Hucknall A. M., Chilkoti A., In pursuit of zero 2.0: Recent developments in nonfouling polymer brushes for immunoassays. Adv. Mater. 32, e1903285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hucknall A., Kim D.-H., Rangarajan S., Hill R. T., Reichert W. M., Chilkoti A., Simple fabrication of antibody microarrays on nonfouling polymer brushes with femtomolar sensitivity for protein analytes in serum and blood. Adv. Mater. 21, 1968–1971 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norman M., Gilboa T., Ogata A. F., Maley A. M., Cohen L., Busch E. L., Lazarovits R., Mao C.-P., Cai Y., Zhang J., Feldman J. E., Hauser B. M., Caradonna T. M., Chen B., Schmidt A. G., Alter G., Charles R. C., Ryan E. T., Walt D. R., Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat. Biomed. Eng. 4, 1180–1118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.J. Seow, C. Graham, B. Merrick, S. Acors, K. J. A. Steel, O. Hemmings, A. O’Bryne, N. Kouphou, S. Pickering, R. P. Galao, G. Betancor, H. D. Wilson, A. W. Signell, H. Winstone, C. Kerridge, N. Temperton, L. Snell, K. Bisnauthsing, A. Moore, A. Green, L. Martinez, B. Stokes, J. Honey, A. Izquierdo-Barras, G. Arbane, A. Patel, L. O’Connell, G. O’Hara, E. M. Mahon, S. Douthwaite, G. Nebbia, R. Batra, R. Martinez-Nunez, J. D. Edgeworth, S. J. D. Neil, M. H. Malim, K. J. Doores, Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv 2020.2007.2009.20148429 (2020).
- 51.Musico A., Frigerio R., Mussida A., Barzon L., Sinigaglia A., Riccetti S., Gobbi F., Piubelli C., Bergamaschi G., Chiari M., Gori A., Cretich M., SARS-CoV-2 epitope mapping on microarrays highlights strong immune-response to N Protein Region. Vaccines (Basel) 9, 35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry J. D., Jones S., Drebot M. A., Andonov A., Sabara M., Yuan X. Y., Weingartl H., Fernando L., Marszal P., Gren J., Nicolas B., Andonova M., Ranada F., Gubbins M. J., Ball T. B., Kitching P., Li Y., Kabani A., Plummer F., Development and characterisation of neutralising monoclonal antibody to the SARS-coronavirus. J. Virol. Methods 120, 87–96 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/26/eabg4901/DC1


