Abstract
SARS-CoV-2 is the culprit causing Coronavirus Disease 2019 (COVID-19). For the study of SARS-CoV-2 infection in a BSL-2 laboratory, a SARS-CoV-2 pseudovirus particle (SARS2pp) production and infection system was constructed by using a lentiviral vector bearing dual-reporter genes eGFP and firefly luciferase (Luc2) for easy observation and analysis. Comparison of SARS2pp different production conditions revealed that the pseudovirus titer could be greatly improved by: 1) removing the last 19 amino acids of the spike protein and replacing the signal peptide with the mouse Igk signal sequence; 2) expressing the spike protein using CMV promoter other than CAG (a hybrid promoter consisting of a CMV enhancer, beta-actin promoter, splice donor, and a beta-globin splice acceptor); 3) screening better optimized spike protein sequences for SARS2pp production; and 4) adding 1 % BSA in the SARS2pp production medium. For infection, this SARS2pp system showed a good linear relationship between MOI 2-0.0002 and then was successfully used to evaluate SARS-CoV-2 infection inhibitors including recombinant human ACE2 proteins and SARS-CoV-2 neutralizing antibodies. The kidney, liver and small intestine-derived cell lines were also found to show different susceptibility to SARSpp and SARS2pp. Given its robustness and good performance, it is believed that this pseudovirus particle production and infection system will greatly promote future research for SARS-CoV-2 entry mechanisms and inhibitors and can be easily applied to study new emerging SARS-CoV-2 variants.
Abbreviations: SARS2pp, SARS-CoV-2 pseudovirus particles; SARSpp, SARS-CoV pseudovirus particles; VSVpp, vesicular stomatitis virus pseudovirus particles; S, SARS-CoV-2 full-length spike protein
Keywords: SARS-CoV-2, Pseudovirus, Viral entry inhibitors, Susceptible cell lines
1. Introduction
Identifying SARS-CoV-2 as a new coronavirus and the pathogen causing the COVID-19 pandemic provides key information for subsequent disease prevention and control (Chan et al., 2020; Wu et al., 2020; Yang et al., 2020; Zhu et al., 2020). Coronaviruses are a group of enveloped single-strand positive-sense RNA viruses that can cause respiratory, gastrointestinal, hepatic and neurological diseases in humans and animals (Weiss and Leibowitz, 2011). Previously, six human coronaviruses have been discovered, including four common coronaviruses (229E, OC43, NL63, and HKU-1) that cause ∼20 % of common cold cases, and two highly pathogenic coronaviruses, namely, SARS-CoV emerging in 2002–2003 and MERS-CoV emerging since 2012 (Cui et al., 2019; Su et al., 2016). Phylogenetic analysis has revealed that SARS-CoV-2 is a new member of the subgenus Sarbecovirus of genus Betacoronavirus and is closely related to several bat coronaviruses and SARS-CoV (Lu et al., 2020; Wu et al., 2020).
The spike glycoproteins of coronaviruses are responsible for their attachment to the host receptor proteins to facilitate viral entry. Similar to SARS-CoV and NL63, SARS-CoV-2 engages angiotensin-converting enzyme 2 (ACE2) as an entry receptor (Glowacka et al., 2009; Hoffmann et al., 2020; Hofmann et al., 2005; Li et al., 2003). Recognition details between the receptor binding domain (RBD) of the spike protein and ACE2 have been analyzed by cryo-EM structure determination of the complex (Benton et al., 2020; Lan et al., 2020; Yan et al., 2020). The spike protein is also a viable and ideal target for the development of vaccines and therapeutics. However, the live virus strains are not easily accessible and must be handled in biosafety level 3 (BSL3) facilities. This condition hampers and delays the development of efficient anti-viral candidates. Pseudovirus particles are useful tools when the live virus strains and specialized biosafety facilities are not accessible such as Ebola virus, Nipah virus, Marburg virus, SARS-CoV and so on (Nie et al., 2019; Simmons et al., 2004; Yonezawa et al., 2005; Zhang et al., 2017). To date, several SARS-CoV-2 pseudovirus particle (SARS2pp) production and infection systems have been successfully generated (Hoffmann et al., 2020; Hu et al., 2020; Huang et al., 2020; Nie et al., 2020; Ou et al., 2020; Tani et al., 2021; Xiong et al., 2020). However, some SARS2pp production systems produce only low titers, and in most reported SARS2pp systems used only a single reporter gene such as eGFP or luciferase at a time, except that one vesicular stomatitis virus (VSV) based SARSpp system was constructed with dual-reporters (Hoffmann et al., 2020).
In this study, a robust dual-reporter pseudovirus system was developed for SARS-CoV-2 entry research based on a newly designed HIV vector that simultaneously expresses eGFP and firefly luciferase (luc2) for easy viral infection observation and analysis. Several important factors affecting pseudovirus particle production were optimized to improve viral titers. Two recombinant ACE2 proteins and two fully human antibodies (REGN10933 and REGN10987) reported previously were also evaluated using the proposed SARS2pp infection system. Finally, several SARS2pp susceptible cell lines were selected to compare their susceptibility to SARSpp.
2. Materials and methods
2.1. Plasmids, cells and reagents
The pCAG and pCMV vectors used for SARS-CoV-2 spike protein expression were derived from pcDNA3 with some modifications. The optimized sequences encoding SARS-CoV-2 spike protein were either designed by the online “GenSmart Codon Optimization” program from the GenScript company (New Jersey, USA) or gifted by Dr. Lu Lu (Fudan University, Shanghai, China). The lentiviral transfer plasmid pLDR bearing dual-reporter genes eGFP and Luc2 was modified from pLNB described by us previously (Xie et al., 2013). The lentiviral packaging plasmid psPAX2 was purchased from Addgene (Cambridge, MA, USA). All these constructs were verified by sequencing. Kidney-derived cell lines HEK293, HEK293 T, 293/hACE2, Vero-E6, and BHK21, the liver-derived cell lines HuH7 and HuH7.5.1, and the small intestine-derived cell line Caco-2 were maintained in Dulbecco’s modified Eagle medium (DMEM, Macgene, Beijing, China) supplemented with 10 % (vol/vol) fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA), 1 mM sodium pyruvate (Macgene), 1×non-essential amino acids (Macgene), and 50 IU/mL penicillin/streptomycin (Macgene) in a humidified 5 % (vol/vol) CO2 incubator at 37 °C. Recombinant ACE2-Fc and ACE2-His were purchased from Sino Biological (Beijing, China) and Bioworld Technology (St. Louis Park, MN, USA) respectively. Recombinant human SARS-CoV-2 neutralizing antibodies (REGN10933 and REGN10987) and IgG1 isotype control were acquired from AtaGenix (Wuhan, China). Recombinant human anti-RBD antibody named CR3022 was gifted by Dr. Jing Wang and Jian-nan Feng (Academy of Military Medical Sciences, Beijing, China). SlowFade® Gold with DAPI were obtained from Life Technologies (Carlsbad, CA, USA).
2.2. Indirect immunofluorescence analysis of spike proteins
Indirect immunofluorescence analysis was performed for cellular spike protein detection. After fixation with 4 % paraformaldehyde and permeabilization with 0.5 % Triton X-100, the HEK293 cells transfected with different kinds of spike protein plasmids were initially stained with anti-RBD antibody CR3022 (1:500 dilution), incubated with 1:1000 diluted Alexa Fluor 488-conjugated goat anti-human IgG secondary antibody (A-11013, Thermo Scientific), washed with PBS for three times, and then stained with SlowFade® Gold with DAPI. Images were captured by FV1000 (Olympus, Tokyo, Japan).
2.3. Production, purification, and infection of SARS2pp, SARSpp, and VSVpp
For SARS2pp, SARSpp, and VSVpp production and purification, the HEK293 T cells were seeded and maintained for 18−24 h until the cell density reached 70 %–90 % confluency at the time of transfection. The spike/pLDR/psPAX2 plasmids were cotransfected with GenJet™ Ver II (SignaGen Laboratories, Rockville, MD, USA) following the manufacturer’s instructions. The medium was changed 10 h post transfection. The cell culture supernatant was collected, pooled, centrifuged at 1000×g for 5 min, filtered through a 0.45-μm filter to further remove cellular debris, aliquoted, and stored at −70 °C until used. For pseudovirus infection, target cells were seeded at a density of 1 × 104 cells per well in 96-well plates at 16 h before infection. The cells in each well were then infected with pseudovirus particles at certain MOI. Culture medium was changed at 9 h post infection. The eGFP was captured using fluorescent microscopy (IX73 microscope, Olympus, Tokyo, Japan), and luciferase activity was detected at 72 h post infection using Steady-Lumi™ II Firefly Luciferase Assay Kit (Beyotime, Shanghai, China). For infection inhibition, pseudovirus particles were cocultured with recombinant ACE2 or antibodies for 30 min at 37 °C prior to SARS2pp infection. Each experiment was repeated at least twice to ensure consistency. Data shown were obtained from three replicates in a single representative experiment.
2.4. Statistical analysis
Data were analyzed using GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA) and expressed as mean ± standard deviation (SD). Statistical significance was determined by the Student’s t test, and P-values are indicated by asterisks (***P < 0.001, **P < 0.01, and *P < 0.05).
3. Results
3.1. Production and infection of four different constructs of SARS2pp
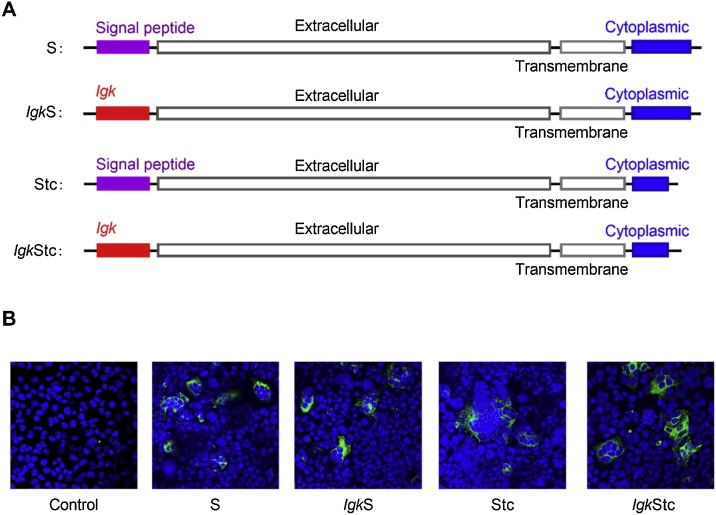
For successful SARS2pp production, the following four different constructs of the SARS-CoV-2 spike protein plasmids were designed and synthesized: 1) full-length spike protein (S), 2) spike protein (IgkS) with the signal peptide substituted by the leader sequence from mouse immunoglobulin κ light chain Igk, 3) spike protein (Stc) with a C-terminal 19 amino acids deletion, and 4) spike protein (IgkStc) with signal peptide substitution and C-terminal 19 amino acids deletion (Fig. 1 A). After transfection into HEK293 cells, the expression of these four different constructs of spike protein was detected by immunofluorescence analysis. All these four constructs were expressed on the cell membrane and showed cell fusion (Fig. 1B).
Fig. 1.
Four different constructs of SARS-CoV-2 spike protein. (A) Schematic of four different constructs of SARS-CoV-2 spike protein. S, full-length spike protein; IgkS, spike protein in which the signal peptide was substituted by Igk; Stc, spike protein with C-terminal 19 amino acids deletion; IgkStc, the spike protein with signal peptide replacement by Igk and C-terminal 19 amino acids deletion. (B) Immunofluorescence analysis of the four different constructs of SARS-CoV-2 spike protein expression at 48 h post transfection of HEK293 cells by confocal microscopy. Control, cells transfected with the empty vector pCMV.
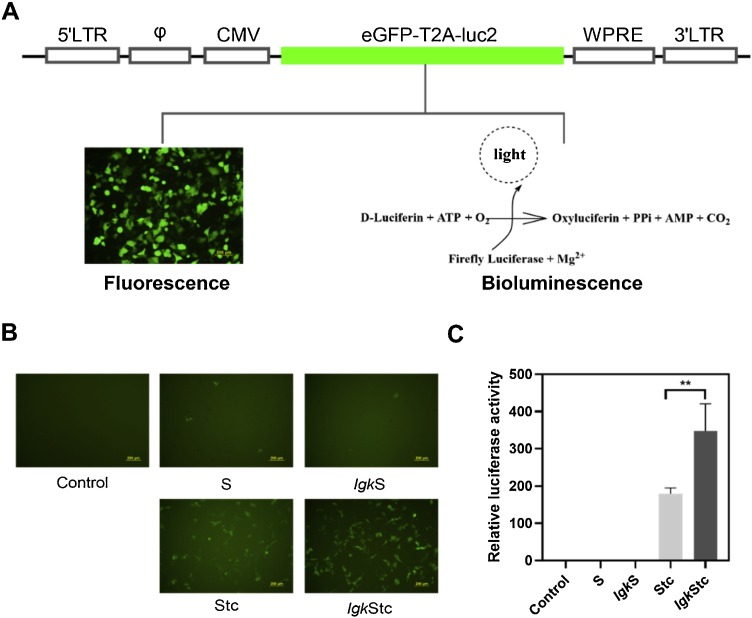
A dual-reporter vector bearing eGFP and Luc2 was designed for simplified observation and analysis (Fig. 2 A). SARS2pps packaged with the four constructs of SARS-CoV-2 spike protein were produced in HEK293 T cells, and its infectivity was determined in 293/hACE2 cells. SARS2pps packaged with S and IgkS showed lower eGFP expression and luciferase activity with that packaged with Stc and IgkStc (Fig. 2B, C), suggesting that the spike C-terminal 19 amino acids mainly restrained the SARS2pp production. Compared with that packaged with Stc, SARS2pp packaged by IgkStc showed approximately twofold increase in eGFP expression and luciferase activity (Fig. 2B, C), indicating that the signal peptide replacement by Igk could further facilitate SARS2pp production.
Fig. 2.
Dual-reporter SARS2pp production and infectivity. (A) Specification of the HIV dual-reporter plasmid for SARS2pp infection indication. eGFP-T2A-luc2, dual-reporter genes eGFP and luc2 connected by the T2A peptide. (B) and (C) Fluorescence and bioluminescence detection of SARS2pp infectivity (293/hACE2 cells) packaged by different constructs of SARS-CoV-2 spike proteins. The luciferase activities were normalized against "S" (set as 1). Control, non-transduced cells. Data were the results from three replicates.
3.2. Three optimized factors to improve SARS2pp titers
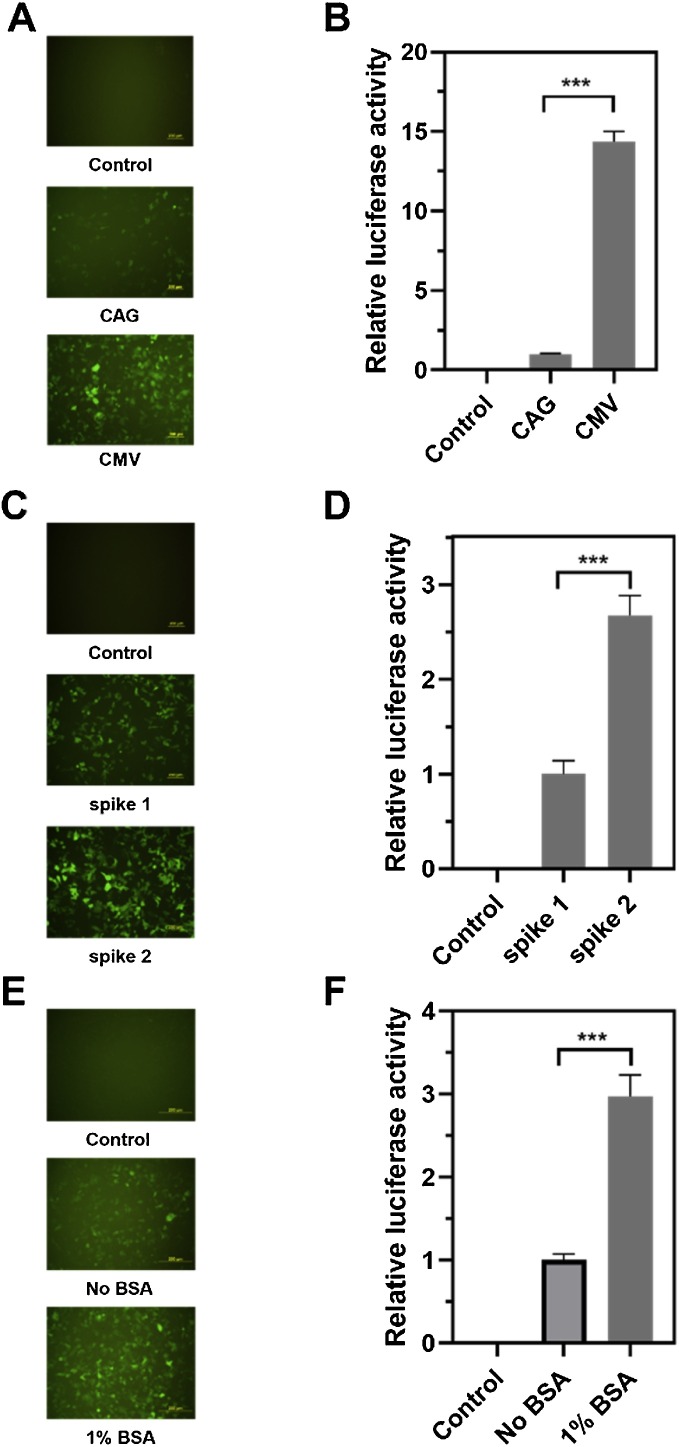
Several factors including the promoters for spike protein expression, different spike optimized sequences, and the production conditions were optimized to obtain high SARS2pp titers. First, SARS2pp titer was increased up to 10 times when the CMV promoter other than CAG was used for spike protein expression (Fig. 3 A). Second, the SARS2pp titer was improved by approximately 2–3 times through comparing different spike optimized sequences (Fig. 3B). Finally, 1 % BSA addition to the medium benefitted SARS2pp production and improved the titer by 2–3 times (Fig. 3C). These three optimized conditions were adopted in the subsequent studies.
Fig. 3.
Three factors to increase SARS2pp titer. (A) and (B) Fluorescence and bioluminescence detection of SARS2pp infectivity (293/hACE2 cells) packaged by SARS-CoV-2 spike protein expressed by different promoters (CAG and CMV). (C) and (D) Two optimized spike protein sequences with SARS2pp production improved by approximately 2.5 times. (E) and (F) 1 % BSA addition to the medium enhanced SARS2pp production about 2-3 times. Control, non-transduced cells. Data were the results from three replicates.
3.3. SARS2pp was a robust system for the evaluation of viral entry inhibitors
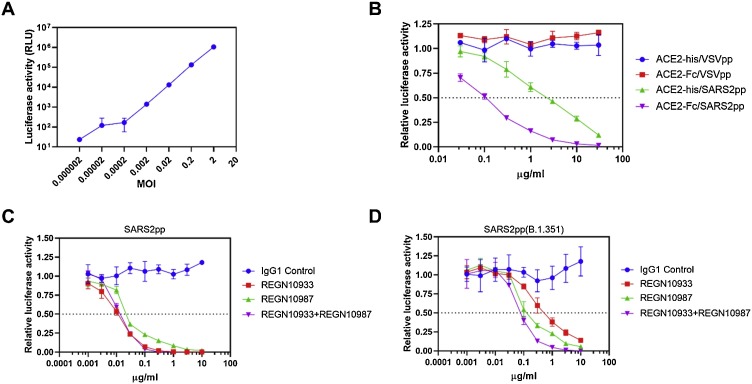
Infection efficiency was firstly detected at different MOIs in 293/hACE2 cells to examine the applicability of this SARS2pp system, and a good linear relationship was observed between MOI 2-0.0002 (Fig. 4 A). Inhibition assay was then performed to test the activity of recombinant ACE2-Fc and ACE2-his proteins (fusion proteins consisting of the extracellular domain (Met 1-Ser 740) of human ACE2 linked to the Fc region of human IgG1 or his tag at the C-terminus) against SARS2pp infection. The results showed that SARS2pp but not VSVpp was inhibited specifically by both recombinant proteins, and the action of ACE2-Fc was superior to that of ACE2-his (Fig. 4B). Two human SARS-CoV-2 neutralizing antibodies (REGN10933 and REGN10987) (Baum et al., 2020) were also re-evaluated against SARS2pp and the Beta variant SARS2pp(B.1.351). REGN10987 maintained about 25 % of its neutralization activity against SARS2pp(B.1.351), but REGN10933 had lost its neutralization activity against SARS2pp(B.1.351) at least 1.5 log units relative to that of SARS2pp. When REGN10933 and REGN10987 were combined into a cocktail (1:1), these two antibodies remained effective against SARS2pp and SARS2pp(B.1.351) (Fig. 4C, D).
Fig. 4.
Detection of neutralizing ability of recombinant ACE2 and human antibodies against SARS2pp infection. (A) SARS2pp showed a good linear relationship between MOI 2-0.0002. (B) ACE2-Fc was superior to ACE2-his for SARS2pp infection inhibition (MOI 0.3). VSVpp containing the spike glycoprotein of the vesicular stomatitis virus (VSV-G) as the control pseudotype particles. (C) and (D) Two SARS-CoV-2 neutralizing antibodies (REGN10933 and REGN10987) re-evaluated against SARS2pp and its South Africa variant SARS2pp(B.1.351) (MOI 0.3). Data were the results from three replicates.
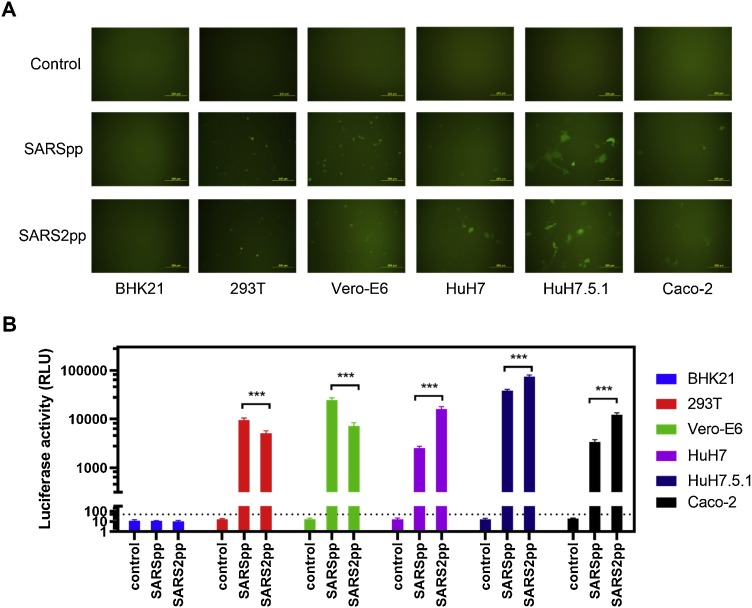
3.4. Different cell lines showed varying susceptibilities to SARSpp and SARS2pp
A panel of SARS2pp susceptible cell lines derived from kidney, liver, and small intestine were selected to compare their susceptibility to SARSpp. Baby Syrian hamster kidney cell line BHK21 served as the unsusceptible control. In accordance with SARS-CoV and SARS-CoV-2 causing multi-organ damage, all the tested cell lines showed susceptibility to SARSpp and SARS2pp but to a different degree, except BHK21. For the kidney-derived cell lines 293 T and Vero-E6, SARSpp showed higher transduction levels than SARS2pp. For liver- and small intestine-derived HuH7, HuH7.5.1, and Caco-2 cell lines, SARS2pp was more infectious than SARSpp (Fig. 5 ).
Fig. 5.
Different cell lines showed different susceptibility to SARSpp and SARS2pp (MOI 2). (A) Fluorescence detection of SARSpp and SARS2pp infectivity to kidney, liver, and small intestine-derived cell lines. (B) Bioluminescence detection of SARSpp and SARS2pp infectivity to kidney, liver, and small intestine-derived cell lines. The luciferase activity above three times of signals obtained from the unsusceptible BHK21 cells was regarded as positive (the dashed line). Data were the results from three replicates.
4. Discussion
The world is still facing the COVID-19 epidemic, and many viral mutations have appeared such as D614 G (Korber et al., 2020), N501Y (Leung et al., 2021), and E484 K (Houriiyah Tegally et al., 2020). E484 K is considered as one of the key mutations for vaccine resistance (Jangra et al., 2021). With the gradual increase in virus mutations, new vaccines and drugs remain urgently needed. However, the live SARS-CoV-2 virus strains must be handled in BSL3 facilities, and this condition greatly limits the research and development of efficient anti-viral candidates.
Pseudovirus is a good choice to mimic the true virus for the study of viral transduction mechanisms and viral infection inhibitors in BSL2 facilities. Several SARS2pp production and infection systems that are based on replication-deficient/-restricted retroviruses or rhabdoviruses (mostly vesicular stomatitis virus) have been proven to be suitable models to mimic SARS-CoV-2 entry (Hu et al., 2020; Huang et al., 2020; Ou et al., 2020). However, the SARS2pp titer was not high in most cases. Here, four different constructs of the SARS-CoV-2 spike proteins were designed and evaluated to construct a robust high-titer SARS2pp system. Results showed that the presence of the spike C-terminal 19 amino acids is the main restraining factor for SARS2pp production. Removing the C-terminal 19 amino acids from the spike protein could remarkably improve SARS2pp titers. This finding is similar to most published studies generating SARS2pp with C-terminal-deleted spike proteins (Hu et al., 2020; Ou et al., 2020). We proposed that similar to SARS-CoV, SARS-CoV-2 spike endodomain may contain a cellular ezrin-binding domain that limits SARS2pp entry and fusion (Millet et al., 2012). Igk is a mouse secretion signal peptide, which is widely used to enhance the expression levels of cytokines, chemokines, or other secreted proteins (Meazza et al., 2000; Peng et al., 2016; Wolschek et al., 2011). In this work, it seemed that the Igk signal peptide could further facilitate SARS2pp production.
Three other optimization conditions were also adopted to further improve SARS2pp titers. First, two promoters used for spike protein expression were compared. The CMV promoter is the most commonly used promoter for the production of recombinant proteins, and the CAG promoter is an artificial promoter composed of the CMV enhancer, chicken β-actin promoter, and a large synthetic intron (Kootstra et al., 2003) and is successfully used for SARSpp production (Simmons et al., 2004). This study showed that expressing the spike protein using the CMV promoter increased the SARS2pp titer by approximately 14 times. Second, two optimized spike protein sequences were screened and were found to improve SARS2pp production by approximately 2.5 times. Finally, 1 % BSA addition to the medium benefitted SARS2pp production. This finding is similar to a previous study stating that the stability of the lentiviral vectors can be increased by adding albumin and lipoproteins as the stabilizing agent (Carmo et al., 2009). Under these optimized conditions, SARS2pp was successfully produced with high titers above 105–106 TU/mL in most cases.
The proposed SARS2pp system showed a good linear relationship with MOI 2-0.0002 when applied for 293/hACE2 cell infection. The potential of recombinant ACE2 and antibodies to block SARS2pp infection was further verified using this pseudovirus system and the different susceptibilities of organ-specific cell lines to SARSpp and SARS2pp infection were also compared. However, over time, more and more SARS-CoV-2 antibody- and vaccine-resistant variants such as the B.1.351 and P.1 have been discovered. The B.1.351 and P.1 variants were first detected in South Africa and Brazil, respectively. Both variants have the E484 K mutation, which decreases the neutralizing antibody response produced by vaccines, monoclonal antibody therapies, and convalescent plasma (Jangra et al., 2021; Li et al., 2021; Zhou et al., 2021). Thus, new SARS-CoV-2 antibody resistance or vaccine escape variants must be carefully monitored.
Briefly, the designed SARS-CoV-2 pseudovirus system SARS2pp provids a safe and convenient tool to study SARS-CoV-2 and its variants for their entry mechanisms and inhibitors when BSL-3 facilities and live virus strains are not accessible.
Author statement
Xuejun Wang and Shengqi Wang: Conceptualization, Funding acquisition, Supervision Xuejun Wang: Methodology Peng Yang, Yuming Wu, Cong Huang and Yanlei Ding: Resources Xuejun Wang, Yang Yang, Peng Yang and Shengqi Wang: Data curation, Formal analysis, Writing- Original draft preparation. Xuejun Wang, Peng Yang, Yang Yang: Visualization, Investigation. Xuejun Wang: Writing- Reviewing and Editing.
Author contributions
Xuejun Wang and Shengqi Wang conceived the project and provided financial support. Xuejun Wang designed the pseudovirus system. Peng Yang, Yuming Wu, Cong Huang and Yanlei Ding constructed and extracted the plasmids. Xuejun Wang, Yang Yang and Peng Yang performed the cellular experiments. Xuejun Wang, Yang Yang, Peng Yang and Shengqi Wang wrote and revised the manuscript. All authors have read and approved the final manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFA0900800), the Medical Innovation Project (17SAZ13), the National Natural Science Foundation of China (81830101), and Beijing Nova Program (Z171100001117119).
References
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature (London) 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo M., Alves A., Rodrigues A.F., Coroadinha A.S., Carrondo M.J., Alves P.M., Cruz P.E. Stabilization of gammaretroviral and lentiviral vectors: from production to gene transfer. J. Gene Med. 2009;11:670–678. doi: 10.1002/jgm.1353. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H., Lo S.K., Chan K., Poon V.K., Chan W., Ip J.D., Cai J., Cheng V.C., Chen H., Hui C.K., Yuen K. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F., Eichler J., Drosten C., Pohlmann S. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2009;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. - PNAS. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houriiyah Tegally E.W.M.G., Fonseca J.G.D.D., Nokukhanya Msomi K.M.A.V., Mushal Allam A.I.T.M., Gert Van Zyl W.P.F.P., Hardie G.M.M.H., Tyers I.M.D.Y., Abrahams O.L.A.A., Godzik C.K.W.B., Junior Alcantara S.L.K.P., J Lessells J.N.B.C. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7:551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Tai C., Hsu Y., Cheng D., Hung S., Chai K.M., Wang Y., Wang J. Assessing the application of a pseudovirus system for emerging SARS-CoV-2 and re-emerging avian influenza virus H5 subtypes in vaccine development. Biomed. J. 2020;43:375–387. doi: 10.1016/j.bj.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Chernet R.L., Eaker L.Q., Ferreri E.D., Floda D.L., Gleason C.R., Kleiner G., Jurczyszak D., Matthews J.C., Mendez W.A., Mulder L.C.F., Russo K.T., Salimbangon A.T., Saksena M., Shin A.S., Sominsky L.A., Srivastava K. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kootstra N.A., Matsumura R., Verma I.M. Efficient production of human FVIII in hemophilic mice using lentiviral vectors. Mol. Ther. 2003;7:623–631. doi: 10.1016/s1525-0016(03)00073-x. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., Zhao C., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Liu J., Liang H., Shi Y., Shen Y., Xie L., Zhang L., Qu X., Xu W., Huang W., Wang Y. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371. doi: 10.1016/j.cell.2021.02.042. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meazza R., Lollini P.L., Nanni P., De Giovanni C., Gaggero A., Comes A., Cilli M., Di Carlo E., Ferrini S., Musiani P. Gene transfer of a secretable form of IL-15 in murine adenocarcinoma cells: effects on tumorigenicity, metastatic potential and immune response. Int. J. Cancer. 2000;87:574–581. [PubMed] [Google Scholar]
- Millet J.K., Kien F., Cheung C.Y., Siu Y.L., Chan W.L., Li H., Leung H.L., Jaume M., Bruzzone R., Peiris J.S., Altmeyer R.M., Nal B. Ezrin interacts with the SARS coronavirus Spike protein and restrains infection at the entry stage. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Liu L., Wang Q., Chen R., Ning T., Liu Q., Huang W., Wang Y. Nipah pseudovirus system enables evaluation of vaccines in vitro and in vivo using non-BSL-4 facilities. Emerg. Microbes Infect. 2019;8:272–281. doi: 10.1080/22221751.2019.1571871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yu X., Li C., Cai Y., Chen Y., He Y., Yang J., Jin J., Li H. Enhanced recombinant factor VII expression in Chinese hamster ovary cells by optimizing signal peptides and fed-batch medium. Bioengineered. 2016;7:189–197. doi: 10.1080/21655979.2016.1176656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. - PNAS. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Kimura M., Tan L., Yoshida Y., Ozawa T., Kishi H., Fukushi S., Saijo M., Sano K., Suzuki T., Kawasuji H., Ueno A., Miyajima Y., Fukui Y., Sakamaki I., Yamamoto Y., Morinaga Y. Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein. Virol. J. 2021;18:16. doi: 10.1186/s12985-021-01490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschek M., Samm E., Seper H., Sturlan S., Kuznetsova I., Schwager C., Khassidov A., Kittel C., Muster T., Egorov A., Bergmann M. Establishment of a chimeric, replication-deficient influenza A virus vector by modulation of splicing efficiency. J. Virol. 2011;85:2469–2473. doi: 10.1128/JVI.01650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y., Wang W., Song Z., Hu Y., Tao Z., Tian J., Pei Y., Yuan M., Zhang Y., Dai F., Liu Y., Wang Q., Zheng J., Xu L., Holmes E.C., Zhang Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P., Xie Y., Zhang X., Huang H., He L., Wang X., Wang S. Inhibition of dengue virus 2 replication by artificial microRNAs targeting the conserved regions. Nucleic Acid Ther. 2013;23:244–252. doi: 10.1089/nat.2012.0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Wu Y., Cao J., Yang R., Liu Y., Ma J., Qiao X., Yao X., Zhang B., Zhang Y., Hou W., Shi Y., Xu J., Zhang L., Wang S., Fu B., Yang T., Ge S., Zhang J., Yuan Q., Huang B., Li Z., Zhang T., Xia N. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa A., Cavrois M., Greene W.C. Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J. Virol. 2005;79:918–926. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li Q., Liu Q., Huang W., Nie J., Wang Y. A bioluminescent imaging mouse model for Marburg virus based on a pseudovirus system. Hum. Vaccin. Immunother. 2017;13:1811–1817. doi: 10.1080/21645515.2017.1325050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Paesen G.C., Lopez-Camacho C., Slon-Campos J., Hallis B., Coombes N., Bewley K., Charlton S., Walter T.S., Skelly D., Lumley S.F., Dold C., Levin R., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S., James W., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China N.C.I.A. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]