Summary
Telomere dysfunction-induced focus (TIF) assay allows efficient profiling of telomere dysfunctions in cells and tissues. Here, we describe the use of the TIF assay to screen synthetic peptides from E3 ubiquitin ligase FBW7, a tumor suppressor gene product, to prevent TIFs caused by environmental radiation stress. We demonstrate peptidomimetic telomere dysfunction inhibitor as a potentially intervening therapeutic drug candidate in aging-related diseases. This work demonstrates a novel utility of the TIF assay protocol in identifying telomere dysfunction inhibitors.
For complete details on the use and execution of this protocol, please refer to Wang et al (2020).
Subject areas: Cell Biology, Cell culture, Cell-based Assays, Cancer, Microscopy, Molecular Biology, Molecular/Chemical Probes, Protein Biochemistry
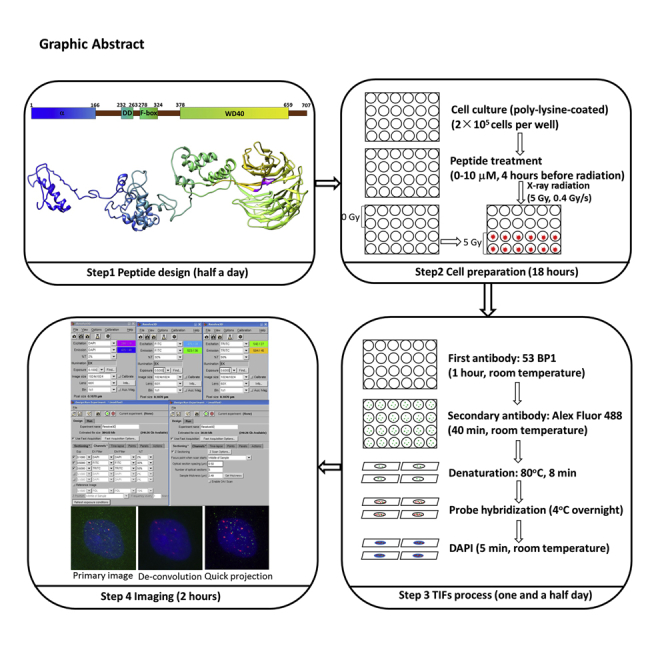
Graphical abstract
Highlights
-
•
Detailed experimental protocol for telomere dysfunction-induced focus (TIF) assay
-
•
Optimized TIF assay for screening inhibitors of telomere dysfunction in cell cultures
-
•
Identification of peptidomimetic telomere dysfunction inhibitor (TELODIN) from FBW7
-
•
Dose-dependent effect of TELODIN on TIFs caused by radiation stress in cancer cells
Telomere dysfunction-induced focus (TIF) assay allows efficient profiling of telomere dysfunctions in cells and tissues. Here, we describe the use of the TIF assay to screen synthetic peptides from E3 ubiquitin ligase FBW7, a tumor suppressor gene product, to prevent TIFs caused by environmental radiation stress. We demonstrate peptidomimetic telomere dysfunction inhibitor as a potentially intervening therapeutic drug candidate in aging-related diseases. This work demonstrates a novel utility of the TIF assay protocol in identifying telomere dysfunction inhibitors.
Before you begin
Telomeres are consisting of highly conserved TTAGGG repetitive sequence. Telomere dysfunction occurs under a variety of conditions including microenvironmental stresses (Wang et al., 2020). Measurement of telomere dysfunction informs telomere status associated with pathophysiological conditions, providing mechanistic insights into relationships between telomere dysfunctions and diseases. Using the nucleotide probes of (TTAGGG)n or (CCCTAA)n that hybridize with telomeric DNAs (Moyzis et al., 1988) provides a specific approach for detection telomere dysfunction foci (TIF) (Cartwright et al., 2019). In a purposely constructed peptide library, linked to an 11 amino acid (aa) residue sequence corresponding to the TAT47-57 (YGRKKRRQRRR) (Nischan et al., 2015), we screened for and identified a specific peptide sequence mimetic to the N terminus of the tumor suppressor FBW7 as an inhibitor to stress-induced mechanism of TIFs, telomere shortening, telomere DNA damage response, stem cell senescence, and pulmonary fibrosis (Wang et al., 2020).
The present protocol details the procedures of identifying TELODIN by using TIF assay in cultured HeLa, A549 and BEAS-2B cells.
Peptide design and synthesis
Timing: 1 week
In choosing synthetic peptide sequences, we focused on the WD40 domain that mediates protein interaction and performed a random selection with preferences of the sequences for two-class properties of (i) surfactant and (ii) cationic charged probabilities, followed by further refining of the positive hits via truncations and mutations for minimal structures.
-
1.Peptide design
- a.
-
b.An 11 aa residue sequence corresponding to the TAT47-57 (YGRKKRRQRRR) (Nischan et al., 2015) was chosen to fuse directly to the N termini of peptides as an alternative mechanism to facilitate peptide entry into cultured cells.
-
c.The aa sequence of control peptide (YK32) was corresponding to the aa 687–707 of FBW7.
-
d.Five short peptides were corresponding to the truncated forms of TELODIN (YK21).
-
2.
Peptide synthesis
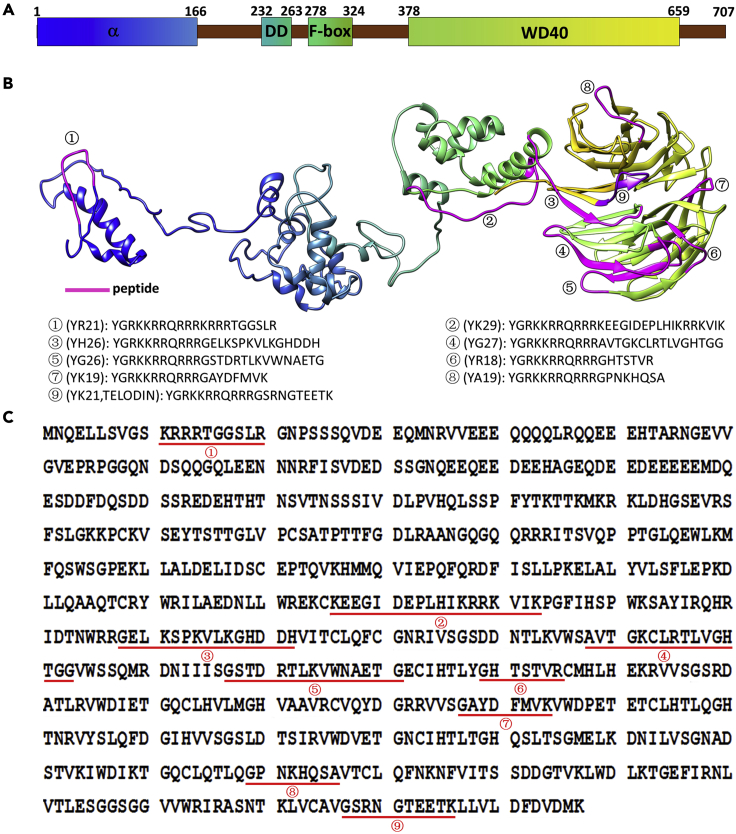
Figure 1.
FBW7 primary structure and synthetic peptide sequences
(A). FBW7 domain structure. α: α helix; DD: dimer domain.
(B) The 3D structure of FBW7. Marked are the sites of primary structures for synthetic peptides to be tested.
(C) Primary sequences of the initial synthetic peptides tested for telomere dysfunction inhibitory activity.
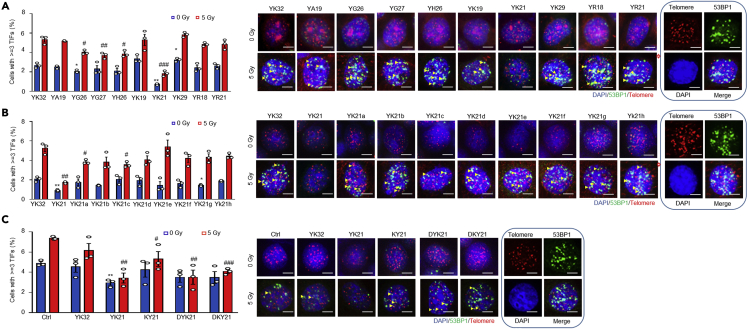
Figure 2.
Screening for effect of synthetic peptides on telomere dysfunction by TIF assay
HeLa seeded onto 12 well plate were incubated with different peptides. After 4 h, cells were exposed to 5 Gy X-ray radiation and TIF was determined by the TIF assay protocol. Similar experiments were repeated three times. Controls included cells treated with the YK32 peptide or cells not treated with any peptide. Scar bar, 2 μm.
(A and B) ∗ vs. YK32 (0 Gy), # vs. YK32 (5 Gy).
(C) ∗ vs. Control (Ctrl, 0 Gy), # vs. Control (Ctrl, 5 Gy). Note: YK21a-h were the mutant forms of YK21. KY21 was with reverse sequence of YK21. DYK21 and DKY21 were the dextrorotatory forms of YK21 and KY21, respectively. GE8, GK10, GT9, SE7 and ST8 were the short forms of YK21.
All peptides, including Cy3-TELODIN used were 95% pure, synthetized by KE Biochem, Shanghai, China. Peptides (1 mg/tube) were resolved in D-PBS to a stock concentration of 1 mM.
Cell culture and treatment
Timing: 2 days
-
3.Sample preparation
-
a.Seed cells on poly-lysine-coated cover slips to 50%–60% confluence (2×105 cells per well for a 12 well plate).
- b.
-
c.Cells were treated with X-ray radiation 4 h after peptide addition. Cells were then removed from incubator and placed onto the insert of X-ray irradiator (Rad Source RS2000XE), the parameter was set by 0.4 Gy/min, total of 750 s (total dose of radiation was 5 Gy). After radiation, cells were incubated for another 1 h.
-
d.Remove culture media from cells after 1 h radiation of TIFs.
-
e.Wash cells with 1 mL 1× PBS for 5 min.
-
a.
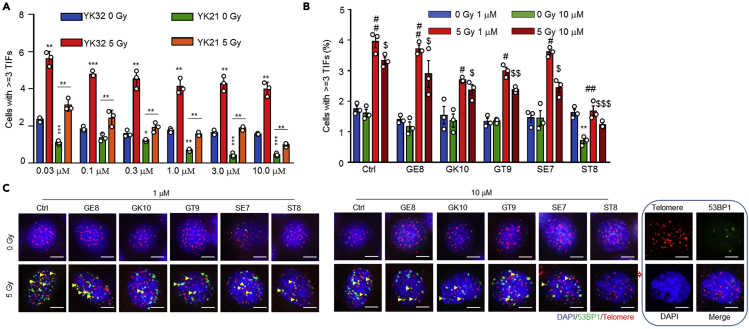
Figure 3.
Concentration-dependent effect of TELODIN on telomere dysfunction by TIF assay
HeLa cells were seeded onto 12 well plate, to which different doses of peptides were added as indicated. After 4 h, cells were exposed to 5 Gy X-ray radiation, followed by TIF assay. Similar experiments were repeated for three times. ∗ vs. Control in the absence of IR. # vs. Control in the presence of 5 Gy IR. $ vs. control at a higher concentration in the presence of 5 Gy IR.
(A) Effects on TIF of different dosages of X-ray radiation plus or minus TELODIN YK21.
(B) Effects on TIF of different dosages of X-ray radiation plus or minus TELODIN ST8.
(C) Representative images of the effect of TELODIN ST8 on radiation-induced TIF.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibody | ||
| 53BP1 | Abcam | Cat#ab36823 |
| Alex Fluor 488-conjugated Goat anti-rabbit IgG (H+L) | Thermo | Cat#A27034 |
| Probe | ||
| PNA probe (Cy3 labeled, CCCTAACCCTAACCCTAA) | Panagene | Cat#F1002 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM (high glucose) | CellMax | CGM101.05 |
| FBS | CellMax | SA311.02 |
| Typsin | CellMax | CPT101.02 |
| D-PBS | Youkang | BS0402 |
| Formamide | Sigma | F9037 |
| Blocking reagent | Roche | Roche # 1196176001 |
| BSA | Sigma | SRE0096 |
| Goat serum | Sigma | G5018 |
| DAPI | Sigma | D8417 |
| Antifade Mounting Medium | Vector Laboratories | H-1000 |
| Fisherbrand Microscope Cover Glass | Thermo Fisher | 12-545-80 |
| DKY21 (TELODIN) | KB Biochem | KTEETGNRSGRRRQRRKKRGY |
| DYK21 (TELODIN) | KB Biochem | YGRKKRRQRRRGSRNGTEETK |
| GE8 | KB Biochem | GSRNGTEE |
| GK10 | KB Biochem | GSRNGTE ETK |
| GT9 | KB Biochem | GSRNGTEET |
| KY21 (TELODIN) | KB Biochem | KTEETGNRSGRRRQRRKKRGY |
| SE7 | KB Biochem | SRNGTEE |
| ST8 (TELODIN) | KB Biochem | SRNGTEET |
| YA19 | KB Biochem | YGRKKRRQRRRGPNKHQSA |
| YG26 | KB Biochem | YGRKKRRQRRRGSTDRTLKVWNAETG |
| YG27 | KB Biochem | YGRKKRRQRRRAVTGKCLRTLVGHTGG |
| YH26 | KB Biochem | YGRKKRRQRRRGELKSPKVLKGHDDH |
| YK19 | KB Biochem | YGRKKRRQRRRGAYDFMVK |
| YK21 (TELODIN) | KB Biochem | YGRKKRRQRRRGSRNGTEETK |
| YK21a (TELODIN) | KB Biochem | YGRKKRRQRRRRNGTEETK |
| YK21b | KB Biochem | YGRKKRRQRRRGSNGTEETK |
| YK21c (TELODIN) | KB Biochem | YGRKKRRQRRRGSRGTEETK |
| YK21d | KB Biochem | YGRKKRRQRRRGSRNGTEET |
| YK21e | KB Biochem | YGRKKRRQRRRGSRNGTEE |
| YK21f | KB Biochem | YGRKKRRQRRRGSRNGTETK |
| YK21g | KB Biochem | YGRKKRRQRRRGDRNGTEETK |
| YK21h | KB Biochem | YGRKKRRQRRRGERNGTEETK |
| YK29 | KB Biochem | YGRKKRRQRRRKEEGIDEPLHIKRRKVIK |
| YK32 | KB Biochem | YGRKKRRQRRRGSRNGTEETKLLVLDFDVDMK |
| YR18 | KB Biochem | YGRKKRRQRRRGHTSTVR |
| YR21 | KB Biochem | YGRKKRRQRRRKRRRTGGSLR |
| Software and algorithms | ||
| ImageJ | ImageJ | https://imagej.net/Downloads |
| GraphPad Prism 7 | GraphPad Prism | http://www.graphpad.com/scientific-siftware/orusm/ |
| softWoRx 6.0 | Cytiva | https://download.cytivalifesciences.com/cellanalysis/download_data/softWoRx/softworx_downloads.htm |
| Other | ||
| DeltaVision® widefield fluorescence microscope | GE Healthcare | N/A |
| Rad Source RS2000XE | Rad Source | N/A |
Materials and equipment
Materials
1×PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 137 mM | 8.0 g |
| KCl | 2.7 mM | 0.2 g |
| KH2PO4 | 2 mM | 0.27 g |
| Na2HPO4·12H2O | 10 mM | 1.42 g |
| ddH2O | Up to 1 L | |
| Total | n/a | 1 L |
Note: PBS was stored at 15°C–25°C for 2 weeks.
Blocking solution
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 1 mg/mL | 50 mg |
| Goat serum | 3% | 1.5 mL |
| Triton X100 | 0.1% | 50 μL |
| EDTA (500 mM) | 1 mM | 100 μL |
| 1×PBS | 48 mL | |
| Total | n/a | 50 mL |
Note: Blocking solution was stored at −20°C in 10 mL aliqutos for 2 months.
Hybridization solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Formamide | 70% | 1.4 mL |
| Blocking reagent (10%) | 0.5% | 100 μL |
| Tris-HCl pH 7.2 (1 M) | 10 mM | 20 μL |
| EDTA (500 mM) | 1 mM | 100 μL |
| Probe (25 μM) | 125 nM | 10 μL |
| ddH2O | 0.37 mL | |
| Total | n/a | 2 mL |
Note: Hybridization solution should be made freshly.
| Blocking reagent | 100 mM maleic acid and 150 mM NaCl, pH 7.5 |
| Washing buffer | 70% formamide and 10 mM Tris-HCl pH 7.2 |
Note: 10× blocking solution stock was prepared according to manufacturer’s instructions (Roche #11096176001)”
Step-by-step method details
TIF analysis
Timing: 2 days
Timing: 3.5 h for antibody incubation
Timing: 20 h for probe hybridization
DNA damage response or DDR at telomere is considered as TIFs (Takai et al., 2003). The TIF assay is performed by the co-localization of DNA damage marker, γH2AX or 53BP1, and telomere markers, such as TRF1 (Takai et al., 2003), TRF2 (Mender and Shay, 2015) or PNA probe (Rai and Chang, 2015).
-
1.Antibody incubation
-
a.Remove the medium of cell culture and wash cells with 1×PBS once.
-
b.Fix cells with 100% cold methanol at −20°C for 10 min.
-
c.Wash cells three times with 1 mL 1× cold PBS for 5 min.
-
d.Incubate cells with blocking solution for 40 min at 15°C–25°C.
-
e.Incubate cells with primary antibody (anti 53BP1 antibody, 1:100 dilution) diluted in blocking solution for 1 h at 15°C–25°C. Coverslips are incubated cell-side-down on antibody solution (10 μL per slide, spotted on parafilm) in a humidified chamber.
-
f.Wash cells three times with 1× PBS, each for 5 min.
-
g.Incubate cells with secondary antibody (1:1000 dilution) diluted in blocking solution for 40 min at 15°C–25°C. Coverslips are incubated cell-side-down on antibody solution (10 μL per slide, spotted on parafilm) in a humidified chamber.
-
h.Wash cells three times with 1 mL 1×PBS, each for 5 min.
-
a.
-
2.Probe hybridization
-
a.Fix the cells for 2% paraformaldehyde for 5 min at 15°C–25°C.
-
b.Wash cells three times with 1× PBS for 5 min.
-
c.Dehydrate the cells in 70%, 95% and 100% ethanol consecutively for 5 min each.
-
d.Dry the cover slips for 5–10 min.
-
e.Place the humidified chamber into 80°C oven for preheating.
-
f.Place 25 μL hybridization solution on the slide and flip the cover slips upside down in hybridization solution.
-
g.Place the slide in preheated humidified chamber.
-
h.Denature with hybridization solution for 8 min at 80°C.
-
i.Take the humidified chamber containing slides out of the 80°C oven
-
j.Put the humidified chamber containing slides in 4°C refrigerator for 16 h.
-
k.Wash cells twice with 1 mL washing solution at 15°C–25°C, each for 15 min.
-
l.Wash cells three times with 1×PBS, each for 5 min.
-
m.Stain the nuclear in DAPI solution (0.1 μg/mL) for 5 min.
-
n.Wash cells once with 1 mL PBS for 5 min.
-
o.Dry the slides for 10 min.
-
p.Place coverslips cell-side-down on slides with one drop of antifade mounting medium being careful to eliminate all air bubbles. Seal the coverslip with clear nail polish and slides are ready for microscopy or can be stored at 4°C and images acquired at a later time.
-
a.
Imaging with a fluorescent microscope
Timing: 1 day
-
3.Imaging
-
a.Acquire images by a DeltaVision® fluorescent microscope with 60x objective.
-
b.The images were taken by softWoRx 6.0 software.
-
c.Set the image size: 1,024 × 1,024.
-
d.Set the parameters: three channels: TRITC (excitation: 555/28 nm, emission: 617/73 nm, for telomere probe), FITC (excitation: 490/20 nm, emission: 528/38 nm, for 53BP1 signal) and DAPI (excitation: 360/40 nm, emission: 457/50 nm, for DNA staining).
-
e.Adjust light source intensity and exposure time for each channels: TRITC, 50% intensity and 0.6 s, FITC, 32% intensity and 0.5 s and DAPI, 2% intensity, 0.1 s.
-
f.Set Z-stack distance: approximately 2 μm, make sure the middle location got the most clear images and take an image every 0.5 μm.
-
g.Take images and label the name the images.
-
h.Perform de-convolution and quick projection processes to improve the quality of images. Particularly, Z-projection using maximum pixel intensity is applied.
-
i.Data collection: A total of 50 images were taken for each condition.
-
a.
Image analysis
Timing: 1 day
-
4.
Co-localization analysis
Images were de-convoluted followed by quick projection process and the co-localization analysis of two different fluorescence signals was performed by Image J software.-
a.Quantitation is performed using Image J software
-
b.Identify images in Photoshop.
-
c.Select images using Image J, and split channels, blue (nuclear, DAPI staining), red (telomere, PNA probe) and green (53 BP1, 488 secondary antibody)
-
d.Choose Plugins: CO-LOCALIZATION threshold, channel 1 (red) and channel 2 (green) will be displayed, choose red and green channels, and pick show scatter button.
-
e.Choose Plugin and use Colocalization Finder, to analyze the co-localization of channel 1 and channel 2, and then press OK button.
-
f.View results in the scatter plot windows.
-
g.Confirm the co-localization: the Pearson’s R value in 0.5–1.0 and the Mander’s value in 0.6–1.0 was considered as co-localization.
-
h.Summarize the results: record the co-localization values in an Excel sheet and get the number of co-localization in cells.
-
a.
In one cell, more than 3 co-localizations of 53BP1 and telomeres were considered to be TIF positive cell. About 300–500 cells were calculated in one condition.
Expected outcomes
TIF assay informs the levels of telomere uncapping in cultured cells in response to ionizing radiation. In this protocol, we treated cells with X-ray irradiation and peptide. TIF assay was performed to determine the inhibitory effects of peptides on telomere DNA damage. The images were taken by a DeltaVision Elite system on an Olympus IX71 inverted microscope, running softWoRx 6.0, fluorescence images were acquired at 60× magnifications, by a CoolSnap HQ2 CCD camera. We expect that several peptides will show protective effects on X-ray irradiation induced TIFs. By testing FBW7 peptides, TIF assay demonstrated effect of the peptides on the basal and radiation-induced levels of telomere uncapping. Our results indicated that TELODIN inhibited both basal and radiation-induced telomere uncapping in cultured human cancer HeLa cells (Figures 2 and 3, YK32 was used as a control).
Quantification and statistical analysis
Each experiment was repeated at least three times. Data were presented as mean ± SD or SEM of at least three independent experiments. Differences among variables were assessed by two-tailed Student’s t-test. A p value of less than 0.05 was considered statistically significant (∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001).
Limitations
A limitation of the protocol would be cellular uptake of the biocompounds of various peptides tested. A valid effect of peptides on TIFs depends on successful delivery of the peptides into the nucleus. The assay protocol would be unavailable or unsuccessful under particular conditions of cellular status where peptides would not reach to telomeres. In addition, the protocol has a limitation of currently unavailable, or yet to be discovered, small molecular positive control, for which TELODIN would be useful in future studies.
Troubleshooting
Problem 1
Weak telomere signal (TIF analysis, probe hybridization)
Potential solution
It would be important that cells were under the log phase proliferative condition and the hybridization buffer was made freshly. The humidified chamber should be preheated at 80°C for 5–10 min of heat denaturation. Additionally, the concentrations of telomere probe can be increased to 250 nM. The hybridization temperature and time can be changed to 37°C for 1 h, or 15°C–25°C for two hours.
Problem 2
Weak DNA damage signals (TIF analysis, antibody incubation)
Potential solution
The cells can be treated with other positive DNA damage conditions, such as X-ray radiation, hydrogen peroxide, or reagent that caused DNA damage. Usage of γH2AX as another maker of DNA damage was recommended.
Problem 3
High background (TIF analysis)
Potential solution
The cover-slips used must be cleaned carefully. The concentrations and incubation conditions (e.g., temperature and time) of the first and secondary antibodies should be tested in advance. Cells were washed by 1×PBS containing 0.1% Triton X100 for 5–10 min each time for five times after antibody incubation. The washing time of telomere probe hybridization can be extend with 40 min every time.
Problem 4
Too little TIF signal of positive cells (TIF analysis)
Potential solution
Cells can be treated by silencing shelterin genes as positive control. Please change the treatment condition, such as increasing the concentration of DNA damage reagent or the incubation time.
Include additional positive controls, including cells transfected with TRF2 siRNA and perform TIFs assay.
Change the treatment condition, such as increasing the concentration of DNA damage reagent or the incubation time.
Problem 5
Difficult to determine the co-localization of DDR signal with telomeres (Image analysis)
Potential solution
Another image analysis software, such as Bitplane Imaris, was recommended to use.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, [Jun-ping Liu] (jun-ping.liu@hznu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate or analyze datasets or code.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2018YFC2000100), the National Natural Science Foundation of China (81530039, 91949207, 91649205, 91849124, and 81871112), the National Basic Research Program of China (2012CB911204), and the Victorian Government’s Operational Infrastructure Support Program of Australia.
Author contributions
L.W. performed all experiments, interpreted data, and wrote the paper. Z.W. assisted bioinformatics analysis. J-.P.L. conceived the study, designed and supervised experiments, interpreted data, and wrote the paper.
Declaration of interests
J-.P.L. is the founder of Hangzhou Duanli Biotechnology Company Limited, Hangzhou, Zhejiang, China.
References
- Cartwright I.M., Haskins J.S., Kato T.A. PNA telomere and centromere FISH staining for accurate analysis of radiation-induced chromosomal aberrations. Methods Mol. Biol. 2019;1984:95–100. doi: 10.1007/978-1-4939-9432-8_11. [DOI] [PubMed] [Google Scholar]
- Mender I., Shay J.W. Telomere dysfunction induced foci (TIF) analysis. Bio Protoc. 2015;5:e1656. doi: 10.21769/bioprotoc.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R.K., Buckingham J.M., Cram L.S., Dani M., Deaven L.L., Jones M.D., Meyne J., Ratliff R.L., Wu J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U S A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nischan N., Herce H.D., Natale F., Bohlke N., Budisa N., Cardoso M.C., Hackenberger C.P.R. Covalent attachment of cyclic TAT peptides to GFP results in protein delivery into live cells with immediate bioavailability. Angew. Chem. Int. Ed. 2015;54:1950–1953. doi: 10.1002/anie.201410006. [DOI] [PubMed] [Google Scholar]
- Rai R., Chang S. Monitoring the DNA damage response at dysfunctional telomeres. Methods Mol. Biol. 2015;1343:175–180. doi: 10.1007/978-1-4939-2963-4_14. [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Wang L., Chen R., Li G., Wang Z., Liu J., Liang Y., Liu J.P. FBW7 mediates senescence and pulmonary fibrosis through telomere uncapping. Cell Metab. 2020;32:860–877 e869. doi: 10.1016/j.cmet.2020.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate or analyze datasets or code.