Abstract
Cellular pyruvate is an essential metabolite at the crossroads of glycolysis and oxidative phosphorylation, capable of supporting fermentative glycolysis by reduction to lactate mediated by lactate dehydrogenase (LDH) among other functions. Several inherited diseases of mitochondrial metabolism impact extracellular (plasma) pyruvate concentrations, and [1-13C]pyruvate infusion is used in isotope-labeled metabolic tracing studies, including hyperpolarized magnetic resonance spectroscopic imaging. However, how these extracellular pyruvate sources impact intracellular metabolism is not clear. Herein, we examined the effects of excess exogenous pyruvate on intracellular LDH activity, extracellular acidification rates (ECARs) as a measure of lactate production, and hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rates across a panel of tumor and normal cells. Combined LDH activity and LDHB/LDHA expression analysis intimated various heterotetrameric isoforms comprising LDHA and LDHB in tumor cells, not only canonical LDHA. Millimolar concentrations of exogenous pyruvate induced substrate inhibition of LDH activity in both enzymatic assays ex vivo and in live cells, abrogated glycolytic ECAR, and inhibited hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rates in cellulo. Of importance, the extent of exogenous pyruvate-induced inhibition of LDH and glycolytic ECAR in live cells was highly dependent on pyruvate influx, functionally mediated by monocarboxylate transporter-1 localized to the plasma membrane. These data provided evidence that highly concentrated bolus injections of pyruvate in vivo may transiently inhibit LDH activity in a tissue type- and monocarboxylate transporter-1–dependent manner. Maintaining plasma pyruvate at submillimolar concentrations could potentially minimize transient metabolic perturbations, improve pyruvate therapy, and enhance quantification of metabolic studies, including hyperpolarized [1-13C]pyruvate magnetic resonance spectroscopic imaging and stable isotope tracer experiments.
Keywords: lactate dehydrogenase, LDH, [1-13C]pyruvate, stable isotope, cancer metabolism, hyperpolarized 13C, 13C magnetic resonance spectroscopy, MRSI
Abbreviations: DMSO, dimethyl sulfoxide; ECAR, extracellular acidification rate; LDH, lactate dehydrogenase; MCT, monocarboxylate transporter; MRSI, magnetic resonance spectroscopic imaging; OxPhos, oxidative phosphorylation; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase; TCA, tricarboxylic acid
Cellular pyruvate is an essential metabolite that resides at the intersection of glycolysis and oxidative phosphorylation (OxPhos). Pyruvate can be oxidized by pyruvate dehydrogenase (PDH) to form acetyl-CoA entering the citric acid (TCA) cycle, further undergoing OxPhos to generate ATP. Alternatively, pyruvate fuels anaplerosis by replenishing oxaloacetate in the TCA cycle, and conversion of pyruvate to alanine facilitates syntheses of various amino acids. In the process of glucose fermentation (anaerobic glycolysis), pyruvate is reduced to lactate via cytoplasmic lactate dehydrogenase (LDH), coupled with NADH to NAD+ oxidation. Pyruvate-to-lactate conversion is an important pathway for ATP generation and homeostasis under physiologically low pO2 (anaerobic glycolysis of exercising muscle) and also supports the reprogramed cancer metabolic state of aerobic glycolysis, known as the Warburg effect (1).
The physiological concentration of pyruvate in plasma ranges from 30 to 260 μM (2). However, two inherited diseases of mitochondrial metabolism impact extracellular (plasma) pyruvate concentration: pyruvate carboxylase (PC) deficiency and PDH deficiency. PC deficiency is a rare genetic disorder present at birth characterized by developmental delay, recurrent seizures, and failure to produce TCA intermediates and neurotransmitters important for brain function (3). PC deficiency is diagnosed by physical symptoms and laboratory analysis, including high plasma levels of pyruvate, ammonia, lactate, acetoacetate, and beta-hydroxybutyrate. The relationships between elevated pyruvate levels and tissue-selective toxicity remain underexplored. In PDH deficiency, wherein pyruvate fails to enter the TCA cycle, systemic increases of lactate result in lactic acidosis with potential profound brain stem and ganglia damage (4). As an alternative energy source, pyruvate therapy can be used to treat patients in whom OxPhos is impaired and an increased NADH/NAD+ ratio limits the activity of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5). The administered exogenous pyruvate is transported into cells, mediated by monocarboxylate transporter-1 (MCT1) (6), a member of the SLC16 family (7). Therein, intracellular pyruvate can then be catalyzed by LDH, producing lactate and NAD+, thereby restoring GAPDH activity. Although highly concentrated pyruvate (0.5–1.0 g/kg/day) is administered to some patients with benefit, the multiorgan metabolic effects of exogenous pyruvate therapy remain under evaluation.
In this context, isotope-labeled pyruvate is a useful biochemical tool compound commonly used in tracing experiments to examine metabolism both in vitro and in humans in vivo (8, 9, 10, 11). One translational application of isotope-labeled substrates is hyperpolarized magnetic resonance spectroscopic imaging (MRSI), which has emerged as a noninvasive metabolic imaging modality with >10,000-fold signal enhancement compared with standard MRI (12, 13, 14, 15, 16, 17, 18, 19, 20). Infused [1-13C]pyruvate is the most well-characterized exogenous hyperpolarization probe for metabolic analysis and clinical translation, wherein intracellular [1-13C]pyruvate is converted to [1-13C]lactate via cytoplasmic LDH, which can be directly monitored by MRSI in living animals and humans (21). Because [1-13C]pyruvate carries a physiological negative charge, transmembrane influx of systemically administered [1-13C]pyruvate from extracellular spaces into intracellular spaces is transporter mediated primarily by MCT1 (6). The conversion of [1-13C]pyruvate to [1-13C]lactate has been used conventionally to inform the state of glycolytic flux of various tumors as well as to assess LDH activity either qualitatively or quantitatively. However, recent data provide a reinterpretation of MRSI signals in terms of MCT1 signatures as the rate-limiting step, pointing to new uses of the technology as a cancer prognostic marker in vivo (6). Technical constraints of hyperpolarized [1-13C]pyruvate MRSI studies require an injection into animals and humans of highly concentrated hyperpolarized [1-13C]pyruvic acid solutions, typically at 80 mM in mice (resulting in ∼8 mM blood pyruvate concentration) or 230 mM in humans at 5 ml/s injection rate in vivo (resulting in ∼2 mM or higher) (14, 22, 23). Studies have reported that bolus injection of pyruvate is not metabolically inert and imposes potential biochemical and physiological effects on biologic systems, including increased cellular oxygen consumption rates and hypoxia, and transient decreases in pO2 (24, 25, 26). However, the biochemical mechanism(s) of the observed phenotypes are not entirely elucidated.
Hence, further analysis in vivo is desired to enhance our understanding of potential metabolic perturbations induced by elevated plasma pyruvate and, more specifically, bolus infusions of hyperpolarized [1-13C]pyruvate during hyperpolarized [1-13C]pyruvate MRSI studies. With this in mind, by evaluating LDH enzyme kinetics quantitatively in vitro and characterizing the effect of concentrated exogenous pyruvate on lactate production semiquantitatively in cellulo using genetic and pharmacologic tools across well-defined cell lines, we found that biomedically relevant millimolar concentrations of pyruvate, typically used during pyruvate therapy and [1-13C]pyruvate MRSI, induced decreased lactate production in live cells, mechanistically related to substrate inhibition of LDH. Furthermore, because intracellular pyruvate accumulation is dependent on pyruvate influx (6), pyruvate-induced LDH substrate inhibition was found to be highly cell-type dependent and mechanistically coupled to MCT1 expression levels.
Results
Excess pyruvate impacts LDH enzymatic activity in vitro
LDH is a tetrameric enzyme comprising LDHA (M subunit primarily expressed in skeletal muscle) and LDHB (H subunit primarily expressed in the heart) (27, 28), wherein various combinations of LDHA and LDHB give rise to homo- and heterotetramers of five different LDH isoforms (LDH1–LDH5) (28). In the cancer metabolism literature, the aerobic glycolytic characteristics of many cancers have been conventionally attributed to LDHA isoforms (27, 29, 30, 31).
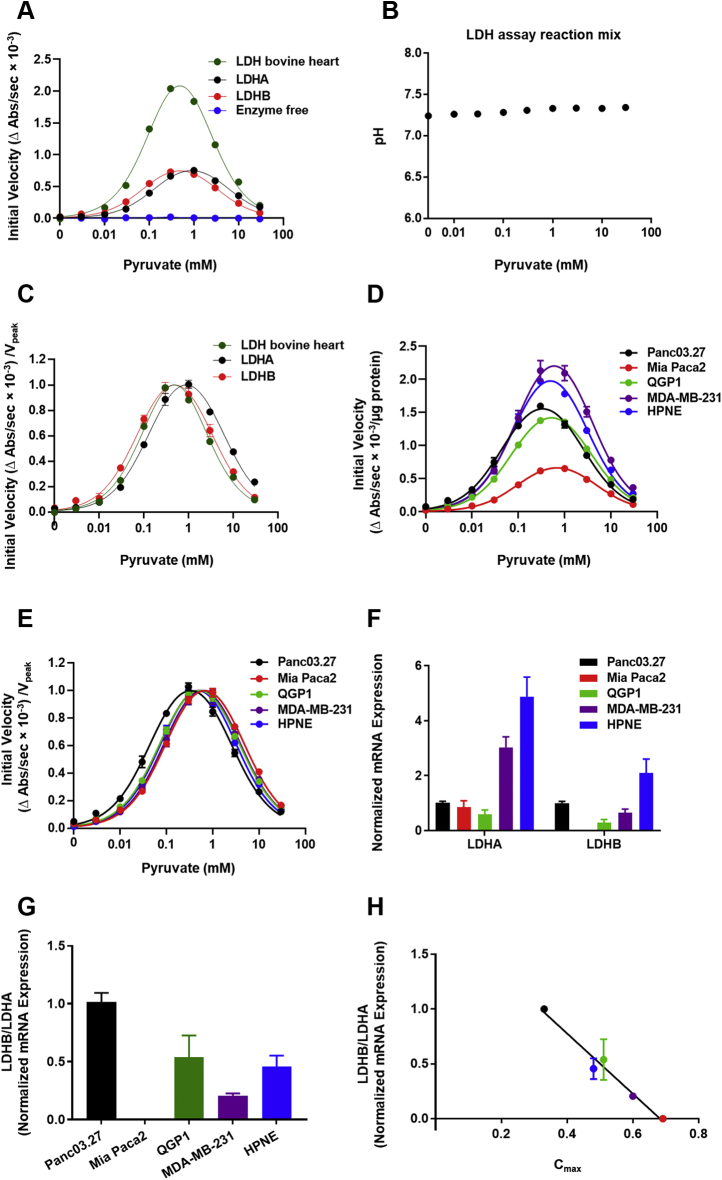
Rather longstanding and perhaps forgotten metabolic studies have suggested that high concentrations of pyruvate can induce substrate inhibition of LDH extracted from various cells, including cardiac cells, erythrocytes, and muscle cells (32, 33). To extend these previous observations to tumor cell-extracted enzymes (32, 33, 34), we first validated a classic coupled-enzyme kinetic assay using commercially available bovine heart LDH, purified human recombinant LDHA, and recombinant LDHB in response to a broad range of pyruvate concentrations. Because initial velocities were evaluated as a change (decrease) in NADH absorbance, which was coupled with pyruvate-to-lactate conversion, it was important to affirm the specificity of initial velocity signals. Indeed, in the absence of LDH enzyme or pyruvate, initial velocities (changes in NADH absorbance) were approximately zero (Fig. 1A). As a further control, pyruvate-induced shifts in buffer pH were negligible across all pyruvate concentrations (±0.1) (Fig. 1B), and thus, any observed pyruvate-induced LDH enzyme effects could not be attributed to pH changes. Strong biphasic pyruvate activation kinetics and substrate inhibition of LDH in response to increasing pyruvate concentrations was observed regardless of LDH isoform or source (R2 bovine LDH = 0.988, R2 LDHA = 0.966, and R2 LDHB = 0.925) (Fig. 1A). Maximal initial velocities of the activation curve fits (upstrokes), peak initial velocities of the biphasic curve fits, pyruvate concentrations at peak initial velocities, and apparent Km and Ki values are summarized in Table 1. Of note, when initial velocities of bovine heart LDH enzyme, recombinant LDHA, and recombinant LDHB were normalized to their corresponding peak velocities, the biphasic substrate inhibition curve of LDHB closely align with the curve of bovine heart LDH, whereas the biphasic substrate inhibition curve of LDHA was right-shifted (Fig. 1C). Thus, the precision of the pyruvate titration curves indicated that bovine heart LDH primarily comprised LDHB, affirming the anticipated results (28, 29) and validating the biochemical sensitivity of the assay.
Figure 1.
Effect of pyruvate on purified and cell-extracted lactate dehydrogenase (LDH) enzyme activity.A, initial velocities of pyruvate-to-lactate conversion for bovine heart LDH, purified human recombinant LDHA and LDHB, as well as enzyme-free control in response to varying concentrations of pyruvate (0–30 mM); R2bovine LDH = 0.988, R2LDHA = 0.966, and R2LDHB = 0.925. B, changes of pH in LDH assay reaction mixtures across various concentrations of pyruvate. C, initial velocities of pyruvate-to-lactate conversion of each enzyme normalized to its corresponding peak initial velocity (Vpeak). D, initial velocities of pyruvate-to-lactate conversion for LDH in cell extracts in response to varying concentrations of pyruvate (0–30 mM); R2Panc03.27 = 0.961, R2Mia Paca2 = 0.976, R2QGP1 = 0.976, R2MDA-MB-231 = 0.917, and R2HPNE = 0.978. E, initial velocities of pyruvate-to-lactate conversion of each cell extract normalized to its corresponding peak initial velocity (Vpeak). F, LDHA and LDHB mRNA expression across the cell panel. All mRNA expression values were normalized to Panc03.27 expression levels. G, ratios of LDHB mRNA to LDHA mRNA across the cell panel. H, correlation between LDHB/LDHA ratios and Cmax, the pyruvate concentration at peak initial velocity, across the cell panel (r = −0.985, p = 0.002). If not shown, error bars are within the size of symbols. All error bars represent SEM. A–D, each experiment was done in triplicate (technical replicate) and F, experiments were done in duplicate (technical replicate); n = 3, independent experiments (biological replicates).
Table 1.
Effect of pyruvate on purified LDH enzyme kinetics
| LDH enzymes | Vmaz (ΔAbs/s∗10−3/μg protein) | Vpeak (ΔAbs/s∗10−3/μg protein) | Cmax (mM) | Km (mM) | Ki (mM) |
|---|---|---|---|---|---|
| LDH bovine heart | 3.23 ± 0.10 | 2.08 | 0.48 | 0.13 ± 0.010 | 1.75 ± 0.11 |
| rLDHA | 1.00 ± 0.04 | 0.75 | 0.90 | 0.15 ± 0.01 | 5.33 ± 0.49 |
| rLDHB | 1.00 ± 0.05 | 0.75 | 0.48 | 0.08 ± 0.01 | 2.88 ± 0.40 |
Abbreviation: LDH, lactate dehydrogenase.
Summary of maximal initial velocities from activation curve fits (Vmax), peak initial velocities of the biphasic curve fits (Vpeak), pyruvate concentrations at peak velocities of the biphasic curve fits (Cmax), and apparent Km and Ki values of commercially available LDH enzymes from bovine heart, human recombinant LDHA and recombinant LDHB. All experiments were done in triplicate (technical replicates); n = 3, independent experiments (biological replicates); mean values with standard errors.
Next, to extend the kinetic analysis to tumor cells, LDH activity was determined in extracts from a multi-cell-line panel comprising four well-established human cancer cell lines and one normal cell line: pancreatic cancer cell lines (Panc03.27 and Mia Paca2), a pancreatic neuroendocrine cancer cell line (QGP1), a triple-negative breast cancer cell line (MDA-MB-231), and an immortalized normal pancreatic epithelial cell line (HPNE). LDH in cell protein extracts, regardless of whether extracts were from cancer or normal cells, demonstrated biphasic pyruvate activation kinetics and substrate inhibition patterns highly similar to commercially available recombinant LDH enzymes (R2 Panc03.27 = 0.961, R2 Mia Paca2 = 0.976, R2 QGP1 = 0.976, R2 MDA-MB-231 = 0.917, and R2 HPNE = 0.978) (Fig. 1D). Furthermore, in the absence of exogenous pyruvate, initial velocity signals were near zero, affirming the LDH specificity of the assay. The highest peak initial velocities of the biphasic curve fits were measured in extracts from MDA-MB-231 cells, then HPNE cells, followed by Panc03.27 and QGP1 cells, and the lowest in Mia Paca-2 cells (Fig. 1D). Pyruvate concentrations at peak initial velocities across all tested cell lines occurred between 0.3 and 0.7 mM pyruvate. Maximal initial velocities of the activation curve fits (upstrokes), peak initial velocities of the biphasic curve fits, pyruvate concentrations at peak initial velocities, and apparent Km and Ki values are summarized in Table 2. Both the Km of activation kinetics and the Ki of substrate inhibition kinetics were highly clustered but the highest in LDH from Mia Paca-2 cells, slightly lower in MDA-MB-231 cells, intermediate in HPNE and QGP1 cells, and the lowest in Panc03.27 cells (Fig. 1E). Across the panel, evidence for inhibition of LDH enzyme activity occurred when pyruvate concentrations were higher than 0.7 mM. The relative abundance of LDHA and LDHB mRNA was determined using real-time quantitative PCR (Fig. 1F), and LDHB/LDHA mRNA ratios for each cell line were calculated (Fig. 1G). Of interest, the LDHB/LDHA mRNA ratios were significantly inversely correlated with Cmax, the pyruvate concentration at the corresponding peak velocity (Vpeak), across the cell panel (r = −0.985, p = 0.002) (Fig. 1H), suggesting that the measured pyruvate-dependent Cmax could be indicative of the LDHA and LDHB heterotetrameric subunit composition within each cell line. Overall, these results indicated that cellular LDH activity in vitro was substrate inhibited uniformly by high concentrations of pyruvate regardless of cell source of enzyme.
Table 2.
Effect of pyruvate on LDH enzyme kinetics in cell extracts
| LDH enzymes | Vmaz (ΔAbs/s∗10−3/μg protein) | Vpeak (ΔAbs/s∗10−3/μg protein) | Cmax (mM) | Km (mM) | Ki (mM) |
|---|---|---|---|---|---|
| Panc03.27 | 2.00 ± 0.07 | 1.56 | 0.33 | 0.05 ± 0.00 | 2.33 ± 0.21 |
| Mia Paca-2 | 0.88 ± 0.03 | 0.66 | 0.69 | 0.11 ± 0.01 | 4.26 ± 0.32 |
| QGP1 | 1.85 ± 0.05 | 1.42 | 0.51 | 0.08 ± 0.01 | 3.42 ± 0.25 |
| MDA-MB-231 | 2.98 ± 0.17 | 2.20 | 0.60 | 0.10 ± 0.02 | 3.39 ± 0.50 |
| HPNE | 2.71 ± 0.08 | 1.98 | 0.48 | 0.09 ± 0.01 | 2.67 ± 0.20 |
Abbreviation: LDH, lactate dehydrogenase.
Summary of maximal initial velocities from activation curve fits (Vmax), peak initial velocities of the biphasic curve fits (Vpeak), pyruvate concentrations at peak velocities of the biphasic curve fits (Cmax), and apparent Km and Ki values of LDH in extracts from Panc03.27, Mia Paca2, QGP1, MDA-MB-231, and HPNE cells. All experiments were done in triplicate (technical replicates); n = 3, independent experiments (biological replicates); mean values with standard errors.
Excess pyruvate impacts lactate production in cellulo
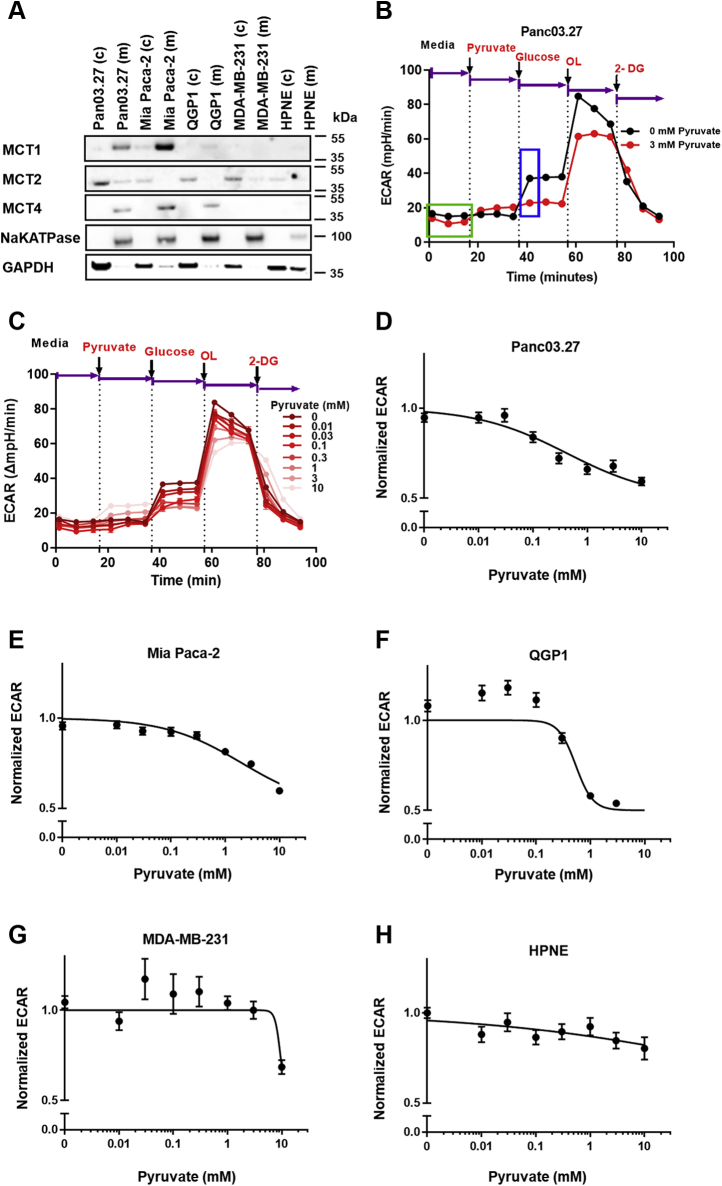
Contrary to kinetics of LDH enzyme extracts in vitro where substrates are freely accessible in the reaction mixture, in intact live cells, only pyruvate that gains access to intracellular pools would be accessible to cytosolic LDH. Because accumulation of intracellular pyruvate is dependent on transmembrane influx, which is primarily mediated by MCT1 (6), the protein expression profiles of MCTs were examined across the cell panel. Because only plasma membrane–bound transporters are involved in pyruvate influx (35), we performed protein fractionation to evaluate plasma membrane-bound transporters. Western blot analysis showed that membrane-localized MCT1, which colocalized with the plasma membrane marker (Na/K-ATPase), was the highest in Mia Paca-2 cells, relatively lower in Panc03.27 cells, minimally detectable in QGP1 cells, and nondetectable in MDA-MB-231 as well as HPNE cells (Fig. 2A). MCT2 was primarily localized to cytosolic fractionations (GAPDH) across the cell panel, with membrane-localized MCT2 minimally detectable in Panc03.27 (Fig. 2A). Membrane-localized MCT4 protein was primarily identified in Panc03.27, Mia Paca-2, and QGP1 cells and nondetectable in MDA-MB-231 and HPNE cells (Fig. 2A).
Figure 2.
Effect of pyruvate on lactate production in cells with different plasma membrane–localized MCT proteins.A, western blot showing membrane and cytosolic fractionations of MCT1, MCT2, and MCT4 protein across the cell line panel. B, the definition of normalized pyruvate-modulated glycolytic ECAR. Seahorse glycolysis stress assays were performed with the addition of substrates in the exact order indicated. Normalized pyruvate-modulated glycolytic ECAR was calculated as . The average of three steady-state ECAR data points (green box) were used to calculated baseline ECAR, and the first glucose-induced ECAR data point (blue box) was used to calculate initial pyruvate-modulated glycolytic ECAR. C, representative glycolytic ECAR plot of Panc03.27 cells pretreated with various concentrations of pyruvate (0–10 mM) and subsequent glycolytic flux measured by the Seahorse assay in live cells. Dotted lines and arrows represent injections of a series of substrates and drugs at different time points during the experiment. The changes in glycolytic ECAR in response to various concentrations of pyruvate pretreatments were measured. Shown is a representation of one experiment performed in quintuplicate. OL represents oligomycin, and 2-DG represents 2-deoxyglucose. Glycolytic ECAR values in response to incubation with various concentrations of pyruvate were normalized to 0 mM pyruvate control for D, Panc 03.27, E, Mia Paca-2, F, QGP1, G, MDA-MB-231, and H, HPNE cells. If not shown, error bars are within the size of symbols and represent SEM. All experiments were done in quintuplicate (technical replicate); n = 3, independent experiments (biological replicate). ECAR, extracellular acidification rate; MCT, monocarboxylate transporter.
Pyruvate-to-lactate conversion rates in live cells were then semiquantitatively monitored in real time using the Seahorse assay to determine the normalized glucose-stimulated extracellular acidification rate (ECAR) as a measure of lactate production (Fig. 2B). Herein, the panel of cells (maintained with DMEM lacking NaHCO3 and glucose; supplemented with 2 mM glutamine) was pretreated with various extracellular pyruvate concentrations for 20 min. As shown with representative Panc03.27 cells, increasing pretreatment pyruvate concentrations decreased subsequent glucose-stimulated ECAR, indicating a decrease in lactate production (Fig. 2, B and C). Of interest, the tested tumor cell lines demonstrated various degrees of decreases in glucose-stimulated ECAR with increasing pyruvate concentration, whereas HPNE did not (Fig. 2, D–H). In contrast to the LDH enzymatic assay, wherein pyruvate concentrations >0.7 mM uniformly produced LDH inhibition in all cell enzyme extracts, in live cells, significant inhibition of glycolysis was observed only in Panc03.27 (IC50 = 0.42 ± 0.09 mM), Mia Paca-2 (IC50 = 2.03 ± 0.30 mM), and QPG1 (IC50 = 0.54 ± 0.08 mM) cells, which were MCT1 positive. By contrast, triple-negative breast cancer cells, MDA-MB-231, were minimally sensitive (IC50 = 9.42 mM, error nonconvergent) and HPNE cells showed no inhibition (IC50 = NA) (Table 3), which were both MCT1 negative. There were no correlations of LDHB/LDHA mRNA ratios with mRNA of MCT1, MCT2, MCT4, or ECAR. Overall, these data demonstrated that, in live cells, high concentrations of exogenous pyruvate inhibited ECAR, consistent with the model of LDH substrate inhibition by excess pyruvate, but only in three of five tested cell lines that expressed MCT1, which would impact transport-mediated pyruvate influx.
Table 3.
Calculated IC50 values of pyruvate-modulated extracellular acidification rate across the cell panel
| Cells | IC50 (mM) ECAR |
|---|---|
| Panc03.27 | 0.42 ± 0.09 |
| Mia Paca2 | 2.03 ± 0.30 |
| QGP1 | 0.54 ± 0.08 |
| MDA-MB-231 | 9.42 ± error nonconvergent |
| HPNE | NA |
IC50 values for pyruvate-modulated glucose-induced extracellular acidification rate (ECAR) for various cell lines as determined by nonlinear regression of log[inhibitor] VS response model using four-parameter variable slope with the constraint that the bottom of the curve fit equals 0.5. Standard errors were included. NA, no inhibition observed. All experiments were done in quintuplicate (technical replicates); n = 3, independent experiments (biological replicate); mean values with standard errors.
The buffering effect of sodium pyruvate
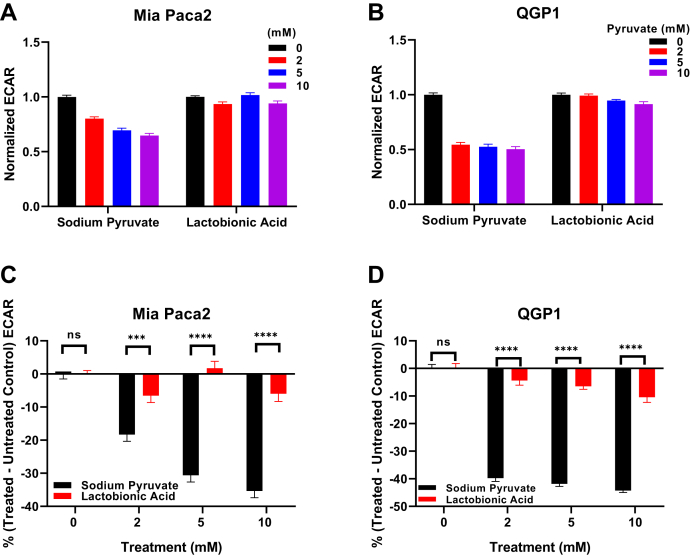
Because Seahorse ECAR assays measure rates of change of extracellular pH in unbuffered media, a control study was performed to determine the potential buffering effect of sodium pyruvate, which could in principle act as a weak base at high concentrations and interfere with the assay. Lactobionic acid, a commonly used cell impermeable antioxidant and osmotic stabilizer with a pKa of 3.6 to 3.8, similar to sodium pyruvate (pKa = 2.93), was used as a pretreatment technical control. Mia Paca-2 and QGP1 cells were treated with various concentrations (0–10 mM) of lactobionic acid or sodium pyruvate (Fig. 3, A and B). Lactate production (measured as glucose-induced ECAR values) was significantly inhibited when cells were pretreated with sodium pyruvate compared with their lactobionic acid pretreated counterparts (Fig. 3, C and D). Thus, we concluded that the observed pyruvate-induced glycolytic ECAR inhibition was the result of sodium pyruvate pretreatment of cells impacting cytosolic LDH rather than buffering artifacts of sodium pyruvate in the reaction medium masking ECAR measurements.
Figure 3.
Glycolytic ECAR modified by sodium pyruvate or lactobionic acid pretreatments (0–10 mM) in Mia Paca-2 and QGP1 cells. Glycolytic ECAR values were normalized to 0 mM of sodium pyruvate or lactobionic acid for (A) Mia Paca-2 and (B) QGP1 cells. C and D, percent changes in normalized treatment-modulated glycolytic ECAR values [% (treated − untreated)] for Mia Paca-2 (C) and QGP1 cells (D). Error bars represent SEM. All experiments were done in quintuplicate (technical replicate); n = 3, independent experiments (biological replicate). ∗∗∗p = 0.0001; ∗∗∗∗p < 0.0001, two-way ANOVA test followed by multiple comparison analyses. ECAR, extracellular acidification rate.
Blockade of pyruvate influx impacts LDH enzyme substrate inhibition in cellulo
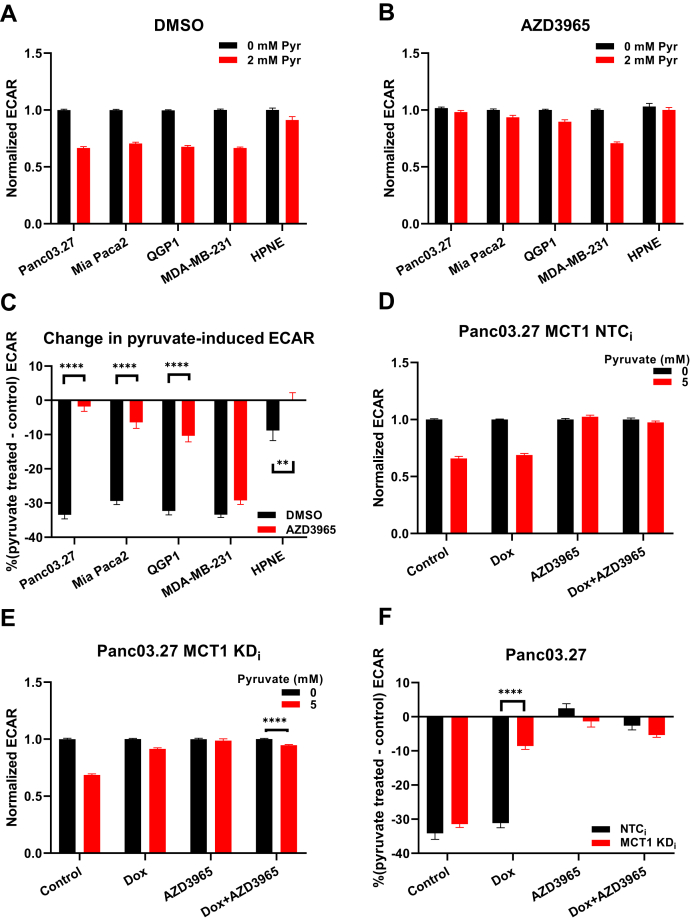
To test if an intervention to limit pyruvate influx impacted the observed inhibition of lactate production, we again used the Seahorse glycolysis assay to measure glycolytic lactate production. For these experiments, basal Seahorse medium was first supplemented with 5 mM of glucose (and dimethyl sulfoxide [DMSO] vehicle control) to ensure cells underwent normal glycolytic activity under a physiologically relevant state. Subsequently, upon addition of 2 mM sodium pyruvate, glucose-stimulated ECAR values then decreased by >30% in Panc03.27 (p < 0.0001), Mia Paca-2 (p < 0.0001), QGP1 (p < 0.0001), and MDA-MB-231 cells (p < 0.0001), but less than 10% in HPNE cells (p = 0.001) (Fig. 4A). By contrast, when cells were pretreated with the MCT1/2 inhibitor AZD3965, thereby limiting MCT1/2-mediated pyruvate influx into cells, the previously observed 2 mM pyruvate-induced inhibition of glucose-stimulated ECAR was blocked in Panc03.27, Mia Paca-2, QGP1, and HPNE cells, but no significant effects in MDA-MB-231 cells were observed (Fig. 4B) compared with their corresponding DMSO-treated control groups (Fig. 4C). These data suggested that, when MCT1/2-mediated pyruvate influx was inhibited pharmacologically, the exogenously delivered high concentrations of pyruvate failed to accumulate intracellularly, and thus, pyruvate-induced substrate inhibition was relieved. However, it is important to note that 2 mM pyruvate inhibited lactate production in MCT1-nondetectable MDA-MB-231 cells regardless of MCT1/2 inhibition, implying the presence of alternative pyruvate transporters other than MCT1/2.
Figure 4.
Dependence of pyruvate-modulated glycolytic ECAR on MCT1-mediated pyruvate influx in cellulo.A, with 5 mM glucose, normalized glycolytic ECAR of the cell panel at 0 mM and 2 mM pyruvate when pretreated with 1 μM DMSO. B, with 5 mM glucose, normalized glycolytic ECAR of the cell panel at 0 and 2 mM pyruvate when pretreated with 0.5 μM AZD3965. C, percent changes in pyruvate-modulated glycolytic ECAR values [% (pyruvate treated − control)] in response to AZD3965-mediated MCT1 inhibition across the cell panel. D, normalized glycolytic ECAR in dox-inducible NTCi shRNA Panc03.27 cells at 0 mM and 5 mM pyruvate pretreatment in response to dox and AZD3965 treatment. E, normalized glycolytic ECAR in dox-inducible MCT1 knockdown Panc03.27 cells at 0 mM and 5 mM pyruvate pretreatment in response to dox and AZD3965 treatment. F, percent changes in pyruvate-modulated glycolytic ECAR values [% (pyruvate treated − control)] in response to MCT1 knockdown and AZD3965 treatment in dox-inducible Panc03.27 cells. Glycolytic ECAR values are normalized to 0 mM pyruvate. Error bars represent SEM. All experiments were done in quintuplicate (technical replicate); n = 3, independent experiments (biological replicate). ∗∗p = 0.001; ∗∗∗∗p < 0.0001, two-way ANOVA test followed by multiple comparison analyses. DMSO, dimethyl sulfoxide; ECAR, extracellular acidification rate; MCT, monocarboxylate transporter; NTC, non-targeting control.
To further examine the hypothesis that MCT1-mediated pyruvate influx altered the intracellular pyruvate concentration, which in turn affected LDH activity, MCT1-expressing Panc03.27 cells transduced with doxycycline-inducible MCT1 knockdown sequences (KDi) or nontargeting control sequences (NTCi) were used to test if the observed inhibition of lactate production was specifically due to MCT1-mediated pyruvate influx. Panc03.27 KDi and NTCi cells were previously generated and validated (6). For control Panc03.27 NTCi cells, pyruvate-induced inhibition of glucose-stimulated ECAR was observed regardless of doxycycline induction (Fig. 4D). By contrast, for Pan03.27 MCT1 KDi cells, the pyruvate-induced inhibition of glucose-stimulated ECAR was mitigated upon doxycycline induction of MCT1 knockdown (Fig. 4, E and F; p < 0.0001). AZD3965 alone and AZD3965 with doxycycline-induced MCT1 knockdown were used as positive controls to compare the efficiency between pharmacological and genetic abrogation of MCT1 (Fig. 4F). Overall, in MCT1-expressing Panc03.27 cells, pyruvate-induced substrate inhibition of LDH was dependent on pyruvate influx mediated by MCT1 transport of exogenous pyruvate into the cells.
Hyperpolarized [1-13C]pyruvate substrate-mediated inhibition in cellulo
Because in vivo hyperpolarized pyruvate MRSI studies require a highly concentrated bolus injection of the metabolite well above physiological pyruvate levels (36), the effects of excess pyruvate on [1-13C]pyruvate-to-[1-13C]lactate conversion rates were therefore evaluated under simulated hyperpolarized MRSI conditions. We thus sought to evaluate the effect of excess pyruvate on initial hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion where the influx of excess hyperpolarized [1-13C]pyruvate was assessable by pyruvate transporters. A previously established cell-encapsulated alginate bead system was utilized to evaluate the initial rate of hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion in isolated live cells (6).
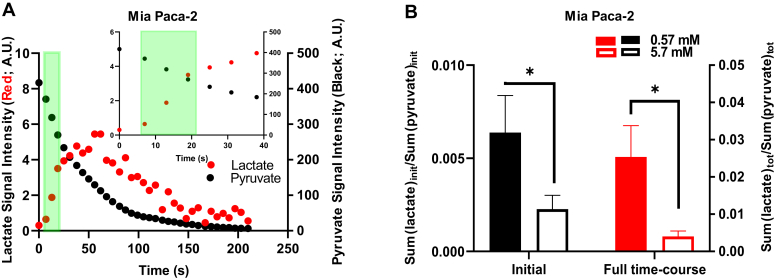
First, we measured the spectra of hyperpolarized [1-13C]pyruvate and [1-13C]lactate under standard conditions over time (Fig. 5A). As previously validated (6), the initial area under the curves of hyperpolarized [1-13C]lactate and [1-13C]pyruvate were integrated over the first 6 to 18 s. For instance, the high MCT1-expressing Mia Paca-2 cells exhibited a monotonic decrease of hyperpolarized [1-13C]pyruvate signal and the production and subsequent decline of converted hyperpolarized [1-13C]lactate signal (Fig. 5A). A [1-13C]lactate to [1-13C]pyruvate ratio over the first 6 to 18 s was used to assess the initial normalized hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rate in Mia Paca-2 cells upon addition of ∼5.7 mM and ∼0.57 mM hyperpolarized [1-13C]pyruvate. The initial hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rate with ∼5.7 mM hyperpolarized [1-13C]pyruvate was significantly lower than that with ∼0.57 mM hyperpolarized [1-13C]pyruvate (p(initial) = 0.04) (Fig. 5B). Conventional analysis of the entire hyperpolarization curves over the 3-min experiment yielded highly similar results (p(full time-course) = 0.03) (Fig. 5B). These data suggested that ∼5.7 mM hyperpolarized [1-13C]pyruvate induced substrate inhibition of LDH in high MCT1-expressing cells within 6 to 18 s.
Figure 5.
The effect of high and low concentrations of exogenous pyruvate on the initial hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rates in cellulo. Using Mia Paca-2 live cell–encapsulated alginate beads, NMR spectra of hyperpolarized [1-13C]pyruvate and [1-13C]lactate were measured over repetitive 6-s time intervals. A, signal intensities of hyperpolarized [1-13C]pyruvate and [1-13C]lactate over time. The light green band demarcates the time intervals for the initial rate calculations (6–18 s; inset, expanded time scale) of intracellular lactate production and pyruvate conversion. B, initial or total hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rates in cellulo. Initial hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion in cellulo was calculated by the sum of converted hyperpolarized [1-13C]lactate during the first 6 to 18 s normalized to the sum of hyperpolarized [1-13C]pyruvate during the first 6 to 18 s in the presence of 0.57 mM or 5.7 mM [1-13C]pyruvate (left). Total hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion in cellulo was calculated by the sum of converted hyperpolarized [1-13C]lactate over the 3-min experiment normalized to the sum of hyperpolarized [1-13C]pyruvate in the presence of 0.57 mM or 5.7 mM [1-13C]pyruvate (right). Error bars represent SEM. All data represent n = 4 independent experiments (biological replicate); ∗p < 0.05; one-tailed t test.
Discussion
For pyruvate therapy and MRSI analysis, boluses of highly concentrated pyruvate are administered (0.5–1.0 g/kg/day and 80 mM, respectively). Although both small animals and humans appear to generally tolerate high bolus doses of pyruvate, the physiological impact of the injected pyruvate remains to be evaluated on a tissue-by-tissue basis for biomedical and clinical translational applications.
We validated that pyruvate itself, at millimolar concentrations, could cause substrate inhibition of cell-extracted LDH regardless of isoform. Furthermore, the close alignment of the biphasic substrate inhibition curves of bovine heart LDH and LDHB functionally revalidated that the LDHB (homotetrameric) isoform is primarily expressed in the heart. It is important to note that the horizontal shift of biphasic substrate inhibition curves across the tumor cell panel strongly implied that cancer cells contain various heterotetrameric isoforms of LDHA and LDHB, not just the canonical LDHA. In addition, in intact cells, pyruvate-induced inhibition of lactate production was also observed among MCT1-expressing cells using the Seahorse glycolytic stress assays, indicating that substrate inhibition of LDH was the underlying mechanism. Because the degree of pyruvate-induced inhibition of lactate production varied with MCT1 protein content of cells, and only intracellular pyruvate is accessible to cytoplasmic LDH, the potential to induce LDH substrate inhibition would be predicted to be a function of MCT1 transporters localized to the plasma membrane of various tissues. By further exploring the role of pyruvate influx, robust pyruvate-induced inhibition of lactate production was observed in MCT1-expressing Panc03.27, Mia Paca-2, and QGP1 cells, but less so in HPNE cells lacking MCT1. Furthermore, such pyruvate-induced inhibition of lactate production was reduced when MCT1-mediated pyruvate influx was inhibited pharmacologically or genetically in MCT1-expressing Panc03.27, Mia Paca-2, and QGP1 cells, but less in MCT1-nondetectable HPNE cells. We also noticed pyruvate-induced inhibition of lactate production in MCT1-nondetectable MDA-MB-231 cells regardless of MCT1/2 inhibition, which indicated the potential for pyruvate influx via alternative pyruvate transporters other than MCT1/2, such as sodium-coupled monocarboxylate transporters (37). Overall, these data suggested that the observed pyruvate-induced substrate inhibition of LDH was dependent on transporter-mediated pyruvate influx in a cell-dependent, or by implication, tissue-dependent manner.
It is important to note that the earliest time point for ECAR measurements in response to pyruvate treatment was 6 min after pyruvate addition. In contrast, for the measurement of the hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion, the apparent rate of the conversion was typically determined in the first 6 to 18 s of the experiment, and the overall hyperpolarization signal normally lasts less than 5 min. Since the Seahorse glycolytic stress assay and in cellulo hyperpolarized [1-13C]pyruvate MRS experiments assessed the lactate conversion at somewhat different time frames, and the accumulation of pyruvate into intracellular spaces was dependent on the rate of pyruvate influx and the duration of exposure, we could not directly and quantitatively compare pyruvate-induced substrate inhibition times between these two assays. The practical net result was nonetheless resolved when we evaluated the effect of different concentrations of [1-13C]pyruvate on initial hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rates in the hyperpolarized MRS setting in live cells. We found that the hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rate at ∼ 0.57 mM hyperpolarized [1-13C]pyruvate was significantly higher than that at ∼ 5.7 mM hyperpolarized [1-13C]pyruvate, the latter well over the apparent Ki, suggesting that ∼5.7 mM hyperpolarized [1-13C]pyruvate induced substrate inhibition of LDH as early as 6 to 18 s.
In addition, millimolar doses of pyruvate have been reported to increase cellular oxygen consumption rates in OxPhos-reliant melanoma cells (24). It is also reported that infusion of hyperpolarized [1-13C]pyruvate could induce increased tumor hypoxia in pancreatic ductal carcinoma and squamous cell carcinoma cells (25, 26). Our findings point to additional mechanisms that explain these previously observed pyruvate-induced phenotypes. First, bolus injections of pyruvate (millimolar concentration) induce substrate inhibition of LDH, which manifests as decreased aerobic glycolysis. Furthermore, cells could compensate for such inhibition by ramping up OxPhos, which would result in decreased pO2 and increased hypoxia. Hence, it may be important to account for the potential for transient metabolic perturbations of bolus infusions of hyperpolarized [1-13C]pyruvate during hyperpolarized [1-13C]pyruvate MRS studies. Technical constraints herein rendered submillimolar hyperpolarized [1-13C]pyruvate MRS experiments difficult to perform, which suffered from poor signal-to-noise ratios in cellulo, and thus, it was challenging to directly examine the potential effect of substrate inhibition on LDH using an even lower range of pyruvate concentrations. Nonetheless, future advancements in detector sensitivity and further enhancement of hyperpolarization technology could potentially reduce the amount of hyperpolarized [1-13C]pyruvate required for MRSI, thus minimizing perturbations induced by isotope-labeled probes on biological systems and physiological processes. In the context of pyruvate therapy, wherein concentrated pyruvate is administrated (0.5–1.0 g/kg/day) over a period of 17 to 66 min (5), it is likely that tissues with moderate to high MCT1 expression will experience sustained pyruvate-induced substrate inhibition during the therapeutic infusion, which could impact the efficacy and patient tolerance of pyruvate therapy. Herein, it may be relevant to monitor plasma pyruvate levels and evaluate the impact on the efficacy of pyruvate therapy in patients with mitochondrial diseases, or perhaps protect sensitive tissues with MCT1 inhibitors.
Overall, using quantitative and semiquantitative orthogonal methods, we demonstrated that millimolar concentrations of pyruvate/[1-13C]pyruvate could induce substrate inhibition of LDH activity at both enzymatic and cellular levels, which provides a mechanism for previously observed pyruvate-induced metabolic alterations (24, 25, 26). Furthermore, in intact cells, pyruvate-induced LDH inhibition was highly dependent on the influx of pyruvate, which in turn was dependent on the expression of MCT1 and related pyruvate transporters. Interrogating time domains from seconds to minutes and systems from non-diffusion-limited enzymatic assays to transporter-mediated cell observations, these experiments clarified links between rates of [1-13C]pyruvate-to-[1-13C]lactate conversion and pyruvate influx as well as the quantitative kinetics of perturbations on biological systems. These data suggested that maintaining plasma pyruvate at submillimolar concentrations could be a potential solution to minimize transient metabolic perturbations, improve pyruvate therapy, and advance quantification of metabolic studies, such as hyperpolarized [1-13C]pyruvate MRSI as well as stable isotope tracer experiments.
Experimental procedures
Cell culture and cell lines
Panc03.27, Mia Paca-2, MDA-MB-231, and hTERT-HPNE cells were obtained from the American Type Culture Collection. QGP1 was purchased from the Japanese Collection of Research Bioresources cell bank. All cells tested negative for mycoplasma infection. Panc03.27 cells were maintained in RPMI-1640 medium supplemented with 10 Units/ml human recombinant insulin and 15% fetal bovine serum (FBS). Mia Paca-2 and MDA 231 cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 2 mM glutamine and 10% FBS. QGP1 cells were cultured in RPMI-1640 medium with 10% FBS. HPNE cells were maintained in 75% DMEM without glucose and 25% Medium M3 Base (Incell Corporation) supplemented with 2 mM L-glutamine, 1.5 g/l sodium bicarbonate, 5.5 mM D-glucose, 750 ng/ml puromycin, 10 ng/ml human recombinant EGF (Thermo Fisher Scientific), and 5% FBS. DMEM, DMEM without glucose, RPMI-1640, sodium bicarbonate, and D-glucose were purchased from Sigma-Aldrich. Cell cultures were maintained at 37 °C in a humidified 5% CO2 atmosphere.
LDH assay
LDH assay experimental procedures have been described in detail previously (6). Bovine heart LDH (Sigma-Aldrich), purified human recombinant LDHA (Sigma-Aldrich), and human recombinant LDHB (Abcam) were purchased commercially. For recombinant enzyme assays, 2 μl of each enzyme stock was diluted in 998 μl of Tris/NaCl buffer as working stocks. For extracts, cells were washed with ice-cold PBS and then lysed in ice-cold LDH Assay Lysis Buffer (Sigma-Aldrich). Protein contents were quantified with the Coomassie Protein Assay. For each well of a 96-well plate, 25 μl of recombinant enzymes or cell extracts, 125 μl of the Tris/NaCl/NADH buffer, and 25 μl of the Tris/NaCl/pyruvate with various pyruvate concentrations were added. The reduced form of NADH absorbs at 340 nm, whereas the oxidized form NAD+ does not. Therefore, to quantify the enzymatic activity of LDH, the change of absorbance at 340 nm due to NADH oxidation was measured using a Synergy H4 microplate reader (BioTek), programmed to shake the microplate for 30 s, and read absorbance at 340 nm five times with 30-s intervals. Controls including enzyme-free assay mixture and pyruvate-free vehicle controls were performed to ensure that changes in absorbance at 340 nm (used to calculate initial velocities of LDH) were LDH and pyruvate specific. Data were analyzed with GraphPad Prism 7.01. Experiments were performed in triplicate and repeated three independent times.
Real-time quantitative PCR assays
RNA was extracted from Mia Paca-2, Panc 03.27, QGP1 cells, MDA-MB-231, and HPNE cells with Aurum total RNA Mini Kit (Bio-Rad). The amount of RNA from each sample was quantified using a Nanodrop 1000 (Thermo Fisher Scientific). Real-time quantitative PCR was performed with primers for LDHA (forward: 5′-TTGAAGGGAGAGATGATGGA-3′ and reverse: 5′-CCAGCCGTGATAATGACCAG-3′), LDHB (forward: 5′-TGCTCTTGTGGATGTTTTGG-3′ and reverse: 5′-CTCTCCCCTTCTTGCTGACG-3′), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward: 5′-ATGGAATCCATCACCATCTT-3′ and reverse: 5′-CGCCCCACTTGATTTTGG-3′) (IDT). RNA was converted to cDNA, amplified, and quantified using iTaq Universal SYBR Green One-Step Kit (Bio-Rad) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Expression of target gene mRNA relative to reference gene mRNA (GAPDH) was calculated using the calculated CT values from the curves, and statistical analysis was done using the GraphPad Prism 7.01. Assays were performed in technical duplicate from three independent experiments.
Western blotting
For whole-cell lysate, cells were lysed in Cell Lysis Buffer (Cell Signaling Technology), supplemented with protease inhibitor cocktail (Thermo Fisher Scientific). Membrane and cytoplasmic subcellular fractions were separated using a commercial Cell Fractionation Kit (Cell Signaling Technology), supplemented with protease inhibitor cocktail (Thermo Fisher Scientific) as per the manufacture protocol. Lysate loading was normalized to protein content quantified with the Coomassie Protein Assay (Thermo Fisher Scientific). Proteins were resolved on 4% to 20% SDS-PAGE gels (Bio-Rad) or NuPAGE 4% to 12% Bio-Tris Protein gels (Thermo Fisher Scientific) and transferred to Trans-blot Turbo nitrocellulose membranes (Bio-Rad). Posttransferred nitrocellulose membranes were stained in Ponceau S staining solution to evaluate total protein loading. For primary antibody–protein hybridization, membranes were probed with the following antibodies at 4 °C overnight: MCT1 (Thermo Fisher Scientific; Lot # TH2617863; TH2585400), MCT2 (Novus Biologicals; Lot # C2E170918), MCT4 (Boster; Lot # 11Q405094), Na/K-ATPase (Abcam), and GAPDH (Abcam; Bio-Rad). Thereafter, secondary anti-mouse or anti-rabbit horseradish peroxidase–conjugated IgG antibodies (Bio-Rad) were incubated for 1 h at room temperature. Protein bands were developed with chemiluminescent reagents (Bio-Rad) and imaged with an Azure c600 (Azure Biosystems).
In vitro analysis of extracellular acidification rate
The Seahorse XF glycolysis stress assay was used to measure the extracellular acidification rate (ECAR) as described in detail (6). The product of glycolysis, pyruvate, when converted into lactate, results in a net production and extrusion of protons into the extracellular environment. Thus, lactate molecules exported to the extracellular environment in the form of lactic acid result in a change of extracellular pH. Five different cell lines were seeded on XF96 Cell Culture Microplates (Seahorse Bioscience) in their original growth medium 48 h before the experiment at the following cell densities: 1.5 × 104 Panc03.27, 1.2 × 104 Mia Paca-2, 2.0 × 104 QGP1, 1.5 × 104 MDA-MB-231, and 1.5 × 104 HPNE. When pretreated with the MCT1 inhibitor AZD3965 (Selleckchem), 0.5 μM of AZD3965 was added to the corresponding group 4 h before the assay started (or DMSO for controls). For the Seahorse assay, cells were washed and maintained with DMEM lacking NaHCO3, supplemented with 2 mM glutamine with/without 5 mM glucose as indicated and no added pyruvate, and adjusted to pH 7.4. Concentrations of sodium pyruvate ranging from 0 to 10 mM, 10 mM of glucose, 2 μM of oligomycin, and 50 mM of 2-deoxyglucose were then sequentially injected. For a technical control, pyruvate was substituted with lactobionic acid (Sigma-Aldrich) to examine the potential buffering effect of sodium pyruvate. The fold-change of glucose-induced ECAR for each cell line was normalized to each cell line’s control group (no treatment). Normalized pyruvate-modulated glycolytic ECAR was calculated as:
where baseline ECAR was the ECAR measurements of cells before the addition of substrates or drugs (Fig. 2B). Experiments were performed with an XF96 analyzer, and raw ECAR values were normalized to cell counts using Hoechst 33342 (Thermo Fisher Scientific) on an Operetta High-Content Imaging System (PerkinElmer). Data were analyzed with GraphPad Prism 7.01. Experiments were performed in triplicate for three independent experiments.
In vitro hyperpolarization experiments using vertical bore NMR
In vitro hyperpolarization experimental procedures have been described in detail (6). Briefly, cell-encapsulated alginate beads (1 ml) made the prior day were washed and resuspended in 2.5 ml of assay medium consisting of phenol red–free DMEM medium supplemented with 2 mM glutamine, 5 mM glucose, 20% (v/v) deuterium oxide (D2O) (Sigma-Aldrich), and 15 mM Hepes buffer. Cell/bead suspensions were then transferred into a 10-mm screw-cap NMR tube (Wilmad-LabGlass) that was fitted with tubing and an injection port for quick delivery of hyperpolarized material. Experiments were conducted in a 7 T Bruker BioSpin NMR spectrometer with a 10-mm broadband probe and running TopSpin 3.5 software. NMR tubes containing cell-encapsulated alginate beads were equilibrated in the NMR spectrometer at 37 °C prior to hyperpolarized [1-13C]pyruvate injection. For analysis, an aliquot of 26.5 mg of a pyruvic acid mix containing [1-13C]pyruvic acid (Isotec, Sigma-Aldrich) with 15 mM Trityl OX063 polarizing radicals (Oxford Instrument) and 1.5 mM Dotarem (Guerbet) was polarized in a HyperSense DNP system (Oxford Instruments) with a 3.35-T magnetic field for ∼30 min at 1.40 K with a microwave irradiation frequency of 96.136 GHz, similar to published procedures (38). The frozen hyperpolarized [1-13C]pyruvic acid was then dissolved in 4 ml of the dissolution medium containing 40 mM Trizma Pre-Set Crystals at pH 7.6 (Sigma-Aldrich), 80 mM sodium hydroxide, 0.1 g/l disodium EDTA dihydrate, and 50 mM sodium chloride at 180 °C. The hyperpolarized [1-13C]pyruvic acid dissolution was further diluted into a specialized hyperpolarization buffer medium comprising modified DMEM supplemented with 2 mM glutamine, 5 mM glucose, and 25 mM Hepes buffer (2 parts:1 part), which was developed to ensure a physiologically balanced osmolality and pH buffering capacity. Thus, the hyperpolarized [1-13C]pyruvic acid stock was 37.7 mM. At time zero of acquisition, 500 μl of the hyperpolarized [1-13C]pyruvic acid stock was injected into the NMR tube (containing 2.5 ml of buffering medium and 1 ml of alginate/cell beads). The final concentration of [1-13C]pyruvic acid in the experiment was ∼5.7 mM or diluted to ∼0.57 mM. Single consecutive 13C scans were taken using Waltz decoupling (zgdc pulse) every 6 s with 15° flip angle for 3 min. The acquired NMR spectra were phased and baseline corrected and referenced, and the signal intensities of hyperpolarized [1-13C]pyruvate (∼173 ± 1 ppm) and [1-13C]lactate (∼185 ± 1 ppm) spectra were integrated using MestReNova 10.0 software (MestreLab Research). After NMR spectra acquisition, samples of cell-encapsulated alginate beads were then subjected to Alamar Blue Assay (Thermo Fisher Scientific). Initial hyperpolarized [1-13C]pyruvate-to-[1-13C]lactate conversion rates = .
Data availability
All relevant data are within the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This study was funded by the National Institutes of Health grant P50 CA94056 to the MD Anderson Cancer Center-Washington University Inter-Institutional Molecular Imaging Center, the Gerald Dewey Dodd, Jr, Endowed Distinguished Chair of the University of Texas MD Anderson Cancer Center, a faculty UT STARs Award, a University of Texas MD Anderson Cancer Center Independent Research Grant (MDACC IRG), and the Erickson Kidney Cancer Research Foundation. We would like to thank the Biostatistics Resource and the Small Animal Imaging Facility (SAIF) of MDACC for animal imaging assistance, especially Charles Kingsley, which obtain support from an NCI Cancer Center Support Grant (P30 CA016672). Financial support was also obtained from P50 CA221707 and U01 CA214263. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
D. P.-W. and Y. R. conceptualization; D. P.-W. funding acquisition; Y. R., M. N. S., N. M. Z., and P. B. investigation; D. P.-W., M. N. S., N. M. Z., and P. B. methodology; D. P.-W. supervision; D. P.-W., Y. R., S. T. G., M. N. S., N. M. Z., and P. B. writing-review and editing; Y. R. data curation; D. P.-W., Y.R., S. T. G., M. N. S., N. M. Z., and P. B. formal analysis; Y. R. writing-original draft.
Edited by John Denu
References
- 1.Liberti M.V., Locasale J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sattler U.G., Walenta S., Mueller-Klieser W. A bioluminescence technique for quantitative and structure-associated imaging of pyruvate. Lab. Invest. 2007;87:84–92. doi: 10.1038/labinvest.3700493. [DOI] [PubMed] [Google Scholar]
- 3.Wang Dong, De Vivo Darryl. GeneReviews® [Internet] University of Washington; Seattle, WA: 2009. Pyruvate carboxylase deficiency; pp. 1993–2020. [Google Scholar]
- 4.Marsac C., Benelli C., Desguerre I., Diry M., Fouque F., De Meirleir L., Ponsot G., Seneca S., Poggi F., Saudubray J.M., Zabot M.T., Fontan D., Lissens W. Biochemical and genetic studies of four patients with pyruvate dehydrogenase E1 alpha deficiency. Hum. Genet. 1997;99:785–792. doi: 10.1007/s004390050449. [DOI] [PubMed] [Google Scholar]
- 5.Fujii T., Nozaki F., Saito K., Hayashi A., Nishigaki Y., Murayama K., Tanaka M., Koga Y., Hiejima I., Kumada T. Efficacy of pyruvate therapy in patients with mitochondrial disease: A semi-quantitative clinical evaluation study. Mol. Genet. Metab. 2014;112:133–138. doi: 10.1016/j.ymgme.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Rao Y., Gammon S., Zacharias N.M., Liu T., Salzillo T., Xi Y., Wang J., Bhattacharya P., Piwnica-Worms D. Hyperpolarized [1-(13)C]pyruvate-to-[1-(13)C]lactate conversion is rate-limited by monocarboxylate transporter-1 in the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22378–22389. doi: 10.1073/pnas.2003537117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Escuredo J., Van Hee V.F., Sboarina M., Falces J., Payen V.L., Pellerin L., Sonveaux P. Monocarboxylate transporters in the brain and in cancer. Biochim. Biophys. Acta. 2016;1863:2481–2497. doi: 10.1016/j.bbamcr.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chokkathukalam A., Kim D.H., Barrett M.P., Breitling R., Creek D.J. Stable isotope-labeling studies in metabolomics: New insights into structure and dynamics of metabolic networks. Bioanalysis. 2014;6:511–524. doi: 10.4155/bio.13.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Garcia J., Altea-Manzano P., Pranzini E., Fendt S.M. Stable isotopes for tracing mammalian-cell metabolism in vivo. Trends Biochem. Sci. 2020;45:185–201. doi: 10.1016/j.tibs.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Jang C., Chen L., Rabinowitz J.D. Metabolomics and isotope tracing. Cell. 2018;173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma E.H., Verway M.J., Johnson R.M., Roy D.G., Steadman M., Hayes S., Williams K.S., Sheldon R.D., Samborska B., Kosinski P.A., Kim H., Griss T., Faubert B., Condotta S.A., Krawczyk C.M. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8(+) T cells. Immunity. 2019;51:856–870.e855. doi: 10.1016/j.immuni.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Ardenkjaer-Larsen J.H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M.H., Servin R., Thaning M., Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brindle K.M., Bohndiek S.E., Gallagher F.A., Kettunen M.I. Tumor imaging using hyperpolarized 13C magnetic resonance spectroscopy. Magn. Reson. Med. 2011;66:505–519. doi: 10.1002/mrm.22999. [DOI] [PubMed] [Google Scholar]
- 14.Guglielmetti C., Najac C., Didonna A., Van der Linden A., Ronen S.M., Chaumeil M.M. Hyperpolarized (13)C MR metabolic imaging can detect neuroinflammation in vivo in a multiple sclerosis murine model. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E6982–E6991. doi: 10.1073/pnas.1613345114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miloushev V.Z., Di Gialleonardo V., Salamanca-Cardona L., Correa F., Granlund K.L., Keshari K.R. Hyperpolarized (13)C pyruvate mouse brain metabolism with absorptive-mode EPSI at 1T. J. Magn. Reson. 2017;275:120–126. doi: 10.1016/j.jmr.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miloushev V.Z., Granlund K.L., Boltyanskiy R., Lyashchenko S.K., DeAngelis L.M., Mellinghoff I.K., Brennan C.W., Tabar V., Yang T.J., Holodny A.I., Sosa R.E., Guo Y.W., Chen A.P., Tropp J., Robb F. Metabolic imaging of the human brain with hyperpolarized (13)C pyruvate demonstrates (13)C lactate production in brain tumor patients. Cancer Res. 2018;78:3755–3760. doi: 10.1158/0008-5472.CAN-18-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues T.B., Serrao E.M., Kennedy B.W., Hu D.E., Kettunen M.I., Brindle K.M. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat. Med. 2014;20:93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrao E.M., Brindle K.M. Potential clinical roles for metabolic imaging with hyperpolarized [1-(13)C]pyruvate. Front. Oncol. 2016;6:59. doi: 10.3389/fonc.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya P., Ross B.D., Bunger R. Cardiovascular applications of hyperpolarized contrast media and metabolic tracers. Exp. Biol. Med. (Maywood) 2009;234:1395–1416. doi: 10.3181/0904-MR-135. [DOI] [PubMed] [Google Scholar]
- 20.Dutta P., Salzillo T.C., Pudakalakatti S., Gammon S.T., Kaipparettu B.A., McAllister F., Wagner S., Frigo D.E., Logothetis C.J., Zacharias N.M., Bhattacharya P.K. Assessing therapeutic efficacy in real-time by hyperpolarized magnetic resonance metabolic imaging. Cells. 2019;8:340. doi: 10.3390/cells8040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurhanewicz J., Vigneron D.B., Ardenkjaer-Larsen J.H., Bankson J.A., Brindle K., Cunningham C.H., Gallagher F.A., Keshari K.R., Kjaer A., Laustsen C., Mankoff D.A., Merritt M.E., Nelson S.J., Pauly J.M., Lee P. Hyperpolarized (13)C MRI: Path to clinical translation in oncology. Neoplasia. 2019;21:1–16. doi: 10.1016/j.neo.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson S.J., Kurhanewicz J., Vigneron D.B., Larson P.E., Harzstark A.L., Ferrone M., van Criekinge M., Chang J.W., Bok R., Park I., Reed G., Carvajal L., Small E.J., Munster P., Weinberg V.K. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-(1)(3)C]pyruvate. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park I., Larson P.E.Z., Gordon J.W., Carvajal L., Chen H.Y., Bok R., Van Criekinge M., Ferrone M., Slater J.B., Xu D., Kurhanewicz J., Vigneron D.B., Chang S., Nelson S.J. Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn. Reson. Med. 2018;80:864–873. doi: 10.1002/mrm.27077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gammon S.T., Pisaneschi F., Bandi M.L., Smith M.G., Sun Y., Rao Y., Muller F., Wong F., De Groot J., Ackroyd J., Mawlawi O., Davies M.A., Gopal Y.N.V., Di Francesco M.E., Marszalek J.R. Mechanism-specific pharmacodynamics of a novel complex-I inhibitor quantified by imaging reversal of consumptive hypoxia with [(18)F]FAZA PET in vivo. Cells. 2019;8:1487. doi: 10.3390/cells8121487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito K., Matsumoto S., Devasahayam N., Subramanian S., Munasinghe J.P., Morris H.D., Lizak M.J., Ardenkjaer-Larsen J.H., Mitchell J.B., Krishna M.C. Transient decrease in tumor oxygenation after intravenous administration of pyruvate. Magn. Reson. Med. 2012;67:801–807. doi: 10.1002/mrm.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojtkowiak J.W., Cornnell H.C., Matsumoto S., Saito K., Takakusagi Y., Dutta P., Kim M., Zhang X., Leos R., Bailey K.M., Martinez G., Lloyd M.C., Weber C., Mitchell J.B., Lynch R.M. Pyruvate sensitizes pancreatic tumors to hypoxia-activated prodrug TH-302. Cancer Metab. 2015;3:2. doi: 10.1186/s40170-014-0026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doherty J.R., Cleveland J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valvona C.J., Fillmore H.L., Nunn P.B., Pilkington G.J. The regulation and function of lactate dehydrogenase A: Therapeutic potential in brain tumor. Brain Pathol. 2016;26:3–17. doi: 10.1111/bpa.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Y., Xiong Y., Qiao T., Li X., Jia L., Han Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018;7:6124–6136. doi: 10.1002/cam4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin L., Chun J., Pan C., Alesi G.N., Li D., Magliocca K.R., Kang Y., Chen Z.G., Shin D.M., Khuri F.R., Fan J., Kang S. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene. 2017;36:3797–3806. doi: 10.1038/onc.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le A., Cooper C.R., Gouw A.M., Dinavahi R., Maitra A., Deck L.M., Royer R.E., Vander Jagt D.L., Semenza G.L., Dang C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutfreund H., Cantwell R., McMurray C.H., Criddle R.S., Hathaway G. The kinetics of the reversible inhibition of heart lactate dehydrogenase through the formation of the enzyme-oxidized nicotinamide-adenine dinucleotide-pyruvate compounds. Biochem. J. 1968;106:683–687. doi: 10.1042/bj1060683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stambaugh R., Post D. Substrate and product inhibition of rabbit muscle lactic dehydrogenase heart (H4) and muscle (M4) isozymes. J. Biol. Chem. 1966;241:1462–1467. [PubMed] [Google Scholar]
- 34.Wang C.S. Inhibition of human erythrocyte lactate dehydrogenase by high concentrations of pyruvate. Evidence for the competitive substrate inhibition. Eur. J. Biochem. 1977;78:569–574. doi: 10.1111/j.1432-1033.1977.tb11770.x. [DOI] [PubMed] [Google Scholar]
- 35.Kirk P., Wilson M.C., Heddle C., Brown M.H., Barclay A.N., Halestrap A.P. CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 2000;19:3896–3904. doi: 10.1093/emboj/19.15.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno K.X., Sabelhaus S.M., Merritt M.E., Sherry A.D., Malloy C.R. Competition of pyruvate with physiological substrates for oxidation by the heart: Implications for studies with hyperpolarized [1-13C]pyruvate. Am. J. Physiol. Heart Circ. Physiol. 2010;298:H1556–H1564. doi: 10.1152/ajpheart.00656.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganapathy V., Thangaraju M., Gopal E., Martin P.M., Itagaki S., Miyauchi S., Prasad P.D. Sodium-coupled monocarboxylate transporters in normal tissues and in cancer. AAPS J. 2008;10:193–199. doi: 10.1208/s12248-008-9022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zacharias N.M., Baran N., Shanmugavelandy S.S., Lee J., Lujan J.V., Dutta P., Millward S.W., Cai T., Wood C.G., Piwnica-Worms D., Konopleva M., Bhattacharya P.K. Assessing metabolic intervention with a glutaminase inhibitor in real-time by hyperpolarized magnetic resonance in acute myeloid leukemia. Mol. Cancer Ther. 2019;18:1937–1946. doi: 10.1158/1535-7163.MCT-18-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the article.