Abstract
Duchenne muscular dystrophy (DMD) is a devastating chromosome X-linked disease that manifests predominantly in progressive skeletal muscle wasting and dysfunctions in the heart and diaphragm. Approximately 1/5000 boys and 1/50,000,000 girls suffer from DMD, and to date, the disease is incurable and leads to premature death. This phenotypic severity is due to mutations in the DMD gene, which result in the absence of functional dystrophin protein. Initially, dystrophin was thought to be a force transducer; however, it is now considered an essential component of the dystrophin-associated protein complex (DAPC), viewed as a multicomponent mechanical scaffold and a signal transduction hub. Modulating signal pathway activation or gene expression through epigenetic modifications has emerged at the forefront of therapeutic approaches as either an adjunct or stand-alone strategy. In this review, we propose a broader perspective by considering DMD to be a disease that affects myofibers and muscle stem (satellite) cells, as well as a disorder in which abrogated communication between different cell types occurs. We believe that by taking this systemic view, we can achieve safe and holistic treatments that can restore correct signal transmission and gene expression in diseased DMD tissues.
Keywords: Duchenne muscular dystrophy, DMD, Signaling pathways, Striated muscles
Introduction
Duchenne muscular dystrophy (DMD) is a chromosome X-linked disease that affects approximately 1/5000 boys and 1/50,000,000 girls [1–3]. DMD manifests in progressive skeletal muscle wasting and heart and diaphragm dysfunctions [4]. Depending on the dystrophin expression profile, some patients also suffer from other phenotypic alterations that include cognitive deficits and psychiatric problems [5, 6]. DMD is typically diagnosed around the age of 5 when the first symptoms of motor delay or abnormal gait become apparent, although preceding delays in independent walking and language development may have also been observed [7]. With progression of the disease, the extent of muscle fiber loss is so vast that patients are forced to use a wheelchair and respiratory system aid to help the diaphragm sustain breathing. To date, DMD is incurable and leads to premature death by the twenties or thirties due to respiratory or cardiac failure [7].
DMD is caused by mutations in the DMD gene that lead to the absence of functional dystrophin protein. A number of research studies have revealed that a lack of functional dystrophin results in aberrant signal transduction, which leads to epigenome modifications and altered gene expression in affected muscle tissues [8–11]. Although still limited, knowledge regarding dystrophin absence-associated disturbed signaling resulted in the first experimental therapies and clinical trials based on gene expression modulation. For example, the use of histone deacetylase inhibitors showed promising therapeutic outcomes in DMD animal models and patients [12, 13]. Nonetheless, due to the systemic and nonspecific action of these therapies, disadvantageous side effects are inevitable [12]. We are only starting to recognize the intricate web of interactions between myofiber and its environment, which consists of various cell types, including muscle stem (satellite) cells, fibro/adipogenic progenitors (FAPs), immune cells, motoneurons, bone cells, and blood vessel cells. Any of the therapeutic approaches based on tweaking gene expression in the diseased tissue would need to take into account the magnitude of these cell-to-cell interactions. In this article, we gather and review the available data on the interdependence of various cell types in the muscle and surrounding tissues in the context of dystrophin deficiency.
Molecular background of Duchenne muscular dystrophy
DMD is localized on the Xp21 locus, which spans ~ 2.5 Mbp and consists of 79 exons. Its activation leads to the synthesis of several mRNA transcripts in a tissue-dependent manner. In particular, a 14 kb mRNA encoding 427 kDa dystrophin isoform (Dp427) is generated in high amounts in skeletal muscles and the heart [14], and all mutations causing DMD result in either the loss or production of highly dysfunctional Dp427 [15]. Dp427 synthesis occurs at three distinct promoters. Dp427m is present in skeletal and cardiac muscles, Dp427c occurs in neurons of the cortex and the CA regions of the hippocampus, and Dp427p is found in cerebellar Purkinje cells [14]. In addition to full-length dystrophin, several shorter isoforms are generated as a result of transcription from several other unique promoters. Isoforms Dp260 and Dp116 are observed mainly in the retina and peripheral nerves, respectively [16, 17], while Dp140 is predominantly synthesized during fetal stages and in the adult brain, similar to the shortest Dp40 [18, 19]. In contrast, Dp71 is ubiquitously present in various tissues, and although previously considered to be a nonmuscle isoform [20, 21], its presence has been recently confirmed in myoblasts and myofibers [21, 22]. Depending on the site of mutation in DMD, the expression of shorter dystrophins might also be compromised [23], which impacts the extent of the observed pathological alterations. For instance, mutations that affect the expression of Dp71, which is especially abundant in the brain, account for intellectual disability in DMD patients [5].
Initially, dystrophin was considered to serve only mechanical functions, particularly in the context of the dystrophin-associated protein complex (DAPC) assembled in the sarcolemma and known to transduce the force during muscle contraction to the extracellular matrix (ECM) [24]. Over time, interactions of either dystrophin or DAPC with several cytoskeletal elements and cell signaling molecules have been reported [25]. Because of its numerous binding domains and interaction partners, dystrophin is now considered part of a multicomponent mechanical scaffold as well as a signal transduction hub [26–29] (Fig. 1).
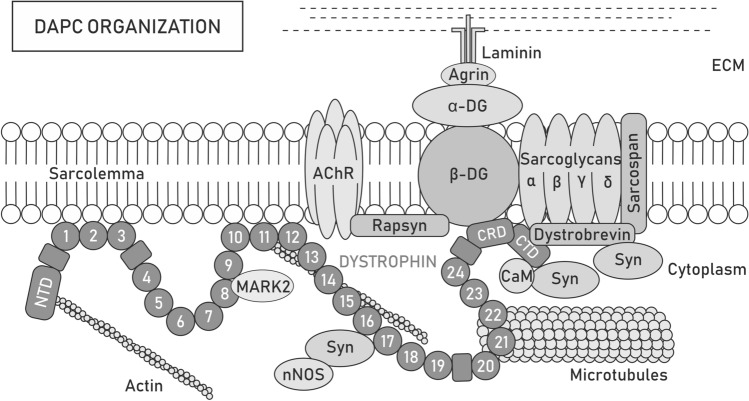
Fig. 1.
Dystrophin-associated protein complex organization. Dystrophin forms a scaffold for the dystrophin-associated protein complex (DAPC) that acts both as a mechanical force transducer and a signaling hub. Depending on the presence of the dystrophin isoform, its paralog utrophin or tissue/cellular localization, the content of DAPC may differ. Full-length dystrophin Dp427 consists of the N-terminal domain (NTD), central rod [with 24 spectrin-like repeats (circles) and 4 hinge modules (rectangles)], cysteine-rich (CRD), and C-terminal (CTD) domains, which provide multiple sites for interactions with proteins. NTD and spectrin-like repeats 11–15 bind costameric actin filaments, while repeats 8 and 9 anchor MARK2 and 20–23 interact with microtubules, which can also contact dystrophin indirectly through ankyrin-B. Spectrin-like repeats 1–3 and 10–12 may additionally participate in stabilizing the complex to the sarcolemma by binding the lipid bilayer. Through CRD, dystrophin interacts with sarcolemma-located β-dystroglycan (β-DG), which is anchored to α-dystroglycan (α-DG). In turn, α-DG binds laminin, a component of the extracellular matrix (ECM) network, which facilitates the transfer of forces during muscle contraction from the cytoskeleton to the ECM and protects the sarcolemma from twich-induced damage. At the neuromuscular junction (NMJ), the interaction of α-DG with agrin and perlecan provides MuSK-induced clustering of acetylcholine receptors (AChRs) and determines the localization of acetylcholine esterase, while the binding of β-DG with rapsyn is involved in the clustering of AChRs. Additionally, β-DG stabilizes α, β, δ- and γ-sarcoglycan and the sarcospan complex at the sarcolemma. The CTD of dystrophin interacts with various cytosolic proteins, such as dystrobrevin or syntrophins (Syn). Syntrophins recruit sodium channels and signaling molecules, such as neuronal nitric oxide synthase (nNOS)
The lack of functional full-length dystrophin (Dp427) inevitably leads to sarcolemma destabilization and muscle degradation. These effects are particularly evident in myofibers that are stretched during contractions, where dystrophin absence results in extensive membrane tearing, influx of Ca2+ through stretch-activated ion channels, a rise in intracellular Ca2+ levels and the associated overproduction of reactive oxygen species (ROS) [30, 31]. This process is considered an important initiating event in dystrophic pathogenesis and a subject of many studies that focus on therapeutic interventions in DMD [32–35]. The absence of dystrophin and the resultant disorganization of DAPC also impair the distribution of proteins at and underneath the sarcolemma, including signaling molecules, additionally contributing to myofiber death [36]. Importantly, dystrophin deficiency seems to be naturally alleviated by expression of the utrophin (UTRN) gene in both humans and animal models [37, 38], which is signified by the structural and functional similarities between utrophin and dystrophin [39, 40] and by the differences in the severity of the phenotype of various DMD mouse models. In particular, mdx mice that lack Dp427 have relatively mild disease symptoms and only slightly affected lifespans [41, 42], while mice missing both dystrophin and utrophin (mdx/utrn−/−) show a much more severe phenotype and usually die before the age of 20 weeks [43, 44]. Despite clear symptomatic differences in the course and severity of the disease in mdx mice and humans, mdx mice continue to be the most commonly used animal model of DMD [41, 45]. The effect of an absence of dystrophin in the muscle is not limited to the myofiber pathology but also pertains to the direct loss of dystrophin-related functions in satellite cells and vasculature [46, 47]. In particular, loss of dystrophin in satellite cells results in alterations in their division kinetics and differentiation into mature fibers [46].
Treatment strategies for DMD target primary defects or attenuate secondary downstream pathologies. The first approach aims to restore functional dystrophin protein. For example, antisense oligonucleotide-mediated exon skipping targets pre-mRNA splicing to restore shortened but functional proteins [48], while the CRISPR/Cas9 system is used to edit the defective DMD gene [49]. Currently, four exon skipping antisense oligonucleotides are FDA-approved for use in the treatment of DMD patients: Amondys 45 (casimersen), Viltepso (viltolarsen), Exondys 51 (eteplirsen) and Vyondys 53 (golodirsen) [50–52]. Moreover, CRISPR/Cas9-based strategies have provided promising therapeutic approaches; however, limitations remain, including the risk of off-target gene editing, and further research is required [50]. The above strategies are mutation-specific and thus are not universal and cannot be used for all patients. In contrast, the sequence of functional dystrophin can be delivered to all cells with recombinant adeno-associated viral (rAAV) vectors, although their capacity is limited to approximately 4.7 kb and thus precludes insertion of a cassette spanning the 11 kb coding fragment of Dp427 mRNA. Importantly, vectors carrying microdystrophins to muscle fibers are in clinical trials with promising initial results [53, 54] and minidystrophins can be generated by dual rAAV expression vectors [55, 56]. Cell-based therapies offer another approach; however, despite promising studies in animal models, the results are not fully transferable to humans, and the limitations include poor survival and limited migration (myoblasts), quick differentiation that limits their regenerative potential (satellite cells; SCs), risk of thalamic stroke (mesangioblasts), and limited ability to differentiate into muscle cells (bone marrow cells and CD133 + cells) [37, 57].
All these therapeutic methods must face the problem of immune rejection of either the restored or delivered dystrophin, as well as other foreign proteins, including viral capsids. Alternative strategies that bypass the problem of potential immune rejection consist of (1) genetically corrected autologous pluripotent stem cells differentiated ex vivo into dedicated muscle stem cells [58, 59] and (2) therapies grounded on either pharmacological induction of the UTRN gene [57] or delivery of vectors encoding micro- and mini-utrophins [40, 60]. Importantly, the Davies and Chamberlain groups revealed the therapeutic potential of full-length [61] and truncated utrophins in the muscles of dystrophic mice [62, 63]. Moreover, increasing the level of utrophin in DMD patients using pharmacological activators also had positive outcomes [40]. Nevertheless, it is important to note that the synthesis of dystrophin and utrophin from several promoters, extensive alternative splicing of the resultant mRNAs and the presence of alternative polyadenylation sites result in the expression of a whole range of specialized protein products in various cell types in a time- and often space-restricted manner [20]. Thus, inadequate dystrophin isoform matching could lead to unwanted side effects and long-term therapy designed for myofibers with one particular protein may expose latent dystrophin-related phenotypes in tissues other than muscle. The current results indicate that utrophin can partially compensate for the lack of functional Dp427, both in mechanical and signaling activities [64]. However, forced body-wide expression of specific utrophins might lead to unexpected pathologies due to, e.g., a lack of neuronal nitric oxide synthase (nNOS)-related signaling [65] or inadequate organization of microtubules [66].
In this review, we focus on DMD pathogenesis from a broader perspective that includes affected myofibers and satellite cells as well as abrogated cell communication and signaling between various cell types in the muscle and surrounding tissues, including inflammatory cells, bones, microvasculature, and innervation; however, signaling-related topics, such as noncoding RNAs or exosomes, are omitted because they have been previously reviewed in-depth [67–69]. The involvement of dystrophins in cell signaling and communication opens new therapeutic avenues as either adjuvants or stand-alone approaches. In addition, because all DMD patients suffer from skeletal muscle weakness as well as diaphragm and heart failure, signaling-based approaches could be a universal method of treatment and contribute to improving their quality of life.
Disrupted signaling in skeletal muscle
Muscle repair is a highly organized sequence of cellular events resulting in reconstruction of damaged tissue [70]. In DMD, this process is abrogated and shows chronically overlapping degeneration and regeneration cycles caused by DAPC loss and the resultant structural and signaling abnormalities in myofibers and satellite cells. On the one hand, the fibers cannot sustain the contraction forces and die, while on the other hand, they cannot be efficiently regenerated due to the affected division and self-renewal of muscle progenitor cells. Recurring inflammation and progressive fibrosis additionally lead to breakdown of the ECM and further change intracellular signaling as well as compromise interactions between different cell types, including myofibers as well as myoblasts and satellite cells, motoneurons, various bone [71] and microvasculature [72] cells (Fig. 2) and recently discovered FAPs [73].
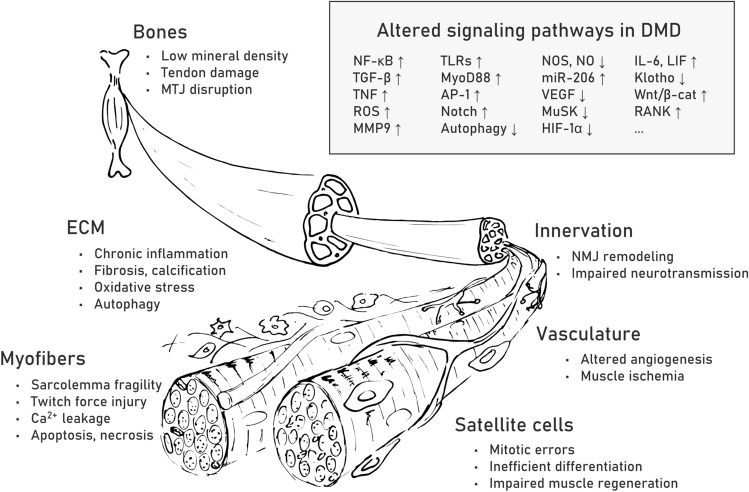
Fig. 2.
Pathological alterations and affected signaling pathways in DMD muscle fibers and surrounding tissues. Pathological alterations are listed for myofibers as well as extracellular matrix (ECM), bones, innervation, vasculature and satellite cells. Examples of upregulated (↑) and downregulated (↓) signaling pathways associated with the observed pathological changes in muscle fibers and surrounding tissues are listed in the box. Abbreviations: nuclear factor-κB (NF-κB), transforming growth factor-β (TGF-β), tissue necrosis factor (TNF), reactive oxygen species (ROS), matrix metalloprotease 9 (MMP9), Toll-like receptors (TLRs), myeloid differentiation primary response 88 (MyoD88), activator protein 1 (AP-1), neuronal nitric oxide synthase (nNOS), nitric oxide (NO), microRNA-206 (miR-206), vascular endothelial growth factor (VEGF), muscle-specific kinase (MuSK), hypoxia-inducible factor-1α (HIF-1α), interleukin 6 (IL-6), leukemia inhibitory factor (LIF), Wnt/β-catenin (Wnt/β-cat), receptor activator of NF-κB (RANK)
Chronic inflammation
Inflammation is a complex biological response in organisms to mitigate harmful stimuli, such as pathogens, irritants, or damaged cells, and initiate tissue repair. It is driven by innate immune response factors (nonspecific, associated with inflammation) and adaptive immune response factors (specialized). The innate immune response is initiated first, and it involves the action of cells, such as neutrophils, monocytes, and macrophages, and soluble factors, including cytokines and complement [74]. The pattern recognition receptors (PRRs) on the membranes of cells of the innate immune response recognize molecules on the pathogen surface as well as released material during cell damage or death, which represent damage-associated molecular patterns (DAMPs). PRRs can be subgrouped depending on the ligand specificity, function, or localization. In particular, Toll-like receptors (TLRs) play a key role in the immune response (Table 1).
Table 1.
Disrupted signaling pathways in DMD muscles and surrounding tissues
| Signaling pathway | Up- (↑) or downregulated (↓) in DMD | Short description | References for further reading | |
|---|---|---|---|---|
| Muscle inflammation | TLRs | ↑ | Toll-like receptors (TLRs) belong to the pattern recognition receptors and play crucial roles in the innate immune response. After interaction with damage-associated molecules patterns (DAMPs) or pathogen-associated molecular patterns (PAMPs), TLRs initiate downstream signaling to activate NF-κB, interferons, or mitogen-activated protein kinases (MAPKs) that regulate the expression of immune defense-related genes | [75, 244] |
| MyD88 | ↑ | Myeloid differentiation primary response 88 (MyD88) is an adaptor protein for inflammatory signaling pathways, downstream of TLRs and IL-1 receptors. MyD88 links TLRs or IL-1 receptors to IL-1R-associated kinase (IRAK), while activation of IRAK activates NF-κB, MAPKs, and activator protein 1, driving immune response | [245] | |
| NF-κB | ↑ | Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a family of inducible transcription factors regulating multiple aspects of immune response and inflammation. NF-κB promotes the expression of proinflammatory genes and regulates the survival, activation, and differentiation of immune cells | [246, 247] | |
| AP-1 | ↑ | Activator protein 1 (AP-1) is a heterodimer transcription factor built of proteins that belong to the Fos, Jun, ATF, and MAF families. In response to growth factor and cytokine signaling, AP-1 controls a wide range of cellular processes, including cell proliferation, death, survival, and differentiation | [248, 271] | |
| MMPs | ↑ | Matrix metalloproteinases (MMPs) are zinc-dependent endopeptidases involved in extracellular matrix remodeling, both during physiological processes and in pathological conditions. MMPs are also important players during inflammation | [249] | |
| TNF | ↑ | Tumor necrosis factor (TNF) is a pro-inflammatory cytokine that regulates a number of signaling pathways with various downstream effects. TNF proteins are predominantly expressed by immune cells. TNF signaling impacts immune response, inflammation, cell proliferation, programmed cell death, and necrosis | [250] | |
| Klotho | ↓ | Klotho proteins are obligate components of endocrine fibroblast growth factor (FGF) receptor complexes and provide the high-affinity binding of FGF19, FGF21, and FGF23 to their receptors. Klotho proteins are known to play a role in aging-related diseases, diabetes, cancer, arteriosclerosis, renal and bone disease, and inflammation processes | [251] | |
| TGF-β | ↑ | Transforming growth factor-β (TGF-β) initiates signaling through the canonical SMAD pathway, regulating the expression of hundreds of genes. TGF-β induces also various noncanonical pathways that are responsible for cytoskeleton organization, cell polarity, and miRNA maturation. The effects of TGF-β signaling depend on the cellular context | [252] | |
| IL | ↑ | Interleukins (ILs) are a group of cytokines with immunomodulatory functions that play an important role in immune cell differentiation and activation. ILs could have pro- and anti-inflammatory effects, depending on the producing and responding cell type or the phase of the immune response | [253] | |
| Muscle – bone | Wnt/β-cat | ↑ | Wnt/β-catenin (Wnt/β-cat) pathway regulates cell fate determination, cell migration, polarity and organogenesis during embryogenesis. Binding of Wnt to its membrane receptor causes translocation of β-cat degradation complex to the cell membrane, effecting in accumulation of β-cat in the cytoplasm and its eventual translocation into the nucleus to act as a transcriptional coactivator | [272] |
| OPN | ↑ | Osteopontin (OPN) is a multifunctional protein involved in physiological processes and the pathogenesis of various diseases (e.g., atherosclerosis, cancer, chronic inflammatory diseases). OPN interacts with several integrins and therefore controls cell migration, adhesion, and survival. Additionally, OPN promotes inflammation and regulates biomineralization | [254] | |
| RANK/RANKL/OPG | ↑ | Receptor activator of NF-κB (RANK), its ligand (RANKL), and osteoprotegerin (OPG) form the triad of the ligand/signaling receptor/decoy receptor. RANKL, RANK, and OPG have pivotal roles in the regulation of bone metabolism and the immune system. The triad is involved in diverse physiological and pathological contexts, including muscle metabolism | [255] | |
| IL-6 | ↑ | Interleukin 6 (IL-6) can act both as a pro-inflammatory cytokine and an anti-inflammatory myokine, depending on the cellular context. Additionally, IL-6 stimulates osteoclastogenesis | [78, 256] | |
| LIF | ↑ | Leukemia inhibitory factor (LIF) is an IL-6 class cytokine involved in controlling stem cell pluripotency, differentiation, bone metabolism, and inflammation. LIF signaling activates the JAK/STAT, MAPK, and PI3K pathways. This pleiotropic cytokine elicits a varied response in different cell types | [257] | |
| POSTN | ↑ | Periostin (POSTN) is an extracellular matrix protein that acts as a structural molecule of the bone matrix and a signaling molecule that stimulates osteoblasts through integrin receptors and the Wnt/β-cat pathway. POSTN is secreted in muscles during regeneration and differentiation | [258] | |
| FGF21 | ↑ | Fibroblast growth factor 21 (FGF21) is a hormone produced by several tissues that controls various metabolic pathways. Muscle-derived FGF21 acts as a stress-induced myokine, found to promote muscle atrophy and bone loss | [259] | |
| Muscle—microvasculature | NOS, NO | ↓ | Nitric oxide synthases (NOSs) catalyze the production of nitric oxide (NO) from L-arginine that controls, among others, vasodilation, and angiogenesis. NO also activates guanylyl cyclases (GC), which synthesize the second messenger cyclic guanosine monophosphate (cGMP), and act on its downstream targets, such as cGMP-activated protein kinase (PKG) or cyclic nucleotide-activated ion channels | [260] |
| VEGF | ↓ | Vascular endothelial growth factors (VEGFs) are key regulators of vascular development and of blood vessel function. Binding of VEGF to the VEGF receptor initiates the downstream signaling cascade and ultimately results in cell proliferation, migration, and the three-dimensional arrangement to form a vascular tube | [261] | |
| miR-206 | ↑ | Micro-RNA miR-206 is expressed specifically in skeletal muscles. miR-206 impedes cell proliferation and promotes SC and myoblast differentiation via posttranscriptional regulation of gene expression, boosting muscle regeneration and growth | [262] | |
| HIF-1α | ↓ | HIF-1α is a regulatory subunit of hypoxia-inducible factor-1 (HIF-1), an oxygen-dependent transcriptional activator. Target genes of HIF-1 are related to angiogenesis, cell proliferation and survival | [263] | |
| Muscle—neuron | MuSK | ↓ | Muscle-specific kinase (MuSK) is a transmembrane tyrosine kinase that forms a multiprotein complex localized in the postsynaptic sarcolemma. In response to neural agrin signaling, autophosphorylation of MuSK drives intracellular signaling cascades to coordinate the local synthesis and assembly of synaptic proteins. It results in the reorganization of the cytoskeleton and the recruitment of AChR-binding scaffolding proteins to aggregate AChRs | [160] |
| AChR | Defects in clustering | Acetylcholine receptors (AChRs) are ligand-gated ion channels that open upon acetylcholine binding and induce postsynaptic depolarization. AChR clustering is necessary for the proper functioning of the neuromuscular junction | [264] | |
| Utrophin | ↑ | Utrophin is a dystrophin homolog. Similar to dystrophin, utrophin presents mechanical functions and forms a signaling hub as a scaffold for various proteins. The upregulation of utrophin gene (UTRN) is one of the potential strategies to treat DMD | [265] | |
| Muscle satellite cells | MARK2 | ↓ | Microtubule affinity regulating kinase 2 (MARK2) is a serine/threonine-protein kinase that is an important regulator of cell polarity. MARK2 modulates microtubule network via phosphorylation and inactivation of microtubule-associating proteins | [46, 266] |
| PARD3 | Loss of polarization | Partitioning defective protein 3 (PARD3) is a part of Par complex built of atypical Protein Kinase C (aPKC)/Bazooka (Baz, PARD3)/Par-6. The Par complex determines cell polarity and asymmetric cell division. Opposite localization of Par complex and MARK2 defines the apicobasal axis | [46, 266] | |
| Autophagy pathways | ↓ | The autophagy pathway is a conserved cellular process of degradation of intracellular components that include soluble or aggregated proteins, organelles, macromolecular complexes, and foreign bodies. The formation of an autophagosome that ultimately fuses with a lysosome is driven by the cooperation of multiple factors | [267] | |
| Notch | ↑ | Notch signaling is a conserved pathway of cell–cell communication. The Notch receptor is localized on the signal-receiving cell, while ligands are located on the neighboring signal-sending cell. The effect of Notch signaling depends on the cellular context and can influence differentiation, proliferation and apoptotic cell fates | [268, 269] | |
| p38γ/Carm1 | Mislocalization | Mitogen-activated protein kinase (MAPK) p38γ regulates SC fate through phosphorylation of Carm1, which further controls epigenetic induction of Myf5 expression during asymmetric SC division | [9] | |
| FGF2 | ↑ | Fibroblast growth factor 2 (FGF2) is one of the FGFs that regulate SC function via activation of ERK MAPK, p38 MAPKs, PI3 kinase, PLCg and STATs. SCs express FGR receptors to detect FGF2 produced by myofibers, fibroblasts and satellite cells | [270] |
TLRs are expressed on the surface of immune cells and cells unrelated to the immune system, including myofibers. Activation of TLR signaling initiates cascades of molecular events that trigger the synthesis and secretion of cytokines and other proinflammatory factors necessary for both innate and adaptive immune responses [75, 76]. Cytokines are small proteins (~ 5–20 kDa) released by a broad range of cells. Their cell-surface receptors are located on various types of cells, and following binding, cytokines activate a cascade of intracellular signaling, resulting in up- or downregulation of a number of genes, although the final effect is cell-type specific. Depending on their influence on the inflammatory process, they can be divided into pro- and anti-inflammatory cytokines. Specifically, families of interleukin (IL)-1, IL-17, tissue necrosis factor-α (TNF-α), and interferons belong to the first group, whereas anti-inflammatory signaling is driven by families of IL-10 and IL-12 cytokines [77] (Table 1). Interestingly, some of them, e.g., IL-6, can have a pleiotropic effect on muscle regeneration, i.e., IL-6 interaction with its soluble receptor mediates pro-inflammatory response while binding to its membrane-bound receptor triggers a cascade of regenerative or anti-inflammatory cytokine action [78, 79] (Table 1). At the time that the mechanisms of innate immunity are already active, the major histocompatibility complex (MHC) proteins present antigens and initiate an adaptive immune response involving the action of T and B cells. Chronic inflammation can adversely affect tissues and organs, and the current data indicate that continuous stimulation of the immune system sustained by the ongoing degeneration and regeneration of myofibers adversely affects the condition of DMD patients.
Membrane instability and leakage of cytoplasmic content into the extracellular matrix initiate chronic activation of the innate immune system in DMD muscles (Fig. 3a). DAMPs, including nucleic acids, heat shock proteins (HSPs), high-mobility group box 1 (HMGB1) proteins, and ROS, are released by destroyed fibers and activate TLRs, e.g., TLR2/4 in myofibers (Fig. 3a). Signaling from TLRs and the IL-1 receptor activates a downstream adaptor protein, myeloid differentiation primary response 88 (MyD88) (Table 1). MyD88 activates IL-1R-associated kinase (IRAK) family kinases, which in turn trigger mitogen-activated protein kinases (MAPKs) and proinflammatory transcription factors, such as nuclear factor-κB (NF-κB; Fig. 3e) and activator protein 1 (AP-1). Importantly, Gallot et al. (2018) showed that inhibition of TLRs or MyD88 in dystrophic mice alleviates the disease symptoms and reduces inflammation [80] (see also “Disrupted signaling in satellite cells”).
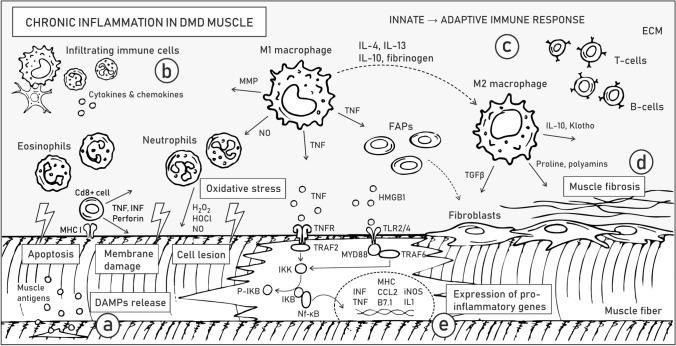
Fig. 3.
Chronic inflammation in DMD skeletal muscles. Pathological alterations related to chronic inflammation are listed in boxes. Arrows illustrate the release of various factors, while dashed line arrows depict the transformation of one type of cell to another. Molecular mechanisms underlying ongoing processes of sarcolemma instability and DAMP release a, activation of the innate immune response b, development of adaptive immune response c, muscle fibrosis d, and inter- and intracellular signaling driving inflammatory response e are described in the text. Abbreviations: extracellular matrix (ECM), damage-associated molecular patterns (DAMPs), major histocompatibility complex I (MHCI), matrix metalloproteases (MMP), fibroadipogenic progenitors (FAPs), interferon γ (INFγ), tissue necrosis factor (TNF), receptor of TNF (TNFR), TNFR Associated Factor 2 (TRAF2), IκB kinase (IKK), IKB, phosphorylated IKB (P-IKB), nuclear factor-κB (NF-κB), high-mobility group box 1 (HMGB1), Toll-like receptors (TLR2/4), TNFR associated factor 6 (TRAF6), myeloid differentiation primary response 88 (MYD88), interleukin (IL), transforming growth factor-β (TGFβ)
After muscle injury, the tissue is infiltrated by innate immune system cells. Among others, (1) CD8 + cytotoxic T lymphocytes induce apoptosis of myofibers through a perforin-mediated pathway, (2) eosinophils release lytic substances and (3) inflammatory M1 macrophages (activated by proinflammatory cytokines, such as IFN-γ) produce nitric oxide (NO) and inflammatory cytokines and participate in direct phagocytosis (Fig. 3b). Macrophage-derived NO further induces muscle fiber lysis and tissue damage, which stimulates neutrophils to release free radicals [81]. Local elevations in oxidative stress aggravate the pathology of DMD muscles, partially through changes in signal transmission [82]. Proinflammatory cytokines trigger constitutive expression of MHC class I and II on muscle cells, which attract T and B cells, consequently inducing an adaptive immune response [83] (Fig. 3c). Meanwhile, promoted by IL-10, IL-4 and IL-13, modulatory M2 macrophages gradually replace M1 macrophages. M2 macrophages release anti-inflammatory cytokines, such as IL-10, IL-4 and transforming growth factor-β (TGF-β) [81] that alleviate muscle injury (Fig. 3b) (Table 1).
The various inflammatory molecules produced by macrophages include matrix metalloproteases (MMPs), which are known to promote inflammation and interstitial fibrosis (Fig. 3d) (Table 1). In particular, high levels of MMP-9 were detected in numerous muscle conditions, including the skeletal muscles of DMD mouse models and patients [84]. Moreover, the level of MMP-9 correlated with disease severity [85]. Hindi et al. showed that inhibition of MMP-9 boosts the promyogenic M2 phenotype while diminishing the percentage of M1 macrophages in mdx mice [84]. Accordingly, MMP-9 downregulation reduced the levels of the inflammatory cytokines IFN-γ and IL-6 and increased the activity of IL-4 (involved in adaptive immunity). The reduced inflammation was also confirmed by the reduced levels of the proinflammatory transcription factors NF-κB and AP-1 (Table 1). Furthermore, enhanced proliferation of SCs and improved regeneration were observed [84].
During DMD muscle degeneration, M1 macrophages and myogenic cells are among the main sources of the proinflammatory factor TNF-α (Fig. 3b) (Table 1), which is suggested to be the major death ligand driving necrosis and programmed necrosis (necroptosis) and is linked to the activation of receptor-interacting protein kinase-1 (RIPK3) signaling [86]. Elevated TNF-α levels also contribute to the aggravation of inflammation via the induction of NF-κB signaling, which triggers the expression of proinflammatory genes and the synthesis of cytokines and chemokines. The Acharyya group demonstrated upregulation of IκB kinase/NF-κB (IKK/NF-κB) signaling in DMD and revealed that reduction of NF-κB or its upstream activator IKK improves the pathology and muscle function in mdx mice [87]. In the same animal model, Yang et al. showed that AAV-mediated shRNA knockdown of the p65 subunit of NF-κB has an anti-inflammatory effect [88]. Genetic reduction of p65 levels also diminished chronic inflammation and improved DMD muscle physiology [89], and by targeting RANKL (the receptor activator of nuclear factor NF-κB ligand), it alleviated the pathology by increasing the proportion of M2 macrophages and reducing muscle edema and fibrosis [90]. Furthermore, anti-RANKL treatment also increased the mechanical properties of bone in dystrophic mice [90]. Overall, inhibition of NF-κB may be a promising therapy for DMD, especially when combined with gene therapy designed to restore dystrophin expression.
The alleviating role of M2 macrophages is also linked to the increased synthesis and secretion of Klotho, a transmembrane protein whose extracellular domain can be cleaved and released (Fig. 3b) (Table 1). Klotho functions as a promyogenic circulating hormone [91] that activates the proliferation and growth of muscle cells. Accordingly, Klotho expressed by leukocytes positively influences the number of SCs in dystrophic muscle [91]. The expression of Klotho is significantly decreased in DMD tissues; the diminished activity of the Klotho pathway in muscle cells is attributed to epigenetic changes associated with oxidative stress in dystrophic muscle; and overexpression of Klotho in macrophages counteracts the expression of profibrotic genes and reduce the pathology of DMD [92]. Under physiological conditions, Klotho diminishes TGF-β expression and thus prevents muscle fibrosis, whereas in DMD patients and mdx mice, TGF-β remains activated and consequently promotes muscle fibrosis.
Muscle–bone relation
Low bone mineral density and fragility in DMD are symptoms that accompany muscle degeneration caused by inflammation and alterations in signaling and cell-to-cell communication [93]. Pathological fractures of long bones and vertebrae significantly impact mobility and decrease the patients’ quality of life [94]; moreover, the standard long-term therapeutics for DMD, i.e., glucocorticosteroids, increase the risk of osteoporosis [95]. DMD mouse models also display a decline in bone biomechanical properties and healing capacity, spinal deformity, and spontaneous ossification in muscles [96–98]. Muscle-derived myokines, osteokines released by the bone, and inflammatory cytokines trigger common signaling pathways, thus providing a functional connection between the cells organized into the muscle–bone unit [71, 93] (Fig. 4).
Fig. 4.
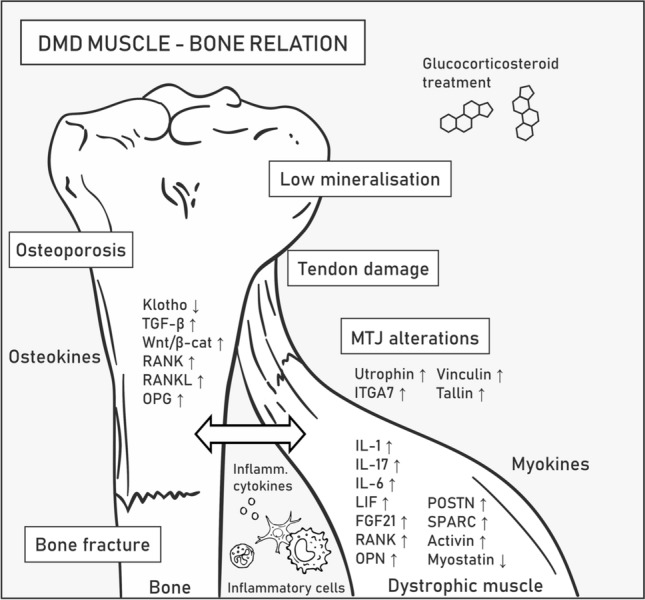
Muscle–bone relation in Duchenne muscular dystrophy. DMD symptoms related to bones and tendons associated with skeletal muscles are listed in boxes. Molecular mechanisms underlying pathogenesis in the muscle–bone relation are described in the text. Abbreviations: transforming growth factor-β (TGF-β), Wnt/β-catenin (Wnt/β-cat), receptor activator of NF-κB (RANK), RANK ligand (RANKL), osteoprotegerin (OPG), interleukin (IL), leukemia inhibitory factor (LIF), fibroblast growth factor 21 (FGF21), periostin (POSTN), osteonectin (SPARC), integrin α7 (ITGA7)
IL-6 is a pro-osteoclastogenic and pro-myogenic cytokine released by tissues with essential functions in bone homeostasis [93] and myogenesis [99]. IL-6 is upregulated in DMD patients and mdx mice compared with healthy controls, and it can mediate bone degradation and elevate the level of IL-10 that switches pro-inflammatory M1 macrophages to anti-inflammatory M2 macrophages, which drives an anti-inflammatory response. The levels of IL-10 and its receptor are also higher in DMD muscles [71]. However, as previously mentioned, IL-6 shows a pleiotropic phenotype during muscle repair, and despite its anti-inflammatory and regenerative effects, it can also aggravate inflammation [100]. Specifically, Pelosi et al. showed that forced expression of IL-6 exacerbates muscle pathology while its receptor blockade has the opposite outcome in mdx mice [79, 100].
LIF is an example of an IL-6 class cytokine involved in bone remodeling and shows higher expression in dystrophic muscles (Table 1). In particular, the study by Matsushita et al. showed that LIF inhibits osteogenesis via the STAT3/SOCS3 signaling pathway [101]. Bone weakness in DMD patients is also driven by osteopontin (OPN), a well-known inhibitor of bone mineralization and a factor that promotes fibrosis in dystrophic muscles [102] (Table 1). This osteokine, among others, is produced by inflammatory cells, e.g., macrophages; therefore, its expression level greatly increases during inflammation. Proinflammatory cytokines engaged in bone homeostasis, such as IL-1 and IL-17, are also elevated in DMD [93, 103, 104]. Thus, potential therapeutic treatments based on reducing the level of various molecules driving inflammation to alleviate inflammation in diseased muscles can be envisioned. Such approaches, however, require in-depth knowledge of the remote effects of these compounds in various organs. For example, IL-15 treatment is potent enough to improve the pathophysiology of dystrophic muscle in mdx mice [105] but was also shown to stimulate osteoclast differentiation, reduce the number of both osteoclasts and osteoblasts in bone marrow, and increase the bone mineral content [71].
Under pathological conditions of dystrophin loss-mediated muscle fiber degeneration and decreased Klotho levels, TGF-β is released in high amounts from the bone matrix, which contributes to muscle weakness. TGF-β promotes myofibroblast differentiation and increases tissue fibrosis by stimulating the canonical Wnt/β-catenin pathway that regulates bone homeostasis and myogenesis (Table 1). In particular, Wnt/β-catenin decreases osteoclast differentiation by promoting the synthesis and secretion of osteoprotegerin (OPG), an osteoclastic inhibitor [106]. Although therapeutic overexpression of Klotho indeed improves the functioning of muscles [92], it was also shown to inhibit mineralization and osteogenic activity in cultured osteoblastic cells, presumably due to decreased Wnt/β-catenin and downregulation of OPG [71]. OPG is produced by osteoblasts and prevents the interaction of receptor activator of NF-κB (RANK), which is located on preosteoclastic cells, with RANKL, thus inhibiting the NF-κB pathway [90] (Table 1). RANK is also present in the sarcolemma of muscle fibers, while RANKL and OPG were found to be expressed in the myoplasm and can be secreted from myofibers, indicative of the bidirectional interaction between both tissues. Moreover, RANK/RANKL signaling is disrupted in mdx/utrn+/−, with significantly upregulated RANKL and RANK protein levels observed in mdx/utrn+/− muscle samples [90]. Overall, the current data indicate that the RANK/RANKL/OPG pathway may be an important platform for muscle–bone crosstalk, which is dysfunctional in DMD patients.
In addition to RANK and RANKL, the expression of many other bone-regulating myokines is altered in DMD, and they include the extracellular matrix proteins periostin (POSTN) and osteonectin (also known as SPARC), which are upregulated in DMD muscles and play important roles in bone remodeling as well as maintaining bone mass and quality by a variety of mechanisms [104] (Table 1). Furthermore, fibroblast growth factor 21 (FGF21) was reported to be elevated in DMD muscles [104] and to negatively regulate bone homeostasis by potentiating peroxisome proliferator-activated receptor gamma (PPAR-γ) activity, resulting in adipogenesis stimulation and osteogenesis inhibition from bone marrow stem cells. Increased FGF21 levels were also found to indirectly promote osteoclastogenesis, presumably by increasing the RANKL/OPG ratio [107] (Table 1).
Activin and myostatin are growth factors that belong to the TGF-β superfamily, and they are known to have negative effects on muscle and bone mass [108, 109]; therefore, their downregulation might be considered a therapeutic approach to prevent muscle wasting and bone degeneration in DMD patients [71]. Follistatin modulates bone metabolism presumably via activin and myostatin signaling, and follistatin-based gene therapy was shown to have positive effects on muscles [110–112]. However, myostatin seems to have a positive impact on tendons. Specifically, in myostatin-deficient mice, tendons have a stiff, brittle and hypocellular phenotype. Stiffness of tendons would worsen the course of DMD and lead to higher sensitivity to contraction-induced injury [113].
Tendons allow movement by providing physical muscle interaction with the skeleton. In DMD patients, the ongoing cycles of myofiber degeneration and regeneration increase fibrosis and results in pseudohypertrophy of certain muscles, especially in the early stages of the disease [114], which results in excessive strain on tendons and in their shortening and contracture (often the case with the Achilles tendon) [115]. The elastic properties of dystrophic tendons are also compromised due to the presence of a higher number of dead cells and collagen concentration as well as the reduction in proteoglycans and infiltration of inflammatory cells [115, 116]. The architecture of myotendinous junctions (MTJs), i.e., specialized structures located on the muscle–tendon interface, is also affected in DMD. Specifically, dystrophic MTJs contain a reduced number of sarcolemmal folds [43, 117], with the compensatory upregulation of a number of proteins, including utrophin, α7 integrin, vinculin, and talin [118, 119].
Muscle–microvasculature relation
Skeletal muscle is a highly vascularized tissue. The presence of necrotic muscle bundles due to local ischemia was one of the first described causes of muscle weakness in DMD before the discovery of the dystrophin gene [120]. Indeed, proper vascularization is obligatory for normal functioning and regeneration of muscle tissue. In addition to myofibers and satellite cells, DMD is expressed in vascular smooth muscle and endothelial cells, and a lack of full-length dystrophin directly affects the formation of blood vessels [47]. Impaired signaling between the muscle tissue and surrounding capillaries also aggravates the phenotype of the patients (Fig. 5).
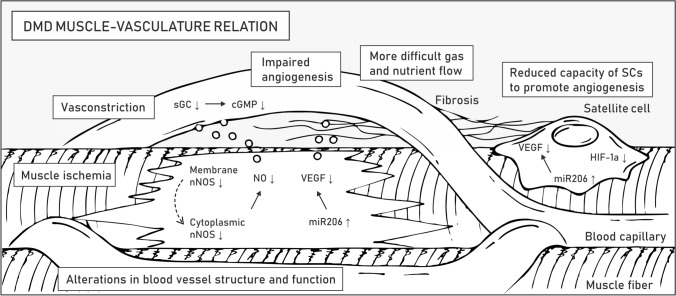
Fig. 5.
Muscle–vasculature relation in DMD. Symptoms related to the microvasculature of striated muscles are listed in boxes. Molecular mechanisms underlying pathogenesis in the muscle-vasculature relation are described in the text. Abbreviations: neuronal nitric oxide synthase (nNOS), soluble guanylyl cyclase (sGC), cyclic guanosine monophosphate (cGMP), vascular endothelial growth factor (VEGF), hypoxia-inducible factor-1α (HIF-1α)
The vascular hypothesis of DMD indicates the critical role of the neuronal nitric oxide synthase (nNOS) pathway [72, 121] (Table 1). In healthy myofibers, the muscle-specific isoform of nNOS (nNOSμ) is anchored to the sarcolemma by binding with dystrophin spectrin-like repeat 17 through α-syntrophin [122] (Fig. 1). nNOSμ-derived nitric oxide (NO) diffuses to neighboring capillaries, where it increases the concentration of cyclic guanosine monophosphate (cGMP) and counteracts vasoconstriction caused by norepinephrine released from sympathetic nerves. Dystrophin deficiency causes displacement of nNOSμ from the sarcolemma to the cytoplasm, where its amount is also greatly reduced [123, 124]. Consequently, DMD myofibers are more susceptible to functional muscle ischemia during exercise and injury due to a reduction in paracrine signaling [123, 124]. The ischemic phenotype inspired multiple studies aimed at targeting the nNOS-NO pathway in DMD patients based either on the expression of dystrophin minigenes to provide the nNOSμ binding site, direct overexpression of the nNOS gene, supplementation with L-arginine (nNOS substrate), delivery of NO-donating drugs, or inhibition of phosphodiesterase 5A (PDE5A) to prolong the half-life of cGMP [72].
Therapeutic strategies grounded on modulation of the nNOS pathway are oriented on increasing vasorelaxation of the vasculature to enable perfusion of diseased muscles. A different approach to obtaining the same effect is to enhance the density of the microvasculature network by boosting angiogenesis [47, 124]. Vascular endothelial growth factor (VEGF) is a glycoprotein synthetized and secreted by myofibers and SCs in muscle tissue, and it plays a crucial role in this process, mainly via the VEGFR-2 pathway [125] (Table 1). VEGF stimulates the proliferation, migration, and survival of endothelial cells upon binding to vascular endothelial growth factor receptors (VEGFRs) [125] and has antiapoptotic properties toward myogenic cells [126]. Importantly, VEGF levels increase during exercise in healthy muscles [47], a process that is abrogated in DMD patients because their physical activity is reduced and associated with tissue damage.
As mdx muscles undergo chronic degeneration–regeneration routes, a strong upregulation of microRNA-206 (miR-206), one of the key regulators of myogenesis synthesized in both muscles and SCs, is observed (Table 1). Apart from its role in muscle repair [127], miR-206 also represses the expression of the VEGF gene in myofibers and therefore negatively impacts angiogenesis [47]. Treatment strategies targeting VEGF signaling include inhibition of VEGFR-1 (a negative regulator of angiogenesis, competing for VEGF with VEGFR-2), direct delivery of VEGF, administration of rAAV vectors carrying the coding sequence of VEGF, or in vivo transplantation of muscle-derived stem cells (MDSCs) with upregulated VEGF. Nevertheless, it is important to note that a high dosage of VEGF can contribute to a profibrotic response and lead to serious adverse effects, e.g., endothelial cell-derived vascular tumors [47]. As an alternative approach, heme oxygenase 1 (HO-1) was found to regulate blood vessel formation and angiogenesis induced by VEGF and stromal cell-derived factor 1 (SDF-1) [128, 129], and such effects in addition to other aspects of HO-1 signaling (see “Disrupted signaling in satellite cells”) underlie the rationale for the therapeutic modulation of HO-1 levels to ameliorate DMD pathology. A treatment based on a different growth factor, angiopoietin 1 (ANG1), combined with VEGF or alone, was also proposed. Notably, administration of ANG1 was shown to enhance muscle perfusion and slow the progression of fibrosis [130].
Signaling crosstalk between capillaries and SCs that are localized in their close proximity may also influence angiogenesis and muscle regeneration. In particular, SCs derived from 12-month-old mdx mice showed a reduced ability to promote angiogenesis in vitro [131], presumably due to the age-related lower levels of proangiogenic VEGF and hypoxia-inducible factor-1α (HIF-1α) [131] (Table 1). Other reports indicate that in young dystrophic animals, showing intensive degeneration and regeneration cycles, angiogenesis is not altered or even enhanced [132, 133]. Interestingly, SCs also seem to be influenced by the muscle vasculature, e.g., Verma et al. showed that their direct interaction may influence the self-renewal and quiescence of SCs via Notch signaling [134] (Table 1). Increased distance between blood vessels and myofibers could also slow gas exchange and impede reciprocal signaling as a result of fibrosis-associated changes in ECM [135]. Latroche et al. compared the microvasculature of 3- and 12-month-old mdx mice that differed in the content of the fibrotic tissue [136], and they identified very severe alterations of the microvascular network structure with reduced perfusion only in older mice. Overall, the current research data clearly indicate that hindered muscle vascularization contributes to the pathophysiology of DMD.
An additional issue associated with muscle-microvasculature interactions is the therapeutic potential of pericytes that localize underneath the basal lamina of small vessels [137]. Pericytes differentiate with high efficiency into skeletal muscle cells in vivo and those isolated from one biopsy can be expanded in vitro to amounts sufficient to treat a pediatric patient [138]. Pericytes are also suitable for systemic delivery because they can cross the vessel wall and are easily transducible with viral vectors [137, 138]. In contrast, SC-derived cells do not efficiently cross the endothelial layer [139, 140] and are challenging to transduce with viral vectors [141, 142].
Muscle–neuron relation
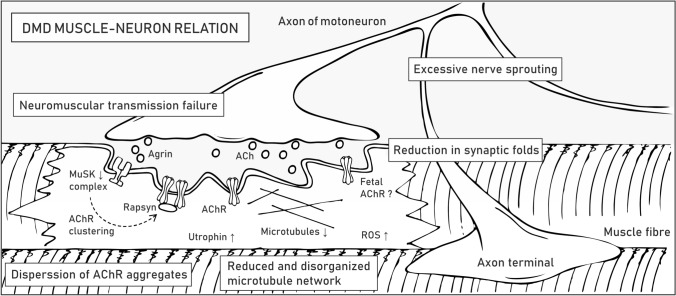
Neuromuscular junctions (NMJs) are specialized regions where terminal buttons of motor neurons contact muscle fibers to form chemical synapses. The signal is transmitted by a small molecule neurotransmitter, acetylcholine (ACh), which, following release from motoneurons, binds ACh receptors (AChRs) on the postsynaptic sarcolemma (called motor end-plate) (Table 1). This interaction enables ion flow across the sarcolemma that leads to local depolarization and induction of an action potential that travels across the myofiber membrane, eventually leading to muscle contraction. The structural organization of the sarcolemma and clustering of AChRs are important factors in efficient signal transmission, signified by the fact that some conditions, such as exercise [143, 144], aging [145], muscle or nerve injury, and lack of associated proteins [146], result in structural and functional alterations.
DAPC accumulates at the postsynaptic membrane of NMJs, where it is required for synaptic homeostasis [29]. Mdx mice show structural abnormalities that include loss in the number and depth of synaptic folds in the motor end-plate, NMJ fragmentation (more scattered AChR clusters), and excessive nerve sprouting [146, 147] (Fig. 6). Such morphological alterations entail profound functional consequences, as revealed by reduced end-plate potential (EPP) and depressed safety factor (the amplitude by which the EPP exceeds depolarization threshold) [148]. Utrophin is also present at the NMJs of healthy individuals and in higher amounts in DMD muscles [149], where it presumably compensates for the absence of dystrophin [64]. Utrn−/− mice have a lower number of AChRs and reduced folding of the postsynaptic membrane; nonetheless, they show no signs of weakness, which suggests that utrophin is not essential but undoubtedly contributes to the organization and maintenance of NMJs [150]. This conclusion is further corroborated by qualitative assessment of NMJs in various DMD mouse models, e.g., mdx, mdx/utrn+/−, and mdx/utrn−/−, which revealed the most severe damage in mice devoid of both dystrophin and utrophin. However, AChR area fragmentation at NMJs was observed for all three DMD mouse models, with no significant difference among them [151].
Fig. 6.
Pathological alterations in dystrophic NMJs. DMD symptoms at NMJs are listed in boxes. The molecular mechanisms underlying the pathogenesis of NMJs are described in the text. Abbreviations: muscle-specific kinase (MuSK), acetylcholine (ACh), acetylcholine receptor (AChR), reactive oxygen species (ROS)
Because disruption of NMJs is one of the main symptoms of DMD, therapeutic strategies are assessed to restore proper communication between nerves and muscles. Although estimates of how much dystrophin is required for the therapeutic effect depend on various factors, e.g., quantification method or reference samples [152, 153], the current results indicate that even a fraction of the normal dystrophin levels could be beneficial in dystrophic muscles. The proper formation and functioning of NMJs might require higher dystrophin amounts, estimated to range between 19 and 50% of the normal content [154]. On the other hand, utrophin upregulation ameliorates muscle pathology in mdx mice and leads to increased AChR clustering and improved morphology of NMJs [155], which shows that either utrophin- or dystrophin-based therapies could be beneficial for the treatment of DMD [64]. In their review, Ng and Ljubicic [148] concisely reported observations regarding the morphology and electrophysiology of NMJs obtained since the 1970s and concisely presented potential therapeutic strategies targeting neuromuscular transmission in dystrophic muscles.
Sequence and structural similarity of dystrophin and utrophin enable the formation of protein complexes characterized by partial functional interchangeability [40]. Both DAPC and utrophin-associated protein complex (UAPC) contain two subunits of dystroglycan that are of special importance at NMJs [156]. α-Dystroglycan is present at the cell periphery on the outer surface of the sarcolemma, where it interacts with a number of proteins, including agrin, a signaling molecule released by the nerve terminal that initiates clustering of AChRs on the postsynaptic membrane, as well as laminin, collagen, entactin, and perlecan. Through the interaction with the latter, α-dystroglycan associates with acetylcholinesterase, which is important in acetylcholine breakdown [157]. Moreover, the transmembrane β subunit of dystroglycan stabilizes the structure of NMJs, thereby facilitating the formation of the cytoskeletal network needed for clustering and stabilization of AChRs at the sarcolemma [156].
α- and β-Dystroglycan are also necessary for the proper organization of other membrane protein assemblies, including the multiprotein complex associated with muscle-specific kinase (MuSK) (Table 1). MuSK is distributed in the postsynaptic sarcolemma, where it plays a pivotal role in AChR clustering and end-plate maintenance [158]. In DMD, loss of dystroglycans leads to reduced levels of MuSK, which presumably directly impacts NMJ morphology and function. Consistent with this finding, Musk−/− mice cannot move and breathe and die at birth due to neuromuscular transmission loss [158]. Additionally, conditional inactivation of MuSK in adult mice resulted in the absence of AChRs, disassembly of the postsynaptic organization, severe muscle weakness and premature death [159]. Other studies have shown that the MuSK complex drives the aggregation of AChRs in response to agrin binding, which initiates the autophosphorylation of MuSK and the formation of a stable and active MuSK complex. This process in turn initiates a downstream signaling cascade that results in recruitment of rapsyn to AChRs and stabilization of the AChR clusters through the DAPC/UAPC linkage [160, 161] (Fig. 6). Rapsyn is also involved in recycling and lifetime regulation of AChRs, a process that is under control of cAMP/PKA signaling, which is shown to be disturbed at dystrophic NMJs [162].
Loss of dystrophin and agrin signaling at the NMJ alters the organization of the microtubule network [66, 163]. Importantly, despite being upregulated at NMJs, utrophin cannot restore the microtubule lattice in dystrophic muscles, as it does not contain the microtubule-binding domain observed for dystrophin (Fig. 1) [66]. Microtubules maintain the right shape of the cell and allow for positioning of organelles and alterations in their network could affect mitochondrial density and function, thereby leading to excessive production of ROS [147, 164]. Another important aspect linked to DMD pathology is the presence of embryonic-type AChRs, as revealed in regenerating muscle fibers of mdx mice [165]. Specifically, Pijl et al. speculated that longer open embryonic-type AChRs in constantly regenerating dystrophic muscles may induce Ca2+ overload and contribute to myofiber focal necrosis by activating Ca2+-dependent proteases [151].
Disrupted signaling in satellite cells
SCs are muscle stem cells that are localized along the myofiber between the sarcolemma and the surrounding ECM (also termed basal lamina) and responsible for postnatal tissue growth and regeneration [166]. In the process of muscle repair, SCs are supported by other cell types that compose the regenerative milieu, including infiltrating immune cells (macrophages, eosinophils, regulatory T-cells, neutrophils), vascular endothelial cells, pericytes, fibroblasts, and FAPs, also called mesenchymal stromal cells [167]. In particular, the latter was recently reported to play crucial roles in muscle repair and maintenance. Experiments in mice revealed that paracrine signaling between SCs and FAPs supports myogenesis by influencing the proliferation and differentiation of SCs [167] while depletion of FAPs results in lessening of the SC pool, regenerative deficit, and muscle atrophy [73]. Some interstitial cells may also have myogenic potential, including myoendothelial [168], CD133 + [169, 170], PW1 interstitial [171], muscle-derived stem cells (MDSCs) [172, 173], Twist2 + progenitor [174] and muscle side population cells [167, 175]. Nonetheless, it is important to note that they are unable to regenerate muscle in the absence of SCs [176].
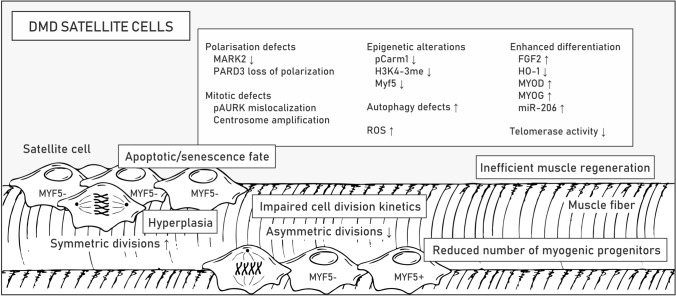
Quiescence of SCs in resting muscle is maintained by expression of the paired box 7 (Pax7) gene controlled by the Notch pathway, which is activated by binding of Notch ligands distributed at the sarcolemma to Notch receptors located on the SC membrane [177] as well as various epigenetic mechanisms [11]. Specifically, nonmethylated Pax7 protein limits the expression of myogenic factor 5 (Myf5) while its methylation enables recruitment of epigenetic machinery to the Myf5 promoter and in turn SC activation, division and differentiation into myotubes [178]. Importantly, SCs can divide in two ways: symmetrically and asymmetrically [179]. While symmetric divisions (occurring parallel to the contiguous myofiber in a planar orientation) preserve the stem cell pool by generating two identical daughter cells, asymmetric divisions (occurring in apicobasal orientation, perpendicular to myofiber) result in two different daughter cells, with one of them returning to the state of quiescence and the other entering the myogenic pathway. The expression of Myf5 is characteristic of myoblasts, while quiescent cells remain Myf5-negative and a direct contact with the basal lamina on one side and the myofiber sarcolemma on the other is a suspected factor determining the polarity of dividing SCs [180]. Notably, during muscle regeneration, SCs can also migrate to interstitial areas of the muscle, where they can proliferate and differentiate into new fibers [181, 182]. A detailed description of the intrinsic and extrinsic mechanisms regulating SC activity can be found elsewhere [178, 179, 183].
In healthy muscles, dystrophin distributes unevenly in activated SCs, thus providing the background for their asymmetric division and myogenic differentiation by anchoring to the plasmalemma microtubule affinity regulating kinase 2 (MARK2) and β1-syntrophin (Fig. 1), with its associated mitogen-activated protein kinase (MAPK) p38γ (also known as MAPK12) [184]. In dystrophic SCs, the greatly reduced content of MARK2 as well as affected polarization of partitioning defective protein 3 (PARD3) and localization of phosphorylated Aurora kinase A (Aurka), impair the apicobasal mitotic spindle orientation, centrosome amplification and division kinetics [46, 184] (Fig. 7; Table 1). Loss of β1-syntrophin and dysregulated p38γ signaling, on the other hand, disable transportation of Carm1 to the nucleus, without which Pax7 cannot be efficiently methylated to trigger Myf5 transcription [185]. Carm1 not only contributes to disturbances in epigenetic control of myogenic gene expression but also induces autophagy-related and lysosomal genes as a coactivator of transcription factor EB (TFEB) via increasing dimethylation of histone H3 Arg17 [186]. Dystrophin loss-mediated Carm1 absence in the nucleus could thus explain the autophagy defects in DMD muscles in addition to persistent activation of Akt and mTOR triggering autophagy-inhibiting pathways [187, 188] (Table 1). Importantly, the failed autophagy pathway may result in the inability to clear damaged mitochondria that could drive the accumulation of ROS and SC senescence [189].
Fig. 7.
Altered functioning of DMD satellite cells. Pathological alterations related to satellite cell function are listed in boxes. Examples of disturbed signaling pathways, altered gene expression and epigenetic changes are shown in the boxes. Molecular mechanisms underlying pathologies in DMD satellite cells are described in the text. Abbreviations: myogenic factor 5 (Myf5), microtubule affinity regulating kinase 2 (MARK2), partitioning defective protein 3 (PARD3), phosphorylated Aurora kinase (pAURK), reactive oxygen species (ROS), fibroblast growth factor 2 (FGF2), heme oxygenase 1 (HO-1), myoblast determination protein (MYOD), myogenin (MYOG)
Although asymmetric division is disrupted, stem cell hyperplasia is observed in muscles of DMD patients and mdx mice, as demonstrated by the elevated numbers of Pax7-positive cells, presumably as a consequence of preferable symmetric SC division [190–192] (Fig. 7). This surplus of SCs, however, does not seem to contribute to muscle repair, and over time, the Pax7 + /Myf5 + muscle progenitor pool declines [46]. Apart from the proliferation deficits, factors that contribute to this reduction include susceptibility to oxidative stress, progressive telomere shortening due to insufficient telomerase activity and high muscular fibroblast growth factor 2 (FGF2) content, which leads to loss of stem cell quiescence [193] (Table 1).
The simultaneous decrease in the level of HO-1 enzyme with the upregulation of myoblast determination protein (MyoD), myogenin (Myog) and miR-206 also implies enhanced differentiation of dystrophic cells that do enter the myogenic differentiation program [194]. Repression of the HO-1-encoding Hmox1 gene in DMD SCs is linked to downregulation of Atf3, MafK, Foxo1 and Klf2 transcription factors as well as attenuation of NO-mediated cGMP-dependent signaling [194]. More intensive differentiation of HO-1-deficient SCs and myoblasts was associated with changes in the activity of miRNA processing enzymes and, consequently, alterations in the miRNA transcriptome [194, 195]. Moreover, diminished HO-1 levels in mdx SCs were linked to increased content of Casp3 [194], which is known to target Pax7 and activate SC differentiation [196]. Interestingly, in myofibers and inflammatory leukocytes, the level of HO-1 is elevated in mdx mice compared to control mice, which presumably has an anti-inflammatory and alleviating effect. Pietraszek-Gremplewicz et al. showed that supplementation of differentiating dystrophic SCs with CO (one of the products of HO-1 enzyme) and NO (induces Hmox1 expression) normalizes the differentiation of mdx SCs [194]. Targeting HO-1 is thus a potentially attractive therapeutic target.
Notch signaling determines many stages of muscle regeneration, including the fate of SCs, proliferation of myoblasts, and the transient inhibition of terminal differentiation of myoblasts into mature myofibers [197]. Moreover, a wide variety of Notch receptors and ligands have to be precisely regulated in a time- and space-restricted manner [197]. Both the quiescent and active states of SCs depend on Notch signaling, and the oscillation of Hes (Notch effector) was found to determine the level of MyoD [198]. In DMD muscles, Notch signaling is dysregulated. Specifically, Mu et al. showed that inhibition of Notch signaling, which is overactivated in DMD muscles and SCs, delays the depletion and senescence of muscle progenitor cells and reduces inflammation and fibrosis in mdx/utrn−/− mice [199]. In contrast, the results from experiments on two mildly affected golden retriever muscular dystrophy (GRMD) dogs [200], which showed a higher content of Jagged 1 (a Notch ligand) due to increased expression of Jag1, indicated the beneficial effects of Notch activation [201]. Compared with the severely affected GRMD dogs, these “escapers” dogs could run, jump and more easily stand on their hindlegs and had a normal lifespan despite the absence of muscle dystrophin [200, 201]. A transcription factor binding site analysis revealed that mildly affected dogs contain a novel myogenin binding site in the Jag1 promoter, which resulted in a myogenin-associated increase during muscle regeneration [201]. Experiments based on the sapje zebrafish, another severe DMD animal model, and an in vitro analysis of GRMD muscle cells from biopsies confirmed the beneficial effects of Jag1 overexpression. Other studies indicate that in addition to the muscle growth and expansion of the satellite cell pool, Jagged-activated Notch signaling promotes angiogenesis [202] and stimulates bone marrow-derived stromal/stem cells (BMSCs) to promote skeletal regeneration [203]. Overall, these findings show the therapeutic potential of either restoring proper Notch signaling or stimulating JAG1 overexpression as a mediator of the regenerative process in DMD muscles independent of the presence of dystrophin.
Increasing the number of asymmetric divisions can be achieved independent from the polarized dystrophin distribution at the membrane in activated SCs, e.g., via activation of epidermal growth factor receptor (EGFR) and Aurka signaling pathways [204]. Promising results were also achieved following Wnt7a overexpression in EDL muscle and Wnt7a protein injection into TA muscles of mdx mice. In addition to an increase in muscle strength and a shift of induction toward slow-twitch fibers, which reduced the level of contractile damage, this treatment also regenerated the SC pool via an enhancement of the symmetric division rate [205]. Presumably, noncanonical Wnt7a signaling stimulates SC symmetric expansion through the planar cell polarity pathway (polarizing distribution of VANGL2, a planar cell polarity effector) and myofiber hypertrophy through the AKT/mTOR pathway [205, 206]. In turn, canonical Wnt signaling, which is elevated in mdx mouse muscles, was shown to activate the TGFβ pathway, resulting in a fibrogenic phenotype in SCs [207]. Importantly, suppression of factors upregulated in DMD that inhibit myogenesis, such as TGFβ, myostatin, and activin, is possible with follistatin treatment [112]. It is important to note, however, that in addition to its beneficial effects on SCs, follistatin can also have detrimental effects on tendons [111, 113].
RNA-seq profiling of SCs from mdx and control mice demonstrated altered expression of over 1000 genes in quiescent SCs and 3000 genes in activated SCs [194]. Interestingly, Boldrin et al. showed that mdx SCs retain their regenerative potential upon transfer to a healthy muscle environment [208], which shows that exploring SC signaling and SC interactions with surrounding tissues can reveal the potential for effective therapies.
Cell signaling in the heart
Cardiac failure is the leading cause of premature mortality in DMD patients [209, 210]. Pathological symptoms develop with age and manifest in progressive fibrosis, left ventricular dilation, and overall reduced systolic function, resulting in cardiomyopathy and atrial arrhythmias [211]. Heart regeneration is particularly challenging because unlike skeletal muscle tissue, this organ lacks resident stem cells/satellite cells that could regenerate the cardiomyocyte population [212]. In healthy tissue, cardiomyocytes, fibroblasts, endothelial and vascular smooth muscle cells are in homeostatic equilibrium [213], whereas in dystrophic muscle, high levels of inflammation and fibrogenic cells in coronary advenitia aggravate the condition of the patients [214–216].
Cardiomyocyte death in DMD patients is associated with the absence of dystrophin-related mechanical functions and signaling. As in skeletal muscle, cardiomyocytes struggle with an overload of Ca2+, the absence of nNOS/NO signaling [217–219] and the overproduction of ROS, which lead to the progression of pathological alterations. Law et al. [220] summarized known mechanisms responsible for mishandling Ca2+ in dilating cardiomyopathy in DMD hearts and reported experimental therapeutic targets that could address this issue. In particular, excess Ca2+ and oxidative stress result in degeneration of mitochondria [221], which cannot be removed effectively in the process of mitophagy [222]. These changes are probably due to dysfunction in the Pink1/Parkin1 pathway, which regulates the process of cleaning cells from nonfunctional mitochondria [222]. Affected cardiomyocytes also release exosomes containing miRNAs, which have been found to aggravate DMD pathogenesis [223]. Furthermore, enhanced Tgfβ1 signaling in coronary adventitial cells was identified as a factor that induces fibrotic changes. In a study by Ieronimakis et al. overexpression of Tgfβ1 ligand in endothelial cells stimulated coronary adventitial cells via the Tgfβ pathway to become fibrotic and produce type I collagen, resulting in perivascular fibrosis [215], which shows that targeting the TGFβ pathway in either skeletal or cardiac muscles could be a viable signaling-based therapeutic strategy in DMD. Therapeutic approaches to treat dystrophic cardiomyopathy that are currently in use or in development have been discussed in-depth in previous works [57, 224].
Conclusions
Although dystrophin and utrophin genes are ubiquitously expressed in various tissues, significant full-length dystrophin synthesis is limited to a few cell types, including myofibers, cardiomyocytes, smooth muscle cells, neurons, endothelial cells, and satellite cells [25, 184, 225]. DMD patients suffer particularly as a consequence of the absence of full-length dystrophin in the striated muscle tissue and succumb to the disease after years of progressive and debilitating symptoms [4]. Some of the dystrophin-related roles pertain to its mechanical functions and widespread interactions with transmembrane as well as cytoskeletal proteins, such as ankyrins, microtubules, plectin, γ-actin and cyotokeratins that enable efficient transmission of forces and structural plasticity [66, 226, 227]. The dystrophin-glycoprotein complex together with its associated proteins, however, is also responsible for the interaction with various signaling molecules [25, 184, 228] that regulate cell proliferation, migration or maintenance via, in some instances, epigenetic and transcriptional changes, such as in the process of myofiber repair [8, 9, 11]. Importantly, in addition to intracellular signaling, cell-to-cell communication is also affected in dystrophin deficiency, as shown, e.g., by vasoconstriction related to nNOS downregulation and displacement from the sarcolemma [121–124], abrogated satellite cell activation due to dysregulated Notch signaling [199], or changes in the production of myokines that affect the bone [71, 93, 98, 104]. Therapeutic approaches should thus focus not only on stopping myofiber degeneration and improving satellite cell division kinetics but also on normalizing abrogated signal transmission. For instance, although warranted by observations of relatively mildly affected Becker muscular dystrophy (BMD) patients, technological shortcomings and methodological feasibility, the rod domain of dystrophin has largely been ignored in either gene therapy- or antisense oligonucleotide-based therapeutic approaches [36]. However, the newly obtained data changed that paradigm and resulted in addition of the nNOS binding domain to rAAV vectors carrying laboratory designed dystrophin coding sequences [229]. The MARK2 binding site might also be included in constructs that could be used in satellite cell therapeutic strategies.
Sodium nitrate (NO donor) as well as inhibitors of myostatin, HDAC and connective tissue growth factor are currently in clinical trials [11, 230–235], with others, including a WNT7a-like compound, in preclinical development [236]. It is also important to note that corticosteroids significantly alleviate tissue damage by downregulating the activity of signaling molecules associated with chronic inflammation, and new treatments based on molecules with less severe side effects are being tested [237–239]. Therapeutic strategies modulating specific signaling pathways could be used either as adjuncts or stand-alone strategies. Importantly, approaches such as stimulation of EGFR-Aurka pathways or Jagged 1 upregulation have been successfully used in animal models and do not seem to require dystrophin to normalize the division kinetics of satellite cells or muscle function. Most of the treatments grounded on dystrophin/utrophin gene delivery or exon skipping aimed at restoring the reading frame do not provide the fully functional dystrophin protein, and signaling approaches might be required as complementary strategies. Disturbances in noncoding RNA levels in DMD also provide an alternative strategy to monitor the progression of the disease or treatment efficacy [67, 240], and noncoding RNAs are under research as potential therapeutic targets [241, 242]. Other approaches would also need to be used in severely affected patients who lack large amounts of functional tissue. Here, therapies based on the reversal of pathological symptoms, such as conversion of fibroblasts to myoblasts by the induction of MYOD, could be employed [243].
In summary, DMD should be understood as a disease of not only affected myofibers and satellite cells but also a disorder in which abrogated communication between different cell types occurs. Therefore, targeting affected signaling pathways in patients is a promising treatment strategy for DMD. We believe that by taking this systemic view, we can achieve safe and holistic treatment that can restore correct signal transmission and gene expression in diseased DMD tissues.
Acknowledgements
This work was supported by the Polish National Science Centre [Grant no. 2018/30/E/NZ5/00065 (to Patryk Konieczny)].
Author contributions
AS and PK wrote the manuscript. AS designed and prepared the figures. Figures were prepared based on freehand sketches, which were further supplemented in CorelDRAW 2019.
Funding
This work was supported by the Polish National Science Centre [Grant no. 2018/30/E/NZ5/00065 (to Patryk Konieczny)].
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Crisafulli S, Sultana J, Fontana A, et al. Global epidemiology of Duchenne muscular dystrophy: an updated systematic review and meta-analysis. Orphanet J Rare Dis. 2020;15:141. doi: 10.1186/s13023-020-01430-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nozoe KT, Akamine RT, Mazzotti DR, et al. Phenotypic contrasts of Duchenne muscular dystrophy in women: two case reports. Sleep Sci. 2016;9:129–133. doi: 10.1016/j.slsci.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishizaki M, Kobayashi M, Adachi K, et al. Female dystrophinopathy: review of current literature. Neuromuscul Disord. 2018;28:572–581. doi: 10.1016/j.nmd.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Birnkrant DJ, Bushby K, Bann CM, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and neuromuscular, rehabilitation, endocrine, and gastrointestinal and nutritional management. Lancet Neurol. 2018;17(3):251–267. doi: 10.1016/S1474-4422(18)30024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naidoo M, Anthony K. Dystrophin Dp71 and the Neuropathophysiology of Duchenne muscular dystrophy. Mol Neurobiol. 2020;57:1748–1767. doi: 10.1007/s12035-019-01845-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rae MG, O’Malley D. Cognitive dysfunction in Duchenne muscular dystrophy: a possible role for neuromodulatory immune molecules. J Neurophysiol. 2016;116:1304–1315. doi: 10.1152/jn.00248.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushby K, Finkel R, Birnkrant DJ, et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 8.Marrone AK, Shcherbata HR. Dystrophin orchestrates the epigenetic profile of muscle cells via miRNAs. Front Genet. 2011;2:64. doi: 10.3389/fgene.2011.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang NC, Sincennes MC, Chevalier FP, Brun CE, Lacaria M, Segalés J, Muñoz-Cánoves P, Ming H, Rudnicki MA. The Dystrophin glycoprotein complex regulates the epigenetic activation of muscle stem cell commitment. Cell Stem Cell. 2018;22(5):755–768.e6. doi: 10.1016/j.stem.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cacchiarelli D, Martone J, Girardi E, et al. MicroRNAs involved in molecular circuitries relevant for the duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Rugowska A, Starosta A, Konieczny P. Epigenetic modifications in muscle regeneration and progression of Duchenne muscular dystrophy. Clin Epigenetics. 2021;13(1):13. doi: 10.1186/s13148-021-01001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consalvi S, Saccone V, Mozzetta C. Histone deacetylase inhibitors : a potential epigenetic treatment for Duchenne muscular dystrophy. Epigenomics. 2014;6:547–560. doi: 10.2217/epi.14.36. [DOI] [PubMed] [Google Scholar]
- 13.Bettica P, Petrini S, D’Oria V, et al. Histological effects of givinostat in boys with Duchenne muscular dystrophy. Neuromuscul Disord. 2016;26:643–649. doi: 10.1016/j.nmd.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 15.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 16.D’souza VN, Man NT, Morris GE, et al. A novel dystrophin isoform is required for normal retinal electrophysiology. Hum Mol Genet. 1995;4:837–842. doi: 10.1093/hmg/4.5.837. [DOI] [PubMed] [Google Scholar]
- 17.Matsuo M, Awano H, Matsumoto M, et al. Dystrophin dp116: a yet to be investigated product of the duchenne muscular dystrophy gene. Genes (Basel) 2017;8:251. doi: 10.3390/genes8100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felisari G, Boneschi FM, Bardoni A, et al. Loss of Dp140 dystrophin isoform and intellectual impairment in Duchenne dystrophy. Neurology. 2000;55:559–564. doi: 10.1212/WNL.55.4.559. [DOI] [PubMed] [Google Scholar]
- 19.Tozawa T, Itoh K, Yaoi T, et al. The shortest isoform of dystrophin (Dp40) interacts with a group of presynaptic proteins to form a presumptive novel complex in the mouse brain. Mol Neurobiol. 2012;45:287–297. doi: 10.1007/s12035-012-8233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doorenweerd N, Mahfouz A, Van Putten M, et al. Timing and localization of human dystrophin isoform expression provide insights into the cognitive phenotype of Duchenne muscular dystrophy. Sci Rep. 2017;7:1–12. doi: 10.1038/s41598-017-12981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi T, Niba ETE, Rani AQM, et al. Detection of dystrophin Dp71 in human skeletal muscle using an automated capillary western assay system. Int J Mol Sci. 2018;19:1546. doi: 10.3390/ijms19061546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapaport D, Lederfein D, den Dunnen JT, et al. Characterization and cell type distribution of a novel, major transcript of the Duchenne Muscular Dystrophy gene. Symp Soc Exp Biol. 1992;49:187–193. doi: 10.1111/j.1432-0436.1992.tb00666.x. [DOI] [PubMed] [Google Scholar]
- 23.Juan-Mateu J, Gonzalez-Quereda L, Rodriguez MJ, et al. DMD mutations in 576 dystrophinopathy families: a step forward in genotype-phenotype correlations. PLoS ONE. 2015;10:e0135189. doi: 10.1371/journal.pone.0135189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deconinck N, Dan B. Pathophysiology of Duchenne muscular dystrophy: current hypotheses. Pediatr Neurol. 2007;36:1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Bhat HF, Mir SS, Dar KB, et al. ABC of multifaceted dystrophin glycoprotein complex (DGC) J Cell Physiol. 2018;233:5142–5159. doi: 10.1002/jcp.25982. [DOI] [PubMed] [Google Scholar]
- 26.Gawor M, Prószyński TJ. The molecular cross talk of the dystrophin–glycoprotein complex. Ann NY Acad Sci. 2018;1412:62–72. doi: 10.1111/nyas.13500. [DOI] [PubMed] [Google Scholar]
- 27.Nichols B, Takeda S, Yokota T. Nonmechanical roles of dystrophin and associated proteins in exercise, neuromuscular junctions, and brains. Brain Sci. 2015;5:275–298. doi: 10.3390/brainsci5030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilgram GSK, Potikanond S, Baines RA, et al. The roles of the dystrophin-associated glycoprotein complex at the synapse. Mol Neurobiol. 2010;41:1–21. doi: 10.1007/s12035-009-8089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, Kodippili K, Yue Y, et al. Dystrophin contains multiple independent membrane-binding domains. Hum Mol Genet. 2016;25(17):3647–3653. doi: 10.1093/hmg/ddw210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feissner RF, Skalska J, Gaum WE, Sheu SS. Crosstalk signaling between mitochondrial Ca2+ and ROS. Front Biosci (Landmark Ed) 2009;14:1197–1218. doi: 10.2741/3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houang EM, Sham YY, Bates FS, Metzger JM. Muscle membrane integrity in Duchenne muscular dystrophy: recent advances in copolymer-based muscle membrane stabilizers. Skelet Muscle. 2018;8(1):31. doi: 10.1186/s13395-018-0177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehead NP, Yeung EW, Allen DG. Muscle damage in mdx (dystrophic) mice: role of calcium and reactive oxygen species. Clin Exp Pharmacol Physiol. 2006;33(7):657–662. doi: 10.1111/j.1440-1681.2006.04394.x. [DOI] [PubMed] [Google Scholar]
- 33.Hughes MC, Ramos SV, Turnbull PC, et al. Early myopathy in Duchenne muscular dystrophy is associated with elevated mitochondrial H2 O2 emission during impaired oxidative phosphorylation. J Cachexia Sarcopenia Muscle. 2019;10(3):643–661. doi: 10.1002/jcsm.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hopf FW, Turner PR, Steinhardt RA. Calcium misregulation and the pathogenesis of muscular dystrophy. Subcell Biochem. 2007;45:429–464. doi: 10.1007/978-1-4020-6191-2_16. [DOI] [PubMed] [Google Scholar]
- 35.Kozakowska M, Pietraszek-Gremplewicz K, Jozkowicz A, Dulak J. The role of oxidative stress in skeletal muscle injury and regeneration: focus on antioxidant enzymes. J Muscle Res Cell Motil. 2015;36(6):377–393. doi: 10.1007/s10974-015-9438-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Judge LM, Haraguchi M, Chamberlain JS. Dissecting the signalling and mechanical functions of the dystrophin-glycoprotein complex. J Cell Sci. 2006;119:1537–1546. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
- 37.Sun C, Serra C, Lee G, Wagner KR. Stem cell-based therapies for Duchenne muscular dystrophy. Exp Neurol. 2020;323:113086. doi: 10.1016/j.expneurol.2019.113086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gramolini AO, Karpati G, Jasmin BJ. Discordant expression of utrophin and its transcript in human and mouse skeletal muscles. J Neuropathol Exp Neurol. 1999;58:235–244. doi: 10.1097/00005072-199903000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Blake DJ, Tinsley JM, Davies KE. Utrophin: a structural and functional comparison to dystrophin. Brain Pathol. 1996;6(1):37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 40.Guiraud S, Roblin D, Kay DE. The potential of utrophin modulators for the treatment of Duchenne muscular dystrophy. Expert Opin Orphan Drugs. 2018;6:179–192. doi: 10.1080/21678707.2018.1438261. [DOI] [Google Scholar]
- 41.Manning J, O'Malley D. What has the mdx mouse model of Duchenne muscular dystrophy contributed to our understanding of this disease? J Muscle Res Cell Motil. 2015;36(2):155–167. doi: 10.1007/s10974-015-9406-4. [DOI] [PubMed] [Google Scholar]
- 42.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21(9):2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 43.Deconinck AE, Rafael JA, Skinner JA, et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90(4):717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 44.Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90(4):729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 45.McGreevy JW, Hakim CH, McIntosh MA, Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech. 2015;8(3):195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang NC, Chevalier FP, Rudnicki MA. Satellite cells in muscular dystrophy - lost in polarity. Trends Mol Med. 2016;22:479–496. doi: 10.1016/j.molmed.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Podkalicka P, Mucha O, Dulak J, Loboda A. Targeting angiogenesis in Duchenne muscular dystrophy. Cell Mol Life Sci. 2019;76:1507–1528. doi: 10.1007/s00018-019-03006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Echevarría L, Aupy P, Goyenvalle A. Exon-skipping advances for Duchenne muscular dystrophy. Hum Mol Genet. 2018;27:R163–R172. doi: 10.1093/hmg/ddy171. [DOI] [PubMed] [Google Scholar]
- 49.Min YL, Bassel-Duby R, Olson EN. CRISPR correction of duchenne muscular dystrophy. Annu Rev Med. 2019;70:239–255. doi: 10.1146/annurev-med-081117-010451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanson B, Wood MJA, Roberts TC. Molecular correction of Duchenne muscular dystrophy by splice modulation and gene editing. RNA Biol. 2021;20:1–15. doi: 10.1080/15476286.2021.1874161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheikh O, Yokota T. Developing DMD therapeutics: a review of the effectiveness of small molecules, stop-codon readthrough, dystrophin gene replacement, and exon-skipping therapies. Expert Opin Investig Drugs. 2021;30(2):167–176. doi: 10.1080/13543784.2021.1868434. [DOI] [PubMed] [Google Scholar]
- 52.Rodrigues M, Yokota T (2018) An overview of recent advances and clinical applications of exon skipping and splice modulation for muscular dystrophy and various genetic diseases. In: Yokota T, Maruyama R (ed) Exon Skipping and Inclusion Therapies. Methods in Molecular Biology, vol 1828. Humana Press, New York [DOI] [PubMed]
- 53.Chamberlain JR, Chamberlain JS. Progress toward gene therapy for Duchenne muscular dystrophy. Mol Ther. 2017;25:1125–1131. doi: 10.1016/j.ymthe.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendell JR, Sahenk Z, Lehman K, et al. Assessment of systemic delivery of rAAVrh74.MHCK7.micro-dystrophin in children with duchenne muscular dystrophy: a nonrandomized controlled trial. JAMA Neurol. 2020;77(9):1122–1131. doi: 10.1001/jamaneurol.2020.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Duan D. Novel mini-dystrophin gene dual adeno-associated virus vectors restore neuronal nitric oxide synthase expression at the sarcolemma. Hum Gene Ther. 2012;23(1):98–103. doi: 10.1089/hum.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kodippili K, Hakim CH, Pan X, et al. AAV gene therapy for duchenne muscular dystrophy with a 7-kb mini-dystrophin gene in the canine model. Hum Gene Ther. 2018;29(3):299–311. doi: 10.1089/hum.2017.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Łoboda A, Dulak J. Muscle and cardiac therapeutic strategies for Duchenne muscular dystrophy: past, present, and future. Pharmacol Rep. 2020;72:1227–1263. doi: 10.1007/s43440-020-00134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Danisovic L, Culenova M, Csobonyeiova M. Induced pluripotent stem cells for duchenne muscular dystrophy modeling and therapy. Cells. 2018;7(12):253. doi: 10.3390/cells7120253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin Y, Shen Y, Su X, Weintraub NL, Tang Y. Effective restoration of dystrophin expression in iPSC Mdx-derived muscle progenitor cells using the CRISPR/Cas9 system and homology-directed repair technology. Comput Struct Biotechnol J. 2020;18:765–773. doi: 10.1016/j.csbj.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy TL, Guiraud S, Edwards B, et al. Micro-utrophin improves cardiac and skeletal muscle function of severely affected D2/mdx mice. Mol Ther Methods Clin Dev. 2018;11:92–105. doi: 10.1016/j.omtm.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4(12):1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 62.Odom GL, Gregorevic P, Allen JM, et al. Microutrophin delivery through rAAV6 increases lifespan and improves muscle function in dystrophic dystrophin/Utrophin-deficient mice. Mol Ther. 2008;16:1539–1545. doi: 10.1038/mt.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19(1):79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- 64.Guiraud S, Edwards B, Babbs A, et al. The potential of utrophin and dystrophin combination therapies for duchenne muscular dystrophy. Hum Mol Genet. 2019;28:2189–2200. doi: 10.1093/hmg/ddz049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li D, Bareja A, Judge L, et al. Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin. J Cell Sci. 2010;123:2008–2013. doi: 10.1242/jcs.064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Belanto JJ, Mader TL, Eckhoff MD, et al. Microtubule binding distinguishes dystrophin from utrophin. Proc Natl Acad Sci USA. 2014;111:5723–5728. doi: 10.1073/pnas.1323842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brusa R, Magri F, Bresolin N, Comi GP, Corti S. Noncoding RNAs in Duchenne and Becker muscular dystrophies: role in pathogenesis and future prognostic and therapeutic perspectives. Cell Mol Life Sci. 2020;77(21):4299–4313. doi: 10.1007/s00018-020-03537-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zanotti S, Gibertini S, Blasevich F, et al. Exosomes and exosomal miRNAs from muscle-derived fibroblasts promote skeletal muscle fibrosis. Matrix Biol. 2018;74:77–100. doi: 10.1016/j.matbio.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Gartz M, Lin CW, Sussman MA, Lawlor MW, Strande JL. Duchenne muscular dystrophy (DMD) cardiomyocyte-secreted exosomes promote the pathogenesis of DMD-associated cardiomyopathy. Dis Model Mech. 2020;13(11):dmm045559. doi: 10.1242/dmm.045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tidball JG. Mechanisms of muscle injury, repair, and regeneration. Compr Physiol. 2011;1:2029–2062. doi: 10.1002/cphy.c100092. [DOI] [PubMed] [Google Scholar]
- 71.Boulanger Piette A, Hamoudi D, Marcadet L, et al. Targeting the muscle-bone unit: filling two needs with one deed in the treatment of Duchenne muscular dystrophy. Curr Osteoporos Rep. 2018;16:541–553. doi: 10.1007/s11914-018-0468-2. [DOI] [PubMed] [Google Scholar]
- 72.Thomas GD. Functional muscle ischemia in Duchenne and Becker muscular dystrophy. Front Physiol. 2013;4:381. doi: 10.3389/fphys.2013.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wosczyna MN, Konishi CT, Carbajal EEP, et al. Mesenchymal stromal cells are required for regeneration and homeostatic maintenance of skeletal muscle. Cell Rep. 2020;27:2029–2035.e5. doi: 10.1016/j.celrep.2019.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 75.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Henriques-Pons A, Yu Q, Rayavarapu S, et al. Role of Toll-like receptors in the pathogenesis of dystrophin-deficient skeletal and heart muscle. Hum Mol Genet. 2014;23(10):2604–2617. doi: 10.1093/hmg/ddt656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta Mol Cell Res. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 78.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8(9):1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pelosi L, Berardinelli MG, De Pasquale L, et al. Functional and morphological improvement of dystrophic muscle by interleukin 6 receptor blockade. EBioMedicine. 2015;2(4):285–293. doi: 10.1016/j.ebiom.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gallot YS, Straughn AR, Bohnert KR, et al. MyD88 is required for satellite cell-mediated myofiber regeneration in dystrophin-deficient mdx mice. Hum Mol Genet. 2018;27:3449–3463. doi: 10.1093/hmg/ddy258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tidball JG, Welc SS, Wehling-Henricks M. Immunobiology of inherited muscular dystrophies. Compr Physiol. 2018;8:1313–1356. doi: 10.1002/cphy.c170052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J-H, Kwak H-B, Thompson LV, Lawler JM. Contribution of oxidative stress to pathology in diaphragm and limb muscles with Duchenne muscular dystrophy. J Muscle Res Cell Motil. 2013;34:1–13. doi: 10.1007/s10974-012-9330-9. [DOI] [PubMed] [Google Scholar]
- 83.Rosenberg AS, Puig M, Nagaraju K, et al. Immune-mediated pathology in Duchenne muscular dystrophy. Sci Transl Med. 2015;7:299rv4. doi: 10.1126/scitranslmed.aaa7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hindi SM, Shin J, Ogura Y, et al. Matrix metalloproteinase-9 inhibition improves proliferation and engraftment of myogenic cells in dystrophic muscle of mdx mice. PLoS ONE. 2013;8:e72121. doi: 10.1371/journal.pone.0072121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nadarajah VD, van Putten M, Chaouch A, et al. Serum matrix metalloproteinase-9 (MMP-9) as a biomarker for monitoring disease progression in Duchenne muscular dystrophy (DMD) Neuromuscul Disord. 2011;21:569–578. doi: 10.1016/j.nmd.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 86.Morgan JE, Prola A, Mariot V, et al. Necroptosis mediates myofibre death in dystrophin-deficient mice. Nat Commun. 2018;9(1):3655. doi: 10.1038/s41467-018-06057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Acharyya S, Villalta SA, Bakkar N, et al. Interplay of IKK/NF- k B signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. J Clin Invest. 2007;117:889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Q, Tang Y, Imbrogno K, et al. AAV-based shRNA silencing of NF-κB ameliorates muscle pathologies in mdx mice. Gene Ther. 2012;19:1196–1204. doi: 10.1038/gt.2011.207. [DOI] [PubMed] [Google Scholar]
- 89.Yin X, Tang Y, Li J, et al. Genetic ablation of P65 subunit of NF-κB in mdx mice to improve muscle physiological function. Muscle Nerve. 2017;56:759–767. doi: 10.1002/mus.25517. [DOI] [PubMed] [Google Scholar]
- 90.Hamoudi D, Marcadet L, Piette AB, et al. An anti-RANKL treatment reduces muscle inflammation and dysfunction and strengthens bone in dystrophic mice. Hum Mol Genet. 2019;28:3101–3112. doi: 10.1093/hmg/ddz124. [DOI] [PubMed] [Google Scholar]
- 91.Wehling-Henricks M, Welc SS, Samengo G, et al. Macrophages escape Klotho gene silencing in the mdx mouse model of Duchenne muscular dystrophy and promote muscle growth and increase satellite cell numbers through a Klotho-mediated pathway. Hum Mol Genet. 2018;27:14–29. doi: 10.1093/hmg/ddx380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wehling-Henricks M, Li Z, Lindsey C, et al. Klotho gene silencing promotes pathology in the mdx mouse model of Duchenne muscular dystrophy. Hum Mol Genet. 2016;25:2465–2482. doi: 10.1093/hmg/ddw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rufo A, Del Fattore A, Capulli M, et al. Mechanisms inducing low bone density in duchenne muscular dystrophy in mice and humans. J Bone Miner Res. 2011;26:1891–1903. doi: 10.1002/jbmr.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morgenroth VH, Hache LP, Clemens PR. Insights into bone health in Duchenne muscular dystrophy. Bonekey Rep. 2012;1:9. doi: 10.1038/bonekey.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King WM, Ruttencutter R, Nagaraja HN, Matkovic V, Landoll J, Hoyle C, Mendell JR, Kissel JT. Orthopedic outcomes of long-term daily corticosteroid treatment in Duchenne muscular dystrophy. Neurology. 2007;68(19):1607–1613. doi: 10.1212/01.wnl.0000260974.41514.83. [DOI] [PubMed] [Google Scholar]
- 96.Isaac C, Wright A, Usas A, et al. Dystrophin and utrophin “double knockout” dystrophic mice exhibit a spectrum of degenerative musculoskeletal abnormalities. J Orthop Res. 2013;31:343–349. doi: 10.1002/jor.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Novotny SA, Warren GL, Lin AS, et al. Bone is functionally impaired in dystrophic mice but less so than skeletal muscle. Neuromuscul Disord. 2011;21:183–193. doi: 10.1016/j.nmd.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mu X, Usas A, Tang Y, et al. RhoA mediates defective stem cell function and heterotopic ossification in dystrophic muscle of mice. FASEB J. 2013;27:3619–3631. doi: 10.1096/fj.13-233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Serrano AL, Baeza-Raja B, Perdiguero E, et al. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7:33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 100.Pelosi L, Berardinelli MG, Forcina L, et al. Increased levels of interleukin-6 exacerbate the dystrophic phenotype in mdx mice. Hum Mol Genet. 2015;24(21):6041–6053. doi: 10.1093/hmg/ddv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matsushita K, Itoh S, Ikeda S, et al. LIF/STAT3/SOCS3 signaling pathway in murine bone marrow stromal cells suppresses osteoblast differentiation. J Cell Biochem. 2014;115:1262–1268. doi: 10.1002/jcb.24777. [DOI] [PubMed] [Google Scholar]
- 102.Vetrone SA, Montecino-Rodriguez E, Kudryashova E, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-β. J Clin Invest. 2009;119:1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.De Pasquale L, D’Amico A, Verardo M, et al. Increased muscle expression of interleukin-17 in Duchenne muscular dystrophy. Neurology. 2012;78:1309–1314. doi: 10.1212/WNL.0b013e3182518302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou S, Qian B, Wang L, et al. Altered bone-regulating myokine expression in skeletal muscle of Duchenne muscular dystrophy mouse models. Muscle Nerve. 2018;58:573–582. doi: 10.1002/mus.26195. [DOI] [PubMed] [Google Scholar]
- 105.Harcourt LJ, Holmes AG, Gregorevic P, et al. Interleukin-15 administration improves diaphragm muscle pathology and function in dystrophic mdx mice. Am J Pathol. 2005;166:1131–1141. doi: 10.1016/S0002-9440(10)62333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Glass DA, II, Bialek P, Ahn JD, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 107.Wei W, Dutchak PA, Wang X, et al. Fibroblast growth factor 21 promotes bone loss by potentiating the effects of peroxisome proliferator-activated receptor γ. Proc Natl Acad Sci USA. 2012;109(8):3143–3148. doi: 10.1073/pnas.1200797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dankbar B, Fennen M, Brunert D, et al. Myostatin is a direct regulator of osteoclast differentiation and its inhibition reduces inflammatory joint destruction in mice. Nat Med. 2015;21:1085–1090. doi: 10.1038/nm.3917. [DOI] [PubMed] [Google Scholar]
- 109.Goh BC, Singhal V, Herrera AJ, et al. Activin receptor type 2A (ACVR2A) functions directly in osteoblasts as a negative regulator of bone mass. J Biol Chem. 2017;292:13809–13822. doi: 10.1074/jbc.M117.782128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haidet AM, Rizo L, Handy C, et al. Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc Natl Acad Sci USA. 2008;105:4318–4322. doi: 10.1073/pnas.0709144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Iskenderian A, Liu N, Deng Q, et al. Myostatin and activin blockade by engineered follistatin results in hypertrophy and improves dystrophic pathology in mdx mouse more than myostatin blockade alone. Skelet Muscle. 2018;8:34. doi: 10.1186/s13395-018-0180-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu J, Li Y, Lu A, et al. Follistatin improves skeletal muscle healing after injury and disease through an interaction with muscle regeneration, angiogenesis, and fibrosis. Am J Pathol. 2011;179:915–930. doi: 10.1016/j.ajpath.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mendias CL, Bakhurin KI, Faulkner JA. Tendons of myostatin-deficient mice are small, brittle, and hypocellular. Proc Natl Acad Sci USA. 2008;105:388–393. doi: 10.1073/pnas.0707069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cros D, Harnden P, Pellissier JF, Serratrice G. Muscle hypertrophy in Duchenne muscular dystrophy. A pathological and morphometric study. J Neurol. 1989;236(1):43–47. doi: 10.1007/BF00314217. [DOI] [PubMed] [Google Scholar]
- 115.De Aro AA, Da Ré GF, Marretto Esquisatto MA, et al. Biochemical and morphological alterations in the Achilles tendon of mdx mice. Microsc Res Tech. 2015;78:85–93. doi: 10.1002/jemt.22448. [DOI] [PubMed] [Google Scholar]
- 116.Rizzuto E, Musarò A, Catizone A, Del Prete Z. Measuring tendon properties in mdx mice: cell viability and viscoelastic characteristics. J Biomech. 2009;42:2243–2248. doi: 10.1016/j.jbiomech.2009.06.041. [DOI] [PubMed] [Google Scholar]
- 117.Ridge JC, Tidball JG, Ahl K, et al. Modifications in myotendinous junction surface morphology in dystrophin-deficient mouse muscle. Exp Mol Pathol. 1994;61:58–68. doi: 10.1006/exmp.1994.1025. [DOI] [PubMed] [Google Scholar]
- 118.Law DJ, Allen DL, Tidball JG. Talin, vinculin and DRP (utrophin) concentrations are increased at mdx myotendinous junctions following onset of necrosis. J Cell Sci. 1994;107:1477–1483. doi: 10.1242/jcs.107.6.1477. [DOI] [PubMed] [Google Scholar]
- 119.Welser JV, Rooney JE, Cohen NC, et al. Myotendinous junction defects and reduced force transmission in mice that lack α7 integrin and utrophin. Am J Pathol. 2009;175:1545–1554. doi: 10.2353/ajpath.2009.090052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fenichel GM. On the pathogenesis of Duchenne muscular dystrophy. Dev Med Child Neurol. 1975;17:527–533. doi: 10.1111/j.1469-8749.1975.tb03511.x. [DOI] [PubMed] [Google Scholar]
- 121.Crosbie RH. NO vascular control in Duchenne muscular dystrophy. Nat Med. 2001;7(1):27–29. doi: 10.1038/83309. [DOI] [PubMed] [Google Scholar]
- 122.Adams ME, Odom GL, Kim MJ, et al. Syntrophin binds directly to multiple spectrin-like repeats in dystrophin and mediates binding of nNOS to repeats 16–17. Hum Mol Genet. 2018;27:2978–2985. doi: 10.1093/hmg/ddy197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kobayashi YM, Rader EP, Crawford RW, et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456:511–515. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ennen JP, Verma M, Asakura A. Vascular-targeted therapies for Duchenne muscular dystrophy. Skelet Muscle. 2013;3:9. doi: 10.1186/2044-5040-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13(1):9–22. doi: 10.1096/fasebj.13.1.9. [DOI] [PubMed] [Google Scholar]
- 126.Arsic N, Zacchigna S, Zentilin L, Ramirez-Correa G, Pattarini L, Salvi A, Sinagra G, Giacca M. Vascular endothelial growth factor stimulates skeletal muscle regeneration in vivo. Mol Ther. 2004;10(5):844–854. doi: 10.1016/j.ymthe.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 127.Liu N, Williams AH, Maxeiner JM, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122(6):2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dulak J, Józkowicz A, Foresti R, et al. Heme oxygenase activity modulates vascular endothelial growth factor synthesis in vascular smooth muscle cells. Antioxid Redox Signal. 2002;4(2):229–240. doi: 10.1089/152308602753666280. [DOI] [PubMed] [Google Scholar]
- 129.Deshane J, Chen S, Caballero S, et al. Stromal cell-derived factor 1 promotes angiogenesis via a heme oxygenase 1-dependent mechanism. J Exp Med. 2007;204(3):605–618. doi: 10.1084/jem.20061609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gutpell KM, Tasevski N, Wong B, et al. ANG1 treatment reduces muscle pathology and prevents a decline in perfusion in DMD mice. PLoS ONE. 2017;12(3):e0174315. doi: 10.1371/journal.pone.0174315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rhoads RP, Flann KL, Cardinal TR, et al. Satellite cells isolated from aged or dystrophic muscle exhibit a reduced capacity to promote angiogenesis in vitro. Biochem Biophys Res Commun. 2013;440:399–404. doi: 10.1016/j.bbrc.2013.09.085. [DOI] [PubMed] [Google Scholar]
- 132.Straino S, Germani A, Di Carlo A, et al. Enhanced arteriogenesis and wound repair in dystrophin-deficient mdx mice. Circulation. 2004;110(21):3341–3348. doi: 10.1161/01.CIR.0000147776.50787.74. [DOI] [PubMed] [Google Scholar]
- 133.Nguyen F, Guigand L, Goubault-Leroux I, Wyers M, Cherel Y. Microvessel density in muscles of dogs with golden retriever muscular dystrophy. Neuromuscul Disord. 2005;15(2):154–163. doi: 10.1016/j.nmd.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 134.Verma M, Asakura Y, Murakonda B, et al. Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and notch signaling. Cell Stem Cell. 2018;23:530–543.e9. doi: 10.1016/j.stem.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Desguerre I, Mayer M, Leturcq F, et al. Endomysial fibrosis in duchenne muscular dystrophy: a marker of poor outcome associated with macrophage alternative activation. J Neuropathol Exp Neurol. 2009;68:762–773. doi: 10.1097/NEN.0b013e3181aa31c2. [DOI] [PubMed] [Google Scholar]
- 136.Latroche C, Matot B, Martins-Bach A, et al. Structural and functional alterations of skeletal muscle microvasculature in Dystrophin-deficient mdx mice. Am J Pathol. 2015;185:2482–2494. doi: 10.1016/j.ajpath.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 137.Birbrair A, Zhang T, Wang ZM, et al. Pericytes: Multitasking cells in the regeneration of injured, diseased, and aged skeletal muscle. Front Aging Neurosci. 2014;6:245. doi: 10.3389/fnagi.2014.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dellavalle A, Sampaolesi M, Tonlorenzi R, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 139.Choi S, Ferrari G, Tedesco FS. Cellular dynamics of myogenic cell migration: molecular mechanisms and implications for skeletal muscle cell therapies. EMBO Mol Med. 2020;12(12):e12357. doi: 10.15252/emmm.202012357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Skuk D, Tremblay JP. First study of intra-arterial delivery of myogenic mononuclear cells to skeletal muscles in primates. Cell Transplant. 2014;23(Suppl 1):S141–S150. doi: 10.3727/096368914X685032. [DOI] [PubMed] [Google Scholar]
- 141.Arnett AL, Konieczny P, Ramos JN, et al. Adeno-associated viral (AAV) vectors do not efficiently target muscle satellite cells. Mol Ther Methods Clin Dev. 2014;1:14038. doi: 10.1038/mtm.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tabebordbar M, Zhu K, Cheng JKW, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deschenes MR. Adaptations of the neuromuscular junction to exercise training. Curr Opin Psychol. 2019;10:10–16. doi: 10.1016/j.cophys.2019.02.004. [DOI] [Google Scholar]
- 144.Nishimune H, Stanford JA, Mori Y. Role of exercise in maintaining the integrity of the neuromuscular junction. Muscle Nerve. 2014;49:315–324. doi: 10.1002/mus.24095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gonzalez-Freire M, de Cabo R, Studenski SA, Ferrucci L. The neuromuscular junction: aging at the crossroad between nerves and muscle. Front Aging Neurosci. 2014;6:208. doi: 10.3389/fnagi.2014.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pratt SJP, Shah SB, Ward CW, et al. Effects of in vivo injury on the neuromuscular junction in healthy and dystrophic muscles. J Physiol. 2013;591:559–570. doi: 10.1113/jphysiol.2012.241679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pratt SJP, Shah SB, Ward CW, et al. Recovery of altered neuromuscular junction morphology and muscle function in mdx mice after injury. Cell Mol Life Sci. 2015;72:153–164. doi: 10.1007/s00018-014-1663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ng SY, Ljubicic V. Recent insights into neuromuscular junction biology in Duchenne muscular dystrophy: impacts, challenges, and opportunities. EBioMedicine. 2020;61:103032. doi: 10.1016/j.ebiom.2020.103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mizuno Y, Nonaka I, Hirai S, Ozawa E. Reciprocal expression of dystrophin and utrophin in muscles of Duchenne muscular dystrophy patients, female DMD-carriers and control subjects. J Neurol Sci. 1993;119:43–52. doi: 10.1016/0022-510x(93)90190-a. [DOI] [PubMed] [Google Scholar]
- 150.Deconinck AE, Potter AC, Tinsley JM, et al. Postsynaptic abnormalities at the neuromuscular junctions of utrophin- deficient mice. J Cell Biol. 1997;136:883–894. doi: 10.1083/jcb.136.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van der Pijl EM, van Putten M, Niks EH, et al. Characterization of neuromuscular synapse function abnormalities in multiple Duchenne muscular dystrophy mouse models. Eur J Neurosci. 2016;43:1623–1635. doi: 10.1111/ejn.13249. [DOI] [PubMed] [Google Scholar]
- 152.Aartsma-Rus A, Morgan J, Lonkar P, et al. Report of a TREAT-NMD/World Duchenne Organisation meeting on Dystrophin quantification methodology. J Neuromuscul Dis. 2019;6(1):147–159. doi: 10.3233/JND-180357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wells DJ. What is the level of dystrophin expression required for effective therapy of Duchenne muscular dystrophy? J Muscle Res Cell Motil. 2019;40(2):141–150. doi: 10.1007/s10974-019-09535-9. [DOI] [PubMed] [Google Scholar]
- 154.van der Pijl EM, van Putten M, Niks EH, et al. Low dystrophin levels are insufficient to normalize the neuromuscular synaptic abnormalities of mdx mice. Neuromuscul Disord. 2018;28:427–442. doi: 10.1016/j.nmd.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 155.Pisani C, Strimpakos G, Gabanella F, et al. Utrophin up-regulation by artificial transcription factors induces muscle rescue and impacts the neuromuscular junction in mdx mice. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1172–1182. doi: 10.1016/j.bbadis.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jacobson C, Côté PD, Rossi SG, et al. The dystroglycan complex is necessary for stabilization of acetylcholine receptor clusters at neuromuscular junctions and formation of the synaptic basement membrane. J Cell Biol. 2001;152:435–450. doi: 10.1083/jcb.152.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Peng HB, Xie H, Rossi SG, Rotundo RL. Acetylcholinesterase clustering at the neuromuscular junction involves perlecan and dystroglycan. J Cell Biol. 1999;145(4):911–921. doi: 10.1083/jcb.145.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.DeChiara TM, Bowen DC, Valenzuela DM, et al. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- 159.Hesser BA, Henschel O, Witzemann V. Synapse disassembly and formation of new synapses in postnatal muscle upon conditional inactivation of MuSK. Mol Cell Neurosci. 2006;31:470–480. doi: 10.1016/j.mcn.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 160.Ghazanfari N, Fernandez KJ, Murata Y, et al. Muscle specific kinase: organiser of synaptic membrane domains. Int J Biochem Cell Biol. 2011;43:295–298. doi: 10.1016/j.biocel.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 161.Banks GB, Fuhrer C, Adams ME, Froehner SC. The postsynaptic submembrane machinery at the neuromuscular junction: requirement for rapsyn and the utrophin/dystrophin-associated complex. J Neurocytol. 2003;32:709–726. doi: 10.1023/B:NEUR.0000020619.24681.2b. [DOI] [PubMed] [Google Scholar]
- 162.Rudolf R, Khan MM, Lustrino D, Labeit S, Kettelhut IC, Navegantes LC. Alterations of cAMP-dependent signaling in dystrophic skeletal muscle. Front Physiol. 2013;4:290. doi: 10.3389/fphys.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schmidt N, Basu S, Sladecek S, et al. Agrin regulates CLASP2-mediated capture of microtubules at the neuromuscular junction synaptic membrane. J Cell Biol. 2012;198(3):421–437. doi: 10.1083/jcb.201111130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Khairallah RJ, Shi G, Sbrana F, et al. Microtubules underlie dysfunction in duchenne muscular dystrophy. Sci Signal. 2012;5:56. doi: 10.1126/scisignal.2002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Koltgen D, Franke C. The coexistence of embryonic and adult acetylcholine receptors in sarcolemma of mdx dystrophic mouse muscle: an effect of regeneration or muscular dystrophy? Neurosci Lett. 1994;173:79–82. doi: 10.1016/0304-3940(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 166.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93(1):23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wosczyna MN, Rando TA. A muscle stem cell support group: coordinated cellular responses in muscle regeneration. Dev Cell. 2018;46:135–143. doi: 10.1016/j.devcel.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zheng B, Cao B, Crisan M, et al. Prospective identification of myogenic endothelial cells in human skeletal muscle. Nat Biotechnol. 2007;25:1025–1034. doi: 10.1038/nbt1334. [DOI] [PubMed] [Google Scholar]
- 169.Torrente Y, Belicchi M, Sampaolesi M, et al. Human circulating AC133+ stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J Clin Invest. 2004;114:182–195. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Negroni E, Riederer I, Chaouch S, et al. In vivo myogenic potential of human CD133+ muscle-derived stem cells: a quantitative study. Mol Ther. 2009;17:1771–1778. doi: 10.1038/mt.2009.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pannérec A, Formicola L, Besson V, et al. Defining skeletal muscle resident progenitors and their cell fate potentials. Dev. 2013;140:2879–2891. doi: 10.1242/dev.089326. [DOI] [PubMed] [Google Scholar]
- 172.Qu-Petersen Z, Deasy B, Jankowski R, et al. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. J Cell Biol. 2002;157:851–864. doi: 10.1083/jcb.200108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huard J. Stem cells, blood vessels, and angiogenesis as major determinants for musculoskeletal tissue repair. J Orthop Res. 2019;37:1212–1220. doi: 10.1002/jor.24058. [DOI] [PubMed] [Google Scholar]
- 174.Liu N, Garry GA, Li S, et al. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol. 2017;19:202–213. doi: 10.1038/ncb3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Majka SM, Jackson KA, Kienstra KA, et al. Distinct progenitor populations in skeletal muscle are bone marrow derived and exhibit different cell fates during vascular regeneration. J Clin Invest. 2003;111:71–79. doi: 10.1172/JCI16157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139(16):2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- 177.Mourikis P, Gopalakrishnan S, Sambasivan R, Tajbakhsh S. Cell-autonomous notch activity maintains the temporal specification potential of skeletal muscle stem cells. Dev. 2012;139:4536–4548. doi: 10.1242/dev.084756. [DOI] [PubMed] [Google Scholar]
- 178.Feige P, Brun CE, Ritso M, Rudnicki MA. Orienting muscle stem cells for regeneration in homeostasis, aging, and disease. Cell Stem Cell. 2018;23:653–664. doi: 10.1016/j.stem.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dumont NA, Wang YX, Rudnicki MA. Intrinsic and extrinsic mechanisms regulating satellite cell function. Dev. 2015;142:1572–1581. doi: 10.1242/dev.114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kuang S, Kuroda K, Le GF, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dumont NA, Bentzinger CF, Sincennes MC, Rudnicki MA. Satellite cells and skeletal muscle regeneration. Compr Physiol. 2015;5(3):1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 182.Ogawa R, Ma Y, Yamaguchi M, et al. Doublecortin marks a new population of transiently amplifying muscle progenitor cells and is required for myofiber maturation during skeletal muscle regeneration. Dev. 2015;142:810. doi: 10.1242/dev.122317. [DOI] [PubMed] [Google Scholar]
- 183.Forcina L, Miano C, Pelosi L, Musarò A. An overview about the muscle satellite cells biology of skeletal. Curr Genomics. 2019;20:24–37. doi: 10.2174/1389202920666190116094736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dumont NA, Wang YX, Von MJ, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kawabe Y, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11(3):333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shin HJR, Kim H, Oh S, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–557. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.De Palma C, Morisi F, Cheli S, et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2014;5:e1363. doi: 10.1038/cddis.2014.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Spitali P, Grumati P, Hiller M, Chrisam M, Aartsma-Rus A, Bonaldo P. Autophagy is impaired in the tibialis anterior of dystrophin null mice. PLoS Curr. 2013;5:ecurrents.md.e1226cefa851a2f079bbc406c0a21e80. doi: 10.1371/currents.md.e1226cefa851a2f079bbc406c0a21e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.García-Prat L, Martínez-Vicente M, Perdiguero E, et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 190.Kottlors M, Kirschner J. Elevated satellite cell number in Duchenne muscular dystrophy. Cell Tissue Res. 2010;340:541–548. doi: 10.1007/s00441-010-0976-6. [DOI] [PubMed] [Google Scholar]
- 191.Bankolé LC, Feasson L, Ponsot E, Kadi F. Fibre type-specific satellite cell content in two models of muscle disease. Histopathology. 2013;63:826–832. doi: 10.1111/his.12231. [DOI] [PubMed] [Google Scholar]
- 192.Keefe AC, Kardon G. A new role for dystrophin in muscle stem cells. Nat Med. 2015;21:1391–1393. doi: 10.1038/nm.4006. [DOI] [PubMed] [Google Scholar]
- 193.Lu A, Poddar M, Tang Y, et al. Rapid depletion of muscle progenitor cells in dystrophic mdx/utrophin-/- mice. Hum Mol Genet. 2014;23:4786–4800. doi: 10.1093/hmg/ddu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pietraszek-Gremplewicz K, Kozakowska M, Bronisz-Budzynska I, et al. Heme oxygenase-1 influences satellite cells and progression of duchenne muscular dystrophy in mice. Antioxid Redox Signal. 2018;29:128–148. doi: 10.1089/ars.2017.7435. [DOI] [PubMed] [Google Scholar]
- 195.Kozakowska M, Ciesla M, Stefanska A, et al. Heme oxygenase-1 inhibits myoblast differentiation by targeting myomirs. Antioxid Redox Signal. 2012;16(2):113–127. doi: 10.1089/ars.2011.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dick SA, Chang NC, Dumont NA, et al. Caspase 3 cleavage of Pax7 inhibits self-renewal of satellite cells. Proc Natl Acad Sci USA. 2015;112(38):E5246–E5252. doi: 10.1073/pnas.1512869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Terragni J, Zhang G, Sun Z, Pradhan S, Song L, Crawford GE, Lacey M, Ehrlich M. Notch signaling genes: myogenic DNA hypomethylation and 5-hydroxymethylcytosine. Epigenetics. 2014;9(6):842–850. doi: 10.4161/epi.28597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sueda R, Kageyama R. Regulation of active and quiescent somatic stem cells by Notch signaling. Dev Growth Differ. 2020;62(1):59–66. doi: 10.1111/dgd.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mu X, Tang Y, Lu A, et al. The role of Notch signaling in muscle progenitor cell depletion and the rapid onset of histopathology in muscular dystrophy. Hum Mol Genet. 2015;24:2923–2937. doi: 10.1093/hmg/ddv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zucconi E, Valadares MC, Vieira NM, et al. Ringo: discordance between the molecular and clinical manifestation in a golden retriever muscular Dystrophy dog. Neuromuscul Disord. 2010;20:64–70. doi: 10.1016/j.nmd.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 201.Vieira NM, Elvers I, Alexander MS, et al. Jagged 1 rescues the Duchenne muscular dystrophy phenotype. Cell. 2015;163:1204–1213. doi: 10.1016/j.cell.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pedrosa AR, Trindade A, Fernandes AC, et al. Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler Thromb Vasc Biol. 2015;35(5):1134–1146. doi: 10.1161/ATVBAHA.114.304741. [DOI] [PubMed] [Google Scholar]
- 203.Dong Y, Long T, Wang C, Mirando AJ, Chen J, O'Keefe RJ, Hilton MJ. NOTCH-mediated maintenance and expansion of human bone marrow stromal/stem cells: a technology designed for orthopedic regenerative medicine. Stem Cells Transl Med. 2014;3(12):1456–1466. doi: 10.5966/sctm.2014-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang YX, Feige P, Brun CE, et al. EGFR-aurka signaling rescues polarity and regeneration defects in dystrophin-deficient muscle stem cells by increasing asymmetric divisions. Cell Stem Cell. 2019;24:419–432. doi: 10.1016/j.stem.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.von Maltzahn J, Renaud JM, Parise G, Rudnicki MA. Wnt7a treatment ameliorates muscular dystrophy. Proc Natl Acad Sci USA. 2012;109:20614–20619. doi: 10.1073/pnas.1215765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Le Grand F, Jones AE, Seale V, et al. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Biressi S, Miyabara EH, Gopinath SD, et al. A Wnt-TGFβ2 axis induces a fibrogenic program in muscle stem cells from dystrophic mice. Sci Transl Med. 2014;6:267. doi: 10.1126/scitranslmed.3008411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Boldrin L, Zammit PS, Morgan JE. Satellite cells from dystrophic muscle retain regenerative capacity. Stem Cell Res. 2015;14:20–29. doi: 10.1016/j.scr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cheeran D, Khan S, Khera R, et al. Predictors of death in adults with duchenne muscular dystrophy-associated cardiomyopathy. J Am Heart Assoc. 2017;6(10):e006340. doi: 10.1161/JAHA.117.006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Birnkrant DJ, Ararat E, Mhanna MJ. Cardiac phenotype determines survival in Duchenne muscular dystrophy. Pediatr Pulmonol. 2016;51(1):70–76. doi: 10.1002/ppul.23215. [DOI] [PubMed] [Google Scholar]
- 211.Kamdar F, Garry DJ. Dystrophin-deficient cardiomyopathy. J Am Coll Cardiol. 2016;67(21):2533–2546. doi: 10.1016/j.jacc.2016.02.081. [DOI] [PubMed] [Google Scholar]
- 212.Vujic A, Natarajan N, Lee RT. Molecular mechanisms of heart regeneration. Semin Cell Dev Biol. 2020;100:20–28. doi: 10.1016/j.semcdb.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ivey MJ, Tallquist MD. Defining the cardiac fibroblast. Circ J. 2016;80(11):2269–2276. doi: 10.1253/circj.CJ-16-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mavrogeni S, Papavasiliou A, Spargias K, et al. Myocardial inflammation in duchenne muscular dystrophy as a precipitating factor for heart failure: a prospective study. BMC Neurol. 2010;10:33. doi: 10.1186/1471-2377-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ieronimakis N, Hays AL, Janebodin K, et al. Coronary adventitial cells are linked to perivascular cardiac fibrosis via TGFβ1 signaling in the mdx mouse model of Duchenne muscular dystrophy. J Mol Cell Cardiol. 2013;63:122–134. doi: 10.1016/j.yjmcc.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Farini A, Gowran A, Bella P, et al. Fibrosis rescue improves cardiac function in dystrophin-deficient mice and duchenne patient-specific cardiomyocytes by immunoproteasome modulation. Am J Pathol. 2019;189(2):339–353. doi: 10.1016/j.ajpath.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 217.Ramachandran J, Schneider JS, Crassous PA, Zheng R, Gonzalez JP, Xie LH, Beuve A, Fraidenraich D, Peluffo RD. Nitric oxide signalling pathway in Duchenne muscular dystrophy mice: up-regulation of L-arginine transporters. Biochem J. 2013;449(1):133–142. doi: 10.1042/BJ20120787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Allen DG, Whitehead NP, Froehner SC. Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol Rev. 2016;96(1):253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Brescia M, Chao YC, Koschinski A, Tomek J, Zaccolo M. Multi-compartment, early disruption of cGMP and cAMP signalling in cardiac myocytes from the mdx model of duchenne muscular dystrophy. Int J Mol Sci. 2020;21(19):7056. doi: 10.3390/ijms21197056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Law ML, Cohen H, Martin AA, Angulski ABB, Metzger JM. Dysregulation of calcium handling in duchenne muscular dystrophy-associated dilated cardiomyopathy: mechanisms and experimental therapeutic strategies. J Clin Med. 2020;9(2):520. doi: 10.3390/jcm9020520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kyrychenko V, Poláková E, Janíček R, Shirokova N. Mitochondrial dysfunctions during progression of dystrophic cardiomyopathy. Cell Calcium. 2015;58(2):186–195. doi: 10.1016/j.ceca.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kang C, Badr MA, Kyrychenko V, Eskelinen EL, Shirokova N. Deficit in PINK1/PARKIN-mediated mitochondrial autophagy at late stages of dystrophic cardiomyopathy. Cardiovasc Res. 2018;114(1):90–102. doi: 10.1093/cvr/cvx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gartz M, Lin CW, Sussman MA, Lawlor MW, Strande JL. Duchenne muscular dystrophy (DMD) cardiomyocyte-secreted exosomes promote the pathogenesis of DMD-associated cardiomyopathy. Dis Model Mech. 2020;13(11):dmm45559. doi: 10.1242/dmm.045559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Meyers TA, Townsend D. Cardiac pathophysiology and the future of cardiac therapies in duchenne muscular dystrophy. Int J Mol Sci. 2019;20(17):4098. doi: 10.3390/ijms20174098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Loufrani L, Matrougui K, Gorny D, Duriez M, Blanc I, Lévy BI, Henrion D. Flow (shear stress)-induced endothelium-dependent dilation is altered in mice lacking the gene encoding for dystrophin. Circulation. 2001;103(6):864–870. doi: 10.1161/01.cir.103.6.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Raith M, Valencia RG, Fischer I, Orthofer M, Penninger JM, Spuler S, Rezniczek GA, Wiche G. Linking cytoarchitecture to metabolism: sarcolemma-associated plectin affects glucose uptake by destabilizing microtubule networks in mdx myofibers. Skelet Muscle. 2013;3(1):14. doi: 10.1186/2044-5040-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Dowling P, Gargan S, Murphy S, Zweyer M, Sabir H, Swandulla D, Ohlendieck K. The dystrophin node as integrator of cytoskeletal organization, lateral force transmission, fiber stability and cellular signaling in skeletal muscle. Proteomes. 2021;9(1):9. doi: 10.3390/proteomes9010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rando TA. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24(12):1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 229.Lai Y, Thomas GD, Yue Y, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009;119(3):624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Nelson MD, Rosenberry R, Barresi R, et al. Sodium nitrate alleviates functional muscle ischaemia in patients with Becker muscular dystrophy. J Physiol. 2015;593(23):5183–5200. doi: 10.1113/JP271252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Timpani CA, Hayes A, Rybalka E. Therapeutic strategies to address neuronal nitric oxide synthase deficiency and the loss of nitric oxide bioavailability in Duchenne Muscular Dystrophy. Orphanet J Rare Dis. 2017;12(1):100. doi: 10.1186/s13023-017-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Suh J, Lee YS. Myostatin Inhibitors: panacea or predicament for musculoskeletal disorders? J Bone Metab. 2020;27(3):151–165. doi: 10.11005/jbm.2020.27.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wagner KR. The elusive promise of myostatin inhibition for muscular dystrophy. Curr Opin Neurol. 2020;33(5):621–628. doi: 10.1097/WCO.0000000000000853. [DOI] [PubMed] [Google Scholar]
- 234.Song Y, Yao S, Liu Y, et al. Expression levels of TGF-β1 and CTGF are associated with the severity of Duchenne muscular dystrophy. Exp Ther Med. 2017;13(4):1209–1214. doi: 10.3892/etm.2017.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chen Z, Zhang N, Chu HY, Yu Y, Zhang ZK, Zhang G, Zhang BT. Connective tissue growth factor: from molecular understandings to drug discovery. Front Cell Dev Biol. 2020;8:593269. doi: 10.3389/fcell.2020.593269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dumont NA, Rudnicki MA. Targeting muscle stem cell intrinsic defects to treat Duchenne muscular dystrophy. NPJ Regen Med. 2016;1:16006. doi: 10.1038/npjregenmed.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Finanger E, Vandenborne K, Finkel RS, et al. Phase 1 study of edasalonexent (CAT-1004), an Oral NF-κB inhibitor, in pediatric patients with duchenne muscular dystrophy. J Neuromuscul Dis. 2019;6(1):43–54. doi: 10.3233/JND-180341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Conklin LS, Damsker JM, Hoffman EP, et al. Phase IIa trial in Duchenne muscular dystrophy shows vamorolone is a first-in-class dissociative steroidal anti-inflammatory drug. Pharmacol Res. 2018;136:140–150. doi: 10.1016/j.phrs.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Donovan JM, Zimmer M, Offman E, Grant T, Jirousek M. A novel NF-κB inhibitor, edasalonexent (CAT-1004), in development as a disease-modifying treatment for patients with duchenne muscular dystrophy: phase 1 safety, pharmacokinetics, and pharmacodynamics in adult subjects. J Clin Pharmacol. 2017;57(5):627–639. doi: 10.1002/jcph.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bronisz-Budzyńska I, Chwalenia K, Mucha O, Podkalicka P, Karolina-Bukowska-Strakova JA, Łoboda A, Kozakowska M, Dulak J. miR-146a deficiency does not aggravate muscular dystrophy in mdx mice. Skelet Muscle. 2019;9(1):22. doi: 10.1186/s13395-019-0207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hrach HC, Mangone M. miRNA profiling for early detection and treatment of duchenne muscular dystrophy. Int J Mol Sci. 2019;20(18):4638. doi: 10.3390/ijms20184638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Podkalicka P, Mucha O, Bronisz-Budzyńska I, Kozakowska M, et al. Lack of miR-378 attenuates muscular dystrophy in mdx mice. JCI Insight. 2020;5(11):e135576. doi: 10.1172/jci.insight.135576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kimura E, Han JJ, Li S, Fall B, Ra J, Haraguchi M, Tapscott SJ, Chamberlain JS. Cell-lineage regulated myogenesis for dystrophin replacement: a novel therapeutic approach for treatment of muscular dystrophy. Hum Mol Genet. 2008;17(16):2507–2517. doi: 10.1093/hmg/ddn151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 245.Deguine J, Barton GM. MyD88: a central player in innate immune signaling. F1000Prime Rep. 2014;7:1–7. doi: 10.12703/P6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 249.Cui N, Hu M, Khalil RA. Biochemical and biological attributes of matrix metalloproteinases. Prog Mol Biol Transl Sci. 2017;147:1–73. doi: 10.1016/bs.pmbts.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Holbrook J, Lara-Reyna S, Jarosz-Griffiths H, McDermott MF. Tumour necrosis factor signalling in health and disease. F1000Research. 2019;8:111. doi: 10.12688/f1000research.17023.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kuro-o M. The Klotho proteins in health and disease. Nat Rev Nephrol. 2019;15:27–44. doi: 10.1038/s41581-018-0078-3. [DOI] [PubMed] [Google Scholar]
- 252.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Brocker C, Thompson D, Matsumoto A, et al. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum Genomics. 2010;5:30–55. doi: 10.1186/1479-7364-5-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Icer MA, Gezmen-Karadag M. The multiple functions and mechanisms of osteopontin. Clin Biochem. 2018;59:17–24. doi: 10.1016/j.clinbiochem.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 255.Ono T, Hayashi M, Sasaki F, Nakashima T. RANKL biology: bone metabolism, the immune system, and beyond. Inflamm Regen. 2020;40:1–16. doi: 10.1186/s41232-019-0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 257.Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF) Cytokine Growth Factor Rev. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bonnet N, Garnero P, Ferrari S. Periostin action in bone. Mol Cell Endocrinol. 2016;432:75–82. doi: 10.1016/j.mce.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 259.Tezze C, Romanello V, Sandri M. FGF21 as modulator of metabolism in health and disease. Front Physiol. 2019;10:1–9. doi: 10.3389/fphys.2019.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Balke JE, Zhang L, Percival JM. Neuronal nitric oxide synthase (nNOS) splice variant function: Insights into nitric oxide signaling from skeletal muscle. Nitric Oxide. 2019;82:35–47. doi: 10.1016/j.niox.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 261.Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Biochem J. 2011;437(2):169–183. doi: 10.1042/BJ20110301. [DOI] [PubMed] [Google Scholar]
- 262.Ma G, Wang Y, Li Y, et al. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. 2015;11:345–352. doi: 10.7150/ijbs.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lee J-W, Bae S-H, Jeong J-W, et al. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 264.Witzemann V, Chevessier F, Pacifici PG, Yampolsky P. The neuromuscular junction: Selective remodeling of synaptic regulators at the nerve/muscle interface. Mech Dev. 2013;130:402–411. doi: 10.1016/j.mod.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 265.Miura P, Jasmin BJ. Utrophin upregulation for treating Duchenne or Becker muscular dystrophy: how close are we? Trends Mol Med. 2006;12:122–129. doi: 10.1016/j.molmed.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 266.Ebnet K. Cell polarity 1 biological role and basic mechanisms. Cham: Springer; 2015. [Google Scholar]
- 267.Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.D'Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Pawlikowski B, Vogler TO, Gadek K, Olwin BB. Regulation of skeletal muscle stem cells by fibroblast growth factors. Dev Dyn. 2017;246:359–367. doi: 10.1002/dvdy.24495. [DOI] [PubMed] [Google Scholar]
- 271.Zumerle S, Alimonti A. In and out from senescence. Nat Cell Biol. 2020;22:753–754. doi: 10.1038/s41556-020-0540-x. [DOI] [PubMed] [Google Scholar]
- 272.Nusse R, Clevers H. Wnt/b-catenin signaling, disease, and emerging therapeutic modalities roel. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]