Abstract
Observed SARS-CoV-2 infections and deaths are low in tropical Africa raising questions about the extent of transmission. We measured SARS-CoV-2 IgG by ELISA in 9,922 blood donors across Kenya and adjusted for sampling bias and test performance. By 1st September 2020, 577 COVID-19 deaths were observed nationwide and seroprevalence was 9.1% (95%CI 7.6-10.8%). Seroprevalence in Nairobi was 22.7% (18.0-27.7%). Although most people remained susceptible, SARS-CoV-2 had spread widely in Kenya with apparently low associated mortality.
Subject terms: Viral infection, SARS-CoV-2, Population screening, Epidemiology
The reported burden of SARS-CoV-2 has been relatively low in tropical Africa compared to Europe and the Americas, but estimating true infection rates is challenging. Here, the authors screen blood donors in Kenya for SARS-CoV-2 antibodies and describe spatiotemporal seroprevalence dynamics.
Introduction
Across tropical Africa, numbers of cases and deaths attributable to COVID-19 have been substantially lower than those in Europe and the Americas1. This could imply reduced transmission, reduced clinical severity or epidemiological under-ascertainment. The first COVID-19 case in Kenya was identified on 12th March 2020. Subsequently, there have been two discrete waves of PCR-detected cases in May–August and October–January separated by a brief nadir in September 2020. At the end of 2020, the government had recorded 96,595 cases and 1792 deaths attributable to SARS-CoV-22. By May 30 2020 when COVID-19 related deaths reached 71, the national anti-SARS-CoV-2 antibody prevalence, estimated in blood donors, was 4.3% (95% confidence interval (CI) 2.9–5.8%)3. Transmission was obviously more widespread than would have been anticipated by reported cases and deaths. In this further study, we examine the dynamics of SARS-CoV-2 seroprevalence among Kenyan blood donors throughout the course of the first epidemic wave.
From 30th April to 30th September 2020, 10,258 samples from blood donors aged 16–64 years were processed at six Kenya National Blood Transfusion Service (KNBTS) regional blood transfusion centres, which serve a countrywide network of satellites and hospitals. We excluded duplicate samples, those from age-ineligible donors and those with missing data, leaving 9922 samples (Supplementary Fig. 1).
The blood donor samples were broadly representative of the Kenyan adult population4 on region of residence and age, although adults aged 55–64 years were under-represented (2.0% vs 7.3%, Supplementary Table 1) and adults aged 25–34 years were over-represented (39.3% vs 27.3%). Males were also over-represented (80.8%).
We tested samples for anti-SARS-CoV-2 IgG antibodies using a previously described ELISA for whole length spike antigen5. Assay sensitivity, estimated in sera from 174 PCR positive Kenyan adults and a panel of sera from the UK National Institute of Biological Standards and Control (NIBSC) was 92.7% (95% CI 87.9–96.1%); specificity, estimated in 910 serum samples from Kilifi drawn in 2018 was 99.0% (95% CI 98.1–99.5%)3. Assays on a subset of test samples were repeated at least once on separate days and reproducibility confirmed. Positive and negative control samples are routinely included in all runs and the results from these were reproducible.
Results and discussion
Of the 9922 samples with complete data 3098 had been reported previously3. In total, 928 were positive for anti-SARS-CoV-2 IgG; crude seroprevalence was 9.4% (95% CI, 8.8–9.9%) with little variation by age or sex (Table 1). We used Bayesian Multi-level Regression with Post-stratification (MRP) to adjust for test sensitivity (93%) and specificity (99%)6, smooth trends over time, and account for the differences in age, sex and residence characteristics of the test sample and the Kenyan population7.
Table 1.
Crude, age/sex standardised and Bayesian-weighted test-adjusted SARS-CoV-2 anti-spike protein IgG seroprevalence across the whole study duration.
| Kenya population | All samples (%) | Sero-positive | Crude seroprevalence | Bayesian weighted, test-adjusted seroprevalencea | |||
|---|---|---|---|---|---|---|---|
| % | 95% CI | % | 95% CI | ||||
| Age | |||||||
| 15–24 years | 9,733,174 | 2763 (27.8) | 241 | 8.7 | 7.7–9.8 | 7.5 | 6.2–8.8 |
| 25–34 years | 7,424,967 | 3902 (39.3) | 379 | 9.7 | 8.8–10.7 | 8.5 | 7.2–9.8 |
| 35–44 years | 4,909,191 | 2261 (22.8) | 224 | 9.9 | 8.7–11.2 | 8.3 | 6.9–9.8 |
| 45–54 years | 3,094,771 | 794 (8.0) | 66 | 8.3 | 6.5–10.5 | 7.3 | 5.5–8.9 |
| 55–64 years | 1,988,062 | 202 (2.1) | 18 | 8.9 | 5.4–13.7 | 7.2 | 5.2–9.1 |
| Sex | |||||||
| Male | 13,388,243 | 8019 (80.8) | 762 | 9.5 | 8.9–10.2 | 8.4 | 7.2–9.5 |
| Female | 13,761,922 | 1903 (19.2) | 166 | 8.7 | 7.5–10.1 | 7.4 | 5.9–8.9 |
| Region | |||||||
| Central | 3,452,213 | 606 (6.1) | 38 | 6.3 | 4.5–8.5 | 5.8 | 3.7–8.0 |
| Coast | 1,671,097 | 1680 (16.9) | 137 | 8.2 | 6.9–9.6 | 7.2 | 5.6–8.9 |
| Eastern/N. Eastern | 5,176,080 | 1482 (14.9) | 108 | 7.3 | 6.0–8.7 | 6.5 | 4.9–8.2 |
| Mombasa | 792,072 | 1654 (16.7) | 239 | 14.4 | 12.8–16.2 | 13.8 | 11.7–16 |
| Nairobi | 3,002,314 | 607 (6.1) | 107 | 17.6 | 14.7–20.9 | 16.7 | 13.4–20.2 |
| Nyanza | 3,363,813 | 1433 (14.4) | 131 | 9.1 | 7.7–10.8 | 8.3 | 6.6–10.2 |
| Rift Valley | 7,035,581 | 2138 (21.6) | 145 | 6.8 | 5.8–7.9 | 5.9 | 4.5–7.4 |
| Western | 2,656,995 | 322 (3.3) | 23 | 7.1 | 4.6–10.5 | 6.6 | 3.9–9.7 |
| National | 27,150,165 | 9922 (100) | 928 | 9.4 | 8.8–9.9 | 7.9 | 6.7–9.0 |
aBayesian Multi-level Regression with Post-stratification (MRP) accounts for differences in the age and sex distribution of blood donors and regional differences in the numbers of samples collected over time. The model also adjusts for sensitivity (93%) and specificity (99%) of the ELISA.
There was marked variation in seroprevalence over time and place with a generally increasing trend over time. Figures 1 and 2 respectively illustrate the cumulative confirmed COVID-19 cases in Kenya during the study period and the crude prevalence and Bayesian model estimates in 10 consecutive periods of ~2 weeks each. In Nairobi, Mombasa and the Coastal Region outside Mombasa, there was a steep rise in seroprevalence across the study period. We divided the observations equally into three consecutive periods (Table 2). In period 1 (30 April–19 June) the adjusted seroprevalence of SARS-CoV-2 was 5.2% (95% CI 3.7–6.7%); in period 2 (20 June–19 August) it had risen to 9.1% (95% CI 7.2–11.3%); and in period 3 (20 August–30 September) it was maintained at 9.1% (95% CI 7.6–10.8%).
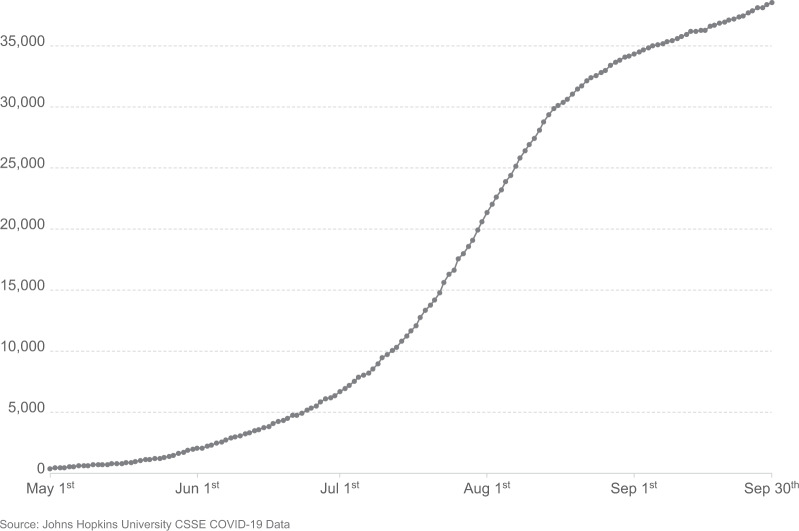
Fig. 1.
Cumulative confirmed COVID-19 cases in Kenya from 1st May - 20th September 2020.
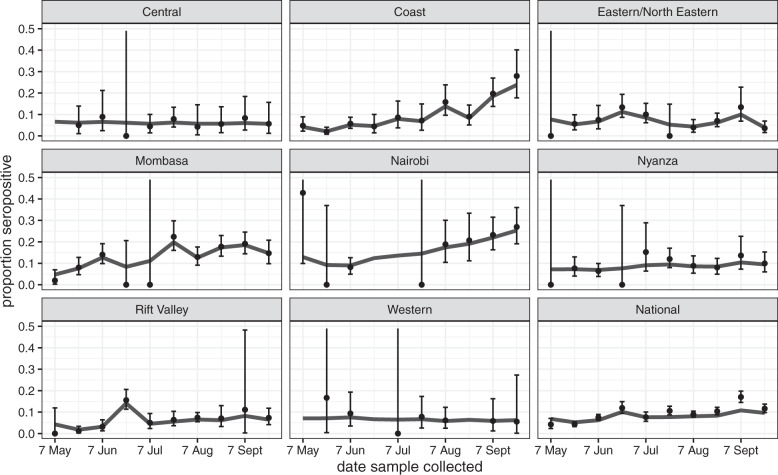
Fig. 2. Seroprevalence positivity across the study period by region.
The figure shows unadjusted estimates (black dots) and Bayesian model estimates (grey line) of seroprevalence in 8 regions of Kenya and overall, by date of sample collection in 10 periods of ~2 weeks each during 2020 (n = 9992). With the exception of the first period in Rift Valley (7 May) and 6th period in Eastern/North Eastern (21 July), all data estimates of zero prevalence are based on small sample sets (<20 samples). Error bars are 95% Confidence Intervals.
Table 2.
Bayesian-weighted test-adjusted SARS-CoV-2 anti-spike protein IgG seroprevalence across three study periods.
| Period 1 (30 Apr–19 Jun) | Period 2 (20 Jun–19 Aug) | Period 3 (20 Aug–30 Sep) | ||||
|---|---|---|---|---|---|---|
| Prevalence | 95% CIa | Prevalence | 95% CI | Prevalence | 95% CI | |
| Age | ||||||
| 15–24 years | 5.2 | 3.4–7.1 | 8.3 | 5.9–10.8 | 8.7 | 6.9–10.6 |
| 25–34 years | 5.2 | 3.6–7.0 | 9.3 | 7.1–12.0 | 10.2 | 8.5–12.4 |
| 35–44 years | 6.1 | 4.1–8.6 | 10.5 | 7.7–13.8 | 8.7 | 6.8–10.7 |
| 45–54 years | 4.1 | 1.6–6.5 | 9.2 | 6.3–12.8 | 8.6 | 6.2–11.0 |
| 55–64 years | 4.1 | 1.1–6.9 | 8.7 | 4.6–12.9 | 8.8 | 6.3–12.9 |
| Sex | ||||||
| Male | 6.1 | 4.6–7.6 | 8.4 | 6.7–10.2 | 10.1 | 8.6–11.7 |
| Female | 4.3 | 2.3–6.5 | 9.7 | 6.9–13.1 | 8.2 | 6.1–10.5 |
| Region | ||||||
| Central | 5.3 | 2.5–9.0 | 6.9 | 3.9–10.4 | 5.9 | 3.0–9.5 |
| Coast | 3.3 | 1.9–4.8 | 11.3 | 7.4–16.0 | 14.1 | 10.6–18.1 |
| Eastern/N. Eastern | 5.4 | 3.2–8.2 | 9.6 | 6.7–12.9 | 5.2 | 3.4–7.3 |
| Mombasa | 7.4 | 5.0–10.1 | 17.2 | 12.5–23.0 | 15.9 | 13.1–19.1 |
| Nairobi | 6.7 | 3.9–10.5 | 10.1 | 3.7–20.1 | 22.7 | 18–27.7 |
| Nyanza | 5.3 | 3.2–7.7 | 11.3 | 8.0–15.2 | 8.5 | 6.1–11.3 |
| Rift Valley | 3.9 | 2.3–5.8 | 7.3 | 5.3–9.6 | 7.3 | 4.9–10.0 |
| Western | 6.3 | 2.9–11.6 | 7.7 | 3.9–12.2 | 6.0 | 2.4–11.1 |
| National | 5.2 | 3.7–6.7 | 9.1 | 7.2–11.3 | 9.1 | 7.6–10.8 |
a95% credible interval.
The results illustrate a heterogeneous pattern of transmission across Kenya and suggest that the seroprevalence first began to rise in Mombasa in May and reached a maximum in July; in Nairobi it increased steadily from June onwards; in the Coastal area seroprevalence began to rise in July and turned up sharply in August and September. Unlike Nairobi and Mombasa this area is mostly rural. Other parts of the country showed less of a temporal trend. These field observations accord closely with epidemic modelling of SARS-CoV-2 across Kenya which integrated early PCR and serological data with mobility trends to describe the transmission pattern nationally8.
Although we used a highly specific and validated assay3,9, and adjusted for biases inherent in the ELISA test performance, we did not control for antibody waning. Given evidence at both individual10 and population11 level that anti-Spike antibodies may decline after an initial immune response, cross-sectional data are likely to underestimate cumulative incidence with increasing error as the epidemic wave declines. Some investigators have adjusted for this effect through modelling12 but as we do not have a clear description of the waning function for these antibodies in our setting, we have not made such an adjustment. We are exploring the application of mixture modelling to account for this challenge13. Therefore, the seroprevalence estimates reported here are likely to underestimate cumulative incidence in Kenya.
The study also relies on convenience sampling of asymptomatic blood transfusion donors which is not representative of the adult population at large and may underestimate seroprevalence because those with a recent history of illness are excluded. Although blood donors are predominantly male in Kenya, we had ~2000 female donor samples and stratified all analyses by sex to ensure that any potential confounding was appropriately adjusted for. We have adjusted for demographic and geographic disparities in our sample set, but we are unable to evaluate whether the behaviour of blood donors increases or reduces their risk of infection by SARS-CoV-2. The exclusion of donors with history of illness in the past 6 months may also contribute to selection bias. A random population sample would overcome these problems, but such studies were difficult to undertake during movement restrictions8. Recruiting household contacts of blood donors was considered but this was beyond the remit of the KNBTS and the movement and other restrictions also made this impractical. The selection bias in KNBTS samples is unlikely to change substantially over time and therefore this survey and the continued surveillance of blood donors will provide valid estimation of trends, which inform the public health management of the epidemic.
The results are also consistent with other surveys in Kenya which have illustrated both high seroprevalence in focal populations and marked geographic variation. For example, seroprevalence was 50% among women attending ante-natal care (ANC) in August 2020 in Nairobi but 1.3%, 1.5% and 11.0% among women attending ANC in Kilifi (Coast) in September, October and November, respectively14. Seroprevalence among truck drivers at two sites (in Coast and Western) was 42% in October 202015 and seroprevalence among health care workers in between August and November 2020 was 43%, 12% and 11% in Nairobi, Busia (Western) and Kilifi (Coast), respectively16.
By 1st September 2020, the first epidemic wave of SARS-CoV-2 in Kenya had declined with a cumulative mortality of 767 COVID-19 deaths and 34,471 cases.2 Our large national blood donor serosurvey illustrates that, at the same point, 1 in 10 donors had antibody evidence of infection with SARS-CoV-2; this rises to 1 in 5 in the two major cities in Kenya. The first epidemic wave rose and fell against a background of constant movement restrictions. The seroprevalence estimates suggest that population immunity alone was inadequate to explain this fall and majority of the population remained susceptible. Nonetheless, they also show that the virus was widely transmitted during the first epidemic wave even though numbers of cases and deaths attributable to SARS-CoV-2 in Kenya were very low by comparison with similar settings in Europe and the Americas at similar seroprevalence17,18. This pattern of widespread SARS-CoV-2 transmission and higher cumulative exposure in general19–22 and targeted populations (including blood donors)23–26 compared to disproportionately lower COVID-19 case numbers and deaths has also been seen across epidemic waves in other parts of Africa. This disparity may be attributable to constraints on morbidity/mortality surveillance and poor availability of PCR testing and, to some degree, to the different age structures of African and European populations.
These and our other estimates of cumulative incidence of SARS-CoV-2 support the need for the SARS-CoV-2 vaccination in Kenya since a significant proportion of the population remains susceptible to infection and COVID-19 disease. In addition, vaccination will interrupt transmission, prevent the development of variants, and ultimately correct the social and economic disruption caused by this pandemic.
Methods
Human samples
The Kenya National Blood Transfusion Service (KNBTS) coordinates and screens blood transfusion donor units at 6 regional centres at Eldoret, Embu, Kisumu, Mombasa, Nairobi and Nakuru, though the units are collected across the whole country and each Regional Centre serves between 5 and 10 of Kenya’s 47 Counties. KNBTS guidelines define eligible blood donors as individuals aged 16–65 years, weighing ≥50 kg, with haemoglobin of 12·5 g/dl, a normal blood pressure (systolic 120–129 mmHg and diastolic BP of 80–89 mmHg), a pulse rate of 60–100 beats per minute and without any history of illness in the past 6 months27. KNBTS generally relies on voluntary non-remunerated blood donors (VNRD) recruited at public blood drives typically located in high schools, colleges and universities. Since September 2019, because of reduced funding, KNBTS has depended increasingly on family replacement donors (FRD) who provide units of blood in compensation for those received by sick relatives. We obtained anonymized residual samples from consecutive donor units submitted to the 6 regional centres for transfusion compatibility-testing and infection screening, as previously described3.
Laboratory analyses
Enzyme linked Immunosorbent Assay (ELISA) IgG antibodies to the SARS-CoV-2 spike protein were measured using a previously described ELISA at the KEMRI-Wellcome Trust Research Programme in Kilifi, Kenya. Following a validation exercise and estimate of sensitivity and specificity, results were expressed as the ratio of test OD to the OD of the plate negative control; samples with OD ratios greater than two were considered positive for SARS-CoV-2 IgG.3,5. In a WHO-sponsored multi-laboratory study of SARS-CoV-2 antibody assays, results from Kilifi were consistent with the majority of the test laboratories9.
Statistical analysis
We estimated crude prevalence based on the proportion of samples with OD ratio > 2. We also used Bayesian Multi-level Regression with Post-stratification (MRP)7 to account for differences in the age and sex distribution of blood donors and regional differences in the numbers of samples collected over time. Data on donor residence were specified at County level. For the purposes of analysis and presentation we collapsed the 47 counties into 8 regions based on the previous administrative provinces of Kenya; as data from two regions (Eastern and North Eastern) was relatively sparse we collapsed these to one stratum. The model was also used to adjust for sensitivity (93%) and specificity (99%) of the chosen cut-off value as previously developed6. Regional and national estimates were produced by combining model predictions with weights from the 2019 Kenyan census4. Two versions of the model were fitted. In the first (Model A), the model included age, sex and region as covariates and was fitted separately to data in three periods (30 Apr–19 Jun, 20 Jun–19 Aug, 20 Aug–30 Sept). In the second (Model B), the model also included a period effect and was fitted to the samples as a whole. A mathematical description of the models and Rstan code28 is provided in the statistical appendix.
Ethical approval
This study was approved by the Scientific and Ethics Review Unit (SERU) of the Kenya Medical Research Institute (Protocol SSC 3426). Blood donors gave individual written consent for the use of their samples for research.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This project was funded by the Wellcome Trust (grant numbers 220991/Z/20/Z, 203077/Z/16/Z), the Bill and Melinda Gates Foundation (INV-017547) and by the Foreign Commonwealth and Development Office (FCDO) through the East Africa Research Fund (EARF/ITT/039). S.U. is funded by DELTAS Africa Initiative [DEL-15-003], L.I.O.-O. is funded by a Wellcome Trust Intermediate Fellowship (107568/Z/15/Z), A.A. is funded by a DFID/MRC/NIHR/Wellcome Trust Joint Global Health Trials Award (MR/R006083/1), J.A.G.S. is funded by a Wellcome Trust Senior Research Fellowship (214320) and the NIHR Health Protection Research Unit in Immunisation, I.A. is funded by the United Kingdom’s Medical Research Council and Department For International Development through a African Research Leader Fellowship (MR/S005293/1) and by the NIHR-MPRU at UCL (grant 2268427 LSHTM). G.M.W. is supported by a fellowship from the Oak Foundation. For the purpose of Open Access, the author has applied a CC-BY public copyright licence to any author accepted manuscript version arising from this submission. This paper has been published with the permission of the Director, Kenya Medical Research Institute. We thank all of the blood donors for their contribution to the research.
Author contributions
I.M.O.A., S.U., A.A., J.A.G.S. and G.W. designed the study and did the literature review. E.W., K.G., A.E., S.V., R.A., M.M., P.A., K.K. and W.N. contributed to the study design, protocol development and implementation. S.U., P.W., C.R., C.Y., K.K.I., E.O., T.R., I.O. and S.K. helped collect the samples and demographic data. T.L., D.W., H.K.K., J.N., J.T., L.I.O.-O. and G.W. developed and validated the ELISA assay. J.N.G., D.M., H.K.K., J.N., J.T. and L.I.O.-O. were responsible for sample preparation and laboratory analyses. E.B., B.T. and P.B. provided coordination and contributed to supervision. M.O., C.B. and J.A.G.S. analysed data and all authors interpreted data. S.U., I.M.O.A., J.A.G.S., A.A. and G.W. wrote the first draft of the report and all authors contributed to editing the final version. A.A., J.A.G.S. and G.W. contributed equally as senior authors.
Data availability
The raw data shown in the manuscript are subject to controlled access because they are the subject of ongoing work and will be made available on request to the corresponding author and approval by the Data Governance Committee at the KEMRI-Wellcome Trust Research Programme. De-identified data has been published on the Harvard dataverse server 10.7910/DVN/FQUNVD.
Code availability
Code related to the Bayesian Multi-level Regression with Post-stratification can be found in the Supplementary note 1: statistical appendix accompanying this article.
Competing interests
R.A., M.M., K.K. and P.A. are from the Ministry of Health, Government of Kenya. All other authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Idris Guessous, Kondwani Jambo, and Helen Ward for their contribution to the peer review of this work. Peer review reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ifedayo M.O. Adetifa, Sophie Uyoga, John N. Gitonga, Daisy Mugo.
Contributor Information
Ifedayo M. O. Adetifa, Email: IAdetifa@kemri-wellcome.org
Sophie Uyoga, Email: SUyoga@kemri-wellcome.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-24062-3.
References
- 1.Maeda JM, Nkengasong JN. The puzzle of the COVID-19 pandemic in Africa. Science. 2021;371:27. doi: 10.1126/science.abf8832. [DOI] [PubMed] [Google Scholar]
- 2.Ministry of Health, K. Kenya Health and Research Observatory (KHRO). COVID-19 Tracker (2021) https://khro.health.go.ke/#/covid_19_tracker?. Accessed 30th March 2021.
- 3.Uyoga, S. et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Kenyan blood donors. Science371, 79–82 (2021). [DOI] [PMC free article] [PubMed]
- 4.Kenya National Bureau of Statistics. 2019Kenya Population and Housing Census. Volume 1. Population by County and sub-County, (Government of Kenya, Nairobi, 2019).
- 5.Amanat, F. et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med.26, 1033–1036 (2020). [DOI] [PMC free article] [PubMed]
- 6.Gelman A, Carpenter R. Bayesian analysis of tests with unknown specificity and sensitivity. J. R. Stat. Soc. Ser. C. 2020;69:1269–1283. doi: 10.1111/rssc.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelman A, Little TC. Poststratification into many categories using hierarchical logistic regression. Surv. Methology. 1997;23:217–135. [Google Scholar]
- 8.Ojal, J. et al. Revealing the extent of the COVID-19 pandemic in Kenya based on serological and PCR-test data. Preprint at https://www.medrxiv.org/content/10.1101/2020.09.02.20186817v1 (2020). [DOI] [PMC free article] [PubMed]
- 9.Mattiuzzo, G. et al. Expert Committee on Biological Standardization. WHO/BS.2020.2403. Establishment of the WHO International Standard and Reference Panel for anti-SARS-CoV-2 antibody https://www.who.int/publications/m/item/WHO-BS-2020.2403 (2020).
- 10.Ibarrondo FJ, et al. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward, H. et al. Prevalence of antibody positivity to SARS-CoV-2 following the first peak of infection in England: Serial cross-sectional studies of 365,000 adults. Lancet Reg Health Eur. 4, 100098 (2021) [DOI] [PMC free article] [PubMed]
- 12.Buss LF, et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371:288–292. doi: 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottomley, C. et al. Improving SARS-CoV-2 cumulative incidence estimation through mixture modelling of antibody levels. Preprint at https://www.medrxiv.org/content/10.1101/2021.04.09.21254250v1 (2021).
- 14.Lucinde, R. et al. Sero-surveillance for IgG to SARS-CoV-2 at antenatal care clinics in two Kenyan referral hospitals. Preprint at https://www.medrxiv.org/content/10.1101/2021.02.05.21250735v1 (2021). [DOI] [PMC free article] [PubMed]
- 15.Kagucia, E. W. et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies among truck drivers and assistants in Kenya. Preprint at https://www.medrxiv.org/content/10.1101/2021.02.12.21251294v1 (2021).
- 16.Etyang, A. O. et al. Seroprevalence of Antibodies to SARS-CoV-2 among Health Care Workers in Kenya. Clin Infect Dis. 24, ciab346 (2021).
- 17.Bajema, K. L. et al. Estimated SARS-CoV-2 Seroprevalence in the US as of September 2020. JAMA Intern. Med. 181, 450–460 (2020). [DOI] [PMC free article] [PubMed]
- 18.Stringhini S, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. 2020;396:313–319. doi: 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulenga, L. B. et al. Prevalence of SARS-CoV-2 in six districts in Zambia in July, 2020: a cross-sectional cluster sample survey. Lancet Glob Health9, e773–81 (2021). [DOI] [PMC free article] [PubMed]
- 20.Kempen, J. H. et al. SARS-CoV-2 serosurvey in Addis Ababa, Ethiopia. Am. J. Tropical Med. Hyg.103, 2022-2023 (2020). [DOI] [PMC free article] [PubMed]
- 21.Aitken, S. et al. COVID19 seroprevalence during the second wave of the pandemic in three districts of South Africa - preliminary findings. 18 (2021) https://www.nicd.ac.za/wp-content/uploads/2021/03/COVID-19-Special-Public-Health-Surveillance-Bulletin-9-12-March-2021_.pdf. Accessed 29th March 2021.
- 22.Wiens KE, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Juba, South Sudan: a population-based study. Emerg Infect Dis. 2021;27:1598–1606. doi: 10.3201/eid2706.210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chibwana, M. G. et al. High SARS-CoV-2 seroprevalence in Health Care Workers but relatively low numbers of deaths in urban Malawi. Preprint at https://www.medrxiv.org/content/10.1101/2020.07.30.20164970v3 (2020).
- 24.Hsiao, M. et al. SARS-CoV-2 Seroprevalence in the Cape Town metropolitan sub-districts after the peak of infections. Natl Inst. Commun. Dis.18, (2020) https://www.nicd.ac.za/wp-content/uploads/2020/09/COVID-19-Special-Public-Health-Surveillance-Bulletin_Issue-5.pdf. Accessed 29th March 2021.
- 25.Milleliri, J. M. et al. SARS-CoV-2 infection in Ivory Coast: a serosurveillance survey among gold mine workers. Am. J. Trop. Med. Hyg.104, 1709–1712 (2021). [DOI] [PMC free article] [PubMed]
- 26.Sykes, W. et al. Prevalence of anti-SARS-CoV-2 antibodies among blood donors in Northern Cape, KwaZulu-Natal, Eastern Cape, and Free State provinces of South Africa in January 2021., 12 February 2021, PREPRINT (Version 1). Research Square10.21203/rs.3.rs-233375/v1 (2021).
- 27.Ministry of Health, K. Policy Guidelines on Blood Transfusion in Kenya. The National Blood Transfusion Service of Kenya. https://nbtskenya.or.ke/wp-content/uploads/2019/02/Policy-Guidelines-on-Blood-Transfusion-in-Kenya.pdf (2001) Accessed 20 July 2020.
- 28.Stan Development Team. RStan: the R interface to Stan. R package version 2.23.2 http://mc-stan.org/ (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data shown in the manuscript are subject to controlled access because they are the subject of ongoing work and will be made available on request to the corresponding author and approval by the Data Governance Committee at the KEMRI-Wellcome Trust Research Programme. De-identified data has been published on the Harvard dataverse server 10.7910/DVN/FQUNVD.
Code related to the Bayesian Multi-level Regression with Post-stratification can be found in the Supplementary note 1: statistical appendix accompanying this article.