Abstract
Lung cancer is one of the deadliest cancers in the world. Two of the most common subtypes, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), have drastically different biological signatures, yet they are often treated similarly and classified together as non-small cell lung cancer (NSCLC). LUAD and LUSC biomarkers are scarce, and their distinct biological mechanisms have yet to be elucidated. To detect biologically relevant markers, many studies have attempted to improve traditional machine learning algorithms or develop novel algorithms for biomarker discovery. However, few have used overlapping machine learning or feature selection methods for cancer classification, biomarker identification, or gene expression analysis. This study proposes to use overlapping traditional feature selection or feature reduction techniques for cancer classification and biomarker discovery. The genes selected by the overlapping method were then verified using random forest. The classification statistics of the overlapping method were compared to those of the traditional feature selection methods. The identified biomarkers were validated in an external dataset using AUC and ROC analysis. Gene expression analysis was then performed to further investigate biological differences between LUAD and LUSC. Overall, our method achieved classification results comparable to, if not better than, the traditional algorithms. It also identified multiple known biomarkers, and five potentially novel biomarkers with high discriminating values between LUAD and LUSC. Many of the biomarkers also exhibit significant prognostic potential, particularly in LUAD. Our study also unraveled distinct biological pathways between LUAD and LUSC.
Subject terms: Medical genomics, Genetic markers
Introduction
Lung cancer is the most commonly diagnosed malignant tumor and is a leading cause of cancer-associated mortality. It is the second highest cause of new cancer cases in both genders in the United States and is the second leading cause of cancer deaths in females globally1,2. The most common subtypes of lung cancers are lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC), classified together as non-small cell lung cancer (NSCLC)3,4. However, recent studies have suggested that LUAD and LUSC should be classified and treated as different cancers5.
Identifying the mechanisms underlying LUAD and LUSC is needed to develop useful biomarkers for better diagnosis and design therapeutic interventions. Multiple gene expression and immunohistochemistry studies have identified biological pathways and biomarkers that differentiate between LUAD and LUSC6–8. Other studies classified cancers using both novel and traditional machine learning or feature selection methods9–12. However, few have investigated cancers by applying multiple feature selection methods and selecting the overlapping features.
In this study, we downloaded LUAD and LUSC RNA-Seq datasets from The Cancer Genome Atlas (TCGA)13 and analyzed them with five feature selection methods with ranking abilities: Differential Gene Expression Analysis (DGE), Principal Component Analysis (PCA), Least absolute shrinkage and selection operator (Lasso), minimal-Redundancy-Maximal Relevance (mRMR), and Extreme Gradient boosting (XGboost). DGE applies a normalization method and uses the negative binomial distribution to detect significant changes in gene expression across samples14,15. Many studies have shown that DGE, though being the most widely used algorithm to detect differentially expressed genes, often yields some false positive results; in addition, it is often sensitive to outliers14–17. On the other hand, XGboost is a tree-based machine learning method that is not sensitive to outliers but is prone to overfitting17,18. To minimize this problem, we chose to use Lasso, a linear regression technique that avoids overfitting but can be influenced by highly correlated features and potentially leading to false discoveries17–20. mRMR is then used to maximize the relevance between the features and the output, and minimize the relevance among the feature themselves, thus, limiting highly correlated features21–23. PCA is another well-known and widely used feature reduction technique in machine learning to reduce high dimensional data into orthogonal principal components, which also removes correlated features17,18. However, amidst other disadvantages, the result of PCA by itself is often not interpretable17,18. These algorithms were also chosen because of their ability to rank features or select a reasonable number of features. In short, overlapping these algorithms is promising because different methods select features using different criteria. Since each method has its strengths and weaknesses, focusing on the overlapping features will optimize the strengths and minimize the weaknesses of each method, thereby reducing the number of false positives and producing reliable results. This study will serve as a proof of concept for the validity of the approach to overlap feature selection methods while investigating NSCLC subtype differences and discovering novel biomarkers.
Results
Study design and overview
We obtained LUAD and LUSC RNA-Seq data from TCGA13 and the summary of their clinical information was provided in Table 1, with more comprehensive details available on TCGA website13. We selected discriminatory genes by overlapping DGE, PCA, mRMR, XGboost, and lasso as depicted in Fig. 1. The genes that were overlapped by two or more algorithms were validated and used for LUAD and LUSC classification as well as gene expression analysis. The genes that were overlapped by three or more algorithms were selected as biomarker candidates, and their diagnostic values were assessed using ROC analysis and AUC value, and then further verified in an external dataset, GSE2858224,25, which is a microarray dataset that includes 50 LUAD and 28 LUSC samples The prognostic values of the biomarker candidates were also assessed using Kaplan Meier Plotter26.
Table 1.
Summary of clinical information from TCGA with each entry indicating number of samples.
| Gender | AJCC pathologic stage | Treatment | Primary diagnosis subtypes | ||||
|---|---|---|---|---|---|---|---|
| Lung adenocarcinoma | |||||||
| Male | 220 | Stage IA | 124 | Pharmacotherapy only | 56 | Adenocarcinoma, NOS | 311 |
| Female | 259 | Stage IB | 131 | Radiotherapy only | 101 | Adenocarcinoma with mixed subtypes | 108 |
| Missing | 50 | Stage IIA | 46 | Both therapies | 70 | Papillary | 22 |
| Stage IIB | 63 | No treatment | 242 | Bronchiolo-alveolar, NOS | 3 | ||
| Stage IIIA | 66 | Missing | 60 | Bronchiolo-alveolar, nonmucinous | 19 | ||
| Stage IIIB | 11 | Brionchio-alviolar Carcinoma, mucinous | 5 | ||||
| Stage IV | 24 | Micropapillary | 3 | ||||
| Stage I | 5 | Clear cell | 2 | ||||
| Stage II | 1 | Solid carcinoma | 6 | ||||
| Missing | 58 | Missing | 50 | ||||
| Lung squamous cell carcinoma | |||||||
| Male | 368 | Stage IA | 89 | Pharmacotherapy only | 57 | Squamous cell carcinoma, NOS | 465 |
| Female | 130 | Stage IB | 150 | Radiotherapy only | 65 | Basaloid | 14 |
| Stage IIA | 64 | Both therapies | 48 | Keratinizing | 13 | ||
| Stage IIB | 94 | No treatment | 265 | Papillary | 3 | ||
| Stage IIIA | 63 | Missing | 63 | Large cell, nonkeratinizing | 2 | ||
| Stage IIIB | 18 | Small cell, nonkeratinizing | 1 | ||||
| Stage IV | 7 | ||||||
| Stage I | 3 | ||||||
| Stage II | 3 | ||||||
| Stage III | 3 | ||||||
| Missing | 4 | ||||||
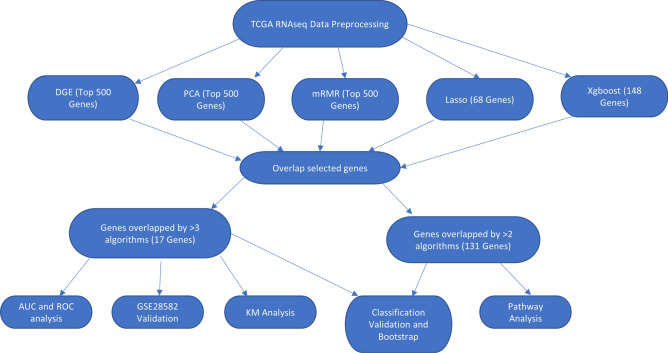
Figure 1.
An overview of the experimental design. A scheme summarizes the selection methods and the numbers of the resulting overlapped genes.
Selection of genes
Top 500 genes from DGE (Table S1) were selected as top features based on their lowest p-values. Similarly, top 500 genes from the first principal component in PCA and the top 500 genes from mRMR (Table S1) were selected based on the ranking of the algorithm. Also, 148 genes in Xgboost (Table S1) and 68 genes in lasso (Table S1) using probability or prediction threshold of 0.5 were identified and selected. The different number of genes selected was due to the nature of the algorithm, with most of the parameters in each algorithm were set to default. The specifics of each metric can be found in the code at the data availability section Since each of these methods has its own selection criteria, the overlapping genes must satisfy multiple selection criteria, making them significant candidate biomarkers that differentiate LUAD and LUSC. Therefore, the five independent sets of top genes were compared with a Venn diagram to identify the overlapping genes detected by multiple algorithms. Venn diagram (Fig. 2) comparison detected 131 genes (Table S2) overlapped by two or more algorithms and 17 genes (Table 2) overlapped by three or more algorithms.
Figure 2.
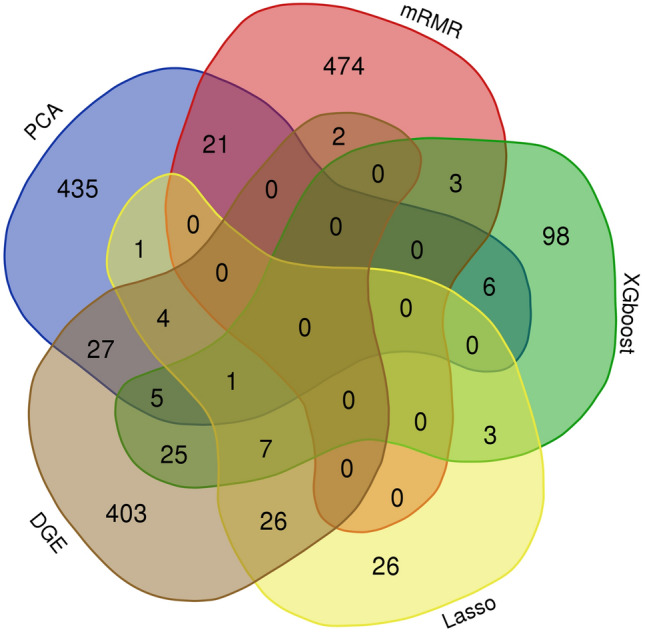
Venn diagram shows overlapping genes selected by each algorithm. Venn diagram of selected genes from PCA, mRMR, DGE, Lasso, and XGboost.
Table 2.
17 Biomarker candidate genes that were selected by three or more.
| Genes | Upregulated or downregulated | Significantly expressed in LUSC or LUAD | Number of algorithms that selected the gene |
|---|---|---|---|
| KRT17 (Keratin 17) | Upregulated | LUSC | DGE, Lasso, PCA, XGBoost |
| KRT14 (Keratin 14) | Upregulated | LUSC | DGE, PCA, XGboost |
| KRT6A (Keratin 6A) | Upregulated | LUSC | DGE, PCA, XGboost |
| KRT5 (Keratin 5) | Upregulated | LUSC | DGE, PCA, XGboost |
| S100A2 (Calcium Binding Protein A2) | Upregulated | LUSC | DGE, PCA, XGboost |
| TUBA1C (Tubulin Alpha 1c) | Upregulated | LUSC | DGE, Lasso, XGboost |
| CELSR2 (Cadherin EGF LAG seven-pass G-type receptor 2) | Upregulated | LUSC | DGE, Lasso, XGboost |
| TRIM29 (Tripartite Motif Containing 29) | Upregulated | LUSC | DGE, Lasso, PCA |
| REPS1 (RALBP1 Associated Eps Domain Containing 1) | Upregulated | LUSC | DGE, Lasso, XGboost |
| PERP (P53 Apoptosis Effector Related To PMP22) | Upregulated | LUSC | DGE, Lasso, PCA |
| NECTIN1 (Nectin Cell Adhesion) Molecule 1 | Upregulated | LUSC | DGE, Lasso XGboost |
| GPC1 (Glypican 1) | Upregulated | LUSC | DGE, PCA, XGBoost |
| MUC1 (Mucin 1, cell surface associated) | Downregulated | LUAD | DGE, Lasso, PCA |
| ELFN2 (Extracellular Leucine Rich Repeat And Fibronectin Type III Domain Containing 2) | Downregulated | LUAD | DGE, Lasso, XGboost |
| ARHGEF38 (Rho Guanine Nucleotide Exchange Factor 38) | Downregulated | LUAD | DGE, Lasso, XGboost |
| ARHGAP12 ( Rho GTPase Activating Protein 12) | Downregulated | LUAD | DGE, Lasso, XGboost |
| QSOX1 (Quiescin Sulfhydryl Oxidase 1) | Downregulated | LUAD | DGE, Lasso, PCA |
Validation of selected genes
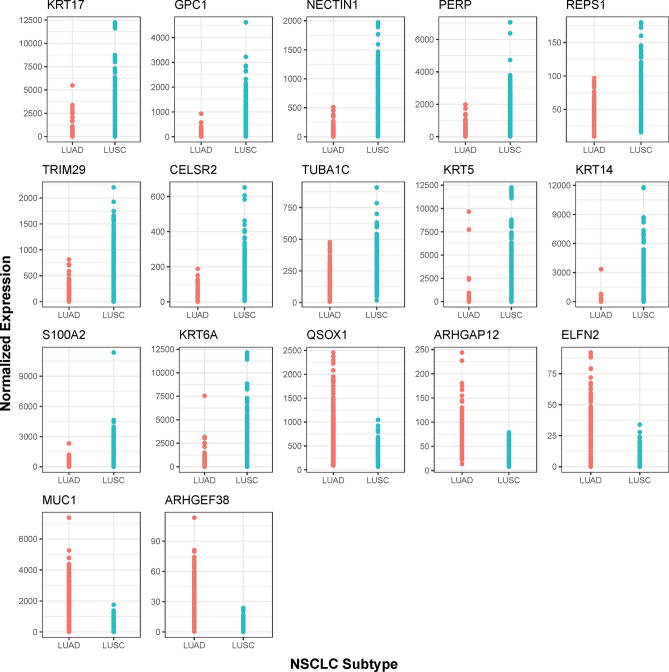
To evaluate how effective the selected genes are in classifying LUAD and LUSC, we used random forest to validate the top 500 genes selected from PCA, mRMR, and DGE, as well as the 148 genes from xgboost and 68 genes from lasso (Table S1). All of the validation results for each feature selection method returned high classification accuracies of over 90% (Table 3). To compare to the previous feature selection methods, the overlapping 131 genes were validated the same way as the other algorithms. The binary classification statistics (Table 3) were calculated using LUAD as ‘positive’ and LUSC as ‘negative’. The overlapping 131 genes showed comparable, if not better, results to the other algorithms (Table 3). The 17 proposed biomarkers also showed to be effective classifiers, having statistics comparable to the other algorithms despite only using 17 genes. Heatmaps for the top 131 and the top 17 genes were also generated (Fig. 3A,B). Both heatmaps, in particular the heatmap with 17 genes, displayed clear borders separating LUAD from LUSC. Dot plots of the gene expression distribution between LUAD and LUSC for each of the 17 genes are displayed in Fig. 4.
Table 3.
LUAD and LUSC Classification Statistics.
| Feature selection method | Accuracy | Specificity | Sensitivity | Precision | F-measure | 95% Bootstrap confidence interval |
|---|---|---|---|---|---|---|
| DGE (Top 500) | 0.932476 | 0.901235 | 0.966443 | 0.9 | 0.932039 | (0.9035, 0.9614) |
| PCA (Top 500) | 0.942122 | 0.901235 | 0.986577 | 0.90184 | 0.942308 | (0.9132, 0.9678) |
| mRMR (Top 500) | 0.916399 | 0.888889 | 0.946309 | 0.886792 | 0.915584 | (0.8842, 0.9453) |
| Lasso (68 Genes) | 0.938907 | 0.907407 | 0.973154 | 0.90625 | 0.938511 | (0.9100, 0.9646) |
| Xgboost (148 Genes) | 0.935691 | 0.901235 | 0.973154 | 0.900621 | 0.935484 | (0.9068, 0.9614) |
| Overlapping 131 Genes | 0.938907 | 0.895062 | 0.986577 | 0.896341 | 0.939297 | (0.9100, 0.9646) |
| 17 Proposed Biomarkers | 0.92926 | 0.889 | 0.9735 | 0.88957 | 0.9296 | ( 0.9003, 0.9550 ) |
Figure 3.
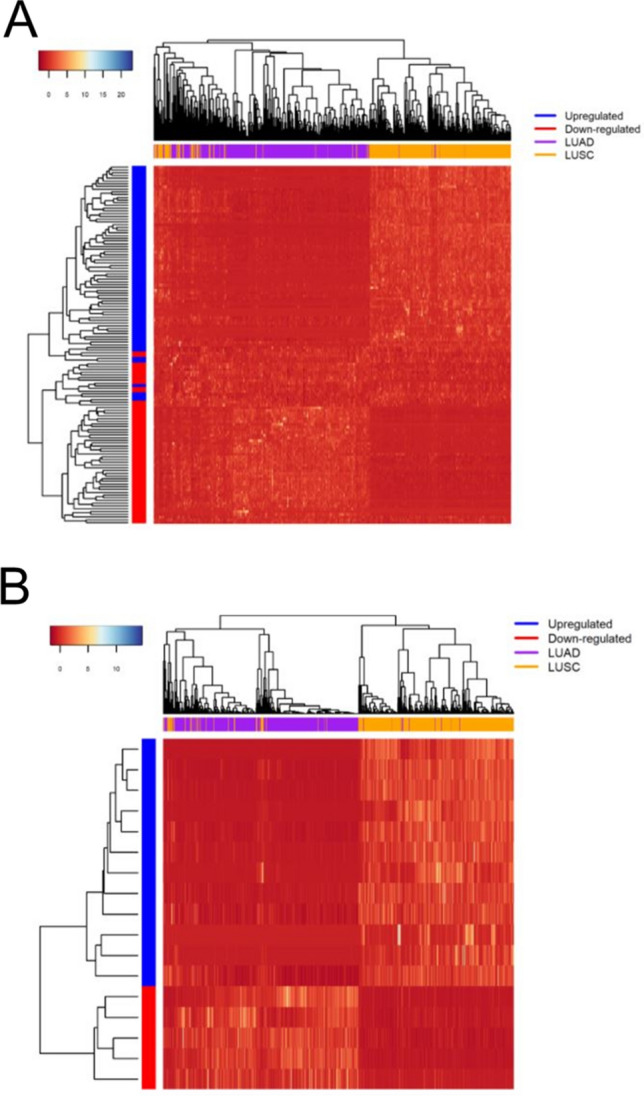
Heatmap shows the 131 selected genes (A) for gene expression analysis and the 17 selected genes (B) as biomarker candidates87. The x-axis represents the samples and the y-axis represents the genes.
Figure 4.
Normalized Gene Expression Distribution Dot Plots for the 17 Biomarker Candidates87. The x-axis represents the NSCLC subtypes and the y-axis represents the normalized gene expression values.
Identification of the 17 potential biomarkers and their ROC analysis
The 17 biomarker candidates (Table 2) were subjected to ROC curve analysis (Fig. 5). Most of the genes had areas under the curve (AUC) of over 0.9, with NECTIN1 (0.9514), PERP (0.9529), KRT5 (0.9731), KRT6A (0.9532), and ARHGEF38 (0.9574) having AUC of over 0.95. Among the upregulated genes (Fig. 5A), KRT5 has the highest AUC of 0.9731, thereby displaying the most significant diagnostic potential in classifying LUAD and LUSC, consistent with the study reported by Jain Xiao et al.6 in which KRT5 also had the highest diagnostic potential. All of the upregulated genes show significant discrimination potential as well (Fig. 5A,B).
Figure 5.
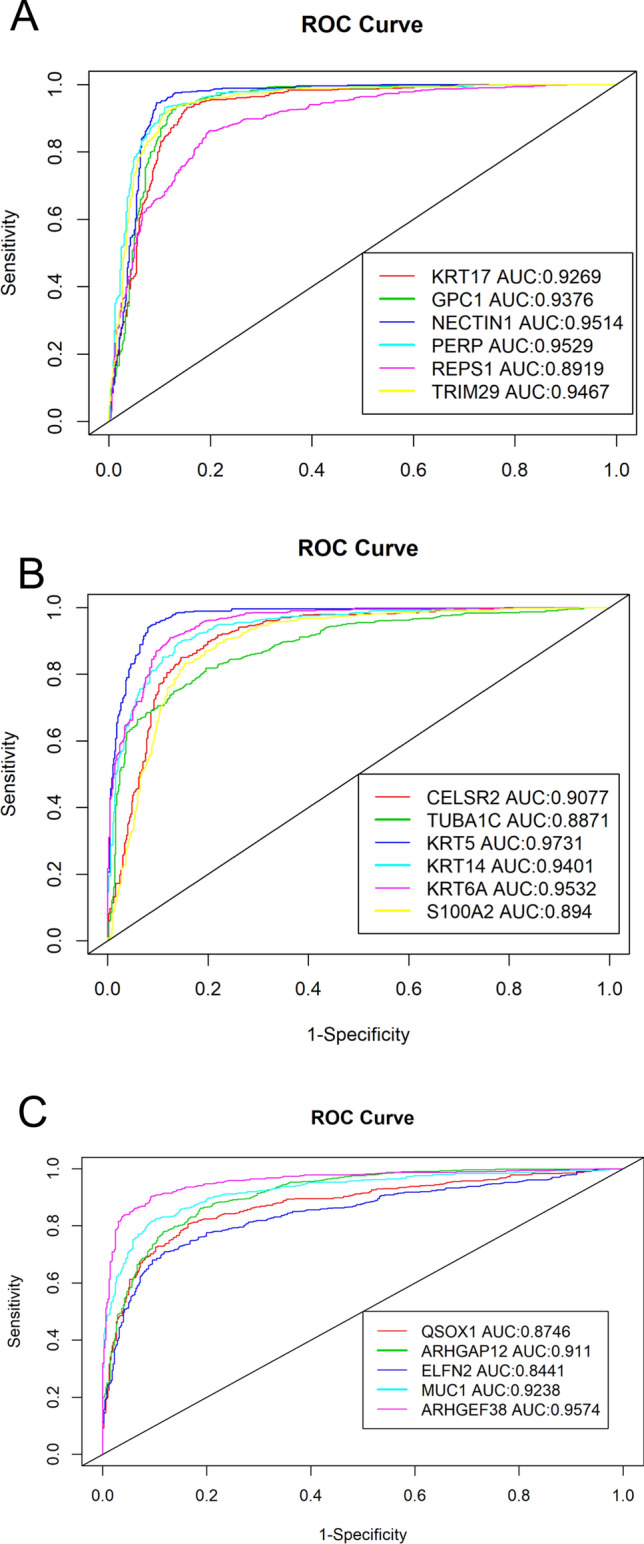
ROC and AUC analysis demonstrate discriminating potential for Upregulated (a,b) and Downregulated (c) Genes in TCGA Dataset87. X-axis is sensitivity, or true positive rate (TPR). The Y-axis is 1-Specificity, or false positive rate (FPR). Higher AUC indicates higher discriminating potential for the gene.
To minimize the inherent RNA expression noise and to ensure that these results are reproducible, an external dataset GSE28582 was used for external validation. AUC and ROC were also used to analyze the 17 genes in GSE28582 validation dataset (Fig. 6). Largely consistent with our result, most of the genes show AUC values well above 0.9; all except one gene, ARHGEF38, have AUC values above 0.8 (Fig. 6).
Figure 6.
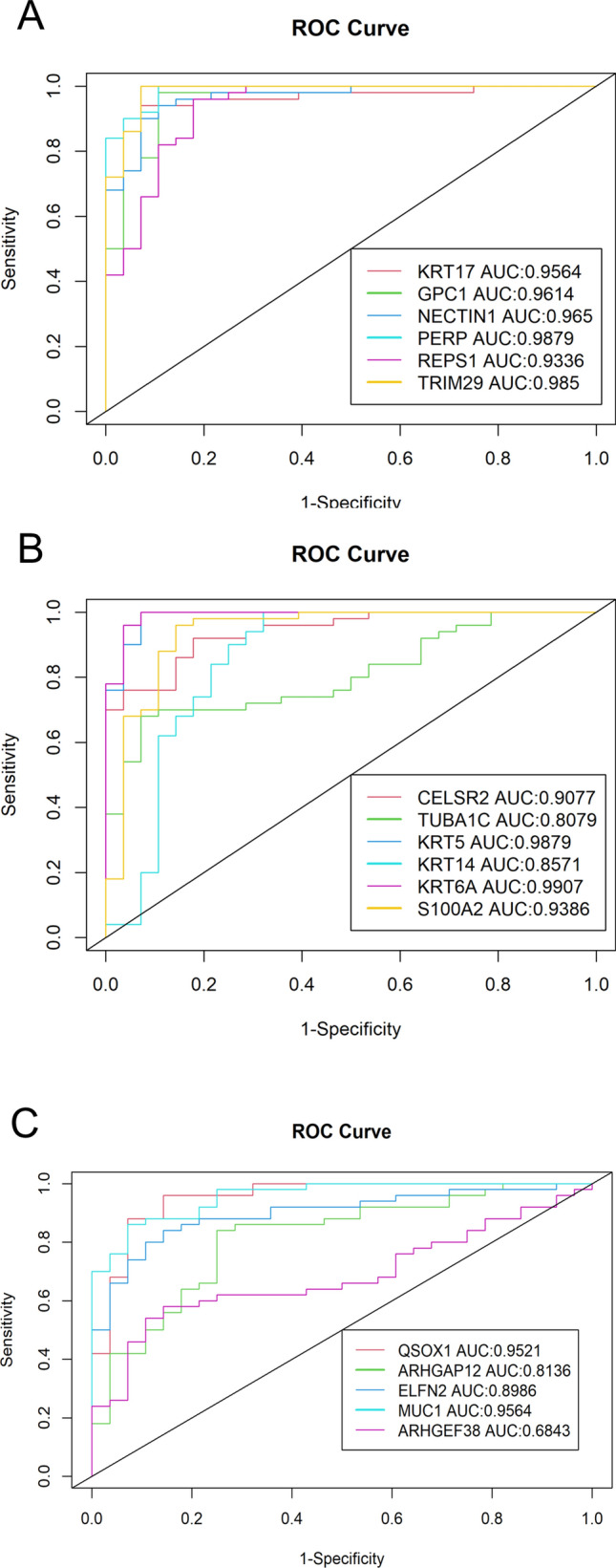
GSE28582 microarray dataset ROC and AUC validation of the 17 candidate biomarkers87. (A,B) The upregulated genes, and (C) shows the downregulated genes. The x-axis represents sensitivity, or true positive rate (TPR). The y-axis is 1 − Specificity, or false positive rate (FPR). Higher AUC indicates higher discriminating potential for the gene.
Kaplan Meier plotter analysis of the 17 potential biomarkers
Of the 17 potential biomarkers (Table 2), only CELSR2 shows a significant prognostic p-value in LUSC, with its higher expression corresponding to a more favorable prognosis in LUSC (Table 4). In contrast, many genes show significant prognostic potential in LUAD. High expressions of KRT17, KRT6A, S100A2, TRIM29, REPS1, and GPC1 correspond to an unfavorable prognosis in LUAD, while high expressions of PERP, ELFN2, ARHGAP12, and QSOX1 correspond to a favorable prognosis in LUAD (Table 4).
Table 4.
Kaplan Meier prognostic values of the 17 biomarker.
| LUAD | LUSC | |||
|---|---|---|---|---|
| HR (95% CIs) | P-value/FDR | HR (95% CIs) | P-value/FDR | |
| KRT17 | 1.28 (1.01–1.61) | 0.037/0.0629 | 1.11 (0.88–1.4) | 0.39/0.947 |
| KRT14 (EBS4) | 1.19 (0.94–1.5) | 0.14/0.2164 | 1.2 (0.95–1.52) | 0.13/1 |
| KRT6A (K6C) | 1.67 (1.32–2.12) | 1.6e−05/0.00014 | 0.99 (0.78–1.25) | 0.92/0.98 |
| KRT5 | 1.14 (0.9–1.43) | 0.28/0.366 | 1 (0.79–1.27) | 1/1 |
| S100A2 | 1.73 (1.36–2.19) | 4.3e−06/7.31E−5 | 1.07 (0.85–1.36) | 0.55/1 |
| TUBA1C | 1.1 (0.87–1.39) | 0.43/0.522 | 1.2 (0.94–1.52) | 0.14/0.793 |
| CELSR2 | 0.92 (0.73–1.16) | 0.47/0.533 | 0.79 (0.62–1) | 0.049/0.833 |
| TRIM29 | 1.31 (1.04–1.66) | 0.022/0.0416 | 0.93 (0.74–1.18) | 0.57/0.969 |
| REPS1 | 1.38 (1.08–1.76) | 0.0093/0.0226 | 0.9 (0.66–1.23) | 0.51/1 |
| PERP | 0.67 (0.52–0.85) | 0.0012/0.0051 | 0.85 (0.62–1.16) | 0.3/0.85 |
| NECTIN1 (PVRL1) | 1.19 (0.94–1.5) | 0.14/0.198 | 0.94 (0.74–1.2) | 0.63/0.974 |
| GPC1 | 1.36 (1.08–1.72) | 0.0091/0.0258 | 0.98 (0.77–1.23) | 0.83/1 |
| MUC1 | 1.02 (0.81–1.29) | 0.84/0.084 | 1.02 (0.8–1.29) | 0.88/1 |
| ELFN2 | 0.72 (0.56–0.92) | 0.0076/0.02584 | 1.07 (0.78–1.47) | 0.67/0.876 |
| ARHGEF38 (FLJ20184) | 0.97 (0.77–1.23) | 0.83/0.882 | 1.16 (0.91–1.47) | 0.22/0.748 |
| ARHGAP12 | 0.61 (0.48–0.77) | 2.3e−05/0.00013 | 1.17 (0.93–1.49) | 0.18/0.765 |
| QSOX1 | 0.76 (0.6–0.96) | 0.021/0.0446 | 0.95 (0.75–1.2) | 0.66/0.935 |
GO term enrichment analysis
To further understand the biological differences between LUAD and LUSC, we performed gene expression analysis by splitting the identified 131 genes into two groups: 57 downregulated and 74 upregulated genes in LUSC compared to LUAD. Functional pathway annotation of these two groups of genes was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID)27 analysis tool with Gene Ontology (GO) biological pathway enrichments. GO terms with P-value < 0.01 were obtained (Tables S3 and S4). The top 10 most significantly upregulated and downregulated GO terms ranked by p-value are shown in Table 5. In addition, DAVID has the functionality to group similar GO terms into clusters of the same biological pathway. To elucidate the potential biological differences between LUAD and LUSC, the top five most significantly upregulated and downregulated clusters ranked by enrichment scores were determined (Table 6 and Tables S5 and S6).
Table 5.
Top 10 Upregulated and Downregulated GO Biological Pathways.
| Top 10 upregulated pathways | Top 10 downregulated pathways | ||||
|---|---|---|---|---|---|
| GO term | Pathway | P-value | GO term | Pathway | P-value |
| GO:0009888 | Tissue development | 4.45E−07 | GO:0002576 | Platelet degranulation | 2.86E−04 |
| GO:0045104 | Intermediate filament cytoskeleton organization | 8.82E−07 | GO:1901575 | Organic substance catabolic process | 8.18E−03 |
| GO:0045103 | Intermediate filament-based process | 9.95E-−07 | GO:0009057 | Macromolecule catabolic process | 8.29E−03 |
| GO:0007155 | Cell adhesion | 4.25E−06 | GO:0045055 | Regulated exocytosis | 1.05E−02 |
| GO:0022610 | Biological adhesion | 4.49E−06 | GO:0009056 | Catabolic process | 1.32E−02 |
| GO:0008544 | Epidermis development | 4.64E−06 | GO:00034613 | Cellular protein localization | 1.80E−02 |
| GO:0098609 | Cell–cell adhesion | 5.07E−06 | GO:0070727 | Cellular macromolecule localization | 1.89E−02 |
| GO:0034330 | Cell junction organization | 9.93E−06 | GO:0043129 | Surfactant homeostasis | 2.36E−02 |
| GO:2001233 | Regulation of apoptotic signaling pathway | 3.06E−05 | GO:0016553 | Base conversion or substitution editing | 2.65E−02 |
| GO:0061436 | Establishment of skin barrier | 5.65E−05 | GO:0048875 | Chemical homeostasis within a tissue | 2.94E−02 |
Table 6.
Top 5 Clusters of Upregulated and Downregulated Biological pathways.
| Top 5 clusters of upregulated biological pathways | Top 5 clusters of downregulated biological pathways | ||
|---|---|---|---|
| Cluster | Enrichment score | Cluster | Enrichment score |
| Cell adhesion | 4.05 | Platelet degranulation and exocytosis | 1.34 |
| Intermediate filament organization | 3.87 | Tyrosine kinase pathways | 0.74 |
| Cell junction organization | 3.42 | Homeostasis | 0.69 |
| Cell component organization | 3.28 | Protein translation and localization | 0.68 |
| Hemidesmosome assembly | 2.67 | Circulatory system regulation | 0.63 |
In the upregulated group, most pathways are concentrated in cell adhesion, intermediate filament organization, and cell junction assembly. In the downregulated group, the most significant cluster is platelet degranulation and cell exocytosis, as well as other pathways such as tyrosine kinase signaling pathway, homeostasis, protein translation and circulatory system. These results suggest that LUSC tends to express more genes related to cell adhesion and cytoskeleton organization, and LUAD tends to express more genes involved in platelet degranulation and exocytosis, along with other signaling pathways.
Reactome gene expression analysis
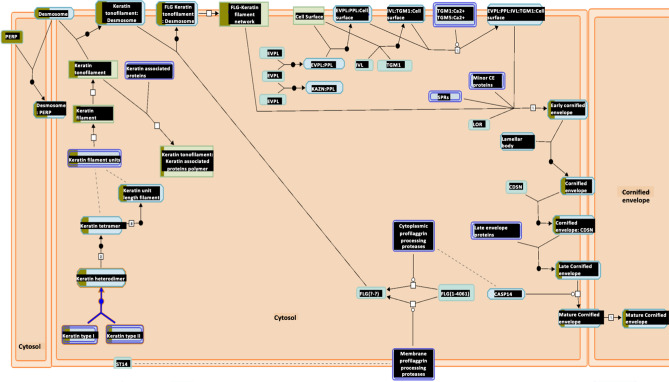
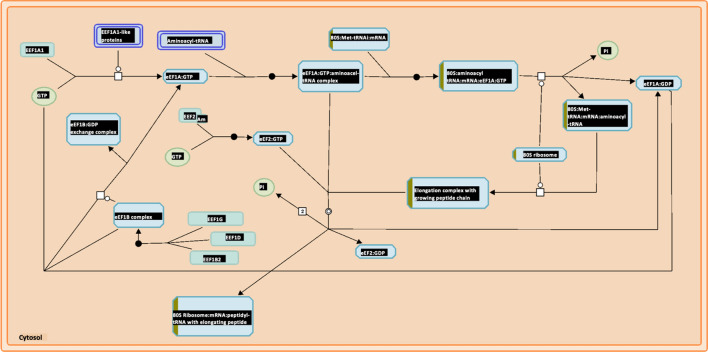
Reactome pathways28 were also generated for both upregulated and downregulated groups. The most significantly upregulated pathway is the cornification, or the keratinization pathway (Fig. 7, Table S7), along with other similar pathways related to cell adhesion, which is consistent with GO term analysis. TP53 regulation pathway, which is often implicated in cancer, is among the top enriched pathways as well (Table S7). For the downregulated group, the most significant pathway is peptide elongation synthesis (Fig. 8, Table S8), which GO term analysis also reveals to be significant.
Figure 7.
Keratinization pathway is upregulated in LUSC28. The Keratinization pathway is the most upregulated pathway according to Reactome analysis with p-value 3.33E−15 and FDR 1.95E−12. The boxes partially highlighted in brown indicate the number of genes identified in the analysis that are associated with each box.
Figure 8.
Peptide elongation pathway is downregulated in LUSC when compared to LUAD28. The peptide elongation pathway is the most down-regulated pathway according to Reactome analysis with p-value 9.72E−6 and FDR 0.00157. The boxes partially highlighted in brown indicate the number of genes identified in the analysis that are associated with each box.
KEGG gene expression analysis
Only the p53 signaling pathway appeared in the upregulated group (Table 7) in Kyoto Encyclopedia of Genes and Genomes (KEGG)29 gene expression analysis. Though it has a p-value of slightly over 0.01, this result is consistent with Reactome analysis which ranks TP53 regulation as the second most upregulated pathway after keratinization and other cell junction related pathways. Only the lysosome seems to be significant in the downregulated group (Table 7). The lysosomal pathway is coherent with platelet degranulation and exocytosis, as reported in GO term analysis. Even though the ribosomal pathway has a p-value slightly greater than 0.05, it is most likely important as it is also shown to be significantly enriched in both GO and Reactome term analyses (Tables S3 and S8).
Table 7.
KEGG Upregulated and Downregulated Pathways.
| KEGG upregulated pathways | KEGG downregulated pathways | ||||
|---|---|---|---|---|---|
| KEGG term | Pathway | P-value | KEGG term | Pathway | P-value |
| Hsa04115 | P53 signaling pathway | 0.0476 | Hsa04142 | Lysosome | 0.00727 |
| NA | NA | NA | Hsa03010 | Ribosome | 0.0749 |
Discussion
Previous studies have utilized traditional feature selection and machine learning methods for cancer diagnosis, detection, and classification10,11,22, but few have extended them to study potential biomarkers and biological pathways to discriminate between LUAD and LUSC. To improve cancer classification accuracy, novel machine learning, and feature selection methods have been developed12,30–32. However, few studies have used overlapping features from different methods for classification, gene expression analysis, and biomarker discovery. To provide a proof of concept for the validity of this method, we took advantage of the capabilities and the strengths of PCA, mRMR, XGboost, DGE, and lasso to select 131 overlapping genes for classification and gene expression analysis, and 17 genes for classification and potential biomarkers. Overall, the overlapping 131 genes showed several high-ranking metrics with lasso and PCA methods. Though the best method may vary depending on the metric, the classification result of using both the overlapping 131 and 17 genes was by many metrics comparable if not better than the other methods that use more genes. The 131 overlapped genes achieved the highest sensitivity with PCA, the second highest accuracy with lasso, and the second highest F-measure overall, indicating that overlapping feature selection methods can be used to perform cancer classification.
Furthermore this method proves to be valuable in biomarker discovery. In agreement with our result, previous studies have reported levels of several genes to be greatly elevated in LUSC compared to LUAD; these genes include KRT66,8,33,34, KRT56,8,35, KRT148,33,34, KRT178,33, PERP8,33, TRIM298,33, GPC18, CELSR28, S100A28, and TUBA1C36. Also, consistent with our result, levels of QSOX133 and MUC18 were reported to be lower in LUSC than in LUAD. Many current biomarkers such as Tumor Protein P63 (TP63), Napsin A Aspartic Peptidase (NAPSA), Melanophilin (MLPH), Desmocollin 3 (DSC3), and others are also part of the top 131 genes selected by our method33,37–40. To our knowledge, ARHGAP12, ARHGEF38, ELFN2, NECTIN1, and REPS1 are among the top 17 genes in this study to be identified as biomarkers for the first time. All 17 candidate biomarkers, except ARHGEF38, are also validated in GSE28582 exhibiting high discriminating potential. Although the selection of ARHGEF38 may be due to bias in the TCGA dataset, it is important to note that there are many more samples in TCGA compared to GSE28582; GSE28582 as a microarray dataset is also significantly worse than RNAseq at detecting gene expression differences when the expression values are low or when the fold change is less than 241–43. Notably, ARHGEF38 has relatively lower fold change and expression value.
Moreover, studies have shown that biomarkers for diagnosis and prognosis are most reliable when they are biologically related to the disease in addition to being statistically significant44,45. Although this study is primarily data-driven, the results reveal biomarkers that would corroborate with a knowledge-based approach. For instance, the most significant candidate biomarkers between LUAD and LUSC are all cytokeratins and cadherins, which is reasonable because they are markers of squamous epithelial cells. In particular, NECTIN1, as a novel cadherin biomarker, consistently demonstrates high discriminating potential both in the TCGA and the external validation dataset; it also directly binds and signals fibroblast growth factor receptor46, a pathological signaling pathway that is more prominent in LUSC47,48. NECTIN1 also serves a key role in herpes simplex virus type 1 (HSV-1) viral entry and is important in oncolytic therapy in squamous cell carcinomas49,50. Similarly, it is logical that MUC1 can be used to identify LUAD, as it is a marker for columnar cells from which LUAD arise. In addition to satisfying the aims of both data-driven and knowledge-based approach, many of the 17 genes identified through this method show significant prognostic importance, particularly in LUAD (Table 4).
The other candidate biomarkers also show strong association with cancers. ARHGEF38 and ARHGAP12 are both part of the Rho family GTPase regulators. Rho GTPases are essential to cell cytoskeletal structure, motility, and morphogenesis, and they have been implicated in many cancer proliferation and metastases51–54. The other upregulated genes ELFN2, QSOX1, and MUC1 have been shown to directly promote metastasis in various cancers55–59, including lung cancer. Furthermore, the loss of certain genes upregulated in LUSC such as TRIM29 and KRT6A is associated with more cellular invasion60,61. Clinical differences between LUAD and LUSC are well known. In particular, LUAD has a higher metastatic rate than LUSC62. Studying these potential biomarkers may provide insight into tumor progression, metastatic, and therapeutic differences between LUAD and LUSC. Overall, these results not only align with known literature, but also provide reasonable and promising biomarkers, suggesting that using overlapping feature selection methods can be used to reliably detect new biomarkers. With the validity of this overlapping method shown both in cancer classification and biomarker identification, we performed gene expression analysis for further investigation.
Aside from cell adhesion or cytoskeleton organization, LUSC demonstrates higher regulation of p53 signaling in both KEGG and Reactome analyses. It is known that TP53 mutation is more common in LUSC than in LUAD63–65, and that such mutation may predominantly be a non-truncated mutation in LUSC leading to higher expression levels of genes involved in the p53 regulation pathway66. Moreover, P53 mutations often lose their tumor suppression function while gaining oncogenic abilities, leading to increased cell growth and proliferation compared to LUAD67.
The most prominent pathway associated with LUAD, compared to LUSC, is platelet degranulation and exocytosis (Tables 5, 6). Interestingly, lung cancer is the most common malignancy to coexist with venous thromboembolism, especially pulmonary embolism68. LUAD, in particular, has been shown to be an independent risk factor for pulmonary embolism even among lung cancers69,70. Because platelet granulation directly causes thrombus formation, the differential enrichment of platelet granulation pathway can therefore help explain a more active and a more common hypercoagulation and thrombotic process in LUAD compared to LUSC71. In addition, platelet degranulation can modulate innate immunity via the release of cytokines, and platelet-leukocyte interactions can lead to leukocyte recruitment and activation in cancer72. In fact, CD63, one of the genes in the platelet degranulation pathway (Tables S3 and S6), is directly involved in leukocyte recruitment through endothelial P-selectin73. LUSC has recently been associated with a relatively more suppressed immune response, implying a more active immune response in LUAD, which supports our result67,74.
There are several limitations of this study. One of them is that this study does not prioritize the RNA expression fold changes, which some groups have used to rank differentially expressed genes75,76. Also, although this study aims to minimize the discovery of false positive biomarkers by overlapping different feature selection methods, the proposed biomarker candidates in this study still lack experimental verification. Nevertheless, these results may shed light into the biological differences between LUAD and LUSC, as well as aid the discovery of better diagnosis and treatment for each77,78.
In conclusion, we designed and implemented a workflow of overlapping five different feature selection methods to perform cancer classification, identify novel biomarkers, and study biological differences in NSCLC. This overlapping method proves to be reliable in both cancer classification and biomarker identification, yielding statistically promising genes that also support our current knowledge. We identified ARHGAP12, ARHGEF38, ELFN2, NECTIN1, and REPS1 as novel biomarkers, along with 12 other strong biomarker candidates. We also provided insight into potential explanations for different clinical findings and biological characteristics between LUSC and LUAD through gene expression analysis. Further validation studies of these biomarkers and biological mechanisms are therefore warranted.
Method
RNA-Seq data processing
The LUAD and LUSC HTSeq read counts data were downloaded from TCGA13 using TCGAbiolinks from R79,80. As of June 2020, there were 529 LUAD and 498 LUSC samples. The samples were normalized using TMM method and standardized using the CPM (read counts per million) function in R. Genes < 1 CPM in over 600 samples were considered noise and discarded to obtain 14,010 genes. The filtered genes were analyzed with different gene selection methods to further narrow down potential gene candidates for biomarkers and pathway analyses.
Gene selection and cancer classification
Gene selection analysis was performed using five different selection methods to generate five independent sets of top genes (Fig. 1). The 5 independent sets were compared, and the resulting overlapped genes were used for cancer classification, biomarker identification, and gene expression analysis. The selection methods used were DGE, PCA, xgboost, lasso, and mRMR. DGE between LUAD and LUSC was performed using the edgeR package81. Though there are other options to perform differential gene expression analysis, edgeR was chosen mostly because of its speed and efficiency in analysis. Also, one of the other popular algorithm, DESeq, has also been shown to yield similar result as edgeR16. After using edgeR analysis and filtering for genes that have FDR < 5E−2 and log(Fold Change) > 0.5, 4702 genes were identified as differentially expressed. Top 500 of the 4702 differentially expressed genes (Table S1) were selected as top features based on their lowest p-values; validation of these genes was performed using random forest with the ranger package82. The top 500 genes from the first principle component in PCA and the top 500 genes ranked from mRMR83 algorithm were selected and validated the same way as the differentially expressed genes. Genes with probability or prediction threshold over 0.5 were selected from Xgboost84 and lasso85 (Table S1), and validated in a similar manner as the other algorithms. For each validation, the data were randomly split into a training set and a testing set in a 7:3 ratio, where the training set was used to construct the model while the testing set was used to evaluate the model’s performance. To compare each selection method more effectively, we split the training sets and testing sets the same way for all validations. We applied fivefold cross validation to decide the optimal parameters for each training model and estimated its accuracy by applying the best determined parameters to the test set. The detailed parameters can be found in the data availability section.
For classification and gene expression analysis, we selected genes that were detected by at least two methods, and they were validated using ranger82. We also used bootstrapping86 with 10,000 replicates to calculate the confidence interval for the accuracy of each method, including the proposed method of classification. The genes that were detected by at least 3 methods were considered candidate biomarkers. Their diagnostic potential was determined and assessed using receiving operating characteristics (ROC) curve analysis. GSE2858224,25, was used as an external dataset to validate the chosen 17-gene classifier.
Prognostic value analysis using Kaplan–Meier plotter
Kaplan–Meier Plotter is an online database that contains comprehensive clinical and microarray data for various cancers, including lung cancer26. Prognostic values of the identified biomarkers in LUAD and LUSC were evaluated using Kaplan–Meier Plotter with each gene used as an univariate analysis. The parameters were set such that the only restricted subtypes were LUAD and LUSC, and the median was used as the cutoff. The rest of the parameters were in the default settings.
Gene expression analysis of selected genes
To further investigate and understand the biological difference between LUAD and LUSC, we performed pathway enrichment analysis using KEGG29, Gene Ontology (GO), and Reactome28. Modified Fisher’s exact tests were performed using DAVID v6.827. Pathways with false discovery rate (FDR) < 5% or p-value less than 0.01 were considered significant. These databases were all accessed in November 2020.
Supplementary Information
Abbreviations
- AUC
Area under curve
- DAVID
The Database for Annotation, Visualization, and Integrated Discovery
- DGE
Differential gene expression
- FPR
False positive rate
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Lasso
Least absolute shrinkage and selection operator
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- mRMR
Minimum redundancy maximum relevance
- NSCLC
Non-small cell lung cancer
- PCA
Principal component analysis
- ROC
Receiving operating characteristics
- TCGA
The Cancer Genome Atlas
- TPR
True positive rate
- Xgboost
Extreme gradient boosting
- QSOX1
Quiescin sulfhydryl oxidase 1
- ARHGAP12
Rho GTPase activating protein 12
- ARHGEF38
Rho guanine nucleotide exchange factor 38
- ELFN2
Extracellular leucine rich repeat and fibronectin type III domain containing 2
- MUC1
Mucin 1, cell surface associated
- GPC1
Glypican 1 GPC1
- NECTIN1
Nectin cell adhesion molecule 1
- PERP
P53 apoptosis effector related to PMP22
- REPS1
RALBP1 associated Eps domain containing 1
- TRIM29
Tripartite motif containing 29
- CELSR2
Cadherin EGF LAG seven-pass G-type receptor 2
- TUBA1C
Tubulin alpha 1c
- S100A2
S100 calcium binding protein A2
- KRT5
Keratin 5
- KRT14
Keratin 14
- KRT6A
Keratin 6A
- TP63
Tumor protein P63
- NAPSA
Napsin A aspartic peptidase
- MLPH
Melanophilin
- DSC3
Desmocollin 3
Author contributions
J.C. proposed the method of overlapping feature selection methods to investigate LUAD and LUSC. J.C. obtained, analyzed, and interpreted the data. J.C. wrote the manuscript and generated the figures and tables. J.D. supervised the study and prepared the figures. J.D. also made substantial suggestions and revisions of the manuscript. All authors read and approved the final manuscript.
Data availability
All data generated and/or analyzed during the current study are included in this published article (and its supplementary information files). The custom code used for data analysis can be accessed at https://github.com/chenjoe569/NSCLC-Research.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92725-8.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N. Engl. J. Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, et al. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relli V, et al. Abandoning the notion of non-small cell lung cancer. Trends Mol. Med. 2019;25(7):585–594. doi: 10.1016/j.molmed.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Xiao J, et al. Eight potential biomarkers for distinguishing between lung adenocarcinoma and squamous cell carcinoma. Oncotarget. 2017;8(42):71759–71771. doi: 10.18632/oncotarget.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu C, et al. Identification of differentially expressed genes between lung adenocarcinoma and lung squamous cell carcinoma by gene expression profiling. Mol. Med. Rep. 2016;14(2):1483–1490. doi: 10.3892/mmr.2016.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhan C, et al. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. J. Thorac. Dis. 2015;7(8):1398–1405. doi: 10.3978/j.issn.2072-1439.2015.07.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian S. Identification of subtype-specific prognostic genes for early-stage lung adenocarcinoma and squamous cell carcinoma patients using an embedded feature selection algorithm. PLoS One. 2015;10(7):e0134630. doi: 10.1371/journal.pone.0134630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhengyan Huang, L. C., Wang, C. Classifying lung adenocarcinoma and squamous cell carcinoma using RNA-Seq data. Cancer Stud. Mol. Med. Open J. 3(2) (2017).
- 11.Cai Z, et al. Classification of lung cancer using ensemble-based feature selection and machine learning methods. Mol. Biosyst. 2015;11(3):791–800. doi: 10.1039/C4MB00659C. [DOI] [PubMed] [Google Scholar]
- 12.Liu XY, et al. Novel regularization method for biomarker selection and cancer classification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020;17(4):1329–1340. doi: 10.1109/TCBB.2019.2897301. [DOI] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rapaport F, et al. Erratum to: Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2015;16:261. doi: 10.1186/s13059-015-0813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapaport F, et al. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol. 2013;14(9):R95. doi: 10.1186/gb-2013-14-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kvam VM, Liu P, Si Y. A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am. J. Bot. 2012;99(2):248–256. doi: 10.3732/ajb.1100340. [DOI] [PubMed] [Google Scholar]
- 17.Hira ZM, Gillies DF. A review of feature selection and feature extraction methods applied on microarray Data. Adv Bioinform. 2015;2015:198363. doi: 10.1155/2015/198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 19.McNeish DM. Using lasso for predictor selection and to assuage overfitting: A method long overlooked in behavioral sciences. Multivar. Behav. Res. 2015;50(5):471–484. doi: 10.1080/00273171.2015.1036965. [DOI] [PubMed] [Google Scholar]
- 20.WeijieSu MB, Candes E. False discoveries occur early on the Lasso path. Ann. Stat. 2017;45(5):2133–2150. [Google Scholar]
- 21.Kalina J, Schlenker A. A robust supervised variable selection for noisy high-dimensional data. Biomed. Res. Int. 2015;2015:320385. doi: 10.1155/2015/320385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 2005;3(2):185–205. doi: 10.1142/S0219720005001004. [DOI] [PubMed] [Google Scholar]
- 23.Peng H, Long F, Ding C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 24.Jabs V, et al. Integrative analysis of genome-wide gene copy number changes and gene expression in non-small cell lung cancer. PLoS One. 2017;12(11):e0187246. doi: 10.1371/journal.pone.0187246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micke P, et al. Gene copy number aberrations are associated with survival in histologic subgroups of non-small cell lung cancer. J. Thorac. Oncol. 2011;6(11):1833–1840. doi: 10.1097/JTO.0b013e3182295917. [DOI] [PubMed] [Google Scholar]
- 26.Gyorffy B, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, et al. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croft D, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(1):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danaee P, Ghaeini R, Hendrix DA. A deep learning approach for cancer detection and relevant gene identification. Pac. Symp. Biocomput. 2017;22:219–229. doi: 10.1142/9789813207813_0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L, et al. Bayesian hyper-LASSO classification for feature selection with application to endometrial cancer RNA-seq data. Sci. Rep. 2020;10(1):9747. doi: 10.1038/s41598-020-66466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang HH, Liu XY, Liang Y. Feature selection and cancer classification via sparse logistic regression with the hybrid L1/2 +2 regularization. PLoS One. 2016;11(5):e0149675. doi: 10.1371/journal.pone.0149675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Relli V, et al. Distinct lung cancer subtypes associate to distinct drivers of tumor progression. Oncotarget. 2018;9(85):35528–35540. doi: 10.18632/oncotarget.26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang HH, Dreyfuss JM, Ramoni MF. A transcriptional network signature characterizes lung cancer subtypes. Cancer. 2011;117(2):353–360. doi: 10.1002/cncr.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miettinen M, Sarlomo-Rikala M. Expression of calretinin, thrombomodulin, keratin 5, and mesothelin in lung carcinomas of different types: An immunohistochemical analysis of 596 tumors in comparison with epithelioid mesotheliomas of the pleura. Am. J. Surg. Pathol. 2003;27(2):150–158. doi: 10.1097/00000478-200302000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Liu S, et al. Transcription factors contribute to differential expression in cellular pathways in lung adenocarcinoma and lung squamous cell carcinoma. Interdiscip. Sci. 2018;10(4):836–847. doi: 10.1007/s12539-018-0300-9. [DOI] [PubMed] [Google Scholar]
- 37.Travis WD, et al. Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: A paradigm shift. J. Thorac. Oncol. 2010;5(4):411–414. doi: 10.1097/JTO.0b013e3181d57f6e. [DOI] [PubMed] [Google Scholar]
- 38.Khayyata S, et al. Value of P63 and CK5/6 in distinguishing squamous cell carcinoma from adenocarcinoma in lung fine-needle aspiration specimens. Diagn. Cytopathol. 2009;37(3):178–183. doi: 10.1002/dc.20975. [DOI] [PubMed] [Google Scholar]
- 39.Ao MH, et al. The utility of a novel triple marker (combination of TTF1, napsin A, and p40) in the subclassification of non-small cell lung cancer. Hum. Pathol. 2014;45(5):926–934. doi: 10.1016/j.humpath.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Travis WD, et al. International association for the study of lung cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011;6(2):244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mantione KJ, et al. Comparing bioinformatic gene expression profiling methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014;20:138–142. doi: 10.12659/MSMBR.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Y, et al. Large scale comparison of gene expression levels by microarrays and RNAseq using TCGA data. PLoS One. 2013;8(8):e71462. doi: 10.1371/journal.pone.0071462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao S, et al. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDermott JE, et al. Challenges in biomarker discovery: Combining expert insights with statistical analysis of complex omics data. Expert Opin. Med. Diagn. 2013;7(1):37–51. doi: 10.1517/17530059.2012.718329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vafaee F, et al. A data-driven, knowledge-based approach to biomarker discovery: Application to circulating microRNA markers of colorectal cancer prognosis. NPJ Syst. Biol. Appl. 2018;4:20. doi: 10.1038/s41540-018-0056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bojesen KB, et al. Nectin-1 binds and signals through the fibroblast growth factor receptor. J. Biol. Chem. 2012;287(44):37420–37433. doi: 10.1074/jbc.M112.345215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schildhaus HU, et al. FGFR1 amplifications in squamous cell carcinomas of the lung: Diagnostic and therapeutic implications. Transl. Lung Cancer Res. 2013;2(2):92–100. doi: 10.3978/j.issn.2218-6751.2013.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salgia R. Fibroblast growth factor signaling and inhibition in non-small cell lung cancer and their role in squamous cell tumors. Cancer Med. 2014;3(3):681–692. doi: 10.1002/cam4.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Z, et al. Nectin-1 expression by squamous cell carcinoma is a predictor of herpes oncolytic sensitivity. Mol. Ther. 2007;15(1):103–113. doi: 10.1038/sj.mt.6300009. [DOI] [PubMed] [Google Scholar]
- 50.Rikitake Y, Mandai K, Takai Y. The role of nectins in different types of cell-cell adhesion. J. Cell Sci. 2012;125(Pt 16):3713–3722. doi: 10.1242/jcs.099572. [DOI] [PubMed] [Google Scholar]
- 51.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: Regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33(31):4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porter AP, Papaioannou A, Malliri A. Deregulation of Rho GTPases in cancer. Small GTPases. 2016;7(3):123–138. doi: 10.1080/21541248.2016.1173767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu K, et al. ARHGEF38 as a novel biomarker to predict aggressive prostate cancer. Genes Dis. 2020;7(2):217–224. doi: 10.1016/j.gendis.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gentile A, et al. Met-driven invasive growth involves transcriptional regulation of Arhgap12. Oncogene. 2008;27(42):5590–5598. doi: 10.1038/onc.2008.173. [DOI] [PubMed] [Google Scholar]
- 55.Zhang YQ, et al. Overexpression of CST4 promotes gastric cancer aggressiveness by activating the ELFN2 signaling pathway. Am. J. Cancer Res. 2017;7(11):2290–2304. [PMC free article] [PubMed] [Google Scholar]
- 56.Knutsvik G, et al. QSOX1 expression is associated with aggressive tumor features and reduced survival in breast carcinomas. Mod. Pathol. 2016;29(12):1485–1491. doi: 10.1038/modpathol.2016.148. [DOI] [PubMed] [Google Scholar]
- 57.Xu T, et al. MUC1 downregulation inhibits non-small cell lung cancer progression in human cell lines. Exp. Ther. Med. 2017;14(5):4443–4447. doi: 10.3892/etm.2017.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohlgraf KG, et al. Contribution of the MUC1 tandem repeat and cytoplasmic tail to invasive and metastatic properties of a pancreatic cancer cell line. Cancer Res. 2003;63(16):5011–5020. [PubMed] [Google Scholar]
- 59.Hollingsworth MA, Swanson BJ. Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer. 2004;4(1):45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 60.Yanagi T, et al. Loss of TRIM29 alters keratin distribution to promote cell invasion in squamous cell carcinoma. Cancer Res. 2018;78(24):6795–6806. doi: 10.1158/0008-5472.CAN-18-1495. [DOI] [PubMed] [Google Scholar]
- 61.Chen C, Shan H. Keratin 6A gene silencing suppresses cell invasion and metastasis of nasopharyngeal carcinoma via the betacatenin cascade. Mol. Med. Rep. 2019;19(5):3477–3484. doi: 10.3892/mmr.2019.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Milovanovic IS, Stjepanovic M, Mitrovic D. Distribution patterns of the metastases of the lung carcinoma in relation to histological type of the primary tumor: An autopsy study. Ann. Thorac. Med. 2017;12(3):191–198. doi: 10.4103/atm.ATM_276_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 64.Petitjean A, et al. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26(15):2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 65.Labbe C, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC) Lung Cancer. 2017;111:23–29. doi: 10.1016/j.lungcan.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Sun Q. TP53 mutations, expression and interaction networks in human cancers. Oncotarget. 2017;8(1):624–643. doi: 10.18632/oncotarget.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen M, et al. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8(1):133–144. doi: 10.18632/oncotarget.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee JE, et al. Clinical characteristics of pulmonary embolism with underlying malignancy. Korean J. Intern. Med. 2010;25(1):66–70. doi: 10.3904/kjim.2010.25.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chew HK, et al. The incidence of venous thromboembolism among patients with primary lung cancer. J. Thromb. Haemost. 2008;6(4):601–608. doi: 10.1111/j.1538-7836.2008.02908.x. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, et al. Prevalence and associations of VTE in patients with newly diagnosed lung cancer. Chest. 2014;146(3):650–658. doi: 10.1378/chest.13-2379. [DOI] [PubMed] [Google Scholar]
- 71.Papageorgiou C, et al. Lobectomy and postoperative thromboprophylaxis with enoxaparin improve blood hypercoagulability in patients with localized primary lung adenocarcinoma. Thromb. Res. 2013;132(5):584–591. doi: 10.1016/j.thromres.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 72.Stoiber D, Assinger A. Platelet–leukocyte interplay in cancer development and progression. Cells. 2020;9(4):855. doi: 10.3390/cells9040855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doyle EL, et al. CD63 is an essential cofactor to leukocyte recruitment by endothelial P-selectin. Blood. 2011;118(15):4265–4273. doi: 10.1182/blood-2010-11-321489. [DOI] [PubMed] [Google Scholar]
- 74.Lucchetta M, et al. Distinct signatures of lung cancer types: Aberrant mucin O-glycosylation and compromised immune response. BMC Cancer. 2019;19(1):824. doi: 10.1186/s12885-019-5965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farztdinov V, McDyer F. Distributional fold change test—A statistical approach for detecting differential expression in microarray experiments. Algorithms Mol. Biol. 2012;7(1):29. doi: 10.1186/1748-7188-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dembele D, Kastner P. Fold change rank ordering statistics: A new method for detecting differentially expressed genes. BMC Bioinform. 2014;15:14. doi: 10.1186/1471-2105-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, et al. Lung cancer and pulmonary embolism: What is the relationship? A review. J. Cancer. 2018;9(17):3046–3057. doi: 10.7150/jca.26008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Z, Liu D. Pathogenesis of molecular signaling pathways changes in smoking-induced lung cancer. Zhongguo Fei Ai Za Zhi. 2009;12(11):1202–1205. doi: 10.3779/j.issn.1009-3419.2009.11.14. [DOI] [PubMed] [Google Scholar]
- 79.Colaprico A, et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silva TC, et al. TCGA Workflow: Analyze cancer genomics and epigenomics data using Bioconductor packages. F1000Res. 2016;5:1542. doi: 10.12688/f1000research.8923.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wright MN, Ziegler A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 2017;77(1):1–17. doi: 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 83.De Jay N, et al. mRMRe: An R package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29(18):2365–2368. doi: 10.1093/bioinformatics/btt383. [DOI] [PubMed] [Google Scholar]
- 84.Tianqi Chen, T. H. et al. xgboost: Extreme Gradient Boosting (2020).
- 85.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33(1):1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Canty, A. & Ripley, B. D. boot: Bootstrap R (S-plus) Functions (2020).
- 87.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and/or analyzed during the current study are included in this published article (and its supplementary information files). The custom code used for data analysis can be accessed at https://github.com/chenjoe569/NSCLC-Research.