Graphical abstract
Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; NLRP3, nucleotide-binding oligomerization domain-like receptor containing pyrin domain 3; S/M/N/E protein, spike/membrane/nucleocapsid/envelope protein; ORF3a/8a/8b, open reading frame 3a/8a/8b; ARDS, acute respiratory distress syndrome; ALI, acute lung injury; IL-18/1β/6/8/10/1RA, interleukin-18/1β/6/8/10/1RA; TNF, tumor necrosis factor; ASC, apoptosis-associated speck-like protein containing a caspase recruitment domain; LRR, , leucine-rich repeat; PYD, pyrin domain; CARD, caspase recruitment domain; PAMPs, pathogen-associated molecular patterns; DAMPs, damage-associated molecular patterns; LPS, lipopolysaccharide; NF-κB, nuclear factor kappa B; PKR, double-stranded RNA-dependent protein kinase; NEK7, never in mitosis A-related kinase 7; ERGIC, endoplasmic reticulum-Golgi apparatus intermediate compartment; ROS, reactive oxygen species; mtDNA, mitochondrial DNA; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane serine protease 2; Ang II, angiotensin II; AT1R, Ang II type 1 receptor; MBL, mannan-binding lectin; MASP2, MBL-associated serine protease 2; MAC, membrane attack complex; ERK, extracellular signal-regulated kinase; P2 × 7, P2X purinergic receptor 7; MEK, mitogen-activated protein kinase; IFN, interferon; CXCL10, C-X-C motif chemokine ligand 10; LDH, lactate dehydrogenase; NETs, neutrophil extracellular traps; BBB, blood-brain barrier; CNS, central nervous system; MERS-CoV, middle east respiratory syndrome coronavirusus
Keywords: NLRP3 inflammasomeome, SARS-CoV-2, COVID-19, Pathogenesis, Inhibitor
Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), exhibits a wide spectrum of clinical presentations, ranging from asymptomatic cases to severe pneumonia or even death. In severe COVID-19 cases, an increased level of proinflammatory cytokines has been observed in the bloodstream, forming the so-called “cytokine storm”. Generally, nucleotide-binding oligomerization domain-like receptor containing pyrin domain 3 (NLRP3) inflammasome activation intensely induces cytokine production as an inflammatory response to viral infection. Therefore, the NLRP3 inflammasome can be a potential target for the treatment of COVID-19. Hence, this review first introduces the canonical NLRP3 inflammasome activation pathway. Second, we review the cellular/molecular mechanisms of NLRP3 inflammasome activation by SARS-CoV-2 infection (e.g., viroporins, ion flux and the complement cascade). Furthermore, we describe the involvement of the NLRP3 inflammasome in the pathogenesis of COVID-19 (e.g., cytokine storm, respiratory manifestations, cardiovascular comorbidity and neurological symptoms). Finally, we also propose several promising inhibitors targeting the NLRP3 inflammasome, cytokine products and neutrophils to provide novel therapeutic strategies for COVID-19.
1. Introduction
Since the first cases of atypical viral pneumonia were reported in December 2019, a global infection has disseminated widely with coronavirus disease 2019 (COVID-19) [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent behind the ongoing COVID-19 pandemic. SARS-CoV-2, an enveloped single-stranded positive-sense RNA virus, belongs to the genus Betacoronavirus within the family Coronaviridae [2]. SARS-CoV-2 is an enveloped virus that is comprised of spike (S), membrane (M), nucleocapsid (N) and envelope (E) proteins [3]. In addition, this virus also encodes a set of accessory proteins, including two ion-channel proteins known as viroporins (open reading frame 3a (ORF3a) and ORF8a) [4]. Most COVID-19-infected individuals present with mild flu-like symptoms (low-grade fever and cough), while 5–10 % are severe cases suffering from a compromised respiratory system and life-threatening pneumonia [1]. In general, severe cases result in acute respiratory distress syndrome (ARDS) and acute lung injury (ALI), which are known to be induced by a storm of inflammatory cytokines, especially interleukin (IL)-1β, IL-6, and tumor necrosis factor-α (TNF-α) [5]. Apart from the typical respiratory symptoms, other cardiovascular and neurological manifestations have also been reported in COVID-19 [6,7].
Generally, the production of inflammatory cytokines in host cells necessitates inflammasomes. Inflammasomes are protein complexes formed in the cytosol responding to different stimuli [8]. Of all the inflammasomes, the nucleotide-binding oligomerization domain-like receptor containing pyrin domain 3 (NLRP3) inflammasome is the best studied inflammasome, comprised of NLRP3 protein, the adapter apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) and pro-caspase-1 [9]. The NLRP3 protein consists mainly of three parts, namely, a leucine-rich repeat (LRR) domain, a central nucleotide-binding domain known as NACHT and a pyrin domain (PYD). ASC is composed of two parts: PYD and a caspase recruitment domain (CARD). In response to danger signals, NLRP3 binds to ASC via interactions between PYDs. Subsequently, ASC recruits pro-caspase-1 through CARD. The homophilic interactions of domains are important for the structural mechanism of NLRP3 inflammasome activation [10].
Activation of the NLRP3 inflammasome leads to proteolytic cleavage of inactive pro-caspase-1 to form caspase-1, which subsequently converts the cytokine precursors pro-IL-1β and pro-IL-18 into mature IL-1β and IL-18 [11]. In general, the activation of the NLRP3 inflammasome occurs in two steps: priming and activation. In the first step (priming), pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), such as lipopolysaccharide (LPS), are recognized by Toll-like receptors, resulting in the activation of nuclear factor kappa B (NF-κB)-mediated signaling. NF-κB upregulates the transcription of inflammasome-related components such as inactive NLRP3, pro-caspase-1, pro-IL-1β and pro-IL-18 [12]. The second step (activation) is mainly triggered by ATP, pore-forming toxins, viral RNA and particulate matter, leading to the assembly of NLRP3, ASC, and pro-caspase-1 into a complex [13]. Since NLRP3-activating factors are chemically diverse, NLRP3 seems to sense some common cellular signals instead of contacting its activators directly. It is worth noting that several molecular and cellular events, including K+ efflux [14], Ca2+ signaling [15], reactive oxygen species (ROS) production [16], mitochondrial dysfunction, and lysosomal rupture [17,18], have been proposed to describe the second step of inflammasome activation. In addition to the signals mentioned above, guanylate-binding protein 5 [19], double-stranded RNA-dependent protein kinase (PKR) [20] and never in mitosis A-related kinase 7 (NEK7) [21] have also been identified as important regulators of NLRP3 inflammasome activation.
As a vital part of the innate immune system, the NLRP3 inflammasome is important for antiviral host defenses, whereas its aberrant activation can lead to pathological tissue injury during infection. Previous studies have characterized the involvement of the NLRP3 inflammasome in the pathogenesis of ARDS and ALI [22,23], which are respiratory symptoms in severe COVID-19 cases [4]. Additionally, cytokine storm, the main cause of inflammation in severe COVID- 19 cases [23], implicates the participation of the NLRP3 inflammasome in the pathogenesis of COVID-19 [24,25]. Accordingly, several studies have reported hyperactivation of the NLRP3 inflammasome in COVID-19 [26,27]. In this context, a better understanding of NLRP3 inflammasome activation during COVID-19 is required for further investigation and better treatment of COVID-19.
Herein, considering the significant involvement of the NLRP3 inflammasome in COVID-19, this review describes the possible pathways of NLRP3 inflammasome activation in response to SARS-CoV-2 infection, the role of the NLRP3 inflammasome in the pathogenesis of COVID-19 and promising treatments based on the inhibition of the NLRP3 inflammasome in COVID-19.
2. Mechanisms of NLRP3 inflammasome activation
The NLRP3 inflammasome, as an important part of the innate immune system, is closely related to inflammation and immunopathology. As mentioned above, the activation of the NLRP3 inflammasome includes 2 steps: priming and activation [9].
In the priming stage, PAMPs or DAMPs (such as LPS) are recognized by Toll-like receptors, initiating the activation of a few transcription factors, such as NF-κB, via intricate signals. The NF-κB pathway subsequently upregulates the expression of NLRP3 inflammasome-related components [12], including NLRP3, proinflammatory cytokines and pro-caspase-1 [28]. The second step (activation) is the oligomerization of NLRP3 and the assembly of NLRP3, ASC, and pro-caspase-1 into a complex. This is typically triggered by ATP, pore-forming toxins, viral RNA and particulate matter [13]. Cellular signaling events can thus be initiated, such as K+ efflux [14], Ca2+ flux [15], ROS production [16], mitochondrial dysfunction [29], lysosomal rupture [17], and chloride intracellular channel-dependent Cl− efflux [30]. Apart from the signaling events, several important regulators of NLRP3 inflammasome activation have also been reported, including PKR [31], guanylate-binding protein 5 [19] and NEK7 [21].
In general, K+ efflux is a crucial step for NLRP3 inflammasome activation. After ATP stimulation, P2X purinergic receptor 7 (P2 × 7), an ATP-gated ion channel, promotes Ca2+ and Na+ influx and coordinates with K+ channels to mediate K+ efflux [32]. The decrease in intracellular K+ concentration thus triggers NLRP3 inflammasome activation in both canonical and noncanonical inflammasome pathways [14,33]. For Ca2+ signaling, either the opening of plasma membrane channels or the release of ER-linked intracellular Ca2+ stores can lead to Ca2+ flux into the cytosol [34], thus promoting NLRP3 complex formation [15]. Additionally, Ca2+ mobilization and mitochondrial Ca2+ overload can induce mitochondrial dysfunction, promoting NLRP3 inflammasome activation [35]. Notably, Ca2+ flux and K+ efflux are often coordinated in NLRP3 inflammasome activation. For instance, ATP induces weak Ca2+ influx via P2 × 7, coordinating K+ efflux [32]. Subsequently, K+ efflux promotes the release of ER-linked Ca2+ stores [36]. Additionally, Cl− efflux via chloride intracellular channel proteins is necessary for NLRP3 inflammasome activation. It acts downstream of the K+ efflux-ROS axis to promote the assembly and activation of the NLRP3 inflammasome [30]. A recent study also proposed that K+ efflux promotes NLRP3 oligomerization, while Cl- efflux facilitates ASC polymerization [37]. Moreover, mitochondrial dysfunction [29], production of ROS [16] and mitochondrial DNA (mtDNA) [38] are also critical mediators of NLRP3 inflammasome activation. In addition, lysosomal rupture and release of particulates into the cytoplasm can activate K+ efflux and Ca2+ influx, thus leading to NLRP3 inflammasome activation [39]. Above all, the mechanisms of NLRP3 inflammasome activation are various. In particular, many NLRP3 inflammasome activation pathways involve either K+ efflux and/or Ca2+ flux. Of note, K+ efflux, ROS and Cl− efflux have been proven to be important for the NEK7-NLRP3 interaction [9], which further implies the intricate correlation of diverse signal events and regulators.
With the activation of the NLRP3 inflammasome, mature caspase-1 leads to the cleavage of pro-IL-1β/18 to form IL-1β/18 and the cleavage of gasdermin D into fragments forming pores on the cell membrane. Cytosolic content is released through these pores, causing the unrestrained dissemination of inflammatory factors such as IL-1β/18, further resulting in a form of cell death termed pyroptosis [40] (Fig. 1 ).
Fig. 1.
Schematic diagram of the canonical NLRP3 inflammasome activation mechanisms. In step 1 (priming), Toll-like receptors are stimulated by PAMPs or DAMPs (such as LPS), upregulating the expression of NLRP3 inflammasome-related components such as inactive NLRP3, NEK7, pro-caspase-1, pro-IL-1β and pro-IL-18 via the NF-κB pathway. Step 2 (activation) is typically triggered by multiple stimuli in the canonical pathway, such as ATP, pore-forming toxins, viral RNA, and particulate matter. A number of molecular/cellular signaling events have been proposed as the key mechanisms of NLRP3 inflammasome activation, including K+ efflux, Ca2+ flux, ROS production, mtDNA, mitochondrial dysfunction, lysosomal rupture and chloride intracellular channel-dependent Cl− efflux. Subsequently, the oligomerization and recruitment of NLRP3, NEK7, ASC and pro-caspase-1 are initiated, leading to the maturation of pro-IL-1β/18 into IL-1β/18 and pyroptosis by the cleavage of gasdermin D.
3. NLRP3 inflammasome activation in response to SARS-CoV-2 infection
The canonical NLRP3 inflammasome activation mechanisms have been described above, which discussed that K+ efflux, Ca2+ flux, ROS, mitochondrial damage and lysosomal rupture are important cellular signals for inflammasome activation. For SARS-CoV-2-induced NLRP3 inflammasome activation, the possible mechanisms are mainly based on canonical activation pathways. SARS-CoV-2 can activate the NLRP3 inflammasome either directly or via diverse cellular/molecular signaling events.
3.1. Ca2+ transport through the SARS-CoV E protein channel activates the NLRP3 inflammasome
SARS-CoV encodes viroporins with ion channel activity, such as protein E, ORF3a and ORF8a. As previously reported, these viroporins oligomerize and form pores during viral infections, which compromise normal physiological homeostasis in host cells and thus lead to viral pathogenicity [41]. Of these viroporins, protein E has been indicated to play a vital role in viral virulence [42]. Notably, deletion of the SARS-CoV E gene can attenuate SARS-CoV infection [43,44], highlighting the importance of the E protein in the high virulence of SARS-CoV.
The role of the E protein in the virulence of SARS-CoV has been widely studied. First, it has been stated that SARS-CoV overstimulates the NF-κB inflammatory pathway in the presence of the E protein [45]. Moreover, the CoV E gene encodes a small transmembrane protein highly expressed during infection. It mainly localizes to the Golgi apparatus and the endoplasmic reticulum-Golgi apparatus intermediate compartment (ERGIC), where it can promote virus production and morphogenesis [46]. Additionally, it is worth noting that the ion channel activity of the E protein allows the CoV E protein to assemble in membranes, forming pentameric protein-lipid pores to induce ion transport [47]. Furthermore, it has been proposed that the SARS-CoV E protein exhibits a slight preference for cations (Na+, K+) over anions (Cl−) when reconstituted in membranes mimicking the charge and composition of the ERGIC/Golgi [47]. These ion channel properties of the E protein and the relevance of the E protein to virulence have further encouraged a recent study, which indicated that the E protein forms a Ca2+-permeable channel in ERGIC/Golgi membranes [5]. As a result, Ca2+ leakage through E protein ion channels in ERGIC/Golgi membranes can induce ionic imbalances within cells, which can exacerbate inflammation and immunopathology.
Increasing evidence has shown that NLRP3 detects RNA viruses by sensing the cellular damage or distress induced by viroporins [48,49]. As mentioned above, Ca2+ flux is a vital signaling event for the activation of the NLRP3 inflammasome. With Ca2+ leakage through the E protein ion channel in ERGIC/Golgi membranes, increased cytosolic Ca2+ may promote NLRP3 inflammasome activation. In accordance, a recent study proved that the SARS-CoV E protein triggers NLRP3 inflammasome activation through its Ca2+ transport ability [5] (Fig. 2 ).
Fig. 2.
Schematic diagram of NLRP3 inflammasome activation in response to SARS-CoV-2 infection. The NLRP3 inflammasome can be activated by SARS-CoV-2 infection through the following possible pathways: 1) the E protein of SARS-CoV can induce Ca2+ transport in the ERGIC/Golgi to trigger NLRP3 inflammasome activation; 2) K+ efflux and ROS production induced by the ORF3a protein of SARS-CoV activate the NLRP3 inflammasome; 3) contact of the SARS-CoV S protein with ACE2 diminishes the degradation of Ang II, resulting in Ang II accumulation, which can activate the NLRP3 inflammasome; 4) the complement cascade induced by the N protein-MBL-MASP2 axis may lead to NLRP3 inflammasome activation via different functions of C3a, C5a and MAC; and 5) the direct interaction of the SARS-CoV ORF8b protein with the NLRP3 inflammasome LRR domain may also cause inflammasome activation.
3.2. K+ efflux and ROS production induced by the ORF3a protein activate the NLRP3 inflammasome
The ORF3a protein, as mentioned above, is also a SARS-CoV viroporin. Similar to the E protein, ORF3a also has ion channel activity and is thought to act as a K+ channel [50]. It has been identified to cause host cell lysosome dysfunction and initiate caspase-1 activation either directly or through increased K+ efflux. Additionally, ORF3a can lead to NF-κB-mediated upregulation of the cytokine IL-1β and pyroptosis, contributing to host inflammatory responses to SARS-CoV infection and tissue damage [51,52].
Considering the indispensable role of NF-κB and K+ efflux in the canonical inflammasome pathway, NLRP3 inflammasome activation by the ORF3a protein was discussed in a recent study [52]. The authors of this study stated that the ion channel activity of the 3a protein is required for inflammasome-mediated IL-1β secretion and that SARS-CoV 3a is sufficient to trigger activation of the NLRP3 inflammasome. In addition, it has been observed that both K+ efflux and ROS production are involved in the IL-1β secretion induced by the SARS-CoV 3a protein [52]. As a result, this suggests a mechanism that the intracellular ionic concentration disruption and mitochondrial damage induced by ORF3a trigger the activation of the NLRP3 inflammasome (Fig. 2).
3.3. Ang II accumulation induced by the S protein-ACE2 pathway activates the NLRP3 inflammasome
SARS-CoV mediates viral membrane fusion via the S protein expressed on the surface of the virus [53]. The S protein can be cleaved into 2 subunits: S1 and S2. S1 directly binds to angiotensin-converting enzyme 2 (ACE2) [54], followed by priming/processing by transmembrane serine protease 2 (TMPRSS2) [55]. Subsequently, S2 is exposed by the cleavage, which facilitates the fusion of the virus and host cell membranes, leading to entry of virus into host cells [56]. It has been shown that the SARS-CoV-2 S protein has almost 80 % amino acid identity with the SARS-CoV S protein. Additionally, the binding affinity of the SARS-CoV-2 S protein to human ACE2 is similar to that of the SARS-CoV S protein [57].
ACE2 is one of the members of the membrane-bound carboxydipeptidase family, which degrades angiotensin II (Ang II) to generate angiotensin 1–7 and negatively regulates various Ang II activities mediated by the Ang II type 1 receptor (AT1R) [58]. Due to the increasing replication of the virus, the S protein-ACE2 contact is enhanced, while the ACE2-Ang II interaction is attenuated. Therefore, the degradation of Ang II is diminished as well. As a result, noncompeting Ang II accumulation occurs, leading to ALI via AT1R activation [59]. Notably, a recent study revealed that Ang II can activate the NLRP3 inflammasome in podocytes, which is partially attributed to mitochondrial dysfunction [60]. In this context, it can be inferred that SARS-CoV-2 can activate the NLRP3 inflammasome through the S protein-ACE2 pathway via Ang II accumulation (Fig. 2).
3.4. The complement cascade induced by SARS-CoV-2 infection activates the NLRP3 inflammasome
The N proteins of SARS-CoV-2 can activate the complement cascade in a mannan-binding lectin (MBL)-dependent manner. The complex formation of MBL and MBL-associated serine protease 2 (MASP2) subsequently induces the release of C3a and C5a anaphylatoxins and the formation of a nonlytic C5b/C9 membrane attack complex (MAC) [[61], [62], [63]]. Accordingly, it has been reported that striking deposition of C5b-9, C4d and MASP2 exists in the microvasculature of 2 organ systems from patients with SARS-CoV-2 infection and severe respiratory failure [63]. This highlights the pathophysiologic importance of complement in COVID-19.
Moreover, several studies have indicated that the initiation of the complement cascade can lead to the activation of the NLRP3 inflammasome. For example, C3a, a potent effector of the complement cascade, engages the C3a receptor and upregulates ATP efflux in an extracellular signal-regulated kinase 1/2 (ERK1/2)-dependent fashion [64]. Increased ATP efflux in turn elevates NLRP3 inflammasome-induced IL-1β/18 secretion via P2 × 7 [64], an ATP-gated ion channel for K+ efflux in NLRP3 inflammasome activation [65]. Furthermore, C5a-C5a receptor 2 signaling has been found to activate the NLRP3 inflammasome by upregulating dsRNA-dependent PKR, an NLRP3 inflammasome activation regulator mentioned above [20], via the mitogen-activated protein kinase (MEK)/ERK pathway and type I interferon (IFN) signaling [66]. In addition, MAC, formed by self-polymerization of terminal components of the complement cascade, contacts the surfaces of target cells and forms transmembrane pores or channels in the cell membrane to influence cytolysis [67]. A recent study revealed that MAC insertion triggers Ca2+ influx and increases cytosolic Ca2+ concentration, inducing mitochondrial damage and NLRP3 inflammasome activation [68].
In conclusion, the complement system contributes to COVID-19 pathology and is closely related to the activation of the NLRP3 inflammasome. In this context, it can be inferred that SARS-CoV-2 infection can activate the NLRP3 inflammasome via the complement cascade pathway (Fig. 2) [61,62].
3.5. ORF8b protein activates the NLRP3 inflammasome by binding to its LRR domain directly
ORF8b, an 84-residue polypeptide, is formed after the cleavage of ORF8 in the late phase of SARS infection. It has been deemed that the cleavage of ORF8 grants an evolutionary advantage to the virus because ORF8b promotes virus replication in the presence of IFN [69,70]. These findings indicate the contribution of ORF8b to SARS-CoV pathology. Regarding the specific cellular mechanism, it has been reported that ORF8b forms intracellular aggregates, which is dependent on a valine at residue 77, leading to lysosomal stress, autophagy, and cell death [71].
Inflammatory monocyte-macrophages in the lungs have been considered vital mediators of SARS-CoV pathology [72] and serve as a critical link between intracellular aggregates and inflammasome activation [73]. In this scenario, the effect of ORF8b on inflammasome activation in macrophages has been studied. It has been suggested that ORF8b induces NLRP3 inflammasome activation in macrophages by binding to the NLRP3 LRR domain directly (Fig. 2) [71].
4. Role of the NLRP3 inflammasome in COVID-19 pathogenesis
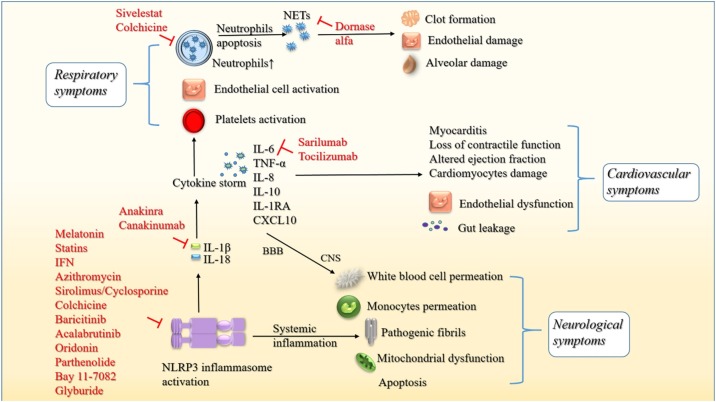
COVID-19 clinical presentations are variable in severity, ranging from asymptomatic cases to severe pneumonia and ARDS, even including death. In symptomatic cases, COVID-19 typically presents as an influenza-like respiratory illness, with fever, cough, dyspnea, and malaise/myalgia. In atypical presentations, it generally manifests as extrapulmonary involvement, such as cardiovascular comorbidity, gastrointestinal symptoms, neurological manifestations and multiorgan failure (liver, kidneys, heart). To date, many studies have provided a better understanding of COVID-19 pathogenesis. Here, this review mainly describes the involvement of the NLRP3 inflammasome in COVID-19-related cytokine storm and respiratory, cardiovascular and neurological manifestations and complications (Fig. 3 ).
Fig. 3.
Schematic diagram of the NLRP3 inflammasome in COVID-19 pathogenesis and promising inhibitors. NLRP3 inflammasome activation during COVID-19 leads to the production of IL-1β/18, facilitating the formation of cytokine storm (IL-6/8/10/1RA, TNF-α and CXCL10). 1) For respiratory symptoms, proinflammatory cytokines further activate platelets, endothelial cells and neutrophils. Higher levels of neutrophils result in the escalated production of NETs, causing clot formation, endothelial damage and alveolar damage. 2) For cardiovascular symptoms, inflammatory cytokines induced by the NLRP3 inflammasome promote endothelial dysfunction, myocarditis and cardiomyocyte damage. Additionally, gut leakage, such as LPS, can exacerbate cardiac inflammation via NLRP3 inflammasome activation. 3) For neurological symptoms, cytokines can cross the BBB to enter the CNS and induce the permeation of peripheral white blood cells and monocytes. Moreover, systemic inflammation induced by the NLRP3 inflammasome leads to the formation of pathogenic fibrils, mitochondrial dysfunction and apoptosis, which can result in neurodegeneration. Promising inhibitors of the NLRP3 inflammasome, cytokine products and the neutrophil-NET axis have been shown.
4.1. NLRP3 inflammasome and cytokine storm in COVID-19
Cytokine release syndrome is a systemic inflammatory response triggered by various stimuli, including drugs and infections [74]. For instance, respiratory viral infection, such as influenza, has been characterized as a stimuli for the development of cytokine release syndrome [25]. Typically, patients undergo a robust cytokine-mediated response to these stimuli, which often is associated with fever, hypotension and hypoxemia [74,75]. The subsequent syndrome can either be mild and relieved spontaneously or develop into persistent high-grade fevers, vasodilatory shock with hemodynamic instability, or severe hypoxemia requiring mechanical ventilation [74,75]. SARS-CoV-2 infection initiates a cytokine storm that has similar characteristics to cytokine release syndrome as stated above [76]. The escalation of IL-6, IL-1β, TNF-α, IL-8, IL-10, IL-1RA, and C-X-C motif chemokine ligand 10 (CXCL10) has been observed in severe COVID-19 patients with cytokine storm [77,78]. These cytokines can produce eosinopenia and lymphocytopenia (low counts of eosinophils, CD8 + T cells, natural killer and naive T-helper cells) and induce naive B-cell activation, T-helper cell 17 lymphocyte differentiation, and the stimulation of monocyte and neutrophil recruitment [[79], [80], [81]].
As mentioned above, the NLRP3 inflammasome is one of the most important innate immune components, which facilitates inflammation by producing IL-1β/18 and causing pyroptosis. With the development of the inflammatory cascade, IL-1β and IL-18 promote the release of additional NLRP3 cytokines, such as IL-6 [82]. Additionally, a recent study indicated that activation of the NLRP3 inflammasome by S. suis leads to the production of IL-1β, resulting in cytokine release syndrome [24]. Accordingly, the positive correlation of IL-18 and caspase-1 with other inflammatory markers, such as C-reactive protein, lactate dehydrogenase (LDH) and IL-6, has been reported with the inflammasome activation in COVID-19 [27]. This further implies the importance of the NLRP3 inflammasome for the formation of cytokine storm and the development of COVID-19 (Fig. 3).
4.2. NLRP3 inflammasome in COVID-19 respiratory symptoms
Based on the clinical data of SARS-CoV-2-infected patients, radiologic evidence has shown that lung edema is quite common. Lung edema is a manifestation of ALI and can develop into hypoxemia and ARDS [83]. COVID-19 ARDS manifests with diffuse alveolar damage, increased epithelial and endothelial cell permeability, leakage of fluid into the pulmonary interstitium, fibrin-rich hyaline membranes, and significant disruption of gas exchange, which finally results in hypoxia and respiratory failure [[84], [85], [86]]. In addition, COVID-19 infection is similar to bacterial and viral pneumonia. Both COVID-19 ARDS and pneumonia are associated with significant morbidity, high levels of mortality and a poor prognosis. Many studies have reported significant levels of immune dysregulation in the lungs of patients with severe COVID-19 pneumonia or ARDS.
Of note, the inflammatory biomarkers of ARDS associated with cytokine storm are IL-6, IL-8, IL-18, and TNF [87]. Additionally, it has recently been reported that increased levels of IL-18 are consistent with increased mortality in ARDS [88]. In this context, it can be inferred that the production of inflammatory cytokines largely contributes to the exacerbation of ARDS [78]. These inflammatory mediators in the pulmonary vasculature promote the development of ARDS by inducing the activation of vascular endothelial cells [89], platelets [90] and neutrophils [91]] and ultimately the formation of platelet neutrophil complexes at the surface of the endothelium [92]. As described above, the NLRP3 inflammasome is closely related to the formation of the severe cytokine storm. Additionally, previous studies have revealed the important role of the NLRP3 inflammasome in the pathogenesis of ARDS and ALI [22,23]. In this scenario, it is obvious that the NLRP3 inflammasome contributes to the development of COVID-19 ARDS. Notably, high mobility group box 1 protein has been proposed to activate the NLRP3 inflammasome, leading to ARDS/ALI in the lungs of COVID-19 patients [93,94]. Furthermore, neutrophil activation has been widely recognized as an amplifier of lung injury in ARDS [95]. In addition, COVID-19 patients exhibit higher neutrophil counts in blood [96]. Neutrophils undergo a particular type of cell death known as NETosis [97]. Subsequently, expiring neutrophils produce thread-like extracellular structures termed neutrophil extracellular traps (NETs) [97]. NETs originally exert a protective function by forming mesh-like structures to trap microbes. However, aberrant NET production has been indicated as a vital factor contributing to ALI and ARDS [98]. Intriguingly, IL-1β secreted by the NLRP3 inflammasome is a key trigger of NETs [99]. In ALI/ARDS caused by infections, due to the chemokines produced by epithelial cells and macrophages, neutrophils migrate massively into the alveoli [98]. With stimulants such as IL-1β in the alveoli, migrated neutrophils thus produce aberrant amounts of NETs, causing potent lung injury [100]. Notably, neutrophil elastase can lead to proinflammatory cytokine production by cleaving cadherins and endothelial cytoskeletal proteins [100], which implies a feedback loop in IL-1β and neutrophils. In conclusion, the elevated level of IL-1β produced by the NLRP3 inflammasome further activates neutrophils, leading to increased levels of NET production. High levels of NETs cause increased clot formation, endothelial damage, and alveolar damage associated with COVID-19.
In summary, the interrelationship of the NLRP3 inflammasome, cytokine storm and NETs emphasizes the vital involvement of the NLRP3 inflammasome in the pathogenesis of COVID-19 ALI/ARDS (Fig. 3).
4.3. NLRP3 inflammasome in COVID-19 cardiovascular symptoms
Many investigations have focused on severe acute respiratory tract distress in COVID-19, yet few are available on the cardiovascular manifestations in COVID-19. It has been observed that patients with coexisting cardiovascular disorders tend to have worse outcomes, including severe cardiac injury and mortality [6]. COVID-19 can exacerbate pre-existing cardiovascular conditions such as ischemic heart disease and chronic heart failure and cause myocardial injury, arrhythmia, acute coronary syndrome, myocarditis, cardiogenic shock, venous thromboembolism, stroke and pulmonary embolism [101].
A recent study stated that SARS-CoV-2 infection may disseminate from the respiratory tract into the cardiovascular system via the circulatory or lymphatic system [102]. With ACE2 receptors expressed in the heart, the COVID-19 virus may trigger a proinflammatory cascade, as it does in the lung. The recruitment of various adaptors, such as mitochondrial antiviral signaling proteins, can increase the expression of the NLRP3 inflammasome and thus the production of inflammatory cytokines (TNF-α, IL-6 and IL-1β) [103,104]. It further leads to myocarditis, loss of contractile function, altered ejection fraction, damage to cardiomyocytes and release of cardiac injury markers [105]. Additionally, endothelial cells are a constitutive and integral part of the vascular system, and endothelial dysfunction can intrinsically cause vascular diseases [106]. Any damage to the endothelium directly results in altered vascular tone and vasoconstriction, leading to ischemia, vascular inflammation, coagulation and thrombosis [107]. A recent study found a high prevalence of hypertension in COVID-19 patients and speculated on the critical role of endothelial dysfunction in COVID-19 [108]. Notably, NLRP3 inflammasome activation has been considered a vital function of endothelial cells in COVID-19 inflammation [106]. Interestingly, another study has suggested that the gut microbiota composition and the gut blood barrier are altered in various forms of cardiovascular disease [109]. Of note, this study reported that the dysfunctional gut mucosal barrier facilitates passive leakage of microbial products such as LPS into the circulation, thus inducing NLRP3 inflammasome activation and contributing to cardiac manifestations in COVID-19 patients [109].
In conclusion, all the findings above highlight the profound participation of the NLRP3 inflammasome in the pathogenesis of COVID-19 cardiovascular symptoms (Fig. 3).
4.4. NLRP3 inflammasome in COVID-19 neurological symptoms
An increasing number of studies have reported the neurological symptoms in patients with COVID-19, including cerebrovascular complications, convulsions, encephalitis, change in mental status, confusion, headaches and febrile seizures, as well as taste (hypogeusia/ageusia) and smell (anosmia/hyposmia) dysfunction [7]. Accordingly, SARS-CoV-2 has been reported to affect human neural progenitor cells and brain organoids, indicating the possibility of direct infection of brain tissue by CoV [110]. SARS-CoV-2 may invade brain tissue in similar ways to other viruses: 1) the hematogenous route, in which viruses reach and invade epithelial cells from the blood-brain barrier (BBB) or the blood cerebrospinal fluid barrier via the bloodstream, or entrance into the central nervous system (CNS) via leukocytes; 2) the neuronal retrograde route, in which viruses invade peripheral neurons to reach the CNS [111]. From this point of view, viruses can cross the BBB and enter the CNS, initiating a neuroinflammatory process [111]. BBB permeability increases in patients with neurodegenerative diseases, facilitating SARS-CoV-2 neuroinvasion.
It has been indicated that inflammasomes are activated in various diseases and injuries affecting the CNS [112]. Therefore, it is obvious that the CNS is able to develop an immune response via inflammasome activation. Additionally, P2 × 7 receptors are ATP-gated ion channels widely expressed in the CNS [65], and their activation initiated by viral infection can result in behavioral alterations and mental disorders [113,114]. As mentioned above, P2 × 7 receptors are important for the indispensable K+ efflux for NLRP3 inflammasome activation. In this context, it has been proposed that neuroinvasion via the BBB and hyperstimulation of neuroimmune responses in COVID-19 may be induced by P2 × 7 receptors in the CNS via NLRP3 inflammasome activation [62]. Additionally, high levels of peripheral cytokines (such as IL-1β and IL-6) can induce neuroinflammation by passing through the BBB directly or diminishing BBB integrity, leading to the permeation of virus-infected peripheral white blood cells and monocytes [115]. Furthermore, systemic inflammation induced by NLRP3 inflammasome activation impairs immune homeostasis in the brain. It facilitates the production of inflammatory cytokines and the aggregation of peptides into pathogenic fibrils and promotes mitochondrial dysfunction and apoptosis. As a result, it ultimately leads to neurodegeneration [116,117]. These findings indicate that the NLRP3 inflammasome may play a key role in the development of COVID-19 neurological manifestations (Fig. 3).
Intriguingly, a recent study [118] summarized the potential similarities of the pathogenesis of COVID-19 and some autoimmune diseases, including rheumatoid arthritis, diabetes, multiple sclerosis, etc. Of note, most of these autoimmune diseases have been characterized by the involvement of the NLRP3 inflammasome [9], which again implies the participation of the NLRP3 inflammasome in COVID-19 pathogenesis. Nonetheless, the specific mechanisms of NLRP3 inflammasome involvement in the development of various COVID-19 symptoms or complications need to be further investigated with more studies.
5. NLRP3 inflammasome-targeted therapeutic strategies for COVID-19
Since the NLRP3 inflammasome, cytokine storm and neutrophil-NET axis all contribute to ALI/ARDS in severe COVID-19 cases, it would be feasible to explore therapies by inhibiting 1) NLRP3 inflammasome activation, 2) cytokine action, or 3) increases in neutrophils and NETs (Fig. 3). This review discusses several inhibitors targeting these three factors. The clinical trials of inhibitors are listed in Table 1 , and the specific characteristics of the clinical trials are listed in Table 2 .
Table 1.
Inhibitors targeting NLRP3 inflammasome-related factors for COVID-19.
| Target | Inhibitor | Clinical triala | Trial phase | Trial status |
|---|---|---|---|---|
| NLRP3 inflammasome | Melatonin | TrialTroveID-375830 | II | Open |
| Statins | TrialTroveID-379443 | III | Open | |
| IFN | TrialTroveID-371689 | IV | Completed | |
| TrialTroveID-370679 | II/III | Completed | ||
| TrialTroveID-392370 | III | Completed | ||
| TrialTroveID-372573 | II | Completed | ||
| Sirolimus | TrialTroveID-378925 | II | Open | |
| TrialTroveID-371558 | II | Open | ||
| TrialTroveID-373755 | II | Planned | ||
| Azithromycin | TrialTroveID-394294 | IV | Completed | |
| TrialTroveID-390593 | IV | Completed | ||
| Cyclosporine | TrialTroveID-383935 | I/II | Completed | |
| Colchicine | TrialTroveID-381747 | II/III | Completed | |
| TrialTroveID-383970 | III | Planned | ||
| Baricitinib | TrialTroveID-393453 | III | Open | |
| TrialTroveID-376590 | III | Completed | ||
| Acalabrutinib | TrialTroveID-374074 | II | Completed | |
| Oridonin | Required | / | / | |
| Parthenolide | Required | / | / | |
| Bay 11-7082 | Required | / | / | |
| Glyburide | Required | / | / | |
| IL-1β | Anakinra | TrialTroveID-387990 | II | Planned |
| TrialTroveID-381687 | II | Completed | ||
| Canakinumab | TrialTroveID-394661 | II | Completed | |
| IL-6 | Sarilumab | TrialTroveID-374439 | II | Open |
| Tocilizumab | TrialTroveID-395500 | IV | Completed | |
| TrialTroveID-379637 | IV | Completed | ||
| Neutrophils | Sivelestat | Required | / | / |
| Colchicine | TrialTroveID-381747 | II/III | Completed | |
| TrialTroveID-383970 | III | Planned | ||
| NETs | Dornase alfa | TrialTroveID-371650 | III | Open |
Clinical trials registered at Citeline.informa.com.
Table 2.
Characteristics of the clinical trials performed on COVID-19 patientsa.
| Agent | Clinical trials | Objective | Intervention | Control | Primary outcome measures |
|---|---|---|---|---|---|
| Melatonin | TrialTroveID-375830 | To evaluate the therapeutic effects of melatonin by inhibition of NLRP3 inflammasome in COVID-19 patients | 9 mg melatonin for 7–10 nights | The usual treatment | The amount of melatonin [Time Frame: Up to 10 days] |
| Statins | TrialTroveID-379443 | To study safety and efficacy of the combination of colchicine and rosuvastatin in addition to standard of care in hospitalized patients with SARS-CoV-2 | 40 mg rosuvastatin daily and 0.6 mg colchicine twice for 3 days and then 0.6 mg daily | The standard care | COVID-19 severity (as defined by World Health Organization Ordinal Scale) [Time Frame: 30 Days] |
| IFN | TrialTroveID-371689 | To investigate the beneficial effects of IFNβ-1a, compared to IFNβ-1b and the base therapeutic regiment in moderate to severe COVID-19 | 44 μg IFNβ-1a on days 1, 3, 6 (for Arm1); 0.25 mg IFNβ-1b on days 1, 3, 6 (for Arm2); 400 mg hydroxychloroquine on day 1 + 400 mg lopinavir/100 mg ritonavir twice a day for 10 days (for Arm1,2) |
400 mg hydroxychloroquine on day 1 + 400 mg lopinavir/100 mg ritonavir twice a day for 10 days | Time to clinical improvement [Time Frame: From date of randomization until 14 days later] |
| TrialTroveID-370679 | To evaluate the efficacy and safety of IFNβ-1b in the treatment of COVID-19 | 250 μg IFNβ-1b subcutaneously every other day for 14 days | Hydroxychloroquine + lopinavir/ritonavir for at least 5 days | Response to the treatment daily; Complications of the treatment daily |
|
| TrialTroveID-392370 | To test whether IFNα-2b can provide additional benefit to these patients in terms of reduced rate of hospitalization and better time to recovery | Single dose of 1 μg/kg pegylated IFNα-2b | The standard care | Change in ordinal scale [Time Frame: From baseline to day 11] | |
| TrialTroveID-372573 | To evaluate peginterferonλ in ambulatory and hospitalized patients with mild to moderate COVID-19 | 180 μg peginterferonλ-1a at baseline and a second dose on day 7 | Placebo | Proportion swab negative [Time Frame: At day 7]; Treatment-emergent and treatment related serious adverse events [Time Frame: Day 0 to day 30]; Time to viral negativity [Time Frame: Day 0 to day 28] |
|
| Sirolimus | TrialTroveID-378925 | To illustrate the efficacy and safety of sirolimus as an adjuvant agent to the standard treatment protocol against COVID-19 infection | 6 mg sirolimus daily on day 1 followed by 2 mg daily for 9 days | The standard care | Time to clinical recovery [Time Frame: 14−28 days]; Viral clearance [Time Frame: 14 days] |
| TrialTroveID-371558 | To determine if treatment with sirolimus can improve clinical outcomes in hospitalized patients with COVID-19 | 6 mg sirolimus daily on day 1 followed by 2 mg daily for the next 13 days | Placebo | Proportion of patients who are alive and free from advanced respiratory support measures at day 28 [Time Frame: 28 days] | |
| TrialTroveID-373755 | To study the effect of hydroxychloroquine in combination with azithromycin or sirolimus for treating COVID-19 patients | 250 mg azithromycin daily for 10 days (for Arm1); 4 mg sirolimus for 1 day then 2 mg daily for 9 days (for Arm2); 600 mg hydroxychloroquine daily for 10 days (for Arm1,2) |
None | Time to clinical improvement | |
| Azithromycin | TrialTroveID-390593 | To assess the clinical effectiveness and safety profile of the COVID-19 treatment protocol (containing both hydroxychloroquine and azithromycin) in an Iraqi specialized hospital | 500 mg azithromycin on day 1, then 250 mg daily for 5 days + 400 mg hydroxychloro twice a day on day 1 then 200 mg twice a day for 5 days (for Arm1,2,3); 75 mg tamiflu twice a day for 5 days (for Arm2,3); 200 mg lopinavir/ 50 mg ritonavir twice a day for 5 days and antibiotic(s) accordingly (for Arm3) |
None | The changes in clinical and biochemical parameters during hospitalization period such as disappearance of clinical symptoms, virologic clearance and occurrence of side effects |
| Cyclosporine | TrialTroveID-383935 | To evaluate low doses of corticosteroids and cyclosporine combined with enoxaparin, in patients with COVID-19 pneumonia | Cyclosporine A at a dose of 1−2 mg /kg / day divided into two doses, for 7 days | The standard treatment: 0.5 mg/kg enoxaparin once, 500 mg clarithromycin twice and metylprednisolone 0.5 mg/kg once | Number of days to clinical improvement until hospital discharge or death. [Time Frame: 28 days] |
| Colchicine | TrialTroveID-381747 | To evaluate whether the addition of colchicine to standard treatment for COVID-19 results in better outcomes | 0.5 mg colchicine three times a day for 5 days, then 0.5 mg twice a day for 5 days (a loading dose of 1 mg if body weight ≥80 kg) | Placebo | Number of days of need of supplemental oxygen by catheter or masks; Number of days from the admission to the discharge; The percentage of individuals who will require admission to the intensive care unit; The percentage of death |
| TrialTroveID-383970 | To evaluate the efficacy and safety of oral colchicine plus standard therapy versus standard therapy in the clinical course of SARS-CoV-2 infection, in a population group with moderate COVID-19 compromise and requiring hospitalization | 1.5 mg colchicine orally on day 1 (initial 1 mg and 0.5 mg at 2 h), then 0.5 mg every 12 h on days 2 to 7, and continuing with 0.5 mg per day until completing 14 ± 1 days | The standard treatment | Number of participants who die or require transfer to intensive care unit [Time Frame: In the first 15 days after randomization] | |
| Baricitinib | TrialTroveID-393453 | To evaluate the efficacy of remdesivir and baricitinib combination therapy for the treatment of ARDS caused by COVID-19; To compare the outcome of the "remdesivir + baricitinib" against "remdesivir + tocilizumab" therapy and find the best option for the management of ARDS in COVID-19 patients |
5 mg/kg (<40 kg) or 200 mg (>40 kg) remdesivir I/V on day 1, then 2.5 mg/kg (< 40 kg) or 100 mg (> 40 kg) daily following randomization (for Arm1,2); 4 mg baricitinib tablets daily for 2–4 weeks (for Arm1); 8 mg/kg tocilizumab I/V (up to 800 mg highest) 12 h apart (for Arm2) |
None | Time to clinical improvement [Time Frame: Following randomization 30 days] |
| TrialTroveID-376590 | To evaluate the efficacy and safety of baricitinib in hospitalized adults with COVID-19 | 4 mg of baricitinib once daily | Placebo | Percentage of participants who die or require non-invasive ventilation/high-flow oxygen or invasive mechanical ventilation (including extracorporeal membrane oxygenation) [Time Frame: Day 1 to day 28] | |
| Acalabrutinib | TrialTroveID-374074 | To investigate the safety, efficacy and pharmacokinetics of acalabrutinib together with best supportive care in the treatment of COVID-19 | 100 mg acalabrutinib twice a day for 10 days | Best supportive care | Occurrence of adverse events and serious adverse events [Time Frame: Day 28]; Proportion of subject alive and free of respiratory failure [Time Frame: Day 28] |
| Anakinra | TrialTroveID-387990 | To determine the efficacy of anakinra, an IL-1 receptor blocker, in reducing the need for mechanical ventilation and/or 28-day mortality among patients with COVID-19 who have features of cytokine storm syndrome and severe respiratory failure | Anakinra IV 4 times a day for 7 days (100 mg anakinra mixed with 100 ml of 0.9 % saline solution for IV administration) | 0.9 % saline | Number of subjects alive without having required mechanical ventilation [Time Frame: 28 days post randomization] |
| TrialTroveID-381687 | To investigate whether anakinra may reduce the need for invasive mechanical ventilation and deaths when compared to standard of care in patients with severe COVID-19 | 100 mg anakinra once daily for 3 days, followed by 100 mg once every other day for 7 days | The standard treatment | Need for invasive mechanical ventilation or admission to the intensive care unit [Time Frame: From patient records between baseline and 14 days]; In-hospital mortality [Time Frame: From patient records between baseline and death or discharge] |
|
| Canakinumab | TrialTroveID-394661 | To evaluate canakinumab to improve respiratory function and laboratory parameters compared with standard therapy (hydroxycloroquine + lopinavir/ritonavir) | 300 mg canakinumab by single subcutaneous administration | Standard therapy: hydroxycloroqui + lopinavir/ritonavir | / |
| Sarilumab | TrialTroveID-374439 | To generate a rapid, still robustly documented, evidence on the potential clinical efficacy and tolerability of a further IL-6R antagonist in COVID-19 pneumonia | Sarilumab prefilled syringe | None | Proportion of patients who show an improvement of the respiratory function [Time Frame: 6 weeks] |
| Tocilizumab | TrialTroveID-395500 | To analyze the effectiveness of tocilizumab in moderate to severe COVID-19 participants on the basis of predefined assessment criteria | Tocilizumab 8 mg/kg (up to 800 mg/dose) over 1 h, followed by up to three additional doses if required as per the response after the first dose with 8 h intervals | 80 mg methylprednisolne daily in two divided doses as per national/local guidelines | Decreased mortality in participants [Time Frame: 30 days]; Hospital & intensive care unit stay in days [Time Frame: 14 days] |
| TrialTroveID-379637 | To assess the effects of tocilizumab, an IL-6 receptor antagonist, on intra-hospital mortality and development of positive cultures in patients with COVID-19 admitted to the intensive care unit | A single dose of 400 mg intravenous tocilizumab twice a day | The standard care | Intra-hospital mortality | |
| Dornase alfa | TrialTroveID-371650 | To evaluate the efficacy and safety of aerosolized intra-tracheal dornase alfa administration in mechanically ventilated patients hospitalized for COVID-19-related ARDS | 2500 IU dornase alfa inhalation solution twice daily, 12 h apart, for 7 consecutive days | The usual care | Efficacy of intra-tracheal administration [Time Frame: Day 7]; Improvement in the ARDS scale severity (Berlin criteria). [Time Frame: Day 7] |
Characteristics of each clinical trial obtained from Citeline.informa.com.
5.1. NLRP3 inflammasome-targeted therapeutic strategies for COVID-19
Several NLRP3 inflammasome inhibitors have been clinically studied for their effectiveness in COVID-19, which implies a promising application of other NLRP3 inflammasome inhibitors.
Melatonin is a hormone of the pineal gland contributing to the regulation of physiological activities and neuroendocrine processes. Recently, it has been proposed that the NLRP3 inflammasome is a new target for melatonin to exert its anti-inflammatory activity [119]. Based on the melatonin inhibitory activity of the NLRP3 inflammasome, a recent clinical trial evaluated the therapeutic effects of melatonin in inhibiting the NLRP3 inflammasome in COVID-19 patients (TrialTroveID-375830). Additionally, statins are a treatment for diverse infections, such as influenza virus or middle east respiratory syndrome coronavirus (MERS-CoV) [120]. It has been shown to limit cytokine storm in severe COVID-19 patients by inhibiting NF-κB and the NLRP3 inflammasome [121]. In addition, colchicine is also capable of inhibiting the NLRP3 inflammasome by preventing caspase activation, inhibiting P2 × 7 receptors and decreasing pyrin gene expression [122]. In accordance, a clinical trial is studying the safety and efficacy of the combination of rosuvastatin and colchicine in addition to standard care in hospitalized patients with SARS-CoV-2 (TrialTroveID-379443). Of note, a completed trial indicated that the use of colchicine reduces the length of supplemental oxygen therapy and hospitalization and increased clinical improvement (TrialTroveID-381747). Therefore, colchicine could be considered a beneficial and inexpensive option for COVID-19 treatment. Additionally, several studies have stated that IFN can compromise the NLRP3 inflammasome via various pathways [123]. A number of trials have been conducted to assess the efficacy of recombinant IFNs in the treatment of COVID-19. The completed clinical trials have shown that IFNβ-1a (TrialTroveID-371689), IFNβ-1b (TrialTroveID-370679), IFNα-2b (TrialTroveID-392370) and peginterferonλ-1a (TrialTroveID-372573) exhibit benefits for clinical improvement in moderate to severe COVID-19. Moreover, azithromycin has been claimed to have an inhibitory effect on NLRP3 inflammasome activation [124]. Accordingly, azithromycin has been studied in several clinical trials (TrialTroveID-394294, TrialTroveID-390593). The results of TrialTroveID-390593 showed that the combination of hydroxychloroquine and azithromycin can help to promote the recovery of most patients and significantly reduce their signs and symptoms. Furthermore, sirolimus and cyclosporine, immunosuppressants that can attenuate NLRP3 inflammasome activation [125,126], are undergoing clinical trials to treat COVID-19 (TrialTroveID-378925, TrialTroveID-371558, TrialTroveID-373755, TrialTroveID-383935). Notably, one trial (TrialTroveID-373755) is investigating the effect of both azithromycin and sirolimus as multiple arms, which further implies the importance of the NLRP3 inflammasome in COVID-19. Additionally, the results of TrialTroveID-383935 have shown that cyclosporine A can improve outcomes and reduce mortality in moderate to severe COVID-19 cases. Likewise, other drugs that can inhibit the NLRP3 inflammasome, such as baricitinib and acalabrutinib [127,128], are being widely studied in clinical trials for COVID-19 (TrialTroveID-376590, TrialTroveID-393453, TrialTroveID-374074). According to the results of TrialTroveID-376590, baricitinib can significantly reduce the mortality of COVID-19 patients.
Apart from NLRP3 inflammasome inhibitors that have been applied in clinical trials of COVID-19, some other potent and effective NLRP3 inflammasome inhibitors deserve more attention. For example, natural product inhibitors such as oridonin and parthenolide and synthetic compound inhibitors such as Bay 11-7082 are closely associated with the attenuation of SARS-CoV infection and the reduction of lung inflammation [45,129,130]. Additionally, glyburide can inhibit the NLRP3 inflammasome in cells infected with RNA viruses [131]. In short, effective NLRP3 inflammasome inhibitors should be discussed further to explore more promising treatments for COVID-19 (Fig. 3).
5.2. Cytokine product-targeted therapeutic strategies for COVID-19
Considering the cytokine-mediated inflammatory response during COVID-19, targeting cytokines directly is a potent therapeutic strategy.
As stated above, IL-1β and IL-18 can facilitate the release of other cytokines, such as IL-6.In addition, IL-1β and IL-6 can promote inflammasome activation in a feedforward mode [132,133]. As a result, the direct inhibition of cytokine products may largely contribute to the blockade of the NLRP3 inflammasome and thus the inflammatory response in COVID-19. For example, the IL-1 receptor antagonists anakinra and canakinumab are undergoing evaluation in several trials for the treatment of COVID-19 (TrialTroveID-381687, TrialTroveID-387990, TrialTroveID-394661). The completed trial TrialTroveID-381687 showed that in patients with severe COVID-19 pneumonia and high oxygen requirements, anakinra could represent an effective treatment option and may confer clinical benefit. Additionally, the results of TrialTroveID-394661 indicated that canakinumab therapy causes rapid and long-lasting improvement in oxygenation levels in the absence of any severe adverse events. Thus, in hospitalized adult patients with mild or severe COVID-19, canakinumab could be a valid therapeutic option. Moreover, the effects on COVID-19 of two anti-IL-6 receptor monoclonal antibodies, sarilumab and tocilizumab [4,[133], [134], [135], [136]], have been evaluated by several clinical trials (TrialTroveID-374439, TrialTroveID-395500, TrialTroveID-379637). The results of TrialTroveID-379637 demonstrated that tocilizumab was associated with rapid improvement in oxygenation and a faster decrease in C-reactive protein and white blood cell counts in patients with COVID-19. Therefore, tocilizumab should be evaluated as rescue therapy for patients with progressive disease. Interestingly, TrialTroveID-393453 mentioned above is evaluating the efficacy of both tocilizumab and baricitinib to determine the best option for the management of ARDS in COVID-19 patients. This further implies the vital role of the NLRP3 inflammasome-cytokine axis in COVID-19 and indicates a novel treatment for COVID-19.
5.3. Neutrophil-NET-targeted therapeutic strategies for COVID-19
As described above, the NLRP3 inflammasome produces IL-1β, which can further recruit and activate neutrophils, resulting in excessive NETs. Additionally, NETs can in turn activate the NLRP3 inflammasome to form a feedback loop [99]. Therefore, considering the participation of neutrophils and NETs in COVID-19 ARDS, inhibition of the neutrophil-NET axis can attenuate inflammation in COVID-19 either directly or via inhibition of the NLRP3 inflammasome.
NET production can be blocked by recombinant dornase alfa, histone deacetylase inhibitors and some IL-6 blockers [99,137]. Of note, dornase alfa has been undergoing clinical trials for COVID-19 (TrialTroveID-371650), which aims to evaluate the effect of dornase alfa aerosol in mechanically ventilated patients hospitalized for COVID-19-related ARDS. In addition, the neutrophil elastase inhibitor sivelestat, approved to treat ARDS, has been studied [138], while clinical trials on COVID-19 are lacking. In addition, the NLRP3 inflammasome inhibitor colchicine mentioned above is also capable of preventing the adhesion and recruitment of neutrophils [139]. The ongoing clinical trials of colchicine are described above (TrialTroveID-379443, TrialTroveID-381747). Clinical improvement through the use of colchicine in COVID-19 implies that the inhibition of both the NLRP3 inflammasome and neutrophils is effective for COVID-19 treatment. All of these inhibitors are safe and FDA-approved drugs, for which more specific clinical trials can be conducted to further assess their efficacy in COVID-19.
Above all, with an enormous number of clinical trials that are planned, ongoing or completed, inhibitors of the NLRP3 inflammasome, cytokines and neutrophil-NETs will be able to provide novel treatments for COVID-19 and help further study the pathogenesis of COVID-19.
6. Conclusion
COVID-19, as a worldwide public health concern, presents with a broad spectrum of symptoms ranging from few or no symptoms to severe pneumonia, which can develop into ARDS or even death. Nevertheless, the specific mechanisms that influence disease severity still remain unclear. Many recent studies have reported that inflammatory mediators such as IL-1β, IL-6 and LDH are closely associated with severe COVID-19 cases. Typically, the inflammatory responses to diverse stimuli in host cells necessitate inflammasomes. Although inflammasome signaling is necessary to defend against viral infection, it contributes to the hyperactivated inflammatory response at the same time. As the best studied inflammasome, the NLRP3 inflammasome has been proven to play a key role in the severity of MERS-CoV, SARS-CoV and SARS-CoV-2 infections [75]. As a result, given the great number of people affected with COVID-19 and the urgent need for drug discovery, a thorough consideration of NLRP3 inflammasome involvement in COVID-19 provides a better understanding of pathogenesis and helps with therapeutic development.
It has been suggested that inflammation signaling pathways are probably better targets than a reduction in viral load for managing COVID-19 [112]. For this reason, this review has highlighted the mechanisms of NLRP3 inflammasome activation by SARS-CoV-2 infection. This finding implies several potential targets for the inhibition of the NLRP3 inflammasome and thus the attenuation of COVID-19. For instance, P2 × 7 receptor blockers can inhibit K+ efflux (e.g., glyburide) [131], and antioxidant/ROS scavengers (e.g., rosuvastatin) can reduce ROS, thus compromising NLRP3 inflammasome activation [9]. In addition, inhibitors targeting the complement cascade or AT1R may also contribute to the inhibition of the NLRP3 inflammasome. Considering the involvement of the NLRP3 inflammasome and its downstream pathways in the pathogenesis of COVID-19 (mainly respiratory, cardiovascular and neurological symptoms), the direct inhibition of NLRP3-related inflammatory cells and cytokine products also provides a potential treatment for COVID-19. This review has proposed several inhibitors targeting the NLRP3 inflammasome, IL-1β, IL-6, neutrophils and NETs. To date, several FDA-approved therapies that interfere with inflammasome activation signaling have been considered for the treatment of COVID-19, including anakinra, tocilizumab and IFN-β. However, there are no FDA-approved drugs that directly target the inflammasome. Therefore, inhibitors directly targeting the NLRP3 inflammasome, such as MCC950 and oridonin, which bind to the NACHT domain of NLRP3 [9,140], are worth investigating further in clinical trials. Additionally, even nutraceutical strategies for the management of COVID-19 via suppression of NLRP3 inflammasome activation have been thoroughly reported [141].
Given that the timing of therapy is important, targeted therapy is urgently needed to protect individuals with mild/moderate disease from severe symptoms and improve patient outcomes in severe cases. Additionally, since CoV infections may become a seasonal infection (e.g., MERS-CoV, SARS-CoV and SARS-CoV-2 infections), it is important to explore more FDA-approved and novel therapeutics that can be applied for the treatment of CoV infections in the future. Therefore, we suggest the NLRP3 inflammasome as a potential target for the treatment of COVID-19.
Funding
This study was supported by projects [81803354] and [81773693] from the National Natural Science Foundation of China, [BK20180564] from the Natural Science Foundation of Jiangsu Province of China, [2018YFC0807402] from the National Key R&D Program of China, [CPU2018GY02] from the Double First Class Innovation Team of China Pharmaceutical University, and [2632021ZD13] from the Fundamental Research Funds for the Central Universities.
CRediT authorship contribution statement
Ni Zhao: Writing - original draft. Bin Di: Writing - review & editing. Li-li Xu: Writing - review & editing.
Declaration of Competing Interest
The authors declare no competing financial interest.
Biographies

Ni Zhao is a postgraduate student at China Pharmaceutical University. She recieved her Bachelor of Science in June 2019 from China Pharmaceutical University. She is currently pursuing a Master in pharmaceutical analysis at China Pharmaceutical University, and plans to graduate in June of 2022. Since September 2019, she has been involved in Dr. Di’s research lab and studied the exploration of metal organic framework. She currently works in the laboratory of Dr. Di and focuses on the exploration of novel inhibitors of NLRP3 inflammasome and the development of therapies for NLRP3 inflammasome-driven diseases. To this end, she has published a review article on NLRP3 inflammasome investigation.

Dr. Bin Di, Ph.D. is a professor of pharmaceutical analysis and vice-dean of School of Pharmacy in the China Pharmaceutical University, Nanjing, China. Dr. Di is currently working on developing novel materials and techniques for pharmaceutical analysis and novel therapies for NLRP3 inflammasome/Nrf2-related diseases. Dr. Di graduated with a Bachelor of Science followed by Master of Science and doctoral degree in China Pharmaceutical University, Nanjing, China. After his Ph.D. he worked in the Department of Pharmaceutical analysis, School of Pharmacy, China Pharmaceutical University till now. He worked as a visiting scholar at the University of Maryland School of Medicine, Institute of Human Virology for one year(2014–2015). To this end, Dr. Di has published over 50 research papers and had 4 patents.

Lili Xu, Ph.D. is a Postdoctoral Fellow in the Department of Pharmaceutical analysis at China Pharmaceutical University. Dr. Xu received her Master of Science and earned her doctorate in Pharmaceutical Chemistry from the China Pharmaceutical University. During her Ph.D. she mainly studied Keap1-Nrf2-ARE signal pathways and investigated therapies for the related diseases. She currently works in the laboratory of Dr. Di and focuses on the development of anti-inflammation medicine such as the exploration of novel inhibitors of NLRP3 inflammasome and agonists of Nrf2. To this end, Dr. Xu has published over 20 SCI research papers.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C.S.G.o.t.I.C.o.T.o. Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan Y.J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antiviral Res. 2005;65(2):69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah A. Novel coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieto-Torres J.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y.Y., Ma Y.T., Zhang J.Y., Xie X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17(5):259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asadi-Pooya A.A., Simani L. Central nervous system manifestations of COVID-19: a systematic review. J. Neurol. Sci. 2020;413:116832. doi: 10.1016/j.jns.2020.116832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broz P., Dixit V.M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016;16(7):407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 9.Zhao N., Li C.C., Di B., Xu L.L. Recent advances in the NEK7-licensed NLRP3 inflammasome activation: mechanisms, role in diseases and related inhibitors. J. Autoimmun. 2020;113:102515. doi: 10.1016/j.jaut.2020.102515. [DOI] [PubMed] [Google Scholar]
- 10.Sharif H., Wang L., Wang W.L., Magupalli V.G., Andreeva L., Qiao Q., Hauenstein A.V., Wu Z., Núñez G., Mao Y., Wu H. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019;570(7761):338–343. doi: 10.1038/s41586-019-1295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis B.K., Wen H., Ting J.P. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauernfeind F.G., Horvath G., Stutz A., Alnemri E.S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B.G., Fitzgerald K.A., Hornung V., Latz E. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009;183(2):787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y., Hara H., Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 2016;41(12):1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14(9):1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 15.Lee G.S., Subramanian N., Kim A.I., Aksentijevich I., Goldbach-Mansky R., Sacks D.B., Germain R.N., Kastner D.L., Chae J.J. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492(7427):123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heid M.E., Keyel P.A., Kamga C., Shiva S., Watkins S.C., Salter R.D. Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. J. Immunol. 2013;191(10):5230–5238. doi: 10.4049/jimmunol.1301490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng W.A., Thein T., Kinnunen K., Lashkari K., Gregory M.S., D’Amore P.A., Ksander B.R. NLRP3 inflammasome activation in retinal pigment epithelial cells by lysosomal destabilization: implications for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2013;54(1):110–120. doi: 10.1167/iovs.12-10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada M., Matsuzawa A., Yoshimura A., Ichijo H. The lysosome rupture-activated TAK1-JNK pathway regulates NLRP3 inflammasome activation. J. Biol. Chem. 2014;289(47):32926–32936. doi: 10.1074/jbc.M114.579961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shenoy A.R., Wellington D.A., Kumar P., Kassa H., Booth C.J., Cresswell P., MacMicking J.D. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336(6080):481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 20.Boriushkin E., Wang J.J., Li J., Bhatta M., Zhang S.X. p58(IPK) suppresses NLRP3 inflammasome activation and IL-1β production via inhibition of PKR in macrophages. Sci. Rep. 2016;6:25013. doi: 10.1038/srep25013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y., Zeng M.Y., Yang D., Motro B., Núñez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530(7590):354–357. doi: 10.1038/nature16959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grailer J.J., Canning B.A., Kalbitz M., Haggadone M.D., Dhond R.M., Andjelkovic A.V., Zetoune F.S., Ward P.A. Critical role for the NLRP3 inflammasome during acute lung injury. J. Immunol. 2014;192(12):5974–5983. doi: 10.4049/jimmunol.1400368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li D., Ren W., Jiang Z., Zhu L. Regulation of the NLRP3 inflammasome and macrophage pyroptosis by the p38 MAPK signaling pathway in a mouse model of acute lung injury. Mol. Med. Rep. 2018;18(5):4399–4409. doi: 10.3892/mmr.2018.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin L., Xu L., Lv W., Han L., Xiang Y., Fu L., Jin M., Zhou R., Chen H., Zhang A. An NLRP3 inflammasome-triggered cytokine storm contributes to Streptococcal toxic shock-like syndrome (STSLS) PLoS Pathog. 2019;15(6) doi: 10.1371/journal.ppat.1007795. e1007795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76(1):16–32. doi: 10.1128/mmbr.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratajczak M.Z., Bujko K., Ciechanowicz A., Sielatycka K., Cymer M., Marlicz W., Kucia M. SARS-CoV-2 entry receptor ACE2 is expressed on very small CD45(-) precursors of hematopoietic and endothelial cells and in response to virus spike protein activates the Nlrp3 inflammasome. Stem Cell Rev. Rep. 2020:1–12. doi: 10.1007/s12015-020-10010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues T.S., de Sá K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Gonçalves A.V., Perucello D.B., Andrade W.A., Castro R., Veras F.P., Toller-Kawahisa J.E., Nascimento D.C., de Lima M.H.F., Silva C.M.S., Caetite D.B., Martins R.B., Castro I.A., Pontelli M.C., de Barros F.C., do Amaral N.B., Giannini M.C., Bonjorno L.P., Lopes M.I.F., Santana R.C., Vilar F.C., Auxiliadora-Martins M., Luppino-Assad R., de Almeida S.C.L., de Oliveira F.R., Batah S.S., Siyuan L., Benatti M.N., Cunha T.M., Alves-Filho J.C., Cunha F.Q., Cunha L.D., Frantz F.G., Kohlsdorf T., Fabro A.T., Arruda E., de Oliveira R.D.R., Louzada-Junior P., Zamboni D.S. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218(3) doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 29.Zhou R., Yazdi A.S., Menu P., Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 30.Tang T., Lang X., Xu C., Wang X., Gong T., Yang Y., Cui J., Bai L., Wang J., Jiang W., Zhou R. CLICs-dependent chloride efflux is an essential and proximal upstream event for NLRP3 inflammasome activation. Nat. Commun. 2017;8(1):202. doi: 10.1038/s41467-017-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu B., Nakamura T., Inouye K., Li J., Tang Y., Lundbäck P., Valdes-Ferrer S.I., Olofsson P.S., Kalb T., Roth J., Zou Y., Erlandsson-Harris H., Yang H., Ting J.P., Wang H., Andersson U., Antoine D.J., Chavan S.S., Hotamisligil G.S., Tracey K.J. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488(7413):670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di A., Xiong S., Ye Z., Malireddi R.K.S., Kometani S., Zhong M., Mittal M., Hong Z., Kanneganti T.D., Rehman J., Malik A.B. The TWIK2 potassium efflux channel in macrophages mediates NLRP3 inflammasome-induced inflammation. Immunity. 2018;49(1):56–65. doi: 10.1016/j.immuni.2018.04.032. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rühl S., Broz P. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K(+) efflux. Eur. J. Immunol. 2015;45(10):2927–2936. doi: 10.1002/eji.201545772. [DOI] [PubMed] [Google Scholar]
- 34.Swanson K.V., Deng M., Ting J.P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19(8):477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami T., Ockinger J., Yu J., Byles V., McColl A., Hofer A.M., Horng T. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. U. S. A. 2012;109(28):11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaron J.R., Gangaraju S., Rao M.Y., Kong X., Zhang L., Su F., Tian Y., Glenn H.L., Meldrum D.R. K(+) regulates Ca(2+) to drive inflammasome signaling: dynamic visualization of ion flux in live cells. Cell Death Dis. 2015;6(10) doi: 10.1038/cddis.2015.277. e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green J.P., Yu S., Martín-Sánchez F., Pelegrin P., Lopez-Castejon G., Lawrence C.B., Brough D. Chloride regulates dynamic NLRP3-dependent ASC oligomerization and inflammasome priming. Proc. Natl. Acad. Sci. U. S. A. 2018;115(40):E9371–e9380. doi: 10.1073/pnas.1812744115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., Fitzgerald K.A., Ryter S.W., Choi A.M. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katsnelson M.A., Lozada-Soto K.M., Russo H.M., Miller B.A., Dubyak G.R. NLRP3 inflammasome signaling is activated by low-level lysosome disruption but inhibited by extensive lysosome disruption: roles for K+ efflux and Ca2+ influx. Am. J. Physiol., Cell Physiol. 2016;311(1):C83–c100. doi: 10.1152/ajpcell.00298.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V.G., Wu H., Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nieva J.L., Madan V., Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10(8):563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins e, 3a, and 8a in replication and pathogenesis. mBio. 2018;9(3) doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Netland J., DeDiego M.L., Zhao J., Fett C., Álvarez E., Nieto-Torres J.L., Enjuanes L., Perlman S. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology. 2010;399(1):120–128. doi: 10.1016/j.virol.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fett C., DeDiego M.L., Regla-Nava J.A., Enjuanes L., Perlman S. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 2013;87(12):6551–6559. doi: 10.1128/jvi.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeño J.M., Fernandez-Delgado R., Fett C., Castaño-Rodriguez C., Perlman S., Enjuanes L. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88(2):913–924. doi: 10.1128/jvi.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieto-Torres J.L., Dediego M.L., Alvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., Kremer L., Shuo S., Enjuanes L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Torres J., Aguilella V.M., Enjuanes L. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432(2):485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito M., Yanagi Y., Ichinohe T. Encephalomyocarditis virus viroporin 2B activates NLRP3 inflammasome. PLoS Pathog. 2012;8(8) doi: 10.1371/journal.ppat.1002857. e1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichinohe T., Pang I.K., Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 2010;11(5):404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K., Xie S., Sun B. Viral proteins function as ion channels. Biochim. Biophys. Acta. 2011;1808(2):510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9(9):904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen I.Y., Moriyama M., Chang M.F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U. S. A. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 55.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santos R.A.S., Sampaio W.O., Alzamora A.C., Motta-Santos D., Alenina N., Bader M., Campagnole-Santos M.J. The ACE2/Angiotensin-(1-7)/MAS Axis of the renin-angiotensin system: focus on angiotensin-(1-7) Physiol. Rev. 2018;98(1):505–553. doi: 10.1152/physrev.00023.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao M., Bai M., Ding G., Zhang Y., Huang S., Jia Z., Zhang A. Angiotensin II stimulates the NLRP3 inflammasome to induce podocyte injury and mitochondrial dysfunction. Kidney Dis. Basel (Basel) 2018;4(2):83–94. doi: 10.1159/000488242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ratajczak M.Z., Kucia M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine "storm" and risk factor for damage of hematopoietic stem cells. Leukemia. 2020;34(7):1726–1729. doi: 10.1038/s41375-020-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro D.E., Oliveira-Giacomelli Á., Glaser T., Arnaud-Sampaio V.F., Andrejew R., Dieckmann L., Baranova J., Lameu C., Ratajczak M.Z., Ulrich H. Hyperactivation of P2X7 receptors as a culprit of COVID-19 neuropathology. Mol. Psychiatry. 2020:1–16. doi: 10.1038/s41380-020-00965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asgari E., Le Friec G., Yamamoto H., Perucha E., Sacks S.S., Köhl J., Cook H.T., Kemper C. C3a modulates IL-1β secretion in human monocytes by regulating ATP efflux and subsequent NLRP3 inflammasome activation. Blood. 2013;122(20):3473–3481. doi: 10.1182/blood-2013-05-502229. [DOI] [PubMed] [Google Scholar]
- 65.Sluyter R. The P2X7 receptor. Adv. Exp. Med. Biol. 2017;1051:17–53. doi: 10.1007/5584_2017_59. [DOI] [PubMed] [Google Scholar]
- 66.Yu S., Wang D., Huang L., Zhang Y., Luo R., Adah D., Tang Y., Zhao K., Lu B. The complement receptor C5aR2 promotes protein kinase R expression and contributes to NLRP3 inflammasome activation and HMGB1 release from macrophages. J. Biol. Chem. 2019;294(21):8384–8394. doi: 10.1074/jbc.RA118.006508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhakdi S., Tranum-Jensen J. Complement lysis: a hole is a hole. Immunol. Today. 1991;12(9):318–320. doi: 10.1016/0167-5699(91)90007-g. discussion 321. [DOI] [PubMed] [Google Scholar]
- 68.Triantafilou K., Hughes T.R., Triantafilou M., Morgan B.P. The complement membrane attack complex triggers intracellular Ca2+ fluxes leading to NLRP3 inflammasome activation. J. Cell. Sci. 2013;126(Pt 13):2903–2913. doi: 10.1242/jcs.124388. [DOI] [PubMed] [Google Scholar]
- 69.McBride R., Fielding B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4(11):2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165–175. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi C.S., Nabar N.R., Huang N.N., Kehrl J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-Infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coll R.C., O’Neill L., Schroder K. Questions and controversies in innate immune research: what is the physiological role of NLRP3? Cell Death Discov. 2016;2:16019. doi: 10.1038/cddiscovery.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimabukuro-Vornhagen A., Gödel P., Subklewe M., Stemmler H.J., Schlößer H.A., Schlaak M., Kochanek M., Böll B., von Bergwelt-Baildon M.S. Cytokine release syndrome. J. Immunother. Cancer. 2018;6(1):56. doi: 10.1186/s40425-018-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freeman T.L., Swartz T.H. Targeting the NLRP3 inflammasome in severe COVID-19. Front. Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/jci137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108(1):17–41. doi: 10.1002/jlb.3covr0520-272r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Root-Bernstein R. Innate receptor activation patterns involving TLR and NLR synergisms in COVID-19, ALI/ARDS and Sepsis cytokine storms: a review and model making novel predictions and therapeutic suggestions. Int. J. Mol. Sci. 2021;22(4) doi: 10.3390/ijms22042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barker B.R., Taxman D.J., Ting J.P. Cross-regulation between the IL-1β/IL-18 processing inflammasome and other inflammatory cytokines. Curr. Opin. Immunol. 2011;23(5):591–597. doi: 10.1016/j.coi.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li L., Huang Q., Wang D.C., Ingbar D.H., Wang X. Acute lung injury in patients with COVID-19 infection. Clin. Transl. Med. 2020;10(1):20–27. doi: 10.1002/ctm2.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tian S., Hu W., Niu L., Liu H., Xu H., Xiao S.Y. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID-19) Pneumonia in Two Patients With Lung Cancer. J. Thorac. Oncol. 2020;15(5):700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carsana L., Sonzogni A., Nasr A., Rossi R.S., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M., Galli M., Catena E., Tosoni A., Gianatti A., Nebuloni M. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 2020;20(10):1135–1140. doi: 10.1016/s1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morris G., Bortolasci C.C., Puri B.K., Olive L., Marx W., O’Neil A., Athan E., Carvalho A., Maes M., Walder K., Berk M. Preventing the development of severe COVID-19 by modifying immunothrombosis. Life Sci. 2021;264:118617. doi: 10.1016/j.lfs.2020.118617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilson J.G., Calfee C.S., Subphenotypes A.R.D.S. Understanding a heterogeneous syndrome. Crit. Care. 2020;24(1):102. doi: 10.1186/s13054-020-2778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogers A.J., Guan J., Trtchounian A., Hunninghake G.M., Kaimal R., Desai M., Kozikowski L.A., DeSouza L., Mogan S., Liu K.D., Matthay M.A., Steingrub J., Wheeler A., Yoon J.H., Nakahira K., Choi A.M., Baron R.M. Association of elevated plasma Interleukin-18 level with increased mortality in a clinical trial of statin treatment for acute respiratory distress syndrome. Crit. Care Med. 2019;47(8):1089–1096. doi: 10.1097/ccm.0000000000003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frantzeskaki F., Armaganidis A., Orfanos S.E. Immunothrombosis in acute respiratory distress syndrome: cross talks between inflammation and coagulation. Respiration. 2017;93(3):212–225. doi: 10.1159/000453002. [DOI] [PubMed] [Google Scholar]
- 90.El Haouari M. Platelet oxidative stress and its relationship with cardiovascular diseases in type 2 diabetes mellitus patients. Curr. Med. Chem. 2019;26(22):4145–4165. doi: 10.2174/0929867324666171005114456. [DOI] [PubMed] [Google Scholar]
- 91.Øynebråten I., Barois N., Bergeland T., Küchler A.M., Bakke O., Haraldsen G. Oligomerized, filamentous surface presentation of RANTES/CCL5 on vascular endothelial cells. Sci. Rep. 2015;5:9261. doi: 10.1038/srep09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sreeramkumar V., Adrover J.M., Ballesteros I., Cuartero M.I., Rossaint J., Bilbao I., Nácher M., Pitaval C., Radovanovic I., Fukui Y., McEver R.P., Filippi M.D., Lizasoain I., Ruiz-Cabello J., Zarbock A., Moro M.A., Hidalgo A. Neutrophils scan for activated platelets to initiate inflammation. Science. 2014;346(6214):1234–1238. doi: 10.1126/science.1256478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kerr N., de Rivero Vaccari J.P., Dietrich W.D., Keane R.W. Neural-respiratory inflammasome axis in traumatic brain injury. Exp. Neurol. 2020;323:113080. doi: 10.1016/j.expneurol.2019.113080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andersson U., Ottestad W., Tracey K.J. Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol. Med. 2020;26(1):42. doi: 10.1186/s10020-020-00172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ueki H., Wang I.H., Fukuyama S., Katsura H., da Silva Lopes T.J., Neumann G., Kawaoka Y. In vivo imaging of the pathophysiological changes and neutrophil dynamics in influenza virus-infected mouse lungs. Proc. Natl. Acad. Sci. U. S. A. 2018;115(28):E6622–e6629. doi: 10.1073/pnas.1806265115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boeltz S., Amini P., Anders H.J., Andrade F., Bilyy R., Chatfield S., Cichon I., Clancy D.M., Desai J., Dumych T., Dwivedi N., Gordon R.A., Hahn J., Hidalgo A., Hoffmann M.H., Kaplan M.J., Knight J.S., Kolaczkowska E., Kubes P., Leppkes M., Manfredi A.A., Martin S.J., Maueröder C., Maugeri N., Mitroulis I., Munoz L.E., Nakazawa D., Neeli I., Nizet V., Pieterse E., Radic M.Z., Reinwald C., Ritis K., Rovere-Querini P., Santocki M., Schauer C., Schett G., Shlomchik M.J., Simon H.U., Skendros P., Stojkov D., Vandenabeele P., Berghe T.V., van der Vlag J., Vitkov L., von Köckritz-Blickwede M., Yousefi S., Zarbock A., Herrmann M. To NET or not to NET:current opinions and state of the science regarding the formation of neutrophil extracellular traps. Cell Death Differ. 2019;26(3):395–408. doi: 10.1038/s41418-018-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schönrich G., Raftery M.J. Neutrophil extracellular traps go viral. Front. Immunol. 2016;7:366. doi: 10.3389/fimmu.2016.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S., Loda M., Looney M.R., McAllister F., Rayes R., Renaud S., Rousseau S., Salvatore S., Schwartz R.E., Spicer J.D., Yost C.C., Weber A., Zuo Y., Egeblad M. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zawrotniak M., Rapala-Kozik M. Neutrophil extracellular traps (NETs) - formation and implications. Acta Biochim. Pol. 2013;60(3):277–284. [PubMed] [Google Scholar]
- 101.Boukhris M., Hillani A., Moroni F., Annabi M.S., Addad F., Ribeiro M.H., Mansour S., Zhao X., Ybarra L.F., Abbate A., Vilca L.M., Azzalini L. Cardiovascular implications of the COVID-19 pandemic: a global perspective. Can. J. Cardiol. 2020;36(7):1068–1080. doi: 10.1016/j.cjca.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 104.Li H., Zhang S., Li F., Qin L. NLRX1 attenuates apoptosis and inflammatory responses in myocardial ischemia by inhibiting MAVS-dependent NLRP3 inflammasome activation. Mol. Immunol. 2016;76:90–97. doi: 10.1016/j.molimm.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 105.Iqubal A., Ahmed M., Ahmad S., Sahoo C.R., Iqubal M.K., Haque S.E. Environmental neurotoxic pollutants: review. Environ. Sci. Pollut. Res. Int. 2020;27(33):41175–41198. doi: 10.1007/s11356-020-10539-z. [DOI] [PubMed] [Google Scholar]
- 106.Jin Y., Ji W., Yang H., Chen S., Zhang W., Duan G. Endothelial activation and dysfunction in COVID-19: from basic mechanisms to potential therapeutic approaches. Signal Transduct. Target. Ther. 2020;5(1):293. doi: 10.1038/s41392-020-00454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Colliva A., Braga L., Giacca M., Zacchigna S. Endothelial cell-cardiomyocyte crosstalk in heart development and disease. J. Physiol. 2020;598(14):2923–2939. doi: 10.1113/jp276758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoel H., Heggelund L., Reikvam D.H., Stiksrud B., Ueland T., Michelsen A.E., Otterdal K., Muller K.E., Lind A., Muller F., Dudman S., Aukrust P., Dyrhol-Riise A.M., Holter J.C., Trøseid M. Elevated markers of gut leakage and inflammasome activation in COVID-19 patients with cardiac involvement. J. Intern. Med. 2021;289(4):523–531. doi: 10.1111/joim.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang B.Z., Chu H., Han S., Shuai H., Deng J., Hu Y.F., Gong H.R., Lee A.C., Zou Z., Yau T., Wu W., Hung I.F., Chan J.F., Yuen K.Y., Huang J.D. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30(10):928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zubair A.S., McAlpine L.S., Gardin T., Farhadian S., Kuruvilla D.E., Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 2020;77(8):1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Rivero Vaccari J.C., Dietrich W.D., Keane R.W., de Rivero Vaccari J.P. The inflammasome in times of COVID-19. Front. Immunol. 2020;11:583373. doi: 10.3389/fimmu.2020.583373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adinolfi E., Giuliani A.L., De Marchi E., Pegoraro A., Orioli E., Di Virgilio F. The P2X7 receptor: a main player in inflammation. Biochem. Pharmacol. 2018;151:234–244. doi: 10.1016/j.bcp.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 114.Sperlágh B., Illes P. P2X7 receptor: an emerging target in central nervous system diseases. Trends Pharmacol. Sci. 2014;35(10):537–547. doi: 10.1016/j.tips.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 115.Mohammadi S., Moosaie F., Aarabi M.H. Understanding the immunologic characteristics of neurologic manifestations of SARS-CoV-2 and potential immunological mechanisms. Mol. Neurobiol. 2020;57(12):5263–5275. doi: 10.1007/s12035-020-02094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heneka M.T., Kummer M.P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T.C., Gelpi E., Halle A., Korte M., Latz E., Golenbock D.T. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493(7434):674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yan Y.Q., Fang Y., Zheng R., Pu J.L., Zhang B.R. NLRP3 inflammasomes in Parkinson’s disease and their regulation by Parkin. Neuroscience. 2020;446:323–334. doi: 10.1016/j.neuroscience.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 118.Najafi S., Rajaei E., Moallemian R., Nokhostin F. The potential similarities of COVID-19 and autoimmune disease pathogenesis and therapeutic options: new insights approach. Clin. Rheumatol. 2020;39(11):3223–3235. doi: 10.1007/s10067-020-05376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ashrafizadeh M., Najafi M., Kavyiani N., Mohammadinejad R., Farkhondeh T., Samarghandian S. Anti-inflammatory activity of melatonin: a focus on the role of NLRP3 inflammasome. Inflammation. 2021 doi: 10.1007/s10753-021-01428-9. [DOI] [PubMed] [Google Scholar]
- 120.Phadke M., Saunik S. COVID-19 treatment by repurposing drugs until the vaccine is in sight. Drug Dev. Res. 2020;81(5):541–543. doi: 10.1002/ddr.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rodrigues-Diez R.R., Tejera-Muñoz A., Marquez-Exposito L., Rayego-Mateos S., Santos Sanchez L., Marchant V., Tejedor Santamaria L., Ramos A.M., Ortiz A., Egido J., Ruiz-Ortega M. Statins: Could an old friend help in the fight against COVID-19? Br. J. Pharmacol. 2020;177(21):4873–4886. doi: 10.1111/bph.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Otani K., Watanabe T., Shimada S., Takeda S., Itani S., Higashimori A., Nadatani Y., Nagami Y., Tanaka F., Kamata N., Yamagami H., Tanigawa T., Shiba M., Tominaga K., Fujiwara Y., Arakawa T. Colchicine prevents NSAID-induced small intestinal injury by inhibiting activation of the NLRP3 inflammasome. Sci. Rep. 2016;6:32587. doi: 10.1038/srep32587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kopitar-Jerala N. The role of interferons in inflammation and inflammasome activation. Front. Immunol. 2017;8:873. doi: 10.3389/fimmu.2017.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lendermon E.A., Coon T.A., Bednash J.S., Weathington N.M., McDyer J.F., Mallampalli R.K. Azithromycin decreases NALP3 mRNA stability in monocytes to limit inflammasome-dependent inflammation. Respir. Res. 2017;18(1):131. doi: 10.1186/s12931-017-0608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ko J.H., Yoon S.O., Lee H.J., Oh J.Y. Rapamycin regulates macrophage activation by inhibiting NLRP3 inflammasome-p38 MAPK-NFκB pathways in autophagy- and p62-dependent manners. Oncotarget. 2017;8(25):40817–40831. doi: 10.18632/oncotarget.17256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iyer S.S., He Q., Janczy J.R., Elliott E.I., Zhong Z., Olivier A.K., Sadler J.J., Knepper-Adrian V., Han R., Qiao L., Eisenbarth S.C., Nauseef W.M., Cassel S.L., Sutterwala F.S. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39(2):311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Collotta D., Hull W., Mastrocola R., Chiazza F., Cento A.S., Murphy C., Verta R., Alves G.F., Gaudioso G., Fava F., Yaqoob M., Aragno M., Tuohy K., Thiemermann C., Collino M. Baricitinib counteracts metaflammation, thus protecting against diet-induced metabolic abnormalities in mice. Mol. Metab. 2020;39:101009. doi: 10.1016/j.molmet.2020.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Purvis G.S.D., Collino M., Aranda-Tavio H., Chiazza F., O’Riordan C.E., Zeboudj L., Mohammad S., Collotta D., Verta R., Guisot N.E.S., Bunyard P., Yaqoob M.M., Greaves D.R., Thiemermann C. Inhibition of Bruton’s TK regulates macrophage NF-κB and NLRP3 inflammasome activation in metabolic inflammation. Br. J. Pharmacol. 2020;177(19):4416–4432. doi: 10.1111/bph.15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.He H., Jiang H., Chen Y., Ye J., Wang A., Wang C., Liu Q., Liang G., Deng X., Jiang W., Zhou R. Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat. Commun. 2018;9(1):2550. doi: 10.1038/s41467-018-04947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Juliana C., Fernandes-Alnemri T., Wu J., Datta P., Solorzano L., Yu J.W., Meng R., Quong A.A., Latz E., Scott C.P., Alnemri E.S. Anti-inflammatory compounds parthenolide and Bay 11-7082 are direct inhibitors of the inflammasome. J. Biol. Chem. 2010;285(13):9792–9802. doi: 10.1074/jbc.M109.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lamkanfi M., Mueller J.L., Vitari A.C., Misaghi S., Fedorova A., Deshayes K., Lee W.P., Hoffman H.M., Dixit V.M. Glyburide inhibits the Cryopyrin/Nalp3 inflammasome. J. Cell Biol. 2009;187(1):61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abbate A., Toldo S., Marchetti C., Kron J., Van Tassell B.W., Dinarello C.A. Interleukin-1 and the inflammasome as therapeutic targets in cardiovascular disease. Circ. Res. 2020;126(9):1260–1280. doi: 10.1161/circresaha.120.315937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yaqinuddin A., Kashir J. Novel therapeutic targets for SARS-CoV-2-induced acute lung injury: targeting a potential IL-1β/neutrophil extracellular traps feedback loop. Med. Hypotheses. 2020;143:109906. doi: 10.1016/j.mehy.2020.109906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bahadoram M., Keikhaei B., Saeedi-Boroujeni A., Mahmoudian-Sani M.R. Chloroquine/hydroxychloroquine: an inflammasome inhibitor in severe COVID-19? Naunyn Schmiedebergs Arch. Pharmacol. 2021:1–5. doi: 10.1007/s00210-020-02034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere-Querini P., Di Lucca G., Martinenghi S., Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2(6):e325–e331. doi: 10.1016/s2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Grebe A., Hoss F., Latz E. NLRP3 inflammasome and the IL-1 pathway in atherosclerosis. Circ. Res. 2018;122(12):1722–1740. doi: 10.1161/circresaha.118.311362. [DOI] [PubMed] [Google Scholar]
- 137.Hamam H.J., Palaniyar N. Post-translational modifications in NETosis and NETs-Mediated diseases. Biomolecules. 2019;9(8) doi: 10.3390/biom9080369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tagami T., Tosa R., Omura M., Fukushima H., Kaneko T., Endo T., Rinka H., Murai A., Yamaguchi J., Yoshikawa K., Saito N., Uzu H., Kase Y., Takatori M., Izumino H., Nakamura T., Seo R., Kitazawa Y., Sugita M., Takahashi H., Kuroki Y., Irahara T., Kanemura T., Yokota H., Kushimoto S. Effect of a selective neutrophil elastase inhibitor on mortality and ventilator-free days in patients with increased extravascular lung water: a post hoc analysis of the PiCCO Pulmonary Edema Study. J. Intensive Care. 2014;2(1):67. doi: 10.1186/s40560-014-0067-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Leung Y.Y., Yao Hui L.L., Kraus V.B. Colchicine--Update on mechanisms of action and therapeutic uses. Semin. Arthritis Rheum. 2015;45(3):341–350. doi: 10.1016/j.semarthrit.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coll R.C., Hill J.R., Day C.J., Zamoshnikova A., Boucher D., Massey N.L., Chitty J.L., Fraser J.A., Jennings M.P., Robertson A.A.B., Schroder K. MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol. 2019;15(6):556–559. doi: 10.1038/s41589-019-0277-7. [DOI] [PubMed] [Google Scholar]
- 141.McCarty M.F., Iloki Assanga S.B., Lewis Luján L., O’Keefe J.H., DiNicolantonio J.J. Nutraceutical strategies for suppressing NLRP3 inflammasome activation: pertinence to the management of COVID-19 and beyond. Nutrients. 2020;13(1) doi: 10.3390/nu13010047. [DOI] [PMC free article] [PubMed] [Google Scholar]