Abstract
Antibodies, and the B cell and plasma cell populations responsible for their production, are key components of the human immune system’s response to SARS-CoV-2, which has caused the coronavirus disease 2019 (COVID-19) pandemic. Here, we review findings addressing the nature of antibody responses against SARS-CoV-2 and their role in protecting from infection or modulating COVID-19 disease severity. In just over a year, much has been learned, and replicated in independent studies, about human immune responses to this pathogen, contributing to the development of effective vaccines. Nevertheless, important questions remain about the duration and effectiveness of antibody responses, differences between immunity derived from infection compared to vaccination, the cellular basis for serological findings, and the extent to which viral variants will escape from current immunity.
Röltgen and Boyd review research in the past year that has analyzed antibody and B cell responses to SARS-CoV-2 infection and vaccination, and their ability to prevent subsequent infection. The duration of immunity and the impact of new SARS-CoV-2 variants are major questions for the pandemic’s next phase.
Introduction
The entry of the novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), into human populations in 2019 set off a global pandemic of coronavirus disease (COVID-19) that has begun to rival the scope and impact of the influenza pandemic of 1918. The zoonotic origins of SARS-CoV-2 are thought to have been from a bat species, potentially followed by other intermediate hosts, and a possible period of cryptic spread in humans (Andersen et al., 2020; Zhou et al., 2020). On encountering largely immunologically naive human populations, the resultant virus has efficiently spread worldwide and caused over 3.8 million deaths (https://covid19.who.int/) at an accelerating pace. New, effective vaccines offer the hope of controlling the pandemic if they can be produced and, equally importantly, distributed on a global scale. In this review, we summarize the main lessons learned from the international effort to understand human antibody and B cell responses to SARS-CoV-2 infection and vaccination and anticipate challenges that lie ahead. The flood of relevant publications has required us to select examples to illustrate the main findings, but these are not meant to represent the first or last word on these topics.
SARS-CoV-2 belongs to the Coronaviridae family of single-stranded RNA viruses that includes four endemic human coronaviruses (HCoVs) that are usually associated with mild respiratory infections: the alphacoronaviruses, NL63 and 229E, and the betacoronaviruses (betaCoVs), HKU1 and OC43, as well as two highly pathogenic betaCoVs, SARS-CoV and MERS-CoV, which have caused deadly epidemics in the past. SARS-CoV-2 can cause a broad spectrum of manifestations in humans, ranging from mild respiratory and/or gastrointestinal symptoms to severe disease with extensive pneumonia and other organ involvement, as well as disorders that include coagulopathies and potential progression to respiratory failure and death. At least one-third of infected individuals remain asymptomatic (Oran and Topol, 2020), but can still be contagious and thus constitute a source of infection. After recovery from acute infection, many individuals have lingering or recurrent symptoms for many months. One of the pressing research imperatives in the COVID-19 pandemic has been the identification of factors that influence the outcome of infection. Apart from viral features, such as the size of the initial viral inoculum and viral mutations that may lead to differential pathogenicity, host determinants including age, sex, co-morbidities, and the state of an individual’s immune system and viral exposure history might also contribute to illness severity. Although humoral immune responses to SARS-CoV-2 infection and vaccination have been the focus of much initial research, evidence is emerging that innate immune mechanisms and T cell responses are also major factors that contribute to protection against SARS-CoV-2 (Sette and Crotty, 2021).
The genome of SARS-CoV-2 encodes four structural proteins, including the highly immunodominant spike (S) and nucleocapsid (N) antigens and 25 putative nonstructural and accessory proteins (Mariano et al., 2020). The S surface glycoprotein plays a major role in viral attachment and entry into host cells. The S receptor binding domain (RBD) varies among different coronaviruses and determines host species range and tissue tropism. SARS-CoV-2 RBD binds to the angiotensin-converting enzyme 2 (ACE2) receptor, which is expressed on a wide range of human cells across a variety of organs. Virus-neutralizing antibodies are thus primarily directed against the S protein and can prevent viral entry into host cells. The N protein is essential for viral replication and the packaging of viral RNA into new virions and is expressed abundantly during infection. Upon infection, SARS-CoV-2 elicits antibody responses mainly directed toward S and N. These antigens, particularly the S protein and its RBD domain, were rapidly and widely adopted for clinical and research serological assays from the early months of the pandemic, aided by the generous and public-spirited sharing of DNA sequences, protocols, and reagents within the scientific community (https://www.ncbi.nlm.nih.gov/nuccore/MN908947) (Stadlbauer et al., 2020; Wu et al., 2020).
More than a year later, and a few months after the rolling out of rapidly developed and highly effective vaccines, questions remain as to the nature and durability of SARS-CoV-2-specific antibody responses after infection and vaccination, and the B cell and plasma cell populations responsible for them. Waning antibody titers after infection or vaccination and reported subsequent infections suggest that immunity may be transient and incomplete, at least in a fraction of individuals. Moreover, some new emerging viral variants show decreased vulnerability to immunity stimulated by infection or vaccination with Wuhan-Hu-1-like antigens.
Pre-existing cross-reactive immunity to SARS-CoV-2
Research on viral infections has shown that established adaptive immunity against closely related viruses or virus variants can protect from infection or severe disease, but in other cases can worsen outcomes (St. John and Rathore, 2019; Welsh et al., 2010). Serological cross-reactivity is commonly observed in the responses to endemic HCoVs, and is associated with transient cross-protection or with attenuated symptoms (Aldridge et al., 2020; Callow et al., 1990; Sariol and Perlman, 2020). SARS-CoV-2 proteins and those of other HCoVs are generally distinct but share specific regions of sequence conservation that might be targeted by cross-reactive antibody responses.
Pre-pandemic specimens, and those from uninfected individuals during the pandemic, have provided interesting insights into pre-existing serum antibodies that bind SARS-CoV-2 antigens, potentially stimulated by prior HCoV infection. Most pre-pandemic/non-infected serum samples contain IgG antibodies to endemic HCoV S proteins (Anderson et al., 2021). In contrast, less than 1% of these samples contain IgG antibodies that bind SARS-CoV-2 S RBD. Low-level cross-reactivity is more commonly observed for the SARS-CoV-2 full-length S (as seen in 4% to 5% of study participants) and against the N protein (∼10% to 16% of study participants) (Anderson et al., 2021; Ng et al., 2020). Whether this pre-pandemic binding activity in polyclonal sera is due to cross-reactive antibodies that bind other HCoV antigens warrants additional study, as relatively few SARS-CoV-2 cross-reactive monoclonal antibodies (mAbs) have been isolated from pre-pandemic specimens (Wec et al., 2020). The S glycoprotein is proteolytically processed into two subunits: S1 (which contains RBD and the N-terminal domain [NTD]) and S2 (which mediates host-viral membrane fusion). S2 is the main target for pre-existing anti-S antibodies, consistent with its greater sequence conservation compared to S1, among HCoV species (Anderson et al., 2021; Jaimes et al., 2020; Ng et al., 2020; Nguyen-Contant et al., 2020). While RBD is the main target for virus-neutralizing antibodies, responses to S epitopes outside of the RBD have been shown to have some neutralizing activity, for example, by binding to the S1 NTD, or by preventing protease cleavage or conformational changes required for entry into cells (Chi et al., 2020; McCallum et al., 2021a; Poh et al., 2020). Antibodies to the N protein show little to no neutralization potential. This is consistent with reports that the pre-existing (predominantly non-RBD binding) SARS-CoV-2-reactive antibodies confer low to undetectable levels of SARS-CoV-2 neutralizing activity (Anderson et al., 2021).
High-resolution, viral linear peptide epitope profiling of antibodies in serum samples from pre-pandemic or non-infected control individuals recapitulates many of the results derived from full SARS-CoV-2 proteins or protein domains, despite not capturing data from conformational or discontinuous epitopes. Many pre-pandemic samples strongly recognize seasonal HCoV peptides, but show only limited reactivity with a few SARS-CoV-2 peptides (Ladner et al., 2021; Shrock et al., 2020). Pre-existing, cross-reactive antibodies preferentially target specific, immunodominant epitopes located in functional sites of the S2 subunit and show greater binding to the homologous endemic HCoV epitopes, compared to SARS-CoV-2 epitopes, while little reactivity is detected for SARS-CoV-2 RBD epitopes (Ladner et al., 2021). Notably, pre-pandemic serum reactivity is also observed for other SARS-CoV-2 peptides that do not show evidence of cross-reactive binding to other HCoVs, suggesting that reactivities might also be generated by non-HCoV antigens (Ladner et al., 2021).
Taken together, these data indicate that some pre-existing antibodies derived from prior exposure to other HCoVs recognize SARS-CoV-2 antigens. Anti-HCoV antibodies are also boosted by SARS-CoV-2 infection, particularly during severe COVID-19 illness (Röltgen et al., 2021). Whether these cross-reactive antibodies confer any protection against infection or whether they modulate disease severity is unclear. One report found that levels of pre-pandemic or pre-infection cross-reactive SARS-CoV-2-binding antibodies did not correlate with protection from SARS-CoV-2 infection and hospitalization (Anderson et al., 2021). In contrast, an epidemiological study based on electronic health records found that recent prior infection with endemic HCoVs was associated with less severe COVID-19 illness (Sagar et al., 2021). It seems possible that some transient immunological protection from SARS-CoV-2 may follow infection with other HCoVs, but if so, it remains to be determined whether this effect is due to antibody responses or to other immune system components such as T cells. Overall, it appears that pre-existing, cross-reactive antibodies form a small fraction of the total humoral response to SARS-CoV-2 infection.
Antibody responses to primary SARS-CoV-2 infection
Systemic antibody responses
While recovery from many acute viral infections, such as from yellow fever, measles, polio, and smallpox, can confer lifelong humoral and cell-mediated immunity, protective immunity to other viruses, including coronaviruses, is comparatively short-lived. Virus-specific antibodies produced by plasma cells may be the most important factor in the long-term prevention of reinfection by most viruses. As discussed in more detail below, durable antibody responses appear to require coordinated T and B lymphocyte interactions within lymphoid tissue germinal centers (GCs) to generate long-lived plasma cells, as well as class-switched memory B cells that can rapidly mount secondary responses after re-encountering antigens (Figures 1 and 2 ).
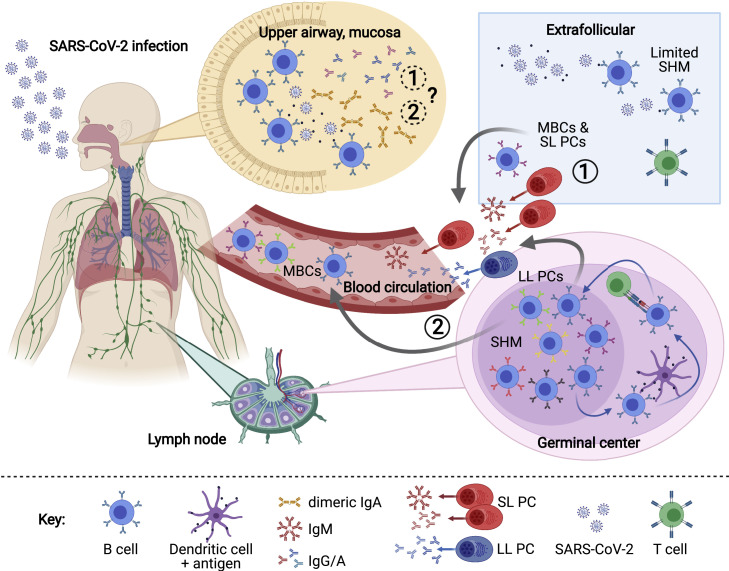
Figure 1.
Overview of the humoral immune response to SARS-CoV-2
SARS-CoV-2 antigen recognition initiates a cascade of immune responses, including the activation of naive B cells. Activated B cells can differentiate rapidly into extrafollicular, short-lived plasma cells (SL PCs) and memory B cells (MBCs) with low rates of somatic hypermutation (SHM) (1), or they can enter germinal centers of secondary lymphoid organs such as lymph nodes, where they undergo rounds of SHM and affinity maturation, resulting in long-lived plasma cells (LL PCs) and MBCs (2). Antibody-secreting plasma cells and MBCs can enter the blood and (potentially) mucosa, where they help to fight viral infection and protect from reinfection. LL PCs also transit to the bone marrow and potentially to other anatomical sites. Schematic created with biorender.com.
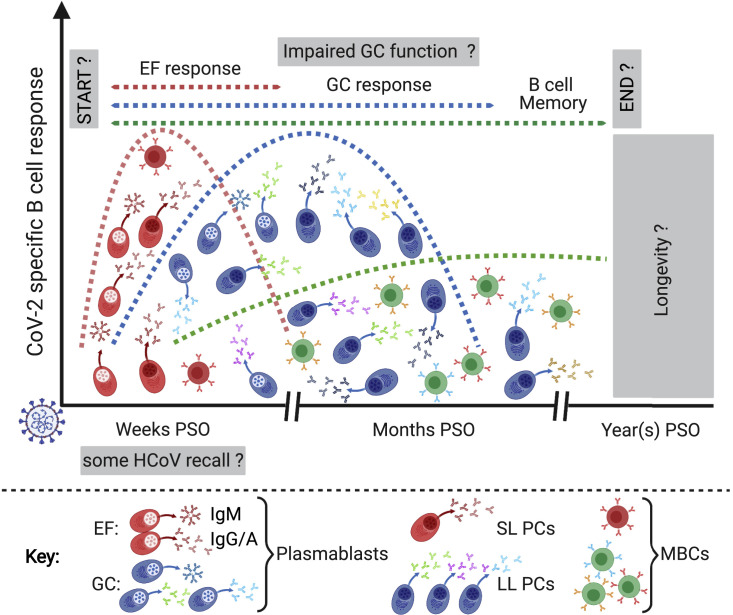
Figure 2.
Anti-SARS-CoV-2 B cell responses
B cell responses to SARS-CoV-2 infection are initiated primarily by the stimulation of naive B cells and potentially some HCoV cross-reactive memory B cells. The time frames for the development of extrafollicular (EF) responses, which produce short-lived antibody-secreting cells (plasmablasts and SL PCs) and memory B cells (MBCs), and germinal center (GC) responses, which provide somatically hypermutated long-lived plasma cells (LL PCs) and class-switched memory B cells, are not well described for human tissues and may take place simultaneously. The duration of these responses, as well as the longevity of memory B cells formed in different microanatomical sites, is still unclear. Signs of impaired GC function have been identified in deceased COVID-19 patients. PSO, post symptom onset. Schematic created with biorender.com.
Upon infection with SARS-CoV-2, naive B cells or potentially pre-existing memory B cells from prior HCoV exposures are activated by antigen recognition and CD4+ T cell help. In many viral infections, such as those caused by Dengue and Zika virus, serum IgM responses precede the appearance of class-switched IgG and IgA antibodies (Ravichandran et al., 2019; Vázquez et al., 2007). In contrast, the serum IgG responses to SARS-CoV-2 S and N appear at approximately the same time as serum IgM and IgA, usually within the first 2 weeks after symptom onset (Iyer et al., 2020; Long et al., 2020a; Röltgen et al., 2020). The median time to seroconversion is between 11 and 13 days post-symptom onset (PSO) for RBD (Iyer et al., 2020; Long et al., 2020a). Seroconversion rates in hospitalized patients for anti-RBD IgM, IgG, and IgA reach their maximum between 4 to 6 weeks PSO (Röltgen et al., 2020). An IgG-based seroconversion rate of >95% that is irrespective of disease severity has been reported for assays relying on the full-length S protein (Wajnberg et al., 2020). IgM and IgA antibodies rapidly decline with median times to seroreversion about 7 and 10 weeks PSO, respectively (Iyer et al., 2020), although more persistent IgA responses have also been reported (Gaebler et al., 2021). The rate at which IgG antibody levels decay remains a topic of some debate and differs between antigens. While anti-S levels appear to be stable for at least 3 months post-infection and show modest decreases after 5 to 8 months (Dan et al., 2021; Wajnberg et al., 2020), anti-RBD and -N antibody responses wane more rapidly (Dan et al., 2021; Ibarrondo et al., 2020; Isho et al., 2020). It is tempting to speculate that anti-S2 antibodies may account for these differences in whole-S and RBD titer persistence, decaying more slowly due to greater contributions from boosted cross-reactive memory clones. The timing of sample collection is a crucial factor to be considered when assessing the decay of antibody titers, as during the initial months after infection there may be a more rapid rate of decrease in IgG produced by transient plasmablasts, followed by a slower long-term rate of decrease that is dependent on fully differentiated plasma cells. Longitudinal studies that are tracking patients over longer periods of time after infection will likely soon provide additional data about the durability and levels of protective titers of anti-SARS-CoV-2 antibodies.
The magnitude of anti-SARS-CoV-2 antibody responses correlates with COVID-19 disease severity, with the highest antibody titers developed by the most severely ill patients, who often also have higher viral loads in nasopharyngeal swabs compared to patients with milder illness (Long et al., 2020b; Röltgen et al., 2020) (Figures 3A and 3B). Broader antibody responses to S and N peptides with more extensive epitope spreading are also seen in patients with severe disease (Shrock et al., 2020). It is possible that individuals whose immune responses cannot fight the infection early, perhaps due to less effective innate or T cell immunity or to antibody targeting of non-neutralizing viral antigens (Atyeo et al., 2020; Röltgen et al., 2020; Sette and Crotty, 2021), develop higher viral antigen loads that contribute to an extended period of antibody evolution and epitope spreading, giving rise to the observed stronger and broader antibody responses to SARS-CoV-2. Heterogeneity in antibody responses is also seen among different age groups. Older adults are at higher risk of developing severe COVID-19 and are therefore also more likely to develop high antibody responses to SARS-CoV-2. Children experience predominantly asymptomatic and mild disease and generate lower antibody responses to the S and N proteins. To date, it is unclear why children are less affected clinically by SARS-CoV-2 and whether specific humoral responses, in addition to factors such as ACE2 expression, might play a role in their protection from more severe COVID-19 disease. One study has reported that a more diverse antibody landscape is present in children compared to adults, featuring a higher proportion of antibodies that target SARS-CoV-2 accessory proteins and open reading frames (Hachim et al., 2021). A few children (∼0.002% of cases) develop severe disease in the form of a multisystem inflammatory syndrome (MIS-C), but similar anti-S and -N antibody profiles are observed in children with and without MIS-C (Weisberg et al., 2021).
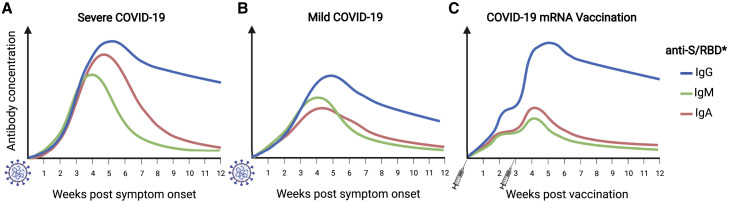
Figure 3.
Anti-SARS-CoV-2 antibody responses
SARS-CoV-2 infection elicits robust antibody responses to the S protein.
(A and B) Patients with severe COVID-19 develop significantly higher anti-S IgM, IgG, and IgA titers than do patients with mild manifestations.
(C) Individuals vaccinated with two doses of S protein-encoding mRNA vaccines develop IgG antibody titers comparable to those of severely ill patients, but lower concentrations of IgM and IgA.
∗Anti-RBD antibody responses show similar patterns but are in general lower than those against S and decline more rapidly. Schematic created with biorender.com.
Neutralizing antibodies are considered a key correlate of immunity to SARS-CoV-2, as demonstrated by passive antibody transfer and viral challenge studies in nonhuman primates (McMahan et al., 2021). Once infection is established, neutralization might play less of a role in controlling SARS-CoV-2, as many severely ill patients (including those who die) develop high neutralizing antibody titers, and lower neutralization titers are observed in most mildly infected adults and children (Robbiani et al., 2020; Weisberg et al., 2021). A threshold for protective neutralizing antibody responses has yet to be defined, and candidate thresholds will likely be affected by viral variants and viral loads encountered during exposures, among other factors.
The finding that a higher titer antibody response does not necessarily correlate with milder disease has focused attention on whether there are differences in the timing or targeting of humoral responses that could contribute to patient outcomes and to disease severity. Differences in the proportions of antibodies that target S compared to N antigens are seen in the early humoral responses in patients who survive compared to those who die of COVID-19, including not only higher anti-S to anti-N antibody ratios but also S-specific phagocytic and complement activity (Atyeo et al., 2020; Röltgen et al., 2020). Despite these associations, there is still considerable heterogeneity in antibody responses within groups defined by disease severity or outcome. A detailed examination of the time course of SARS-CoV-2 antibody responses in individual patients has revealed several response patterns. Among patient groups who were admitted to hospital but not requiring ICU care, who were admitted to ICU, or who were dying of their illness, a range of antibody responses were observed—from undetectable to high neutralizing titers—in different individuals by the time of their recovery or death (Röltgen et al., 2020). These observations indicate that some patients are able to resolve their illness prior to the appearance of antibodies in their blood, while others suffer severe disease despite mounting strong antibody responses. Notably, although some patients who died of COVID-19 had delayed seroconversion, this was not generally the case, and time to seroconversion was not significantly associated with patient outcomes. Causation is difficult to establish in human studies, but these discordant relationships between antibody responses and patient outcomes strongly suggest that innate immune mechanisms and T cell responses play major roles in determining the disease course in the primary infection of individual patients.
Structural variations in the antibody Fc domain and in the IgG glycome, previously associated with age, sex, and autoimmune diseases (Gudelj et al., 2018), might also modulate the course of SARS-CoV-2 infection. Even minor changes in the composition of Fc-associated glycans can significantly alter Fc conformations and interaction with Fcγ receptor (FcγR) family members on leukocytes to modulate effector responses. Patients with severe COVID-19 have elevated concentrations of anti-SARS-CoV-2 IgG lacking glycan fucosylation, compared to patients with milder illness (Chakraborty et al., 2021; Larsen et al., 2021). Afucosylated IgG responses formed against some enveloped viruses have been reported to have increased binding affinity to the activating FcγRIIIa, potentially promoting cytokine storms and immune-mediated pathologies (Larsen et al., 2021). The importance of this association for COVID-19 disease severity warrants further evaluation.
In summary, consistent associations between greater disease severity and higher antibody responses have been reported in many studies, while antibody responses that target the S compared to the N antigen are greater in patients with mild disease. Within patient groups, however, considerable heterogeneity in the antibody response is seen.
Mucosal antibody responses
In contrast to systemic antibody responses, less is known about the role of mucosal immunity to SARS-CoV-2. Mucosal surfaces of the respiratory tract are the entry point for respiratory pathogens such as SARS-CoV-2 and likely represent an important site for the initiation of antibody responses (Figure 1). Secretory IgA is the principal isotype on mucosal surfaces and may thus play a central role in early SARS-CoV-2 defense.
Antibody kinetics in saliva samples obtained from SARS-CoV-2-infected individuals are reported to be similar to those of serum samples, in that anti-S and anti-RBD IgG levels are stable over several months PSO, whereas IgM and IgA antibodies wane more rapidly (Cervia et al., 2021; Isho et al., 2020). The sensitivity with which antibodies could be detected in saliva samples was lower than in serum samples, for example, the detection of anti-SARS-CoV-2 S antibodies in 90 saliva samples from COVID-19 patients was 57% for IgM, 89% for IgG, and 51% for IgA (Isho et al., 2020). IgM and IgG antibody levels in saliva showed a significant positive correlation with those in serum, while the positive correlation of saliva and plasma IgA was more limited (Cervia et al., 2021; Isho et al., 2020). Elevated functional IgG-related responses were apparent in nasal samples from COVID-19 convalescent donors who had experienced severe disease, while donors with mild-to-moderate COVID-19 had elevated nasal IgA-related responses (Butler et al., 2021). In contrast to data from serum samples, which show pseudovirus neutralization activity to correlate positively with disease severity, nasal wash samples from subjects with severe disease show little to no viral neutralization, whereas individuals with mild symptoms have elevated mucosal neutralization activity (Butler et al., 2021). Intriguingly, neutralization activity detected in nasal samples, but not in serum samples, showed a positive correlation with IgA responses.
Although cohorts in all of the mucosal studies discussed here were not large enough to enable a robust examination of trends, the described associations between mucosal IgA binding, virus neutralization responses, and less severe disease together suggest that mucosal IgA might play an important role in protective immune mechanisms and might also be a factor that contributes to disease outcome. One study of neutralizing mAbs has highlighted the greater potency of dimeric IgA, the predominant secretory form at mucosal sites (Wang et al., 2021a). A protective role for mucosal IgA might also have important implications for vaccine development. Vaccines that are administered intramuscularly, as opposed to intranasally, are expected to induce mainly systemic IgG, and not mucosal IgA, although this has not yet been determined for approved SARS-CoV-2 vaccines. Mucosal immunity might contribute, or even be required, to achieve protection from infection and to prevent onward transmission of SARS-CoV-2 in humans.
B-lineage cells in SARS-CoV-2 infection
The convenience and widespread availability of serological testing for SARS-CoV-2 antibodies has meant that it is the primary source of data for large cohorts and for epidemiological studies analyzing humoral immune responses to SARS-CoV-2 infection in human populations. However, detailed study of the clonal populations of antigen-specific B cells and plasma cells that are stimulated by infection or vaccination (and the immunoglobulin gene rearrangements they express) is required to arrive at a more complete understanding of the cellular mechanisms that give rise to the serological data. As viral variants have arisen in human populations, it has become increasingly important to understand how structural changes in S, particularly in the RBD and NTD (Figure 4 ) domains, affect the binding of important classes of neutralizing antibodies. The complementary and supporting roles of innate immunity, T cell immunity, and humoral immunity, and the interactions of B cells with other immune cell types, particularly CD4+ T cells, all likely need to be considered to explain the disease course and outcomes of individual patients. Initial findings related to these interactions have been surveyed recently (Sette and Crotty, 2021). Here, we focus on patient responses through the lens of B cell biology to highlight areas of growing consensus and remaining gaps in our knowledge.
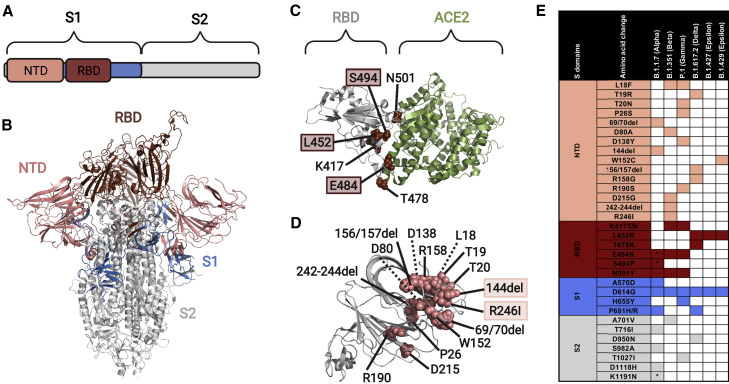
Figure 4.
S variant emergence and impact on antibody recognition
(A) The S protein consists of two subdomains: S1 (shaded blue, contains the NTD and RBD) and S2 (shaded gray).
(B) S protein structure (PDB: 6ZGG; Wrobel et al., 2020), showing the RBD (dark red) and NTD (salmon) domains.
(C) S protein structure (PDB: 6MOJ; Lan et al., 2020) showing key surface residues in RBD (sphere representation, dark red) that have been reported in variants of concern. Amino acid residues highlighted with a colored box decreased neutralization by polyclonal sera from COVID-19 convalescent patients (Greaney et al., 2021).
(D) S protein structure (PDB: 6ZGG) showing key surface residues in NTD (sphere representation, salmon) reported in variants of concern. Amino acid residues highlighted with a colored box are part of the NTD “supersite,” a region that is involved in binding by at least eight reported neutralizing antibodies (Cerutti et al., 2021).
(E) Table of amino acid changes reported in currently circulating viral variants of concern (source: CDC, WHO).
∗Detected in some sequences but not all. The protein and domain structures were visualized with PyMOL.
B cells in acute infection
Models of human B cell responses to viral infection, combining extrapolation of results from mouse data and actual observations from humans, include extrafollicular B cell responses that provide a rapid, anti-viral function by generating short-lived antibody-secreting cells and non-class-switched memory B cells. In parallel to these events, other B cells enter the GCs of secondary lymphoid tissues, where they undergo cell division, somatic hypermutation (SHM) of antibody genes, and selection for improved antigen binding, giving rise to long-lived plasma cells and class-switched memory B cells. Thus, in the initial weeks of an infection, rapidly expanding plasmablasts produce high antibody titers, which decay after the acute infection has resolved and are followed by the more sustained production of antibodies at lower levels due to distinct populations of longer-lived plasma cells (Figures 1 and 2).
B cells and plasma cells from the extrafollicular and GC reactions can enter the blood, which has been the most common source of these cells in human immunological studies. Relatively few studies of human immune responses have directly examined lymph nodes, spleen, or other potent organizing sites of adaptive immunity, although fine-needle aspiration of lymph node cells offers a strategy to address this limitation in the future (Havenar-Daughton et al., 2020).
Descriptions of peripheral blood B cells in the initial weeks of an acute SARS-CoV-2 infection have reported modest relative B cell lymphopenia, and variable increases in plasmablast frequencies, in some cases exceeding 30% of total B cells, reminiscent of findings in other severe acute viral illnesses such as Ebola virus infection (Kaneko et al., 2020; Mathew et al., 2020; McElroy et al., 2015). These B cell subset changes are consistent but transient, as reported in a longer follow-up study of COVID-19 patients that analyzed plasmablast frequencies within 7 days of hospitalization, at discharge, and after 3 to 6 months of convalescence (Shuwa et al., 2021). This study confirmed the finding of elevated plasmablasts in acute COVID-19 but reported that plasmablasts returned to baseline frequencies in convalescent samples 3 to 6 months after infection. Other B cell phenotypes have been reported in the acute phase of infection; for example, one study performed a single-B cell transcriptomic analysis and identified non-plasmablast CD71+-activated B cells, CD11c+ FcRL5+ “atypical” B cells, and a cell phenotype with intermediate characteristics between these two (Sokal et al., 2021).
The sequencing of immunoglobulin heavy chain genes obtained from peripheral blood B cells during acute COVID-19 has revealed the presence of highly polyclonal B cell populations that have class switched, usually to IgG subtypes with a lesser contribution of IgA subtypes, but with little or no SHM (Nielsen et al., 2020). These cells appear in the blood at approximately the same time that patients seroconvert and when plasmablast frequencies are elevated. Another analysis of total immunoglobulin gene repertoires in acute COVID-19 patients’ blood reported the oligoclonal expansion of certain clones within the repertoires (Kuri-Cervantes et al., 2020). Isotype-specific deep sequencing of heavy chain gene repertoires in a third study confirmed the abundance of low-SHM IgG3 or IgG1-expressing B cells in the initial weeks following the onset of COVID-19 symptoms (Kim et al., 2021).
Together, these data from acute COVID-19 patients suggest that the early B cell response to infection derives from the stimulation of naive B cells that class switch and differentiate to plasmablast and potentially to other activated phenotypes without accumulating substantial SHM prior to their appearance in the blood. But is there any evidence from acute COVID-19 patients of a cross-reactive secondary response to epitopes that are shared by SARS-CoV-2 and one or more of the endemic HCoVs? Such a response might be expected, given reports of pre-pandemic serum antibodies in many children and some adults that cross-react with SARS-CoV-2 antigens, and given evidence of pre-pandemic B cell clones that express heavy chain genes with high sequence similarity to known SARS-CoV-2 specific antibodies, with higher frequencies in children (Anderson et al., 2021; Ng et al., 2020; Yang et al., 2021). A study of SARS-CoV-2 S-binding mAbs isolated from a pre-pandemic donor that had survived a SARS-CoV infection in the 2003 outbreak demonstrated that a small proportion of these mAbs were cross-reactive with HCoV S proteins (Wec et al., 2020). However, the unusual circumstance of this donor’s SARS-CoV infection might have stimulated clones that would not be present in human populations whose prior coronavirus exposures were limited to endemic HCoVs. In another study, testing of the antigen specificity and cross-reactivity of large numbers of SARS-CoV-2 S-binding memory B cells from samples collected 3 and 6 months after SARS-CoV-2 infection showed that a significant fraction (approximately 12% at 3 months) of clones were cross-reactive and bound to the S proteins of HCoVs OC43, HKU1, or both (Sokal et al., 2021). Evidence that these clones were derived from preexisting cross-reactive memory B cells was provided by the presence of substantially higher levels of SHM in the cross-reactive clones compared to those that bound SARS-CoV-2 RBD alone.
Germinal center versus extrafollicular B cell responses in infection
In an effort to address the question of whether acute serological responses to SARS-CoV-2 infection derive from GC-dependent B cell populations, one study analyzed post-mortem lymph node and spleen specimens obtained from patients who had died of COVID-19. This study found GCs and the numbers of BCL-6+ GC B cells to be markedly decreased in the follicles of these secondary lymphoid tissues (Kaneko et al., 2020). These observations are suggestive of impaired GC function and of the lack of formation of long-lived memory B cells, high-affinity antigen-specific B cells, and, presumably, long-lived plasma cells. As the authors note, these findings are based on the most severely ill patients, and those with milder disease may not exhibit a comparable impairment of GC structures or function. Furthermore, for severely ill patients who survive COVID-19, accumulated evidence now indicates that their titers of SARS-CoV-2-specific antibodies persist at higher levels than those of patients with milder illness. For recovered severely ill or mildly ill patients, class-switched memory B cells with accumulating SHM appear to increase in frequency in their blood for several months after the onset of symptoms and persist stably for at least 6 to 8 months, suggesting that some GC-derived clones contribute to the responses (Dan et al., 2021; Hartley et al., 2020; Rodda et al., 2021; Sokal et al., 2021).
A further exploration of B cell responses in COVID-19 patients used flow cytometry to examine peripheral blood B cell phenotypes associated with extrafollicular B cell proliferations. These included CD11c+ “activated naïve” B cells that differentiate into double-negative-type-2 (DN2) B cells classified as “double-negative” due to their lack of CD27 and IgD expression, and as “type-2” due to expression of CD11c but lack of CD21 (Woodruff et al., 2020). Indeed, patients with severe COVID-19 showed higher frequencies of the DN2 B cells compared to those with mild illness and also had higher plasmablast frequencies. The patients in this cohort did not show the overall lymphopenia or B cell lymphopenia noted in some other studies (Kaneko et al., 2020; Mathew et al., 2020). A third and novel B cell population designated by Woodruff et al. as DN3, characterized by the lack of CD27, IgD, CD21, and CD11c expression, has also been identified in COVID-19 patients. Consistent with other reports, most of the antibody-secreting cells in the peripheral blood showed low SHM frequencies in class-switched cells.
B cell memory
Data from other coronavirus infections indicate that, for most patients with mild or moderate disease, antibody titers wane relatively rapidly, becoming negative within 2 to 3 years for most SARS and MERS patients, and decreasing to the point where reinfection is common in the case of endemic HCoVs (Edridge et al., 2020; Sariol and Perlman, 2020; Tang et al., 2011; Wu et al., 2007). However, data from limited numbers of patients who have survived severe SARS or MERS show longer-lasting serological responses. Similar patterns have begun to emerge for SARS-CoV-2 serology after infection, as described above. These data indicate that most coronavirus infections do not efficiently produce long-lived plasma cells, but whether they generate long-lived memory B cell populations that can mount a rapid, secondary response upon re-exposure is less clear. A study of SARS patients 6 years after their infection found that their serum antibody levels had decreased to undetectable levels in 21 of 23 patients and that none of the patients had detectable specific memory B cells in culture ELISPOT experiments (Tang et al., 2011).
Data obtained from the memory B cells of convalescent SARS-CoV-2 patients in the initial 16 months of the pandemic appear to be somewhat more encouraging. One study detected RBD or N-specific memory B cells in all 25 patients of a cohort convalescing from mild, moderate, or severe COVID-19 and found that the frequency of these cells appeared to increase from the early weeks of infection to approximately 150 days PSO (Hartley et al., 2020). Whether the increasing affinity of the B cells for antigen at later time points contributed to their detection was not evaluated. IgM+ memory B cells in this study formed the largest fraction of total memory B cells in the first month PSO, but declined in frequency at later time points, while IgG1+ memory B cells predominated at later time points and showed more stable frequencies. Notably, the absolute number of memory B cells per ml of blood varied by up to 10-fold between different participants convalescent from mild disease, indicating that severe disease may not be necessary to form high frequencies of memory B cells in the months after acute infection. An additional in-depth analysis of adaptive immune memory confirmed that memory B cells specific for S, RBD, or N increased in frequency in the blood for the first 3 to 4 months and remained stable for up to 8 months PSO (Dan et al., 2021). These reports of stable memory B cell frequencies are further supported by a study that compared S-specific memory B cell frequencies in acute infection and at 3 and 6 months post infection (Sokal et al., 2021), and an analysis of RBD-specific memory B cells at 3 months (Rodda et al., 2021). As noted above, the Sokal et al. study identified cross-reactive memory clones that bound SARS-CoV-2 RBD, as well as the S proteins of HCoVs OC43, HKU1, or both at 3 months, but the frequencies of these cross-reactive memory B cell clones decreased by 6 months post-infection, whereas the frequency of non-cross-reactive RBD binders increased in proportion, suggesting that the cross-reactive clones had shorter half-lives or were otherwise disfavored in the memory pool after longer times. The memory B cells that bound SARS-CoV-2 S or RBD also showed progressive increases in SHM during the months after infection (Sokal et al., 2021).
BCR repertoires and mAbs
An intense effort by many laboratories around the world to identify neutralizing mAbs to SARS-CoV-2 has resulted in a rich body of knowledge about the antibodies that bind the S protein, and the genomic rearrangements that encode them. Current databases of SARS-CoV-2-specific antibody sequences are heavily biased toward RBD binders, as these comprise most of the validated neutralizing mAbs, and were expected and subsequently shown to be responsible for most of the neutralizing activity in polyclonal sera (http://opig.stats.ox.ac.uk/webapps/covabdab/) (Greaney et al., 2021; Piccoli et al., 2020).
Several strategies used to isolate neutralizing antibodies have helped to characterize the specificities of B cells in acute and convalescent patient samples. One illustrative study evaluating the frequency of neutralizing antibodies in clonally expanded B cells from convalescent patient memory B cell populations sequenced using single-cell transcriptomic methods found only one neutralizing mAb of 130 tested. By contrast, mAbs from B cells meeting the criteria of binding to RBD in flow cytometric sorting, expressing IgG1, not being in clones containing IgG2+ members, having clone members with at least 2% SHM in the heavy chain, and not being in clones whose members all had an exhausted B cell or naive B cell phenotype, yielded a 25% success rate in identifying neutralizing antibodies (Cao et al., 2020). Notably, sequences with high similarity to known SARS-CoV neutralizers were also validated as being neutralizing in pseudotyped SARS-CoV-2 assays. Other cross-neutralizing antibodies for SARS-CoV and SARS-CoV-2 have been isolated from SARS and COVID-19 patient repertoires (Ju et al., 2020).
Several key epitope regions that overlap with the ACE2 binding site on RBD and appear to be the most common sites targeted by neutralizing antibodies have also been identified in numerous studies that have characterized mAbs isolated from SARS-CoV-2 RBD or from S-binding B cells (Brouwer et al., 2020; Dejnirattisai et al., 2021a; Ju et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Shi et al., 2020; Tortorici et al., 2020; Zost et al., 2020). Notable in this extensive body of work are the structural biology studies, particularly those using cryo-electron microscopy and X-ray crystallography, which have accelerated the process of determining the paratope:epitope interactions of large numbers of mAbs and antigens (Barnes et al., 2020; Ju et al., 2020; Robbiani et al., 2020; Shi et al., 2020; Tortorici et al., 2020; Yuan et al., 2020). A particular tour de force in this regard is a paper reporting the structures of 19 Fabs bound to SARS-CoV-2 RBD (Dejnirattisai et al., 2021a).
Some studies have also identified SARS-CoV-2 neutralizing antibodies that bind the NTD of S1, the domain encoded by amino acid residues upstream of the RBD (Brouwer et al., 2020; Chi et al., 2020). Other antibodies with similar activity appear to recognize a common glycan-free region on the NTD surface (Cerutti et al., 2021; Dejnirattisai et al., 2021a; McCallum et al., 2021a; Suryadevara et al., 2021). The precise neutralization mechanism employed by these antibodies remains to be elucidated, but it appears to involve a post-attachment step in the virus’s infection cycle, because antibodies that are added after the virus has been allowed to adhere to cells still prevent infection (Suryadevara et al., 2021).
Convergent antibodies to SARS-CoV-2
A surprising feature of SARS-CoV-2 antibody responses that has emerged in the past year is the frequency of convergent or public antibody gene rearrangements that have high sequence similarity among different individuals (Nielsen et al., 2020; Robbiani et al., 2020). Heavy chain gene sequencing without antibody binding characterization has revealed that many convergent sequence types exist among COVID-19 patients (Nielsen et al., 2020); other convergent B cell receptors (BCRs) have been revealed by single-B cell cloning and mAb expression studies (Andreano et al., 2021; Dejnirattisai et al., 2021a; Robbiani et al., 2020; Yuan et al., 2020). Although each patient’s repertoire of SARS-CoV-2-binding antibodies is predominantly composed of clonotypes not observed in other individuals, certain common gene rearrangements in human naive repertoires have been found to bind SARS-CoV-2 S antigen and even to have strong neutralizing activity (Andreano et al., 2021; Robbiani et al., 2020; Yuan et al., 2020). Some of the convergent neutralizing antibodies for SARS-CoV-2 do not appear to require many, or any, SHM changes from the germline gene segments that encode the heavy and light chain variable regions (Andreano et al., 2021; Robbiani et al., 2020; Yuan et al., 2020). These antibodies provide informative counter-examples to the idea that high-affinity neutralizing antibodies must be the product of GC reactions and of rounds of SHM and affinity maturation. This is not to say that the total pool of convergent antibodies is particularly enriched for functionally important binders; rather, it seems most likely that the interplay between the frequencies of particular kinds of rearrangements in the primary human B cell repertoire, and the constraints enforced by selection for binding to particular epitopes on the viral antigens, determine the fraction of convergent sequences that have a protective role. Over time, as SARS-CoV-2 persists in human populations, selective pressure on the virus to escape common human neutralizing antibody types will likely also play a major role in the observed B cell responses.
Antibody and B cell responses to SARS-CoV-2 vaccination
The rapid and prolonged spread of SARS-CoV-2, as well as waning humoral immune responses, particularly after mild infection, suggest that vaccination will be required to end the COVID-19 pandemic while minimizing deaths and the long-term consequences of infection. Recent regulatory authorization and the beginning of mass vaccination programs in the US and elsewhere, with lipid nanoparticle mRNA vaccines and adenoviral-vectored vaccines, each encoding the SARS-CoV-2 S, have led to enormous interest in the magnitude, neutralizing titers, and duration of vaccine-stimulated antibody responses, as well as in the effects on other immune cell types such as T cells. COVID-19 vaccines currently authorized by the US Food and Drug Administration (FDA) for emergency use include the BNT162b2 (Pfizer-BioNTech) and mRNA-1273 (Moderna) mRNA vaccines, which are administered intramuscularly in two doses, and the adenoviral vector Ad26.COV2.S (Janssen) vaccine, which is administered intramuscularly in one dose. Phase III trials showed greater than 90% efficacy at preventing COVID-19 after two doses of the mRNA vaccines (Baden et al., 2021; Polack et al., 2020) and ∼67% efficacy for Ad26.COV2.S (Sadoff et al., 2021). Other countries have authorized additional vaccines such as the adenoviral vector ChAdOx1 nCoV-19 (AZD1222, AstraZeneca) vaccine, which is ∼70% efficacious (Voysey et al., 2021). While correlates of vaccine-mediated protection are not yet defined quantitatively, all of these vaccines collectively target the SARS-CoV-2 S protein, with the aim of inducing high viral neutralization titers.
The very high efficacy of novel mRNA-based vaccines has been one of the most important insights from research efforts targeting SARS-CoV-2. Both the Pfizer-BioNTech and Moderna vaccines induce high levels of anti-S and anti-RBD IgG binding and neutralizing antibodies that decline slightly by the studied time points, 6 weeks and 4 months after the initial dose (Röltgen et al., 2021; Widge et al., 2021). Peak antibody responses are comparable to those of severely ill COVID-19 patients (Röltgen et al., 2021). Interestingly, serological responses to the Pfizer-BioNTech mRNA vaccine, compared to severe SARS-CoV-2 infection, show a greater dominance of IgG over IgM and IgA isotypes, indicating efficient IgG class switching (Figure 3). Antibodies induced by this vaccine also showed decreased breadth of binding to the S proteins of other HCoVs. We speculate that dissimilar inflammatory environments during infection versus vaccination, as well as the different anatomical compartments where immune responses are initiated, might lead to the narrower antibody response observed after vaccination (Röltgen et al., 2021). A single dose of the BNT162b2 or the mRNA-1273 vaccine in individuals that were seropositive (due to a previous SARS-CoV-2 infection) elicited post-vaccination IgG titers that were at least as high as those of seronegative individuals after two vaccine doses (Krammer et al., 2021). Most previously uninfected single-dose recipients mount moderate titers by day 21, prompting discussions of dose-sparing strategies and the fastest routes to herd immunity, in light of the limited supply of vaccines in most countries. Initial data on the B cell populations stimulated by vaccination indicate that the frequencies of memory B cells generated are approximately equivalent to those seen in survivors of severe COVID-19 (Wang et al., 2021b).
Key questions about the duration of vaccine-induced antibody titers for all of the SARS-CoV-2 vaccines await further data in the coming months. Initial observations of decreases in titers in the first months following vaccination suggest that booster doses may be required to maintain sufficiently high titers for protection over the long term.
Antibody responses to viral variants
A major concern from the beginning of the SARS-CoV-2 pandemic has been the possibility that new viral variants would evolve with increased pathogenicity, transmissibility, or escape from human immune responses. To balance the capacity for evolution, adaptation, and host-response escape with the need to maintain a long and complex genome, coronaviruses replicate their RNA with an error-prone RNA polymerase, but also encode a 3′ to 5′ exonuclease with proofreading function. Unfortunately, SARS-CoV-2 has encountered an extremely favorable environment (a large immunologically naive reservoir) to explore its sequence space. A comparison of 3,823 representative viral genomes (https://nextstrain.org/ncov/global) to the early Wuhan-Hu-1 strain identified that SARS-CoV-2 viruses had accumulated around 75 heritable non-synonymous nucleotide mutations as of January 2021 and 24 non-synonymous mutations that potentially arose independently in different viral isolates (Wu et al., 2021a). A viral variant with a D614G substitution in the S protein appeared early in the pandemic, rapidly replacing the early Wuhan-Hu-1 due to its enhanced viral replication and infectivity (Yurkovetskiy et al., 2020). Since then, several amino acid changes in the S protein and particularly in the RBD have been identified with increasing frequency in viral isolates, showing improved receptor binding activities and viral escape from both therapeutic mAbs and plasma antibodies (Plante et al., 2021; Wu et al., 2021a) (Figure 4).
Deep mutational scanning using yeast display of RBD variants and isolation of those with reduced binding by COVID-19 patient plasma has revealed the importance of a few immunodominant RBD epitopes that are targeted by neutralizing antibodies (Greaney et al., 2021). This prescient work has successfully anticipated viral variants that have appeared singly, or recurrently, in primary viral isolates from patients in different parts of the world. Interestingly, E484 (and to a lesser extent, nearby residues such as L452, L455, F456, G485, F486, and F490) was identified as the amino acid position at which changes had the largest effect on binding and neutralization by plasma antibodies. One potential explanation for this observation is that this site is often targeted by antibodies that are common in human populations. Indeed, two of the four predominant SARS-CoV-2 variants of concern, as defined by the Centers for Disease Control and Prevention (CDC), currently co-circulating globally, P.1 (Gamma, first detected Brazil) and B.1.351 (Beta, emerging from South Africa), share amino acid substitution E484K (in addition to K417N/T) and are associated with resistance to antibody neutralization in sera from infected and vaccinated individuals (Chen et al., 2021; Edara et al., 2021; Wu et al., 2021b). IgG titers stimulated by Wuhan-Hu-1-like virus infection or mRNA vaccination show remarkably consistent decreases of IgG binding to the S and RBD antigens of major viral variants of concern, with the greatest decreases seen for B.1.351, then P.1, and relatively minimal decreases in binding to a third variant, the B.1.1.7 (Alpha) virus first reported in the UK (Röltgen et al., 2021). The amino acid change N501Y, found in the B.1.1.7 variant, is shared with variants P.1 and B.1.351 and has likely been selected because it enhances the binding affinity of the virus to ACE2, rather than because it plays a major role in the evasion of humoral immune responses (Muik et al., 2021; Starr et al., 2020; Wu et al., 2021b). Immune sera also showed reduced neutralizing potency against a fourth variant of concern, B.1.427/B.1.429 (Epsilon, first identified in California), characterized by amino acid substitution L452R (McCallum et al., 2021b). The World Health Organization (WHO) has classified another variant, B.1.617.2 (Delta, first detected in India) containing RBD substitutions L452R and T478K, as being of global concern; this variant also has decreased neutralization by Pfizer-BioNTech vaccinated sera (Wall et al., 2021).
As would be predicted, the viral variants that contain mutations in RBD amino acids in the epitopes of mAbs, including therapeutic mAbs, can show partial or complete ablation of binding. Examples of such immune escape, including loss of neutralizing mAb activity, have been shown for B.1.1.7 (Graham et al., 2021), B.1.351 (Hoffmann et al., 2021; Li et al., 2021), P.1 (Dejnirattisai et al., 2021b; Hoffmann et al., 2021), B.1.427/B.1.429 (McCallum et al., 2021b), and B.1.617.2 (Planas et al., 2021). We expect more data on the efficacy of vaccines and antibody-based therapeutics against these and other newly emerging SARS-CoV-2 variants to become available in the coming months.
Summary and outlook
A remarkable scientific effort during the past 16 months has rapidly addressed many major questions about humoral immunity to SARS-CoV-2 and about the B cell and plasma cell populations that generate protective antibody responses. Key questions, such as the duration of antibody titers, the thresholds at which protective immunity will begin to be impaired, and the severity of illness in individuals who do become reinfected, will require additional time and data to answer. The relative importance of systemic, as compared to mucosal, antibody responses remains in need of more study. Without doubt, current and new viral variants will have a major impact on the fitness and spread of SARS-CoV-2, particularly with regard to its ability to evade previously established antibody responses from infection or from vaccination with antigens derived from earlier virus sequences. While vaccine efficacy data for SARS-CoV-2 variants have yet to be systematically analyzed and published, vaccines may have to be updated periodically to match relevant variants, as they are for influenza. Whether updated vaccines for viral variants will actually elicit a good immune response to the modified epitopes, or simply boost older responses, will be a critically important topic of research in coming years. Similarly, the ability of cross-reactive antibody responses elicited by SARS-CoV-2 vaccination or infection to potentially protect against other more divergent coronaviruses that could cause future pandemics will be an important area of investigation (Saunders et al., 2021).
Acknowledgments
This work was supported by NIH/NIAID R01AI127877 (S.D.B.), NIH/NIAID R01AI130398 (S.D.B.), NIH 1U54CA260517 (S.D.B.), and an endowment to S.D.B. from the Crown Family Foundation.
Declaration of interests
S.D.B. has consulted for Regeneron, Sanofi, and Novartis on topics unrelated to this manuscript, and owns stock in AbCellera Biologics.
References
- Aldridge R.W., Lewer D., Beale S., Johnson A.M., Zambon M., Hayward A.C., Fragaszy E.B., Flu Watch Group Seasonality and immunity to laboratory-confirmed seasonal coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): results from the Flu Watch cohort study. Wellcome Open Res. 2020;5:52. doi: 10.12688/wellcomeopenres.15812.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. UPenn COVID Processing Unit Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., et al. Distinct Early Serological Signatures Track with SARS-CoV-2 Survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S.E., Crowley A.R., Natarajan H., Xu S., Weiner J.A., Bobak C.A., Mattox D.E., Lee J., Wieland-Alter W., Connor R.I., et al. Distinct Features and Functions of Systemic and Mucosal Humoral Immunity Among SARS-CoV-2 Convalescent Individuals. Front. Immunol. 2021;11:618685. doi: 10.3389/fimmu.2020.618685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervia C., Nilsson J., Zurbuchen Y., Valaperti A., Schreiner J., Wolfensberger A., Raeber M.E., Adamo S., Weigang S., Emmenegger M., et al. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J. Allergy Clin. Immunol. 2021;147:545–557.e9. doi: 10.1016/j.jaci.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e22. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.V., Norwood C., Floyd K., Lai L., Davis-Gardner M.E., Hudson W.H., Mantus G., Nyhoff L.E., Adelman M.W., Fineman R., et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 2021;29:516–521.e3. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D., Kaczorowska J., Hoste A.C.R., Bakker M., Klein M., Loens K., Jebbink M.F., Matser A., Kinsella C.M., Rueda P., et al. Seasonal coronavirus protective immunity is short-lasting. Nature Medicine. 2020;26:1691–1693. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham C., Seow J., Huettner I., Khan H., Kouphou N., Acors S., Winstone H., Pickering S., Galao R.P., Dupont L., et al. Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant. Immunity. 2021;54:1276–1289.e6. doi: 10.1016/j.immuni.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudelj I., Lauc G., Pezer M. Immunoglobulin G glycosylation in aging and diseases. Cell. Immunol. 2018;333:65–79. doi: 10.1016/j.cellimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Hachim A., Gu H., Kavian O., Kwan M.Y., Chan W.-H., Yau Y.S., Chiu S.S., Tsang O.T., Hui D.S., Ma F., et al. MedRxiv; 2021. The SARS-CoV-2 antibody landscape is lower in magnitude for structural proteins, diversified for accessory proteins and stable long-term in children. [Google Scholar]
- Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., Peleg A.Y., Boo I., Drummer H.E., Hogarth P.M., et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Newton I.G., Zare S.Y., Reiss S.M., Schwan B., Suh M.J., Hasteh F., Levi G., Crotty S. Normal human lymph node T follicular helper cells and germinal center B cells accessed via fine needle aspirations. J. Immunol. Methods. 2020;479:112746. doi: 10.1016/j.jim.2020.112746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid Decay of Anti-SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5:eabe0367. doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Chappie J.S., Millet J.K., Whittaker G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Massachusetts Consortium on Pathogen Readiness Specimen Working Group Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., Lee H., Jung J., Kang C.K., Song K.-H., et al. Stereotypic neutralizing VH antibodies against SARS-CoV-2 spike protein receptor binding domain in patients with COVID-19 and healthy individuals. Sci. Transl. Med. 2021;13:eabd6990. doi: 10.1126/scitranslmed.abd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermúdez-González M.C., Bielak D.A., Carreño J.M., Chernet R.L., et al. Antibody Responses in Seropositive Persons after a Single Dose of SARS-CoV-2 mRNA Vaccine. N. Engl. J. Med. 2021;384:1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D’ambrozio J., Schaecher K.E., Stone M., Dong W., et al. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep. Med. 2021;2:100189. doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Nouta J., Hoepel W., Chen H.-J., Linty F., Visser R., Brinkhaus M., et al. Amsterdam UMC COVID-19. biobank study group Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371:eabc8378. doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front. Mol. Biosci. 2020;7:605236. doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Bassi J., Marco A.D., Chen A., Walls A.C., Iulio J.D., Tortorici M.A., Navarro M.-J., Silacci-Fregni C., Saliba C., et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. BioRxiv. 2021 doi: 10.1101/2021.03.31.437925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy A.K., Akondy R.S., Davis C.W., Ellebedy A.H., Mehta A.K., Kraft C.S., Lyon G.M., Ribner B.S., Varkey J., Sidney J., et al. Human Ebola virus infection results in substantial immune activation. Proc. Natl. Acad. Sci. USA. 2015;112:4719–4724. doi: 10.1073/pnas.1502619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio. 2020;11 doi: 10.1128/mBio.01991-20. e01991–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.-Y., Rustagi A., Rogers A.J., et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host & Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oran D.P., Topol E.J. Prevalence of Asymptomatic SARS-CoV-2 Infection : A Narrative Review. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. ). Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. BioRxiv. 2021 doi: 10.1101/2021.05.26.445838. [DOI] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19's next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y.-P., Chee R.S.-L., Fong S.-W., Yeo N.K.-W., Lee W.-H., Torres-Ruesta A., et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020;11:2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran S., Hahn M., Belaunzarán-Zamudio P.F., Ramos-Castañeda J., Nájera-Cancino G., Caballero-Sosa S., Navarro-Fuentes K.R., Ruiz-Palacios G., Golding H., Beigel J.H., Khurana S. Differential human antibody repertoires following Zika infection and the implications for serodiagnostics and disease outcome. Nat. Commun. 2019;10:1943. doi: 10.1038/s41467-019-09914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Arunachalam P.S., Yang F., Hoh R.A., Wirz O.F., Lee A.S., Gao F., Mallajosyula V., Li C., et al. mRNA vaccination compared to infection elicits an IgG-predominant response with greater SARS-CoV-2 specificity and similar decrease in variant spike recognition. MedRxiv. 2021 doi: 10.1101/2021.04.05.21254952. [DOI] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. New England Journal of Medicine. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. American Society for Clinical Investigation; 2021. Recent endemic coronavirus infection is associated with less-severe COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K.O., Lee E., Parks R., Martinez D.R., Li D., Chen H., Edwards R.J., Gobeil S., Barr M., Mansouri K., et al. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021 doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. MGH COVID-19 Collection & Processing Team Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuwa H.A., Shaw T.N., Knight S.B., Wemyss K., McClure F.A., Pearmain L., Prise I., Jagger C., Morgan D.J., Khan S., et al. Alterations in T and B cell function persist in convalescent COVID-19 patients. Med. 2021;2:720–735.e4. doi: 10.1016/j.medj.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John A.L., Rathore A.P.S. Adaptive immune responses to primary and secondary dengue virus infections. Nat. Rev. Immunol. 2019;19:218–230. doi: 10.1038/s41577-019-0123-x. [DOI] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez S., Cabezas S., Pérez A.B., Pupo M., Ruiz D., Calzada N., Bernardo L., Castro O., González D., Serrano T., et al. Kinetics of antibodies in sera, saliva, and urine samples from adult patients with primary or secondary dengue 3 virus infections. Int. J. Infect. Dis. 2007;11:256–262. doi: 10.1016/j.ijid.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall E.C., Wu M., Harvey R., Kelly G., Warchal S., Sawyer C., Daniels R., Hobson P., Hatipoglu E., Ngai Y., et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. The Lancet. 2021;397:2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.-H., Oliveira T.Y., Oren D.A., et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.-H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.M., Che J.W., Brehm M.A., Selin L.K. Heterologous immunity between viruses. Immunol. Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2021;384:80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-P., Wang N.-C., Chang Y.-H., Tian X.-Y., Na D.-Y., Zhang L.-Y., Zheng L., Lan T., Wang L.-F., Liang G.-D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Wang L., Zhou H.-Y., Ji C.-Y., Xia S.Z., Cao Y., Meng J., Ding X., Gold S., Jiang T., Cheng G. One year of SARS-CoV-2 evolution. Cell Host Microbe. 2021;29:503–507. doi: 10.1016/j.chom.2021.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine - Preliminary Report. N Engl J Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Nielsen S.C.A., Hoh R.A., Röltgen K., Wirz O.F., Haraguchi E., Jean G.H., Lee J.-Y., Pham T.D., Jackson K.J.L., et al. Shared B cell memory to coronaviruses and other pathogens varies in human age groups and tissues. Science. 2021;372:738–741. doi: 10.1126/science.abf6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]