Summary
Increasing evidence suggests infants develop unique neutralizing antibody (nAb) responses to HIV compared to adults. Here, we dissected the nAb response of an infant whose virus is in clinical trials as a vaccine immunogen, with a goal of characterizing the broad responses in the infant to this antigen. We isolated 73 nAbs from infant BG505 and identified a large number of clonal families. Twenty-six antibodies neutralized tier 2 viruses—in some cases, viruses from the same clade as BG505, and in others, a different clade, although none showed notable breadth. Several nAbs demonstrated antibody-dependent cellular cytotoxicity activity and targeted the V3 loop. These findings suggest an impressive polyclonal response to HIV infection in infant BG505, adding to the growing evidence that the nAb response to HIV in infants is polyclonal—a desirable vaccine response to a rapidly evolving virus like HIV.
Keywords: HIV, neutralizing antibody, infant, polyclonal, ADCC, V3, BG505
Graphical abstract
Highlights
Infant BG505 developed a broad and polyclonal antibody response to HIV-1
We isolate 73 neutralizing antibodies spanning 19 clonal lineages from BG505
Many antibodies mediate cell killing but show limited neutralization breadth
V3 is a common epitope target of the BG505 antibodies
Although BG505 Envelope trimer is reputable in the HIV field, this infant’s antibody response was unknown. Simonich et al. isolated 73 neutralizing antibodies encompassing 19 clonal lineages from BG505 at 27 months of age. BG505 developed a vast polyclonal antibody response to HIV, an advantageous response to elicit by vaccination.
Introduction
Massive efforts have been made to understand broadly neutralizing antibody (nAb) responses in HIV-infected individuals in an attempt to inform their induction by vaccination.1,2 Broad nAb (bnAb) responses capable of neutralizing viruses from diverse clades of HIV have high levels of somatic hypermutation (SHM) and take years to develop in HIV-infected adults.3 In contrast, cross-clade nAb responses were achieved in a majority of infants by a median of 20 months of age; remarkably, bnAb responses were detectable within a year after infection for a subset of infants.4 A subsequent study identified bnAb responses in 75% of children over 5 years of age compared to 19% of infected adults.5 Together, these studies suggest a unique nAb response in infants and children compared to adults that leads to breadth. Dozens of adult-derived bnAbs have been studied in detail,1,2,6 whereas until recently, nAbs from infants have been largely ignored. We isolated the first HIV nAbs from an infant, BF520, and demonstrated that infant HIV-specific nAbs and bnAbs have unique characteristics compared to adult bnAbs, including much lower levels of SHM.7,8 Similar antibody characteristics have also been observed for another V3-specific bnAb isolated from a pediatric elite neutralizer infected with a clade C virus.9 Thus, infant HIV-specific nAbs may provide unique insights into productive and potentially protective antibody responses to HIV.
Infant BG505 developed an impressive nAb response detected at approximately 2 years after HIV infection, neutralizing 91% of Tier 2 viruses in an expanded panel, where HIV was first detected at 6 weeks of life.4 The transmitted Env variant from infant BG50510 is something of a household name in the HIV field because it was used to generate the first native-like soluble Env trimer, BG505.SOSIP.664.11 The BG505.SOSIP.664 trimer has been the focus of numerous detailed studies of the HIV Env trimer, including the first high-resolution structural characterization of a native-like Env trimer.12,13 The BG505 trimer has facilitated greater understanding of the conformational states of Env and its function in receptor binding,12, 13, 14, 15, 16, 17, 18 trimer glycosylation,17,19, 20, 21, 22, 23 the antigenic landscape of Env, and the epitope targets of bnAbs.12,14,17,24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 The BG505 SOSIP has also been used to generate more biologically relevant simian-human immunodeficiency virus chimeras (SHIVs) for macaque models of infection.39, 40, 41 Additionally, this trimer has been tested in a number of pre-clinical vaccine trials, eliciting tier 2 nAbs in rabbits, guinea pigs, and macaques and bnAbs in cows.14,35,42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53 Human clinical trials using the BG505 Env trimer are currently underway. For these reasons, the details of the nAb response to the BG505 Env in the infected infant is of high interest to the field.
A single vial of peripheral blood mononuclear cells (PMBCs), taken approximately 2 years post-infection, was available from infant BG505. To identify HIV-specific B cells contributing to the broadly neutralizing plasma response of infant BG505, we utilized several available tools for nAb isolation by combining two methods commonly used to identify HIV-specific B cells.2 We used both the autologous clade A BG505 SOSIP trimer and a heterologous clade C SOSIP trimer from a virus neutralized by contemporaneous plasma antibodies as baits to selectively isolate B cells that bind these antigens. We coupled this with large-scale culture and functional screening of all remaining immunoglobulin G (IgG) B cells. From these efforts, we isolated 73 nAbs from infant BG505. Many of these monoclonal antibodies (mAbs) mediated antibody-dependent cellular cytotoxicity (ADCC), which has been associated with improved infant outcomes.54,55 Characterization of the mAbs that developed in response to this unique transmitted HIV variant complements studies using the BG505 Env as an immunogen and adds to our understanding of cross-clade nAb responses.
Results
HIV-specific nAbs isolated from infant BG505
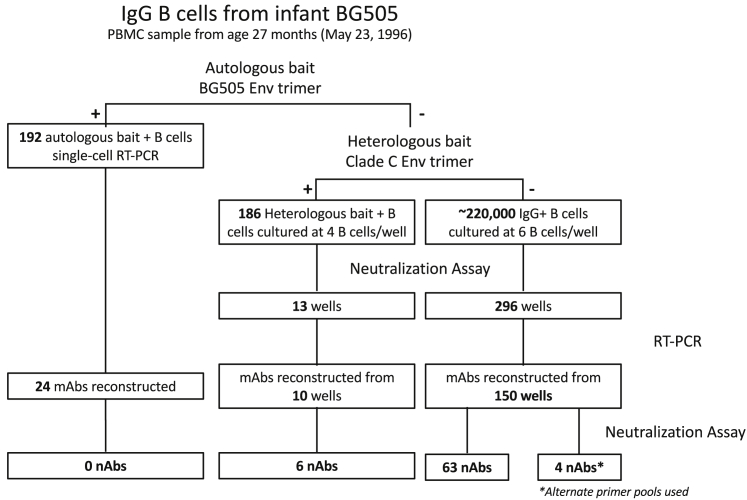
The BG505 PBMC sample from 27 months of age (M27) collected in 1996 was ∼90% viable and contained ∼6 million total live cells. The autologous BG505.SOSIP.664 Env trimer bait was bound by 192 IgG B cells, and attempts were made to amplify heavy- and light-chain sequences from all 192 cells. Paired heavy and light chains were amplified from 24 wells. None of the 24 reconstructed mAbs demonstrated neutralizing activity against SF162 virus (no detectable half maximal inhibitory concentration (IC50) at ≤20 μg/mL), which is an easy-to-neutralize tier 1 virus that should detect even low-level neutralizing activity. These mAbs also did not neutralize the autologous BG505.W6M.C2 transmitted founder (T/F) virus expressing the same Env on which the BG505.SOSIP.664 trimer protein is based, nor did the subset tested by ELISA (N = 14) bind trimer or monomer.
The heterologous subtype C DU422.SOSIP.664 Env trimer bait was bound by 186 B cells. These bait-isolated B cells were subsequently cultured to promote antibody secretion and screened for neutralizing activity as a second screen to avoid labor-intensive attempts at reconstruction of such a large number of antibodies. Additionally, ∼220,000 IgG-expressing B cells that did not bind either Env trimer were cultured and screened for neutralizing activity (Figure 1). IgG was detected in 58% of randomly sampled wells at day 12. For the B cells isolated using the heterologous DU422.SOSIP.664 bait, 13/46 of the culture wells showed evidence of HIV-specific neutralizing activity. Ten of the thirteen mAbs were successfully amplified and produced, and six demonstrated neutralization of SF162 (Figure 1). Of the ∼220,000 cultured IgG B cells, an additional 296 wells were identified by neutralization assay as potentially containing HIV-specific nAbs. Antibodies were reconstructed from 150/296 wells and 63 mAbs neutralized SF162 (Figure 1). Fifty representative mAbs that did not neutralize SF162 were tested for neutralization of autologous BG505 T/F virus, and none showed activity.
Figure 1.
Identification of HIV-neutralizing antibodies
Schematic of the BG505 B cell sort and numbers of HIV-specific neutralizing antibodies identified by each method, representing the single biological and technical replicate performed. The four nAbs isolated using alternative amplification methods are discussed separately and the data shown in Figure S3.
The 69 nAbs isolated from BG505 at M27 using this three-pronged approach belong to 18 clonal lineages with as many as 12 nAbs in a single lineage. The clonal lineages were assigned using deep sequencing data of the BG505 antibody repertoire at 14 weeks (W14) of life. Interestingly, BG505 nAbs utilized only 4 heavy-chain germline V-genes, VH1-18, VH1-69, VH3-21, and VH5-51, with a single VH3-21 antibody and >50% that utilized VH1-69. Heavy-chain complementarity-determining region 3 (CDR3) lengths ranged from 10 to 28 amino acid residues, and heavy-chain SHM ranged from 3.9% to 10.8% at the nucleotide level (Figure 2).
Figure 2.
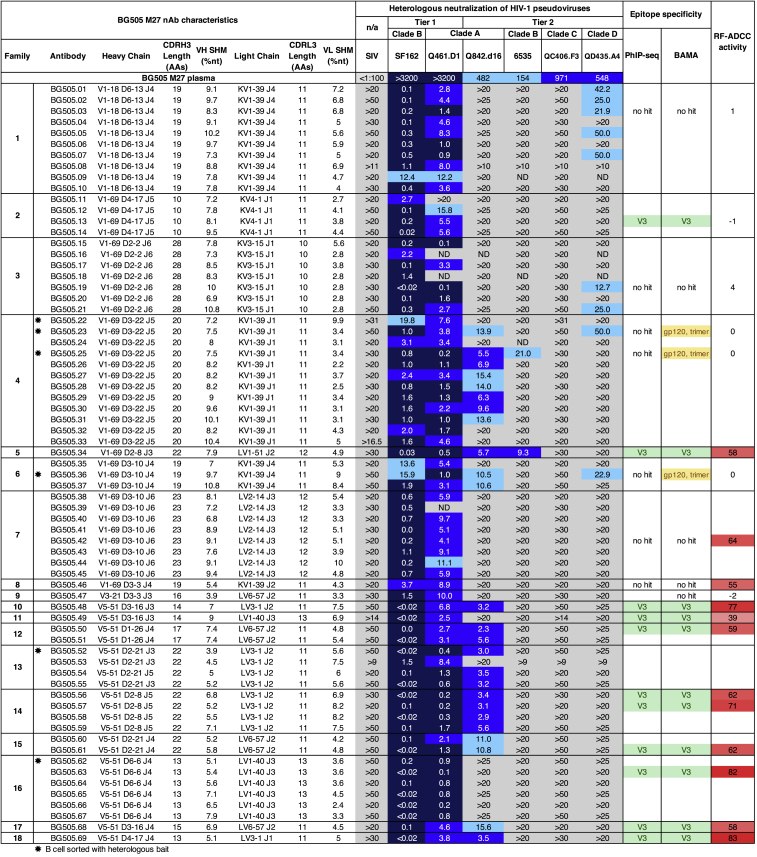
Heterologous neutralization by BG505 antibodies
Neutralization of panel viruses by BG505 M27 plasma and nAbs. Plasma-neutralizing activity against this heterologous virus panel is shown at the top. Antibodies are in rows with names and sequence characteristics of each nAb shown on the left. Stars indicate antibodies identified using the heterologous bait to bind B cells prior to culturing. Heterologous viruses are in columns to the right, with the first two rows indicating virus tier and clade. SIV was included as a negative control. IC50 values (μg/mL) represent an average of two independent experiments performed in duplicate. Darker blue shading indicates more potent neutralization. Gray indicates that 50% neutralization was not achieved at the highest mAb concentration tested. A second clade C pseudovirus was tested (CAP210.E8) and was not neutralized by any of the BG505 nAbs at 20 μg/mL. RF-ADCC activity indicates the percentage of target cells killed, normalized to polyclonal anti-HIV immunoglobulin from pooled serum (HIVIG) activity, with darker shades of red indicating more potent activity; data are representative of at least two technical replicates. PhIP-seq epitope mapping indicates V3 linear peptide specificity; data are representative of at least two technical replicates. See also Figure S5.
Heterologous neutralizing activity of BG505 antibodies
Heterologous neutralizing activity of the 69 isolated BG505 nAbs is shown in Figure 2. Viruses in the panel included tier 1 viruses and tier 2 viruses representing multiple clades based on their susceptibility to BG505 M27 plasma neutralization.4 The majority of mAbs neutralized only tier 1 viruses. The tier 2 clade A virus Q842.d16, the same clade as the infecting virus (92% nucleotide identity), was neutralized by 25 of the isolated antibodies. Eleven BG505 nAbs demonstrated low potency neutralization of tier 2 viruses from clades B and D, with different antibodies mediating neutralization of viruses from different clades, suggestive of a polyclonal response. Two antibodies demonstrated tier 2 clade A and B neutralizing activity, and two antibodies demonstrated tier 2 clade A and D neutralizing activity. Three of the eleven cross-clade nAbs (BG505.23, 25, and 36) were identified using the subtype C trimer bait combined with functional screening, and the other eight (BG505.1-3, 5, 7, 19, 21, and 34) were isolated from cultured IgG-expressing B cells that were not pre-selected for HIV binding. Five of the clade D QD435-neutralizing antibodies belong to the second largest clonal family identified, defined by VH1-18 and CDRH3 length of 19 amino acids. The largest clonal family identified with 12 members (VH1-69 and CDRH3 length of 20 amino acids) included nAbs that showed different cross-clade neutralizing activity, with one member (BG505.25) neutralizing a tier 2 clade B virus and the other (BG505.23) a tier 2 clade D virus, both of which were identified using the heterologous bait. None of the antibodies were able to recapitulate the full breadth of the plasma, and none neutralized the tier 2 clade C viruses despite using a clade C SOSIP trimer as bait.
BG505 nAbs bind to but do not neutralize BG505 autologous Env variants
To determine whether the diverse HIV-specific nAb lineages were able to bind or neutralize autologous Env isolates, Env variants from the time point when HIV was detected, week 6 (W6), were previously cloned10 and used to generate the BG505 SOSIP trimer.11 Additional Env sequences from early infection (W14), and later (M27), were isolated and cloned into an expression vector to generate pseudoviruses. Samples were not available from this infant between W14 and M27. The sequences isolated from BG505 at W6 and W14 cluster together, whereas the later variants from M27 show more variation, including those from maternal sequences isolated near the time of transmission (Figure S1).
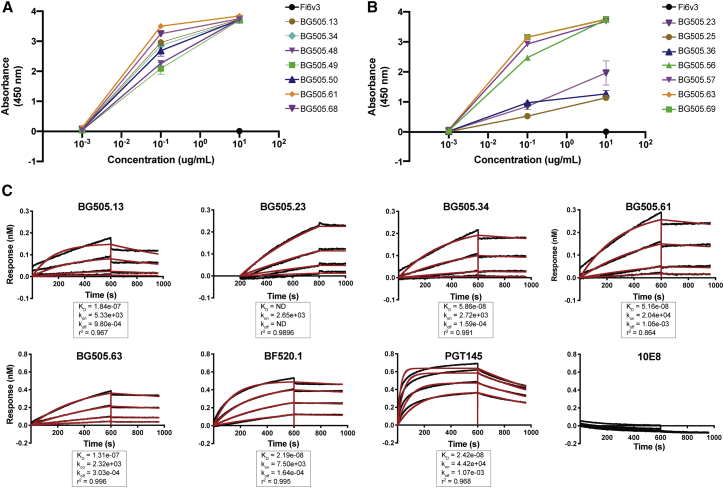
A subset of BG505 nAbs (BG505.13, 23, 25, 34, 36, 48–50, 56, 57, 61, 63, 68, and 69) were tested for their ability to bind to autologous envelope by ELISA (Figures 3A and 3B). All of the tested mAbs were found to bind BG505 SOSIP trimer, and this was further supported by biolayer interferometry (BLI) demonstrating that binding to the BG505 SOSIP trimer was detected with dissociation constants ranging from 51.6 to 184 nM (Figure 3C).
Figure 3.
BG505 mAbs bind to autologous Env
(A and B) BG505 mAbs were tested by ELISA for binding to native Env trimer BG505.SOSIP.664 (1 μg/mL) at a concentration series of 10, 0.1, and 0.001 μg/mL. Mean absorbance at 450 nm ± the standard deviation between at least two technical replicate measurements is shown for each antibody relative to the negative control influenza mAb Fi6v3 at 10 μg/mL.
(C) BG505 mAbs binding to autologous HIV Env trimer as measured by biolayer interferometry. Multiple BG505 mAbs (ligand) at 8 μg mL−1 bind to BG505.SOSIP.664 (analyte). Response is measured as nanometer shift. PGT145 mAb confirms the presence of native trimer conformation, while 10E8 mAb serves as a negative control, because the Env trimer lacks the MPER epitope that 10E8 targets. BF520.1 mAb was included as a positive control based on previously published findings.7 Data are representative of two technical replicates. Binding-affinity constants (KD, kon, and kdis) for each BG505 mAb binding to BG505 SOSIP are derived from the global best fit (red lines) using a 1:1 model of ligand:analyte binding. No KD could be calculated for BG505.23, as there was no measurable dissociation.
See also Figures S1 and S2.
Earlier Env variants isolated from BG505 at W6 and W14 were potently neutralized by plasma antibodies from M27. Plasma from BG505 at M27 neutralizes contemporaneous viruses to varying degrees. A few M27 Env variants were potently neutralized by the M27 plasma, with IC50 values as high as 1,875, but most were neutralized to a lesser extent (IC50 200–500; Figure S2A). By contrast, none of the 51/69 representative monoclonal nAbs isolated from BG505 neutralized autologous virus. This was true when tested against early (W6 and W14) or contemporaneous viruses (M27; Figure S2B for representative data and data not shown).
Alternative approaches to isolate additional HIV-specific antibodies from BG505
The large collection of isolated antibodies did not recapitulate the breadth of plasma antibodies (Figure 2). To rule out the possibility that primers used for antibody chain amplification were biased and did not amplify certain HIV-specific antibodies, two alternative primer sets were used28,56 to probe for additional antibody chains from the 309 wells that exhibited HIV specificity by microneutralization screening (Figure 1). From these efforts, 119 additional antibody chains were identified by PCR and 45 mAbs were synthesized, focusing on those that generated novel antibody chain pairings and had characteristics suggestive of HIV bnAbs—including more SHM and longer CDRH3 regions.3 Of the 45 additional mAbs, four neutralized HIV (Figure S3A). Consistent with the 69 nAbs in Figure 2, these additional nAbs clustered into the previously identified clonal families and one additional clonal family, with shared characteristics, such as overall percent SHM and CDR3 lengths, and none were broadly neutralizing, nor did they neutralize autologous virus (Figure S3B). Despite using an exhaustive collection of primer pools to identify antibody chains from BG505, the isolated nAbs from these combined efforts had similar HIV neutralization profiles and shared clonal family characteristics (Figures 2 and S3).
Epitope mapping for BG505 nAbs
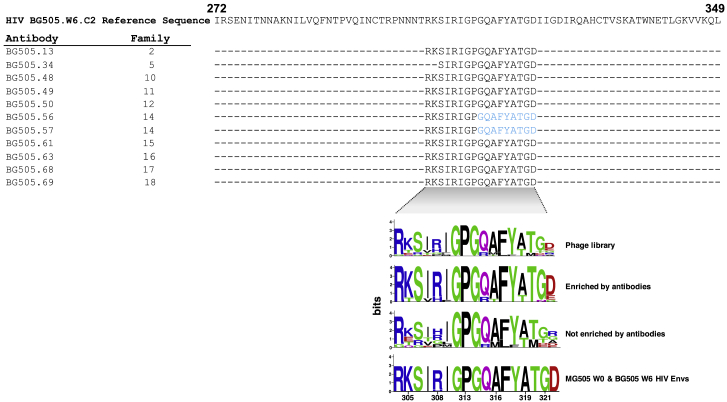
Phage immunoprecipitation with deep sequencing (PhIP-seq) was used to identify linear peptides that bind the mAbs, as performed previously.57 The phage library contained multiple HIV Env sequences, including consensus sequences for clades A, B, C, and D; specific sequences circulating in Kenya; and the T/F BG505.W6.C2 virus. For ten of the antibody families, phage-displayed peptides were significantly enriched in the V3 region of HIV Env (spanning positions 304–322, based on HXB2 numbering), suggesting this region comprises a key part of the epitope of these isolated nAbs (Figures 4 and S4). This included one mAb (BG505.34) with tier 2 clade B neutralizing activity and several nAbs (BG505.48, 50, 56, 57, 61, 68, and 69) that showed clade A tier 2 activity (Figures 2 and 4). For two of the antibodies tested (BG505.56 and BG505.57), we observed weak but significant enrichment of a smaller peptide that truncated the minimal epitope sequence, suggesting this is a core part of the epitope (Figure 4, blue residues). The remaining representative antibodies did not demonstrate binding to the linear epitopes present in the library.
Figure 4.
PhIP-seq analysis of BG505 antibody family representatives
Sequence alignment of the minimal consensus epitopes identified by PhIP-seq for each tested antibody. See Figure S4 for all peptides significantly enriched and not enriched for a subset of tested antibodies. Residues in blue signify where the minimal epitope was extended in cases where there was weak but significant enrichment of a peptide that truncated the minimal epitope sequence. Logo plot of sequences corresponds to the minimal epitope region (HIV Env V3) in the phage library, sequences that were significantly enriched or not enriched by tested antibodies, MG505 W0 (Envs from the mother of BG505 near the time of transmission), and BG505 W6 Envs.
Because the library included sequences of several different HIV-1 variants, our PhIP-seq data gave us insight into which residues were preferred at highly variable sites within the library sequences. For example, K at 305, R at 308, and GD at 321 to 322 are preferentially found in the enriched peptides, and these residues are also present in the BG505 sequence (Figure 4).
Binding antibody multiplex assay (BAMA) using a variety of HIV Env antigens, including variants of gp120, gp41, trimer, and peptide epitopes,58 was used to broadly define the targets of the BG505 nAbs that could not be determined by PhIP-seq. Antibodies BG505.23, 25, and 36 were not mapped by PhIP-seq but demonstrated binding to gp120 and trimer (Figure S5). We also included select nAbs identified as V3 specific by PhIP-seq (BG505.13, 34, 48, 49, 50, 56, 57, 61, 63, 68, and 69) and confirmed their specificity for V3 linear peptides by BAMA. The BAMA data (Figure S5) also confirmed that nAbs from 12 of the clonal families bound to the autologous gp120 and Env trimer (BG505 SOSIP trimer), as we observed by ELISA and BLI (Figure 3). BG505.03, 19, 42, 46, and 47 did not bind any of the proteins tested, suggesting the targeted epitopes were not represented in the panel.
ADCC activity of representative BG505 nAbs
Given that passively acquired ADCC antibodies have been associated with improved outcomes in infants,54 we also determined the capacity of a set of representative BG505 nAbs to mediate ADCC. Eleven clonal families demonstrated rapid fluorometric antibody-dependent cellular cytotoxicity (RF-ADCC) activity using a clade A envelope protein, BL035 (Figure 2). Of the antibodies with ADCC activity, most were V3 specific, including one (BG505.34) that demonstrated cross-clade activity. Only one V3-specific antibody (BG505.13) did not mediate ADCC. Two antibodies that were not V3-specific, BG505.42 and 46, showed potent ADCC, but the remainder did not (Figure 2). None of the tier 2 clade D nAbs demonstrated ADCC.
Discussion
Infant BG505 developed a broad and potent nAb response by approximately 2 years post-infection. Here, we isolated 73 nAbs from 19 clonal lineages from infant BG505 at this time point, suggesting the antibody response in this infant was due to a diverse collection of B cell responses. This is consistent with plasma-mapping studies of infant responses, which did not show evidence of a dominant epitope specificity, though there were smaller contributions by known bnAb specificities in some infants, including BG505; this also is consistent with a detailed study of one infant,8 collectively suggesting polyclonal responses drive the broad nAb responses detected in HIV-infected infants. Despite isolating a large number of unique nAbs using several different approaches to both identify B cells and amplify antibody sequences, we did not fully recapitulate the nAb response of infant BG505 plasma, indicating that the response may be even larger than captured here.
Isolation of adult-derived bnAbs has been achieved using either single B cell culture coupled to high-throughput neutralization screening or sorting of single B cells that bind HIV Env bait.2 The latter approach has been successful in isolating antibodies of known specificity but is not as amenable to the more challenging problem of identifying the collection of nAbs contributing to a polyclonal response. Selecting Env variants and/or appropriate viruses that have the potential to identify any B cell expressing a nAb also presents challenges. For BG505, a single vial of cells was available at a time when there was HIV-specific nAb breadth and potency. To maximize our chances of identifying as many HIV-specific nAbs as possible, we combined methodologies for HIV-specific B cell identification, which included using both autologous and heterologous Env trimers and two heterologous virus variants for microneutralization screening. Surprisingly, the autologous SOSIP trimer did not capture any HIV-specific nAbs. Bulk culture screening techniques were successful in that twenty-six of the isolated nAbs demonstrated limited tier 2 neutralization restricted to the clade of the infecting virus and eleven nAbs had low-potency, cross-clade neutralization. The cross-clade nAbs isolated were primarily found in the larger clonal families; two of these families had multiple members that targeted a tier 2 clade D virus with low potency. In family 4, there were different antibodies targeting tier 2 clade B and tier 2 clade D viruses, suggesting there may be other members of this family with the potential to neutralize both and possibly have greater breadth; perhaps we simply did not capture the broadest member of this clonal family.
Overall, our findings suggest that no one method nor a combined approach was sufficient to capture antibodies that recapitulate BG505’s plasma activity, though a combined approach may help maximize identification of diverse nAbs contributing to a polyclonal response. It is possible bnAbs were simply not sampled, given the extremely limited sample availability as well as the nature of sampling PBMCs, which may not contain the long-lived plasma cells contributing to plasma breadth.
Several strain-specific nAbs have been isolated and characterized from rabbits, guinea pigs, and macaques that were immunized with the BG505 SOSIP trimer.45,52,59 Many isolated rabbit antibodies target holes in the glycan shield that are easily filled by the virus as an escape mechanism.45 Unlike the rabbit nAbs, the nAbs from infant BG505 bind but do not neutralize the transmitted variant. The isolated nAbs were also unable to neutralize other autologous viruses from W6 when infection was first detected and shortly thereafter (W14). Sample availability precluded studies to determine which, if any, of the evolving viruses were recognized by intermediates in these antibody lineages.
The V3-glycan loop is commonly targeted by nAbs and bnAbs and is considered a potential target for vaccine-elicited antibodies. In the setting of mother-to-child transmission, V3-specific antibody responses are correlated with a reduced risk of transmission.60 Antibodies from 10 of 19 clonal families isolated from BG505 target V3, suggesting it was a major target of the antibody response. We defined the epitope of these nAbs using PhIP-seq and, in the process, detected evidence for preferences at certain positions, including K305, R308, G321, and D322, and these nAbs recognized the GPGQ motif. Interestingly, these residues were also present in the T/F virus, which was a virus that had escaped maternal antibodies, suggesting these V3-specific nAbs did not play a major role in the selection that occurred during transmission. The majority of these V3-specific nAbs mediated ADCC, which in infants has been associated with improved outcomes in the context of passive antibodies.54
Overall, BG505 mounted an impressive neutralizing antibody response from which we isolated and characterized a substantial diversity of nAbs, both in terms of the number of clonal families identified and the number of related variants in the families. Our data suggest that, despite isolating a large collection of HIV-specific nAbs, there may be additional antibody families that contribute to breadth and/or that there are members of the families we isolated that have more breadth and potency.
Limitations of the study
This study focuses on characterizing the antibody response of infant BG505 to HIV infection at 27 months of age. Due to extremely limited sample availability, we were unable to comprehensively analyze this infant’s antibody response dynamics. Moreover, the sample volume was also limited, which may have influenced sampling depth. The nature of sorting HIV-specific memory B cells—whether single cell or bulk sorted—is biased based on which antigens are used, whether they be for bait or for functional screening following culture. We took a multi-faceted approach to mitigate some of these biases and used both diverse antigen bait constructs as well as bulk sorting and screening techniques with multiple pseudoviruses. However, it is quite plausible that we missed HIV-specific antibodies that were circulating in infant BG505 at the time of sampling despite using this combination of techniques. Following rescue and purification of 73 mAbs from infant BG505, we used several different methods to test for antibody function and HIV specificity, some of which have limitations. Although the RF-ADCC assay is widely used in the field,61 there are also other ADCC assays that have different endpoint measurements of cytotoxicity that we could have implemented to corroborate our findings.62 The peptides displayed in the PhiP-seq library are linear and do not undergo post-translational modification, resulting in the inability to identify conformational and/or glycan-specific antibody epitopes. Thus, we may not have obtained a comprehensive profile of epitopes for the isolated nAbs from infant BG505.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human monoclonal antibodies from subject BG505 at month 27 of age | This paper | GenBank: MW602658 - Genbank: MW602803 |
| 2F5 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-1475 |
| PG9 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-12149; RRID:AB_2491030 |
| PGT121 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-12343; RRID:AB_2491041 |
| 447-52D | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-4030; RRID:AB_2491016 |
| HIVIG | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-3957; RRID:AB_2890264 |
| VRC01 | Williams et al.57 | RRID:AB_2491019 |
| anti-p24 | Williams et al.57 | N/A |
| Fi6v3 | Jesse Bloom | N/A |
| Bacterial and virus strains | ||
| HIV Env clones from BG505.W14 | This paper | GenBank: MW650605- Genbank: MW650618 |
| HIV Env clones from BG505.M27 | This paper | GenBank: MW650619-Genbank: MW650637 |
| HIV Env clone BG505.W6M.C2 | Wu et al.10 | GenBank: DQ208458.1 |
| HIV Env clone BG505.W6M.B1 | Wu et al.10 | GenBank: DQ208457.1 |
| HIV Env clone BG505.W6M.A5 | Wu et al.10 | N/A |
| HIV Env clones from MG505.W0 | Wu et al.10 | GenBank: DQ208449.1- Genbank: DQ208455.1 |
| HIV Env clone BL035.W6M.C1 | Immune-Tech | Cat#IT-001-115p; GenBank: DQ208480.1 |
| HIV Env clone C.ZA.1197MB | Rousseau, 200663 | GenBank: AAR27754.1 |
| HIV Env clone SF162 | Cheng-Mayer, 199764 | GenBank: EU123924.1 |
| HIV Env clone QC406.70M.F3 | Blish, 200965 | GenBank: FJ866133.1 |
| Biological samples | ||
| Human PBMC sample from subject BG505 | Nairobi Breastfeeding Clinical Trial, Nduati et al.66 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Q5 High-Fidelity Master Mix | New England BioLabs | Cat#M0492S |
| FreeStyle MAX | Thermo Fisher | Cat#16447500 |
| Protein G agarose | Pierce | Cat#20397 |
| 293F FreeStyle Expression media | Invitrogen | Cat#12338-026 |
| BG505.SOSIP.664 | Sanders et al.11 | N/A |
| BG505.SOSIP.664-aviB Envelope trimer | Sok et al., 201467 | N/A |
| DU422.SOSIP.664-aviB Envelope trimer | Sok et al., 201467 | N/A |
| HIV PhIP-Seq bacteriophage library | Williams et al.57 | N/A |
| HIV Mn gp41 | ImmunoDX, LLC through the AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-12027 |
| IIIB recombinant p24 | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-12028 |
| BAMA HIV antigen panel | Ronen et al.68 | N/A |
| Critical commercial assays | ||
| Gal-Screen | Thermo Fisher | Cat#T1028 |
| AllPrep DNA/RNA Mini Kit | QIAGEN | Cat#80204 |
| SMARTer RACE 5′/3′ Kit | Takara Bio USA | Cat#634858 |
| KAPA library quantification kit | Kapa Biosystems | Cat#KK4824 |
| 600-cycle MiSeq Reagent Kit v3 | Illumina | Cat#MS-102-3003 |
| Nextera XT 96-well index kit | Illumina | Cat#FC-131-1001 |
| Deposited data | ||
| BG505 W14 antibody variable gene sequencing data | This paper | BioProject SRA: PRJNA588318 |
| Experimental models: cell lines | ||
| Human: FreeStyle 293F | Invitrogen | Cat#R790-07; RRID:CVCL_D603 |
| Human: CEM.NKR | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#ARP-458; RRID:CVCL_X622 |
| Human: HEK293T | ATCC® | Cat# CRL-3216; RRID:CVCL_0063 |
| Human: TZM-bl | AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat# ARP-8129; RRID:CVCL_B478 |
| Mouse: 3T3/CD40L | Kershaw 2001; AIDS Reagent Program, Division of AIDS, NIAID, NIH. | Cat#12535-444; RRID:CVCL_1H10 |
| Oligonucleotides | ||
| Primers for antibody variable gene amplification | Tiller et al.69; Liao et al.56; Doria-Rose, 201670 | N/A |
| Primers for Env amplification and cloning | Wu et al.10; Milligan et al.71 | N/A |
| Recombinant DNA | ||
| Human Igγ1 expression vector | Tiller et al.69 | Addgene: 80795 |
| Human Igκ expression vector | Tiller et al.69 | Addgene: 80796 |
| Human Igλ expression vector | Tiller et al.69 | Addgene: 99575 |
| Software and algorithms | ||
| FLASH v1.2.11 | Magoc, 201172 | http://ccb.jhu.edu/software/FLASH/ |
| Cutadapt 1.14 with Python 2.7.9 | Martin, 201173 | https://cutadapt.readthedocs.io/en/stable/ |
| FASTX toolkit 0.0.14 | Hannon Lab, Cold Spring Harbor | http://hannonlab.cshl.edu/fastx_toolkit/ |
| Partis | Ralph and Matsen74 | https://github.com/psathyrella/partis |
| Clustal Omega | Sievers, 201175 | RRID:SCR_001591 |
| WebLogo | Crooks et al.76 | RRID:SCR_010236 |
| Geneious v11.1.2 | Kearse et al. 201277 | RRID:SCR_010519 |
| FlowJo v10 | TreeStar | RRID:SCR_008520 |
| Excel | Microsoft | RRID:SCR_016137 |
| ForteBio’s Octet Software “Data Analysis 7.0” | Pall ForteBio | N/A |
| Prism 8.0c | GraphPad | RRID:SCR_002798 |
| Other | ||
| Anti-human IgG Fc capture biosensors | Pall ForteBio | Cat#18-5063 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Julie Overbaugh (joverbau@fredhutch.org).
Materials availability
Materials used or generated in this manuscript are available upon request, without restriction, though we may require a completed Materials Transfer Agreement.
Data and code availability
The accession number for the antibody gene sequencing data of BG505’s B cell repertoire at week 14 reported in this paper can be found at BioProject SRA: accession PRJNA588318. GenBank accession numbers for BG505 antibodies isolated from month 27 are Genbank: MW602658 - Genbank: MW602803.
GenBank accession numbers for HIV Env variants isolated from BG505 at week 14 and month 27 are Genbank: MW650605 - Genbank: MW650637. GenBank accession numbers for HIV Env variants used in this study are Genbank: DQ208458.1 (BG505.W6M.C2), Genbank: DQ208457.1 (BG505.W6M.B1), Genbank: DQ208456.1 (BG505.W6M.A5), Genkbank: DQ208449.1 - Genbank: DQ208455.1 (MG505.W0M Env clones), Genbank: DQ208480.1 (BL035.W6M.C1), Genbank: AAR27754.1 (C.ZA.1197MB), Genbank: EU123924 (SF162), and Genbank: FJ866133 (QC406.70M.F3).
The code used in this study for partis analysis is publicly available on GitHub: (https://github.com/psathyrella/partis).
Experimental model and subject details
Ethics statement
Plasma and peripheral blood mononuclear cell (PBMC) samples from male infant BG505 at 27 months of age were collected when the subject was enrolled in the Nairobi Breastfeeding Clinical Trial, 66 which was conducted prior to the use of antiretrovirals for the prevention of mother-to-child transmission. The protocols for the original trial were approved by the Institutional Review Boards of the University of Washington and University of Nairobi, including the approach to consent. All women gave oral consent, which was deemed the best approach because of a high rate of illiteracy in the population. This trial was conducted prior to when trial registries were used. The current study was approved by the Institutional Review Boards of Fred Hutchinson Cancer Research Center, University of Nairobi, and University of Washington.
Cell lines
For antibody production: HEK293-F cells (RRID:CVCL_D603; originally derived from female human embryonic kidney cells) were obtained from Invitrogen (Thermo Fisher Scientific, Waltham, MA, catalog #R790-07) and grown at 37°C in Freestyle 293 Expression Medium (Thermo Fisher Scientific, catalog #12338002) in baffle-bottomed flasks orbiting at 135 rpm. These cells were not further authenticated in our hands.
For pseudovirus production: 293T cells (RRID:CVCL_0063; transformed cell line originally derived from female human embryonic kidney cells) were obtained from ATCC (Manassas, VA, catalog #CRL-3216) and grown at 37°C in DMEM media with added fetal bovine serum (10%), penicillin (10,000 units/mL), streptomycin 10,000 μg/mL, and Amphotericin B (250 ng/mL) (all from Thermo Fisher Scientific). These cells were not further authenticated in our hands.
For neutralization assays: TZM-bl cells (RRID:CVCL_B478; originally derived from female cancerous human cervical tissue) were obtained from NIH AIDS Reagent Program (Germantown, MD, catalog #ARP-8129) and grown at 37°C in DMEM media with added fetal bovine serum (10%), penicillin (10,000 units/mL), streptomycin 10,000 μg/mL, and Amphotericin B (250 ng/mL) (all from Thermo Fisher Scientific). These cells were not further authenticated in our hands.
For RFADCC: CEM.NKR cells (RRID:CVCL_X622; originally derived from female human T-lymphoblastoid cells) were obtained from NIH AIDS Reagent Program (Germantown, MD, catalog #ARP-458) and grown at 37°C in RPMI 1640 media with added penicillin (100 U/mL), streptomycin (100 μg/mL), Amphotericin B (250 ng/mL), L-glutamine (2mM), and fetal bovine serum (10%) (all from Thermo Fisher Scientific). These cells were not further authenticated in our hands.
Method details
B cell sorting
BG505 was HIV-1 DNA-negative by PCR and HIV-1 RNA-negative using the Gen-Probe Viral Load assay at birth and was HIV-1 DNA- and RNA-positive at 6 weeks of age. The infecting virus was clade A based on envelope sequence.10 A PBMC sample from BG505 from 27 months of age (M27) (sample date 05/23/1996), which was ∼2 years after first detection of HIV infection and a time-point at which the plasma demonstrated broad neutralizing activity,4 was thawed at 37°C and re-suspended in 10 mL B cell media (IMDM medium, GIBCO; 10% low IgG FBS, Life Technologies; 5 mL GlutaMAX, Life Technologies; 1 mL MycoZap plus PR, Lonza) plus 20 μL/ml benzonase followed by centrifugation at 300 x g for 10 minutes. Biotinylated Env SOSIP.664-aviB trimers67 kindly provided by Marit van Gils were conjugated to streptavidin-fluorophores (3.75 μg trimer and 1.5 μL streptavidin-fluorophore) at 4°C for 1 hour. BG505.SOSIP.664-aviB was conjugated to streptavidin-PE (Premium Grade, Life Sciences Technologies) and streptavidin-APC (BD PharMingen), separately. DU422.SOSIP.664-aviB was conjugated to streptavidin-PECy7 (eBioscience) and streptavidin-BV21 (BD Horizon). Cells were stained on ice for 30 minutes using a cocktail of the four trimer conjugates, anti-CD19-BV510, anti-IgD-FITC, anti-IgM-FITC, anti-IgA-FITC, anti-CD3-BV711, anti-CD14-BV711, and anti-CD16-BV711. Cells were then washed once and resuspended in fluorescence-activated cell sorting (FACS) wash buffer (1X phosphate buffered saline (PBS), 2% FBS). Cells were loaded onto a BD FACS Aria II cell sorter. The gating strategy was set up such that IgG expressing B cells (CD3-CD14-CD16-CD19+IgD-IgM-IgA- cells) that were positive for the BG505.SOSIP.664-aviB were sorted into FACS wash buffer. The remaining IgG B cells that were positive for the DU422.SOSIP.664-aviB were sorted into B cell collection media (IMDM medium, GIBCO; 10% low IgG FBS, Life Technologies; 5 mL GlutaMAX, Life Technologies; 1 mL MycoZap plus PR, Lonza). All remaining IgG B cells that were not positive for IgG and either bait were bulk sorted into B cell collection media. Immediately following the sorting of B cells, cells that were sorted for binding to the BG505.SOSIP.664 trimer were diluted in RNA storage buffer (15 mM Tris and 10 U murine RNase inhibitor, NEB) to a final concentration of one B cell per 20 μL and then 20 μL aliquots were made and frozen at −80°C. Cells that bound the DU422.SOSIP.664 trimer were plated at a density of four B cells in 60 μL per well into 46 culture wells and IgG-expressing B cells that did not bind bait were plated at six B cells in 60 μL per well into 130 × 384-well plates in B cell growth media (100 U mL-1 IL-2, Roche; 0.05 μg/ml IL-21, Invitrogen, and irradiated 3T3/CD40L feeder cells, 8.85x105/mL). Cultured B cells were incubated for 12 days at 37°C in a 5% CO2 incubator based on the protocol by Huang et al. 78
B cell culture supernatant IgG ELISA
IgG was measured by ELISA on culture days 9 through 12 from a random sample of wells from multiple culture plates. Immunolon 2HB ELISA plates were coated with 2500 ng/well Sigma goat anti-human Ab in 0.1 M sodium bicarbonate, pH 9.4 overnight at 4°C. Plates were washed three times with 200 μL wash buffer (1X PBS, 0.05% Tween-20) and blocked for 1 hour at room temperature (RT) in blocking buffer (1X PBS with 10% non-fat milk and 0.05% Tween-20). IgG at a known concentration (3 μg/mL) was serially diluted four-fold a total of eight times for a standard curve. Five μL of B cell culture supernatant was diluted 1:10 into blocking buffer. Fifty microliters of the standard curve and diluted supernatants were added to each well and incubated at 37°C for 1 hour. Plates were washed and incubated with secondary antibody (goat anti-human IgG-HRP, Sigma diluted 1:2500 in blocking buffer) for 1 hour at RT. After washing, 50 μL of TMB-ELISA solution (Pierce) was added for 10 minutes at RT and stopped with 50 μL 1N sulfuric acid. Absorption was read at 450 nM, and concentrations were interpolated based on the standard curve.
B cell culture harvest and microneutralization
On day 12, B cell culture supernatants were divided into 2 × 384-well plates at 20 μL each for neutralization assays using a Tecan automated liquid handling system. B cells were frozen at −80°C in 20 μL RNA storage buffer per well. Microneutralization assays were performed by incubating 20 μL of B cell culture supernatant with ∼325 infectious pseudovirus particles in 20 μL for 1 hr at 37°C. Screening was done using two viruses (Tier 1 clade B SF162 and Tier 2 clade C QC406.F3) to maximize isolation of HIV-specific antibodies. Following incubation, 3,000 TZM-bl cells in 20 μl DMEM plus 10% FBS and 1% PSF, GIBCO (1.5 × 105 cells mL-1) and diethylamino ethyl (DEAE) dextran (10 μg mL-1 final concentration) were added to each well and cultured at 37°C in a CO2 incubator for 48 hr Luminescence levels were measured using the Gal-Screen system (ThermoScientific). Briefly, 30 μl was removed from each well, 25 μl of substrate diluted 1:25 was added and incubated for 40 min at RT, and read using a luminometer. Wells demonstrating > 40% neutralization of one or both viruses were selected for antibody cloning (n = 309).
Reconstruction of antibodies
RT-PCR amplification of the IgG heavy and light chain variable regions was performed for B cells identified by the functional assay, which included the B cells that bound the heterologous bait as well as for B cells that were sorted based on binding to BG505.SOSIP.664. Briefly, cDNA was generated using Superscript III with random hexamers (Invitrogen) and then two rounds of five independent, semi-nested PCRs were performed to amplify Igγ, Igκ, and Igλ variable genes using previously described primer sets.69 Products from positive PCRs were sequenced and functional heavy and light chain variable region sequences were determined using IMGT V-QUEST79 before cloning into the corresponding Igγ1, Igκ, and Igλ expression vectors.
In parallel, for the B cells identified by the functional assay, an aliquot of B cell RNA was sent to Atreca (https://www.atreca.com) for deep sequencing of the antibody heavy and light chain variable regions from each well. In cases where Atreca identified additional heavy and/or light chains not amplified by our methods (N = 32 wells), those were synthesized as gene fragments by Integrated DNA Technologies (IDT) and subsequently cloned into Igγ1 expression vectors.
In addition, wells that exhibited HIV-specificity by microneutralization were further screened using alternate primer sets28,56 designed to amplify heavy and light chains that may have been missed by the original primer pools used following memory B cell sorting and RNA isolation.69 Carrier RNA (500 ng, Thermo Fisher) was added to each PCR reaction to improve DNA yields. From the pool of 119 additional antibody chains that were amplified from these efforts, those that yielded novel antibody pairings and had intriguing characteristics such as higher percent SHM (≥9%) and longer CDRH3 regions (≥17 amino acids) were synthesized as gene fragments from IDT and cloned into Igγ1 expression vectors.
The Freestyle MAX system (Invitrogen) was used to co-transfect HEK293-F cells with equal amounts of paired heavy and light chain plasmids cloned from the same well; transfections proceeded at 37°C with 8% CO2 in baffled-bottom flasks, rotating at 135 rpm and were harvested after 3-6 days. Supernatants were collected following centrifugation of cells at 1,900 rpm for 10 minutes. Antibody supernatants were passed three times over columns packed with immobilized Protein G resin (Thermo Fisher, catalog #20397) as previously described (Scherer et al., 201480). For each well, all possible heavy and light chain pairs were generated when multiple antibody sequences were identified. The 45 mAbs generated using alternate primer pools were screened for HIV-specificity using a 1:2 dilution of unpurified supernatant after three days of transfection (Figure S3). Antibody supernatant from transfection of the heavy and light chains of bnAb QA013.2 was used in a 1:2 dilution as a positive control for screening.81
Pseudovirus production and neutralization assays
To generate pseudoviruses as previously described by Goo et al.,82 we co-transfected plasmids containing Envelope chimeras with an Env-deficient sub-type A proviral plasmid (Q23ΔEnv) at a 1:2 mass ratio into 3 × 106 293T cells plated in a T-75 flask 24 hr prior to transfection. Four micrograms of total DNA was mixed with 12 μL of the transfection reagent Fugene6 (Roche). Forty-eight hours after transfection, supernatant was collected and sterile filtered through a 0.2-μm filter and subsequently used to infect TZM-bl cells in the presence of DEAE-dextran (10 μg/mL). Viral titer was determined by counting blue (infected) cells present at 48 hr post-infection after staining fixed cells for β-galactosidase activity.82
Neutralization assays were performed using approximately 500 infectious pseudovirus particles as determined by TZM-bl titer diluted to a final volume of 25 μL and incubated with an equal volume of serial dilutions of either heat-inactivated BG505 plasma or isolated antibodies in duplicate at 37°C for 60 min TZM-bl cells (1x104 in 100 μL DMEM) were then added to each well containing plasma/antibody and pseudovirus. Forty-eight hours post-infection, luminescence levels were measured using the Gal-Screen system (Thermo Fisher Scientific). Percent neutralization was calculated as the percent reduction in luminescence activity of pseudovirus incubated with each dilution of plasma or antibody compared to the same virus incubated with media and TZM-bl cells only. Plasma IC50 values are the reciprocal plasma dilution resulting in 50% reduction of virus infectivity. Monoclonal antibody (mAb) IC50 values represent the mAb concentration (μg/mL) at which 50% of the virus was neutralized. Reported IC50 values are an average of two independent experiments performed in duplicate.
BG505 neutralizing antibody sequence analysis
In order to assign gene families to antibodies, we sequenced the antibody repertoire of BG505 at 14 weeks of life using methods described.68,83,83 Briefly, RNA was extracted and prepared for antibody gene sequencing from PBMCs collected from infant BG505 14 weeks after birth, which was ∼8 weeks after HIV-1 infection. Antibody sequencing was performed using the SMARTer RACE 5′/3′ Kit (Takara Bio). RACE-ready cDNA synthesis was performed using primers specific for IgM, IgG, IgK, and IgL (Simonich et al.7). Following synthesis, cDNA was diluted in 10mM Tricine-EDTA according to the manufacturer’s recommended protocol. First-round PCR was performed using Q5 High-Fidelity Master Mix (New England BioLabs) and nested gene-specific primers at 20 μM. First-round amplicons were used as templates for second-round PCR amplification, which added MiSeq adaptors. Second-round products were gel purified and indexed using Nextera XT P5 and P7 indices (Illumina). Gel-purified indexed libraries were quantified using the KAPA library quantification kit (Kapa Biosystems) using an Applied Biosystems 7500 real-time PCR machine. Libraries were denatured and loaded onto an Illumina MiSeq using 600-cycle V3 cartridges, according to the manufacturer’s instructions.
HIV-1 env amplification and cloning
Infant BG505 week 6 (W6) and mother MG505 week 0 (W0) Env variants were previously isolated by Wu et al. 10 BG505 week 14 (W14) and month 27 (M27) full-length Env variants were cloned from total RNA that was extracted from plasma as described.84 To isolate RNA, plasma samples were centrifuged at 16,000 x g for 1 hr at 4°C and following centrifugation, the virion pellet was treated with 50 μL of 5 mM Tris-Cl (pH 8) with 200 μg proteinase K for 30 min at 55°C. Then the pellet was treated with 200 μL of 5.8 M guanidinium isothiocyanate containing 200 μg glycogen as a carrier. Next, 270 μL of 100% isopropanol was added and the lysate was centrifuged at 21,000 x g for 15 min at 4°C. The supernatant was decanted and the resulting pellet washed with 70% ethanol and resuspended in 40 μL of 5 mM Tris-Cl (pH 8) containing dithiothreitol (DTT) and 1,000 units of RNaseOUT (Thermo Fisher). cDNA synthesis and nested PCR of envelope was performed as previously described using all of the isolated RNA.10,71 The primer used for cDNA synthesis was nef50a (5′ AGAGCTCCCTTGTAAGTCATTGG 3′). Round one nested PCR primers were vpr9 (5′ TGTCGACATAGCAGAATAGG 3′), and nef24 (5′ TACTTGTGATTGCTCCAT GT 3′) mixed in an equal molar ratio with nef34 (5′ TACTTGTGACTGCTCCATGT 3′). Second round primers were vpr21a1 (5′ TAACCTAGACGCGTGGAATCACCCGGGAAGTCAGCCTACAACACCTTGTA 3′) and vpr21a2 (5′ TAACCTAGACGCGTGGAATCACCCGGGAAGCCGGCCTACAACACCTTGTA 3′) and primers nef60a1 (5′CTTGTGGCGGCCGCATGTTTATCTAAATCTCGAGATACTGCTCCTACTCCTGGTGCTG 3′) and nef60a2 (5′ CTTGTGGCGGCCGCATGTTTAGCTAAATCTCGAGATACTGCTCCTACTCCTGGTGCT 3′) in equal molar ratios. The cycling parameters for PCR amplification were 1 cycle of 94°C for 4 min, 35 cycles of 94°C for 30 s, 58°C or 60°C for 30 s, and 68°C for 4 min, and 1 cycle of 72°C for 10 min PCR products were cloned into pCI-Neo (Invitrogen).
Phylogenetic tree construction
MG505 (mother) and BG505 (infant) Env sequences were aligned using Geneious version 11.1.2. A maximum likelihood phylogenetic tree was constructed using the LANL HIV tools database PHYML interface.85
Rapid and fluorometric ADCC (RF-ADCC) assay
The RF-ADCC assay was performed as described57,61 using gp120 (BL035.W6M.Env.C1, Immuntech) -coated CEM-NKr target cells (AIDS Research and Reference Reagent Program, NIAID, NIH from Dr. Alexandra Trkola) stained with PKH-26-cell membrane dye (Sigma-Aldrich) and cytoplasmic dye (Vybrant CFDA SE. Cell Tracer Kit, Life Technologies). Peripheral blood mononuclear effector cells (Bloodworks Northwest) from an HIV-negative donor were then added at a ratio of 50 effector cells (250,000) per target cell (5,000).
PhIP-seq
Phage display immunoprecipitation sequencing (PhIP-seq) was performed as previously described using a library that encodes sequences from several full-length HIV viruses from clades A, B, C, and D.57 The sequences of the HIV genomes were encoded as 39 amino acid peptides that tile across the entire genome with 20 amino acid overlap. Two concentrations (2 ng and 20 ng) of each monoclonal antibody were tested in technical replicates. Deep-well plates were blocked overnight at 4°C while rocking with a 3% Tris-Base solution containing 1% Tween-20 (TBST) solution. The following day, 1 mL of PhIP-seq library diluted to a final concentration that yielded sufficient representation of each peptide as determined by Larman et al. was added to each well along with 10 ng of each antibody.86 Samples and PhIP-seq library were incubated at 4°C for 20 hours while rocking. After overnight incubation, equal volumes of Protein A and Protein G Dynabeads (Thermo Fisher) were added to all wells and incubated at 4°C for 4 hours to immunoprecipitate enriched antibodies and bound peptides + phage. Enriched phage were lysed at 95°C for 10 min and subsequent PCR was performed to prepare sample libraries for deep sequencing. Thirty rounds of round 1 PCR were performed using Q5 mastermix (New England Biolabs) and 10 μM primers as previously described.86 The first round PCR products were used as the template for the second round PCR, which was comprised of 8 cycles and again used Q5 mastermix. Sample libraries were quantified using the PicoGreen assay kit for detection of dsDNA (Thermo Fisher). Equal masses of each sample library were pooled, gel extracted, and denatured prior to deep sequencing on an Illumina MiSeq using v3 chemistry.
Biolayer Interferometry
BG505 nAb binding to autologous HIV Env SOSIP trimer was measured using biolayer interferometry on an Octet RED instrument (ForteBio). Antibodies diluted to 8 μg mL-1 in a filtered buffer solution of 1X PBS containing 1% BSA, 0.03% Tween-20, and 0.02% sodium azide were immobilized onto anti-human IgG Fc capture biosensors (AHC). BG505.SOSIP.664 was diluted to 1 μM in the same buffer as above and a series of four, two-fold dilutions of Env trimer were tested as analyte in solution at a shake speed of 600 rpm at 30°C. The kinetics of mAb binding were measured as follows: association was monitored for 10 minutes, dissociation was monitored for 6 minutes, and regeneration was performed in 10mM Glycine HCl (pH 1.5).
Env trimer, gp120, and gp41 ELISAs
Immunolon 2-HB plates were coated with 100 μL of HIV antigen of interest diluted to 1 μg mL-1 in 0.1 M sodium bicarbonate buffer (pH 9.4) overnight at 4°C. Plates were washed 3-4 times using a Tecan HydroFlex plate washer with 200 μL wash buffer (PBS-0.05% Tween-20) and then blocked with a 10% non-fat dry milk (NFDM) in wash buffer solution for 1 hour at RT. Following blocking, the plates were incubated with 100 μL of primary antibody diluted in NFDM solution for 1 hour at 37°C. The plates were washed a second time prior to the addition of 100 μL of anti-IgG-HRP (Sigma) diluted 1:2500 in NFDM solution. Plates were incubated in secondary antibody for 1 hour at RT and then washed a third time. One hundred microliters of Ultra TMB solution (Thermo) was added to each plate and incubated in the dark for 10 minutes at RT. The reaction was stopped by adding equal volume of 0.1 M H2SO4. Absorbance was measured at 450 nm. Influenza-specific antibody Fi6v3 was included as a negative control.
BG505.SOSIP.664 trimer was obtained from Kelly Lee, BL035.W6M.ENV.C1 gp120 peptide was purchased from ImmuneTech (IT-001-115p), and MN gp41 recombinant protein was obtained from ImmunoDX, LLC through the AIDS Reagent Program, Division of AIDS, NIAID, NIH. FI6V3 was generated in stably producing 293F cells (courtesy of Jesse Bloom, Fred Hutchinson Cancer Research Center).
Binding antibody multiplex assay (BAMA)
BAMA87,88 was utilized to measure IgG binding against a customized panel of HIV antigens. Briefly, carboxylated fluorescent beads (Luminex) were covalently coupled to the previously employed HIV antigen panel68 that included gp140 clade A consensus protein; several gp120 proteins including BG505.W6M.ENV.C1, BL035.W6M.ENV.C1, ZM109F.PB4, SF162, and SIV; v1v2 scaffolds MuLVgp70-caseA2_v1v2; 2J9C-ZM53_v1v2; 1FD6-Fc-ZM109_v1v2; v3 consensus peptides for clades A1, B, C, and D; SOSIP Env trimer; gp41 protein from MN and the gp41 ectodomain from ZA.1197/MB; C1 consensus peptide for clade A1; the resurfaced Env core protein (RSC3) and CD4-binding site defective mutant (RSC3Δ371I); with the addition of IIIB recombinant p24 (AIDS Reagent Program). Coupled beads were incubated with antibodies at 25 μg/ml for 30 min at RT. Following sample incubation, the coupled beads were washed with 1X PBS containing 1% BSA and 0.05% Tween-20. Biotin-conjugated mouse anti-human IgG (Southern Biotec) or goat anti-human IgA (Jackson Immunoresearch) secondary antibodies were used at 4 μg/ml in a 30 min incubation at RT for detection of binding. Following incubation in secondary antibody, the beads were washed again and then incubated with a 1:100 dilution of Streptavidin-PE (BD Pharmigen) for 30 min at RT. VRC01, 2F5, anti-p24, PG9, PGT121, 447-52D, and influenza-specific FI6V3 were tested at 25 μg/ml as control antibodies. VRC01 and anti-p24 were produced as previously described57, whereas 2F5, PG9, PGT121, 447-52D, and polyclonal anti-HIV immunoglobulin from pooled serum (HIVIG) were obtained through the AIDS Reagent Program. Binding was measured on a Bio-Plex 200 instrument (Bio-Rad Laboratories) and is reported as the average median fluorescent intensity (MFI) of background-subtracted technical duplicates.
Quantification and statistical analysis
Antibody chain clonal family clustering
Antibody variable gene library preparation and sequencing was performed in technical duplicate to increase depth of coverage using the same isolated sample of BG505 W14 RNA. Following next-generation sequencing, sequence reads were pre-processed into amplicons using FLASH, primers were trimmed using cutadapt, and the FASTX-toolkit was used to remove sequence reads containing low-confidence base calls (N’s) as previously described.7,83 Sequence reads were annotated and deduplicated using partis (https://github.com/psathyrella/partis) with the default parameters. Sequences that were out of frame or contained internal stop codons were removed, while singletons, or sequences that were observed only once in the sampled repertoire were included in an attempt to retain undersampled or rare sequences. Heavy and light chain nAb sequences were analyzed and clustered into clonal families using the seeded clustering method of partis, which is designed specifically for clonal family clustering of B cell repertoires, including antibody gene family annotation. NAb clusters were delineated using IgH chain clustering information.74,89 Percent SHM was calculated as the mutation frequency at the nucleotide level compared to the predicted naive allele, as determined by partis using BG505-specific germline inference based on antibody deep sequencing of BG505 W14 early-infection PBMCs.
RF-ADCC analysis
Effector and target cells were analyzed by flow cytometry (LSR II, BD) and ADCC activity was defined as the percent of PKH-26+ CFDA- cells with background subtracted, where background (antibody-mediated killing of uncoated cells) was between 3%–5% as analyzed using FlowJo software (Tree Star). All values were normalized to HIVIG (positive control) activity.
PhIP-seq analysis
Enriched peptides were identified using a zero-inflated generalized Poisson significant-enrichment assignment algorithm to generate a -log10(p value) for enrichment of each phage clone across all samples, as previously described.90 Of note, the -log10(p value) reproducibility threshold when testing these antibodies in PhIP-seq was 2.3. Thus, we considered a phage-displayed peptide as significantly enriched if its -log10(p value) was ≥ 2.3 in both technical replicates. A phage-displayed peptide was considered to be part of the antibody’s epitope sequence only if it was significantly enriched in both conditions tested (2 ng and 20 ng). Fold-enrichment of each phage-displayed peptide was also calculated across all monoclonal antibodies tested.
Phage that were incubated without any monoclonal antibody served as a negative control for non-specific binding and were used to identify and eliminate background hits. For each monoclonal antibody tested, enriched and unenriched peptides were aligned using Clustal Omega. The minimal epitope of an antibody was defined as the shortest amino acid sequence present in all of the enriched peptides. Logo plots representing peptides in the phage library, peptides that were enriched by BG505 antibodies, peptides that were not enriched by BG505 antibodies, MG505 W0, and BG505 W6 HIV Env sequences were generated using WebLogo.76 For the “phage library” and “not enriched by antibodies” logo plots, only peptides that spanned the full length of the minimal epitope (at least from positions 308-322) were included.
Biolayer interferometry analysis
Binding-affinity constants (KD; on-rate, kon; off-rate, kdis) were calculated using ForteBio’s Data Analysis Software 7.0. Responses (nanometer shift) were calculated using data that were background-subtracted from reference wells and processed by Savitzky-Golay filtering, prior to fitting using a 1:1 global model of binding kinetics.
Acknowledgments
We acknowledge Guy Cavet and the Atreca, Inc. team for their help with sequencing of B cell RNA; Marit vanGils, James Williams, Adam Nguyen, and Kelly Lee for providing trimer proteins; and Jesse Bloom for providing FI6V3 mAb. This work was supported by NIH grant NIH R01 AI12096 and by training awards T32 AI07140, F30 AI122866, and F30 AI142870.
Author contributions
J.O. conceived the study. J.O. and C.S. contributed to the design of the study. C.S., M.M.S., L.D., T.G., M.G., E.M.C., B.H., H.I., and V.C. performed experiments. R.N. developed the cohort and collected samples. All authors contributed to the analysis and interpretation of data. C.S., M.M.S., and J.O. wrote the manuscript with input from all authors.
Declaration of interests
The authors declare no competing interests.
Published: June 15, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100314.
Contributor Information
Cassandra Simonich, Email: abelca@uw.edu.
Julie Overbaugh, Email: joverbau@fredhutch.org.
Supplemental information
References
- 1.Bonsignori M., Liao H.X., Gao F., Williams W.B., Alam S.M., Montefiori D.C., Haynes B.F. Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol. Rev. 2017;275:145–160. doi: 10.1111/imr.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCoy L.E., Burton D.R. Identification and specificity of broadly neutralizing antibodies against HIV. Immunol. Rev. 2017;275:11–20. doi: 10.1111/imr.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hraber P., Seaman M.S., Bailer R.T., Mascola J.R., Montefiori D.C., Korber B.T. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goo L., Chohan V., Nduati R., Overbaugh J. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat. Med. 2014;20:655–658. doi: 10.1038/nm.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muenchhoff M., Adland E., Karimanzira O., Crowther C., Pace M., Csala A., Leitman E., Moonsamy A., McGregor C., Hurst J. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci. Transl. Med. 2016;8:358ra125. doi: 10.1126/scitranslmed.aag1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton D.R., Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonich C.A., Doepker L., Ralph D., Williams J.A., Dhar A., Yaffe Z., Gentles L., Small C.T., Oliver B., Vigdorovich V. Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing antibody lineage. Nat. Commun. 2019;10:2190. doi: 10.1038/s41467-019-09481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonich C.A., Williams K.L., Verkerke H.P., Williams J.A., Nduati R., Lee K.K., Overbaugh J. HIV-1 neutralizing antibodies with limited hypermutation from an infant. Cell. 2016;166:77–87. doi: 10.1016/j.cell.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Panda H., Makhdoomi M.A., Mishra N., Safdari H.A., Chawla H., Aggarwal H., Reddy E.S., Lodha R., Kumar Kabra S. An HIV-1 broadly neutralizing antibody from a clade C-infected pediatric elite neutralizer potently neutralizes the contemporaneous and autologous evolving viruses. J. Virol. 2019;93:e01495-18. doi: 10.1128/JVI.01495-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X., Parast A.B., Richardson B.A., Nduati R., John-Stewart G., Mbori-Ngacha D., Rainwater S.M., Overbaugh J. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julien J.P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyumkis D., Julien J.P., de Val N., Cupo A., Potter C.S., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., Carragher B. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Taeye S.W., Ozorowski G., Torrents de la Peña A., Guttman M., Julien J.P., van den Kerkhof T.L., Burger J.A., Pritchard L.K., Pugach P., Yasmeen A. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M., Garcia N.K., Cupo A., Matsui T., Julien J.P., Sanders R.W., Wilson I.A., Moore J.P., Lee K.K. CD4-induced activation in a soluble HIV-1 Env trimer. Structure. 2014;22:974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon Y.D., Pancera M., Acharya P., Georgiev I.S., Crooks E.T., Gorman J., Joyce M.G., Guttman M., Ma X., Narpala S. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pancera M., Zhou T., Druz A., Georgiev I.S., Soto C., Gorman J., Huang J., Acharya P., Chuang G.Y., Ofek G. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders R.W., Moore J.P. HIV: a stamp on the envelope. Nature. 2014;514:437–438. doi: 10.1038/nature13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behrens A.J., Crispin M. Structural principles controlling HIV envelope glycosylation. Curr. Opin. Struct. Biol. 2017;44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behrens A.J., Vasiljevic S., Pritchard L.K., Harvey D.J., Andev R.S., Krumm S.A., Struwe W.B., Cupo A., Kumar A., Zitzmann N. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao L., Diedrich J.K., Kulp D.W., Pauthner M., He L., Park S.R., Sok D., Su C.Y., Delahunty C.M., Menis S. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doores K.J. The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J. 2015;282:4679–4691. doi: 10.1111/febs.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard L.K., Vasiljevic S., Ozorowski G., Seabright G.E., Cupo A., Ringe R., Kim H.J., Sanders R.W., Doores K.J., Burton D.R. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep. 2015;11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blattner C., Lee J.H., Sliepen K., Derking R., Falkowska E., de la Peña A.T., Cupo A., Julien J.P., van Gils M., Lee P.S. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derking R., Ozorowski G., Sliepen K., Yasmeen A., Cupo A., Torres J.L., Julien J.P., Lee J.H., van Montfort T., de Taeye S.W. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog. 2015;11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dingens A.S., Haddox H.K., Overbaugh J., Bloom J.D. Comprehensive mapping of HIV-1 escape from a broadly neutralizing antibody. Cell Host Microbe. 2017;21:777–787.e4. doi: 10.1016/j.chom.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doria-Rose N.A., Schramm C.A., Gorman J., Moore P.L., Bhiman J.N., DeKosky B.J., Ernandes M.J., Georgiev I.S., Kim H.J., Pancera M., NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doria-Rose N.A., Bhiman J.N., Roark R.S., Schramm C.A., Gorman J., Chuang G.-Y., Pancera M., Cale E.M., Ernandes M.J., Louder M.K. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J. Virol. 2015;90:76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkowska E., Le K.M., Ramos A., Doores K.J., Lee J.H., Blattner C., Ramirez A., Derking R., van Gils M.J., Liang C.H. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freund N.T., Wang H., Scharf L., Nogueira L., Horwitz J.A., Bar-On Y., Golijanin J., Sievers S.A., Sok D., Cai H. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 2017;9:eaal2144. doi: 10.1126/scitranslmed.aal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garces F., Lee J.H., de Val N., de la Pena A.T., Kong L., Puchades C., Hua Y., Stanfield R.L., Burton D.R., Moore J.P. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garces F., Sok D., Kong L., McBride R., Kim H.J., Saye-Francisco K.F., Julien J.P., Hua Y., Cupo A., Moore J.P. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J., Kang B.H., Pancera M., Lee J.H., Tong T., Feng Y., Imamichi H., Georgiev I.S., Chuang G.Y., Druz A. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong L., Ju B., Chen Y., He L., Ren L., Liu J., Hong K., Su B., Wang Z., Ozorowski G. Key gp120 glycans pose roadblocks to the rapid development of VRC01-class antibodies in an HIV-1-infected Chinese donor. Immunity. 2016;44:939–950. doi: 10.1016/j.immuni.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders R.W., Moore J.P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 2017;275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharf L., Scheid J.F., Lee J.H., West A.P., Jr., Chen C., Gao H., Gnanapragasam P.N., Mares R., Seaman M.S., Ward A.B. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sok D., Doores K.J., Briney B., Le K.M., Saye-Francisco K.L., Ramos A., Kulp D.W., Julien J.P., Menis S., Wickramasinghe L. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci. Transl. Med. 2014;6:236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Gils M.J., van den Kerkhof T.L., Ozorowski G., Cottrell C.A., Sok D., Pauthner M., Pallesen J., de Val N., Yasmeen A., de Taeye S.W. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2016;2:16199. doi: 10.1038/nmicrobiol.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyd D.F., Sharma A., Humes D., Cheng-Mayer C., Overbaugh J. Adapting SHIVs in vivo selects for envelope-mediated interferon-α resistance. PLoS Pathog. 2016;12:e1005727. doi: 10.1371/journal.ppat.1005727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Wang S., Kong R., Ding W., Lee F.H., Parker Z., Kim E., Learn G.H., Hahn P., Policicchio B. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2016;113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma A., Boyd D.F., Overbaugh J. Development of SHIVs with circulating, transmitted HIV-1 variants. J. Med. Primatol. 2015;44:296–300. doi: 10.1111/jmp.12179. [DOI] [PubMed] [Google Scholar]
- 42.Cheng C., Pancera M., Bossert A., Schmidt S.D., Chen R.E., Chen X., Druz A., Narpala S., Doria-Rose N.A., McDermott A.B. Immunogenicity of a prefusion HIV-1 envelope trimer in complex with a quaternary-structure-specific antibody. J. Virol. 2015;90:2740–2755. doi: 10.1128/JVI.02380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y., Tran K., Bale S., Kumar S., Guenaga J., Wilson R., de Val N., Arendt H., DeStefano J., Ward A.B., Wyatt R.T. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog. 2016;12:e1005767. doi: 10.1371/journal.ppat.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Havenar-Daughton C., Carnathan D.G., Torrents de la Peña A., Pauthner M., Briney B., Reiss S.M., Wood J.S., Kaushik K., van Gils M.J., Rosales S.L. Direct probing of germinal center responses reveals immunological features and bottlenecks for neutralizing antibody responses to HIV Env trimer. Cell Rep. 2016;17:2195–2209. doi: 10.1016/j.celrep.2016.10.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy L.E., van Gils M.J., Ozorowski G., Messmer T., Briney B., Voss J.E., Kulp D.W., Macauley M.S., Sok D., Pauthner M. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016;16:2327–2338. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pauthner M., Havenar-Daughton C., Sok D., Nkolola J.P., Bastidas R., Boopathy A.V., Carnathan D.G., Chandrashekar A., Cirelli K.M., Cottrell C.A. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity. 2017;46:1073–1088.e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanders R.W., van Gils M.J., Derking R., Sok D., Ketas T.J., Burger J.A., Ozorowski G., Cupo A., Simonich C., Goo L. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sok D., Le K.M., Vadnais M., Saye-Francisco K.L., Jardine J.G., Torres J.L., Berndsen Z.T., Kong L., Stanfield R., Ruiz J. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature. 2017;548:108–111. doi: 10.1038/nature23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torrents de la Peña A., de Taeye S.W., Sliepen K., LaBranche C.C., Burger J.A., Schermer E.E., Montefiori D.C., Moore J.P., Klasse P.J., Sanders R.W. Immunogenicity in rabbits of HIV-1 SOSIP trimers from clades A, B, and C, given individually, sequentially, or in combination. J. Virol. 2018;92:e01957-17. doi: 10.1128/JVI.01957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torrents de la Peña A., Julien J.P., de Taeye S.W., Garces F., Guttman M., Ozorowski G., Pritchard L.K., Behrens A.J., Go E.P., Burger J.A. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 2017;20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gach J.S., Mara K.J.V., LaBranche C.C., van Gils M.J., McCoy L.E., Klasse P.J., Montefiori D.C., Sanders R.W., Moore J.P., Forthal D.N. Antibody responses elicited by immunization with BG505 trimer immune complexes. J. Virol. 2019;93:e01188-19. doi: 10.1128/JVI.01188-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klasse P.J., Ketas T.J., Cottrell C.A., Ozorowski G., Debnath G., Camara D., Francomano E., Pugach P., Ringe R.P., LaBranche C.C. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog. 2018;14:e1006913. doi: 10.1371/journal.ppat.1006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ringe R.P., Pugach P., Cottrell C.A., LaBranche C.C., Seabright G.E., Ketas T.J., Ozorowski G., Kumar S., Schorcht A., van Gils M.J. Closing and opening holes in the glycan shield of HIV-1 envelope glycoprotein SOSIP trimers can redirect the neutralizing antibody response to the newly unmasked epitopes. J. Virol. 2019;93:e01656-18. doi: 10.1128/JVI.01656-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milligan C., Richardson B.A., John-Stewart G., Nduati R., Overbaugh J. Passively acquired antibody-dependent cellular cytotoxicity (ADCC) activity in HIV-infected infants is associated with reduced mortality. Cell Host Microbe. 2015;17:500–506. doi: 10.1016/j.chom.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mabuka J., Nduati R., Odem-Davis K., Peterson D., Overbaugh J. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog. 2012;8:e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao H.X., Levesque M.C., Nagel A., Dixon A., Zhang R., Walter E., Parks R., Whitesides J., Marshall D.J., Hwang K.K. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams K.L., Stumpf M., Naiman N.E., Ding S., Garrett M., Gobillot T., Vézina D., Dusenbury K., Ramadoss N.S., Basom R. Identification of HIV gp41-specific antibodies that mediate killing of infected cells. PLoS Pathog. 2019;15:e1007572. doi: 10.1371/journal.ppat.1007572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naiman N.E., Slyker J., Nduati R., Overbaugh J.M. Maternal envelope gp41 ectodomain-specific antibodies are associated with increased mother-to-child transmission of human immunodeficiency virus-1. J. Infect. Dis. 2020;221:232–237. doi: 10.1093/infdis/jiz444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lei L., Yang Y.R., Tran K., Wang Y., Chiang C.I., Ozorowski G., Xiao Y., Ward A.B., Wyatt R.T., Li Y. The HIV-1 envelope glycoprotein C3/V4 region defines a prevalent neutralization epitope following immunization. Cell Rep. 2019;27:586–598.e6. doi: 10.1016/j.celrep.2019.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Permar S.R., Fong Y., Vandergrift N., Fouda G.G., Gilbert P., Parks R., Jaeger F.H., Pollara J., Martelli A., Liebl B.E. Maternal HIV-1 envelope-specific antibody responses and reduced risk of perinatal transmission. J. Clin. Invest. 2015;125:2702–2706. doi: 10.1172/JCI81593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gómez-Román V.R., Florese R.H., Patterson L.J., Peng B., Venzon D., Aldrich K., Robert-Guroff M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods. 2006;308:53–67. doi: 10.1016/j.jim.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 62.Lewis G.K., Ackerman M.E., Scarlatti G., Moog C., Robert-Guroff M., Kent S.J., Overbaugh J., Reeves R.K., Ferrari G., Thyagarajan B. Knowns and unknowns of assaying antibody-dependent cell-mediated cytotoxicity against HIV-1. Front. Immunol. 2019;10:1025. doi: 10.3389/fimmu.2019.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rousseau Christine, Birditt Brian, McKay Angela, Stoddard Julia, Lee Tsan Chun, McLaughlin Sherry. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. Journal of Virological Methods. 2006;136:118–125. doi: 10.1016/j.jviromet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Cheng-Mayer C., Liu R., Landau N.R., Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. Journal of Virology. 1997;71(2):1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blish Catherine, Jalalian-Lechak Zahra, Rainwater Stephanie, Nguyen Minh-An, Dogan Ozge, Overbaugh Julie. Cross-subtype neutralization sensitivity despite monoclonal antibody resistance among early subtype A, C, and D envelope variants of human immunodeficiency virus type 1. Journal of Virology. 2009;83(15):7783–7788. doi: 10.1128/JVI.00673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nduati R., John G., Mbori-Ngacha D., Richardson B., Overbaugh J., Mwatha A., Ndinya-Achola J., Bwayo J., Onyango F.E., Hughes J., Kreiss J. Effect of breastfeeding and formula feeding on transmission of HIV-1: a randomized clinical trial. JAMA. 2000;283:1167–1174. doi: 10.1001/jama.283.9.1167. [DOI] [PubMed] [Google Scholar]
- 67.Sok D., van Gils M.J., Pauthner M., Julien J.P., Saye-Francisco K.L., Hsueh J., Briney B., Lee J.H., Le K.M., Lee P.S. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. USA. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ronen K., Dingens A.S., Graham S.M., Jaoko W., Mandaliya K., McClelland R.S., Overbaugh J. Comprehensive characterization of humoral correlates of human immunodeficiency virus 1 superinfection acquisition in high-risk Kenyan women. EBioMedicine. 2017;18:216–224. doi: 10.1016/j.ebiom.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doria-Rose Nicole, Bhiman Jinal, Roark Ryan, Schramm Chaim, Gorman Jason, Chuang Gwo-Yu. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. Journal of Virology. 2016;90(1):76–91. doi: 10.1128/JVI.01791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milligan C., Omenda M.M., Chohan V., Odem-Davis K., Richardson B.A., Nduati R., Overbaugh J. Maternal neutralization-resistant virus variants do not predict infant HIV infection risk. MBio. 2016;7 doi: 10.1128/mBio.02221-15. e02221–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Magoč Tanja, Salzberg Steven. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin Marcel. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 74.Ralph D.K., Matsen F.A., 4th Consistency of VDJ rearrangement and substitution parameters enables accurate B cell receptor sequence annotation. PLoS Comput. Biol. 2016;12:e1004409. doi: 10.1371/journal.pcbi.1004409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sievers Fabian, Wilm Andreas, Dineen David, Gibson Toby, Karplus Kevin, Li Weizhong. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7(1):539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kearse Matthew, Moir Richard, Wilson Amy, Stones-Havas Steven, Cheung Matthew, Sturrock Shane. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang J., Doria-Rose N.A., Longo N.S., Laub L., Lin C.L., Turk E., Kang B.H., Migueles S.A., Bailer R.T., Mascola J.R., Connors M. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat. Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lefranc M.P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scherer Erin, Smith Robin, Simonich Cassandra, Niyonzima Nixon, Carter Joseph, Galloway Denise. Characteristics of Memory B Cells Elicited by a Highly Efficacious HPV Vaccine in Subjects with No Pre-existing Immunity. PLoS Pathogens. 2014;10(12) doi: 10.1371/journal.ppat.1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams K.L., Wang B., Arenz D., Williams J.A., Dingens A.S., Cortez V., Simonich C.A., Rainwater S., Lehman D.A., Lee K.K., Overbaugh J. Superinfection drives HIV neutralizing antibody responses from several B cell lineages that contribute to a polyclonal repertoire. Cell Rep. 2018;23:682–691. doi: 10.1016/j.celrep.2018.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goo L., Milligan C., Simonich C.A., Nduati R., Overbaugh J. Neutralizing antibody escape during HIV-1 mother-to-child transmission involves conformational masking of distal epitopes in envelope. J. Virol. 2012;86:9566–9582. doi: 10.1128/JVI.00953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vigdorovich V., Oliver B.G., Carbonetti S., Dambrauskas N., Lange M.D., Yacoob C., Leahy W., Callahan J., Stamatatos L., Sather D.N. Repertoire comparison of the B-cell receptor-encoding loci in humans and rhesus macaques by next-generation sequencing. Clin. Transl. Immunology. 2016;5:e93. doi: 10.1038/cti.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Palmer S., Kearney M., Maldarelli F., Halvas E.K., Bixby C.J., Bazmi H., Rock D., Falloon J., Davey R.T., Jr., Dewar R.L. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 86.Larman H.B., Zhao Z., Laserson U., Li M.Z., Ciccia A., Gakidis M.A., Church G.M., Kesari S., LeProust E.M., Solimini N.L., Elledge S.J. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haynes B.F., Gilbert P.B., McElrath M.J., Zolla-Pazner S., Tomaras G.D., Alam S.M., Evans D.T., Montefiori D.C., Karnasuta C., Sutthent R. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tomaras G.D., Yates N.L., Liu P., Qin L., Fouda G.G., Chavez L.L., Decamp A.C., Parks R.J., Ashley V.C., Lucas J.T. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ralph D.K., Matsen F.A., 4th Per-sample immunoglobulin germline inference from B cell receptor deep sequencing data. PLoS Comput. Biol. 2019;15:e1007133. doi: 10.1371/journal.pcbi.1007133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu G.J., Kula T., Xu Q., Li M.Z., Vernon S.D., Ndung’u T., Ruxrungtham K., Sanchez J., Brander C., Chung R.T. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the antibody gene sequencing data of BG505’s B cell repertoire at week 14 reported in this paper can be found at BioProject SRA: accession PRJNA588318. GenBank accession numbers for BG505 antibodies isolated from month 27 are Genbank: MW602658 - Genbank: MW602803.
GenBank accession numbers for HIV Env variants isolated from BG505 at week 14 and month 27 are Genbank: MW650605 - Genbank: MW650637. GenBank accession numbers for HIV Env variants used in this study are Genbank: DQ208458.1 (BG505.W6M.C2), Genbank: DQ208457.1 (BG505.W6M.B1), Genbank: DQ208456.1 (BG505.W6M.A5), Genkbank: DQ208449.1 - Genbank: DQ208455.1 (MG505.W0M Env clones), Genbank: DQ208480.1 (BL035.W6M.C1), Genbank: AAR27754.1 (C.ZA.1197MB), Genbank: EU123924 (SF162), and Genbank: FJ866133 (QC406.70M.F3).
The code used in this study for partis analysis is publicly available on GitHub: (https://github.com/psathyrella/partis).