Abstract
DNA hypermethylation is an important epigenetic mechanism for gene expression inactivation in head and neck cancer (HNC). Saliva has emerged as a novel liquid biopsy representing a potential source of biomarkers. We performed a comprehensive meta-analysis to evaluate the overall diagnostic accuracy of salivary DNA methylation for detecting HNC. PubMed EMBASE, Web of Science, LILACS, and the Cochrane Library were searched. Study quality was assessed by the Quality Assessment for Studies of Diagnostic Accuracy-2, and sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (dOR), and their corresponding 95% confidence intervals (CIs) were calculated using a bivariate random-effect meta-analysis model. Meta-regression and subgroup analyses were performed to assess heterogeneity. Eighty-four study units from 18 articles with 8368 subjects were included. The pooled sensitivity and specificity of salivary DNA methylation were 0.39 and 0.87, respectively, while PLR and NLR were 3.68 and 0.63, respectively. The overall area under the curve (AUC) was 0.81 and the dOR was 8.34. The combination of methylated genes showed higher diagnostic accuracy (AUC, 0.92 and dOR, 36.97) than individual gene analysis (AUC, 0.77 and dOR, 6.02). These findings provide evidence regarding the potential clinical application of salivary DNA methylation for HNC diagnosis.
Keywords: DNA methylation, epigenetics, head and neck cancer, saliva, biomarkers, liquid biopsy, meta-analysis
1. Introduction
Head and neck cancer (HNC) comprises a heterogenous group of epithelial malignancies arising from mucosal linings of the oral cavity, oropharynx, larynx, and hypopharynx. According to data from the World Health Organization’s GLOBOCAN network, HNC is highly prevalent worldwide, accounting for an estimated 890,000 new cases and 450,000 deaths in 2018 [1]. Despite improvements in diagnosis and therapeutic strategies, the 5-year survival rate for HNC has remained around 50% for the last decade. Unfortunately, HNC patients are frequently diagnosed in advanced stages involving a high risk of locoregional recurrence and distant metastasis [2,3]. Therefore, it is necessary to identify new biomarkers for diagnosis and prognosis to allow early detection and improved overall survival. HNC arises through multistep carcinogenic pathways as a result of cumulative genetic and epigenetic aberrations resulting from risk factors including alcohol and tobacco consumption, human papillomavirus infection, chronic inflammation, and genetic predisposition, and leading to reduced tumor suppressor gene function as well as oncogene activation [2,4]. In recent years, accumulating scientific evidence has highlighted the important role in tumorigenesis of epigenetic mechanisms, which represent a cancer hallmark [5,6]. Epigenetic alterations such as DNA methylation, histone covalent modifications, chromatin remodeling, and non-coding RNAs have been implicated in the landscape of phenotypical changes occurring in a wide variety of malignancies, including HNC [7]. DNA hypermethylation has been shown to be an important epigenetic mechanism for gene expression inactivation. The hypermethylation of cytosine–phosphodiester bond–guanine (CpG) islands within promoter regions plays an important role in carcinogenesis through the transcriptional silencing of different tumor suppressor genes or dysfunction in DNA repair genes [7,8]. Several studies have focused on the identification of aberrant promoter methylation patterns in HNC tissue and liquid biopsies [9,10]. Moreover, a number of investigations have detected promoter hypermethylation in various genes using saliva from HNC patients [10,11,12]. Therefore, salivary DNA hypermethylation represents a promising biomarker for non-invasively diagnosing HNC.
DNA methylation is a heritable and stable epigenetic mechanism implicated in the regulation of gene expression that plays an important role during normal development, regulating X chromosome inactivation, genomic imprinting, and preventing the transcription of DNA repetitive sequences, inserted viral sequences, and transposons [8,13]. DNA methylation is characterized by the covalent addition of a methyl group to the 5’-position of the pyrimidine ring of cytosines by DNA methyltransferases, giving rise to 5-methylcytosine. This enzymatic process occurs predominantly within CpG dinucleotides which are concentrated at CpG-rich DNA stretches named CpG islands (CGIs), which overlap the promoter region of 60–70% of protein-coding genes [14]. In the human genome, approximately 80% of CpG dinucleotides are heavily methylated whereas CGIs in gene promoters are mostly unmethylated, allowing active gene transcription [13]. Dysregulation of DNA methylation has been found to be involved in several diseases, representing an early epigenetic event in carcinogenesis. These alterations of normal DNA methylation patterns in cancer have been characterized as global hypomethylation and gene-specific hypermethylation. The global hypomethylation of repetitive sequences and transposable elements within the genome induces genomic instability and mutagenesis. In this line, the loss of DNA methylation may also activate latent viral sequences, promoting carcinogenesis. By contrast, in addition to global hypomethylation, aberrant promoter hypermethylation can drive the inactivation of key tumor suppressor genes, which are unmethylated in non-malignant tissues [15]. In this sense, although silencing by DNA hypermethylation of some genes is common in many types of tumors, the methylation profile of gene promoters is different for each human cancer, allowing the identification of cancer-specific hypermethylation patterns [16]. Although tissue biopsy of the primary tumor or metastatic lesions remains the gold standard method for diagnosis, DNA methylation biomarkers can be assessed in different liquid biopsies, representing a non-invasive alternative for early cancer detection [10,17,18].
Recently, saliva has emerged as an attractive liquid biopsy for genomic and epigenomic analysis. Saliva-based liquid biopsy is a fast, reliable, cost-effective, and non-invasive approach to analyze epigenetic alterations involved in the onset and course of the disease. Some researchers have found comparable methylation profiles between saliva and tissue, representing a non-invasive alternative for epigenomic profiling [19,20]. Additionally, although similar methylation DNA patterns have been reported between saliva and blood [21,22], methylation differences between both biofluids can be identified due to tumor shedding and tissue-specific methylation [23,24]. Focusing on promoter hypermethylation, a number of investigations have detected various methylated genes in saliva, representing a promising biomarker for non-invasive HNC detection [10,11,12].
The purpose of this systematic review and meta-analysis was to summarize the results of published clinical studies to assess the overall diagnostic accuracy of salivary DNA hypermethylation for discriminating HNC.
2. Materials and Methods
2.1. Protocol and Registration
This study was conducted according to Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) guidelines [25], and the protocol was registered with the International Prospective Register of Systematic Reviews (reference No. CRD42020199114).
2.2. Search Strategy and Study Selection
The systematic literature search of eligible articles published up to 27 August 2020 was carried out without language restrictions using PubMed, EMBASE, Web of Science, LILACS, and the Cochrane Library. The search strategy was based on the following combinations of keywords and medical subject headings: (methylation OR hypermethylation OR epigenomics) AND (saliva OR oral rinse OR mouthwash) AND (head and neck cancer OR head and neck neoplasm OR head and neck carcinoma OR head and neck squamous cell carcinoma OR HNSCC OR oral cancer OR pharyngeal cancer OR laryngeal cancer). Studies were screened based on title and abstract, and eligible manuscripts were retrieved for full-text review. In addition, reference lists from each original and review article were searched manually in order to find further relevant studies. The literature search was performed independently by two investigators (ORG and MMSC), and disagreements during the selection process were resolved by consensus. The studies selected by means of the search strategy and other references were managed using RefWorks software (https://www.refworks.com/content/path_learn/faqs.asp, accessed 28 October 2020), and duplicate items were removed using the associated tools.
2.3. Selection Criteria
The inclusion criteria were as follows: (1) studies that evaluated the diagnostic accuracy of gene promoter hypermethylation in saliva samples from HNC patients; (2) inclusion of a control group consisting of healthy controls; (3) sufficient data for generating a two-by-two (2 × 2) contingency table containing true positive (TP), false positive (FP), true negative (TN), and false negative (FN) values. The exclusion criteria were as follows: (1) reviews, letters, personal opinions, book chapters, case reports, conference abstracts, and meetings; (2) duplicate publications; (3) in vitro and in vivo animal experiments.
2.4. Data Extraction
All eligible studies were assessed independently by two investigators (ORG and MMSC) and data were extracted using a pre-established form designed on a Microsoft Excel spreadsheet (Microsoft Corp. Redmond, WA, USA). Any disagreement among reviewers was resolved by consensus. The following information was extracted from each study: name of first author, year of publication, country, anatomic tumor location, number of cases and controls, positive methylated cases, positive methylated controls, method for DNA methylation detection, type of saliva sample (saliva or oral rinse), methylated gene names, and statistical analysis outcomes, including diagnostic accuracy and cut-off values. If the required data were incomplete, attempts were made to contact the authors to obtain the missing information. We defined “study unit” as the analysis of a relationship between gene promoter hypermethylation and HNC. Therefore, a single publication could potentially include more than one study unit as a result of reporting promoter hypermethylation for multiple genes.
2.5. Quality Assessment of Individual Studies
Following Healthcare Research and Quality Agency recommendations, two independent researchers (ADL and LMR) applied the Quality Assessment of Diagnostic Accuracy Studies-2 checklist (QUADAS-2) [26]. Any discrepancies were resolved by a third reviewer (MMSC). The QUADAS-2 checklist assesses study quality by analyzing four key domains: (1) patient selection, (2) index tests, (3) reference tests, and (4) flow and times. Risk of bias and applicability concerns for each domain were assessed as “low”, “high”, or “unclear”. One point was assigned to each item assessed as “low”. Thus, articles were grouped into the following quality categories based on their cumulative score: “high” quality (6–7 points), “moderate” quality (4–5 points), and “low” quality (0–3 points).
2.6. Statistical Analysis
MetaDiSc software (v.1.4) [27], free R software (v.3.4.4; https://www.r-project.org, accessed 30 November 2020), and STATA (v.14.0; https://www.stata.com, accessed 30 November 2020) were used to carry out statistical analysis. The numbers of TP, FP, FN, and TN in each study unit in the diagnostic meta-analysis were extracted to calculate pooled sensitivity [TP/(TP + FN)], specificity [TN/(TN + FP)], positive likelihood ratio (PLR) [(sensitivity/(1 − sensitivity)], negative likelihood ratio (NLR), [(1 − specificity)/specificity)], diagnostic odds ratio (dOR), and their corresponding 95% confidence intervals (CIs) using a bivariate random or fixed effect meta-analysis model. The pooled diagnostic performance of salivary DNA promoter hypermethylation for HNC detection was determined by plotting the summary receiver operator characteristic (SROC) curve and calculating the area under the SROC curve (AUC). Heterogeneity analysis was used to identify factors influencing accuracy indicators and the statistical model applied [27]. Spearman’s correlation analysis and ROC plane plots were used to assess heterogeneity due to the threshold effect. Cochran’s Q statistic test-based chi-squared test and I2 statistics were used to assess non-threshold heterogeneity. When I2 > 50% and/or p < 0.05 for the Cochran’s Q test, heterogeneity was considered to be significant. The DerSimonian and Laird random effects model was applied when heterogeneity was significant; otherwise, we applied the Mantel–Haenszel fixed effects model. Potential sources of non-threshold heterogeneity were explored by meta-regression and subgroup analyses. In addition, the predictive value of post-salivary DNA promoter hypermethylation for HNC diagnosis was evaluated by Fagan’s nomogram. Post-test probability was calculated using Bayes theorem under the assumption of prior probabilities of 25%, 50%, and 75%, respectively [28]. Deeks’ funnel plot asymmetry test [29] was used to ascertain publication bias (statistical significance: p < 0.05).
3. Results
3.1. Study Selection
A PRISMA flowchart for the literature identification and selection process is shown in Figure 1. A total of 576 studies were identified based on the search strategy across the five electronic databases, which was reduced to 470 after removing duplicates. After title and abstract review, 27 articles were submitted for full-text reading, of which nine were excluded for the following reasons: non-independent cancer group (two articles); reviews, letters, personal opinions, book chapters, case reports, conference abstracts, and meetings (three articles); absence of a healthy control group (one article); saliva enriched with brush oral cytology (two articles); and insufficient information for meta-analysis (one article). In the end, 18 articles met the inclusion criteria for final analysis [10,11,12,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44].
Figure 1.
PRISMA flow diagram.
3.2. Characteristics of Included Studies
The characteristics of the included studies are shown in Table 1. The 18 articles comprised a total of 84 study units, including 4758 HNC patients and 3605 healthy individuals (the sample size ranged from 13 [31] to 210 [41]). All articles were published between 2001 and 2020, and studies were conducted in the following geographical regions: the United States (5), Australia (3), Thailand (2), Japan (2), France (1), Brazil (1), India (1), Taiwan (1), Colombia (1), and Italy (1). Saliva samples included oral rinses (12) and whole saliva (6). Salivary DNA methylation was detected by different methods, including methylation-specific polymerase chain reaction (MSP) (11) and quantitative MSP (7). A total of 34 different genes were identified in the studies. Five studies evaluated the methylation status of a single gene [31,34,36,39,41] and 13 studies evaluated two or more genes [10,11,12,30,32,33,35,37,38,40,42,43,44]. Ten studies focused on gene promoter methylation panels combining two to four genes, whereas eight studies evaluated only single genes.
Table 1.
Summary of descriptive characteristics of included studies.
| First Author | Anatomic Tumor Location | Type of Sample | Method | Biomarker | Cancer Group N (M+) |
Control Group N (M+) |
|---|---|---|---|---|---|---|
| Rosas 2001 | HNC | Oral rinse (NaCl) | MSP | p16 | 30 (11+) | 30 (1+) |
| DAPK | 30 (6+) | 30 (0+) | ||||
| MGMT | 30 (4+) | 30 (1+) | ||||
| Righini 2007 | HNC | Oral rinse (NaCl) | MSP | TIMP3 | 60 (17+) | 30 (0+) |
| ECAD | 60 (12+) | |||||
| p16 | 60 (16+) | |||||
| MGMT | 60 (13+) | |||||
| DAPK | 60 (9+) | |||||
| RASSF1A | 60 (10+) | |||||
| p15 | 60 (7+) | |||||
| p14 | 60 (2+) | |||||
| APC | 60 (4+) | |||||
| FHIT | 60 (2+) | |||||
| hMLH1 | 60 (0+) | |||||
| Franzmann 2007 | HNC | Oral rinse (NaCl) | MSP | CD44 | 11 (9+) | 10 (0+) |
| Guerrero-Preston 2011 | HNC | Oral rinse (NaCl) | qMSP | HOXA9 | 32 (20+) | 19 (9+) |
| NID2 | 32 (23+) | 19 (12+) | ||||
| HOXA9+NID2 | 32 (25+) | 19 (6+) | ||||
| OC | HOXA9 | 16 (11+) | 19 (9+) | |||
| NID2 | 16 (14+) | 19 (12+) | ||||
| HOXA9+NID2 | 16 (14+) | 19 (6+) | ||||
| OPC | HOXA9 | 16 (9+) | 19 (9+) | |||
| NID2 | 16 (9+) | 19 (12+) | ||||
| HOXA9+NID2 | 16 (11+) | 19 (6+) | ||||
| Nagata 2011 | OC | Oral rinse (NaCl) | MSP | ECAD | 34 (32+) | 24 (5+) |
| TMEFF2 | 34 (29+) | 24 (3+) | ||||
| RARβ | 34 (28+) | 24 (2+) | ||||
| MGMT | 34 (26+) | 24 (5+) | ||||
| FHIT | 34 (27+) | 24 (8+) | ||||
| WIF1 | 34 (24+) | 24 (5+) | ||||
| DAPK | 34 (19+) | 24 (6+) | ||||
| p16 | 34 (13+) | 24 (2+) | ||||
| HIN | 34 (10+) | 24 (2+) | ||||
| TIMP3 | 34 (8+) | 24 (1+) | ||||
| p15 | 34 (22+) | 24 (9+) | ||||
| APC | 34 (18+) | 24 (9+) | ||||
| SPARC | 34 (14+) | 24 (8+) | ||||
| ECAD+TMEFF2+RARβ+MGMT | 34 (34+) | 24 (3+) | ||||
| ECAD+TMEFF2+MGMT | 34 (33+) | 24 (2+) | ||||
| ECAD+TMEFF2+RARβ | 34 (32+) | 24 (1+) | ||||
| ECAD+RARβ+MGMT | 34 (31+) | 24 (2+) | ||||
| Ovchinnikov 2012 | OC | Saliva | Nested MSP | p16+RASSF1A+DAPK1 | 143 (117+) | 46 (6+) |
| Rettori 2012 | HNC | Oral rinse (NaCl) | qMSP | DCC | 143 (75+) | 50 (5+) |
| CCNA1 | 146 (17+) | 60 (2+) | ||||
| DAPK | 146 (12+) | 39 (1+) | ||||
| MGMT | 146 (11+) | 57 (2+) | ||||
| TIMP3 | 146 (7+) | 60 (2+) | ||||
| MINT31 | 68 (3+) | 20 (0+) | ||||
| AIM1 | 71 (2+) | 41 (0+) | ||||
| SFRP1 | 71 (2+) | 20 (0+) | ||||
| APC | 62 (2+) | 20 (0+) | ||||
| CDKN2A | 69 (1+) | 20 (0+) | ||||
| HIN1 | 134 (16+) | 57 (11+) | ||||
| CCNA1+DAPK+DCC+MGMT+TIMP3 | NA | NA | ||||
| CCNA1+DAPK+MGMT+TIMP3 | NA | NA | ||||
| CCNA1+DAPK+MGMT | NA | NA | ||||
| CCNA1+MGMT+TIMP3 | NA | NA | ||||
| CCNA1+DAPK+TIMP3 | NA | NA | ||||
| DAPK+MGMT+TIMP3 | NA | NA | ||||
| CCNA1+MGMT | NA | NA | ||||
| CCNA1+DAPK | NA | NA | ||||
| CCNA1+TIMP3 | NA | NA | ||||
| Ksumoto 2012 | OC | Oral rinse | MSP | p16 | 10 (4+) | 3 (0+) |
| Ovchinnikov 2014 | HNC | Saliva | MSP | PCQAP5’ | 62 (42+) | 49 (17+) |
| PCQAP3’ | 60 (41+) | 45 (19+) | ||||
| Gaykalova 2015 | HNC | Oral rinse | qMSP | ZNF14 | 59 (5+) | 35 (0+) |
| ZNF160 | 59 (10+) | 35 (0+) | ||||
| ZNF420 | 59 (8+) | 35 (0+) | ||||
| ZNF14+ZNF160+ZNF420 | 59 (13+) | 35 (0+) | ||||
| Lim 2016 | HNC | Saliva | MSP | RASSF1α | 88 (36+) | 122 (10+) |
| p16 | 88 (41+) | 122 (38+) | ||||
| TIMP3 | 88 (33+) | 122 (22+) | ||||
| PCQAP5’ | 88 (72+) | 122 (66+) | ||||
| PCQAP3’ | 88 (30+) | 122 (18+) | ||||
| RASSF1α+p16+TIMP3+PCQAP5’+PCQAP3’ | 88 (62+) | 122 (24+) | ||||
| Ferlazzo 2017 | OC | Saliva (Oragene DNA kit) | MSP | P16 | 58 (10+) | 90 (5+) |
| MGMT | 58 (16+) | 90 (7+) | ||||
| P16 + MGMT | 58 (12+) | 90 (0+) | ||||
| Cheng 2017 | OC | Oral rinse (0.12% clorhexidine) | qMSP | ZNF582 | 94 (62+) | 65 (10+) |
| PAX1 | 94 (64+) | 65 (7+) | ||||
| ZNF582+PAX1 | 94 (75+) | 65 (14+) | ||||
| Puttipanyalears 2018 | HNC | Oral rinse (NaCl) | qMSP | TRH | 66 (57+) | 54 (4+) |
| OC | 42 (37+) | |||||
| OPC | 24 (20+) | |||||
| Liyanage 2020 | HNC | Saliva | MSP | p16 | 88 (62+) | NA |
| RASSF1 α | 88 (59+) | NA | ||||
| TIMP3 | 88 (68+) | NA | ||||
| PCQAP/MED15 | 88 (66+) | NA | ||||
| p16+RASSF1α+TIMP3+PCQAP | 84 (80+) | 60 (5+) | ||||
| OC | p16 | 54 (39+) | NA | |||
| RASSF1α | 54 (37+) | NA | ||||
| TIMP3 | 54 (43+) | NA | ||||
| PCQAP/MED15 | 54 (43+) | NA | ||||
| p16+RASSF1α+TIMP3+PCQAP | 54 (46+) | 60 (5+) | ||||
| OPC | p16 | 34 (23+) | NA | |||
| RASSF1α | 34 (22+) | NA | ||||
| TIMP3 | 34 (25+) | NA | ||||
| PCQAP/MED15 | 34 (23+) | NA | ||||
| p16+RASSF1α+TIMP3+PCQAP | 34 (34+) | 60 (5+) | ||||
| Srisuttee 2020 | OC | Oral rinse (NaCl) | qMSP | NID2 | 43 (34+) | 90 (0+) |
| Shen 2020 | OPC | Oral rinse (NaCl) | qMSP | EDNRB | 21 (15+) | 40 (2+) |
| PAX5 | 21 (15+) | 40 (4+) | ||||
| p16 | 21 (3+) | 40 (0+) | ||||
| González-Pérez 2020 | OC | Saliva | MSP | p16 | 43 (19+) | 40 (4+) |
| RASSF1A | 43 (10+) | 40 (2+) | ||||
| p16+RASSF1A | 43 (23+) | 40 (5+) |
Abbreviations: HNC = head and neck cancer; OC = oral cancer; OPC = oropharyngeal cancer; MSP = methylation-specific polymerase chain reaction; qMSP = quantitative MSP; NA = not available.
3.3. Quality Assessment of the Included Studies
All included articles were evaluated for risk of bias using the QUADAS-2 checklist (Figure S1). The major risk of bias in this study was patient selection domain, as 13 out of 18 publications were unclear or lacked detail on whether the patient sample was consecutive or random. Additionally, 14 out of 18 studies did not provide a detailed description of the inclusion/exclusion criteria. Moreover, there was high risk of bias in the index test, since some studies lacked a setting threshold. All domains were considered to have a low risk of bias in terms of applicability concern. All studies were of moderate-to-high quality, with an average QUADAS-2 score of 5.6.
3.4. Diagnostic Accuracy of Salivary DNA Promoter Hypermethylation
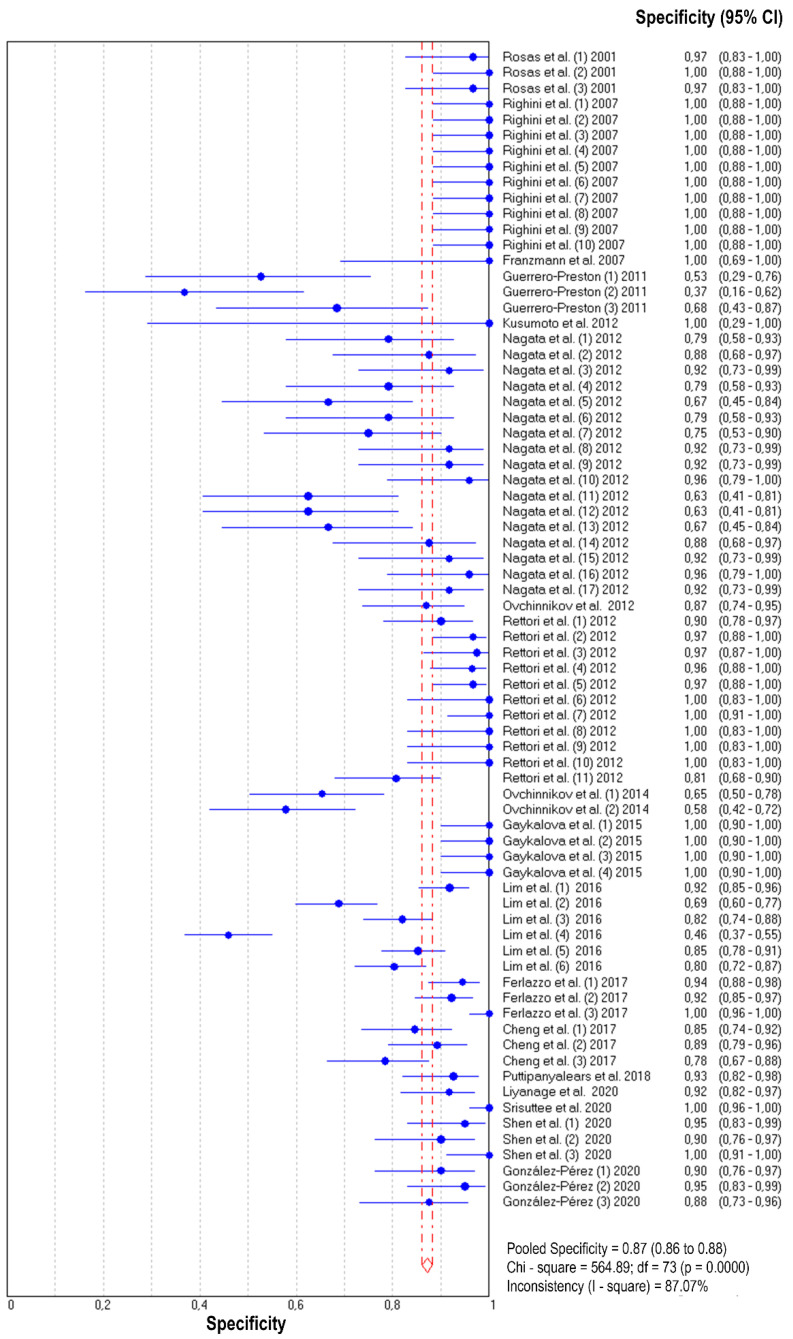
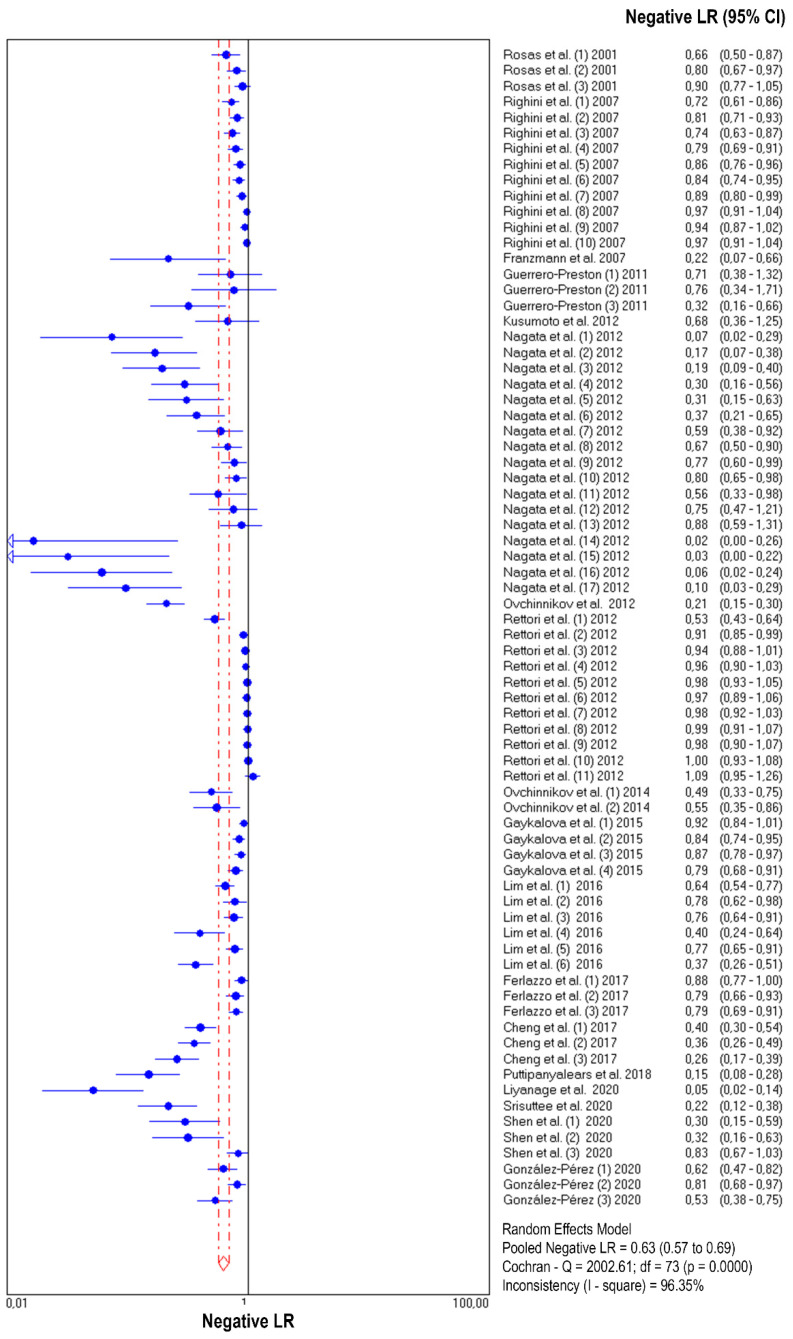
The diagnostic accuracy (sensitivity, specificity, and 95% confidence interval) of each study unit (n = 74) included in this meta-analysis is shown in Figure 2 and Figure 3. The pooled sensitivity and specificity of salivary DNA hypermethylation genes in the diagnosis of HNC were 0.39 (95% CI: 0.38–0.41) and 0.87 (95% CI: 0.86–0.88), respectively. The PLR and NLR were 3.68 (95% CI: 2.97–4.57) and 0.63 (95% CI: 0.57–0.69) (Figure 4 and Figure 5), respectively; the summary dOR was 8.34 (95% CI: 6.10–11.39) (Figure S2); and the area under the SROC was 0.81 (95% CI 0.77–0.84) (Figure 6). As shown in Fagan’s nomogram (Figure S3), given a pre-test probability of 27.8%, a positive measurement leads to a post-test cancer probability of 59%, whereas a negative measurement leads to a post-test probability of 20%.
Figure 2.
Forest plot of sensitivities from test accuracy studies of salivary DNA methylation for predicting HNC diagnosis.
Figure 3.
Forest plot of specificities from test accuracy studies of salivary DNA methylation for predicting HNC diagnosis.
Figure 4.
Forest plot of likelihood ratios for positive test results from salivary DNA methylation studies for predicting HNC diagnosis.
Figure 5.
Forest plot of likelihood ratios for negative test results from salivary DNA methylation studies for predicting HNC diagnosis.
Figure 6.
SROC curve with pooled estimates of sensitivity, specificity, and the AUC for all included studies of the salivary DNA methylation studies for detecting HNC.
3.5. Heterogeneity and Subgroup Analysis
As shown in Figure 2, Figure 3, Figure 4 and Figure 5 and Figure S2, significant heterogeneity was observed regarding the pooled sensitivity (I2 = 96.33%; p < 0.001), specificity (I2 = 87.07%; p < 0.001), PLR (I2 = 73.99%; p < 0.001), NLR (I2 = 96.35%; p < 0.001), and dOR (I2 = 71.83%; p < 0.001). The representation of accuracy estimates from each study in the SROC space revealed a typical pattern of a “shoulder arm”, suggesting the presence of a threshold effect (Figure S4). Moreover, Spearman’s correlation coefficient between the logit of the true positive rate and the logit of the false positive rate was 0.633 (p = 0.000), which showed further indication of a threshold effect. In addition to the variations due to the threshold effect, meta-regression was performed to determine the possible sources of heterogeneity using the following covariates as predictor variables: sample type, sample size, anatomic tumor location, DNA methylation methods, and methylation gene profiling. The results indicated that anatomic tumor location (p = 0.002) and gene profiling (p < 0.001) were potential sources of heterogeneity in this study (Table S1). Consequently, subgroup analysis based on anatomic tumor location (HNC vs. oral cancer vs. oropharyngeal cancer) and gene profiling (single vs. combination of genes) was performed. As shown in Table S2, the results indicated similar accuracy for salivary methylated genes in oral cancer (sensitivity, 0.63; specificity, 0.87; PLR, 4.02; NLR, 0.40; dOR, 13.07; AUC, 0.88) and oropharyngeal cancer (sensitivity, 0.70; specificity, 0.86; PLR, 3.67; NLR, 0.41; dOR, 13.26; AUC, 0.87). However, differences in diagnostic accuracy were observed in the HNC group (sensitivity, 0.31; specificity, 0.86; PLR, 3.03; NLR, 0.75; dOR, 5.78; AUC, 0.81). When basing the meta-analysis on gene profile, the combination of methylated genes showed higher diagnostic accuracy (sensitivity, 0.73; specificity, 0.88; PLR, 5.76; NLR, 0.22; dOR, 36.97; AUC, 0.92) compared to individual genes (sensitivity, 0.32; specificity, 0.87; PLR, 3.17; NLR, 0.71; dOR, 6.02; AUC, 0.77). Although the meta-regression results were negative for other covariates, we conducted subgroup analyses based on these factors to further explore the diagnostic potential of salivary DNA methylated genes (Table S2).
3.6. Publication Bias
The potential publication bias in each salivary DNA methylation study was explored by Deeks’ funnel plot asymmetry test, which yielded a slope coefficient p-value of 0.711 overall. This indication of a symmetric data pattern suggests the absence of publication bias (Figure S5).
4. Discussion
Over the last few years, saliva has aroused great interest in the scientific community due to its potential as a non-invasive liquid biopsy in cancer. Several studies have evidenced the diagnostic capability of salivary biomarkers for diagnosing both HNC [45] and tumors distant from the oral cavity [46]. In this sense, a wide variety of biomolecules have been assessed as tumor biomarkers using saliva-omics approaches, including genomic, epigenomic, transcriptomic, metabolomic, proteomic, and microbiomic technologies [47]. In the field of epigenomics, DNA promoter hypermethylation represents one of the most intensively studied epigenetic alterations in human cancer. Promoter hypermethylation of critical pathway genes has been recognized as an important epigenetic mechanism of carcinogenesis [13]. Its potential role as an early diagnostic biomarker stems from the fact that gene promoter hypermethylation is an early event in cancer development [13]. DNA hypermethylation as a common event in cancer plays an important role in HNC development and progression [7]. The detection of DNA methylation in body fluids has emerged as an opportunity to assess the methylation status non-invasively and cost-effectively. In this line, several studies have investigated the promoter methylation of different tumor-suppressor genes in saliva from HNC patients [10,11,12,38]. Therefore, salivary DNA methylation biomarkers could be potentially used in the screening and early detection of HNC.
To the best of our knowledge, this study represents the first meta-analysis evaluating the diagnostic accuracy of promoter hypermethylation genes in saliva for differentiating HNC patients from healthy individuals. The present analysis included a total of 18 articles (84 study units) involving 4758 HNC patients and 3605 healthy individuals. According to QUADAS-2 quality evaluation, most of the included studies were of moderate quality. As for the overall accuracy of salivary hypermethylated genes for discriminating HNC from healthy individuals, the pooled diagnostic parameters of sensitivity, specificity, and AUC values were 0.39, 0.87, and 0.81, respectively. The summary dOR of 8.34 reflects the diagnostic capacity of salivary hypermethylated genes for HNC. The pooled PLR value of 3.68 indicates that a person testing positive had approximately 3.68 times higher probability of having cancer than a healthy individual. On the other hand, the pooled NLR indicated that a person testing negative had a 63% probability of not having cancer. Additionally, given a pre-test probability of 27.8% in Fagan’s nomogram, correct HNC diagnosis increased to 59% after a positive test, and reduced to 20% after a negative test. Overall, these results show a low sensitivity for HNC detection, indicating a high false negative rate. Therefore, the salivary gene methylation evaluated in this meta-analysis presented limitations as a screening biomarker for HNC. However, the diagnostic specificity of gene methylation for HNC was very high, suggesting that detection in saliva may aid assessment in HNC diagnosis. New molecular biology techniques such as next-generation sequencing platforms and digital PCR represent an opportunity for improving the sensitivity of methylation assays and for discovering new methylation patterns.
Due to the fact that heterogeneity is inherent to any diagnostic accuracy meta-analysis, an evaluation of the reasons contributing to inconsistencies across studies should be carried out. In the present research, overall heterogeneity among studies was high, so a bivariate random effects model was applied. This significant heterogeneity was reflected numerically in Cochran’s Q test and the I2 statistic. Further exploration of the heterogeneity revealed the presence of a threshold effect. The threshold effect is a major source of heterogeneity in meta-analyses of diagnostic tests. It arises from differences in sensitivities and specificities or likelihood ratios due to different cut-offs among studies for defining positive (or negative) test results [27]. In the present meta-analysis, the threshold effect was suggested by the visual inspection of accuracy estimates in forest plots where increasing specificities with decreasing sensitivities were observed. Later, the ROC plane and the Spearman correlation test also indicated the presence of the threshold effect. The variations in accuracy estimates among different studies could be due to a number of reasons other than the threshold, such as study population (anatomic tumor location, TNM staging), index test (differences in technology, assays), reference standard, and study design. Therefore, heterogeneity should be explored by relating study level co-variates to an accuracy measure by meta-regression techniques [27]. In the present study, meta-regression was performed to test the effect of sample type, sample size, anatomic tumor location, DNA methylation method, and methylated gene profiling. The results point to anatomic tumor location and gene profiling strategy as possible causes of heterogeneity. Stratified analysis by anatomic tumor location showed that salivary gene methylation had a higher diagnostic accuracy for discriminating oral (AUC = 0.88) and oropharyngeal (AUC = 0.87) tumors than overall HNC (AUC = 0.81). The explanation for these findings may be that tumors located in the oral cavity and oropharynx release more tumor cells directly into saliva. With respect to gene profiling strategy, subgroup analysis showed that single genes presented low sensitivity, but were highly specific to cancer tissue. The combination of salivary methylated genes had better diagnostic accuracy than single gene-based tests, with a dOR of 36.97 vs. 6.02 and AUC of 0.92 vs. 0.77, respectively, demonstrating that the use of salivary methylated gene panels as biomarkers may increase HNC detection accuracy without decreasing specificity. We also conducted subgroup analyses based on sample type, sample size, and DNA methylation method but no significant differences in diagnostic accuracy were observed. Future studies should be conducted to clarify the impact of these factors on the diagnostic potential of salivary DNA methylation.
The current meta-analysis is not free of limitations. Firstly, this meta-analysis included case–control studies, but none was multicenter. Moreover, no randomized controlled trials exist on this topic. Secondly, considerable heterogeneity was observed among the included studies. Although we examined the sources of heterogeneity in five variables, we were not able to explore other demographic and clinicopathological factors due to lack of information. Thirdly, studies involving saliva samples only evaluated some of the genes methylated in HNC tissue. The remaining genes should also be tested in saliva to determine their diagnostic potential. Lastly, confounding variables such as gender, age, lifestyle (tobacco and alcohol), and diet were not considered in most of the included studies. Due to these limitations, future research based on large-scale prospective diagnostic studies involving multiple health centers would contribute to further evaluating the clinical utility of salivary gene promoter methylation for HNC diagnosis. Furthermore, better comparison among future studies would benefit from standardization of analytic strategies and cut-off selection.
5. Conclusions
Our meta-analysis suggests that the detection of DNA promoter hypermethylation in saliva is a promising biomarker for HNC diagnosis, mainly in oral and oropharyngeal tumors. The use of salivary hypermethylated gene panels improves diagnostic accuracy with respect to single-gene analysis. This meta-analysis could provide valuable insights into methodology design for further research studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jpm11060568/s1, Figure S1: Quality assessment of the included studies according to Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria, Figure S2: Forest plot of pooled dOR salivary DNA methylation for the diagnosis of HNC, Figure S3: Fagan’s monogram evaluating the clinical utility of salivary DNA methylation for differentiating HNC patients, Figure S4: Representation of sensitivity against (1-specificity) in ROC space for each study of salivary methylation in the diagnosis of HNC, Figure S5: Deeks’ funnel plot asymmetry test for the assessment of potential bias of included studies, Table S1: Results of meta-regression analysis, Table S2: Subgroup analysis of salivary DNA methylation for HNC detection based on different covariates.
Author Contributions
Conceptualization, Ó.R.-G., L.M.-R., and M.M.S.-C.; methodology, Ó.R.-G. and M.M.S.-C.; software, C.M.-R. and Á.S.-B.; formal analysis, C.M.-R. and Á.S.-B.; investigation, Ó.R.-G., Á.D.-L., and M.M.S.-C.; data curation, Ó.R.-G., Á.D.-L., and M.M.S.-C.; writing—original draft preparation, Ó.R.-G. and M.M.S.-C.; writing—review and editing, L.M.-R., Á.D.-L., and R.L.-L.; visualization, Ó.R.-G. and J.M.-L.; supervision, M.M.S.-C.; project administration, M.M.S.-C.; funding acquisition, M.M.S.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was co-funded by the Instituto de Salud Carlos III (ISCIII) (PI20/01449) and the European Regional Development Fund (FEDER). A.D.-L. is funded by a “Juan Rodés” contract from ISCIII (JR17/00016). L.M.-R. is funded by a “Miguel Servet” contract from ISCIII (CP20/00129).
Conflicts of Interest
R.L.-L. reports other from Nasasbiotech, during the study; grants and personal fees from Roche, grants and personal fees from Merck, personal fees from AstraZeneca, personal fees from Bayer, personal fees and non-financial support from BMS, personal fees from Pharmamar, personal fees from Leo, outside the submitted work. The rest of the authors have nothing to disclose. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Leemans C.R., Snijders P.J.F., Brakenhoff R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer. 2018;18:269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 3.Sacco A.G., Cohen E.E. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J. Clin. Oncol. 2015;33:3305–3313. doi: 10.1200/JCO.2015.62.0963. [DOI] [PubMed] [Google Scholar]
- 4.Feller L., Altini M., Lemmer J. Inflammation in the context of oral cancer. Oral Oncol. 2013;49:887–892. doi: 10.1016/j.oraloncology.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 6.Darwiche N. Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. Am. J. Cancer Res. 2020;10:1954–1978. [PMC free article] [PubMed] [Google Scholar]
- 7.Castilho R.M., Squarize C.H., Almeida L.O. Epigenetic modifications and head and neck cancer: Implications for tumor progression and resistance to therapy. Int. J. Mol. Sci. 2017;18:1506. doi: 10.3390/ijms18071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garinis G.A., Patrinos G.P., Spanakis N.E., Menounos P.G. DNA hypermethylation: When tumour suppressor genes go silent. Hum. Genet. 2002;111:115–127. doi: 10.1007/s00439-002-0783-6. [DOI] [PubMed] [Google Scholar]
- 9.Misawa K., Imai A., Matsui H., Kanai A., Misawa Y., Mochizuki D., Mima M., Yamada S., Kurokawa T., Nakagawa T., et al. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene. 2020;39:4741–4755. doi: 10.1038/s41388-020-1327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liyanage C., Wathupola A., Muraleetharan S., Perera K., Punyadeera C., Udagama P. Promoter hypermethylation of tumor-suppressor genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in salivary DNA as a quadruple biomarker panel for early detection of oral and oropharyngeal cancers. Biomolecules. 2019;9:148. doi: 10.3390/biom9040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim Y., Wan Y., Vagenas D., Ovchinnikov D.A., Perry C.F.L., Davis M.J., Punyadeera C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer. 2016;16:749. doi: 10.1186/s12885-016-2785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrero-Preston R., Soudry E., Acero J., Orera M., Moreno-López L., Macía-Colón G., Jaffe A., Berdasco M., Ili-Gangas C., Brebi-Mieville P., et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev. Res. 2011;4:1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 14.Borchiellini M., Ummarino S., Di Ruscio A. The bright and dark side of DNA methylation: A matter of balance. Cells. 2019;8:1243. doi: 10.3390/cells8101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulis M., Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M., Corn P.G., Baylin S.B., Herman J.G. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 17.O’Reilly E., Tuzova A.V., Walsh A.L., Russell N.M., O’Brien O., Kelly S., Dhomhnallain O.N., DeBarra L., Dale C.M., Brugman R., et al. epiCaPture: A urine DNA methylation test for early detection of aggressive prostate cancer. JCO Precis. Oncol. 2019:1–18. doi: 10.1200/PO.18.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang W., Zhao Y., Huang W., Gao Y., Xu W., Tao J., Yang M., Li L., Ping W., Shen H., et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA) Theranostics. 2019;9:2056–2070. doi: 10.7150/thno.28119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith A.K., Kilaru V., Klengel T., Mercer K.B., Bradley B., Conneely K.N., Ressler K.J., Binder E.B. DNA extracted from saliva for methylation studies of psychiatric traits: Evidence tissue specificity and relatedness to brain. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2015;168:36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hearn N.L., Coleman A.S., Ho V., Chiu C.L., Lind J.M. Comparing DNA methylation profiles in saliva and intestinal mucosa. BMC Genomics. 2019;20:163. doi: 10.1186/s12864-019-5553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H.C., Wang Q., Chung W.K., Andrulis I.L., Daly M.B., John E.M., Keegan T.H., Knight J., Bradbury A.R., Kappil M.A., et al. Correlation of DNA methylation levels in blood and saliva DNA in young girls of the LEGACY girls study. Epigenetics. 2014;9:929–933. doi: 10.4161/epi.28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson T.M., Sharfi D., Lee M., Yrigollen C.M., Naumova O.Y., Grigorenko E.L. Comparison of whole-genome DNA methylation patterns in whole blood, saliva, and lymphoblastoid cell lines. Behav. Genet. 2013;43:168–176. doi: 10.1007/s10519-012-9579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langie S.A.S., Szarc Vel Szic K., Declerck K., Traen S., Koppen G., Van Camp G., Schoeters G., Vanden Berghe W., De Boever P. Whole-genome saliva and blood DNA methylation profiling in individuals with a respiratory allergy. PLoS ONE. 2016;11:e0151109. doi: 10.1371/journal.pone.0151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho A.L., Jeronimo C., Kim M.M., Henrique R., Zhang Z., Hoque M.O., Chang S., Brait M., Nayak C.S., Jiang W.W., et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin. Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 25.McInnes M.D.F., Moher D., Thombs B.D., McGrath T.A., Bossuyt P.M., the PRISMA-DTA Group. Clifford T., Cohen J.F., Deeks J.J., Gatsonis C., et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA. 2018;319:388–396. doi: 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 26.Whiting P.F., Rutjes A.W.S., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M.G., Sterne J.A.C., Bossuyt P.M.M. Quadas-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 27.Zamora J., Abraira V., Muriel A., Khan K., Coomarasamy A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellmich M., Lehmacher W. A ruler for interpreting diagnostic test results. Methods Inf. Med. 2005;44:124–126. [PubMed] [Google Scholar]
- 29.Deeks J.J., Macaskill P., Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Rosas S.L., Koch W., da Costa Carvalho M.G., Wu L., Califano J., Westra W., Jen J., Sidransky D. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and Death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–942. [PubMed] [Google Scholar]
- 31.Kusumoto T., Hamada T., Yamada N., Nagata S., Kanmura Y., Houjou I., Kamikawa Y., Yonezawa S., Sugihara K. Comprehensive Epigenetic Analysis Using Oral Rinse Samples: A Pilot Study. J. Oral Maxillofac. Surg. 2012;70:1486–1494. doi: 10.1016/j.joms.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 32.Gaykalova D.A., Vatapalli R., Wei Y., Tsai H.L., Wang H., Zhang C., Hennessey P.T., Guo T., Tan M., Li R., et al. Outlier Analysis Defines Zinc Finger Gene Family DNA Methylation in Tumors and Saliva of Head and Neck Cancer Patients. PLoS ONE. 2015;10:e0142148. doi: 10.1371/journal.pone.0142148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng S.J., Chang C.F., Ko H.H., Lee J.J., Chen H.M., Wang H.J., Lin H.S., Chiang C.P. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck. 2018;40:355–368. doi: 10.1002/hed.24958. [DOI] [PubMed] [Google Scholar]
- 34.Srisuttee R., Arayataweegool A., Mahattanasakul P., Tangjaturonrasme N., Kerekhanjanarong V., Keelawat S., Mutirangura A., Kitkumthorn N. Evaluation of NID2 promoter methylation for screening of Oral squamous cell carcinoma. BMC Cancer. 2020;20:218. doi: 10.1186/s12885-020-6692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagata S., Hamada T., Yamada N., Yokoyama S., Kitamoto S., Kanmura Y., Nomura M., Kamikawa Y., Yonezawa S., Sugihara K. Aberrant DNA methylation of tumor-related genes in oral rinse. Cancer. 2012;118:4298–4308. doi: 10.1002/cncr.27417. [DOI] [PubMed] [Google Scholar]
- 36.Franzmann E.J., Reategui E.P., Pedroso F., Pernas F.G., Karakullukcu B.M., Carraway K.L., Hamilton K., Singal R., Goodwin W.J. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:1348–1355. doi: 10.1158/1055-9965.EPI-06-0011. [DOI] [PubMed] [Google Scholar]
- 37.Righini C.A., de Fraipont F., Timsit J.F., Faure C., Brambilla E., Reyt E., Favrot M.C. Tumor-specific methylation in saliva: A promising biomarker for early detection of head and neck cancer recurrence. Clin. Cancer Res. 2007;13:1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 38.Ovchinnikov D.A., Cooper M.A., Pandit P., Coman W.B., Cooper-White J.J., Keith P., Wolvetang E.J., Slowey P.D., Punyadeera C. Tumor-suppressor gene promoter hypermethylation in saliva of head and neck cancer patients. Transl. Oncol. 2012;5:321–326. doi: 10.1593/tlo.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ovchinnikov D.A., Wan Y., Coman W.B., Pandit P., Cooper-White J.J., Herman J.G., Punyadeera C. DNA methylation at the novel CpG sites in the promoter of MED15/PCQAP gene as a biomarker for head and neck cancers. Biomark. Insights. 2014;9:53–60. doi: 10.4137/BMI.S16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Pérez L., Isaza-Guzmán D., Arango-Pérez E., Tobón-Arroyave S. Analysis of salivary detection of P16INK4A and RASSF1A promoter gene methylation and its association with oral squamous cell carcinoma in a Colombian population. J. Clin. Exp. Dent. 2020:e452–e460. doi: 10.4317/jced.56647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puttipanyalears C., Arayataweegool A., Chalertpet K., Rattanachayoto P., Mahattanasakul P., Tangjaturonsasme N., Kerekhanjanarong V., Mutirangura A., Kitkumthorn N. TRH site-specific methylation in oral and oropharyngeal squamous cell carcinoma. BMC Cancer. 2018;18:786. doi: 10.1186/s12885-018-4706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferlazzo N., Currò M., Zinellu A., Caccamo D., Isola G., Ventura V., Carru C., Matarese G., Ientile R. Influence of MTHFR genetic background on p16 and MGMT methylation in oral squamous cell cancer. Int. J. Mol. Sci. 2017;18:724. doi: 10.3390/ijms18040724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen S., Saito Y., Ren S., Liu C., Guo T., Qualliotine J., Khan Z., Sadat S., Califano J.A. Targeting viral DNA and promoter hypermethylation in salivary rinses for recurrent HPV-positive oropharyngeal cancer. Otolaryngol. Neck Surg. 2020;162:512–519. doi: 10.1177/0194599820903031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rettori M.M., de Carvalho A.C., Bomfim Longo A.L., de Oliveira C.Z., Kowalski L.P., Carvalho A.L., Vettore A.L. Prognostic significance of TIMP3 hypermethylation in post-treatment salivary rinse from head and neck squamous cell carcinoma patients. Carcinogenesis. 2013;34:20–27. doi: 10.1093/carcin/bgs311. [DOI] [PubMed] [Google Scholar]
- 45.Guerra E.N.S., Acevedo A.C., Leite A.F., Gozal D., Chardin H., De Luca Canto G. Diagnostic capability of salivary biomarkers in the assessment of head and neck cancer: A systematic review and meta-analysis. Oral Oncol. 2015;51:805–818. doi: 10.1016/j.oraloncology.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Rapado-González Ó., Martínez-Reglero C., Salgado-Barreira Á., Takkouche B., López-López R., Suárez-Cunqueiro M.M., Muinelo-Romay L. Salivary biomarkers for cancer diagnosis: A meta-analysis. Ann. Med. 2020;52:131–144. doi: 10.1080/07853890.2020.1730431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshizawa J.M., Schafer C.A., Schafer J.J., Farrell J.J., Paster B.J., Wong D.T.W. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013;26:781–791. doi: 10.1128/CMR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.