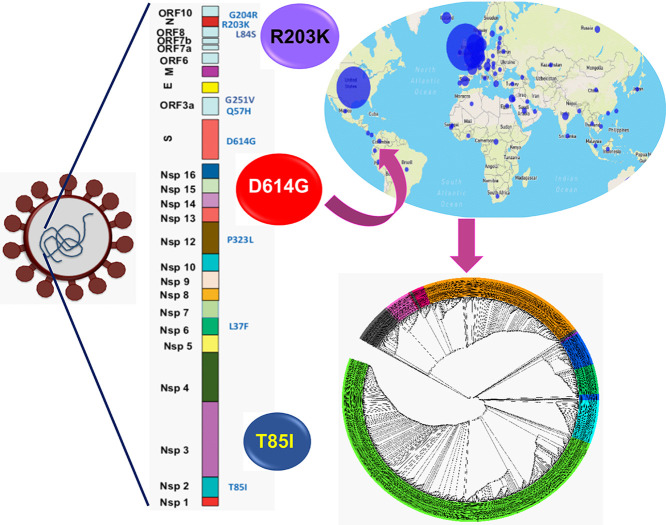
Abstract
SARS-CoV-2 is currently causing major havoc worldwide with its efficient transmission and propagation. To track the emergence as well as the persistence of mutations during the early stage of the pandemic, a comparative analysis of SARS-CoV-2 whole proteome sequences has been performed by considering manually curated 31,389 whole genome sequences from 84 countries. Among the 7 highly recurring (percentage frequency≥10%) mutations (Nsp2:T85I, Nsp6:L37F, Nsp12:P323L, Spike:D614G, ORF3a:Q57H, N protein:R203K and N protein:G204R), N protein:R203K and N protein: G204R are co-occurring (dependent) mutations. Nsp12:P323L and Spike:D614G often appear simultaneously. The highly recurring Spike:D614G, Nsp12:P323L and Nsp6:L37F as well as moderately recurring (percentage frequency between ≥1 and <10%) ORF3a:G251V and ORF8:L84S mutations have led to4 major clades in addition to a clade that lacks high recurring mutations. Further, the occurrence of ORF3a:Q57H&Nsp2:T85I, ORF3a:Q57H and N protein:R203K&G204R along with Nsp12:P323L&Spike:D614G has led to 3 additional sub-clades. Similarly, occurrence of Nsp6:L37F and ORF3a:G251V together has led to the emergence of a sub-clade. Nonetheless, ORF8:L84S does not occur along with ORF3a:G251V or Nsp6:L37F. Intriguingly, ORF3a:G251V and ORF8:L84S are found to occur independent of Nsp12:P323L and Spike:D614G mutations. These clades have evolved during the early stage of the pandemic and have disseminated across several countries. Further, Nsp10 is found to be highly resistant to mutations, thus, it can be exploited for drug/vaccine development and the corresponding gene sequence can be used for the diagnosis. Concisely, the study reports the SARS-CoV-2 antigens diversity across the globe during the early stage of the pandemic and facilitates the understanding of viral evolution.
Keywords: SARS-CoV-2 viromics, Highly recurring mutations, Moderately recurring mutations, Phyloproteomics, Proteome analysis, Mutational susceptibility
Graphical abstract
1. Introduction
The severe acute respiratory syndrome virus 2 (SARS-CoV-2) pandemic outbreak has so far claimed several millions of lives. Treating the SARS-CoV-2 pandemic is the biggest challenge being faced by the world today. Although SARS-CoV-2 is expected to have a slow mutation rate due to its sophisticated proof reading mechanism (Pachetti et al., 2020), it can incorporate changes in the genome (Catanzaro et al., 2020; Garcia, 2020; Lucas et al., 2001) to escape from the host immune defences. A comparative analysis of 31,389 clinically relevant SARS-CoV-2 whole genome sequences obtained from 84 countries are exploited here to accelerate the identification of universal vaccine candidates and drug targets. A country wise local proteome repository has been created and subjected to multiple sequence alignment to analyze the position specific recurrence of each amino acid variation, henceforth referred to as percentage frequency (see Methods), in the SARS-CoV-2 whole proteome.
The SARS-CoV-2 genome has 10 open reading frames (ORFs) flanked by 5′UTR and 3′UTR (Kim et al., 2020; Wu et al., 2020). Upon entering into the host by either direct membrane fusion or receptor-mediated endocytosis, the ORF1ab is translated into two large polyproteins (PP1a and PP1ab) with the utilization of the host translational machinery. PP1a consists of non-structural proteins (Nsps) 1 to 11, whereas, the PP1ab comprises of Nsps 1 to 16 (except Nsp11) and are translated from the genomic RNA. The matured Nsp1 to Nsp16 are produced with the help of two viral encoded cysteine proteases, namely, the papain-like protease (PLpro) (proteolyzes Nsps 1–3) and the main protease (Mpro or 3CLpro) (proteolyzes Nsps 4–16). The Nsps 7–16 act as the replication machinery that enables the synthesis of the viral genome. The subgenomic RNAs from S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8, N and ORF10 coding regions are involved in the translation of virion structural and accessory proteins. While the proteins synthesized by ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10 are designated as accessory proteins, the proteins from S, E, M and N are designated as structural proteins (Kim et al., 2020). ORF10 is being utilized in the diagnostics to differentiate SARS-CoV from SARS-CoV-2 (Khailany et al., 2020; Takahiko Koyama DPaLP, 2020).
SARS-CoV-2 whole proteome analysis encompassing the above proteins indicates that 7, 28 and 2081 unique mutations exhibit high (≥10%), moderate (≥1–<10%) and low (≥0.01–<1%) percentage frequency respectively during the early stage of the pandemic (December 2019-May 17, 2020). 5 major clades have evolved encompassing these mutations. Among the mutations, Spike:D614G and Nsp12:P323L are occurring in nearly 75% of the sequences soon after their first appearance in January 2020 (GISAID ID: EPI_ISL_422425) indicating their evolutionary advantage. Interestingly, R203K and G204R present in nucleoprotein are found to be co-occurring (dependent) mutations. The study also reports the presence of several deletions and co-occurring mutations that recur at a moderate percentage frequency. As the envelope protein, Nsp4, Nsp9 and Nsp10 are less heterogeneous compared with the rest of the proteins, they can be good vaccine/drug targets. Thus, the results presented here would help in identifying the potential therapeutic targets. It may further have an impact on correlating the mutations with the viral evolution and virulence.
2. Methods
The whole genome sequences (WGS) obtained from the clinical samples collected from the diverse geographical background were downloaded from the NCBI (Benson et al., 2011) (3,730 sequences) (TableSD1A.xlsx) and GISAID (Shu and McCauley, 2017) (27,659 sequences) (TableSD1B.xlsx) (deposited on or before May 17, 2020). 31,389 sequences were considered to analyze SARS-CoV-2 proteomic diversity (in the above-mentioned proteins) across 84 countries. Subsequently, the dataset was manually curated to create the local database. The viral genome sequences with more than 15,000 bases (half of the entire viral genome) were alone considered in the creation of the database. Note that as there is no link between the WGS deposited in the NCBI and GISAID, there is a possibility of sequence(s) duplication in the local database. The genomic sequences were then translated into individual proteins (Kim et al., 2020; Wu et al., 2020) (as mentioned in the Introduction) by considering the first published SARS-CoV-2 proteome as the reference (herein onwards, reference sequence) using an in-house script. Note that if any translated region contained an undefined amino acid (due to the presence of one or more undefined nucleotides (“N”) in the coding region), the region was not considered for the analysis. However, the remaining coding regions were considered for the analysis (Table SD2.xlsx).
The country wise mutational analysis specific to each protein was analyzed through multiple sequence alignment (MSA) using CLUSTAL OMEGA (Sievers and Higgins, 2014). It is worth mentioning that the sequences were available for the following countries: England, Wales, Scotland, France, Luxembourg, Netherlands, Spain, Belgium, Germany, Italy, Czech, Austria, Ireland, Denmark, Latvia, Greece, Hungary, Sweden, Switzerland, Poland, Turkey, China, Japan, Malaysia, Russia, Saudi Arabia, Singapore, South Korea, Taiwan, Hong Kong, Thailand, Vietnam, Georgia, India, Iran, Israel, USA, Canada, South Africa, Congo, Australia, Argentina, Brazil, Chile, Uruguay, Colombia, Iceland, Portugal, Norway, Finland, Estonia, Slovenia, Lithuania, Slovakia, Belarus, Kuwait, Nepal, Pakistan, Mexico, Ghana, Algeria, Senegal, New Zealand, Peru, Ecuador, Panama, Cambodia, Tunisia, Serbia, Romania, Croatia, Philippines, Indonesia, UAE, Jordan, Qatar, Bangladesh, Brunei, Kazakhstan, Sri Lanka, Costa Rica, Guam, Gambia and Egypt.
Prior to the MSA, the 26 proteins (non-structural (Nsps 1 to 16), structural (Spike, Envelope, Membrane and Nucleocapsid) and accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10)) (Kim et al., 2020) encoded by the genomic and subgenomic RNAs of SARS-CoV-2 were segregated individually and stored country wise in the database.
2.1. Calculation of the percentage frequency
The country wise MSA alignments were manually verified for each of the 26 proteins and subjected to position wise amino acid variation analysis using in-house scripts. When an amino acid at a particular position changes to a different amino acid, the number of such changes was counted to calculate the frequency of mutation in each country. A mutation was considered as recurrent (viz., the mutation that does not occur by chance) only if the summation of the country wise percentage frequency (PF) (Eq. (1)) of that particular mutation is (occurs at least 3 times) 0.01%.
| (1) |
The recurrent mutations were further classified into three categories: highly recurring (HR), moderately recurring (MR) and low recurring(LR). A mutation was considered as highly (dominant) or moderately recurring when it occurs at the percentage frequency ≥10% or ≥1 and <10% respectively. On the other hand, a mutation was considered as a low recurrent when it occurs at the percentage frequency between 0.01 and <1% (frequency in the range of 3-314).
Further, the mutation susceptibility of a protein in terms of number of recurrences and number of positions undergoing mutations are quantified using the following equations:
| (2) |
| (3) |
2.2. Construction of phyloproteomic tree
Initially, the whole genome sequences that do not have undefined nucleotides (‘N') were translated to 11,198 single sequences with each containing 26 translated proteins (5’UTR, 3’UTR, ORF1ab stemloop 1, ORF1ab stemloop 2, ORF10 stemloop 1 and ORF10 stemloop 2 were excluded). The phyloproteomic trees corresponding to the countries that occupy top 10 positions in terms of number of SARS-CoV-2 sequences were constructed: USA (number of sequences = 1372), Australia (219), Belgium (142), China (135), Denmark (156), France (180), Iceland (118), India (107), Netherlands (173) and UK (981). The sequences were subsequently subjected to alignment using MAFFT (Katoh and Standley, 2013) and the phyloproteomic tree was constructed using maximum likelihood method in IQ-TREE software (Nguyen et al., 2015). Itol tool (Letunic and Bork, 2019) was used to visualize and analyze the phylogram.
3. Results
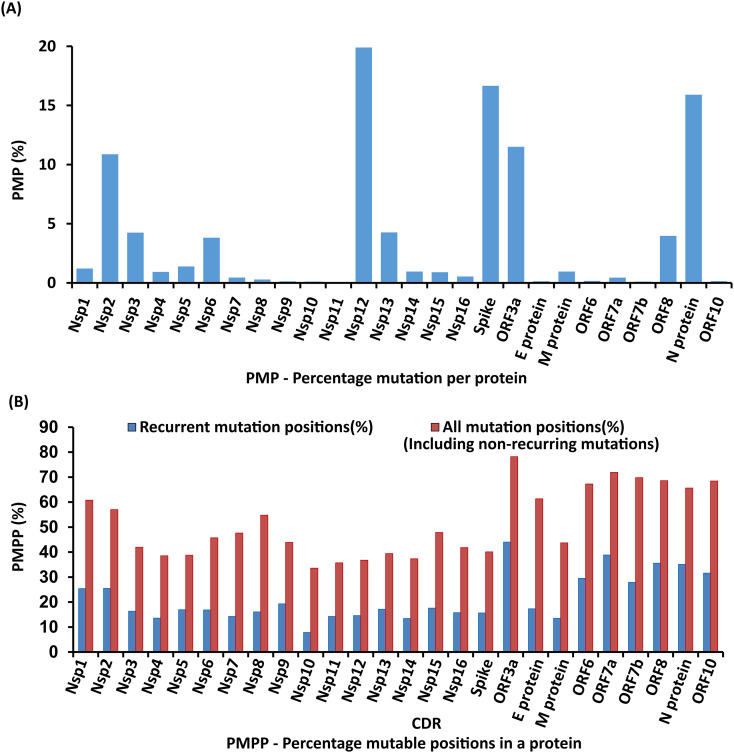
31,389 sequences have been considered to analyze SARS-CoV-2 proteome diversity across 84 countries. Functions of all the twenty-six SARS-CoV-2 proteins are provided in Table S1. Further, the PMP of the 26 proteins falls in the following order: Nsp12 (19.900%) > Spike (16.657%) > N protein (15.911%) > ORF3a (11.505%) > Nsp2 (10.867%) > Nsp13 (4.265%) > Nsp3 (4.242%) > ORF8 (3.969%) > Nsp6 (3.816%) > Nsp5 (1.387%) > Nsp1 (1.210%) > Nsp14 (0.963%) > M protein (0.961%) > Nsp4 (0.930%) > Nsp15 (0.895%) > Nsp16 (0.543%) > Nsp7 (0.452%) > ORF7a (0.437%) > Nsp8 (0.289%) > ORF6 (0.169%) > ORF10 (0.148%) > E protein (0.136%) > Nsp9 (0.124%) > ORF7b (0.108%) > Nsp10 (0.106%) > Nsp11 (0.008%). Similarly, PMPP falls in the following order: ORF3a (44%) > ORF7a (38.842%) > ORF8 (35.537%) > N protein (35.083%) > ORF10 (31.578%) > ORF6 (29.508%) > ORF7b (27.906%) > Nsp2 (25.508%) > Nsp1 (25.414%) > Nsp9 (19.298%) > Nsp15 (17.579%) > E protein (17.333%) > Nsp13 (17.109%) > Nsp5 (16.938%) > Nsp6 (16.838%) > Nsp3 (16.341%) > Nsp8 (16.08%) > Nsp16 (15.719%) > Spike (15.632%) > Nsp12 (14.576%) > Nsp7 (14.285%)≥ Nsp11 (14.285%) > Nsp4 (13.572%) > M protein (13.513%) > Nsp14 (13.446%) > Nsp10 (7.857%). Taking into account the mutation recurrence frequency as well as the number of positions exhibiting variations in each protein, it can be suggested that ORF3a and Nsp10 are the proteins that have the highest and lowest susceptibility to the mutation respectively (Fig. 1 ).
Fig. 1.
Bar diagram illustrating the mutation susceptibility (%) of 26 proteins (See Supplementary Table S1 for functions). (A) Considering the mutation count from individual proteins, the mutation susceptibility (Eq. (2)) falls in the following order: Nsp12 > Spike >N protein > ORF3a > Nsp2 > Nsp13 > Nsp3 > ORF8 > Nsp6 > Nsp5 > Nsp1 > Nsp14 > M protein > Nsp4 > Nsp15 > Nsp16 > Nsp7 > ORF7a > Nsp8 > ORF6 > ORF10 > E protein > Nsp9 > ORF7b > Nsp10 > Nsp11. (B) When the mutation position count is considered, the mutation susceptibility (Eq. (3)) falls in the following order: ORF3a > ORF7a > ORF8 > N protein> ORF10 > ORF6 > ORF7b > Nsp2 > Nsp1 > Nsp9 > Nsp15 > E protein > Nsp13 > Nsp5 > Nsp6 > Nsp3 > Nsp8 > Nsp16 > Spike > Nsp12 > Nsp7 > Nsp11 > Nsp4 > M protein > Nsp14 > Nsp10.
The Tables SD3-SD28.xlsx provide country wise percentage frequency of SARS-CoV-2 amino acid mutations.
3.1. Mutations with high recurrence
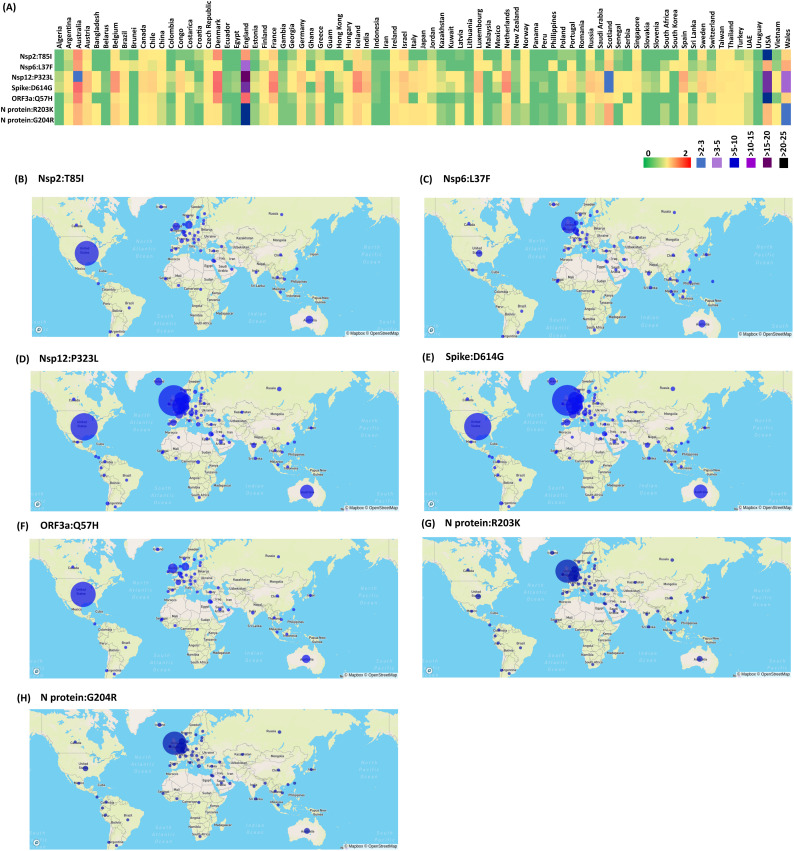
In Nsp2, T85I mutation recurs at the percentage frequency of 20.43% in 47 countries (Fig. 2A & B) indicating the pre-lockdown global spread of I85 (54% occupancy among the mutations in Nsp2). This mutation is found at the highest recurring level in USA with nearly 56% of its sequences having this mutation. Similarly, L37F in Nsp6 has a percentage frequency > 11.98% (Fig. 2A). 59 countries possess these mutations (Fig. 2C). Further, P323L in Nsp12 that is located in the proximity of the Nsp8-Nsp12 binding site (Fig. S1, PDB ID: 6YYT) is found in 71 countries (except Brunei, Croatia, Iran, Malaysia, South Korea, Nepal, Philippines, Turkey, Pakistan, Tunisia, Guam and Uruguay) out of 84 countries considered here with a high recurrence (percentage frequency of 69.47%) (Fig. 2A & D). Prevalence of this mutation is also reported earlier (Pachetti et al., 2020; Chand et al., 2020; Ugurel et al., 2020; Zhao et al., 2020).
Fig. 2.
Emergence and dissemination of the mutations that highly recur during pre-lockdown period: A) Heat map representing the country wise percentage frequency of 7 highly recurring mutations. B-H) Global distribution of (represented by percentage frequency) highly recurring mutations: B) Nsp2:T85I, C) Nsp6:L37F, D) Nsp12:P323L, E) Spike:D614G, F) ORF3a:Q57H, G) N protein:R203K and H) N protein:G204R.
In spike protein, D614G (present in the S1 subunit flanked by the receptor binding site) (Fig. 2A & E) mutation has become the most dominant mutation as reported earlier (Daniloski et al., 2020; Klumpp-Thomas et al., 2020; Korber et al., 2020; Zhang et al., 2020a) with the percentage frequency of 69.20%. Totally, 70 countries (except 14 countries, namely, Brunei, Panama, Cambodia, Tunisia, Malaysia, Nepal, South Korea, Philippines, Iran, Pakistan, Ghana, Uruguay, Qatar and Guam) have this mutation (Fig. 2A & E). In addition to above, Q57H in ORF3a is found to be the highly recurring mutation with the percentage frequency of 24.43% in 51 countries (Fig. 2A & F). N protein co-mutations (dependent), R203K and G204R are also found to occur with a high percentage frequency (~23.6%). Both of them co-exist in 63 countries and occur at a high percentage frequency of 12.5% in England followed by Wales (3.34%) and Scotland (1.01%) (Fig. 2A, G & H).
Thus, Nsp2:T85I, Nsp6:L37F, Nsp12:P323L, Spike:D614G, ORF3a:Q57H, N protein:R203K and G204R have occurred at a high recurrence during the early stage of the pandemic.
3.2. Evolution of new lineages with moderate and low recurring mutations
3.2.1. Protein encoded by ORF1ab
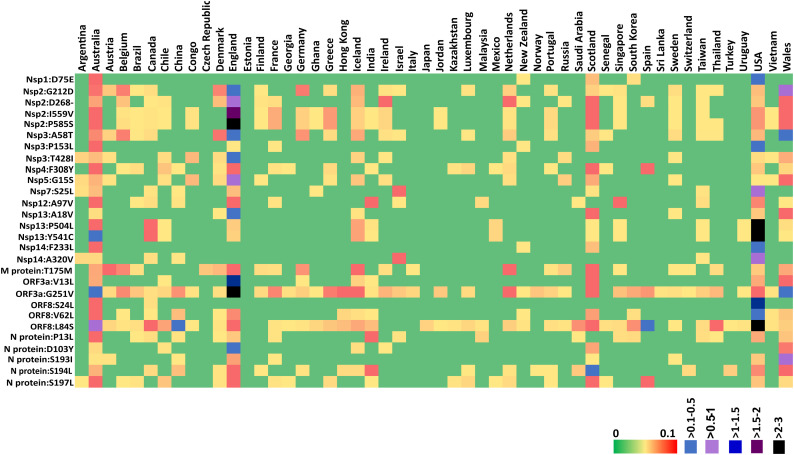
In addition to the abovementioned 7 highly recurring mutations, 28 moderately recurring mutations have been observed during the early stage of the pandemic. For instance, the exchange between the negatively charged amino acids D75 and E75 in Nsp1 occurs at a moderate percentage frequency of ~1% (Fig. 3 ). Further, co-occurring (dependent) K141, S142 and F143 deletions in Nsp1 are found to recur above 0.35% PF. Strikingly, G82-V86 is yet another stretch in Nsp1 that is prone to undergo mutations. For instance, the deletion of M85 (~0.34%) is found with the highest occurrence in England. Similar to K141, S142 and F143 deletions, G82 deletion is also found at the low recurrence (PF = 0.1%). Independent deletion of V84 and V86 is also found in certain countries (TableSD3.xlsx).
Fig. 3.
Heat map representing the country wise percentage frequency of 28 moderately recurring (MR) mutations. Note that, 52 countries have at least one of the MR mutations (percentage frequency between ≥1 and <10%). Among the 52 countries, Australia is found to have 28 out of the 28 MR mutations, followed by USA (27), England (24), Scotland (23) and Wales (22).
Similarly, P585S (~4.1%), I559V (~3.8%), deletion of D268 (~2.87%) and G212D (~2.64%) in Nsp2 are found at a moderate percentage frequency. A total of 26 countries (England, Wales, Scotland, France, Belgium, Germany, Ireland, Greece, Sweden, Singapore, Taiwan, Thailand, Vietnam, India, Jordan, USA, Australia, Chile, Iceland, Netherlands, Portugal, Finland, Canada, Ghana, Congo and Brazil) undergo P585 to S585 mutation (Fig. 3) indicating the spread of the SARS-CoV-2 variant having this mutation during pre-lockdown period. Very interestingly, the same countries/sequences undergo I559 to V559 mutation with a nearly similar percentage frequency. These two mutations occur at the highest percentage frequency in England (~2.9–3%) followed by Wales (0.3%) and Australia (0.2–0.3%) (Fig. 3).
Nsp3 mutations that are found with a moderate percentage frequency are: A58T which is present in the UBL1 domain of Nsp3 is the moderately recurring mutation with the percentage frequency of 1.8%, P153L (occurs at the percentage frequency in the range of 0.7% and 0.26% respectively in USA and Australia) and T428I (England possesses the highest PF of 0.86%). Notably, these mutations do not occur in the PL-pro domain (PDB ID: 6WUU), which makes this domain a potential drug target in line with the earlier attempts (Zhou et al., 2020) (Fig. S1, TableSD5.xlsx).
In Nsp4, the exchange between the aromatic amino acids F308 and Y308 is the highest among the (Nsp4) mutations, which occurs with a moderate percentage frequency (1.2%). Nsp5 or main protease is an attractive target to treat SARS-CoV-2 as it plays a major role in producing the functional forms of Nsp4 to Nsp16 (Zhang et al., 2020b). Although the catalytic site and N-terminal finger of Nsp5 are conserved, G15S located in the chymotrypsin-like domain is recurring at a moderate PF of 1.6% (Fig. 3). Luckily, the structure of Mpro is available (PDB ID: 6LU7) which indicates that this mutation does not occur either at the catalytic site or at the N-terminal finger (important for the recognition of the target protein) or at the dimeric interface indicating the conservation of its functional mechanism (Fig. S1). Since this mutation occurs on the surface exposed region of Nsp5, it may have an influence on the interaction with other virion protein(s) as well as the host protein(s).
3.3. Mutations in the replication machinery
Nsp7 to Nsp16 are the replicase polyproteins and crucial for the protein replication. S25L is the only recurring mutation in Nsp7 (Fig. 3 and TableSD9.xlsx). S25L of Nsp7 occurs at a moderate percentage frequency of 1.33%. Notably, this mutation occurs at the turn region of the alpha helical bundle of Nsp7, which interacts with Nsp8 (Fig. S1). Next, A97V of Nsp12 is a moderately recurring mutation (1.2%) (Fig. 3). In Nsp13, the exchange between the aliphatic A18 and V18 (PF = 1.1%), P504L (PF = 5.6%) and Y541C (PF = 5.8%) are found with the moderate recurrence (Fig. 3). F233L (~1.26%) and A320V of Nsp14 (~1.26%) are moderately recurring mutation (Fig. 3) which are found in the exoribonuclease (ExoN) and (guanine-N7)-methyl transferase (N7-Mtase) domains of Nsp14 respectively (Ma et al., 2015) (as seen in the Nsp10-Nsp14 SWISS-MODEL complex structure based on SARS-CoV which has the sequence identity more than 90% (PDB ID: 5C8S)) (Fig. S1). Interestingly, Nsp8, Nsp9, Nsp10, Nsp11, Nsp15 and Nsp16 of the replication machinery do not have moderately or highly recurring mutations.
3.4. Moderately and low recurring mutations in the proteins encoded by SARS-CoV-2 subgenomic mRNA
G251V of ORF3a protein is the moderately recurring mutation with the second highest percentage frequency (~9%) among the moderately recurring mutations and is highly occurring in England (PF = 5.18%) (Fig. 3).V13L of ORF3a is a moderately recurring mutation (PF = 2.6%) with England holding the top position followed by Wales and Scotland (Fig. 3). Additionally, H93Y and G196V in ORF3a (PF = 0.76%) are found to be low recurring mutations (Table S2). In M protein, T175M is a moderately recurring mutation with the percentage frequency of ~1.6%. The Envelope protein, ORF6, ORF7a, ORF7b and ORF10 do not have any mutations that recur at a moderate PF. L84S in ORF8 is the moderately recurring mutation with the upmost percentage frequency (~9.8%) among the moderately recurring mutations and is highly recurring in USA (PF = 5.93%). Additionally, S24L (PF = 2.2%) and V62L (PF = 1.2%) are the moderately recurring mutations in ORF8 (Fig. 3).
Notably, in N protein, S180, S183, S187, S188, S190, S193, S194, S197 and S202 present in a serine rich stretch (SR rich linker domain, residue number 180–202) change primarily to aliphatic/aromatic amino acids I, Y, L, L/P, I, I, L, L and N respectively. Among these, S193I, S194L and S197L recur with the percentage frequency in the range of 1 to 1.6% (moderate recurrence) (Fig. 3), whereas, S188L and S190I occur with the percentage frequency of 0.45% and 0.23% respectively (TableSD27.xlsx). Indeed, certain countries have percentage frequency more than 1.5% for these mutations: S193I (the highest in Wales followed by England), S194I (the highest in Scotland followed by England, India and Wales) and S197L (the highest Australia followed by Scotland, England and Spain). Apart from these, P13L (N-terminal disordered region, the highest in Australia) (PF = 1.03%) and D103Y (PF = 1.05%) also occur at a moderate percentage frequency (Fig. 3). Notably, 11 recurring mutations (including R203K and G204R co-mutations) occur in the S180-R209 stretch of SR rich linker region, for which, the structural information is unknown (Fig. S1).
Some notable substitutions, co-occurring mutations, deletions and insertions which occur with a low to moderate recurrence and may have a significant influence on the viral pathogenic mechanism are: co-deletion (dependent) of K141, S142 and F143, co-deletion (dependent) of G82 and H83, D75E, co-occurring (dependent) S135N, Y136 deletion and M85 deletion in Nsp1, D268 deletion, G212D, co-occurring (dependent) I559V and P585S, F10L, V198I, P91S, T166I, H237R, T371I, S211F and G339S in Nsp2, A58T, P153L and T428I in Nsp3, F308Y, T295I (0.3%) and M33I in Nsp4, G15S and K90R in Nsp5, S25L and S26F in Nsp7, A97V and A449V in Nsp12, A18V and co-mutation P504L & Y541C in Nsp13, F233L and A320V in Nsp14, V13L in ORF3a, S24L and A62L in ORF8, D936Y in spike and P13L, D103Y, S193I, S194L and S197L with a percentage frequency above 1% in nucleoprotein. Very interestingly, a deletion in ORF8 region is found in Singapore and Taiwan. In these sequences, the ORF8 encodes only 9 amino acid peptide instead of the 121 long amino acid protein. This mutation is suggested to be associated with SARS-CoV-2 infection found in Taiwan and the Middle East countries (Gong et al., 2020).
The Table S2 contains the list of above discussed low, moderate and high recurrent mutations found in the structural and accessory proteins of SARS-CoV-2.
3.5. Evolution of five major clades of SARS-CoV-2
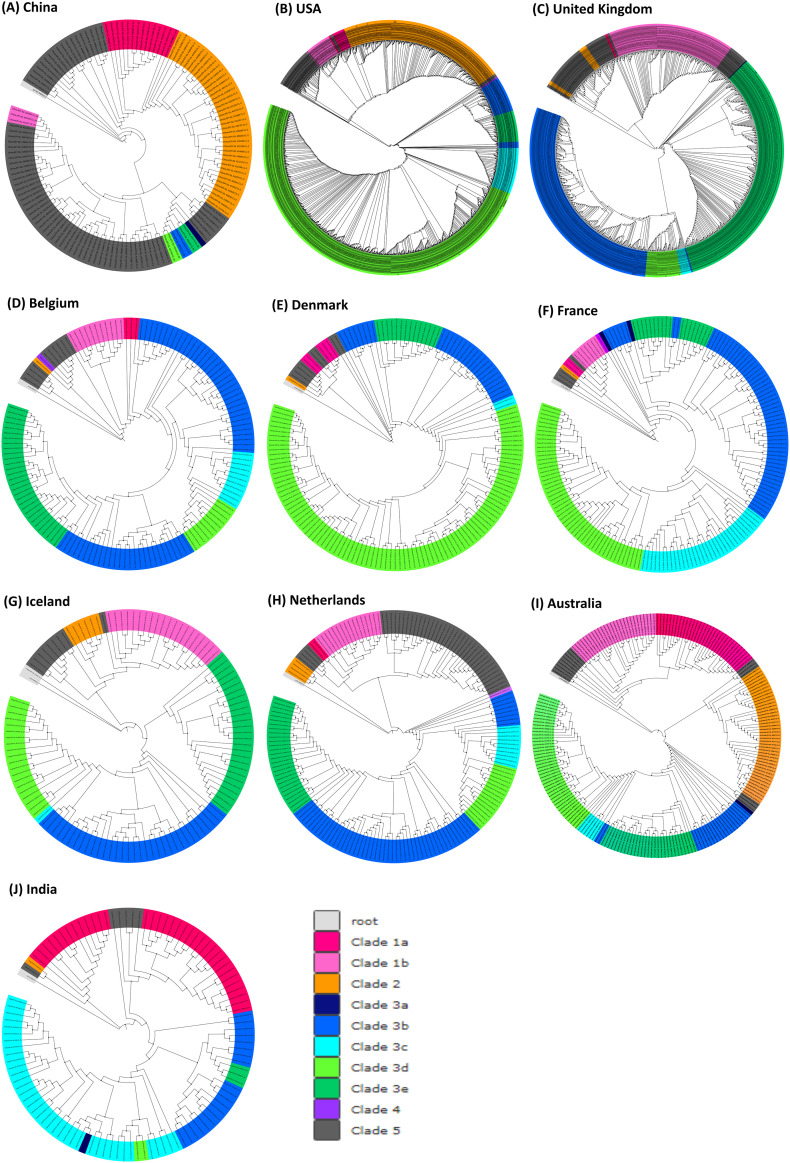
Individual phyloprotemic trees generated here for top 10 countries (based on the sequences deposited) by considering the reference proteome sequence (Genbank ID: NC_045512.2) as the root and the non-redundant proteome sequences which vary at least in one position of the SARS-CoV-2 proteome with respect to the reference sequence indicate the region wise evolution of 5 major clades (Fig. 4 (A-J)) during the early stage of the pandemic. 3 of the clades are due to the high recurrent mutations [Nsp6:L37F (Clade 1), Spike:D614G (Clade 3) and Nsp12:P323L (Clade 4)] and one of the clade is due to the moderately recurring mutation [ORF8:L84S (Clade 2)]. The sequences devoid of the highly recurring mutations form a separate clade (Clade 5). Among these clades, the spike protein clade has the highest number of sub-clades as it is in the phase of overtaking the wild-type during the pre-lockdown time. Further, 4 major sub-clades of Clade 3 (Spike:D614G) have also emerged during the pre-lockdown. For instance, additional mutations in Spike:D614G (Clade 3a) have led to: Clade 3b (Nsp12:P323L), Clade 3c (Nsp12:P323L and ORF3a:Q57H), Clade 3d (Nsp12:P323L, ORF3a:Q57H & Nsp2:T85I) and Clade 3e (Nsp12:P323L & N protein:R203K & G204R). Simultaneous occurrence of Nsp6:L37F and ORF3a:G251V has led to Clade 1b.
Fig. 4.
(A-J) Phyloproteome analyses represent the pre-lockdown emergence and the dissemination of 5 major clades of SARS-CoV-2 across the countries that are top 10 in terms of their SARS-CoV-2 sequence data.
Intriguingly, individual phyloproteomic trees provide information about the clades that are prevalent in certain countries. The Clade 5 (that lacks high recurrent mutations) followed by Clade 2 (which are deficient of Spike:D614G mutation) are found to be prevalent in China which is the epicenter of the pandemic. Similarly, Clades 3d & 2 and Clades 3b & 3e are prominent in USA and Europe respectively. However the Clade 3d is dominant in Denmark. In Australia, all the clades are found to occur with nearly similar frequency. The Clade 3c followed by Clade 1a are found to be highly recurring in India.
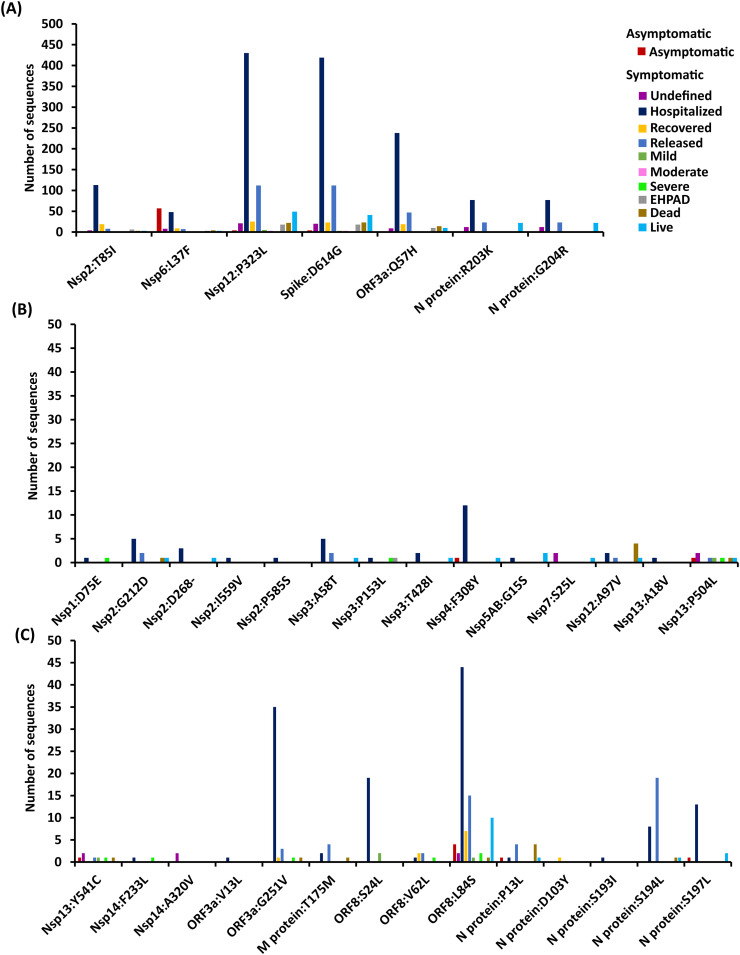
3.6. Spike:D614G and Nsp12:P323L mutations are highly found in the hospitalized patients
Not surprisingly, the available patient condition data for 1040 sequences (as on May 17, 2020) indicates that Spike:D614G and Nsp12:P323L are dominantly found in the hospitalized patients due to their surge after January 2020 (Fig. 5 ). Followed by these, ORF3a:Q57H is found in the hospitalized patients. In comparison to highly recurring mutations, moderately recurring mutations are found in relatively lesser frequency (lesser than 10-fold) in the hospitalized patients. Among the moderately recurring mutations, following mutations are found in the hospitalized patients in the following order of frequency: ORF8:L84S > ORF3a:G251V > ORF8:S24L > N protein: S197L > Nsp4:F308Y. It is noteworthy that only a few sequences in GISAID have information about the mild, moderate or severe symptom of the patients. Thus, this point is not discussed in detail here.
Fig. 5.
Bar chart illustrating the relationship between patients' condition with the (A) highly recurring and (B, C) moderately recurring mutations. Note that the color associated with the patients' conditions are indicated alongside graph.
4. Discussion
The SARS-CoV-2 pandemic is a major threat to the public health. Several attempts are being made to obtain a potential drug molecule or vaccine candidate with a broad coverage (Martin and Cheng, 2020; Poran et al., 2020; Shyr et al., 2020). Although the reasons behind variations in the vulnerability to SARS-CoV-2 infection between individuals are still unclear, the variations in the human (Hou et al., 2020) as well as the viral genome may have an important role in the degree of severity of SARS-CoV-2 infection between individuals. To this end, a comparative analysis of SARS-CoV-2 whole proteome has been performed here by considering 31,389 whole genome sequences from a diverse clinical and geographical (84 countries) origin to identify the mutations that occur in 26 SARS-CoV-2 proteins. Prior to the analyses, a manually curated database has been created for the whole genome protein sequences (country wise and protein wise) and is subjected to the mutation analyses.
The results indicate that 2116 mutations occur recurrently in SARS-CoV-2 proteome in 84 nations (Table S2). Among these, 7, 28 and 2081 are highly, moderately and low recurring mutations respectively (Fig. 2, Fig. 3 and Table S2). The highly recurring mutations include Nsp2:T85I, Nsp6:L37F, Nsp12:P323L, Spike:D614G, ORF3a:Q57H, N protein:R203K and N protein:G204R which are found across 47, 59, 71, 70, 51, 63 and 63 countries respectively (Fig. 2). Interestingly, R203K and G204R are co-occurring (dependent) mutations as they occur in nearly similar frequency in same countries. Among all the highly recurring mutations, Nsp12:P323L and Spike:D614G are found in majority of the countries (~69%). Intriguingly, some of these mutations are highly prevalent in certain countries compared to the rest of the mutations: P323L, D614G, R203K and G204R in England and P323L, D614G, Q57H and T85I in USA. The high recurrence of the above mutations clearly indicates that these mutations might have evolved very early and might have a positive selection pressure.
Further, 28 mutations are found in moderate recurrence (Fig. 3) and are found in 78 countries. Australia has the highest number of moderate mutations (28 out of 28) followed by USA (27) and United Kingdom (England, Wales and Scotland carry these moderately recurring mutations in the range of 22–24) (Fig. 3). Denmark also has 14 of these moderately recurring mutations. Among the Asian countries, India and Taiwan carry the highest number (viz.,13) of moderate mutations. Chile has 13 moderate mutations. Most of the moderately recurring mutations are absent in African countries, perhaps due to the limited availability of the data. In addition to the moderately and highly recurring mutations, 2081 mutations are found at a low recurrence (1%< PF ≥0.01%).
Six SARS-CoV-2 proteins (Nsp2, Nsp6, Nsp12, Spike, ORF3a and N protein) have at least one of the highly recurring mutations and 10 proteins (Nsp2, Nsp3, Nsp4, Nsp5, Nsp7, Nsp13, ORF3a, M protein, ORF8 and N protein) have at least one of the moderately recurring mutations. The rest of the proteins have only low recurrent mutations (Table S2).
Seven HR (Fig. 2A), 2 MR (ORF8:L84S and ORF3a:G251V, Fig. 3) and sequences devoid of HR mutations have led to the emergence of 5 major clades and 5 sub-clades of SARS-CoV-2 during the early stage of the pandemic which have disseminated across many countries during the pre-lockdown period. Among them, Clade 2 has originated from the moderately recurring mutation ORF8:L84S and Clade 5 mainly possesses low recurring mutations. However, Nsp6:L37F has led to Clade 1a and its sub-clade 1b (ORF3a:G251V). Interestingly, 70% of proteome sequences are found under Clade 3 which encompasses 4 sub-clades emerging from Nsp12:P323L, ORF3a:Q57H, N protein:R203K & G204R and Nsp6:T85I mutations indicating their evolutionary advantage.
Further, the structural (Fig. S1) and functional perspective of the SARS-CoV-2 protein mutations reported here provide some useful information for the therapeutics. The PL-pro domain and macrodomain of Nsp3 can be potential drug targets as they are less prone to mutations (Virdi et al., 2020). Occurrence of S25L mutation in Nsp7, which is in the proximity to Nsp7-Nsp8 binding interface and the proximity of Nsp12:P323L to the Nsp12-Nsp8 binding site indicate their possible role in modulating the viral replication mechanism. Further, majority of the amino acids in Nsp6 mutates either to F or to a more hydrophobic amino acid implicating the role of these mutations in host membrane manipulations (Fig. S1). Similarly, Nsp16:P134S located in the cavity that is in proximity to the SAM binding site may have a role in replication modulation (PDB ID: 7BQ7) (Fig. S1). In general, the mutations in the replicase machinery, except Nsp12:P323L, occur with a low recurrence indicating their role in functional conservation and thus, could be potential therapeutic targets. Interestingly, Spike:D614G, Spike:D936Y and ORF3a:V13L mutations are observed in potential B-cell or T-cell epitope regions (Grifoni et al., 2020), thus, may have an influence on the host defence mechanisms. Yet another notable point is a high mutability found in the SR stretch of the nucleoprotein which may have a role in host immune evasion (Gong et al., 2020; Li et al., 2020) (Fig. S1).
Since the interaction between SARS-CoV-2 spike protein and ACE2 receptor of the host is important for its entry into the host, one can envisage that ACE2 receptor polymorphism could influence the susceptibility and severity of the infection across different host population. Although a few studies have been carried out to address the role of expression and polymorphism of ACE2 receptor in SARS-CoV-2 infection susceptibility and severity (Hou et al., 2020; Cao et al., 2020; Suryamohan et al., 2021; Hussain et al., 2020; Devaux et al., 2020; Benetti et al., 2020), the unavailability of ACE2 receptor sequence information limits the establishment of one-to-one correlation between the ACE2 receptor polymorphism and spike protein variation and its implication in susceptibility and severity of the infection.
As SARS-CoV-2 variants having Spike:D614G mutation (which has enhanced infectivity and transmissibility (Korber et al., 2020; Zhang et al., 2020c; Plante et al., 2021; Gobeil et al., 2021; Raghav et al., 2020)) have conquered the wild-type across the globe, it is important to analyze the occurrence of high and moderate recurrent mutations along with Spike:D614G. Luckily, the availability of the mutations present in the SARS-CoV-2 proteome enables the understanding of the prevalence of certain mutations along with Spike:D614G (TableSD29.xlsx). For instance, Nsp2:T85I and ORF3a:Q57H (mostly occur together) along with Spike:D614G are found to be highly prevalent in USA (Nsp2:T85I and Spike:D614G = ~75% and ORF3a:Q57H and Spike:D614G = ~85%) and Denmark (Nsp2:T85I and Spike:D614G = ~72% and ORF3a:Q57H and Spike:D614G = ~76%) population. Intriguingly, these combinations are found with low prevalence in UK. Similarly, Spike:D614G and the co-mutations N protein:R203K and N protein:G204R are highly recurring in countries like Greece (78%), Portugal (68%), Russia (65%), UK (52%), Netherlands (35%) etc. Not surprisingly, Spike:D614G and Nsp12:P323L are found together in all the countries with above 95%. Among the moderately recurring mutations, M protein:T175M along with Spike:D614G occur above 25% in the Netherlands. Similarly, N protein:S194L and Spike:D614G occur ~25% in India.
In summary, the present investigation reveals the emergence of 5 major divergence from the reference sequence (Genbank ID: NC_045512.2) quite early during the pandemic based on the recurring (high, moderate and low recurrence) mutations in the proteome of SARS-CoV-2 for the dataset considered in this investigation. The high and moderate recurrent mutations which are found ≥10% and ≥1-<10% frequency respectively indicate that the virus has picked-up them as its choice quite early as they may be evolutionarily advantageous. Although the variations in SARS-CoV-2 proteins reported here (based on the comparative proteome analysis) would not directly reflect the viral transmissibility, pathogenicity, virulence and immunogenicity mechanisms, it clearly pinpoints the effective drug and vaccine targets.
Author contribution
PPU collected and organized the whole genome sequence data of SARS-CoV-2. LPPP and CS wrote the codes to fetch the information from the data. LPPP, PPU and CS analysed the data. LPPP, PPU, CS and TR wrote the manuscript. TR designed and supervised the entire project.
Funding
None.
Declaration of Competing Interest
None.
Acknowledgements
The authors thank all the researchers who have deposited the SARS-CoV-2 genome sequences (used in this study) to GISAID (Acknowledgement Table SD30) and GISAID for providing the sequences. The authors thank Indian Institute of Technology Hyderabad for the computational resources and financial support from BIRAC-SRISTI GYTI (PMU_2017_010) and BIRAC-SRISTI (PMU2019/007).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104973.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct Target Ther. 2020;5:84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand G.B., Banerjee A., Azad G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ. 2020;8 doi: 10.7717/peerj.9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Guo X., Sanjana N.E. The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. bioRxiv. 2020 doi: 10.1101/2020.06.14.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Rolain J.M., Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020;53:425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia L.F. Immune response, inflammation, and the clinical spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S.M., Janowska K., McDowell S., Mansouri K., Parks R., Manne K., et al. D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 2021;34:108630. doi: 10.1016/j.celrep.2020.108630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.N., Tsao K.C., Hsiao M.J., Huang C.G., Huang P.N., Huang P.W., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg Microbes Infect. 2020;9:1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680. doi: 10.1016/j.chom.2020.03.002. (e2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18:216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020;92:1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;100682 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. (e10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp-Thomas C., Kalish H., Hicks J., Mehalko J., Drew M., Memoli M.J., et al. D614G spike variant does not alter IgG, IgM, or IgA spike seroassay performance. medRxiv. 2020 doi: 10.1101/2020.07.08.20147371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;183:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz239. W256-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M., Karrer U., Lucas A., Klenerman P. Viral escape mechanisms--escapology taught by viruses. Int. J. Exp. Pathol. 2001;82:269–286. doi: 10.1046/j.1365-2613.2001.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., et al. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W.R., Cheng F. A rational design of a multi-epitope vaccine against SARS-CoV-2 which accounts for the glycan shield of the spike glycoprotein. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12770225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poran A., Harjanto D., Malloy M., Arieta C.M., Rothenberg D.A., Lenkala D., et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020;12:70. doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghav S., Ghosh A., Turuk J., Kumar S., Jha A., Madhulika S., et al. Analysis of Indian SARS-CoV-2 genomes reveals prevalence of D614G mutation in spike protein predicting an increase in interaction with TMPRSS2 and virus infectivity. Front. Microbiol. 2020;11:594928. doi: 10.3389/fmicb.2020.594928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr Z.A., Gorshkov K., Chen C.Z., Zheng W. Drug discovery strategies for SARS-CoV-2. J. Pharmacol. Exp. Ther. 2020;375:127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D.G. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Suryamohan K., Diwanji D., Stawiski E.W., Gupta R., Miersch S., Liu J., et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun. Biol. 2021;4:475. doi: 10.1038/s42003-021-02030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahiko Koyama DPaLP . In: Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ, editor. 2020. pp. 498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel O.M., Ata O., Turgut-Balik D. An updated analysis of variations in SARS-CoV-2 genome. Turk. J. Biol. 2020;44:157–167. doi: 10.3906/biy-2005-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdi R.S., Bavisotto R.V., Hopper N.C., Frick D.N. Discovery of drug-like ligands for the Mac1 domain of SARS-CoV-2 Nsp3. bioRxiv. 2020 doi: 10.1101/2020.07.06.190413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhai X., Zhou J. Snapshot of the evolution and mutation patterns of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.07.04.187435. [DOI] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2