Abstract
The immunogenicity of the SARS-CoV-2 proteome is largely unknown, especially for non-structural proteins and accessory proteins. In this study, we collect 2,360 COVID-19 sera and 601 control sera. We analyze these sera on a protein microarray with 20 proteins of SARS-CoV-2, building an antibody response landscape for immunoglobulin (Ig)G and IgM. Non-structural proteins and accessory proteins NSP1, NSP7, NSP8, RdRp, ORF3b, and ORF9b elicit prevalent IgG responses. The IgG patterns and dynamics of non-structural/accessory proteins are different from those of the S and N proteins. The IgG responses against these six proteins are associated with disease severity and clinical outcome, and they decline sharply about 20 days after symptom onset. In non-survivors, a sharp decrease of IgG antibodies against S1 and N proteins before death is observed. The global antibody responses to non-structural/accessory proteins revealed here may facilitate a deeper understanding of SARS-CoV-2 immunology.
Keywords: humoral immunity, non-structural/accessory proteins, SARS-CoV-2, COVID-19, proteome microarray, immune response
Graphical abstract
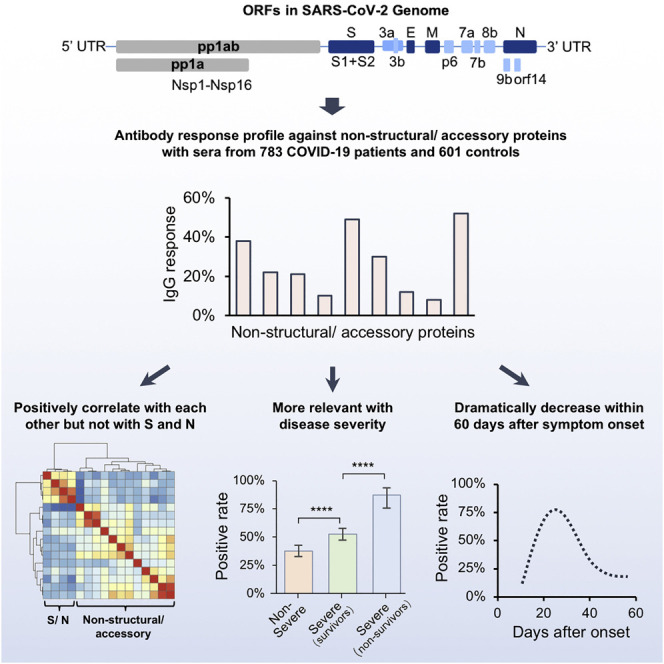
Li et al. profile antibody responses against SASR-CoV-2 non-structural/accessory proteins with sera from 783 COVID-19 patients and 601 controls, and they demonstrate that the responses against the non-structural/accessory proteins are distinct from those of the structural proteins.
Introduction
COVID-19, caused by SARS-CoV-2 (Wu et al., 2020c; Zhou et al., 2020), has become one of the most threatening crises to global public health. By November 4, 2020, 47,328,401 cases were diagnosed and 1,212,070 lives were claimed (https://coronavirus.jhu.edu/map.html) (Dong et al., 2020). SARS-CoV-2 belongs to the betacoronavirus genus, and its genome encodes 4 major structural proteins, i.e., spike (S), envelope (E), membrane (M), and nucleocapsid (N), and 16 non-structural proteins (Nsp1–16) and 9 accessory proteins (Gordon et al., 2020; Wu et al., 2020a). Among them, the S protein consists of an N-terminal S1 fragment and a C-terminal S2 fragment, and it plays an essential role in viral attachment, fusion, and entry into the target cells that express the viral receptor, i.e., angiotensin-converting enzyme 2 (ACE2) (Ge et al., 2013; Hoffmann et al., 2020; Lan et al., 2020; Wrapp et al., 2020; Yan et al., 2020). However, the function, including immunogenicity, of most of the non-structural proteins and accessory proteins is still elusive.
One of the major features of patients with COVID-19 is the extreme variability of clinical severity from asymptomatic to death (Shrock et al., 2020). However, the factors that cause this variability are still largely unknown. Humoral immune responses elicited by SARS-CoV-2 play essential roles, especially in diagnosis, neutralizing antibody production, and vaccine development (Jiang et al., 2020c; Long et al., 2020a; Vabret et al., 2020). Among all the SARS-CoV-2 proteins, the S protein and N protein exhibit high immunogenicity. Antibodies against the S protein and N protein are elicited in most patients, with higher titers in severe patients, demonstrating the association between severity and humoral immune responses (Jiang et al., 2020a, 2020b). It was reported that the antibodies against peptides derived from non-structural and accessory proteins were also detectable in patients (Li et al., 2020; Shrock et al., 2020; Wang et al., 2020). However, the prevalence, clinical relevance, and the dynamics of non-structural proteins and accessory proteins in patients are still largely unknown.
Recently, we constructed a SARS-CoV-2 proteome microarray, containing S protein, N protein, and most of the non-structural and accessory proteins (Jiang et al., 2020b). The microarray is a powerful tool to systematically study the humoral immune response, especially the immunoglobulin (Ig)G and IgM responses against the SARS-CoV-2 proteome. Based on this platform, we have successfully characterized the humoral immune in convalescent patients (Jiang et al., 2020b) and asymptomatic patients (Lei et al., 2021).
In this study, we adopted an updated SARS-CoV-2 proteome microarray that contains 20 proteins, and we profiled 2,360 sera from 783 patients with COVID-19 and 601 control sera. We identified that NSP1, NSP7, NSP8, RdRp, ORF3b, and ORF9b can strongly elicit antibodies in patients with COVID-19. Further analysis revealed that the patterns of humoral immunity of these non-structural/accessory proteins were distinct from those of S and N proteins. The global antibody responses to non-structural proteins and accessory proteins revealed in this study will facilitate the comprehensive understanding of SARS-CoV-2 humoral immunity, and they may provide potential biomarkers for precise monitoring of COVID-19 progression.
Results
COVID-19 severity and clinical outcome are associated with a set of clinical parameters
To systematically analyze the clinical characteristics of SARS-CoV-2 infection, we first analyzed the correlations between severity and each of the available laboratory parameters of 783 patients with COVID-19 monitored when admitted (Table S1). According to the severity and clinical outcome, the patients were divided into three groups, i.e., non-severe patients (all of whom were recovered), severe but survived patients, and non-survivors. Statistical comparisons among these three groups enable us to investigate the features either related to the severity for survivors or to the outcome under similar severity (Table 1 ). Because for some patients some laboratory examinations were missing, for each clinical parameter only the effective patient numbers were given. As expected, sex, age, and the comorbidities of hypertension and diabetes are associated factors of severity; however, only age is significantly associated with clinical outcome for severe patients. In addition, consistent with many previous studies (García, 2020; Huang et al., 2020; Wu et al., 2020b), we identified a set of clinical and laboratory parameters that are highly related to severity or outcome, such as lymphopenia, increased CRP (C reaction protein), and factors associated with blood coagulation, cardiac injury, liver injury, and kidney injury. Most of these factors are associated both with severity and outcome, while some are likely associated either with severity or outcome. For instance, thrombocytopenia and some kidney injury-related factors are more common in non-survivors as compared to the other two groups, while most liver injury-related factors are only associated with severity but not outcome.
Table 1.
Clinical parameters related to severity of patients with COVID-19
| Related functions | Clinical parametersa | Non-severe |
Severe (survivors) |
Severe (non-survivors) |
p value 1b | p value 2b | |||
|---|---|---|---|---|---|---|---|---|---|
| Samples (n) | n, % (95% CI) | Samples (n) | n, % (95% CI) | Samples (n) | n, % (95% CI) | ||||
| Gender | sex (male) ↑→ | 369 | 156, 42.3% (37.3%–47.4%) | 354 | 193, 56.2% (51.6%–60.7%) | 60 | 38, 58.2% (47.2%–68.5%) | <0.0001 | 0.733 |
| Complication |
hypertension ↑→ | 369 | 106, 28.7% (23.3%–33.5%) | 354 | 176, 49.7% (42.6%–51.7%) | 60 | 24, 40.0% (28.6–52.6%) | <0.0001 | 0.164 |
| diabetes ↑→ | 369 | 49, 13.3% (10.2%–17.1%) | 354 | 82, 23.2% (19.1%–27.8%) | 60 | 15, 25.0% (15.8%–37.2%) | 0.0006 | 0.756 | |
| Related functions | Clinical parameters | Samples (n) | Median (IQR) | Samples (n) | Median (IQR) | Samples (n) | Median (IQR) | p value 1 | p value 2 |
| Age | age ↑↑ | 369 | 58 (47, 67.5) | 354 | 65 (56, 72) | 60 | 68 (59.5, 78) | 2.74E−14 | 2.27E−03 |
| Infection | lymphocyte (no.) ↓↓ | 246 | 1.115 (0.7875, 1.5525) | 305 | 0.85 (0.5825, 1.17) | 48 | 0.64 (0.45, 0.86) | 7.39E−14 | 1.86E−04 |
| lymphocyte (%) ↓↓ | 246 | 22.5 (15.25, 29.625) | 305 | 13.4 (9.1, 20.275) | 48 | 8.4 (4.9, 12.2) | 4.07E−25 | 3.42E−07 | |
| neutrophils (no.) ↑↑ | 246 | 3.57 (2.49, 4.94) | 305 | 4.795 (3.4175, 6.7825) | 48 | 6.77 (4.5, 11.38) | 6.63E−15 | 3.33E−05 | |
| neutrophils (%) ↑↑ | 246 | 66.9 (58.6, 76.1) | 305 | 77.75 (70.05, 85) | 48 | 86.4 (80.8, 90.1) | 8.47E−24 | 3.42E−07 | |
| eosinophils (no.) ↓↓ | 246 | 0.03 (0, 0.08) | 305 | 0.01 (0, 0.0575) | 48 | 0 (0, 0.01) | 3.27E−04 | 3.30E−04 | |
| eosinophils (%) ↓↓ | 246 | 0.5 (0, 1.4) | 305 | 0.2 (0, 0.9) | 48 | 0 (0, 0.2) | 2.51E−05 | 5.20E−05 | |
| basophils (%) ↓↓ | 246 | 0.2 (0.1, 0.4) | 305 | 0.2 (0.1, 0.2) | 48 | 0.1 (0, 0.2) | 2.39E−06 | 0.01 | |
| monocytes (%) ↓↓ | 246 | 8.4 (6.6, 10.1) | 305 | 7.3 (4.825, 9.6) | 48 | 4.9 (2.7, 7.2) | 1.98E−06 | 3.89E−05 | |
| white blood cells ↑↑ | 246 | 5.375 (4.1775, 6.95) | 305 | 6.29 (4.8625, 8.405) | 43 | 8.03 (5.49, 13.08) | 1.19E−08 | 1.27E−04 | |
| CRP ↑↑ | 228 | 20.15 (5.3, 57.95) | 277 | 55.5 (20.1, 102.4) | 38 | 86.25 (50.85, 170.15) | 2.42E−14 | 5.26E−05 | |
| procalcitonin ↑↑ | 197 | 0.05 (0.03, 0.09) | 236 | 0.07 (0.04, 0.18) | 43 | 0.18 (0.09, 0.9) | 2.18E−09 | 2.20E−07 | |
| globulin ↑→ | 228 | 33.3 (30.1, 36.8) | 284 | 35.05 (32.025, 38.7) | 40 | 35.8 (31.55, 40.05) | 7.61E−06 | 0.45 | |
| Blood coagulation | d-dimer ↑↑ | 216 | 0.68 (0.36, 1.55) | 251 | 1.29 (0.68, 2.47) | 41 | 2.82 (1.41, 13.545) | 1.87E−11 | 1.84E−07 |
| prothrombin activity ↓↓ | 218 | 92 (85, 101) | 256 | 88 (80, 96.25) | 41 | 78 (69, 89) | 7.64E−06 | 4.51E−06 | |
| prothrombin time ↑↑ | 218 | 13.8 (13.3, 14.375) | 256 | 14.1 (13.5, 14.825) | 36 | 15 (14.1, 15.9) | 1.23E−06 | 8.27E−06 | |
| fibrinogen ↑→ | 200 | 4.935 (3.975, 5.76) | 211 | 5.47 (4.64, 6.47) | 23 | 5.215 (3.4, 6.525) | 1.91E−06 | 0.12 | |
| FDP ↑↑ | 102 | 4 (4, 5.3) | 99 | 4.4 (4, 7.8) | 48 | 13.6 (6.8, 51.55) | 1.10E−03 | 1.66E−06 | |
| platelet count → ↓ | 246 | 219 (162, 278) | 305 | 222 (165, 295) | 46 | 179 (119, 247) | 0.51 | 1.88E−04 | |
| Plateletcrit → | 239 | 0.23 (0.18, 0.29) | 300 | 0.24 (0.18, 0.31) | 43 | 0.2 (0.14, 0.26) | 0.29 | 6.67E−04 | |
| Cardiac injury | LDH ↑↑ | 229 | 251 (210, 306) | 284 | 330.5 (270.25, 443) | 41 | 480 (364, 552.5) | 7.19E−23 | 1.95E−07 |
| hs-cTnI ↑↑ | 167 | 4 (1.9, 8.525) | 211 | 7.25 (3.8, 15.025) | 37 | 24.4 (8.1, 103.7) | 4.00E-09 | 2.35E-08 | |
| NT-proBNP ↑↑ | 142 | 99.5 (33.75, 281.25) | 189 | 269 (113.75, 569.25) | 23 | 942.5 (349.25, 2,794.25) | 1.82E-11 | 7.08E-08 | |
| myoglobin ↑↑ | 66 | 40.1 (29.825, 76.625) | 93 | 75.3 (46.625, 124.325) | 43 | 157.7 (85.5, 344.85) | 4.71E−06 | 2.50E−04 | |
| Liver Injury | aspartate aminotransferase ↑↑ | 229 | 25 (18, 37) | 284 | 31 (22, 48) | 43 | 38 (25.5, 55.5) | 4.57E−07 | 0.02 |
| alanine aminotransferase ↑→ | 229 | 21 (13, 39.5) | 284 | 28 (18, 44) | 43 | 29 (18, 39) | 3.10E−04 | 0.56 | |
| γ-GT ↑→ | 228 | 29 (18, 53) | 284 | 35 (22, 71) | 43 | 40 (27, 87.5) | 8.74E−04 | 0.11 | |
| total bilirubin ↑↑ | 228 | 8.6 (6.4, 11.7) | 284 | 10.05 (8, 13.875) | 43 | 12 (9.4, 18.1) | 1.39E−07 | 5.85E−03 | |
| direct bilirubin ↑↑ | 228 | 3.7 (2.7, 5) | 284 | 4.65 (3.5, 6.475) | 41 | 5.4 (4.1, 9.7) | 5.39E−10 | 5.39E−03 | |
| indirect bilirubin ↑→ | 224 | 4.8 (3.5, 6.425) | 283 | 5.2 (4, 7.6) | 43 | 5.8 (4.325, 7.475) | 9.45E−04 | 0.34 | |
| albumin ↓↓ | 228 | 35.7 (32.8, 38.7) | 284 | 32.6 (30.025, 35.4) | 43 | 32 (29.3, 34.3) | 3.13E−16 | 0.07 | |
| albumin-to-globulin ratio ↓↓ | 228 | 1.06 (0.92, 1.26) | 284 | 0.92 (0.8, 1.0775) | 43 | 0.87 (0.775, 0.995) | 2.51E−15 | 0.12 | |
| total protein ↓→ | 228 | 69.3 (65.8, 72.9) | 284 | 67.8 (64.325, 72.125) | 43 | 68.5 (62.9, 72.65) | 8.14E−03 | 0.72 | |
| total cholesterol ↓→ | 228 | 3.81 (3.24, 4.41) | 284 | 3.55 (3.0625, 4.04) | 30 | 3.48 (2.975, 4.1) | 6.30E−05 | 0.84 | |
| ferritin ↑↑ | 119 | 488.5 (303.85, 745.95) | 121 | 852.85 (525.5, 1,542.975) | 43 | 1,340.4 (953.3, 2,005.45) | 3.38E−10 | 6.39E−03 | |
| alkaline phosphatase →↑ | 228 | 65 (54, 82) | 284 | 67 (55.25, 86) | 43 | 80 (60, 102) | 0.30 | 3.46E−03 | |
| Kidney injury | creatinine →↑ | 228 | 67 (57, 82) | 282 | 69 (56.75, 86) | 23 | 82 (66.5, 102.5) | 0.15 | 3.25E−04 |
| creatine kinase ↑↑ | 61 | 0.6 (0.4, 1.425) | 89 | 1 (0.675, 1.9) | 41 | 1.8 (1, 2.95) | 1.77E−03 | 2.70E−03 | |
| eGFR ↓↓ | 225 | 92.7 (79.1, 104.45) | 272 | 89.7 (76.3, 99.3) | 43 | 69.9 (47.7, 91) | 7.70E−03 | 1.96E−05 | |
| bicarbonate →↓ | 228 | 24.3 (22.9, 26.1) | 282 | 24.1 (22.4, 26.1) | 43 | 21.9 (19.5, 24) | 0.35 | 1.95E−06 | |
| urea ↑↑ | 228 | 4.2 (3.3, 5.4) | 282 | 4.9 (3.575, 6.325) | 43 | 8 (5.75, 12.2) | 6.21E−05 | 2.02E−11 | |
| uric acid →↑ | 228 | 253 (198.1, 309) | 282 | 236 (181.75, 298.125) | 11 | 270 (184.5, 388) | 0.05 | 0.03 | |
| Thyroid function | free T3 ↓↓ | 56 | 4.38 (3.57, 4.82) | 62 | 3.67 (3.3, 4.32) | 11 | 2.94 (2.6475, 3.22) | 2.66E−03 | 6.31E−05 |
| free T4 ↑→ | 56 | 17.5 (15.07, 20.08) | 62 | 19.63 (16.89, 21.35) | 13 | 17.25 (16.25, 17.985) | 1.49E−03 | 0.10 | |
| Cytokines | IL-6 ↑↑ | 47 | 6.025 (2.6225, 28) | 32 | 24.785 (5.5575, 59.565) | 12 | 89.47 (16.82, 283.7) | 1.17E−03 | 8.65E−03 |
| IL-8↑↑ | 40 | 7.5 (5, 13.4) | 29 | 10.9 (6.275, 21.9) | 12 | 30.95 (16.975, 156.175) | 0.02 | 7.67E−04 | |
| IL-10 →↑ | 40 | 5 (5, 5.7) | 29 | 5 (5, 9.2) | 12 | 10.8 (6.9, 22.6) | 0.09 | 8.60E−04 | |
| IL-2R ↑↑ | 40 | 395 (243, 676) | 29 | 731.5 (413, 1,007) | 12 | 1,321.5 (811.5, 2,083.5) | 4.77E-04 | 7.05E-04 | |
| TNF-α ↑↑ | 40 | 7.3 (5.3, 9.2) | 29 | 8.55 (6.375, 12.25) | 42 | 14.15 (10.15, 21.15) | 0.04 | 9.69E−04 | |
| Others | Glucose ↑↑ | 215 | 5.9 (5.1975, 7.02) | 275 | 6.69 (5.6575, 8.8275) | 47 | 7.42 (6.4375, 9.1475) | 1.78E−08 | 0.01 |
| RDW (CV) →↑ | 242 | 12.3 (11.9, 13) | 303 | 12.4 (11.9, 13) | 47 | 12.85 (12.2, 14.1) | 0.39 | 7.13E−05 | |
| RDW (SD) →↑ | 242 | 40 (38, 42.1) | 303 | 40 (38.4, 42.125) | 43 | 41.55 (39.8, 44.225) | 0.46 | 3.63E−04 | |
| potassium →↑ | 228 | 4.18 (3.82, 4.51) | 282 | 4.17 (3.78, 4.54) | 43 | 4.42 (3.865, 4.79) | 0.97 | 0.02 | |
| chlorine ↓→ | 228 | 100.8 (98.225, 103) | 282 | 100 (97.1, 102.725) | 43 | 100.6 (97.75, 103.8) | 9.61E−03 | 0.18 | |
| calcium ↓→ | 228 | 2.17 (2.11, 2.27) | 282 | 2.11 (2.05, 2.2) | 43 | 2.11 (2.04, 2.17) | 1.19E−10 | 0.50 | |
| sodium ↓→ | 228 | 139.8 (137.5, 141.875) | 282 | 138.9 (136.3, 141.225) | 59 | 139.2 (136.25, 141.8) | 3.01E−03 | 0.54 | |
↓, significantly decrease; ↑, significantly increase; →, no significant change. CPR, C-reactve protein; FDP, fibrinogen degradation product; LDH, lactate dehydrogenase; hs-cTnI, high-sensitivity cardiac troponin I; NT-proBNP, N-terminal pro-B-type natriuretic peptide; γ-GT, γ-glutamyl transpeptidase; eGFR, epidermal growth factor receptor; RDW (CV), red cell volume distribution width (coefficient of variation); RDW (SD), red cell volume distribution width (standard deviation).
The two arrows immediately after each clinical parameter indicate the change of severe survivors compared with non-severe patients and severe non-survivors compared with severe survivors, respectively.
p value 1: comparison between non-severe and severe survivors; p value 2: comparison between severe survivors and non-survivors.
Several non-structural and accessory proteins elicit highly prevalent antibody responses
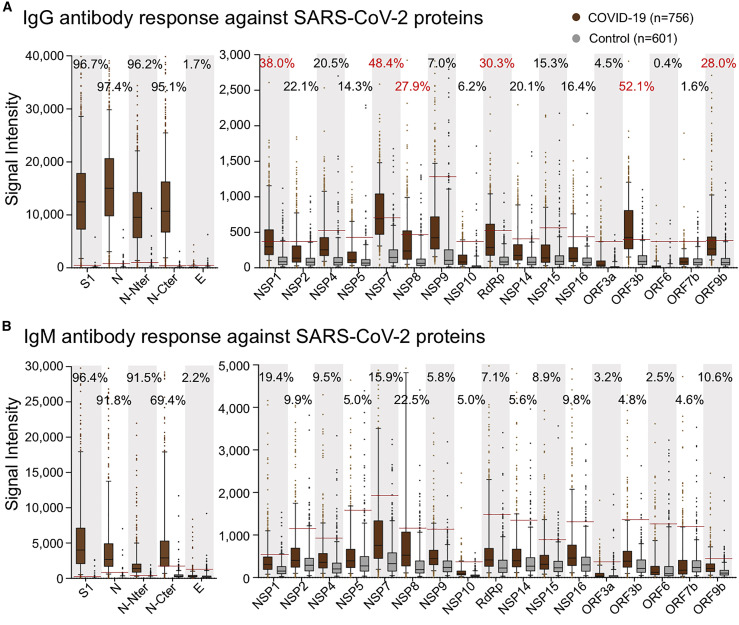
Previously, we constructed a SARS-CoV-2 proteome microarray (Figures S1A and S1B; Table S2) and screened a small cohort of convalescent patients (Jiang et al., 2020b). In this study, we aimed to systematically analyze the immune responses and their dynamic changes against SARS-CoV-2 proteins with a much larger cohort of samples. In total, we collected 2,360 sera from 783 laboratory-confirmed patients with COVID-19 as well as 601 control sera (Table S1). All of these sera were analyzed on the SARS-CoV-2 protein microarray. To acquire high-quality data for the microarray experiments, we prepared a positive control by mixing 50 randomly selected COVID-19 sera. This control was then probed on each microarray to assess and normalize the data. It turns out that high reproducibility is achieved in our assay (Figures S1C and S1D). To construct the antibody response landscape against the whole proteome, a subset of the sera collection was selected. Since seroprevalence would gradually increase to a plateau until about 2 weeks after symptom onset (Long et al., 2020a), the samples collected within 14 days were excluded. For patients with more than one sample, the serum collected at the earliest time point after 2 weeks of symptom onset was selected. A total of 756 serum samples were selected to construct the landscape (Figure 1 ). Immune response frequency was calculated for each protein with the cutoff value set by mean + 2 × SD of the control group. Except for S1 and N, which are known to be highly antigenic, we found that several non-structural and accessory proteins elicited prevalent antibody responses, especially for IgG, including NSP1, NSP7, NSP8, RdRp, ORF3b, and ORF9b, for which the positive rates were 38%, 48.4%, 27.9%, 30.3%, 52.1%, and 28%, respectively. Although the IgM responses were high in some cases, the overall responses were much lower than those of IgG. We then decided to focus on IgG for in-depth analysis.
Figure 1.
Antibody response landscape against SARS-CoV-2 proteins
(A and B) IgG (A) and IgM (B) responses against each SARS-CoV-2 protein are depicted as boxplots according to the signal intensity of each sample on the proteome microarray. The data are presented as median with quintiles and the hinges (n = 756). Cutoff values (the red line) for each protein were set as mean + 2 × SD of the control group (n = 601). The positive rates of the patient group were labeled for each protein; positive rates >25% are labeled as red.
The IgG pattern of non-structural and accessory proteins is distinct from that of S1 and N protein
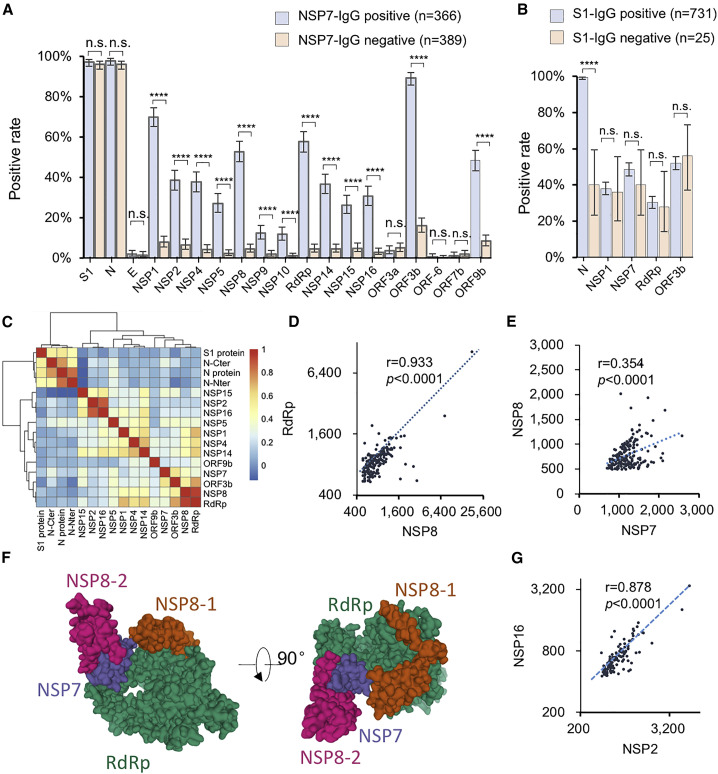
We next asked whether the IgG responses to these proteins were associated with each other. We first chose NSP7 as a reference. The samples were divided into two groups depending on positive or negative NSP7 IgG. Positive rates of the rest of the proteins were calculated for the two groups. Unexpectedly, for all of the non-structural and accessory proteins, except for ORF3a, ORF6, and ORF7a, which barely elicit antibodies, the positive rates in the NSP7-IgG-positive group were significantly higher than those in the NSP7-IgG negative group, demonstrating high correlations (Figure 2 A). Interestingly, there is no clear difference for the IgG responses of S1 and N. Similar observations were shown for NSP1, RaRp, and ORF3b as references (Figure S2). To further confirm our observation, we reversely compared the positive rates of IgG to non-structural/accessory proteins between the groups of S1-IgG-positive and -negative (Figure 2B). The positive rates of the IgG response to N protein are significantly different between these two groups, while no clear difference was shown for the non-structural/accessory proteins. These observations demonstrate that the structural proteins elicit antibodies with a distinct pattern to that of the non-structural and accessory proteins, suggesting that the underlying mechanisms by which the antibodies are triggered are different for these two groups of proteins. To further study the correlations of IgG signal intensity among the proteins, Pearson correlation coefficients between any two of these proteins were calculated and then clustered. The proteins with positive rate less than 10% were not included due to statistical limits (Figure 2C).
Figure 2.
Antibodies against structural proteins and other proteins are in different patterns
(A) Antibody-positive rates for the SARS-CoV-2 proteins in two patient groups divided according to NSP7 IgG signal, either positive or negative.
(B) Antibody-positive rates for selected proteins in two patient groups; the patient groups were divided according to the S1 IgG signal, either positive or negative.
(C) The Pearson correlation coefficients of the IgG responses among the proteins were calculated and clustered.
(D and E) Correlations of the IgG responses against RdRp and NSP8 (D) and NSP8 and NSP7 (E).
(F) Location and accessibility of NSP7, NSP8, and RdRp in the SARS-CoV-2 RNA polymerase complex (PDB: 7BV1).
(G) Correlations of the IgG responses against NSP2 and NSP16.
For (A) and (B), the error bar is given as the 95% confidential interval. p values were calculated by a two-sided χ2 test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n.s., not significant.
Consistently, the S and N proteins have lower correlations with the non-structural/accessory proteins, while the non-structural/accessory proteins were clustered together. In addition, several subclusters were shown among the non-structural/accessory proteins. Interestingly, NSP8 and RdRp have a high correlation (Figure 2D). It is known that RdRp, NSP8, and NSP7 could form a tight complex (Gao et al., 2020; Yin et al., 2020), which might contribute to the high correlation. However, the correlation between NSP8 and NSP7 is less significant (Figures 2C and 2E). The structure of the complex shows NSP7 physically connect to RdRp and NSP8, but with most of the protein surface blocked (Figure 2F), while NSP8 and RdRp are more accessible. However, NSP7 elicits antibody in a higher rate (Figure 1A), suggesting that NSP7 might mainly exist in other forms rather than in a complex with NSP8 and RdRp, and thus it has other biological functions to be discovered. In addition, the IgG responses of NSP2 and NSP16 also have a high correlation (Figures 2C–2G). It was reported that PPI (protein-protein interaction) was detected between NSP2 and NSP16 (Li et al., 2021). However, we did not detect any direct binding signals between NSP2 and NSP16 in vitro (data not shown), suggesting that NSP2 and NSP16 might form a complex through the bridging of other proteins. In addition, differences of the positive rates among the proteins are not associated with the in vivo protein expression level (Finkel et al., 2021) and the protein length (Figure S3).
IgG responses are associated with severity
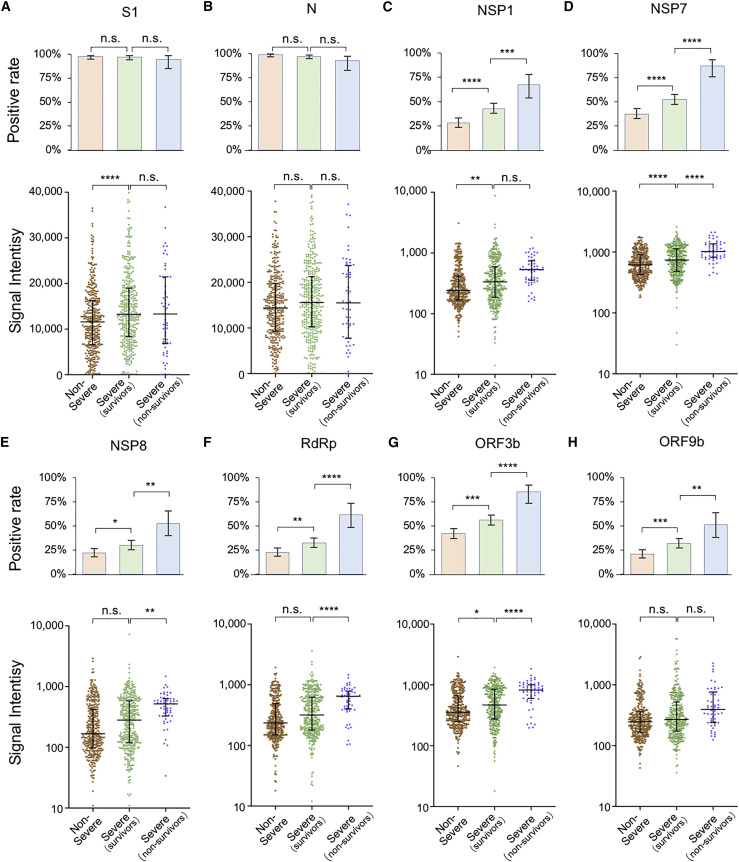
It is known that IgG responses against S and N proteins are associated with disease severity and clinical features (Jiang et al., 2020b; Long et al., 2020a), however, the correlations to other SARS-CoV-2 proteins, especially non-structural and accessory proteins, have not been revealed before. As described above, we divided the patient population into three groups, i.e., non-severe, severe survivors, and severe non-survivors. Two statistical methods were applied to assess the correlations. One is to analyze the positive rate of IgG against each protein, and the other is to compare the signal intensity distribution among groups (Figure 3 ). For the N protein, no significant difference was found among the three groups, whereas for the S1 protein, the only statistically significant difference was observed between the severe and non-severe groups (Figures 3A and 3B). In contrast, the non-structural/accessory proteins with high positive rates/signal intensities are more significantly correlated with severity. It is noteworthy that for the six non-structural and accessory proteins, both the positive rates and signal intensities are significantly higher in more severe groups (Figures 3C–H). These results indicate that the IgG responses against non-structural/accessory proteins have higher correlations with disease severity, and they may serve as a better predictor of COVID-19 severity than those of S1 and N proteins.
Figure 3.
IgG responses are associated with disease severity
(A–H) IgG-positive rate and signal intensity distribution among three patient groups, i.e., non-severe, severe (survivors), and severe (non-survivors) patients for S1 (A), N protein (B), NSP1 (C), NSP7 (D), NSP8 (E), RdRp (F), ORF3b (G), and ORF9b (H). For positive rate analyses, the error bar is given as the 95% confidential interval. The p values were calculated by a two-sided χ2 test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n.s., not significant. For signal intensity analysis, the middle line is the median value; the upper and lower hinges are the values of the 75% and 25% percentile. The p value was calculated by a two-sided t test.
We next analyzed the correlation between antibody response and clinical parameter. The clinical parameters, which have statistical correlations with IgG against S1, N, NSP7, NSP8, RdRp, ORF3b, and ORF9b, were determined (Table S3; Figure S4). All of these parameters are related to severity, suggesting that severity is a major factor and confounder that contributes to the correlations. Interestingly, thrombocytopenia is related with clinical outcome but not severity (Table 1), and it is significantly correlated with NSP7 and ORF3b but not S1 and N, further confirming the higher correlations among antibodies of non-structural/accessory proteins and clinical outcome.
IgG responses of S1 and N proteins decrease several days before death in non-survivors
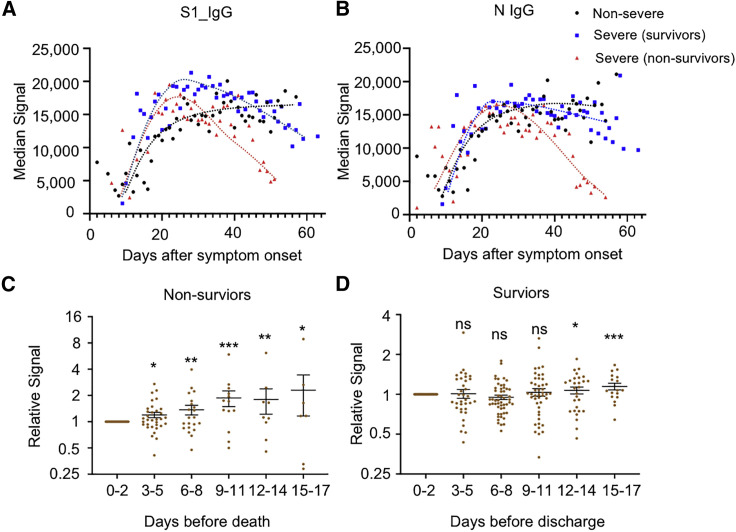
The preservation of high-level neutralizing antibodies is essential for protecting patients from re-infection. One critical question concerns how long the antibodies against SARS-CoV-2 can last. A recent study found that antibody titers did not decline within 4 months after diagnosis (Gudbjartsson et al., 2020), while other studies observed rapid decay of antibodies in patients with mild cases (Ibarrondo et al., 2020; Long et al., 2020b). Accordingly, we analyzed the antibody dynamics with our sample set in which the sera were collected from 0 to about 60 days after initial symptom onset. The seroprevalence or positive rates for both S1 and N reached a plateau at about 20 days after symptom onset and were maintained afterward for at least 2 months for all three groups (Figures 4A and 4B). However, with regard to signal intensity, a dramatic decrease was observed for the non-survivor group, but not for other two groups, although a slight decline was observed for the severe survivor group (Figures 4C and 4D). The sharp decline in non-survivors might be related to death. To confirm this possibility, we analyzed 35 patients with serum available 0–2 days before death, and 108 survivors with serum available 0–2 days before discharge served as control. For each patient, we defined the relative signal for each sample to the sample immediately before death or discharge. Overall, the relative signals declined gradually during the disease progression from about 10 days before death (Figure 4E) for non-survivors, although the trend differed among individuals. In contrast, there was no significant change for the survivors (Figure 4F). These observations might imply a collapse in SARS-CoV-2-related humoral immunity in most patients before death, and further studies are needed to confirm this.
Figure 4.
S1 and N IgG decrease several days before death in non-survivors
(A and B) The trends of median signal intensities of IgG at different time points for S1 (A) and N (B), among three sample groups, i.e., non-severe, severe (survivors), and severe (non-survivors). Samples were grouped per day, and the time points with a sample number less than four were excluded due to lack of statistical significance.
(C and D) Relative S1 IgG signal levels were calculated for each patient by dividing the signal intensity of the samples collected at other time points versus samples collected at 0–2 days before the death of non-survivors (C, n = 35) or the discharge of survivors (D, n = 108). The samples were grouped per 3 days. For each patient, the signals were averaged when there was more than one sample during each 3-day period. The p values were calculated by a two-sided t test between the indicated group and the first group (0–2 days). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., not significant.
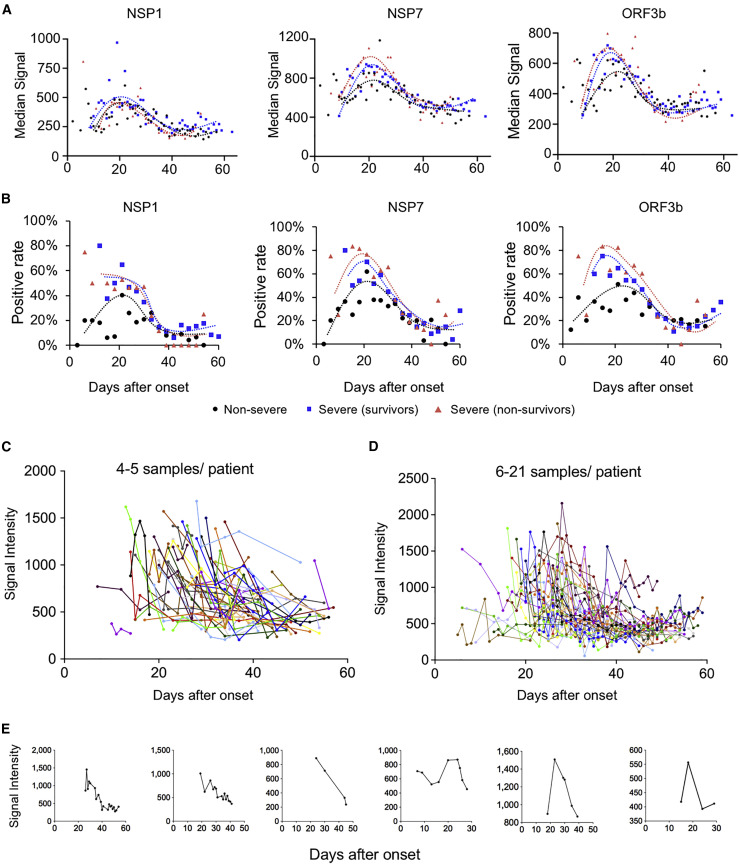
IgG against the six non-structural/accessory proteins decline rapidly during COVID-19 progression
We next analyzed the dynamic of IgG responses for the six non-structural/accessory proteins. Surprisingly, the IgG responses, i.e., the signal intensities and positive rates, against the six proteins reached a plateau in all the three groups at about 20 days after the symptom onset, and then they decreased rapidly for all three groups (Figures 5A and 5B); this is largely different from S1 and N protein (Figure 4). We next selected NSP7 IgG as an example for further analysis and depicted the change for each patient (Figures 5C–5E). A continuous and dramatic decline of IgG against NSP7 for most patients was observed (Figures 5D and 5E). These results imply that the B cells that produce IgG antibodies against non-structural/accessory proteins might be short-lived and/or the underlying mechanism of generating IgG antibodies against non-structural/accessory proteins may differ from that of S1 and N proteins.
Figure 5.
Antibody responses against non-structural proteins and accessory proteins decrease rapidly after 20 days of symptom onset
(A) Trends of median signal intensities of IgG at different time points for NSP1, NSP7, and ORF3b among three samples groups, i.e., non-severe, severe (survivors), and severe (non-survivors). Samples were grouped per day, and the points with sample number less than four were excluded.
(B) Trends of positive rate of IgG at different time points for NSP1, NSP7, and ORF3b among three samples groups, i.e., non-severe, severe (survivors), and severe (non-survivors). Samples were grouped per 3 days.
(C–E) NSP7-IgG signal dynamic changes for the patients with four to five samples (C) or more samples (D) or for some representative individuals (E). Each line represents one person.
Discussion
In this study, we profiled 2,360 sera from 783 patients with COVID-19 and 601 control sera using a SARS-CoV-2 proteome microarray. We found that six non-structural/accessory proteins elicit strong antibody responses in patients with COVID-19, including NSP1, NSP7, NSP8, RdRp, ORF3b, and ORF9b.
It is broadly reported that a batch of clinical laboratory parameters are associated with disease severity of patients with COVID-19, such as lymphopenia, neutrophilia, increased value of CRP, lactate dehydrogenase (LDH), and d-dimer (García, 2020; Huang et al., 2020; Wu et al., 2020b). What we observed is mostly consistent with these reports. We also found some parameters that were more related with clinical outcome. For example, there is no clear difference in the levels of thrombocytopenia between non-severe and severe survivors, but there was a significant decline in levels in non-survivors, suggesting that the issue of blood coagulation in patients with COVID-19 should be carefully monitored during therapy and fully evaluated for its contribution to death.
NSP1 is a major virulence factor that binds with the small subunit of host ribosome to suppress host gene expression (Thoms et al., 2020). NSP7, NSP8, and RdRp (NSP12) form a complex that is involved in the replication and transcription of the SARS-CoV-2 genome, playing an essential role for virus replication. RdRp is the target of a promising drug, remdesivir (Yin et al., 2020). ORF3b is reported to be a potent interferon (IFN) antagonist (Konno et al., 2020) and was identified to elicit antibodies in patients with COVID-19 (Hachim et al., 2020). ORF9b can suppress type I IFN production by targeting host protein TOM70 (Jiang et al., 2020a). Previous studies have found the presence of antibodies against SARS ORF9b (Guo et al., 2004; Qiu et al., 2005), and the ORF9b IgG antibody was identified in sera from convalescents, although from a small cohort of sera (Jiang et al., 2020b). However, to our knowledge, all of these findings are based on small cohorts of samples. In the present study, by analyzing a large cohort of samples, we actually constructed an antibody response landscape of the SARS-CoV-2 proteome. This landscape extends our knowledge of the interaction between SARS-CoV-2 and the immune system. However, due to the difficulty of protein preparation, there are still some proteins that are missing on the SARS-CoV-2 proteome microarray, such as ORF8, which has recently been reported to be able to elicit a strong antibody response (Hachim et al., 2020). Some of these missing proteins will be added when we update the microarray.
Comparison of the IgG responses among the antigenic proteins revealed the possibility that the antibodies generated against the non-structural/accessory proteins are not independent of one another, which means for a patient who is positive for one protein tends to have a significant chance to be positive for other proteins. This is expected since these proteins may be simultaneously exposed or not to the immune system. Interestingly, high correlations were shown between some particular proteins, likely revealing the high associations of these proteins during infection and disease progression. For instance, it is known that RdRp, NSP7, and NSP8 form a complex for replication and translation of the viral genome. Correspondingly, high correlations of their elicited antibody responses, particular for NSP8 and RdRp, were observed. We also observed high correlations between NSP2 and NSP16, suggesting that the two proteins might associate in vivo.
The functions of the antibodies to non-structural/accessory proteins are still largely unknown. Our results reveal that the antibody levels are more associated with disease severity, particularly with the final outcome. These findings imply that the antibodies against non-structural/accessory proteins may play more important roles, which are worth further and in-depth investigation. One concern about the antibody against S protein is the possible ADE (antibody-dependent enhancement) (Wu et al., 2020d), which causes uncontrolled release of proinflammatory cytokines, such as interleukin (IL)-1, IL-6, IFN-γ, and tumor necrosis factor (TNF)-α (Liu et al., 2019; Ragab et al., 2020). Meanwhile, the severity of COVID-19 is highly associated with cytokine release syndrome or cytokine storm (Mangalmurti and Hunter, 2020; Ragab et al., 2020). One possible role of these antibodies against the non-structural/accessory proteins might be to trigger production of more cytokines when they bind the released antigens from the infected cells.
The duration of the protective antibodies in patients is still controversial (Gudbjartsson et al., 2020; Ibarrondo et al., 2020). Our data reveal that there is no significant decline of IgG antibodies against S or N protein for mild and severe survivors within 60 days after symptom onset. Also, the IgG antibodies against non-structural/accessory proteins rapidly decline when they reach the plateau about 20 days after symptom onset. This may be explained by the gradual decline of virus load, which is usually detectable about 20 days after symptom onset (Azkur et al., 2020; He et al., 2020). A recently published study showed that ORF3b can elicit antibodies and that the antibodies can last for 2–3 months (Hachim et al., 2020). In fact, for most of the patients, a clear trend of decline is observed. The decline of the antibody response might be associated with a decrease of the protein expression level of these proteins when the viral replication is inhibited; however, it cannot fully explain the distinct patterns of antibody responses between the structural and non-structural/accessory proteins. The duration of an antibody is largely dependent on the type of the corresponding B cells or antibody-secreting plasma cells, either long-lived or short-lived (Cyster and Allen, 2019; Nutt et al., 2015). The clear short lifetime of the antibodies against non-structural/accessory proteins might due to the suppressed production of long-lived B cells or the tendency to generate short-lived B cells with an unknown mechanism (Nutt et al., 2015). It seems that there are two distinct mechanisms through which the proteins of SARS-CoV-2 elicit host humoral immune responses. (1) The viral particle is involved as an antigen resource, specifically S and N proteins, which elicit potent antibody responses and tend to generate long-lived B cells. These antibodies mainly play a protective role. (2) The infected cell is involved as an antigen resource with non-structural/accessory proteins, which elicit weaker antibody responses and tend to be suppressed to generate long-lived B cells. These antibodies might be stronger to induce cytokines to contribute to more severe outcomes. However, this hypothesis should be confirmed by further studies.
In contrast to the antibody levels of S and N proteins that are stable for survivors, we observed the overall antibody levels start to decline in non-survivors at about 10 days before death. This observation implies the possible protective function of these antibodies in patients, and a collapse of humoral immunity might occur for most patients immediately before death. Further studies are needed to confirm this observation with more samples.
Taken together, we revealed a comprehensive antibody landscape against the SARS-CoV-2 proteome. The results were assured by a large cohort of 2,360 COVID-19 sera. Distinct characteristics of the antibodies against non-structural/accessory proteins and structural proteins were shown for the first time, with regard to patterns of antibody responses, associations with severity/outcome, and the dynamics. We strongly think that the antibody landscape revealed in this study will facilitate a deeper understanding of immunity related to SARS-CoV-2, predict the final outcome, may provide potential biomarkers for precise monitoring of COVID-19 progression, and may guide the development of effective vaccines.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cy3-Goat Anti-Human IgG | Jackson ImmunoResearch | Cat# 109-165-008 |
| Alexa Fluor 647 Donkey Anti-Human IgM | Jackson ImmunoResearch | Cat# 709-605-073 |
| Anti-Human IgG | Sigma | Cat# I2136 |
| Anti-Human IgM | Sigma | Cat# I2386 |
| Anti-His Antibody | Merck Millipore | Cat# 05-949 |
| Bacterial and virus strains | ||
| E. coli BL21 orf3a expression strain | This paper | N/A |
| E. coli BL21 orf3b expression strain | This paper | N/A |
| E. coli BL21 orf7b expression strain | This paper | N/A |
| Biological samples | ||
| Human Sera | This paper | Table S1 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike S1 subunit | Hangzhou Bioeast | Cat# SC2S302 |
| SARS-CoV-2 Nucleocapsid protein | VACURE Biotechnology | Cat# AG-PL-2101 |
| SARS-CoV-2 Nucleocapsid protein | Our Lab | N/A |
| SARS-CoV-2 Nucleocapsid protein (C terminus) | Healthcode | Cat# PROTN_nCoVN-CterHG01000 |
| SARS-CoV-2 Nucleocapsid protein (N terminus) | Healthcode | Cat# PROTN_nCoVN-NterHG01000 |
| SARS-CoV-2 NSP1 | Our Lab | N/A |
| SARS-CoV-2 NSP2 | Healthcode | Cat# PROTN_nCoVNSP2HG01000 |
| SARS-CoV-2 NSP4 | Our Lab | N/A |
| SARS-CoV-2 NSP5 | Healthcode | Cat# PROTN_nCoV3ClpHG01000 |
| SARS-CoV-2 NSP7 | Our Lab | N/A |
| SARS-CoV-2 NSP8 | Our Lab | N/A |
| SARS-CoV-2 NSP9 | Our Lab | N/A |
| SARS-CoV-2 NSP10 | Our Lab | N/A |
| SARS-CoV-2 RdRp | A gift from H. Eric Xu, Shanghai Institute of Materia Medica | N/A |
| SARS-CoV-2 NSP14 | Healthcode | Cat# PROTN_nCoVNSP14HG01000 |
| SARS-CoV-2 NSP15 | Healthcode | Cat# PROTN_nCoVNdUHG01000 |
| SARS-CoV-2 NSP16 | Healthcode | Cat# PROTN_nCoVOMTHG01000 |
| SARS-CoV-2 ORF-3a | Our Lab | N/A |
| SARS-CoV-2 ORF-3b | Our Lab | N/A |
| SARS-CoV-2 ORF6 | Our Lab | N/A |
| SARS-CoV-2 ORF-7b | Our Lab | N/A |
| SARS-CoV-2 ORF-9b | Our Lab | N/A |
| SARS-CoV-2 E-protein | Healthcode | Cat# PROTN_nCoVEHG01000 |
| BSA | Yeasen Biotech | Cat# B27371 |
| Ni2+ Sepharose beads | Senhui Microsphere Technology | Cat# 11-0010 |
| GST-Sepharose beads | Senhui Microsphere Technology | Cat# 12-0010 |
| Recombinant DNA | ||
| Plasmid: SARS-CoV-2 ORF3a | This paper | N/A |
| Plasmid: SARS-CoV-2 ORF3b | This paper | N/A |
| Plasmid: SARS-CoV-2 ORF7b | This paper | N/A |
| Deposited data | ||
| Raw data of the protein microarray | This paper | http://www.proteinmicroarray.cn (PMD: PMDE259) |
| Structure of RdRp, NSP8 and NSP7 complex | Yin et al., 2020 | PDB: 7BV1 |
| SARS-CoV-2 gene sequences | GenBank | GenBank: MN_908947.3 |
| Software and algorithms | ||
| SPSS | IBM | https://www.ibm.com/cn-zh/analytics/spss-statistics-software |
| Pymol | Pymol | https://pymol.org/2/ |
| Pheatmap package | R | https://cran.r-project.org/web/packages/pheatmap/index.html |
| Other | ||
| PATH protein microarray slides | Grace Bio-Labs | Cat# 805025 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact, Sheng-ce Tao (taosc@sjtu.edu.cn).
Materials availability
All unique/stable reagents generated in this study are available from the Lead contact with a completed Materials Transfer Agreement.
Data and code availability
The SARS-CoV-2 proteome microarray data are deposited on Protein Microarray Database (PMD) under the accession number PMDE259 (PMD: PMDE259) (http://www.proteinmicroarray.cn/index.php/experiment/detail?experiment_id=259). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.
Experimental model and subject details
Patients and samples
The study was approved by the Ethical Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (ITJ-C20200128). Written informed consent was obtained from all participants enrolled in this study. All patients with COVID-19 were laboratory confirmed and hospitalized during the period from 25 January 2020 and 28 April 2020. A total of 2,360 sera from 783 patients with COVID-19 were collected (Table S1). For the 756 patients whose samples were used to build the antibody landscape, 377 are males and 379 are females and the average age is 61.4. Sera of the control group from healthy donors, patients with lung cancer, patients with autoimmune diseases or other diseases were collected from Ruijin Hospital, Shanghai, China or Tongren Hospital, Shanghai, China. The negative reference samples were from National Institutes for Food and Drug Control. All the samples were stored at −80°C until use.
Cell lines
E.coli BL21 strain was used both for vector construction and protein expression. The bacterial strain was cultured in LB (Luria-Bertani) medium at 37°C. More information can be found in the following Method details.
Method details
Construction of expression vectors and protein preparation
In addition to the protein library of SARS-CoV-2 we previously constructed (Jiang et al., 2020b), three more proteins, i. e., ORF3a, ORF3b and ORF7b, were prepared according to the same methods. Briefly, the original sequences of the proteins were downloaded from GenBank (Accession number: MN908947.3). The genes were optimized and synthesized by Sangon Biotech. (Shanghai, China) followed by being cloned into pET32a or pGEX-4T-1 vector and transformed into E. coli BL21 strain to construct the transformants.
The recombinant proteins were expressed in E. coli BL21 by growing cells in 200 mL LB medium to an A600 of 0.6 at 37°C. Protein expression was induced by the addition of 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) before incubating cells overnight at 16°C. For the purification of 6xHis-tagged proteins, cell pellets were re-suspended in lysis buffer containing 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole (pH 8.0), then lysed by a high-pressure cell cracker (Union-biotech, Shanghai, China). Cell lysates were centrifuged at 12,000 × g for 20 min at 4°C. Supernatants were purified with Ni2+ Sepharose beads (Senhui Microsphere Technology, Suzhou, China), then washed with lysis buffer and eluted with buffer containing 50 mM Tris-HCl pH 8.0, 500 mM NaCl and 300 mM imidazole pH 8.0. For the purification of GST-tagged proteins, cells were harvested and lysed by a high-pressure cell cracker in lysis buffer containing 50mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM DTT. After centrifugation, the supernatant was incubated with GST-Sepharose beads (Senhui Microsphere Technology, Suzhou, china). The target proteins were washed with lysis buffer and eluted with 50mM Tris-HCl, pH 8.0, 500 mM NaCl, 1mM DTT, 40 mM glutathione. The purified proteins were analyzed by SDS-PAGE followed by western blotting using an anti-His antibody (Merck Millipore, USA, Cat#05-949) and Coomassie brilliant blue staining.
Protein microarray fabrication
The SARS-CoV-2 proteome microarray used this study is a updated version of the original one (Jiang et al., 2020b). Four more proteins were added. Except for ORF3a, ORF3b and ORF7bthe 4th protein, RdRp was provided by H. Eric Xu (Yin et al., 2020). The protein microarray was fabricated as previously described (Jiang et al., 2020b). Briefly, the proteins with indicated concentrations, along with the negative (GST, Biotin-control and eGFP) and positive controls (Human IgG, Human IgM and ACE2-Fc), were printed in quadruplicate on PATH substrate slide (Grace Bio-Labs, Oregon, USA) to generate identical arrays in a 2 × 7 subarray format using Super Marathon printer (Arrayjet, UK). The microarrays were used for serum profiling as described previously with minor modifications19. Protein microarrays were stored at −80°C until use.
Microarray-based serum analysis
A 14-chamber rubber gasket was mounted onto each slide to create individual chambers for the 14 identical subarrays. The microarray was used for serum profiling as previously described (Li et al., 2020) with minor modifications. Briefly, the arrays stored at −80°C were warmed to room temperature and then incubated in blocking buffer (3% BSA in 1 × PBS buffer with 0.1% Tween 20) for 3 h. Serum samples were diluted 1: 200 in PBS containing 0.1% Tween 20, added with 0.5 mg/mL-1 total E. coli lysate. A total of 200 μL of diluted serum or buffer only was incubated with each subarray overnight at 4°C. The arrays were washed with 1 × PBST and the signals were readout by incubating with Cy3-conjugated goat anti-human IgG and Alexa Fluor 647-conjugated donkey anti-human IgM (Jackson ImmunoResearch, PA, USA). These two fluorescent conjugated antibodies were diluted 1: 1,000 in 1 × PBST and incubated at room temperature for 1 h. The microarrays were then washed with 1 × PBST, dried by centrifugation at room temperature, scanned by LuxScan 10K-A (CapitalBio Corporation, Beijing, China) with the parameters set as 95% laser power/ PMT 550 and 95% laser power/ PMT 480 for IgM and IgG, respectively. The fluorescent intensity data was extracted by GenePix Pro 6.0 software (Molecular Devices, CA, USA). A pool of 50 randomly selected patient sera was used as a standard positive control. Block #14 of each slide was incubated with the positive control. Data normalization among slides was performed by a linear method with the data from the positive control. Specifically, a normalization factor for each slide was calculated by a liner regression function of the signals of the positive control of the given slide with the averaged signals of all slides, and then the signals of all the proteins from the slide were divided by the factor.
Quantification and statistical analysis
Signal Intensity was defined as the median of the foreground subtracted by the median of background for each spot and then the quadruplicate spots were averaged for each protein. IgG and IgM data were analyzed separately. Pearson correlation coefficient between two proteins or indicators and the corresponding p-value was calculated by SPSS software under the default parameters. Cluster analysis was performed by pheatmap package of R (Kolde, 2015). To calculate the positive rate of antibody response for each protein, mean + 2 x standard deviation (SD) of the control sera were used to set the threshold. The statistical details of experiments as well as the p-values can be found in the figures or in the figure legends.
Acknowledgments
We thank Dr. Daniel M. Czajkowsky for English editing and critical comments. We thank Prof. H. Eric Xu (Shanghai Institute of Materia Medica) for providing RdRp protein and Dawei Shi from the National Institutes for Food and Drug Control (Beijing, China) for providing negative reference control samples. We also thank Healthcode Co., Ltd., Hangzhou Bioeast Biotech Co., Ltd., and VACURE Biotechnology Co., Ltd. for providing reagents. This work was partially supported by the National Key Research and Development Program of China (grant no. 2016YFA0500600) and the National Natural Science Foundation of China (31970130, 31600672, 31670831, 31370813, 31900112, and 21907065).
Author contributions
S.-c.T. and X.F. developed the conceptual ideas and designed the study. Y.L., Z.X., Q.L., D.-y.L., M.-l.M., B.Z., H.C., C.Y., J.-b.X., X.-n.W., Y.-x.Z., H.-n.Z., H.-w.J., H.Q., Y.Z., X.L., Z.Y., and S.-j.G. performed the experiments and data analysis. Z.S., F.W., H.H., Y.Z., X.L., Z.Y., H.S., J.W., and H.S. collected the sera samples. S.-C.T. and Y.L. wrote the manuscript with suggestions from other authors.
Declaration of interests
The authors declare no competing interests.
Published: June 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109391.
Supplemental information
References
- Azkur A.K., Akdis M., Azkur D., Sokolowska M., van de Veen W., Brüggen M.C., O’Mahony L., Gao Y., Nadeau K., Akdis C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster J.G., Allen C.D.C. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177:524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García L.F. Immune response, inflammation, and the clinical spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Norddahl G.L., Melsted P., Gunnarsdottir K., Holm H., Eythorsson E., Arnthorsson A.O., Helgason D., Bjarnadottir K., Ingvarsson R.F. Humoral immune response to SARS-CoV-2 in Iceland. N. Engl. J. Med. 2020;383:1724–1734. doi: 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.-P., Petric M., Campbell W., McGeer P.L. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324:251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.-W., Zhang H.-N., Meng Q.-F., Xie J., Li Y., Chen H., Zheng Y.-X., Wang X.-N., Qi H., Zhang J. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.W., Li Y., Zhang H.N., Wang W., Yang X., Qi H., Li H., Men D., Zhou J., Tao S.C. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020;11:3581. doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2015. pheatmap: Pretty heatmaps. R package version 1.07.https://rdrr.io/cran/pheatmap/ [Google Scholar]
- Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., Sauter D., Gifford R.J., Nakagawa S., Sato K., USFQ-COVID19 Consortium SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lei Q., Li Y., Hou H., Wang F., Ouyang Z., Zhang Y., Lai D. Antibody dynamics to SARS-CoV-2 in asymptomatic COVID-19 infections. Allergy. 2021;76:551–561. doi: 10.1111/all.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li C.-Q., Guo S.-J., Guo W., Jiang H.-W., Li H.-C., Tao S.-C. Longitudinal serum autoantibody repertoire profiling identifies surgery-associated biomarkers in lung adenocarcinoma. EBioMedicine. 2020;53:102674. doi: 10.1016/j.ebiom.2020.102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y. Vol. 2. 2021. pp. 99–112.e7. (Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:e123158. doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., Hu J.L., Xu W., Zhang Y., Lv F.J. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Mangalmurti N., Hunter C.A. Cytokine storms: Understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- Qiu M., Shi Y., Guo Z., Chen Z., He R., Chen R., Zhou D., Dai E., Wang X., Si B. Antibody responses to individual proteins of SARS coronavirus and their neutralization activities. Microbes Infect. 2005;7:882–889. doi: 10.1016/j.micinf.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., Sinai Immunology Review Project Immunology of COVID-19: Current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Yan R., Liu M., Liu Z., Wang Y., Luan D., Wu K., Song Z., Sun T., Ma Y. Antibody-dependent enhancement (ADE) of SARS-CoV-2 infection in recovered COVID-19 patients: Studies based on cellular and structural biology analysis. MedRxiv. 2020 doi: 10.1101/2020.10.08.20209114. [DOI] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SARS-CoV-2 proteome microarray data are deposited on Protein Microarray Database (PMD) under the accession number PMDE259 (PMD: PMDE259) (http://www.proteinmicroarray.cn/index.php/experiment/detail?experiment_id=259). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request. This paper does not report original code.