Abstract
Staphylococcus pseudintermedius is a zoonotic pathogen that can cause life-threatening infections in animals and humans. The study of methicillin-resistant S. pseudintermedius (MRSP) and its ability to produce biofilms is important to select the most suitable treatment. The prevalence and characteristics of S. pseudintermedius isolated from dogs admitted at the Veterinary Teaching Hospital, Prince of Songkla University, Thailand were assessed. Results showed that 28.30% (15/53) of the isolates were MRSP. Amplification of the mecA gene was observed in 93.33% (14/15) MRSP. Methicillin-resistant strains revealed co-resistant patterns against other antibiotics, including chloramphenicol, clindamycin, tetracycline, clarithromycin, ciprofloxacin, and trimethoprim. In this study, all bacterial isolates produced biofilms, while 90.55% of S. pseudintermedius isolates were strong or moderate biofilm producers. Most (45–60%) of the resistant strains were strong biofilm producers, while the correlation between biofilm production and antibiotic resistance was not statistically significant. This is the first study in southern Thailand to investigate the drug-resistant profile of S. pseudintermedius and its ability to form biofilm. The results will contribute to a better understanding of the emergence and prevalence of antimicrobial resistance in S. pseudintermedius.
Keywords: Staphylococcus pseudintermedius, methicillin, antibiotic resistance, biofilm
1. Introduction
Staphylococcus pseudintermedius is an opportunistic pathogen that causes wound infections, pyoderma, otitis externa, and endometritis in canines. The Staphylococcus intermedius group (SIG) includes Staphylococcus intermedius, S. pseudintermedius, and Staphylococcus delphini. S. pseudintermedius, found as a novel species in 2005, colonizes the skin, nose, and anus of domestic pets. SIG isolates from canine infections should be identified as S. pseudintermedius when tested by conventional methods [1,2]. Recently, S. pseudintermedius has gained increased attention, with many reports of serious invasive and opportunistic infections in humans [3]. In addition, antimicrobial resistance in S. pseudintermedius has also been observed, with increasing emergence of methicillin-resistant S. pseudintermedius (MRSP). A case report from a tertiary hospital in Sweden showed that elders were infected by MRSP, while in Thailand, S. pseudintermedius and MRSP were reported in pets and people who had been in contact with animals [4,5,6]. MRSP was also cultured from the grounds and exposed areas of a hospital environment [7]. S. pseudintermedius and MRSP were reported to show resistance to antibiotics [8,9,10], while resistance against tetracyclines, macrolides and lincosamides, chloramphenicol, aminoglycosides trimethoprim, fluoroquinolones, rifampicin, and fusidic acid was reported in S. pseudintermedius, with limited treatment options [11].
Biofilm is a virulence factor of S. pseudintermedius that promotes the adherence of bacteria to host surfaces. Biofilm formation boosts bacterial survival and growth [12,13,14]. Biofilm-related infections have been reported and investigated because the bacteria could be more tolerant to antibiotics compared to the equivalent planktonic forms and resistant to host immune responses [15,16]. Antibiotics often fail to penetrate biofilms due to the extracellular matrix, which limits the transport of antimicrobial agents. The ability of S. pseudintermedius to produce biofilms has been previously reported. In Japan, all S. pseudintermedius isolates were able to produce biofilm [17], while in South Korea, a biofilm-forming quantitative assay showed that all S. pseudintermedius isolates produced either strong or moderate biofilm [18].
In Thailand, the molecular epidemiology of S. pseudintermedius has been studied in different regions but not in southern areas [6,19]. This research assessed the prevalence and characteristics of S. pseudintermedius isolated from dogs admitted at the Veterinary Teaching Hospital, Prince of Songkla University, Thailand. S. pseudintermedius isolates were identified using a conventional biochemical method, while a genotypic method targeted the spsL gene. Antibiotic resistance patterns were also phenotypically and genotypically characterized, and all isolates were assessed for quantitative biofilm production. The correlation between antibiotic resistance patterns and the ability of S. pseudintermedius to produce biofilms was also evaluated.
2. Results
In this study, 53 isolates were collected from dogs admitted at the Veterinary Teaching Hospital, Prince of Songkla University. All isolates were identified using a conventional biochemical method, including a coagulase test, urease test, acetoin production test, polymyxin B resistance, and acid production from mannitol and glucose. For genotypic characterization, the PCR method was employed to confirm species of S. pseudintermedius by the detection of the gene encoding surface protein L (spsL) as a cell wall-anchored protein. A total of 53 isolates showed a positive band of 512 bp product size using gel electrophoresis.
Detection of methicillin resistance in S. pseudintermedius was conducted by the Kirby–Bauer disk diffusion method. Oxacillin was chosen as a representative of all β-lactam agents owing to its stability and no longer commercially available methicillin. All S. pseudintermedius isolates were tested for susceptibility to oxacillin and interpretation was performed based on CLSI guidelines [20]. Results showed that 15 (28.30%) isolates were resistant to oxacillin; these were characterized as MRSP. Amplification of the mecA gene was observed in 14 of 15 (93.33%) MRSP isolates, while 7 (18.42%) mecA gene-positive isolates were found in methicillin-susceptible S. pseudintermedius (MSSP) (Table 1).
Table 1.
Comparison of methicillin susceptibility obtained by the oxacillin disk diffusion method and mecA gene detection.
| Results Obtained by Oxacillin Disk Diffusion Method | mecA Gene Detection |
|---|---|
| MSSP (38/53; 71.70%) | 7/38 (18.42%) |
| MRSP (15/53; 28.30%) | 14/15 (93.33%) |
MSSP: methicillin-susceptible S. pseudintermedius; MRSP: methicillin-resistant S. pseudintermedius.
All isolates were tested for susceptibility to six antibiotic categories, including chloramphenicol, clindamycin, tetracycline, clarithromycin, ciprofloxacin, and trimethoprim-sulfamethoxazole, using the Kirby–Bauer disk diffusion method. Antimicrobial susceptibility testing of S. pseudintermedius determined that 11 (20.75%), 15 (28.30%), 15 (28.30%), 16 (30.19%), 17 (32.08%), and 20 (37.74%) isolates were resistant to chloramphenicol, trimethoprim, clarithromycin, clindamycin, ciprofloxacin, and tetracycline, respectively. Results from the disk diffusion method showed that all MRSP isolates were resistant to β-lactams and also to other antibiotics, including 8 (53.33%), 11 (73.33%), 11 (73.33%), 12 (80.00%), 14 (93.33%), and 15 (100.00%) isolates that were resistant to chloramphenicol, trimethoprim, clindamycin, clarithromycin, ciprofloxacin, and tetracycline, respectively. The oxacillin-resistant isolates were classified into seven drug-resistance patterns, as shown in Table 2.
Table 2.
Antibiotic resistance profile of methicillin-resistant and methicillin-susceptible S. pseudintermedius isolates.
| Antibiotic Resistance Profile | MRSP (n = 15 isolates) | |
| % Resistant | Number of Isolates | |
| C-DA-TE-CRT-CIP-SXT | 20 | 3 |
| C-DA-TE-CRT-CIP | 6.67 | 1 |
| C-DA-TE-CRT-SXT | 6.67 | 1 |
| C-TE-CRT-CIP | 20 | 3 |
| DA-TE-CRT-CIP-SXT | 26.67 | 4 |
| DA-TE-CIP-SXT | 13.33 | 2 |
| TE-CIP-SXT | 6.67 | 1 |
| Total | 100 | 15 |
| Antibiotic Resistance Profile | MSSP (n = 38 isolates) | |
| % Resistant | Number of Isolates | |
| C-DA-TE-CRT-CIP-SXT | 2.63 | 1 |
| C-DA-TE-CRT-SXT | 2.63 | 1 |
| C | 2.63 | 1 |
| DA-TE-CRT-CIP-SXT | 2.63 | 1 |
| DA-TE-CIP | 2.63 | 1 |
| DA | 2.63 | 1 |
| TE-SXT | 2.63 | 1 |
| Total | 18.41 | 7 |
C: Chloramphenicol; DA: Clindamycin; TE: Tetracycline; CRT: Clarithromycin; CIP: Ciprofloxacin; SXT: Trimethoprim-sulfamethoxazole.
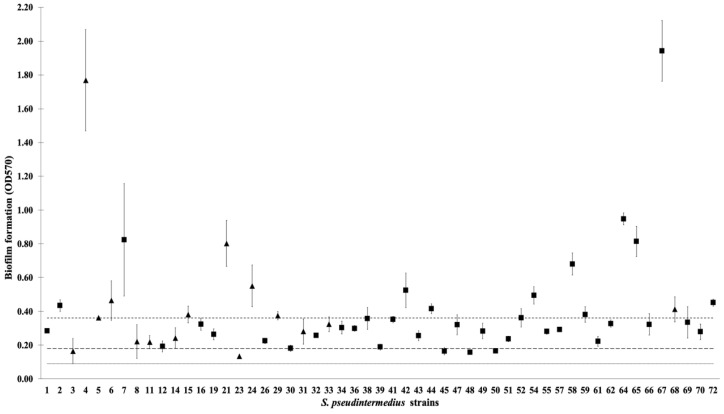
In this study, all S. pseudintermedius isolates produced biofilms. The mean OD values obtained by a quantitative biofilm-production assay are plotted in Figure 1. Most S. pseudintermedius isolates showed strong and moderate production of biofilms (Table 3). Twenty-two (41.50%) isolates were identified as strong biofilm producers, 26 (49.05%) were identified as moderate biofilm producers, while only 5 (9.43%) isolates were identified as weak biofilm producers. Based on antibiotic resistance, 34.21% (13/38) MSSP isolates were classified as strong biofilm producers, 57.89% (22/38) as moderate biofilm producers, and 7.89% (3/38) as weak biofilm producers. Out of the 15 MRSP isolates, 60.00% (9/15), 26.67% (4/15), and 13.33% (2/15) were strong, moderate, and weak biofilm producers, respectively.
Figure 1.
Biofilm formation values (OD570) of S. pseudintermedius isolates (n = 53) obtained by quantitative biofilm production assay. The OD cut-off used to distinguish weak and moderate biofilm producers from strong biofilm producers is 0.09 (dashed line). Categories: non-biofilm producers (OD ≤ 0.09), weak biofilm producers (0.09 < OD ≤ 0.18 (dashed line)), moderate biofilm producers (0.18 < OD ≤ 0.36 (dashed line)), and strong biofilm producers (0.36 < OD). (▲) MRSP; (■) MSSP.
Table 3.
Correlation between antibiotic resistant/susceptible strains and biofilm formation ability by the phenotypic method.
| Antibiotics | % Biofilm Producer (Isolates) | p Value | |||||
|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | ||||||
| Weak | Moderate | Strong | Weak | Moderate | Strong | ||
| OX | 13.33% (2) | 26.67% (4) | 60.00% (9) | 7.89% (3) | 57.89% (22) | 34.21% (13) | 0.12 |
| C | 9.09% (1) | 45.45% (5) | 45.45% (5) | 9.52% (4) | 50.00% (21) | 40.48% (17) | 0.96 |
| DA | 6.25% (1) | 43.75% (7) | 50.00% (8) | 10.81% (4) | 51.35% (19) | 37.84% (14) | 0.68 |
| TE | 10.00% (2) | 40.00% (8) | 50.00% (10) | 9.09% (3) | 54.55% (18) | 36.36% (12) | 0.58 |
| CRT | 13.33% (2) | 40.00% (6) | 46.67% (7) | 7.89% (3) | 52.63% (20) | 39.47% (15) | 0.66 |
| CIP | 11.76% (2) | 35.29% (6) | 52.94% (9) | 8.33% (3) | 55.56% (20) | 36.11% (13) | 0.39 |
| SXT | 6.67% (1) | 40.00% (6) | 53.33% (8) | 10.53% (4) | 52.63% (20) | 36.84% (14) | 0.54 |
OX: Oxacillin; C: Chloramphenicol; DA: Clindamycin; TE: Tetracycline; CRT: Clarithromycin; CIP: Ciprofloxacin; SXT: Trimethoprim-sulfamethoxazole.
Most (45–60%) of the resistant strains were strong biofilm producers, 26–45% were moderate biofilm producers, while 34–40% of susceptible strains were identified as strong biofilm producers and 50–57% were moderate biofilm producers (Table 3).
3. Discussion
S. pseudintermedius is an opportunistic pathogen that causes wound infections, pyoderma, otitis externa, and endometritis in canines. This pathogen has attracted increasing attention in recent years because invasive S. pseudintermedius infections have been reported in humans [3,21,22]. In this study, 53 isolates collected from dogs admitted at the Veterinary Teaching Hospital, Prince of Songkla University were identified by a conventional method. Confirmation of the strains was conducted by identifying genomic characteristics and the cell wall-anchored protein gene encoding S. pseudintermedius (spsL). Methicillin resistance is concerning because it confers resistance to all β-lactam groups. In Thailand, MRSP isolates have emerged with increasing incidence in clinical isolates, in particular, in pets and people who have been in contact with animals. A report from the Faculty of Veterinary Science Chulalongkorn University stated that 45% of S. pseudintermedius strains isolated from dogs between 2010 and 2012 were MRSP, while MRSP cultured from veterinarians who work there was confirmed at 76%, with 23% cultured from dog owners [6]. Phumthanakorn et al. reported 93 MRSP isolates collected from dogs (n = 43), humans (n = 18), and the environment (n = 32) [9]. Our study showed that MRSP was detected at 28.30%, similar to Kadlec et al., who recorded 28% MRSP isolated from dogs admitted at a small animal hospital between July 2006 and April 2013 [7]. Moreover, the Department of Dermatology at the Veterinary Hospital of Universidade Federal de Minas Gerais (Belo Horizonte, Brazil) indicated 29.55%, similar to a study at the Department of Veterinary Medicine and Animal Production, University of Naples (Naples, Italy) indicating 30.17% [23,24].
In mecA detection, 93.33% of the methicillin-resistant isolates contained the mecA gene, while 1 out of 15 isolates (6.67%) recorded an absence of the mecA gene. This result concurred with a previous report showing that 98.53% of oxacillin-resistant isolates were identified as carrying the mecA gene [25], while 9.76% of methicillin-resistant S. aureus (MRSA) isolates failed to detect the expression of the mecA gene [26]. In addition, 18.4% of methicillin-susceptible isolates contained the mecA gene. Chen et al. reported that 52/91 (57.1%) and 6/180 (3.3%) isolates classified as methicillin-susceptible S. aureus (MSSA) with oxacillin MICs were mecA positive [27]. In Korea, Cho et al. detected the mecA gene in 25 S. pseudintermedius isolates (19 in MRSP and 6 in MSSP isolates) [28]. Detection of the mecA gene was shown to be important when classifying methicillin-susceptibility testing in staphylococci, with the disk diffusion method giving high accuracy for methicillin-resistant strains. The results here suggest that antimicrobial susceptibility testing was important as well as screening for the mecA gene.
The antibiotic resistance patterns were classified into seven categories by the resistant profiles. According to antibiotic classification mechanisms, including cell wall synthesis, protein synthesis inhibitors, and nucleic acid inhibitors, all resistant organisms were resistant to protein synthesis inhibitors, including chloramphenicol, clindamycin, tetracycline, and clarithromycin. Most isolates were resistant to tetracycline and protein (30s subunit) synthesis inhibitors. In S. pseudintermedius, tetracycline resistance was thought to be associated with producing ribosome protective protein to pump out antibiotics by an efflux system [11]. Ciprofloxacin and trimethoprim were the second and third highest resistant antibiotics. In ciprofloxacin, DNA synthesis inhibitor resistance has been reported in S. pseudintermedius with mutations in the gyrA, gyrB, grlA, and grlB genes, and considered resistant to all fluoroquinolones [11], while in trimethoprim, nucleic acid inhibitors were not commonly used for therapeutic interventions. The resistance was associated with an alternative insensitive target (dihydrofolate reductase) in S. pseudintermedius [29], probably due to the use of alternative antibiotics without knowing the need for use. Therefore, antibiotics were used excessively. Our study demonstrated antimicrobial resistance patterns of S. pseudintermedius isolates cultured from domestic pets. Although rarely isolated from humans, many reports detailed serious invasive and opportunistic infections in patients that may lead to cases of drug resistance S. pseudintermedius infection.
Biofilms produced by S. pseudintermedius play an important role in the pathophysiology of infection and colonization [14,15]. In this study, all bacterial isolates produced biofilms, and more than 90% were classified as either strong or moderate biofilm producers. Meroni et al. showed that 94.5% of S. pseudintermedius isolates were biofilm producers, with 21 isolates strong biofilm producers, 29 moderate biofilm producers, and 19 weak biofilm producers [30]. In another study, S. pseudintermedius classified as strong, moderate, weak, and non-biofilm producers were 61%, 34%, 3%, and 2 %, respectively [14]. The ability to produce biofilms was different in each isolate because of several factors, such as the biofilm gene genotypic characteristic, physical interaction, and type of bacteria attachment surface. The association between biofilm production and antibiotic resistance has not been identified and correlated clearly with clonal types of drug-resistant bacteria [31,32,33]. In this study, the majority (45–60%) of the resistant strains were strong biofilm producers, with second place as moderate biofilm producers. By contrast, in susceptible strains, the majority (50–57%) were moderate biofilm producers, while strong biofilm producers were ranked below. However, variation between the OD values of resistant and susceptible strains was not statistically significant. In our study, a clear correlation was not found between antibiotic resistance and the ability to produce biofilms.
4. Materials and Methods
4.1. Isolation of Staphylococcus pseudintermedius
Fifty-three isolates were collected from dogs admitted at the Veterinary Teaching Hospital, Prince of Songkla University (Songkhla, Thailand). All animal protocols were approved by the Institutional Animal Care and Use Committee, Prince of Songkla University (EC 2562-05-041, 7 October 2019). The bacterial strains were kept in cryogenic vials (Biologix, Shandong, China) via the glycerol (VMR International, Radnor, PA, USA) method in Luria-Bertani (LB) broth (Becton Dickinson, Le Pont de Claix Cedex, France) and stored at −20 °C. All isolates were identified using a conventional biochemical method, including a coagulase test, urease test, acetoin production test, polymyxin B resistance, and acid production from mannitol and glucose.
4.2. Antimicrobial Susceptibility Testing
All isolates were grown on LB agar at 37 °C and shaken overnight. The bacteria were prepared by suspending colonies in LB broth. The log-phase bacteria were adjusted to an optical density 0.5 McFarland standard. All isolates were tested by the Kirby–Bauer disk diffusion method on Mueller-Hinton agar (Becton Dickinson, Le Pont de Claix Cedex, France) plates using oxacillin (30 μg), clarithromycin (15 μg), ciprofloxacin (5 μg), clindamycin (2 μg), trimethoprim-sulfamethoxazole (5 μg), tetracycline (30 μg), and chloramphenicol (30 μg) (OXOID, Thermo Fisher, Hampshire, UK). In this study, S. aureus ATCC 25923 was used as the reference strain, while all isolates were interpreted based on the Guidelines of the Clinical Laboratory and Standards Institute (CLSI, 2020) [20].
4.3. DNA Extraction
Pure colonies were picked from LB agar plates into 1 mL of LB broth and shaken at 37 °C overnight. The bacteria were pelleted using centrifugation at 8000× g for 3 min and washed three times in TE buffer. The cells were re-suspended in 500 µL of TE buffer and boiled using a heating box for 15 min. After immediate cooling on ice for 10 min, centrifugation was performed at 14,500× g for 10 min. The supernatant was transferred into a 1.5-mL PCR tube (Axygen, Union City, CA, USA). DNA was evaluated for purity with a NanoDrop 2000c Spectrophotometer at A260/280 (Thermo Scientific, Wilmington, DE, USA) and kept at −20 °C until required for use.
4.4. Gene Detection
The expression levels of spsL and mecA genes were evaluated by the polymerase chain reaction (PCR) method using a thermal cycler machine (Bio-Rad T100™ Thermal Cycler, Hercules, CA, USA). Sequences of the used primers are shown in Table 4. The reaction mixture for the PCR consisted of 2 µL of DNA extract in a total volume of 20 µL composed of 5 U/µL of Taq DNA polymerase (Invitrogen, San Diego, CA, USA), 10 µM of each primer, 2 mM dNTPs, 50 mM MgCl2, and 10X PCR buffer (Invitrogen, San Diego, CA, USA). The reaction mixture of spsL was thermally cycled under the following conditions: pre-incubation at 94 °C for 5 min, 25 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, extension for 1 min at 72 °C, and final extension for 5 min at 72 °C. The PCR condition of mecA was set as follows: pre-incubation at 94 °C for 15 min, 30 cycles of denaturation for 30 s at 94 °C, annealing for 1 min at 59 °C, extension for 1 min at 72 °C, and final extension for 10 min at 72 °C. The purified PCR products were separated by gel electrophoresis (1.5% agarose gel (SBIO by SmartScience, Pathum Thani, Thailand) in TBE buffer (VMR International, Radnor, PA, USA)). The gel was subsequently stained with GelRed® and visualized under ultraviolet light (Uvitec Ltd., Cambridge, UK).
Table 4.
Primer sequences of the spsL and mecA genes used for polymerase chain reaction.
4.5. Quantitative Biofilm Production Assay
For each bacterial strain, 20 µL of 0.5 McFarland standard adjusted-bacteria were filled in 96-well flat-bottom microplates with 180 µL of tryptic soy broth (TSB) containing 1% glucose and incubated at 37 °C for 24 h. Sterile TSB containing 1% glucose was used as a negative control, while S. aureus ATCC 29213 was selected as a positive control. After incubation, the wells were washed three times with 300 µL of phosphate buffer saline (PBS, pH 7.2) to remove free-floating or planktonic bacteria. The wells were fixed by absolute methanol for 20 min and dried overnight. Adherence of bacteria to the culture plate was stained with 2% Hucker’s crystal violet for 15 min and excess stain was washed with distilled water. After drying, 33% (v/v) glacial acetic acid was added to the wells and the absorbance of stained adherent bacteria was measured at 570 nm using a microplate reader. The optical density (OD) value was considered as the formation of biofilm mass on the surface of the culture plate. This assay was carried out for three independent experiments in triplicate. The following criteria were used for biofilm gradation in clinical isolates. The OD cut-off (ODc) was used for biofilm gradation as three standard deviations (SD) above the mean OD of the negative control (ODc = OD average of negative control + 3×SD of OD of negative control). All strains were classified based on adherence capabilities into non-biofilm producers (OD ≤ ODc), weak biofilm producers (ODc < OD ≤ 2ODc), moderate biofilm producers (2ODc < OD ≤ 4ODc), and strong biofilm producers (4ODc < OD) [36].
4.6. Statistical Analysis
Association between the antibiotic-resistant categories and quantitative biofilm production of S. pseudintermedius was assessed by the Chi-square test, with results expressed as means and SD of three independent experiments. Differences were considered statistically significant when p < 0.05.
5. Conclusions
Our findings showed that S. pseudintermedius isolates from dogs displayed MRSP, which showed co-resistance to antibiotics. All isolates evaluated in this study could produce biofilms and this may be one of the important virulence factors in the rapidly increasing emergence of S. pseudintermedius. Despite the majority of resistance strains being strong biofilm producers, the correlation between antibiotic resistance and biofilm production was not statistically significant. This is the first time S. pseudintermedius, its drug-resistant profile, and its ability to form biofilm has been investigated in southern Thailand. The results allow us to understand more about the emergence and prevalence of antimicrobial-resistant S. pseudintermedius.
Acknowledgments
The project was supported by Prince of Songkla University (Grant No. MET6302171S).
Author Contributions
Conceptualization, P.J. and D.S.; methodology, P.J. and D.S., validation, P.J. and D.S.; formal analysis, P.J. and D.S.; investigation, P.J., H.H. and D.S.; resources, T.S., S.I., J.S., W.W. and D.S.; data curation, P.J., J.C. and D.S.; writing—original draft preparation, P.J. and D.S.; writing—review and editing, V.T. and D.S.; supervision, D.S.; project administration, D.S.; funding acquisition, D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Prince of Songkla University (Grant No. MET6302171S) for D.S.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Devriese L.A., Vancanneyt M., Baele M., Vaneechoutte M., De Graef E., Snauwaert C., Cleenwerck I., Dawyndt P., Swings J., Decostere A., et al. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 2005;55:1569–1573. doi: 10.1099/ijs.0.63413-0. [DOI] [PubMed] [Google Scholar]
- 2.Becker K., Skov R.L., von Eiff C. Manual of Clinical Microbiology. ASM Press; Washington, DC, USA: 2015. Staphylococcus, Micrococcus, and other catalase-positive cocci; pp. 354–382. [Google Scholar]
- 3.Somayaji R., Priyantha M.A., Rubin J.E., Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: Report of 24 cases. Diagn. Microbiol. Infect. Dis. 2016;85:471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Starlander G., Börjesson S., Grönlund-Andersson U., Tellgren-Roth C., Melhus A. Cluster of infections caused by methicillin-resistant Staphylococcus pseudintermedius in humans in a tertiary hospital. J. Clin. Microbiol. 2014;52:3118–3120. doi: 10.1128/JCM.00703-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadlec K., Weiß S., Wendlandt S., Schwarz S., Tonpitak W. Characterization of canine and feline methicillin-resistant Staphylococcus pseudintermedius (MRSP) from Thailand. Vet. Microbiol. 2016;194:93–97. doi: 10.1016/j.vetmic.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Chanchaithong P., Perreten V., Schwendener S., Tribuddharat C., Chongthaleong A., Niyomtham W., Prapasarakul N. Strain typing and antimicrobial susceptibility of methicillin-resistant coagulase-positive staphylococcal species in dogs and people associated with dogs in Thailand. J. Appl. Microbiol. 2014;117:572–586. doi: 10.1111/jam.12545. [DOI] [PubMed] [Google Scholar]
- 7.Phumthanakorn N., Fungwithaya P., Chanchaithong P., Prapasarakul N. Enterotoxin gene profile of methicillin-resistant Staphylococcus pseudintermedius isolates from dogs, humans and the environment. J. Med. Microbiol. 2018;67:866–873. doi: 10.1099/jmm.0.000748. [DOI] [PubMed] [Google Scholar]
- 8.Zur G., Gurevich B., Elad D. Prior antimicrobial use as a risk factor for resistance in selected Staphylococcus pseudintermedius isolates from the skin and ears of dogs. Vet. Dermatol. 2016;27:468–e125. doi: 10.1111/vde.12382. [DOI] [PubMed] [Google Scholar]
- 9.Grönthal T., Eklund M., Thomson K., Piiparinen H., Sironen T., Rantala M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J. Antimicrob. Chemother. 2017;72:1021–1030. doi: 10.1093/jac/dkx086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva V., Oliveira A., Manageiro V., Caniça M., Contente D., Capita R., Alonso-Calleja C., Carvalho I., Capelo J.L., Igrejas G., et al. Clonal diversity and antimicrobial resistance of methicillin-resistant Staphylococcus pseudintermedius isolated from canine pyoderma. Microorganisms. 2021;9:482. doi: 10.3390/microorganisms9030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadlec K., Schwarz S. Antimicrobial resistance of Staphylococcus pseudintermedius. Vet. Dermatol. 2012;23:276–282, e255. doi: 10.1111/j.1365-3164.2012.01056.x. [DOI] [PubMed] [Google Scholar]
- 12.Flemming H.C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 13.Otto M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A., Walker M., Rousseau J., Weese J.S. Characterization of the biofilm forming ability of Staphylococcus pseudintermedius from dogs. BMC Vet. Res. 2013;9:93. doi: 10.1186/1746-6148-9-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain A., Agarwal A. Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J. Microbiol. Methods. 2009;76:88–92. doi: 10.1016/j.mimet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Pompilio A., De Nicola S., Crocetta V., Guarnieri S., Savini V., Carretto E., Di Bonaventura G. New insights in Staphylococcus pseudintermedius pathogenicity: Antibiotic-resistant biofilm formation by a human wound-associated strain. BMC Microbiol. 2015;15:109. doi: 10.1186/s12866-015-0449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arima S., Ochi H., Mitsuhashi M., Kibe R., Takahashi K., Kataoka Y. Staphylococcus pseudintermedius biofilms secrete factors that induce inflammatory reactions in vitro. Lett. Appl. Microbiol. 2018;67:214–219. doi: 10.1111/lam.13018. [DOI] [PubMed] [Google Scholar]
- 18.Han J.I., Yang C.H., Park H.M. Emergence of biofilm-producing Staphylococcus pseudintermedius isolated from healthy dogs in South Korea. Vet. Q. 2015;35:207–210. doi: 10.1080/01652176.2015.1096429. [DOI] [PubMed] [Google Scholar]
- 19.Tonpitak W., Sornklien C. Prevalence of methicillin-resistant Staphylococcus pseudintermedius isolates from healthy dogs and healthy cats in a small animal teaching hospital. Vet. Integr. Sci. 2014;12:95–105. [Google Scholar]
- 20.Clinical and Laboratory Standards Institute . M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI; Wayne, PA, USA: 2020. [Google Scholar]
- 21.Bhooshan S., Negi V., Khatri P.K. Staphylococcus pseudintermedius: An undocumented, emerging pathogen in humans. GMS Hyg. Infect. Control. 2020;15:1–11. doi: 10.3205/dgkh000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darlow C.A., Paidakakos N., Sikander M., Atkins B. A spinal infection with Staphylococcus pseudintermedius. BMJ Case Rep. 2017;2017 doi: 10.1136/bcr-2017-221260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Botoni L.S., Scherer C.B., Silva R.O., Coura F.M., Heinemann M.B., Paes-Leme F.O., Costa-Val A.P. Prevalence and in vitro susceptibility of methicillin-resistant Staphylococcus pseudintermedius (MRSP) from skin and nostrils of dogs with superficial pyoderma. Pesq. Vet. Bras. 2016;36:1178–1180. doi: 10.1590/s0100-736x2016001200006. [DOI] [Google Scholar]
- 24.Nocera F.P., Meroni G., Fiorito F., De Martino L., Martino P.A. Occurrence and antimicrobial susceptibility patterns of canine Staphylococcus pseudintermedius strains isolated from two different Italian university veterinary hospitals. Vet. Ital. 2020;56:263–269. doi: 10.12834/VetIt.2195.12897.1. [DOI] [PubMed] [Google Scholar]
- 25.Nisa S., Bercker C., Midwinter A.C., Bruce I., Graham C.F., Venter P., Bell A., French N.P., Benschop J., Bailey K.M., et al. Combining MALDI-TOF and genomics in the study of methicillin resistant and multidrug resistant Staphylococcus pseudintermedius in New Zealand. Sci. Rep. 2019;9:1271–1273. doi: 10.1038/s41598-018-37503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elhassan M.M., Ozbak H.A., Hemeg H.A., Elmekki M.A., Ahmed L.M. Absence of the mecA gene in methicillin resistant Staphylococcus aureus isolated from different clinical specimens in Shendi City, Sudan. Biomed. Res. Int. 2015;2015:895860. doi: 10.1155/2015/895860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F.J., Huang I.W., Wang C.H., Chen P.C., Wang H.Y., Lai J.F., Shiau Y.R., Lauderdale T.L. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J. Clin. Microbiol. 2012;50:1679–1683. doi: 10.1128/JCM.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho J.-K., Lee M.-R., Kim J.-M., Kim H.-D. Methicillin-resistant or susceptible Staphylococcus pseudintermedius isolates from dogs and cats. Korean J. Vet. Serv. 2016;39:175–181. doi: 10.7853/kjvs.2016.39.3.175. [DOI] [Google Scholar]
- 29.Sekiguchi J., Tharavichitkul P., Miyoshi-Akiyama T., Chupia V., Fujino T., Araake M., Irie A., Morita K., Kuratsuji T., Kirikae T. Cloning and characterization of a novel trimethoprim-resistant dihydrofolate reductase from a nosocomial isolate of Staphylococcus aureus CM.S2 (IMCJ1454) Antimicrob. Agents. Chemother. 2005;49:3948–3951. doi: 10.1128/AAC.49.9.3948-3951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meroni G., Soares Filipe J.F., Drago L., Martino P.A. Investigation on antibiotic-resistance, biofilm formation and virulence factors in multi drug resistant and non multi drug resistant Staphylococcus pseudintermedius. Microorganisms. 2019;7:702. doi: 10.3390/microorganisms7120702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senobar Tahaei S.A., Stájer A., Barrak I., Ostorházi E., Szabó D., Gajdács M. Correlation between biofilm-formation and the antibiotic resistant phenotype in Staphylococcus aureus isolates: A laboratory-based study in Hungary and a review of the literature. Infect. Drug Resist. 2021;14:1155–1168. doi: 10.2147/IDR.S303992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behzadi P., Urbán E., Gajdács M. Association between biofilm-production and antibiotic resistance in uropathogenic Escherichia coli (UPEC): An in vitro study. Diseases. 2020;8:17. doi: 10.3390/diseases8020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena P., Joshi Y., Rawat K., Bisht R. Biofilms: Architecture, resistance, quorum sensing and control mechanisms. Indian J. Microbiol. 2019;59:3–12. doi: 10.1007/s12088-018-0757-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maali Y., Martins-Simões P., Valour F., Bouvard D., Rasigade J.P., Bes M., Haenni M., Ferry T., Laurent F., Trouillet-Assant S. Pathophysiological mechanisms of Staphylococcus non-aureus bone and joint infection: Interspecies homogeneity and specific behavior of S. pseudintermedius. Front. Microbiol. 2016;7:1063–1072. doi: 10.3389/fmicb.2016.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegger Á., Andersen P., Kearns A., Pichon B., Holmes M., Edwards G., Laurent F., Teale C., Skov R., Larsen A. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin. Microbiol. Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 36.Stepanović S., Vuković D., Hola V., Di Bonaventura G., Djukić S., Cirković I., Ruzicka F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in article.