Abstract
Listeria monocytogenes is an increasing food safety concern throughout the produce supply chain as it has been linked to produce associated outbreaks and recalls. To our knowledge, this is the first systematic literature review to investigate Listeria species and L. monocytogenes prevalence, persistence, and diversity at each stage along the supply chain. This review identified 64 articles of 4863 candidate articles obtained from four Boolean search queries in six databases. Included studies examined naturally detected/isolated Listeria species and L. monocytogenes in fresh produce-related environments, and/or from past fresh produce associated outbreaks or from produce directly. Listeria species and L. monocytogenes were detected in each stage of the fresh produce supply chain. The greatest prevalence of Listeria species was observed in natural environments and outdoor production, with prevalence generally decreasing with each progression of the supply chain (e.g., packinghouse to distribution to retail). L. monocytogenes prevalence ranged from 61.1% to not detected (0.00%) across the entire supply chain for included studies. Listeria persistence and diversity were also investigated more in natural, production, and processing environments, compared to other supply chain environments (e.g., retail). Data gaps were identified for future produce safety research, for example, in the transportation and distribution center environment.
Keywords: food safety, environment, detection, identification, foodborne pathogen, contamination
1. Introduction
The genus Listeria contains 17 species, but Listeria monocytogenes (Lm) is the only known pathogenic strain [1]. Lm is a bacterial, intracellular parasite that causes approximately 1600 illnesses and 260 deaths annually within the United States [2]. Regulation from the U.S. Food and Drug Administration (FDA) has a zero tolerance for Lm in food due to the high mortality rate associated with listeriosis, and its undetermined infectious dose [3,4]. Lm is able to penetrate the blood-brain and placental barriers, increasing severity of disease, compared to other foodborne pathogens that predominantly infect the gastrointestinal tract [5]. Pregnant women, elderly, infants, and immunocompromised individuals are the most at risk populations for Listeria infections and complications [3].
Lm has historically been a concern in ready-to-eat (RTE) foods and soft cheeses, due largely to its ability to survive and grow at refrigeration temperatures, low pH, and high salinity. Lm can outcompete other organisms under these less hospitable conditions [6], and has resulted in outbreaks linked to hard-boiled eggs, deli meats, raw milk, and ice cream [7]. Fresh produce is increasingly being recognized as a potential vehicle for Lm contamination as it often does not undergo a complete microbial kill step (i.e., often consumed raw). Mitigation of Lm on fresh produce can be challenging as Lm is naturally found and survives in soils [8,9]. Additionally, Lm can adhere to surfaces and form biofilms, which allow Lm to be resistant to desiccation, acid, heat, and/or sanitizers/disinfectants [10,11]. The combination of Lm’s ability to tolerate a wide array of environmental conditions, combined with its relatively high mortality rate, make Lm a concern throughout each stage of the produce supply chain (e.g., production, retail).
There have been at least ten recognized outbreaks of listeriosis related to fresh produce, sprouts, and mushrooms since 1979 (Table 1). Lm has been linked to fresh produce-associated outbreaks from products including caramel apples, cantaloupes, stone fruits, and minimally processed vegetables [12,13,14,15,16,17]. Notably, one of the most severe listeriosis outbreaks in the U.S. was from contaminated whole cantaloupe [13,14]. This outbreak led to 147 cases across 28 states with 143 hospitalizations and 33 deaths, making it the deadliest produce-related foodborne illness outbreak in the U.S. [13,14]. Thus, the goal of this systematic literature review was to synthesize the published data on Lm and Listeria spp. prevalence, persistence, and diversity throughout each stage of the fresh produce supply chain. Objectives include (1) identifying sites and associated factors (e.g., meteorological, geographical) of greatest Listeria and Lm prevalence, (2) further clarifying the relationship between Listeria spp. as index organisms for Lm, and (3) determining research needs regarding fresh produce supply chain stages.
Table 1.
U.S. listeriosis outbreaks associated with fresh produce.
| Associated Food | Source of Contamination | Year | Number of States Affected | Cases | Hospitalizations | Deaths | Reference |
|---|---|---|---|---|---|---|---|
| Enoki mushrooms | TBD | 2020 | 17 | 36 | 30 | 4 | [18] |
| Frozen vegetables | Processing environment | 2016 | 4 | 9 | 9 | 3 | [19] |
| Packaged salad | Processing environment | 2016 | 9 | 19 | 19 | 1 | [20] |
| Caramel apples | Processing environment | 2014 | 12 | 35 | 34 | 7 | [16] |
| Mung bean sprouts | Production environment | 2014 | 2 | 5 | 5 | 2 | [21] |
| Peaches and nectarines | Packinghouse environment | 2014 | 4 | 2 | 2 | 1 | [17,22] |
| Cantaloupes | Processing environment | 2011 | 28 | 147 | 143 | 33 | [14] |
| Chopped celery | Processing environment | 2010 | 1 | 10 | 10 | 5 | [15] |
| Sprouts | Production environment | 2008 | Unknown | 20 | 16 | 0 | [23] |
| Raw celery, tomatoes, and lettuce | Unknown | 1979 | 1 | 20 | 20 | 5 | [23,24] |
2. Materials and Methods
2.1. Definitions and Scope
The broader genus of Listeria was included in this review as Listeria spp. are often used as index organisms for Lm [25]. Listeria spp. are defined as those species excluding Lm, with Lm reported separately unless otherwise noted. Here, in the reported review, we considered prevalence to be the occurrence of culturable Listeria in the natural or farm environment (e.g., soil, water), on non-food contact surfaces (NFCS) or food contact surfaces (FCS), or on fresh produce. Persistence was classified as prolonged or repeated detection of Listeria in an environment, or on fomites or fresh produce. Diversity was defined as species abundance and or genetic difference(s) within the Listeria genus and between species of Listeria. This review presents a panoramic view of the prevalence, persistence, and diversity of Listeria spp. and Lm throughout each stage of the produce supply chain. Stages of the produce supply chain were partitioned into seven main stages: (1) natural and outdoor production (e.g., farm), (2) packinghouse, indoor production, and processing, (3) transportation and distribution, (4) retail, (5) farmers’ market, (6) restaurant, and (7) domestic.
2.2. Literature Search
Searches were completed using Web of Science (beginning 1900), PubMed (beginning 1966), Food Science Source (beginning in 1941), PubAg (beginning in 1895), AGRICOLA (beginning in the 15th century), and CAB Abstracts and CAB Archives (beginning in 1973) through August 2020. Four separate Boolean searches were used: (1) Listeria AND prevalen* AND presen*, (2) Listeria AND persist*, (3) Listeria AND divers*, and (4) Listeria AND fresh AND produce. Boolean terminology was used for selected second and third terms to encompass truncations of each word. For example, “prevalen*” includes prevalence and prevalent. Searches were also restricted to English only and studies originating from U.S. based institutions or study locations. Book chapters, conference materials, and reports were excluded.
2.3. Study Selection
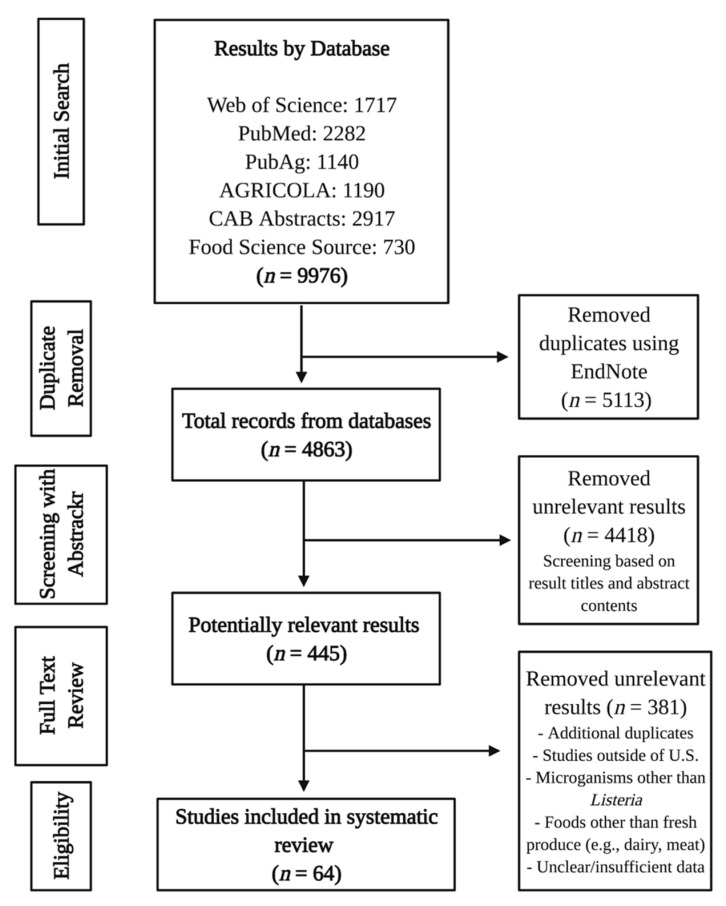
The study selection process was conducted by co-author Townsend. Results from database searches were exported to EndNote (Version X9, Clarivate, Philadelphia, PA, USA). Duplicate results were removed in EndNote using the “Find Duplicates” function after importing search results from each individual database. The remaining results were screened for relevance using a free, open-source web tool Abstrackr [26], which maintains a digital paper trail on screening decisions. Abstracts were rejected if they did not contain specified review terms (e.g., “Listeria,” “produce,” “fresh,” “fruit,” “prevalen”). Screening results from Abstrackr were downloaded as a CSV file containing the assigned Abstrackr (internal) ID, source ID, PubMed ID, keywords, abstract, title, journal, authors, tags, and screening decision for each study. Abstrackr assigned a score of -1 (rejected), 0 (unsure), or 1 (accepted) for the reviewer’s screening decisions. Studies were re-screened in Excel (Microsoft Corp., Redmond, WA, USA) to verify correct screening decisions through Abstrackr by examining titles and abstracts. Duplicates not identified through EndNote were also removed manually during re-screening. Studies receiving a score of 0 or “unsure” were reviewed using the full text if the title and abstract were not sufficient to gauge relevance. After re-screening, results were filtered to those with a score of 1 and were examined using the full text. Full text review evaluated whether the study included fresh produce or was directly related to the fresh produce supply chain stages. Exclusion characteristics included studies completed outside of the U.S. and focus on microorganisms other than Listeria. Experimental studies evaluating isolation and/or detection methods were not included if samples did not contain naturally occurring Listeria species from produce-related environments or Listeria isolates from fresh produce or from previous fresh produce-associated outbreaks. An overview of the systematic review process can be seen in Figure 1.
Figure 1.
Flow diagram illustrating the systematic literature review process.
2.4. Data Extraction and Synthesis
Data extraction and synthesis were completed by co-author Townsend. Studies were assessed individually, grouped by supply chain stage, and discussed by U.S. Department of Agriculture (USDA) Agriculture Research Service (ARS) region and chronological order. USDA ARS regions were chosen because of their distinction between climate and geography, and available natural resources and agricultural commodities and products within each region. Data regarding the study location(s), sampling site(s), sample types, duration, organisms of interest (Listeria spp. and/or Lm), prevalence (e.g., number of Listeria positive samples over total samples), and strain/serotype identification were extracted from the full texts. These data were entered manually into Excel. During full text review, studies were flagged for discussion if they contained data or evidence on Listeria diversity (e.g., strain/serotype data) and persistence (e.g., longitudinal studies).
3. Results
3.1. Included Study Characteristics
The initial primary search identified 9976 results. After duplicate removal, 4863 studies remained for screening. After analyzing result titles and abstracts in Abstrackr, 4418 results were removed, leaving 445 results for selection criteria evaluation. Sixty-four studies were considered relevant for review based on criteria. Of the 64 studies, 54 included prevalence data for Listeria spp. and Lm based on detection of these microorganisms from environmental samples. Included studies examined a broad variety of sample types. Sample types from natural and outdoor production environments included surface water (non-irrigation and irrigation), soil, compost (raw and treated), feces (wild and domestic animals), fresh produce, and drag swabs, as well as man-made materials, such as sidewalks and doors, in urban environments. Sample types from packinghouse, indoor production, and processing environments included NFCS and produce (e.g., leafy greens, stone fruits). All retail and farmers’ market environments included produce, such as leafy greens, sprouts, packaged salads, and mushrooms, with one study also examining NFCS and FCS in retail grocery departments. The only domestic-related study examined NFCS and FCS (e.g., refrigerators, sinks, dishcloths) in consumer homes. Most identified studies performed a microbial survey of various fresh produce-related locations, while a handful of studies examined methods validation, and modeling.
Relevant studies were organized by fresh produce supply chain and then by geographic region. Geographic regions approximately follow the USDA Agricultural Research Service (ARS) regions (Northeast, Southeast, Midwest, Plains, and Pacific West) with some deviation due to multiregional studies or studies with unspecified locations. Studies with sampling locations in states or areas within two or more geographical regions were classified as multiregional. Studies not pertaining to a specific geographic region, such as laboratory-based experiments, were included in a separate “Additional Relevant Studies” section. Summary information from identified studies that assessed the prevalence of Listeria spp. and Lm throughout the fresh produce supply chain are provided in Table 2. In Table 2, those studies with only Listeria spp. as target organisms examined all Listeria species prevalence (including Lm). However, those studies with Listeria spp. and Lm as target organisms exclude Lm from the Listeria spp. prevalence. The overall range of Listeria spp. and Lm prevalence in each supply chain stage can be found in Table 3.
Table 2.
Summary of Listeria spp. and L. monocytogenes prevalence throughout the fresh produce supply chain. All Listeria species (including L. monocytogenes) are considered when Listeria spp. are the only target organisms within a study. However, Listeria spp. exclude L. monocytogenes when both are included in target organisms within a study. Prevalence is given in number of positive samples divided by the total number of samples with corresponding percentage in parentheses.
| Geographical Location(s) | Sampling Site(s) | Study Duration | Type of Sample(s) | Target Organism(s) | Presence (Yes/Not Detected) | Prevalence | Positive Sample Type(s) | Strain/Serotype | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Natural Environment and Outdoor Production | |||||||||
| Colorado | Wilderness areas (n = 5) | Two years | Soil, water, sediment, surface soil, and wildlife feces (n = 572) | Listeria spp. (excludes Lm) | Yes | 19/572 (3.32) | All sample types | L. welshimeri and undetermined strains | [27] |
| Listeria monocytogenes | Yes | 3/572 (0.52) | Feces and water | 1/2a, 1/2b, 3a, 3b, or 7 | |||||
| Washington, Oregon, and California | Compost facilities (n = 94) | Three weeks | Market-ready, organic compost (n = 94) | Listeria spp. (all) | Yes | 22/47 (46.81) | Compost from OR and CA | Not identified | [28] |
| Arizona, California, Georgia, Kentucky, Maryland, New York, North Carolina, South Carolina, Tennessee | Farms, outside locations, commercial operations | Four years | Organic fertilizers (n = 103) | Listeria monocytogenes | ND | N/A | N/A | N/A | [29] |
| New York | Natural environments (n = 5) | Two years | Composite soil, drag swab, water, and wildlife/domestic animal feces (n = 1322) | Listeria spp. (excludes Lm) | Yes | 186/734 (25.34) | All sample types | L. innocua, L. seeligeri, L. welshimeri, and L. marthii | [25] |
| Listeria monocytogenes | Yes | 59/734 (8.04) | Not specified | Not identified | |||||
| California | Watershed areas near produce field (n = 30) | Two years | Surface water (n = 1405) | Listeria monocytogenes | Yes | ~ 604/1405 (43) | Surface water | 1/2a, 1/2b, 3a, 4d, and 4e | [30] |
| California | Watershed areas near produce field (n = 14) | Ten months | Surface water | Listeria monocytogenes | Yes | Culture positive: 22/36 (61.1) PCR positive: 21/36 (58.3) |
Surface water | Not identified | [31] |
| California | Watershed areas near produce field (n = 30) | Three months | Surface water (n = 206) | Listeria monocytogenes | Yes | 62/206 (30.1) | Surface water | 1/2a, 1/2b, and 4b | [32] |
| California | Watershed areas (n > 30) | Two years | Surface water (n = 860) | Listeria monocytogenes | Yes | 381/860 (44.3) | Surface water | Not identified | [33] |
| Ohio | Experimental treatments (n = 5) with varying amendments (sawdust, straw, and water) to manure | Sampling occurred over 57-day period | Compost testing occurred on days 0, 14, 28, and 56 | Listeria spp. (all) | Yes | Not specified | Day 0: Five compost treatments. Day 3: Three compost treatments. Day 14: One compost treatment |
Not identified | [34] |
| Pennsylvania | Six parallel plots (5 m × 2 m) | Not specified | Irrigation water and spinach | Listeria spp. (all) | ND | N/A | N/A | N/A | [35] |
| Maine | Randomized complete block split plot design with five replications completed in duplicate | Two years | Liquid dairy, pig, chicken, cow manure, and potatoes grown with and without liquid dairy manure | Listeria monocytogenes | Yes | Total: 3/24 (12.5) Liquid dairy: 2/6 (33.3) Cow: 1/6 (16.7) |
Liquid dairy and cow manure | N/A | [36] |
| Maryland | Mixed produce and dairy farm; 12 sampling sites | 14 months | Cow feces, cow feed, cow drinking water, bird feces, bird gathering areas, raw liquid manure, water from lagoon, raw separated solids, partially composted material, fully composed material, surface water, and soil from vegetable production area and cow pasture (n = 159) | Listeria spp. (excludes Lm) | Yes | 8/159 (5.03) | Cow feed, cow feces, raw separated solid, windrow compost, finished compost, bird feces, pasture soil | Not identified | [37] |
| Listeria monocytogenes | Yes | 2/159 (1.26) | Cow feed and pasture soil | Not identified | |||||
| Rio Grande Valley and Texas | Cabbage farms with packing sheds and separate packing sheds (n = 6) | Seven months | Cabbage (n = 425), water (n = 205), and environmental (n = 225) | Listeria spp. (excludes Lm) | Yes | 24/855 (2.81) | All sample types | L. grayi, L. innocua, L. ivanoii, and L. welshimeri | [38] |
| Listeria monocytogenes | Yes | 26/855 (3.04) | All sample types | Not identified | |||||
| Maryland | Non-tidal freshwater creek | One year | Creek water, soil, radishes, and kale | Listeria monocytogenes | Yes | 0.04 and 0.07 MPN/L | Creek water | Not identified | [39] |
| New York | Spinach fields (n = 2) | Seven weeks | Feces, leaves, soil, and surface water (n = 1492) | Listeria spp. (excludes Lm) | Yes | 74/1492 (4.96) | All sample types | Not identified | [40] |
| Listeria monocytogenes | Yes | 130/1492 (8.71) | Not identified | ||||||
| New York | Natural (n = 4) and urban (n = 4) sites | Two years | Soil, vegetation, surface water, floors, sidewalks, and human contact surfaces (n = 1805) | Listeria monocytogenes | Yes | Urban: 67/898 (7.5) Natural: 13/907 (1.4) |
Urban: soil, vegetation, water, sidewalk/floor, and human contact surfaces Natural: soil, vegetation, and water |
Not identified | [41] |
| New York | Natural (n = 4) and urban (n = 4) sites | Two years | Soil, vegetation, surface water, floors, sidewalks, and human contact surfaces (n = 1805) | Listeria spp. (excludes Lm) | Yes | 362/1805 (20.1) | Variety from natural and urban sites | L. marthii, L. innocua, L. seeligeri, and L. welshimeri | [42] |
| Listeria monocytogenes | Yes | 80/1805 (4.43) | Not identified | ||||||
| Arizona and New York | Watershed areas (n = 9) | Eleven months | Surface water (n = 1053) | Listeria spp. (excludes Lm) | Yes | AZ: 0/76 (0) NY: 58/257 (22.57) |
Streams in NY | L. booriae, L. innocua, L. marthii, L. seeligeri, and L. welshimeri | [43] |
| Listeria monocytogenes | Yes | AZ: 3/76 (3.95) NY: 30/257 (11.67) |
Canals in AZ and streams in NY | Not identified | |||||
| Arizona | Five sites in two wastewater treatment plants | 20 months | Reclaimed and return flow water (n = 28) | Listeria monocytogenes | ND | N/A | N/A | N/A | [44] |
| Mid-Atlantic U.S. | Convention water sources (n = 6) | Three years | Tidal freshwater river (n = 34), non-tidal freshwater creek (n = 32), reclaimed water holding pond (n = 25), pond water sites (n = 69), and produce wash water (n = 10) | Listeria monocytogenes | Yes | 53/170 (31.18) | All sample types | Not identified | [45] |
| Washington | Red raspberry field with individual plots (22.86 m × 3.05 m; n = 4) with buffer rows between treatment plots; completed in duplicate | Two years | Fertilizer, soil, foliar, and raspberry fruit | Listeria monocytogenes | ND | N/A | N/A | N/A | [46] |
| New York | Produce farms (n = 5) | 27 months | Soil, water (engineered and surface), feces, and drag swabs (n = 588) | Listeria monocytogenes | Yes | 88/588 (15.0) | All sample types except for engineered water | Not identified | [47] |
| New York | Produce farms (n = 21) | Five weeks | Fields (n = 263) and environmental samples (soil, drag swab, and water; n = 600) | Listeria monocytogenes | Yes | Field: 46/263 (17.5) Soil: 30/263 (11) Drag: 21/263 (8) Water: 22/74 (30) |
All sample types | Nine allelic types representing lineages I, II, and IIIa | [48] |
| New York | Produce farms (n = 10) | Six weeks | Terrestrial, water, and fecal (n = 124) | Listeria spp. (excludes Lm) | Yes | 24/124 (19.35) | All sample types | L. seeligeri, L. welshimeri, and L. innocua | [49] |
| Listeria monocytogenes | Yes | 28/124 (22.58) | All sample types | Not identified | |||||
| Packinghouse, Indoor Production, and Processing | |||||||||
| Pennsylvania | Small scale mushroom production facility (n = 1) |
Two months | NFCS, such as shovels, drains, doors, floors, conveyor belts, brooms, dust pans, etc. (n = 184) | Listeria spp. (excludes Lm) | Yes | 26/184 (14.13) | Phase I composting, phase II composting, tray filling line, and growing rooms | L. innocua, L. welshimeri, and L. grayi | [50] |
| Listeria monocytogenes | Yes | 3/184 (1.63) | Phase I composting | Not identified | |||||
| Maryland | Organic farms (n = 7) | Two years | Produce (tomatoes, leafy greens, peppers, cucumbers, etc.), well water, and surface water (n = 206) | Listeria monocytogenes | ND | N/A | N/A | N/A | [51] |
| Southern U.S. | Farms (n = 13) and packing sheds (n = 5) | 19 months | Produce (leafy greens, herbs, and cantaloupe; n = 398) | Listeria monocytogenes | ND | N/A | N/A | N/A | [52] |
| Northeast U.S. | Apple and other tree fruit packinghouses (n = 3) | Six months | NFCS (n = 117) | Listeria monocytogenes | Yes | 66/117 (56.41) | Washing, drying, and waxing areas of all facilities | Not identified | [53] |
| Southern U.S. | Packinghouses (n = 8) | 14 months | Leafy greens (n = 109), herbs (n = 165), and cantaloupe (n = 36) | Listeria monocytogenes | Yes | Leafy greens: 3/43 (6.98) | Cabbage | Not identified | [54] |
| Southeastern U.S. | Packinghouses (n = 11) | Nine months | NFCS, such as forklift wheels, drains, dump tank legs, cold room floors, etc. | Listeria spp. (excludes Lm) | Yes | 52/1588 (3.27) | Drains, cold storage rooms, wet NFCS, mobile NFCS, dry NFCS, and outside packing/handling area | L. innouca, L. marthii, L. seeligeri, and L. welshimeri | [55] |
| Listeria monocytogenes | Yes | 60/1588 (3.78) | Not identified | ||||||
| California | Stone fruits (n = 105; from seven lots) | Not specified | White nectarines (n = 30), yellow nectarines (n = 30), white peaches (n = 30), and yellow peaches (n = 15) | Listeria monocytogenes | Yes; 11.3 CFU/fruit (geometric mean) | Total: 56/105 (53.3) Nectarines: 15/60 (25) Peaches: 41/45 (91.1) | All sample types | IVb-v1 and 1/2b | [17] |
| Four U.S. states | Packinghouses (n = 3) and fresh-cut facilities (n = 5) | One year | Sponge samples (n = 2014) | Listeria spp. (excludes Lm) | Yes | Packinghouse: 5/252 (2) to 8/171 (4.7) Fresh-cut: 0/249 (0) to 5/325 (1.5) |
Zones 2 and 3 for packinghouse and only zone 3 for fresh-cut | Not identified | [56] |
| Listeria monocytogenes | Packinghouse: 2/252 (0.8) to 10/171 (5.8) Fresh-cut: 0/249 (0) to 4/246 (1.6) |
Zones 2 and 3 for packinghouse and only zone 3 for fresh-cut | Not identified | ||||||
| Multiregional | Processing plants (n = 2) | 14 months | Baby spinach (n = 409) | Listeria spp. (excludes Lm) | Yes | 2/409 (0.49) | Processed samples | L. seeligeri | [57] |
| Listeria monocytogenes | Yes | 3/409 (0.73) | One processed and two minimally processed baby spinach samples | Not identified | |||||
| Pacific Northwest U.S. | Produce handling and processing facilities (n = 7) | One year | Environmental sponge samples (n = 350) | Listeria spp. (excludes Lm) | Yes | 11/350 (3.14) | Drain, entry point, floor, forklift tire, forklift traffic area, equipment leg | L. innocua, L. ivanoii, and L. welshimeri | [58] |
| Listeria monocytogenes | Yes | 15/350 (4.29) | 1/2a, 3a, 4b, 4d, 4e | ||||||
| California | Grower (n = 1) | Four months | Conventional and organic spring mix | Listeria monocytogenes | ND | N/A | N/A | N/A | [59] |
| Not specified | Fresh mushroom slicing and packaging operation; 98 sampling sites within the facility | 14 months | NFCS, such as loading dock doors, floors, walls, pallets, drains, squeegees, electrical utility boxes, forklifts, plastic curtains, etc. (n = 255) | Listeria spp. (excludes Lm) | Yes | 16/255 (6.27) | Receiving and staging, washing and slicing, packaging, and shipping sites | L. innocua and L. grayi | [60] |
| Listeria monocytogenes | Yes | 48/255 (18.8) | Receiving and staging, washing and slicing, and packaging sites | 1/2a, 1/2b, and 1/2c | |||||
| Retail | |||||||||
| Michigan and New Jersey | Distribution (cilantro), retail (cilantro and mung bean sprouts) and farm (cucumber) | Not specified | Cilantro (pre-retail and retail), cucumbers, and mung bean sprouts | Listeria spp. and Listeria monocytogenes | No live isolates obtained; however, species level proportional abundances illustrate presence of Listeria DNA | L. monocytogenes DNA present in two cilantro samples | Not identified | [61] | |
| South America, North America, Europe, Africa, and Asia | Published studies (n = 25) | Not specified | Packaged salads (n = 20,904), including packaged greens (n = 1212), packaged RTE (n = 11,978), unsure if packaged (n = 2637), and packaged with meat (n = 5077) | Listeria monocytogenes | Yes | 543/20,904 (2.60) | All sample types, except for some unsure if packaged samples | Not identified | [62] |
| Maryland | Retail stores (n = 3) | One year | Basil, cilantro, lettuce, scallion, spinach, and parsley (n = 414) | Listeria monocytogenes | Yes | Not specified | Spinach | Not identified | [63] |
| California, Texas, Iowa, Minnesota, Ohio, Massachusetts, and Florida | Retail grocery produce departments (n = 30) | Eight months | FCS and NFCS | Listeria monocytogenes | Yes | Total: 226/5112 (4.42) NFCS: 178/2205 (8.1) FCS: 48/2907 (1.7) |
Drain (cold room storage), standing water, drain (produce area), squeegee/floor cleaners, floor (cold room storage), etc. | Not identified | [64] |
| Maryland and California | Retail markets | Over 14 to 23 months | Bagged salads (n = 2966) | Listeria monocytogenes | Yes | Total: 22/2966 (0.74) MD: 8/1465 (0.55) CA: 14/1501 (0.93) |
Bagged salads | Not identified | [65] |
| California, Maryland, Connecticut, and Georgia | Retail stores (n = 1042) | Two years | Produce, including cut vegetables (raw), low-acid cut fruit, and sprouts (n = 6749) | Listeria monocytogenes | Yes | 36/6749 (0.53) | All sample types | Not identified | [66] |
| Virginia | Retail markets | Two months | Whole (n = 20) and sliced (n = 8) shiitake mushrooms | Listeria spp. (excludes Lm) | Yes | Total: 3/28 (10.71) Whole: 1/20 (5) Sliced: 2/8 (25) |
Whole and sliced mushrooms | Not identified | [67] |
| Listeria monocytogenes | ND | N/A | N/A | N/A | |||||
| Delaware | Grocery stores (number not specified) | 15 months | Mushrooms (n = 202) and alfalfa sprouts (n = 206) | Listeria spp. (excludes Lm) | Yes | Total: 24/408 (5.88) Mushroom: 17/202 (8.42) Sprouts: 7/206 (3.40) |
Mushroom and sprouts | L. welshimeri, L. innocua, and L. seeligeri | [68] |
| Listeria monocytogenes | Total: 1/408 (0.25) Mushroom: 0/202 Sprouts: 1/206 (0.49) |
Only sprouts | Not identified | ||||||
| Seattle, Washington | Retail stores | One year | Sprouts (n = 200) and mushrooms (n = 100) | Listeria monocytogenes | Yes | 1/100 (1) | Mushroom | Not identified | [69] |
| Colorado, Connecticut, Georgia, Maryland, Minnesota, California, Texas, and Washington | Retail locations | Six years | Leafy greens (n = 14,183), sprouts (2652), and melons (3411) | Listeria monocytogenes | Yes | Leafy greens: (0.11) Sprouts: (0.11) Melons: (0.23) |
Spinach, romaine, alfalfa sprouts, broccoli sprouts, cucumber, and mango | Not identified | [70] |
| Philadelphia, Pennsylvania | Retail food establishments (n = 60) | Two years | RTE fresh fruit, greens, and herbs | Listeria monocytogenes | ND | N/A | N/A | N/A | [71] |
| Minnesota | Supermarket | Not specified | Produce (lettuce, potato peels, corn husks, broccoli stems, cabbage outer leaves, carrot peels, cauliflower stems, mushroom stems, spinach, beet peels, and frozen green beans, pea pods, green peas, and spinach) | Listeria monocytogenes | ND | N/A | N/A | N/A | [72] |
| Minneapolis, Minnesota | Supermarkets (n = 2) | One year | Produce (broccoli, cabbage, carrots, cauliflower, cucumbers, lettuce, mushrooms, potatoes, radishes, and tomatoes; n = 1000) | Listeria spp. and Listeria monocytogenes | Yes | 97/1000 (9.7) | Lettuce, cabbage, cucumbers, mushrooms, potatoes, radishes | L. monocytogenes, L. innocua, L. welshimeri, and L. seeligeri | [73] |
| Farmers’ Markets | |||||||||
| West Virginia and Kentucky | Farmers’ markets (n = 2) | Four months | Produce (tomatoes, peppers, cucumber, cantaloupe, and spinach; n = 212) | Listeria spp. (excludes Lm) | Yes | 4/212 (1.89) | Peppers and cantaloupes | Not identified | [74] |
| Listeria monocytogenes | Yes | 4/212 (1.89) | Tomatoes, cucumbers, and cantaloupes | Not identified | |||||
| Pennsylvania | Farmers’ markets (n = 25) and vendors (n = 58) | 8 months | Leafy greens (n = 50 each of lettuce, spinach, and kale) | Listeria spp. (excludes Lm) | Yes | 5/152 (3.30) | Kale, lettuce, and spinach | Not identified | [75] |
| Listeria monocytogenes | Yes | 1/152 (0.66) | Spinach | ||||||
| Washington D.C. | Farmers’ markets and supermarkets | Not specified | Produce (alfalfa sprouts, beets, broccoli, broccoli sprouts, cauliflower, celery, cilantro, cucumbers, field cress, green peppers, lettuce, mung bean sprouts, potatoes, soybean sprouts, watercress, yams; n = 127) | Listeria spp. (excludes Lm) | Yes | 19/127 (14.96) | Celery, field cress, lettuce, mung bean sprouts, potatoes, soybean sprouts, watercress, yams | L. innocua, L. welshimeri, and L. grayi | [76] |
| Listeria monocytogenes | Yes | 6/127 (4.72) | Field cress and potatoes | N/A | |||||
| Florida | Farmers’ markets (n = 9) and supermarkets (n = 12) | 10 months | Leafy greens (n = 103), berries (n = 106), spinach (n = 77), and tomatoes (n = 115) | Listeria monocytogenes | Yes | 4/401 (1) | Leafy greens and spinach from farmer’s markets | Not identified | [77] |
| Domestic | |||||||||
| Philadelphia, Pennsylvania | Homes (n = 100) | One year | Refrigerator door handle, bottom shelf, meat drawer; kitchen counter near sink; used kitchen sponge or dishcloth | Listeria spp. (excludes Lm) | Yes | 12/557 (2.15) | Meat drawer | L. innocua, L. welshimeri, L. grayi, and L. seeligeri | [78] |
| Listeria monocytogenes | Yes | 4/557 (0.72) | Refrigerator door handle, refrigerator drawer, kitchen sink, and dishcloth/sponge | Not identified | |||||
Table 3.
Range of Listeria spp. and Lm prevalence and identified Listeria spp. and Lm serovars by supply chain stage.
| Stage of Supply Chain | Listeria spp. Prevalence (%) a,b | Identified Listeria Species | Lm Prevalence (%) | Identified Lm Serovars c | ||
|---|---|---|---|---|---|---|
| Low | High | Low | High | |||
| Natural environment and outdoor production | ND | 46.81 [28] | L. welshimeri, L. innocua, L. seeligeri, L. grayi, L. ivanoii, L. marthii, L. booriae | ND | 61.1 [31] | 1/2a, 1/2b, 3a, 3b, 4d, 4e, 7 |
| Packinghouse, indoor production, and processing | 0.08 | 14.13 [50] | L. welshimeri, L. innocua, L. seeligeri, L. grayi, L. ivanoii, L. marthii | ND | 56.41 [53] | 1/2a, 1/2b, 1/2c, 3a, 4b, 4c, 4d, IVb-v1 |
| Retail | 6.13 [68] | 10.71 [67] | L. innocua, L. welshimeri, L. seeligeri | ND | 4.42 [64] | NI |
| Farmers’ markets | ND | 14.96 [76] | L. welshimeri, L. innocua, L. seeligeri, L. grayi | ND | 4.72 [76] | NI |
| Domestic | 2.15 [78] | L. welshimeri, L. innocua, L. seeligeri, L. grayi | 0.72 [78] | NI | ||
aListeria spp. exclude Lm except for the high Listeria spp. prevalence at the natural environment and outdoor production stage which includes Lm. b Not detected (ND) c Not identified (NI).
3.2. Natural Environment and Outdoor Production
Twenty-five studies evaluated the prevalence, persistence, and/or diversity of Listeria species in the natural and/or outdoor production (e.g., farm) environment. Among the included natural environment and outdoor production studies, Listeria spp. prevalence ranged between undetected and 46.81%, while Lm prevalence ranged from undetected to 61.1% (Table 3). The largest reported Listeria spp. prevalence (including Lm) was in market-ready organic composts [28], while the largest reported Lm prevalence was in surface water in California [31]. Three of the relevant studies [35,44,46] in natural environment and outdoor production did not detect Listeria spp. (including Lm). No relevant studies were identified in the U.S. southeastern region.
3.2.1. Northeast Studies
In 2003, different types of compostable manures in Maryland were evaluated for gastrointestinal pathogens [36]. Overall, 12.5% of manures contained Lm; of these, two samples were liquid dairy manure, and one sample was cow manure [36]. These manures were also applied to potato fields, and there was no detectable Lm on potatoes grown in the manure-applied fields [36].
In 2005, a study [41] characterized eighty Lm isolates from urban and natural environments and observed the majority of natural isolates in lineage II, while urban isolates were evenly split between lineages I and II [41]. Seven and twenty-six EcoRI ribotypes were also differentiated between urban and natural environments, respectively [41]. Additionally, this study examined an additional 921 isolates from farm, food and food processing facilities, and clinical samples in combination with the previous 80 isolates to determine lineage disparities between sample categories [41]. Isolates from human clinical samples were more commonly identified as lineage I, while those from the natural environment, farms, and food were more often characterized as lineage II [41]. Ribotype diversity was also highest for isolates from farms and lowest for those in natural environments; therefore, isolates from different locations appear to be diverse [41].
A separate study [42] examined 1805 samples (soil, water, and environment) in both natural and urban environments. While prevalence of Listeria spp. was similar between natural and urban environments (23.4% and 22.3%, respectively), there were statistically significant differences in the species and allelic types between natural and urban areas [42]. There was also evidence of persistent species and allelic types during the course of the study in specific sampling sites [42]. In another study [47], allelic diversities were exhibited among Lm isolates from soil, water, fecal, and drag swab samples from fruit and vegetable farms. Three allelic types of sigB (57, 58, and 61) were determined from isolates identified from water samples [47]. Lm isolates from this study also represented nine allelic types from lineages I, II, and IIIa [48]. Soil available water storage, temperature, proximity to water, roads and urban development, and pasture/hay grass had an effect on the likelihood of detecting Lm [47]. Recent cultivation of fields and application of manure were significantly associated with an increased likelihood of isolating both Lm and Salmonella from produce fields [48].
In 2010, a study [49] identified meteorological and landscape factors and management practices associated with prevalence of several foodborne pathogens, such as Lm. Terrestrial samples from recently irrigated fields had greater likelihood of Lm isolation [49]. In 2014, prevalence and diversity of Listeria spp. were determined from environmental samples from produce production and natural environments [25]. Additionally, detection of Listeria was associated with identified geographical and meteorological factors [25] Random forest models suggested that soil moisture and proximity to water and pastures were highly associated with Listeria spp. isolation from produce production [25]. Alternatively, elevation, study site, and proximity to pastures were highly associated with Listeria spp. isolation from natural environments [25]. In 2015, a study examining spatial and temporal factors associated with Lm prevalence in spinach fields also found that Listeria spp. and Lm isolates that were associated with irrigation events showed significantly lower sigB allele type diversity than those isolates associated with rain events [40]. Based on this result, it was suggested that irrigation water may be a source of contamination as isolates were more similar to one another [40].
A study examining meteorological risk factors associated with a mixed produce and dairy farm in Maryland assessed 159 samples of various animal-related and environmental samples (e.g., cow feces, cow drinking water, cow and bird feces, surface water, partially and fully composted material, water, and soil from vegetable production area) over 14 months [37]. Listeria spp. were found in 5.03% of samples, while Lm isolates were obtained from 1.26% of samples [37].
Surface water from a non-tidal freshwater creek in Maryland was evaluated for bacterial pathogens and applied to produce fields containing kale and radishes to evaluate pathogen prevalence in soil and produce [39]. Lm was detected in and enumerated from creek water at 0.04 and 0.07 MPN/L, but prevalence regarding total sample number was not specified [39]. Conventional water sources, including tidal freshwater rivers, non-tidal freshwater creeks, reclaimed water holding ponds, pond water sites, and produce wash water in the mid-Atlantic U.S. were examined for Lm prevalence [45]. Lm was found in 31.18% of samples in all conventional water sources [45].
3.2.2. Midwest Studies
Various experimental treatments were applied to varying compost amendments (sawdust, straw, and water) to monitor the change in pathogen prevalence during simulated composting [34]. Compost was sampled on days 0, 3, 7, 14, 28, and 56 for pathogen presence, and Listeria species (including Lm) were found in all three treated compost types on days 0 and 3; however, their prevalence was not reported [34].
3.2.3. Plains Studies
In 2002, a study [38] examining cabbage farms and packinghouses in the Rio Grande Valley and Texas found 2.81% Listeria spp. and 3.04% Lm prevalence among cabbage, water, and environmental samples.
A study performed in five Colorado pristine wilderness areas detected Listeria spp. in all tested sample types (soil, water, sediment, surface soil, and wildlife feces) and Lm from only water and feces [27]. Additionally, this study observed approximately 75% of Listeria spp. positive samples from a single wilderness location [27]. It is suggested that some Lm strains may be persistent, as one unique Lm strain was found in both the summer of the first sampling year and in the fall of the second sampling year [27]. Overall, this study indicated that Listeria incidence was rare at the study site and isolates tended to be genetically distinct based on wilderness area [27].
3.2.4. Pacific West Studies
Four separate studies in California evaluated the microbial composition of watershed areas and all found Lm in surface water samples [30,31,32,33]. Lm prevalence ranged from 30.1 to 61.1% across the four studies. Two studies [30,32] found a range of serotypes, 1/2a, 1/2b, 3a, 4b, 4d, and 4e, indicating strain-level diversity in and among watershed sites in California. One study [79] characterized over 100 Lm isolates from watersheds near leafy green production sites using MLVA and found 49 different Lm strains, illustrating the diversity of strains within these sites. In Arizona, Lm was not detected in reclaimed and return flow water in two wastewater treatment plants [44].
One study examined market-ready organic compost from 94 compost facilities across Washington, Oregon, and California and detected a Listeria species prevalence of 46.81% [28]. Another study did not detect Lm in fertilizer, soil, foliar, and raspberry fruit from raspberry fields in Washington [46].
3.2.5. Multiregional Studies
Two studies contain sampling regions across U.S. geographic regions. A study over nine states (AZ, CA, GA, KY, MD, NY, NC, SC, and TN) examined over 100 organic fertilizers (e.g., spent-mushroom compost, vermicompost, mixed animal waste) and did not detect Lm [29]. Another multiregional study [43] examined interactions between weather and microbial and physiochemical water quality to determine the likelihood of pathogens in agricultural water in Arizona and New York. Two types of sampling methods were used: Moore swabs (MS), which use sterile gauze or cheesecloth to continuously filter microorganisms from water, and grab swabs (GS), which collect a single volume of water for analysis [43]. The odds of isolating Lm using MS was significantly lower than that of isolating Lm from paired GS [43]. However, more competitive microflora and fewer Listeria-like colonies were observed after plating MS enrichments compared to GS enrichments [43]. Highly ranked factors associated with Lm isolation from both MS and GS included flow rate of agricultural water, weather conditions 0–4 days and 0–1 day before sample collection [43].
3.3. Packinghouse, Indoor Production, and Processing
Eighteen identified studies included microbial sampling at indoor production, packinghouse, or processing environments. These studies primarily focused on non-food contact surface areas, while some studies performed microbiological analysis of produce. Five studies evaluated environmental and some produce samples from packinghouses.
3.3.1. Northeast Studies
Two studies focused on mushroom production and processing facilities. The first sampled NFCS in a small-scale mushroom production facility in Pennsylvania and detected Listeria spp. and Lm in 14.13% and 1.63% of all samples [50]. Listeria species positive samples included phase I composting, phase II composting, tray filling line, and growing rooms, while Lm positive samples were only found in phase I composting [50]. Identified Listeria species other than Lm included L. innocua, L. welshimeri, and L. grayi [50]. The second study examined NFCS in a fresh mushroom slicing and packaging operation [60]. The location of the packaging facility is not specified in this study; however, based on the sampling methodology it is reasonable to assume the facility is in Pennsylvania. Listeria species were found in 6.27% of samples, where positive samples included those in receiving and staging, washing and slicing, packaging, and shipping sites [60]. Lm was detected in 18.8% of samples, with positive samples in receiving and staging, washing and slicing, and packaging sites [60].
NFCS samples were taken at apple and other tree fruit packinghouses in the northeastern U.S. [53]. Lm was found in 56.41% of NFCS samples [53]. Positive samples included washing drying, and waxing areas of the packinghouse facilities [53].
One northeast-based studies failed to detect any Listeria spp. or Lm in produce samples. Lm was not detected in tomatoes, leafy greens, peppers, cucumbers, and other produce (n = 177) from seven organic farms in Maryland [51]. Well water and surface water (n = 29) were also tested at these organic farms and Lm was not detected [51].
3.3.2. Southern/Southeastern Studies
The microbiological quality of leafy greens, herbs, and cantaloupes was evaluated, and Lm was detected in three cabbage samples—6.98% prevalence out of all samples (n = 43) [54]. Another study examining leafy greens, herbs, and cantaloupe from thirteen farms and five packing sheds in the southern U.S. did not detect any Lm [52].
A study based in the southeastern U.S. evaluated environmental samples collected from eleven fresh produce packinghouses and found 2.64% of samples were positive for Listeria species and 3.15% for Lm [55]. Positive samples included drains, cold storage rooms, wet NFCS, mobile NFCS, dry NFCS, and outside packing/handling areas [55]. Non-Lm Listeria species isolated included L. innocua, L. marthii, L. seeligeri, and L. welshimeri [55].
3.3.3. Pacific West Studies
In the Pacific northwest U.S., seven produce handling and processing facilities were environmentally sampled for both Listeria spp. and Lm [58]. The detectable Lm strains represented serotypes 1/2a, 3a, 4b, 4d, and 4e [58]. In addition to Lm, researchers also detected L. innocua, L. ivanovii, and L. welshimeri [58]. Positive sample locations included drains, entry points, floors, forklift tires, and equipment legs [58].
Stone fruits, such as white and yellow nectarines and white and yellow peaches, were acquired from a packinghouse environment in California from lots associated with the 2014 stone fruit-associated listeriosis outbreak [17]. Implicated stone fruits from were examined for listeria prevalence, and 53.3% of all samples were positive for Lm [17]. Of all nectarines sampled, 25% were positive for Lm and of all peaches, 91.1% were positive for Lm. Serotypes of Lm were determined to be IVb-v1 and 1/2b [17]. SNP-based whole-genome sequencing demonstrated the outbreak-associated isolates differed by up to 42 SNPs compared to unrelated clinical isolates [17]. Isolates belonging to serotype Ivb-v1 belonged to the singleton ST382, which is an emerging clonal group of Lm and was associated with the 2011 cantaloupe-related outbreak [17]. However, a core genome MLST study to identify globally distributed clonal groups and outbreak strains of Lm did not associate ST382 with the cantaloupe-associated outbreak [80]. ST382 was only associated with caramel apple, stone fruit, and packaged leafy green salad outbreaks [80]. Otherwise, both studies emphasize ST382 as an emerging clonal group associated with produce [17,80].
Lastly, an analysis of microflora on organically and conventionally grown spring mix from a California processor did not yield any Lm isolates [59].
3.3.4. Multiregional Studies
Baby spinach samples from farms in TX, AZ, CA, CO, MD, and NJ and Ontario and Quebec, Canada were examined from two processing plants (unspecified location) for Listeria spp. and Lm [57]. Lm was detected in one processed and two minimally processed spinach samples (3/409; 0.73%), while L. seeligeri was found in two processed spinach samples (2/409; 0.49%) [57].
A study in three packinghouses and five fresh-cut facilities on both the East and West coasts of the U.S. environmentally sampled zones 2 and 3 for Listeria. Zone 2 consisted of areas that are in close proximity to food or FCS, while zone 3 included areas in the processing and packing area that are not adjacent to FCS [56]. Listeria spp. and Lm were found in zones 2 and 3 in packinghouses and only in zone 3 for fresh-cut facilities [56].
3.3.5. Other Relevant Studies
In 2017, researchers examined the genetic relatedness of 387 Lm serotype 4b strains, with some strains previously isolated from caramel apples, lettuce, collard greens, curly parsley, nectarines, peaches, spinach, and walnuts [81]. While there were clinical and non-clinical samples in each of the seven clades identified by the CFSAN SNP Pipeline, nearly all of the strains were organized into one clade by the multi-virulence-locus sequencing type (MVLST) analysis [81]. These findings suggest that 4bV strains may be under different selection pressure at varied geographic regions, which may conserve diversity across virulence loci but permit variability across less highly conserved areas of the genome [81].
From a diversity perspective, copper stress in Lm isolates and mutants from the 2011 cantaloupe-associated outbreak can be mediated by the penicillin-bind protein, pbp4 [82]. A transposon mutant of pbp4 demonstrated reduced copper tolerance and minimal inhibitory concentrations for selected cell wall-active antibiotics [82]. Genomic characterization of the cantaloupe-associated outbreak strains found that 1/2b isolates were closely related, while 1/2a isolates comprised two genomic groups [83]. An analysis of non-hemolytic mutants of a 1/2b strain that contributed to the cantaloupe-associated outbreak illustrated that prfA and hly contribute to biofilm formation and aggregation of Lm [84]. PrfA and hyl are well-known Lm virulence genes [41].
Genetic diversity of Listeria isolates from food production facilities, including farms and packinghouses, was investigated to assess the relationship among isolates [85]. In addition to whole genome sequencing (WGS) data, metadata from the FDA Field Accomplishments and Compliance Tracking System (FACTS) database was also included in the analysis to link isolates to facilities [85]. SNP data indicated that isolates from different facilities, as well as isolates within the same facility, have fairly large genetic distances (>20 SNPs) [85].
3.4. Transportation and Distribution
There were no studies identified that investigated Listeria spp. and Lm presence, persistence, and/or diversity in regard to fresh produce transport and distribution.
3.5. Retail
Fifteen relevant studies involve microbiological analysis of NFCS, FCS, or fresh produce acquired in retail environments. Only one study examined FCS and NFCS [64], while all other identified studies microbially examined produce from retail locations. The range of Listeria prevalence at the retail level was 6.13 to 10.71, while that of Lm was not detected to 4.42% (Table 3).
3.5.1. Northeast Studies
In Delaware, mushrooms and alfalfa sprouts (n = 408) were acquired from grocery stores over a period of 15 months [68]. Listeria spp. were more frequently detected in mushrooms (8.42%) than sprouts (3.88%), while Lm was only detected in sprouts (0.49%) [68].
One study that investigated food safety risk differences in RTE fresh fruit, greens, and herbs acquired from retail locations in Philadelphia, PA did not detect Lm [71].
Basil, cilantro, lettuce, scallions, spinach, and parsley from three retail chain groceries in Maryland were sampled bi-weekly for Lm presence [63]. Only spinach samples contained Lm, but the number of positive samples was not reported [63].
A third study out of Virginia sampled whole and sliced shiitake mushrooms purchased from retail markets for both Listeria spp. and Lm [67]. Listeria spp. were only found on whole mushrooms (5%), while Lm was not detected [67].
3.5.2. Midwest Studies
One study examining both fresh (lettuce, potato peels, corn husks, broccoli stems, cabbage outer leaves, carrot peels, cauliflower stems, mushroom stems, spinach, and beet peels) and frozen (green beans, pea pods, green peas, and spinach) produce from a supermarket in Minnesota did not detect Lm [72]. Broccoli, cabbage, carrots, cauliflower, cucumbers, lettuce, mushrooms, potatoes, radishes, and tomatoes were sampled from two supermarkets and assessed for Listeria spp. [73]. Listeria spp. were isolated from 9.7% of samples, including lettuce, cabbage, cucumbers, mushrooms, potatoes, and radishes [73].
3.5.3. Pacific West Studies
In Washington state, a study [69] examined sprouts and mushrooms from retail stores in Seattle for the presence of Lm. Lm was not detected in any sprout samples but was detected in 1% of mushroom samples [69].
3.5.4. Multiregional Studies
A 2007 study characterizing Lm strains isolated from retails foods in Florida and the greater Washington, D.C. area identified three isolates from conventionally grown fresh produce and two from organically grown fresh produce [86]. Serotype designations for these isolates are not explicitly stated, but may include 1/2b, 1/2c, 3b, 3c, 4a, or 4c [86]. Isolates from conventional produce were all resistant to sulfonamide, while those from organic produce were resistant to either ciprofloxacin or sulfonamide [86].
Fresh cut vegetables, low-acid fruit, and sprouts were acquired from 1,042 retail stores across California, Maryland, Connecticut, and Georgia [66]. Lm was detected in 0.53% (36/6,749) of samples [66]. Another multistate investigation of fresh produce from retail locations over six years found Lm in leafy greens (0.11%), sprouts (0.11%), and melons (0.23%) [70].
In Michigan and New Jersey, several produce items were sampled from the farm (cucumbers), prior to retail distribution (cilantro), and in retail (cilantro and mung bean sprouts) to evaluate their microbiomes [61]. Listeria spp. and Lm isolates were not obtained; however, species level proportional abundances identified Listeria DNA in two cilantro samples [61].
A systematic review of the prevalence of Listeria in RTE foods incorporated retail-acquired produce from South America, North America, Europe, Africa, and Asia [62]. Packaged salad samples had a Lm prevalence of 2.60% [62]. Another study [65] analyzed approximately 3000 bagged salad samples in Maryland and California and isolated Lm from 0.75% of samples.
Thirty retail grocery produce departments across CA, TX, IA, MN, OH, MA, and FL were environmentally sampled for Lm [64]. Positive sample types included areas such as cold room storage drains, standing water, produce area drains, squeegee/floor cleaners, and the cold room storage floor [64]. Lm was isolated from 4.42% of all samples (both FCS and NFCS) [64].
3.5.5. Other Relevant Studies
Heavy metal and disinfectant resistance were assessed in Lm strains from foods and food processing plants with unspecified locations [87]. Isolates from fresh avocados and other avocado products (e.g., paste, pulp, chunks) demonstrated little resistance to heavy metals (cadmium, benzalkonium chloride, and arsenic) [87]. Only two 4b serotypes, one from avocado and the other from frozen avocado pulps, demonstrated resistance to cadmium and arsenic [87]. None of the avocado and processed avocado isolates contained cadmium resistance determinants (cadA1, cadA2, and cadA3) [87].
3.6. Farmers’ Market
Five studies examined produce from farmers’ markets for Listeria. The range of Listeria spp. prevalence was not detected to approximately 15.0%, while that of Lm was not detected to 4.72%. No Lm isolate serovars were identified in relevant studies at the farmers’ market level.
3.6.1. Northeast Studies
A variety of produce was sampled from both farmers’ markets and supermarkets in Washington, D.C. [76]. Listeria spp., including L. innocua, L. welshimeri, and L. grayi, were isolated from celery, field cress, lettuce, mung bean sprouts, potatoes, soybean sprouts, watercress, and yams [76]. Lm was isolated from field cress and potatoes [76]. This study observed the greatest Listeria spp. and Lm prevalence at the farmers’ market level compared to that from the other relevant farmers’ market studies (Table 3). Additionally, this study isolated Listeria from produce acquired from both supermarkets and farmers’ markets [76]. Listeria positive samples obtained from farmer’s markets included celery, field cress, and potatoes, while those from supermarkets were celery, lettuce, mung bean sprouts, soybean sprouts, watercress, and yams [76].
Leafy greens (lettuce, spinach, and kale) were acquired from 58 vendors at 25 farmers’ markets in Pennsylvania [75]. Both Listeria spp. and Lm were isolated from kale, lettuce, and spinach, and spinach, respectively [75].
3.6.2. Southeast Studies
In Florida, leafy greens, berries, spinach, and tomatoes acquired from nine farmers’ markets and twelve supermarkets were evaluated for Lm [77]. Lm was found on leafy greens and spinach from farmers’ markets; no Lm was detected in any supermarket-acquired produce [77].
3.6.3. Multiregional Studies
Listeria spp. and Lm were recovered from produce collected at two farmers’ markets, one of which was in Kentucky and the other in West Virginia [74]. Listeria spp. were isolated from peppers and cantaloupes, while Lm was isolated from tomatoes, cucumbers, and cantaloupes [74]. Additionally, the research group investigated farmers’ market vendors’ handling of produce containers and the survival of Lm on plastic, pressed card, and wood container surfaces [88]. Two Lm isolates with serotype 1/2b from the cantaloupe-associated outbreak were used for inoculation of container surfaces [88]. The initial population of Lm recovered from each material surface after inoculation ranged from 6.39 to 6.93 log CFU/cm2 at 3.2 and 22.5 °C, respectively [88]. After the first day of storage, Lm populations were fairly constant or only slightly decreased [88]. By day 21, the Lm population remaining on all surfaces ranged from 4.89 to 5.73 log CFU/cm2 [88]. Lm persisted longer and at a greater population level on pressed card surfaces compared to on plastic and wood surfaces [88].
3.7. Restaurant
There were no relevant studies that investigated Listeria spp. and Lm prevalence, persistence, and/or diversity in restaurant environments.
3.8. Domestic
One relevant study was identified examining consumer kitchens and homes. One hundred homes in Philadelphia, PA were sampled over one year for Listeria spp. and Lm [78]. Types of samples included kitchen areas, such as refrigerator door handles and interiors, counters, and used sponges and dishcloths [78]. Listeria spp. were found in the meat drawer of refrigerators, which accounted for 2.15% (12/557) of samples [78]. Lm was present on refrigerator door handles, refrigerator drawers, kitchen sinks, and on dishcloths/sponges, which accounted for 0.72% (4/557) of samples [78].
4. Discussion
4.1. Natural Environments and Outdoor Production
Detection of Listeria spp. and Lm from natural environments and outdoor production provides insight into identifying potential sources and routes for cross-contamination onto produce. The extent of Listeria prevalence and persistence may also depend on the environment. The ubiquity of Listeria in natural environments presents a baseline level of risk for fresh produce at the farm level. Persistence of Listeria in natural or farm environments is especially challenging as harborage sites can be difficult to determine, access, or transplanted from other sources. Therefore, Listeria prevention at the farm level will remain a concern for producers, produce handlers, and processors. In 2016, the Produce Safety Rule (PSR) within the Food Safety Modernization Act was finalized and established science-based standards for the safe production of fresh produce for human consumption. Based on the identified studies focusing on Listeria prevalence at the natural environment and outdoor production level, surface water samples have a consistently high percentage of Listeria-positive samples. Quality and safety of surface water should be prioritized, as it is under the PSR, to mitigate possible cross-contamination of Listeria onto fresh produce. Furthermore, natural events or production activities including irrigation, precipitation, and other on-farm management practices or inputs impact Listeria isolation; these events can be observed and when possible, modified to limit the prevalence of Listeria in natural and farm environments [25,40,43,47,48,49]. As Listeria has demonstrated to be prevalent, persistent, and diverse at the natural environment and outdoor production level, it is crucial for producers to adhere to Good Agricultural Practices (GAPs) and Good Handling Practices (GHPs) to prevent cross-contamination of Listeria onto produce from agricultural water, soil, workers, and equipment during harvest and packaging. Research areas of need include those completed in the southeast region, as this review did not identify any relevant studies in that region.
These studies also support molecular source tracking and fingerprinting of Listeria species that naturally occur in the environment, which is useful for foodborne illness outbreak traceback investigations and evidence of persistent species or strains [42]. The longitudinal study data reported in this review also suggests persistence of strains within certain environments. A range of Listeria diversity was exhibited in relevant studies at the natural environment and outdoor production level, with ten studies identifying Listeria species other than Lm and/or serotyping Lm strains [25,27,30,32,38,42,43,48,49,50]. Therefore, not only may certain Listeria species be persistent in natural and farm environments, but they may also be diverse. This evidence also supports the use of Listeria spp. as index organisms for Lm, as many studies at this supply chain stage isolated both Listeria spp. and Lm from the same sample types and locations. Generally, these studies found a greater prevalence of Listeria spp. compared to Lm, which is a criterion of an acceptable index organism [25]. However, the Listeria spp. isolated at this supply chain stage include both Listeria sensu stricto (i.e., isolated from animal hosts), as well as Listeria sensu lato (i.e., inability to colonize animal hosts) [89]. Therefore, it may not be appropriate to classify certain Listeria spp. as index organisms for Lm due to differences in metabolic activity and pathogenicity. This is consistent with conclusions in Chapin et al. (2014), where the strength of association between isolation of Listeria spp. and Lm may depend on the Listeria species.
4.2. Packinghouse, Indoor Production, and Processing
Across these studies, both Listeria spp. and Lm were found in all food contact zones (i.e., ranging from food contact surfaces to remote areas of a facility). There was also evidence that some Lm strains are from the same source or are persistent in this environment [50]. Contamination within several facility zones may indicate movement of personnel and/or equipment between zones that could contribute to contamination [50]. Therefore, limitation of movement between zones and review of cleaning and sanitation operations may reduce Listeria prevalence throughout the facility [50]. Cleaning and sanitation operations within processing facilities should focus on specific surfaces (e.g., porous, difficult to clean) that may offer greater harborage or support of Listeria populations.
One identified study investigated strain-level diversity of Listeria in stone fruit in response to the 2014 stone fruit-associated listeriosis outbreak [17]. This outbreak is an example of a polyclonal outbreak where several strains of the bacterial pathogen are linked to the outbreak. Polyclonal outbreaks may be indicative of single or multiple persistent Listeria strains that may establish in a facility and undergo selection over time, or strains that may transplant to other locations [90]. Strain diversity can complicate traceback investigations by increasing the difficulty of matching clinical isolates with implicated food isolates. Incidence of polyclonal outbreaks has further bolstered the necessity of WGS for molecular surveillance and fingerprinting of foodborne pathogens [90]. Therefore, more studies are needed that focus on strain-level identification and fingerprinting of Listeria species at this supply chain stage.
Listeria is also prevalent and diverse in packinghouse and processing environments; however, more research is needed to address persistence of strains within these environments. Attention should also be placed toward relevant environments in the Midwest and plains regions, as no relevant studies were identified in those areas for this review. While some studies suggest persistence of isolates, another suggests a reintroduction of Listeria species within produce packinghouses, rather than persistence [55]. Therefore, it is critical for studies to sample locations frequently and use high-resolution subtyping to more accurately assess strain persistence. Verifying that cleaning and sanitation procedures are preventing establishment and colonization of Listeria will assist in minimizing Listeria prevalence and possible persistence or reintroduction within a facility.
4.3. Transportation and Distribution
Transportation vehicles and distribution centers can be bottlenecks within the food supply chain. Within this supply chain stage, large quantities and varieties of products move into a singular location and are then dispersed to many retail, foodservice, or secondary storage areas. As the majority of products in transport and distribution are in their final packaging and typically in secondary or even tertiary containers, they are considered to be at low risk of contamination. Therefore, few best practices are implemented with food safety or current Good Manufacturing Practices in mind. Additionally, as most products sold at major grocers, restaurants, or food service facilities pass through at least one distribution center before reaching the consumer, it is reasonable to assume that many foods associated with foodborne illness outbreaks transit through this type of facility. Without data identifying the hazards associated with environmental contamination in this supply chain stage and quantifying the likelihood for microbial hazards to penetrate and subsequently contaminate fresh produce in unsealed containers, it is difficult to assess the degree of risk associated with transit of these products. Because no studies were identified that assess Listeria spp. and Lm prevalence, persistence, and diversity at the transportation and distribution stage, research is needed in this area.
4.4. Retail
Many retail studies focusing on Listeria have occurred in delicatessens, as RTE salads and deli meats are foods traditionally associated with Listeria contamination. Additional studies on Listeria prevalence, and especially regarding persistence and diversity, are needed in retail produce departments. This review did not identify any relevant studies in the southeastern and plains regions either. Nearly all (14/17, 82.4%) identified retail-based studies, with publications ranging between 1988 and 2020, detected Listeria and/or Lm in fresh produce samples. Therefore, Listeria has been found in retail fresh produce prior to and after the implementation of the “zero-tolerance” policy. This is an indication that Listeria is a consistent issue in retail fresh produce with contributing factors not yet identified or addressed through food safety regulations. RTE foods, including many types of fresh produce, do not undergo additional “kill steps,” such as cooking, so it is critical that no opportunity exists for retail produce items to become contaminated with foodborne pathogens. Frequent and thorough cleaning and sanitation of retail produce departments, as well as good handling practices, should be implemented to avoid cross-contamination and establishment of Listeria species and other pathogenic microorganisms.
While many relevant retail-based studies occurred over long durations of time, these studies did not characterize Listeria or Lm isolates at the species or strain level. Therefore, persistence of specific isolates is unable to be determined. While there are legal hurdles, future research should focus on high-resolution identification of isolates from frequently sampled retail fresh produce environments to determine Listeria persistence. Additionally, more prevalence data between Listeria spp. and Lm is needed to designate Listeria spp. as index organisms at the retail level.
4.5. Farmers’ Market
Supermarkets are subject to governmental regulations and have numerous resources to implement food safety practices, while farmers’ markets typically do not have strict, defined food safety guidelines. Additionally, relevant studies completed at farmers’ market locations illustrate a greater prevalence of Listeria compared to that of supermarkets. Farmers’ markets vendors are generally exempt from federal regulations and may have little oversight. Knowledge and implementation of food safety practices of farmers’ market vendors and managers have been previously investigated [91,92,93]. Some practices may put consumers at risk of contracting foodborne illness as some markets do not have food safety plans nor provide food safety guidance [93]. Lack of facilities, equipment, and resources with food safety guidelines can be barriers to implementing food safety practices [91]. Therefore, farmers’ market-specific training may be able to improve food safety practices, as generalized food safety trainings may not lead to behavioral changes [91,92,93].
Only one identified study [76] characterized the diversity of Listeria species in farmers’ market produce. Therefore, future studies should also examine the genetic diversity of Listeria spp. in farmers’ market environments and produce. This review identified research gaps in geographic regions, where there were no relevant studies identified in the upper Midwest, plains area, and Pacific west. Additionally, more sample types focusing on NFCS are needed at the farmers’ market level, as the identified studies only focused on fresh produce sampling. Isolate characterization is also needed to characterize the diversity of Listeria and validate Listeria spp. as index organisms for Lm at the farmers’ market level and assist in isolate tracking from farm to market.
4.6. Restaurant
While research evaluating restaurant food preparation practices and employee behaviors has been completed [94,95,96], there appear to be no studies on Listeria spp. and/or Lm prevalence, persistence, or diversity at the restaurant level. This particular research gap may not be able to be addressed due to legal implications and other reputational consequences restaurants may face if included in research studies on environmental foodborne bacteria.
4.7. Domestic
Consumer behavior may lead to persistence of Lm and/or cross-contamination of Lm onto other foods. For example, poor cleaning and sanitation of home refrigerators can lead to transfer of bacteria on surfaces to foods. Improper separation of meats, soft cheeses, and other Listeria-associated foods and RTE foods, such as produce, can also lead to cross-contamination if Listeria is present. Reusable bags can also be a route of cross-contamination, especially if they are not washed often or are reused for other purposes that may pose a food safety risk [97]. Temperature abuse is a known contributor to foodborne pathogen, including Lm, persistence and proliferation on fresh produce [98,99,100,101,102,103]. Therefore, ensuring that foods are not held at abusive temperatures after purchase (i.e., for prolonged periods of time in a vehicle) and are properly placed in a clean, properly operating refrigerator will help prevent food safety issues. Readily available and widespread consumer-friendly food safety materials, such as internet-accessible and printed resources, may help improve food safety at home [97].
Additional studies evaluating Listeria spp. and Lm prevalence at the domestic level is needed to determine sources of Listeria (i.e., from the produce itself or via cross contamination). Furthermore, one relevant study does not provide enough evidence to support Listeria spp. as index organisms for Lm in the domestic environment. At this stage, Listeria spp. may be more appropriately considered as indicator organisms in relation to hygienic status of consumer kitchens.
5. Conclusions
Based on these studies, potential Listeria contamination is a risk at each stage along the fresh produce supply chain. The largest prevalence of Listeria spp. and Lm was observed at the natural and outdoor production environments, with prevalence generally decreasing with each progression in the supply chain (e.g., packinghouse to distribution to retail). Generally, both Listeria spp. and Lm were isolated together in many studies at the natural environment and outdoor production, as well as at the packinghouse, indoor production, and processing stages. Therefore, Listeria spp. many serve as index organisms for Lm at these stages; however, more research is needed to address Listeria spp. as index organisms at subsequent supply chain stages (e.g., retail). Another research deficit includes work in the Midwest and plains regions as these areas had the fewest relevant studies. Research is also needed to examine Lm prevalence, diversity, and persistence in the produce transport and distribution environment. Additional strain-level studies focusing on isolates obtained from environments closer to the supply endpoint, as well as across smaller food systems (i.e., on farms that supply farmers’ markets, markets themselves, and consumer homes), will also provide evidence of Listeria diversity and persistence.
Acknowledgments
We would like to thank Kelsey Forester for library assistance and Claire M. Marik for editorial suggestions. All figures created with BioRender.com.
Author Contributions
Conceptualization, A.T. and L.L.D.; methodology, A.T. and L.K.S.; formal analysis, A.T.; investigation, A.T.; data curation, A.T.; writing—original draft preparation, A.T.; writing—review and editing, L.K.S., B.J.C., L.L.D.; visualization, A.T.; supervision, L.L.D.; project administration, L.L.D.; funding acquisition, L.L.D., L.K.S., B.J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Center for Produce Safety.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Orsi R.H., Wiedmann M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016;100:5273–5287. doi: 10.1007/s00253-016-7552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Listeria (Listeriosis) [(accessed on 6 January 2019)]; Available online: https://www.cdc.gov/listeria/index.html.
- 3.Archer D.L. The evolution of FDA’s policy on Listeria monocytogenes in ready-to-eat foods in the United States. Curr. Opin. Food Sci. 2018;20:64–68. doi: 10.1016/j.cofs.2018.03.007. [DOI] [Google Scholar]
- 4.U.S. Food Drug Administration (FDA) The Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins Handbook. [(accessed on 8 January 2020)]; Available online: https://www.fda.gov/food/foodborne-pathogens/bad-bug-book-second-edition.
- 5.Farber J.M., Peterkin P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radoshevich L., Cossart P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018;16:32. doi: 10.1038/nrmicro.2017.126. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC) Listeria Outbreaks. [(accessed on 25 April 2020)]; Available online: https://www.cdc.gov/listeria/outbreaks/index.html.
- 8.Welshimer H.J. Survival of Listeria monocytogenes in soil. J. Bacteriol. 1960;80:316–320. doi: 10.1128/jb.80.3.316-320.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dowe M.J., Jackson E.D., Mori J.G., Bell C.R. Listeria monocytogenes survival in soil and incidence in agricultural soils. J. Food Prot. 1997;60:1201–1207. doi: 10.4315/0362-028X-60.10.1201. [DOI] [PubMed] [Google Scholar]
- 10.Srey S., Jahid I.K., Ha S.D. Biofilm formation in food industries: A food safety concern. Food Control. 2013;31:572–585. doi: 10.1016/j.foodcont.2012.12.001. [DOI] [Google Scholar]
- 11.Carpentier B., Cerf O. Review-Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011;145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Marik C.M., Zuchel J., Schaffner D.W., Strawn L.K. Growth and Survival of Listeria monocytogenes on Intact Fruit and Vegetable Surfaces during Postharvest Handling: A Systematic Literature Review. J. Food Prot. 2020;83:108–128. doi: 10.4315/0362-028X.JFP-19-283. [DOI] [PubMed] [Google Scholar]
- 13.Cosgrove S., Cronquist A., Wright G., Ghosh T., Vogt R., Teitell P., Gelfius A., Spires C., Duvernoy T., Merriweather S., et al. Multistate Outbreak of Listeriosis Associated With Jensen Farms Cantaloupe-United States, August–September 2011 (Reprinted from MMWR, volume 60, pp. 1357–1358, 2011) JAMA. 2011;306:2321. [Google Scholar]
- 14.Centers for Disease Control and Prevention (CDC) Multistate Outbreak of Listeriosis Linked to Whole Cantaloupes from Jensen Farms, Colorado (Final Update) [(accessed on 22 April 2020)]; Available online: https://www.cdc.gov/listeria/outbreaks/cantaloupes-jensen-farms/index.html.
- 15.Gaul L.K., Farag N.H., Shim T., Kingsley M.A., Silk B.J., Hyytia-Trees E. Hospital-acquired listeriosis outbreak caused by contaminated diced celery—Texas, 2010. Clin. Infect. Dis. 2013;56:20–26. doi: 10.1093/cid/cis817. [DOI] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention (CDC) Multistate Outbreak of Listeriosis Linked to Commercially Produced, Prepackaged Caramel Apples Made from Bidart Bros. Apples (Final Update) [(accessed on 22 April 2020)]; Available online: https://www.cdc.gov/listeria/outbreaks/caramel-apples-12-14/index.html.
- 17.Chen Y., Burall L.S., Luo Y., Timme R., Melka D., Muruvanda T., Payne J., Wang C., Kastanis G., Maounounen-Laasri A., et al. Listeria monocytogenes in stone fruits linked to a multistate outbreak: Enumeration of cells and whole-genome sequencing. Appl. Environ. Microbiol. 2016;82:7030–7040. doi: 10.1128/AEM.01486-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention (CDC) Outbreak of Listeria Infections Linked to Enoki Mushrooms. [(accessed on 22 April 2020)]; Available online: https://www.cdc.gov/listeria/outbreaks/enoki-mushrooms-03-20/index.html.
- 19.Centers for Disease Control and Prevention (CDC) Multistate Outbreak of Listeriosis Linked to Frozen Vegetables (Final Update) [(accessed on 22 April 2020)]; Available online: https://www.cdc.gov/listeria/outbreaks/frozen-vegetables-05-16/index.html.
- 20.Centers for Disease Control and Prevention (CDC) Multistate Outbreak of Listeriosis Linked to Packaged Salads Produced at Springfield, Ohio Dole Processing Facility (Final Update) [(accessed on 22 April 2020)]; Available online: https://www.cdc.gov/listeria/outbreaks/bagged-salads-01-16/index.html.
- 21.U.S. Food and Drug Administration (FDA) FDA Investigated Listeria monocytogenes in Sprouts from Wholesome Soy Products, Inc. [(accessed on 22 April 2020)]; Available online: http://wayback.archive-it.org/7993/20171114154907/https://www.fda.gov/Food/RecallsOutbreaksEmergencies/Outbreaks/ucm422562.htm.
- 22.Centers for Disease Control and Prevention (CDC) National Outbreak Reporting System (NORS) [(accessed on 22 April 2020)]; Available online: www.cdc.gov/norsdashboard.
- 23.Garner D., Kathariou S. Fresh produce–associated listeriosis outbreaks, sources of concern, teachable moments, and insights. J. Food Prot. 2016;79:337–344. doi: 10.4315/0362-028X.JFP-15-387. [DOI] [PubMed] [Google Scholar]
- 24.Ho J.L., Shands K.N., Friedland G., Eckind P., Fraser D.W. An outbreak of type-4B Listeria monocytogenes infection involving patients from 8 Boston hospitals. Arch. Intern. Med. 1986;146:520–524. doi: 10.1001/archinte.1986.00360150134016. [DOI] [PubMed] [Google Scholar]
- 25.Chapin T.K., Nightingale K.K., Worobo R.W., Wiedmann M., Strawn L.K. Geographical and Meteorological Factors Associated with Isolation of Listeria Species in New York State Produce Production and Natural Environments. J. Food Prot. 2014;77:1919–1928. doi: 10.4315/0362-028X.JFP-14-132. [DOI] [PubMed] [Google Scholar]
- 26.Wallace B.C., Small K., Brodley C.E., Lau J., Trikalinos T.A. Deploying an interactive machine learning system in an evidence-based practice center: Abstrackr; Proceedings of the 2nd ACM SIGHIT international health informatics symposium; Miami, FL, USA. 28–30 January 2012; pp. 819–824. [Google Scholar]
- 27.Ahlstrom C.A., Manuel C.S., Den Bakker H.C., Wiedmann M., Nightingale K.K. Molecular ecology of Listeria spp., Salmonella, Escherichia coli O157:H7 and non-O157 Shiga toxin-producing E. coli in pristine natural environments in Northern Colorado. J. Appl. Microbiol. 2018;124:511–521. doi: 10.1111/jam.13657. [DOI] [PubMed] [Google Scholar]
- 28.Brinton W.F., Storms P., Blewett T.C. Occurrence and Levels of Fecal Indicators and Pathogenic Bacteria in Market-Ready Recycled Organic Matter Composts. J. Food Prot. 2009;72:332–339. doi: 10.4315/0362-028X-72.2.332. [DOI] [PubMed] [Google Scholar]
- 29.Miller C., Heringa S., Kim J., Jiang X.P. Analyzing Indicator Microorganisms, Antibiotic Resistant Escherichia coli, and Regrowth Potential of Foodborne Pathogens in Various Organic Fertilizers. Foodborne Pathog. Dis. 2013;10:520–527. doi: 10.1089/fpd.2012.1403. [DOI] [PubMed] [Google Scholar]
- 30.Cooley M.B., Quinones B., Oryang D., Mandrell R.E., Gorski L. Prevalence of Shiga-toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front. Cell. Infect. Microbiol. 2014;4:13. doi: 10.3389/fcimb.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooley M.B., Carychao D., Gorski L. Optimized Co-extraction and Quantification of DNA From Enteric Pathogens in Surface Water Samples Near Produce Fields in California. Front. Microbiol. 2018;9:9. doi: 10.3389/fmicb.2018.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorski L., Walker S., Liang A.S., Nguyen K.M., Govoni J., Carychao D., Cooley M.B., Mandrell R.E. Comparison of Subtypes of Listeria monocytogenes Isolates from Naturally Contaminated Watershed Samples with and without a Selective Secondary Enrichment. PLoS ONE. 2014;9:e92467. doi: 10.1371/journal.pone.0092467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian P., Yang D., Shan L., Wang D.P., Li Q.Q., Gorski L., Lee B.G., Quinones B., Cooley M.B. Concurrent Detection of Human Norovirus and Bacterial Pathogens in Water Samples from an Agricultural Region in Central California Coast. Front. Microbiol. 2017;8:11. doi: 10.3389/fmicb.2017.01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grewal S.K., Rajeev S., Sreevatsan S., Michel F.C. Persistence of Mycobacterium avium subsp. paratuberculosis and other zoonotic pathogens during simulated composting, manure packing, and liquid storage of dairy manure. Appl. Environ. Microbiol. 2006;72:565–574. doi: 10.1128/aem.72.1.565-574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu G.Y., Yin H.B., Ottesen A., Bolten S., Patel J., Rideout S., Nou X.W. Microbiomes in Ground Water and Alternative Irrigation Water, and Spinach Microbiomes Impacted by Irrigation with Different Types of Water. Phytobiomes J. 2019;3:137–147. doi: 10.1094/PBIOMES-09-18-0037-R. [DOI] [Google Scholar]
- 36.Liao C.H., Honeycutt C.W., Griffin T.S., Jemison J.M. Occurrence of gastrointestinal pathogens in soil of potato field treated with liquid dairy manure. J. Food Agric. Environ. 2003;1:224–228. [Google Scholar]
- 37.Pang H., McEgan R., Mishra A., Micallef S.A., Pradhan A.K. Identifying and modeling meteorological risk factors associated with pre-harvest contamination of Listeria species in a mixed produce and dairy farm. Food Res. Int. 2017;102:355–363. doi: 10.1016/j.foodres.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 38.Prazak A.M., Murano E.A., Mercado I., Acuff G.R. Prevalence of Listeria monocytogenes during production and postharvest processing of cabbage. J. Food Prot. 2002;65:1728–1734. doi: 10.4315/0362-028X-65.11.1728. [DOI] [PubMed] [Google Scholar]
- 39.Allard S.M., Callahan M.T., Bui A., Ferelli A.M.C., Chopyk J., Chattopadhyay S., Mongodin E.F., Micallef S.A., Sapkota A.R. Creek to Table: Tracking fecal indicator bacteria, bacterial pathogens, and total bacterial communities from irrigation water to kale and radish crops. Sci. Total Environ. 2019;666:461–471. doi: 10.1016/j.scitotenv.2019.02.179. [DOI] [PubMed] [Google Scholar]
- 40.Weller D., Wiedmann M., Strawn L.K. Spatial and Temporal Factors Associated with an Increased Prevalence of Listeria monocytogenes in Spinach Fields in New York State. Appl. Environ. Microbiol. 2015;81:6059–6069. doi: 10.1128/AEM.01286-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauders B.D., Durak M.Z., Fortes E., Windham K., Schukken Y., Lembo A.J., Akey B., Nightingale K.K., Wiedmann M. Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 2006;69:93–105. doi: 10.4315/0362-028X-69.1.93. [DOI] [PubMed] [Google Scholar]
- 42.Sauders B.D., Overdevest J., Fortes E., Windham K., Schukken Y., Lembo A., Wiedmann M. Diversity of Listeria Species in Urban and Natural Environments. Appl. Environ. Microbiol. 2012;78:4420–4433. doi: 10.1128/AEM.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weller D., Brassill N., Rock C., Ivanek R., Mudrak E., Roof S., Ganda E., Wiedmann M. Complex Interactions between Weather, and Microbial and Physicochemical Water Quality Impact the Likelihood of Detecting Foodborne Pathogens in Agricultural Water. Front. Microbiol. 2020;11:20. doi: 10.3389/fmicb.2020.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu L., Torres M., Betancourt W.Q., Sharma M., Micallef S.A., Gerba C., Sapkota A.R., Sapkota A., Parveen S., Hashem F., et al. Incidence of fecal indicator and pathogenic bacteria in reclaimed and return flow waters in Arizona, USA. Environ. Res. 2019;170:122–127. doi: 10.1016/j.envres.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 45.Sharma M., Handy E.T., East C.L., Kim S.O.Y., Jiang C.S., Callahan M.T., Allard S.M., Micallef S., Craighead S., Anderson-Coughlin B., et al. Prevalence of Salmonella and Listeria monocytogenes in non-traditional irrigation waters in the Mid-Atlantic United States is affected by water type, season, and recovery method. PLoS ONE. 2020;15:e229365. doi: 10.1371/journal.pone.0229365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheng L.N., Shen X.Y., Benedict C., Su Y., Tsai H.C., Schacht E., Kruger C.E., Drennan M., Zhu M.J. Microbial Safety of Dairy Manure Fertilizer Application in Raspberry Production. Front. Microbiol. 2019;10:2276. doi: 10.3389/fmicb.2019.02276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strawn L.K., Fortes E.D., Bihn E.A., Nightingale K.K., Grohn Y.T., Worobo R.W., Wiedmann M., Bergholz P.W. Landscape and Meteorological Factors Affecting Prevalence of Three Food-Borne Pathogens in Fruit and Vegetable Farms. Appl. Environ. Microbiol. 2013;79:588–600. doi: 10.1128/AEM.02491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strawn L.K., Grohn Y.T., Warchocki S., Worobo R.W., Bihn E.A., Wiedmann M. Risk Factors Associated with Salmonella and Listeria monocytogenes Contamination of Produce Fields. Appl. Environ. Microbiol. 2013;79:7618–7627. doi: 10.1128/AEM.02831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weller D., Wiedmann M., Strawn L.K. Irrigation Is Significantly Associated with an Increased Prevalence of Listeria monocytogenes in Produce Production Environments in New York State. J. Food Prot. 2015;78:1132–1141. doi: 10.4315/0362-028X.JFP-14-584. [DOI] [PubMed] [Google Scholar]
- 50.Viswanath P., Murugesan L., Knabel S.J., Verghese B., Chikthimmah N., LaBorde L.F. Incidence of Listeria monocytogenes and Listeria spp. in a Small-Scale Mushroom Production Facility. J. Food Prot. 2013;76:608–615. doi: 10.4315/0362-028X.JFP-12-292. [DOI] [PubMed] [Google Scholar]
- 51.Xu A.X., Pahl D.M., Buchanan R.L., Micallef S.A. Comparing the Microbiological Status of Pre- and Postharvest Produce from Small Organic Production. J. Food Prot. 2015;78:1072–1080. doi: 10.4315/0362-028X.JFP-14-548. [DOI] [PubMed] [Google Scholar]
- 52.Johnston L.M., Jaykus L.A., Moll D., Martinez M.C., Anciso J., Mora B., Moe C.L. A field study of the microbiological quality of fresh produce. J. Food Prot. 2005;68:1840–1847. doi: 10.4315/0362-028X-68.9.1840. [DOI] [PubMed] [Google Scholar]
- 53.Tan X.Q., Chung T., Chen Y., Macarisin D., LaBorde L., Kovac J. The occurrence of Listeria monocytogenes is associated with built environment microbiota in three tree fruit processing facilities. Microbiome. 2019;7:18. doi: 10.1186/s40168-019-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnston L.M., Jaykus L.A., Moll D., Anciso J., Mora B., Moe C.L. A field study of the microbiological quality of fresh produce of domestic and Mexican origin. Int. J. Food Microbiol. 2006;112:83–95. doi: 10.1016/j.ijfoodmicro.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 55.Estrada E.M., Hamilton A.M., Sullivan G.B., Wiedmann M., Critzer F.J., Strawn L.K. Prevalence, Persistence, and Diversity of Listeria monocytogenes and Listeria Species in Produce Packinghouses in Three US States. J. Food Prot. 2020;83:277–286. doi: 10.4315/0362-028X.JFP-19-411. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan G., Wiedmann M. Detection and prevalence of Listeria in US produce packinghouses and fresh-cut facilities. J. Food Prot. 2020 doi: 10.4315/JFP-20-094. [DOI] [PubMed] [Google Scholar]
- 57.Ilic S., Odomeru J., LeJeune J.T. Coliforms and Prevalence of Escherichia coli and Foodborne Pathogens on Minimally Processed Spinach in Two Packing Plants. J. Food Prot. 2008;71:2398–2403. doi: 10.4315/0362-028X-71.12.2398. [DOI] [PubMed] [Google Scholar]
- 58.John J., Joy W.C., Jovana K. Prevalence of Listeria spp. in produce handling and processing facilities in the Pacific Northwest. Food Microbiol. 2020;90:103468. doi: 10.1016/j.fm.2020.103468. [DOI] [PubMed] [Google Scholar]
- 59.Phillips C.A., Harrison M.A. Comparison of the microflora on organically and conventionally grown spring mix from a California processor. J. Food Prot. 2005;68:1143–1146. doi: 10.4315/0362-028X-68.6.1143. [DOI] [PubMed] [Google Scholar]
- 60.Murugesan L., Kucerova Z., Knabel S.J., LaBorde L.F. Predominance and Distribution of a Persistent Listeria monocytogenes Clone in a Commercial Fresh Mushroom Processing Environment. J. Food Prot. 2015;78:1988–1998. doi: 10.4315/0362-028X.JFP-15-195. [DOI] [PubMed] [Google Scholar]
- 61.Jarvis K.G., Daquigan N., White J.R., Morin P.M., Howard L.M., Manetas J.E., Ottesen A., Ramachandran P., Grim C.J. Microbiomes Associated With Foods From Plant and Animal Sources. Front. Microbiol. 2018;9:2540. doi: 10.3389/fmicb.2018.02540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Churchill K.J., Sargeant J.M., Farber J.M., O’Connor A.M. Prevalence of Listeria monocytogenes in Select Ready-to-Eat Foods-Deli Meat, Soft Cheese, and Packaged Salad: A Systematic Review and Meta-Analysis. J. Food Prot. 2019;82:344–357. doi: 10.4315/0362-028X.JFP-18-158. [DOI] [PubMed] [Google Scholar]
- 63.Korir R.C., Parveen S., Hashem F., Bowers J. Microbiological quality of fresh produce obtained from retail stores on the Eastern Shore of Maryland, United States of America. Food Microbiol. 2016;56:29–34. doi: 10.1016/j.fm.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Burnett J., Wu S.T., den Bakker H.C., Cook P.W., Veenhuizen D.R., Hammons S.R., Singh M., Oliver H.F. Listeria monocytogenes is prevalent in retail produce environments but Salmonella enterica is rare. Food Control. 2020;113:11. doi: 10.1016/j.foodcont.2020.107173. [DOI] [Google Scholar]
- 65.Gombas D.E., Chen Y.H., Clavero R.S., Scott V.N. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 2003;66:559–569. doi: 10.4315/0362-028X-66.4.559. [DOI] [PubMed] [Google Scholar]
- 66.Luchansky J.B., Chen Y.H., Porto-Fett A.C.S., Pouillot R., Shoyer B.A., Johnson-DeRycke R., Eblen D.R., Hoelzer K., Shaw W.K., Van Doren J.M., et al. Survey for Listeria monocytogenes in and on Ready-to-Eat Foods from Retail Establishments in the United States (2010 through 2013): Assessing Potential Changes of Pathogen Prevalence and Levels in a Decade. J. Food Prot. 2017;80:903–921. doi: 10.4315/0362-028X.JFP-16-420. [DOI] [PubMed] [Google Scholar]
- 67.Kim C., Nartea T.J., Pao S., Li H., Jordan K.L., Xu Y., Stein R.A., Sismour E.N. Evaluation of Microbial Loads on Dried and Fresh Shiitake Mushrooms (Lentinula edodes) as Obtained from Internet and Local Retail Markets, Respectively. Food Saf. 2016;4:45–51. doi: 10.14252/foodsafetyfscj.2016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strapp C.M., Shearer A.E.H., Joerger R.D. Survey of retail alfalfa sprouts and mushrooms for the presence of Escherichia coli O157:H7, Salmonella, and Listeria with BAX, and evaluation of this polymerase chain reaction-based system with experimentally contaminated samples. J. Food Prot. 2003;66:182–187. doi: 10.4315/0362-028X-66.2.182. [DOI] [PubMed] [Google Scholar]
- 69.Samadpour M., Barbour M.W., Nguyen T., Cao T.M., Buck F., Depavia G.A., Mazengia E., Yang P., Alfi D., Lopes M., et al. Incidence of enterohemorrhagic Escherichia coli, Escherichia coli O157, Salmonella, and Listeria monocytogenes in retail fresh ground beef, sprouts, and mushrooms. J. Food Prot. 2006;69:441–443. doi: 10.4315/0362-028X-69.2.441. [DOI] [PubMed] [Google Scholar]
- 70.Zhang G.D., Chen Y., Hu L.J., Melka D., Wang H., Laasri A., Brown E.W., Strain E., Allard M., Bunning V.K., et al. Survey of Foodborne Pathogens, Aerobic Plate Counts, Total Coliform Counts, and Escherichia coli Counts in Leafy Greens, Sprouts, and Melons Marketed in the United States. J. Food Prot. 2018;81:400–411. doi: 10.4315/0362-028X.JFP-17-253. [DOI] [PubMed] [Google Scholar]
- 71.Signs R.J., Darcey V.L., Carney T.A., Evans A.A., Quinlan J.J. Retail Food Safety Risks for Populations of Different Races, Ethnicities, and Income Levels. J. Food Prot. 2011;74:1717–1723. doi: 10.4315/0362-028X.JFP-11-059. [DOI] [PubMed] [Google Scholar]
- 72.Petran R.L., Zottola E.A., Gravani R.B. Incidence of Listeria monocytogenes in Market Samples of Fresh and Frozen Vegetables. J. Food Sci. 1988;53:1238–1240. doi: 10.1111/j.1365-2621.1988.tb13576.x. [DOI] [Google Scholar]
- 73.Heisick J.E., Wagner D.E., Nierman M.L., Peeler J.T. Listeria spp. found on fresh market produce. Appl. Environ. Microbiol. 1989;55:1925–1927. doi: 10.1128/aem.55.8.1925-1927.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li K.W., Weidhaas J., Lemonakis L., Khouryieh H., Stone M., Jones L., Shen C.L. Microbiological quality and safety of fresh produce in West Virginia and Kentucky farmers’ markets and validation of a post-harvest washing practice with antimicrobials to inactivate Salmonella and Listeria monocytogenes. Food Control. 2017;79:101–108. doi: 10.1016/j.foodcont.2017.03.031. [DOI] [Google Scholar]
- 75.Scheinberg J.A., Dudley E.G., Campbell J., Roberts B., DiMarzio M., DebRoy C., Cutter C.N. Prevalence and Phylogenetic Characterization of Escherichia coli and Hygiene Indicator Bacteria Isolated from Leafy Green Produce, Beef, and Pork Obtained from Farmers’ Markets in Pennsylvania. J. Food Prot. 2017;80:237–244. doi: 10.4315/0362-028X.JFP-16-282. [DOI] [PubMed] [Google Scholar]
- 76.Thunberg R.L., Tran T.T., Bennett R.W., Matthews R.N., Belay N. Microbial evaluation of selected fresh produce obtained at retail markets. J. Food Prot. 2002;65:677–682. doi: 10.4315/0362-028X-65.4.677. [DOI] [PubMed] [Google Scholar]
- 77.Roth L., Simonne A., House L., Ahn S. Microbiological analysis of fresh produce sold at Florida farmers’ markets. Food Control. 2018;92:444–449. doi: 10.1016/j.foodcont.2018.05.030. [DOI] [Google Scholar]
- 78.Borrusso P.A., Quinlan J.J. Prevalence of Pathogens and Indicator Organisms in Home Kitchens and Correlation with Unsafe Food Handling Practices and Conditions. J. Food Prot. 2017;80:590–597. doi: 10.4315/0362-028X.JFP-16-354. [DOI] [PubMed] [Google Scholar]
- 79.Gorski L., Parker C.T., Liang A.S., Walker S., Romanolo K.F. The Majority of Genotypes of the Virulence Gene inlA Are Intact among Natural Watershed Isolates of Listeria monocytogenes from the Central California Coast. PLoS ONE. 2016;11:e167566. doi: 10.1371/journal.pone.0167566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Y., Gonzalez-Escalona N., Hammack T.S., Allard M.W., Strain E.A., Brown E.W. Core Genome Multilocus Sequence Typing for Identification of Globally Distributed Clonal Groups and Differentiation of Outbreak Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2016;82:6258–6272. doi: 10.1128/AEM.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burall L.S., Grim C.J., Mammel M.K., Datta A.R. A Comprehensive Evaluation of the Genetic Relatedness of Listeria monocytogenes Serotype 4b Variant Strains. Front. Public Health. 2017;5:241. doi: 10.3389/fpubh.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parsons C., Costolo B., Brown P., Kathariou S. Penicillin-binding protein encoded by pbp4 is involved in mediating copper stress in Listeria monocytogenes. FEMS Microbiol. Lett. 2017;364:fnx207. doi: 10.1093/femsle/fnx207. [DOI] [PubMed] [Google Scholar]
- 83.Laksanalamai P., Joseph L.A., Silk B.J., Burall L.S., Tarr C.L., Gerner-Smidt P., Datta A.R. Genomic Characterization of Listeria monocytogenes Strains Involved in a Multistate Listeriosis Outbreak Associated with Cantaloupe in US. PLoS ONE. 2012;7:e42448. doi: 10.1371/journal.pone.0042448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Price R., Jayeola V., Niedermeyer J., Parsons C., Kathariou S. The Listeria monocytogenes Key Virulence Determinants hly and prfA are involved in Biofilm Formation and Aggregation but not Colonization of Fresh Produce. Pathogens. 2018;7:18. doi: 10.3390/pathogens7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y., Pettengill J.B., Pightling A., Timme R., Allard M., Strain E., Rand H. Genetic Diversity of Salmonella and Listeria Isolates from Food Facilities. J. Food Prot. 2018;81:2082–2089. doi: 10.4315/0362-028X.JFP-18-093. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y.F., Yeh E., Hall G., Cripe J., Bhagwat A.A., Meng J.H. Characterization of Listeria monocytogenes isolated from retail foods. Int. J. Food Microbiol. 2007;113:47–53. doi: 10.1016/j.ijfoodmicro.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 87.Ratani S.S., Siletzky R.M., Dutta V., Yildirim S., Osborne J.A., Lin W., Hitchins A.D., Ward T.J., Kathariou S. Heavy Metal and Disinfectant Resistance of Listeria monocytogenes from Foods and Food Processing Plants. Appl. Environ. Microbiol. 2012;78:6938–6945. doi: 10.1128/AEM.01553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li K.W., Khouryieh H., Jones L., Etienne X., Shen C.L. Assessing farmers market produce vendors’ handling of containers and evaluation of the survival of Salmonella and Listeria monocytogenes on plastic, pressed-card, and wood container surfaces at refrigerated and room temperature. Food Control. 2018;94:116–122. doi: 10.1016/j.foodcont.2018.06.036. [DOI] [Google Scholar]
- 89.Schardt J., Jones G., Müller-Herbst S., Schauer K., D’Orazio S.E., Fuchs T.M. Comparison between Listeria sensu stricto and Listeria sensu lato strains identifies novel determinants involved in infection. Sci. Rep. 2017;7:1–14. doi: 10.1038/s41598-017-17570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerner-Smidt P., Besser J., Concepcion-Acevedo J., Folster J.P., Huffman J., Joseph L.A., Kucerova Z., Nichols M.C., Schwensohn C.A., Tolar B. Whole Genome Sequencing: Bridging One-Health Surveillance of Foodborne Diseases. Front. Public Health. 2019;7:172. doi: 10.3389/fpubh.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mohammad Z.H., Yu H.Y., Neal J.A., Gibson K.E., Sirsat S.A. Food Safety Challenges and Barriers in Southern United States Farmers Markets. Foods. 2020;9:12. doi: 10.3390/foods9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pollard S., Boyer R., Chapman B., di Stefano J., Archibald T., Ponder M.A., Rideout S. Identification of risky food safety practices at southwest Virginia farmers’ markets. Food Prot. Trends. 2016;36:168–175. [Google Scholar]
- 93.Harrison J.A., Gaskin J.W., Harrison M.A., Cannon J.L., Boyer R.R., Zehnder G.W. Survey of Food Safety Practices on Small to Medium-Sized Farms and in Farmers Markets. J. Food Prot. 2013;76:1989–1993. doi: 10.4315/0362-028X.JFP-13-158. [DOI] [PubMed] [Google Scholar]
- 94.Jones T.F., Angulo F.J. Eating in restaurants: A risk factor for foodborne disease? Clin. Infect. Dis. 2006;43:1324–1328. doi: 10.1086/508540. [DOI] [PubMed] [Google Scholar]
- 95.Gould L.H., Rosenblum I., Nicholas D., Phan Q., Jones T.F. Contributing Factors in Restaurant- Associated Foodborne Disease Outbreaks, FoodNet Sites, 2006 and 2007. J. Food Prot. 2013;76:1824–1828. doi: 10.4315/0362-028X.JFP-13-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.US Food and Drug Administration FDA Report on the Occurrence of Foodborne Illness Risk Factors in Fast Food and Full Service Restaurants, 2013–2014. [(accessed on 22 April 2020)];2018 Available online: https://www.fda.gov/food/cfsan-constituent-updates/fda-releases-report-occurrence-foodborne-illness-risk-factors-fast-food-and-full-service-restaurants.
- 97.Byrd-Bredbenner C., Berning J., Martin-Biggers J., Quick V. Food Safety in Home Kitchens: A Synthesis of the Literature. Int. J. Environ. Res. Public Health. 2013;10:4060–4085. doi: 10.3390/ijerph10094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Beirne D., Gomez-Lopez V., Tudela J.A., Allende A., Gil M.I. Effects of oxygen-depleted atmospheres on survival and growth of Listeria monocytogenes on fresh-cut Iceberg lettuce stored at mild abuse commercial temperatures. Food Microbiol. 2015;48:17–21. doi: 10.1016/j.fm.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 99.Thomas C., Prior O., O’Beirne D. Survival and growth of Listeria species in a model ready-to-use vegetable product containing raw and cooked ingredients as affected by storage temperature and acidification. Int. J. Food Sci. Technol. 1999;34:317–324. doi: 10.1046/j.1365-2621.1999.00273.x. [DOI] [Google Scholar]
- 100.Ndraha N., Hsiao H.I., Vlajic J., Yang M.F., Lin H.T.V. Time-temperature abuse in the food cold chain: Review of issues, challenges, and recommendations. Food Control. 2018;89:12–21. doi: 10.1016/j.foodcont.2018.01.027. [DOI] [Google Scholar]
- 101.Jayeola V., Jeong S., Almenar E., Marks B.P., Vorst K.L., Brown J.W., Ryser E.T. Predicting the Growth of Listeria monocytogenes and Salmonella Typhimurium in Diced Celery, Onions, and Tomatoes during Simulated Commercial Transport, Retail Storage, and Display. J. Food Prot. 2019;82:287–300. doi: 10.4315/0362-028X.JFP-18-277. [DOI] [PubMed] [Google Scholar]
- 102.Huang J.W., Luo Y.G., Zhou B., Zheng J., Nou X.W. Growth and survival of Salmonella enterica and Listeria monocytogenes on fresh-cut produce and their juice extracts: Impacts and interactions of food matrices and temperature abuse conditions. Food Control. 2019;100:300–304. doi: 10.1016/j.foodcont.2018.12.035. [DOI] [Google Scholar]
- 103.Huang J.W., Luo Y.G., Nou X.W. Growth of Salmonella enterica and Listeria monocytogenes on Fresh-Cut Cantaloupe under Different Temperature Abuse Scenarios. J. Food Prot. 2015;78:1125–1131. doi: 10.4315/0362-028X.JFP-14-468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.