Abstract
Lung cancer has been a leading cause of cancer-related death for decades and therapeutic strategies for non-driver mutation lung cancer are still lacking. A novel approach for this type of lung cancer is an emergent requirement. Here we find that loss of LSAMP (Limbic System Associated Membrane Protein), compared to other IgLON family of proteins NTM (Neurotrimin) and OPCML (OPioid-binding Cell adhesion MoLecule), exhibits the strongest prognostic and therapeutic significance in predicting lung adenocarcinoma (LUAD) progression. Lower expression of LSAMP and NTM, but not OPCML, were found in tumor parts compared with normal parts in six LUAD patients, and this was validated by public datasets, Oncomine® and TCGA. The lower expression of LSAMP, but not NTM, was correlated to shorter overall survival. Two epigenetic regulations, including hypermethylation and miR-143-3p upregulation but not copy number variation, were associated with downregulation of LSAMP in LUAD patients. Pathway network analysis showed that NEGR1 (Neuronal Growth Regulator 1) was involved in the regulatory loop of LSAMP. The biologic functions by LSMAP knockdown in lung cancer cells revealed LSMAP was linked to cancer cell migration via epithelial-mesenchymal transition (EMT) but not proliferation nor stemness of LUAD. Our result showed for the first time that LSAMP acts as a potential tumor suppressor in regulating lung cancer. A further deep investigation into the role of LSAMP in lung cancer tumorigenesis would provide therapeutic hope for such affected patients.
Keywords: EMT, lung adenocarcinoma, LSAMP
1. Introduction
Lung cancer is still the most deadly cancer worldwide [1,2]. In Taiwan, lung cancer has been responsible for the leading cause of cancer-related death for decades [3]. The most common causes that lead to lung cancer are tobacco smoking, air pollution either indoor or outdoor, asbestos, etc. [4]. Furthermore, LUAD harbors distinctive features of gene alterations such as epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase-echinoderm microtubule-associated protein-like4 (ALK-EML4), c-ros oncogene 1 (ROS1), REarranged during Transfection (RET), and neurotrophic tropomyosin-related kinases (NTRK) [5]. Nowadays, the existence of these driver mutations provides therapeutic benefits via specific tyrosine kinase inhibitors (TKIs) with the greatest efforts focusing on them. However, the critical issue lies in that the LUAD patients of indistinctive gene alternations only benefit from platinum doublets and/or immunotherapy with a demise hope [6]. To overcome this difficulty, the novel mechanism and relevant treatment strategy are still necessary to pursue higher therapeutic rates.
Avoidance of metastasis is a hot target for lung cancer research. Tumors of metastatic entities contribute to more than 90% of cancer-related deaths and the aggressiveness of lung cancer enables them to metastasize by enhancing mobility, invasion, and resistance to apoptotic stimuli [7] and regains their stem-like trait [8]. The epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) mechanisms confer tumor aggressiveness and metastasis with EMT is associated with loss of cell-cell adhesion and the gain of invasiveness [9]. At the same time, the MET causes morphologic and functional changes and the acquisition of epithelial characters to facilitate metastasis. In addition, EMT tightly contributes to the cancer stem cell (CSC) theory, which implies that only a few subpopulations of “stem-like” cells can lead to metastatic disease [9]. CSCs are cells with the ability to self-renew and generate a heterogeneous population within the tumor. The roles of EMT/MET and CSC are keys to elucidate the mechanisms of cancer metastasis.
IgLONs are an immunoglobulin (Ig) subfamily, including opioid binding protein/cell adhesion molecule–like (OPCML/OBCAM) [10], limbic system–associated membrane protein (LSAMP/LAMP) [10], neurotrimin (NTM) [11], neuronal growth regulator 1 (NEGR1/Kilon) [12], and IgLON family member 5 (IgLON5) [13]. The IgLON family consists of a group of cell adhesion molecules to regulate neurite outgrowth [14], dendritic arborization [15], and synapse formation [16]. They execute their functions by forming homophilic and heterophilic complexes along the cell surface. The functional specificity and divergence of the IgLONs are to control neuronal development and psychiatric disorders such as schizophrenia and major depression [17,18]. The behavioral changes and structural brain endophenotypes related to IgLONs are not limited to humans but also affect other mammals such as mice [18]. IgLON neural adhesion molecules not only regulate neuronal events, but also function as tumor-suppressor genes in a number of non-neural organs and tissue types [19]. IgLONs have been linked to sporadic ovarian tumors [20], gastric cancer [21], and osteosarcoma [22]; however, the IgLONs have not been well investigated in lung cancer, and elucidation of this family might be of great help in curing this disease.
To overcome the demise of hope for non-driver mutation, the advancement in genome-wide profiling technology has brought about a substantial improvement of the understanding of the genetic landscape and pathways leading to lung cancer [23,24]. Global analysis of gene expression by transcriptomic sequencing has generated a complete set of RNA transcripts (messenger RNA, micro RNA, etc.) of the genome of lung cancer in-depth [25,26]. In this study, we took advantage of the powerful next-generation sequencing (NGS) to investigate the expression of the three members of the IgLON family from six-paired adjacent normal versus tumor lung tissue samples from patients with lung cancer. This study analyzed the three members of the IgLON family—OPCML, LSAMP, and NTM—which revealed decreased LSAMP expression in patient samples and public databases, such as The Cancer Genome Atlas (TCGA) and Oncomine datasets. Furthermore, the lower the LSAMP was, the more aggressive lung cancer behaved. The low level of LSAMP conferred a shorter survival time and under the epigenetic regulation by miR-141-3p and DNA methylation. These results provided evidence that LSAMP regulates tumorigenesis and brings treatment hope for lung cancer. The advanced exploration of LSAMP in tumorigenesis might offer therapeutic hope in lung cancer.
2. Materials and Methods
2.1. Cell Lines and Cell Culture
Human lung adenocarcinoma cell lines A549 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). A549 cells were cultured in F-12K Medium (ATCC) with the supplement of 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin (Thermo Fisher Scientific, Boston, MA, USA). A549 was authenticated by the short tandem repeat analysis (Promega, Madison, WI, USA) and ascertained negative for mycoplasma contamination by MycoAlert™ mycoplasma detection kit (Lonza, Switzerland) every 3 months.
2.2. Next-Generation Sequencing
Six pairs of adjacent lung non-tumor and tumor tissues were acquired from the Division of Thoracic Surgery and Division of Pulmonary and Critical Care Medicine, Kaohsiung Medical University Hospital (Kaohsiung, Taiwan). The study protocol was reviewed and approved by the Institutional Review Board of Kaohsiung Medical University Hospital (KMUH-IRB-20130054, 24 May 2013). The freshly isolated and unfixed surgical tissues were immediately extracted for total RNA preparation. The mRNA profiles were further examined utilizing NGS using the Illumina platform (Welgene Biotechnology Company, Taipei, Taiwan) [27]. RNA and small RNA library construction were carried out using an Illumina sample preparation kit, following the protocol of the TruSeq RNA or Small RNA Sample Preparation Guide. The criteria for differentially expressed mRNA by NGS analysis were fold change >2 and fragments per kilobase million (FPKM) >0.3. Furthermore, the IgLON family, OPCML, LSAMP, and NTM were extracted for analysis.
2.3. Bioinformatics
The mRNA expression of IgLONs was derived from the TCGA of UALCAN (accessed on 1 January 2021; ualcan.path.uab.edu) [28]. According to the data from this website, the expression levels of IgLONs were reclassified under the criteria of lymph node metastasis (N0-N3) and tumor stage (pathologic stages 1 to 4) based on AJCC (American Joint Committee on Cancer) 8th edition. The ratio of mRNA expression in lung cancer and normal specimens (cancer vs. normal) was derived from Oncomine database (accessed on 1 January 2021; http://www.oncomine.org), Compendia biosciences, Ann Arbor, MI, USA). The criteria for analysis were >2 fold change and p-value < 0.0001 through the independent t-test. The protein expression of IgLONs was retrieved from the CPTAC of UALCAN (accessed on January 2021; ualcan.path.uab.edu) [28]. The expression levels of LSAMP, NTM but lacking OPCML, were reclassified under the criteria of lymph node metastasis (N0–N3) and tumor stage (stage 1 to 4). The relationship between IgLONs and overall survival rates in lung adenocarcinoma was assessed by either the RNA-seq or the RNA gene chip cohort in the KM plotter (accessed on 1 August 2020; http://kmplot.com/analysis/). Patients were divided into 2 groups (high and low expression) based on the best cut-off in RNA-seq cohort and by the median value in the RNA gene chip cohort. The probability of survival between the two groups was computed. The hazard ratios (95% confidence intervals) were calculated using the Cox proportional model.
2.4. DNA Methylation and Copy Number
The extent of LSAMP DNA methylation was compared according to the tissue source, the tumor stages, and the stages of lymph node metastasis on the UALCAN website (accessed on 1 January 2021; ualcan.path.uab.edu). The expression profiles of gene-specific DNA methylation data, copy number (gene level) and clinical information of patients with LUAD were extracted from the dataset TCGA Pan-Cancer (PANCAN) from the UCSC Xena website. Pearson’s correlation between LSAMP mRNA expression level and the copy number and/or DNA methylation was calculated using the metadata.
2.5. miRNAs and LSAMP Correlation
Pearson’s correlation between LSAMP and all microRNAs in the TCGA LUAD dataset was calculated. The miRNAs were ranked by r value and validated by TargetScan (accessed on 1 August 2020; http://www.targetscan.org/vert_72/) from the top-ranked miRNAs. Excluding the miRNAs with context++ score percentile <90, only miR-141-3p remained with a good context++ score 98th percentile.
2.6. Gene and Protein Interaction of LSAMP
Genes interacting with LSAMP are predicted by the Pathway Common website (accessed on 1 January 2021; https://www.pathwaycommons.org/; Version 12), whereas the proteins that interact with LSAMP are predicted by String Website (accessed on 1 January 2021; https://string-db.org/; Version 11.0).
2.7. LSAMP Knockdown
Knockdown of LSAMP in A549 cells was performed using an shRNA expression system obtained from the National RNAi Core Facility (Taipei, Taiwan). The knockdown efficacy of LSAMP shRNA plasmid was determined by Immunoblot.
2.8. Cell Proliferation, Transwell Migration, Wound Healing, and Tumor Spheroid Formation
Cell proliferation was assessed by BrdU (Bromodeoxyuridine/5-bromo-2′-deoxyuridine) incorporation analysis for 72 h (EMD Millipore, Burlington, MA, USA), according to the manufacturer’s protocol. For migration, cells were seeded into inserts with polyester membranes with a pore size of 8 μm (EMD Millipore, Burlington, MA, USA). Complete cell culture medium was then added to the bottom wells for 48 h as a chemo-attractant. Migratory cells were visualized using crystal violet staining. Alternatively, cells were placed into a 12 well-plate at 100% confluence, and cell movement was measured by determining the migration of cells into the acellular area formatted by a sterile tip. The quantitative results of transwell migration and spheroid formation were performed using the counting method. For tumor spheroid formation, cells were seeded in ultralow-attachment plates (Corning Life Sciences, Tewksbury, MA, USA) for 7 days and the tumor spheres were assessed using an ImageXpress Micro system (Molecular Devices LLC, Sunnyvale, CA, USA).
2.9. Immunoblot
Cellular total protein was extracted using RIPA lysis buffer (EMD Millipore, Billerica, MA, USA) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). An equal amount of cellular protein was denatured by heating, and then separated by SDS-PAGE. Proteins were transferred to PVDF membranes (EMD Millipore, Burlington, MA, USA) and probed with various primary antibodies for 4–16 h, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell-Signaling Technology, Danvers, MA, USA). Signals of specific proteins were detected using a chemiluminescence kit (EMD Millipore, Burlington, MA, USA). The primary antibodies used included LSAMP (PA5-69373, PA5-51223 for IHC, ThermoFisher, Waltham, MA, USA), those not characteristic of cancer stem cells, c-Myc (Catalog # 18583, Cell-Signaling Technology, Danvers, MA, USA), OCT4 (Catalog#2890, Cell-Signaling Technology, Danvers, MA, USA), Nanog (Catalog#4903, Cell-Signaling Technology, Danvers, MA, USA), SOX2 (Catalog#3579, Cell-Signaling Technology, Danvers, MA, USA), and GAPDH (Catalog#5174, Cell-Signaling Technology, Danvers, MA, USA) and EMT markers, N-cadherin (Catalog #610920, BD Biosciences (San Jose, CA, USA)), E-cadherin (Catalog #610404, BD Biosciences, Franklin Lakes, NJ, USA), Vimentin (Catalog#550513, BD Biosciences, Franklin Lakes, NJ, USA), α-SMA (Catalog #A5228, Sigma-Aldrich, St. Louis, MO, USA), Snail (Catalog t# 3879, Cell-Signaling Technology, Danvers, MA, USA), and Slug (Catalog # 9585, Cell-Signaling Technology, Danvers, MA, USA).
2.10. Statistical Analysis
Results of the cell experiments are presented as mean ± standard deviation (SD). Differences between the two tested groups were compared using the Student’s t-test with GraphPad Prism software (7.04 version, Graphpad Software, San Diego, CA, USA). Statistical significance was defined as p-value < 0.05.
3. Results
3.1. The Downregulated Expressions of LSAMP and NTM Genes in Lung Adenocarcinoma
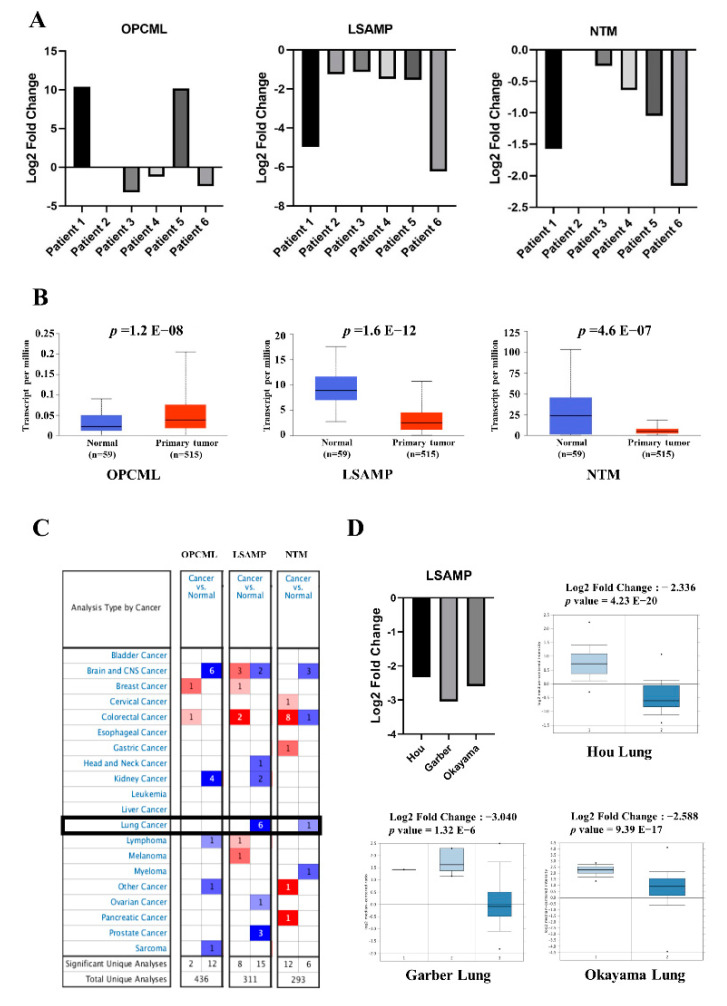
To identify the family of IgLON genes including OPCML, LSAMP, and NTM that regulate lung tumorigenesis, six paired normal and adenocarcinoma lung tissues were collected for analysis (Table 1). The expression levels of LSAMP and NTM in tumor tissues were lower than that in normal tissue samples of lung adenocarcinoma (LUAD) (middle and right panel, Figure 1A); however, the levels of OPCML were relatively inconsistent (left panel, Figure 1A). To further validate the gene expression of OPCML, LSAMP, and NTM, we retrieved data from Oncomine and TCGA cohorts with analysis showing higher expression of OPCML and lower expressions of LSAMP and NTM in tumor tissues compared with normal tissue with statistical significance in LUAD (Figure 1B). In the Oncomine database, there were 6 (3 from LUAD, 2 LUSC and large cell carcinoma) and 1 LUAD cohorts showing downregulation of LSAMP and NTM genes, respectively. Based on these expression profiles, LSAMP was the most significant gene linked to lung cancer (Figure 1C). The available three cohorts focusing on LUAD patients were listed in Figure 1D. Taking these data into consideration, LSAMP and NTM, but not OPCML, in the IgLON family acts as a tumor suppressor in lung cancer.
Table 1.
Patient characteristics.
| Number | Gender | Age | Stage | AJCC, 8th Ed. | Comorbidity |
|---|---|---|---|---|---|
| 01 | M | 70 | 2B | T3N0M0 | Old stroke |
| 02 | M | 66 | 4B | T2aN0M1c | Hypertension |
| 03 | F | 51 | 1B | T2aN0M0 | Hepatitis B carrier |
| 04 | M | 53 | 3A | T3N2M0 | Hypertension |
| 05 | F | 60 | 1A | T1bN0M0 | Hepatitis B carrier |
| 06 | M | 67 | 1A | T1aN0M0 | UTUC, 2016 |
UTUC, Upper Tract Urothelial Carcinoma.
Figure 1.
LSAMP mediated lung cancer tumorigenesis disclosed by transcriptomic data and bioinformatic analysis: The specimens from six human subjects were collected for IgLON family analysis. The expression levels of OPCML, LSAMP and NTM of six patients with LUAD (A). The comparative expression levels of OPCML, LSAMP, and NTM in tumor versus normal tissue samples were retrieved from TCGA database (B). The expression of OPCML, LSAMP, and NTM from datasets of the Oncomine was summarized (C), and the fold change of LSAMP in three different cohorts, specifically on adenocarcinoma, was extracted from the Oncomine database (D).
3.2. LSAMP Expression Is Negatively Correlated with Tumor Aggressiveness
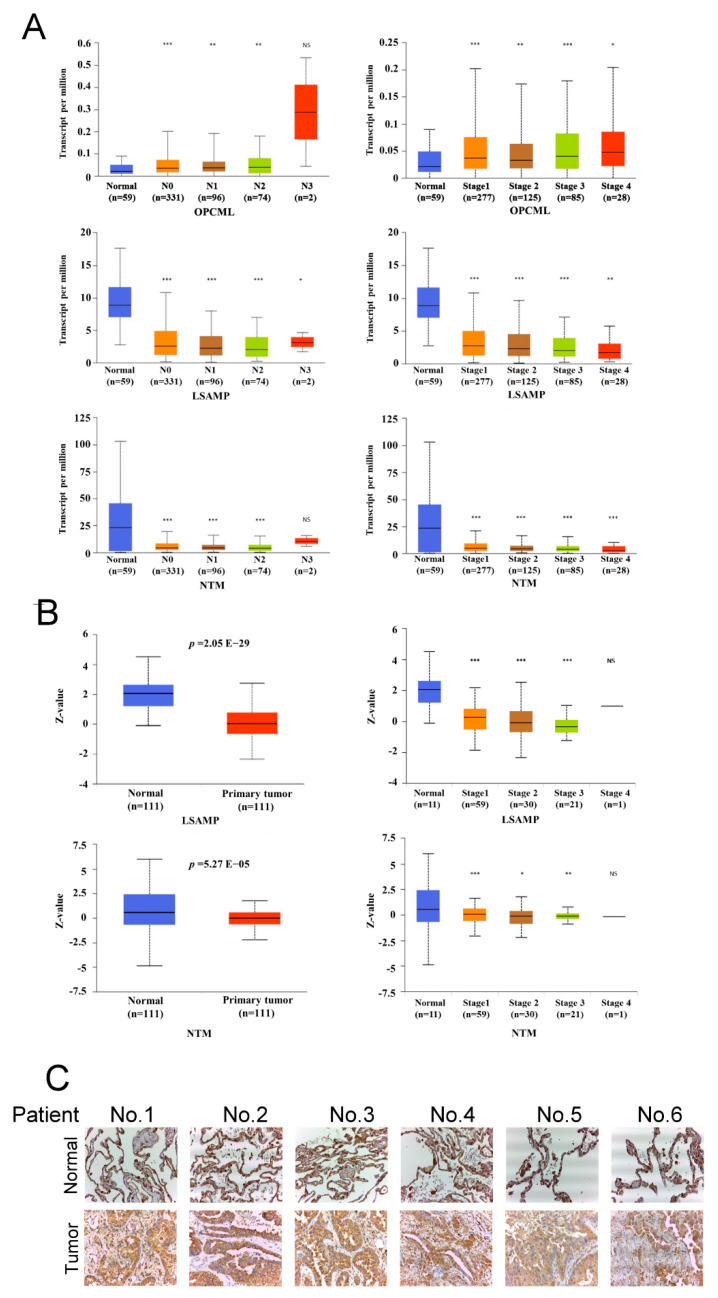
Concerning the function as suppressors of IgLONs in tumor aggressiveness, nodal metastasis and tumor staging were adopted for analysis. The expression of OPCML had no significant difference in nodal metastasis and tumor stages in the TCGA cohort (upper panel, Figure 2A). On the contrary, the expression of LSAMP and NTM correlated negatively with nodal metastasis and tumor stage (middle and lower panels, Figure 2A). The encoded genes are translated into specific proteins, which execute their specific physiologic function. Consistently, the protein expression of LSAMP and NTM of the CPTAC cohort revealed that tumor parts expressed lower levels of LSAMP and NTM protein in LUAD (left panel, Figure 2B); furthermore, the levels of LSAMP and NTM protein were correlated negatively with tumor stage (right panel, Figure 2B). However, the difference in LSAMP was more significant than that of NTM. LSAMP expression was lower in lung tumor parts than in lung normal parts from six lung cancer patients (Figure 3C). In summary, the expression of LSAMP was negatively correlated with tumor aggressiveness in both RNA and protein levels, which suggests that low expression of LSAMP is involved in tumor progression in LUAD.
Figure 2.
Low expression of LSAMP conferred lung tumor aggressiveness. The TCGA cohort showed the relationship between mRNA levels of OPCML (upper panel, (A)), LSAMP (middle panel, (A)), NTM (lower panel, (A)) and nodal metastasis and tumor stages. Furthermore, the CPTAC cohort showed the protein levels, LSAMP, and NTM (left panel, (B)) in both normal and tumor tissues. The correlation between the protein expression levels of LSAMP and NTM and tumor stages (right panel, B). The immunohistochemical staining of LSAMP from six lung cancer patients (C). * p < 0.05; ** p < 0.01; *** p < 0.005; ns, not significant.
Figure 3.
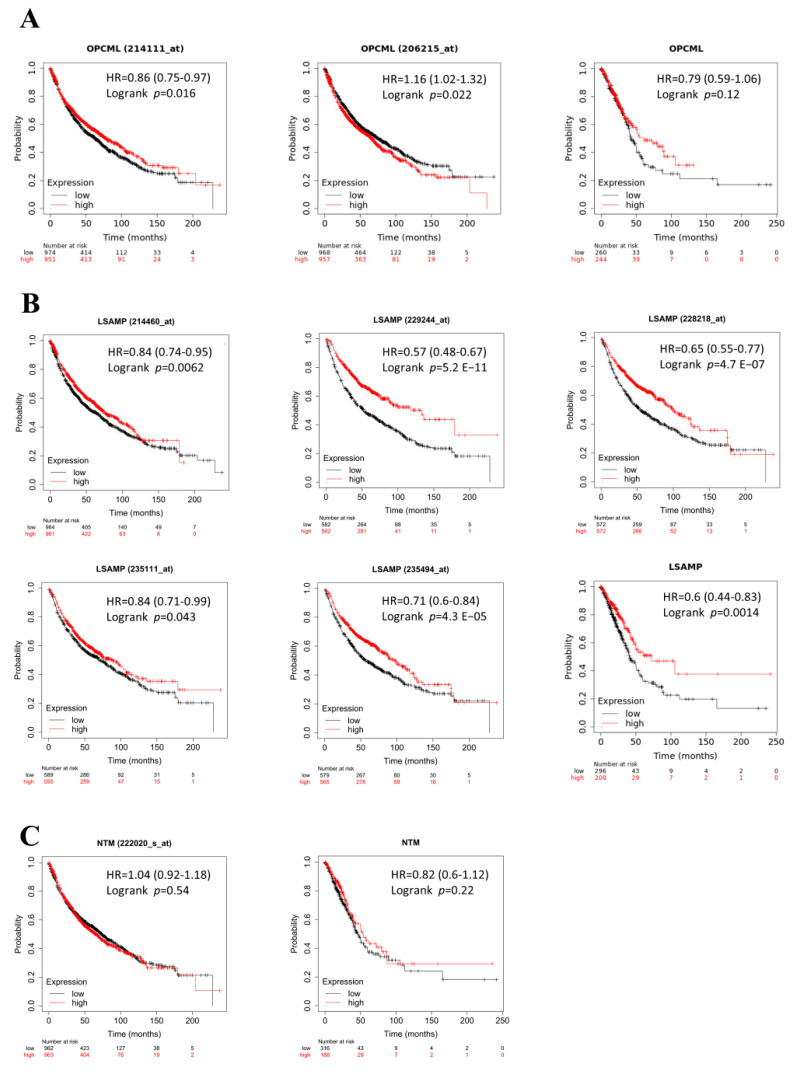
The survival analysis of IgLONs. Survival advantage or disadvantage should signify the role of each gene in lung cancer. Each gene of IgLONs and its K-M survival analyses were listed as OPCML (A), LSAMP (B), and NTM (C), respectively. HR, hazard ratio.
3.3. The Lower Levels of LSAMP Conferred Poor Survival Prognosis Clinically
To further verify the prognostic value of IgLONs in LUAD, the survival analyses were retrieved from the K-M plotter. The expression of OPCML (Figure 3A) did not correlate with survival and neither did NTM, but the survival analyses of LSAMP illustrated survival disadvantage from low LSAMP expression in LUAD extracted from the KM plotter. (Figure 3A–C). The aggressiveness of cancer shortens survival time in cancer patients and based on the survival data, lower expression of LSAMP but not NTM conferred shorter survival time in lung cancer. This suggests that LSAMP exhibits survival benefits in LUAD patients.
3.4. The Epigenetic Regulatory Mechanisms for LSAMP Expression
The expression of each gene is tightly controlled by epigenetic mechanisms as a DNA transcription step to posttranscriptional regulations. To elucidate the dysregulating mechanisms of LSAMP, possible regulatory mechanisms such as DNA methylation, copy number variation, and miRNAs were explored. Hypermethylation in the promotor of LSAMP was found in the tumor parts of LUAD, which correlated negatively with LSAMP mRNA. In addition, the status of hypermethylation of LSAMP correlated with nodal metastasis and higher tumor stages (Figure 4A). However, the correlation between copy number variation and expression of LSAMP was irrelevant (Figure 4B). To further elucidate the post-transcriptional control by microRNA, the TCGA cohort was utilized to find possible candidates, as validated by 3’UTR binding ability of microRNAs predicted by TargetScan. With the aid of bioinformatics, miR-141-3p was determined to be a regulator for LSAMP (Figure 4C). The binding prediction showed miR-141-3p has the ability to bind to 3’UTR of LSAMP from TargetScan (Figure 4E). The negative correlation between miR-141-3p and LSAMP was also proven from the TCGA cohort (Figure 4D). Moreover, the expression of miR-141-3p was higher in tumors compared with normal tissue parts, and its expression was higher in the advanced stage of LUAD, despite being not stage-dependent (Figure 4F). According to these data, both transcriptional and post-transcriptional modifications regulate LSAMP expression in LUAD.
Figure 4.
The regulatory mechanisms for LSAMP expression. The possible regulatory mechanisms included DNA methylation and nodal metastasis and tumor stage (A) and the correlation between copy number variation and LSAMP expression (B). The flow chart for the prediction of negative correlation between miRNAs and LSAMP from TCGA showed in Panel (C) with its r-value of 0.32 (D). The high binding score predicted from TargetScan was 98% (E). The correlation between miR-141-3p expression and tumor, nodal metastasis, and stage (F). * p < 0.05; *** p < 0.005; ns, not significant.
3.5. The NEGR1 Interacts with LSAMP Expression
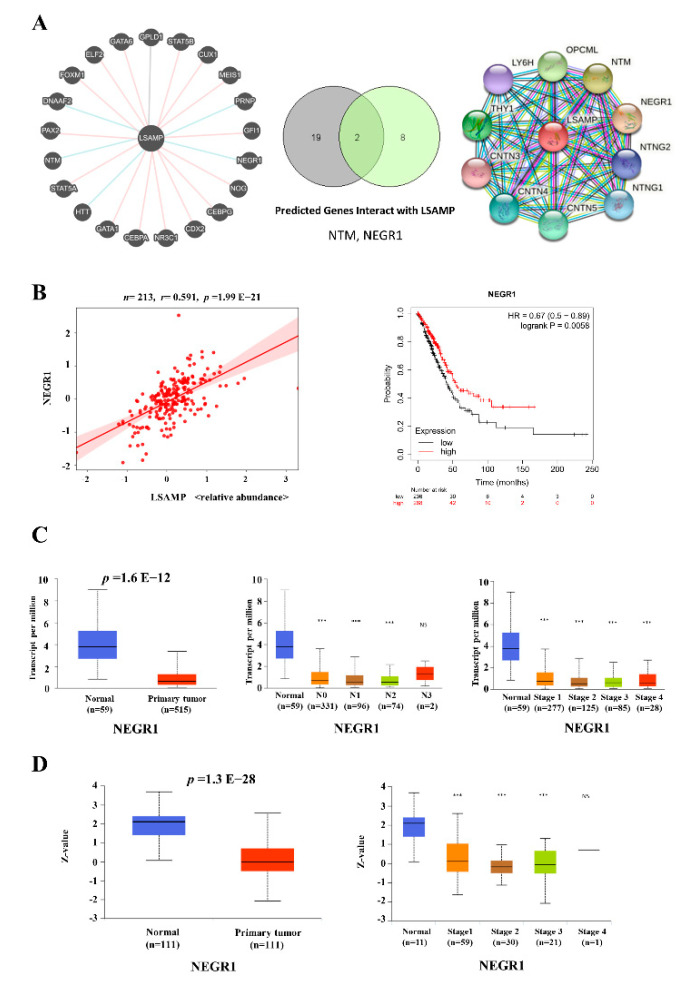
Signaling transduction pathways mediate the underlying mechanism of cell growth, cell apoptosis, organismal development, and pathways-aberrant diseases [29]. To elucidate the signaling transduction pathway, the Pathway Commons (gene, version 12) and STRING (protein, version 11.0b) were utilized to predict the protein interaction of LSAMP. There were two common genes/proteins, including NTM and NEGR1 predicted by the intersection of Pathway Commons and STRING (Figure 5A). In addition, a positive correlation (r = 0.591) between LSAMP and NEGR1 further confirmed the interaction between these two genes/proteins (left panel, Figure 5B). In addition, higher NEGR1 expression conferred longer survival time in LUAD patients (right panel, Figure 5B). Furthermore, the mRNA expression of NEGR1 was lower in tumor parts compared with normal parts (left panel, Figure 5C), and its expression was correlated negatively in nodal metastasis and staging from TCGA (right two panels, Figure 5C). The protein expression of NEGR1 was lower in tumor parts compared with normal parts (left panel, Figure 5D) and its expression was correlated negatively in staging from the CPTAC cohort (right panel, Figure 5D). Based on these data, NEGR1 might interact with LSAMP, which affects the clinical outcomes of LUAD.
Figure 5.
NEGR1 has an interaction with LSAMP. NTM and NEGR1 were the predicted genes interacting with LSAMP by intersecting the results from websites, Pathway Commons (left plot) and STRING (right plot) (A). NEGR1 had a good correlation with LSAMP and positively linked to better survival (B). NEGR1 mRNA expression was suppressed in LUAD, and associated with lymph node metastasis and advanced tumor stages (C). NEGR1 protein has lower expression in tumor parts in LUAD and correlates with advanced tumor stages (D). *** p < 0.005.
3.6. LSAMP Interferes with Epithelial-Mesenchymal Transition
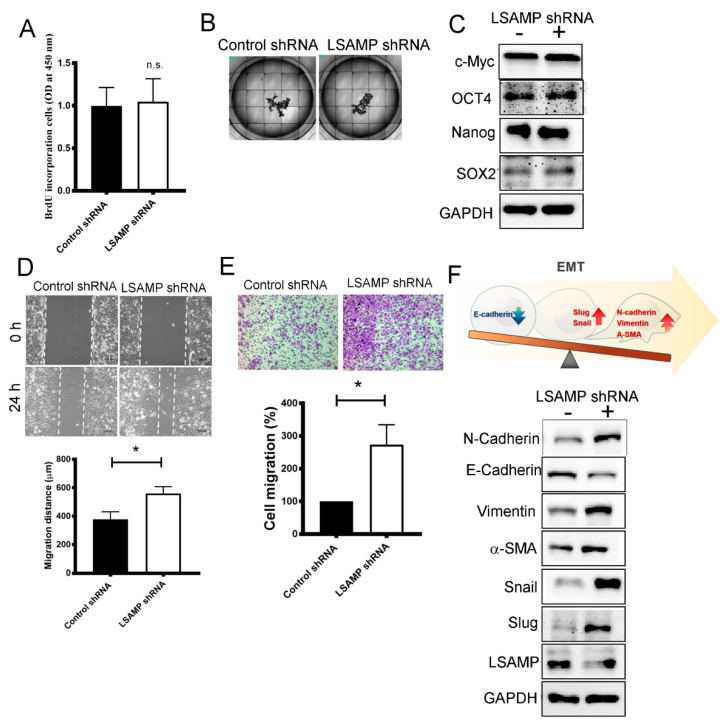
To validate our previous data showing that LSAMP acts as a suppressor in tumorigenesis, functional assays were performed through an LSAMP shRNA knockdown model. With the aid of shRNA knockdown in a lung cancer cell line, A549, LSAMP did not affect cell proliferation through BrdU incorporation (Figure 6A), tumor spheroid formation (Figure 6B), nor stem cell characteristics (Figure 6C). On the contrary, cell migration, measured by both wound-healing and transwell migration assays, was enhanced after the loss of function in LSAMP (Figure 6D,E). The relevant mesenchymal markers, N-cadherin, vimentin, α-SMA, snail and slug strengthened after the knockdown of LSAMP via shRNA technique but an epithelial marker, E-cadherin, was attenuated (Figure 6F). These results reveal that the functional loss of LSAMP enables cancer cell migration via the trigging of EMT.
Figure 6.
Loss of function in LSAMP enhances lung cancer progression. Cellular functions changed after knocking down LSAMP. Several cellular behaviors were investigated as proliferation, as determined by BrdU after 72 h incubation (A), tumor spheroid formation after 7-day culture (B), stemness (C), wound-healing assay (D), transwell migration after 48 h movement (E) and EMT markers and the illustration of EMT change. (F). All experiments were performed independently at least three times. The results were presented as mean ± SD. * Significant difference between the two test groups (p < 0.05).
4. Discussion
Lung cancer has been one of the most deadly cancer for decades worldwide. However, the present treatment strategy is imperfect, especially in non-driver-mutation-type lung cancer [30]. In this study, advanced techniques, NGS and bioinformatics were utilized to discover and validate LSAMP in lung cancer tumorigenesis. Clinically, LUAD patients with low levels of LSAMP carried the worst clinical outcomes in several public datasets. Furthermore, the loss of function of LSAMP in lung cancer enhanced cellular migration via EMT under the epigenetic regulation of DNA hypermethylation and miR-141-3p. Our study indicated that LSAMP acts as a suppressor in lung cancer tumorigenesis. This study provides evidence that LSAMP would be a good candidate target for developing a treatment strategy in non-driver mutation lung cancer.
LSAMP has been known as a tumor suppressor gene in osteosarcoma and ovarian cancers [20,31]; however, the evidence of LSAMP functions for lung cancer tumorigenesis is still lacking. By utilizing the NGS data from six paired lung normal/tumor tissues and public datasets, this study found that tumor tissues expressed low-level LSAMP compared with normal tissues in LUAD. To verify its role in LUAD, several public datasets confirmed that low levels of LSAMP conferred tumor invasiveness, nodal metastasis, and advanced tumor stages as well as poor clinical outcomes with short survival time. To elucidate the role of the loss of function in LSAMP in tumorigenesis, several molecular mechanisms have been explored through shRNA knockdown. The wound-healing assay revealed enhanced wound closure as increased migration through EMT but not proliferation, tumor spheroid formation, or stemness in this study. LSMAP belongs to the superfamily of cell adhesion molecules (CAMs), which are the most abundantly expressed glycosylphosphatidylinositol (GPI)-anchored cell surface glycoprotein [32]. CAMs act as a subset of adhesion proteins on the cell surface and might affect cellular mechanisms of growth, contact inhibition, and apoptosis [33]. The specific characteristic of LSAMP as a CAM might explain its function in lung cancer progression.
To investigate the regulation over LSAMP, several proposed regulatory mechanisms as DNA methylation of the promoter region and copy number might regulate LSAMP expression [22,31]. In this study, the DNA methylation and the miR-141-3p negatively correlated with LSAMP expression, but failed to identify the correlation with copy number variation of LSAMP through the TCGA cohort. In addition, the protein-protein interaction would affect the expression of candidate protein. Through the Pathway Commons and STRING open website, strong interaction in LSAMP-NTM and LSAMP-NEGR1 was discovered. Reports illustrate the number of potential interactions between IgLONs due to their ability to form homophilic and heterophilic bonds, both in cis and trans orientations [32]. The interaction among IgLONs might be worthy of further investigation.
In this study, a series of investigations in vitro suggested that loss of function of LSAMP enhanced migratory ability via EMT but not proliferation or stemness in LUAD. However, this study has some limitations. Firstly, with only six paired samples, a definite conclusion for the role of LSAMP in lung cancer might not be reachable. This limitation might be improved by using public datasets. Secondly, the regulatory mechanisms for the downregulation of LSAMP were predicted via public databases. However, with this solid molecular evidence, this study provides the role of LSAMP in LUAD tumorigenesis and might offer hope for lung cancer patients.
5. Conclusions
Based on our study results, LSAMP acts as a suppressor in lung cancer tumorigenesis and LSAMP might be of great potential in developing a therapeutic strategy to overcome lung cancer.
Acknowledgments
We thank the National RNAi Core Facility at Academia Sinica in Taiwan for providing shRNA reagents and related services. The authors thank the Center for Research Resources and Development of Kaohsiung Medical University.
Author Contributions
Conceptualization, C.-Y.C., Y.-M.T. and Y.-L.H.; methodology, Y.-Y.C., Y.-W.L., Y.-C.H., S.-F.J., Y.-S.L. and P.-H.T.; software, C.-Y.C.; validation, K.-L.W., C.-Y.C., Y.-M.T. and Y.-L.H.; formal analysis, Y.-M.T. and Y.-L.H.; investigation, Y.-L.H.; resources, Y.-L.H.; data curation, C.-Y.C., Y.-M.T. and Y.-L.H.; writing—original draft preparation, C.-Y.C. and Y.-M.T.; writing—review and editing, Y.-M.T.; visualization, J.-Y.H.; supervision, Y.-L.H.; project administration, Y.-L.H.; funding acquisition, Y.-M.T., J.-Y.H. and Y.-L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the Ministry of Science and Technology (Grant No. MOST 109-2314-B-037-091-) and Kaohsiung Medical University Hospital (Grant No. KMU109-9R12).
Institutional Review Board Statement
The present study was approved by the Institutional Review Board of Kaohsiung Medical University Hospital (KMUH-IRB-20130054, 24 May 2013).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from the patient(s), if applicable, to publish this paper.
Data Availability Statement
All data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Pineros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Hsu J.C., Wei C.-F., Yang S.-C., Lin P.-C., Lee Y.-C., Lu C.Y. Lung cancer survival and mortality in Taiwan following the initial launch of targeted therapies: An interrupted time series study. BMJ Open. 2020;10:e033427. doi: 10.1136/bmjopen-2019-033427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 5.Gristina V., Malapelle U., Galvano A., Pisapia P., Pepe F., Rolfo C., Tortorici S., Bazan V., Troncone G., Russo A. The significance of epidermal growth factor receptor uncommon mutations in non-small cell lung cancer: A systematic review and critical appraisal. Cancer Treat. Rev. 2020;85:101994. doi: 10.1016/j.ctrv.2020.101994. [DOI] [PubMed] [Google Scholar]
- 6.Corrales L., Scilla K., Caglevic C., Miller K., Oliveira J., Rolfo C. Immunotherapy in Lung Cancer: A New Age in Cancer Treatment. Adv. Exp. Med. Biol. 2018;995:65–95. doi: 10.1007/978-3-030-02505-2_3. [DOI] [PubMed] [Google Scholar]
- 7.Mittal V. Epithelial Mesenchymal Transition in Aggressive Lung Cancers. Adv. Exp. Med. Biol. 2016;890:37–56. doi: 10.1007/978-3-319-24932-2_3. [DOI] [PubMed] [Google Scholar]
- 8.Zou H., Wang S., Wang S., Wu H., Yu J., Chen Q., Cui W., Yuan Y., Wen X., He J., et al. SOX5 interacts with YAP1 to drive malignant potential of non-small cell lung cancer cells. Am. J. Cancer Res. 2018;8:866–878. [PMC free article] [PubMed] [Google Scholar]
- 9.Banyard J., Bielenberg D.R. The role of EMT and MET in cancer dissemination. Connect. Tissue Res. 2015;56:403–413. doi: 10.3109/03008207.2015.1060970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levitt P. A monoclonal antibody to limbic system neurons. Science. 1984;223:299–301. doi: 10.1126/science.6199842. [DOI] [PubMed] [Google Scholar]
- 11.Struyk A., Canoll P., Wolfgang M., Rosen C., D’Eustachio P., Salzer J. Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. J. Neurosci. 1995;15:2141–2156. doi: 10.1523/JNEUROSCI.15-03-02141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funatsu N., Miyata S., Kumanogoh H., Shigeta M., Hamada K., Endo Y., Sokawa Y., Maekawa S. Characterization of a novel rat brain glycosylphosphatidylinositol-anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J. Biol. Chem. 1999;274:8224–8230. doi: 10.1074/jbc.274.12.8224. [DOI] [PubMed] [Google Scholar]
- 13.Nissen M.S., Blaabjerg M. Anti-IgLON5 Disease: A Case With 11-Year Clinical Course and Review of the Literature. Front. Neurol. 2019;10:1056. doi: 10.3389/fneur.2019.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akeel M., McNamee C.J., Youssef S., Moss D. DIgLONs inhibit initiation of neurite outgrowth from forebrain neurons via an IgLON-containing receptor complex. Brain Res. 2011;1374:27–35. doi: 10.1016/j.brainres.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 15.Pischedda F., Piccoli G. The IgLON Family Member Negr1 Promotes Neuronal Arborization Acting as Soluble Factor via FGFR2. Front. Mol. Neurosci. 2015;8:89. doi: 10.3389/fnmol.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto T., Maekawa S., Miyata S. IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem. Funct. 2009;27:496–498. doi: 10.1002/cbf.1600. [DOI] [PubMed] [Google Scholar]
- 17.Karis K., Eskla K.-L., Kaare M., Täht K., Tuusov J., Visnapuu T., Innos J., Jayaram M., Timmusk T., Weickert C.S., et al. Altered Expression Profile of IgLON Family of Neural Cell Adhesion Molecules in the Dorsolateral Prefrontal Cortex of Schizophrenic Patients. Front. Mol. Neurosci. 2018;11:8. doi: 10.3389/fnmol.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh K., Jayaram M., Kaare M., Leidmaa E., Jagomäe T., Heinla I., Hickey M.A., Kaasik A., Schäfer M.K., Innos J., et al. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci. Rep. 2019;9:5457. doi: 10.1038/s41598-019-41991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanaveski T., Singh K., Narvik J., Eskla K.-L., Visnapuu T., Heinla I., Jayaram M., Innos J., Lilleväli K., Philips M.-A., et al. Promoter-Specific Expression and Genomic Structure of IgLON Family Genes in Mouse. Front. Neurosci. 2017;11:38. doi: 10.3389/fnins.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ntougkos E., Rush R., Scott D., Frankenberg T., Gabra H., Smyth J.F., Sellar G.C. The IgLON Family in Epithelial Ovarian Cancer: Expression Profiles and Clinicopathologic Correlates. Clin. Cancer Res. 2005;11:5764–5768. doi: 10.1158/1078-0432.CCR-04-2388. [DOI] [PubMed] [Google Scholar]
- 21.Zhang N., Xu J., Wang Y., Heng X., Yang L., Xing X. Loss of opioid binding protein/cell adhesion molecule-like gene expression in gastric cancer. Oncol. Lett. 2018;15:9973–9977. doi: 10.3892/ol.2018.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barøy T., Kresse S.H., Skårn M., Stabell M., Castro R., Lauvrak S., Llombart-Bosch A., Myklebost O., Meza-Zepeda L.A. Reexpression of LSAMP inhibits tumor growth in a preclinical osteosarcoma model. Mol. Cancer. 2014;13:93. doi: 10.1186/1476-4598-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pros E., Saigi M., Alameda D., Gomez-Mariano G., Martinez-Delgado B., Alburquerque-Bejar J.J., Carretero J., Tonda R., Esteve-Codina A., Catala I., et al. Genome-wide profiling of non-smoking-related lung cancer cells reveals common RB1 rearrangements associated with histopathologic transformation in EGFR-mutant tumors. Ann. Oncol. 2020;31:274–282. doi: 10.1016/j.annonc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Ocak S., Sos M.L., Thomas R.K., Massion P.P. High-throughput molecular analysis in lung cancer: Insights into biology and potential clinical applications. Eur. Respir. J. 2009;34:489–506. doi: 10.1183/09031936.00042409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakai T., Prasoon P., Hirose Y., Shimada Y., Ichikawa H., Nagahashi M. Next-generation sequencing-based clinical sequencing: Toward precision medicine in solid tumors. Int. J. Clin. Oncol. 2019;24:115–122. doi: 10.1007/s10147-018-1375-3. [DOI] [PubMed] [Google Scholar]
- 26.Nagahashi M., Shimada Y., Ichikawa H., Kameyama H., Takabe K., Okuda S., Wakai T. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci. 2019;110:6–15. doi: 10.1111/cas.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu Y.L., Hung J.Y., Lee Y.L., Chen F.W., Chang K.F., Chang W.A., Tsai Y.M., Chong I.W., Kuo P.L. Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget. 2017;8:104831–104854. doi: 10.18632/oncotarget.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B., Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devreotes P., Horwitz A.R. Signaling Networks that Regulate Cell Migration. Cold Spring Harbor Perspect. Biol. 2015;7:a005959. doi: 10.1101/cshperspect.a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu Q.S. Targeting non-small cell lung cancer: Driver mutation beyond epidermal growth factor mutation and anaplastic lymphoma kinase fusion. Ther. Adv. Med. Oncol. 2020;12:1758835919895756. doi: 10.1177/1758835919895756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kresse S.H., Ohnstad H.O., Paulsen E.B., Bjerkehagen B., Szuhai K., Serra M., Schaefer K.L., Myklebost O., Meza-Zepeda L.A. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2009;48:679–693. doi: 10.1002/gcc.20675. [DOI] [PubMed] [Google Scholar]
- 32.Singh K., Lilleväli K., Gilbert S.F., Bregin A., Narvik J., Jayaram M., Rahi M., Innos J., Kaasik A., Vasar E., et al. The combined impact of IgLON family proteins Lsamp and Neurotrimin on developing neurons and behavioral profiles in mouse. Brain Res. Bull. 2018;140:5–18. doi: 10.1016/j.brainresbull.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Harjunpää H., Llort Asens M., Guenther C., Fagerholm S.C. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front. Immunol. 2019;10:1078. doi: 10.3389/fimmu.2019.01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available upon request.