Abstract
Snakebite envenoming is a neglected tropical disease that may claim over 100,000 human lives annually worldwide. Snakebite occurs as the result of an interaction between a human and a snake that elicits either a defensive response from the snake or, more rarely, a feeding response as the result of mistaken identity. Snakebite envenoming is therefore a biological and, more specifically, an ecological problem. Snake venom itself is often described as a “cocktail”, as it is a heterogenous mixture of molecules including the toxins (which are typically proteinaceous) responsible for the pathophysiological consequences of envenoming. The primary function of venom in snake ecology is pre-subjugation, with defensive deployment of the secretion typically considered a secondary function. The particular composition of any given venom cocktail is shaped by evolutionary forces that include phylogenetic constraints associated with the snake's lineage and adaptive responses to the snake's ecological context, including the taxa it preys upon and by which it is predated upon. In the present article, we describe how conceptual frameworks from ecology and evolutionary biology can enter into a mutually enlightening relationship with clinical toxinology by enabling the consideration of snakebite envenoming from an “ecological stance”. We detail the insights that may emerge from such a perspective and highlight the ways in which the high-fidelity descriptive knowledge emerging from applications of -omics era technologies – “venomics” and “antivenomics” – can combine with evolutionary explanations to deliver a detailed understanding of this multifactorial health crisis.
Graphical abstract
Highlights
-
•
The conceptual frameworks of ecology and evolutionary biology have a mutually enriching relationship with clinical toxinology.
-
•
Omics technologies combined with evolutionary explanations deliver a detailed understanding of snakebite envenoming.
-
•
Integrating and contextualising eco-evolutionary and clinical toxinology is a powerful approach to tackle snake envenoming.
-
•
A deep understanding of how to mitigate the multifactorial snakebite health crisis requires a biological lens.
-
•
Understanding the problem will help us to solve it.
1. Evolutionary ecology and clinical impact of snake venoms—a brief introduction
Venomous snakes share a long co-evolutionary history with primates, including our ancestors: their continuous co-existence across Africa (since ~6.7 My) and more recently (last ~2.5 My) in Asia, has shaped selection pressures for both lineages (Harris et al., 2021; Kazandjian et al., 2021). In our world today, humans and venomous snakes coexist in tropical and subtropical rural regions. While a certain level of human ophidian accidents is inevitable, habitat reshaping due to agricultural and other practices, and climate change, contribute to increasing the level of human-snake interactions and, thus, the incidence of snakebites (Gutiérrez, 2020). Legitimate snakebite envenomings inflicted on humans by any of the approximately 110 species included in the World Health Organisation's (WHO) category 1 (highest medical importance) are an occupational hazard and a disease of poverty in many tropical regions (Harrison et al., 2009). The majority of envenoming from these snakes, which either belong to the Viperidae or Elapidae family (WHO, 2017), occur during capricious encounters with people engaged in rural activities in tropical and subtropical regions, especially in Asia and Africa. Such encounters can be described as ecological interactions between snakes and humans (Jackson et al., 2019; see below). Literature reports estimate that 400,000–1,200,000 snakebite envenomings occur annually, causing 81,000–138,000 deaths and many more injuries, such as physical sequelae (stigmatising disfigurements and amputations) and chronic mental morbidity (Gutiérrez et al., 2017a).
Snakebite envenoming affects not only the victims but often their entire families, which may enter a cycle of generational poverty that is difficult to break. In spite of its magnitude, the problem of snakebite envenoming has historically faced a neglect: therapeutic antivenoms are scarce or even unavailable in many regions (Longbottom et al., 2018); knowledge regarding the venoms/toxins and their pathophysiological actions is relatively limited; and modern biotechnological tools such as humanised or fully human antibodies still seem far from being introduced into active therapeutic deployment. Recent international efforts fostered by the WHO, science-funding agencies, and other stakeholders including Médecins Sans Frontières, the Wellcome Trust, the Kofi Annan Foundation, and the Global Snakebite Initiative (Chippaux, 2017; Longbottom et al., 2018; Harrison et al., 2009), are attempting to reverse this neglect and reduce the mortality and disability from snakebite envenoming worldwide by 50% before 2030 (WHO, 2019).
Snakebite envenoming represents a One Health challenge requiring clinical, ecological, and public health expertise (Longbottom et al., 2018). Hence, to comprehensively address this disease burden, the clinical challenge of appropriate case diagnosis needs to be considered within an ecological evolutionary context. Throughout this manuscript we reflect on the meaning, experimental methodologies, application and interpretation of shared or specific concepts of the partly overlapping domains of ecological and clinical toxinology. We argue that increased knowledge of snake behavior and life-history traits are crucial for understanding and predicting the frequency and geographical distribution of snakebites (Jackson et al., 2019). Furthermore, knowledge of the evolutionary ecology of venomous snakes and the identity of the most relevant toxin molecules in the context of prey capture and human envenoming, are both key for improving our understanding of both the underlying pharmacology and the ability to improve the efficacy of antivenoms—the only effective antidote for envenoming. This remains a major challenge, as nearly half of all snake species capable of inflicting a life-threatening bite do not have antivenoms on the market, and distribution pipelines in the most afflicted countries are often substandard (WHO, 2017).
Venoms are intrinsically ecological traits (Jackson et al., 2019) and their natural history should thus be understood in the context of a network of chance and selection events. Identifying the specific pressures that tailored the composition of extant medically relevant venoms, e.g. by inferring influential nodes across phylogeny (Calvete, 2013, 2019; Pla et al., 2013, 2017a; Gibbs et al., 2013; Lomonte et al., 2014; Blanchet et al., 2017; Calvete et al., 2017; Ainsworth et al., 2018; Sanz et al., 2019a; Zaher et al., 2019; Jackson and Koludarov, 2020; Kazandjian et al., 2021; Holding et al., 2021), may have implications for the clinical treatment of human envenomings. The thesis advocated in this essay is that integration and contextualisation of complementary evolutionary, ecological, and clinical toxinological information represents a powerful holistic approach to tackle the global challenge of snake envenoming.
2. Evolutionary and contemporary contexts of snake venoms
Venoms are bioactive secretions typically consisting of cocktails of secretory proteins collectively referred to as toxins. Venoms have evolved independently in over a hundred animal lineages, where they are key evolutionary innovations that play a wide range of important ecological roles that predominantly involve predation and/or defense (Casewell et al., 2013; Schendel et al., 2019). One such lineage is caenophidian snakes, which explosive radiation in the wake of the Cretaceous–Paleogene mass extinction approximately 60–50 million years ago (Hsiang et al., 2015), was perhaps a consequence of the evolution of venom (Fry et al., 2006, 2012). Snake venom is thought to have evolved primarily as a trophic adaptation, that is for predatory purposes (Davies and Arbuckle, 2019; Ward-Smith et al., 2020), and among the ca. 3700 extant snakes, at least 800 species (within Elapidae, Viperidae, Colubridae and Atractaspididae) use venom to immobilize their invertebrate or vertebrate prey before swallowing it whole. Venom is also used defensively by many snakes, and several lineages of venomous snakes have evolved specialised defensive venom adaptations (Arbuckle et al., 2017; Calvete, 2017), such as the blindingly cytotoxic venoms of the three lineages of spitting cobra (Kazandjian et al., 2021) and the hyperalgesic venoms of micrurine snakes (Bohlen et al., 2011; Lomonte et al., 2016; Sanz et al., 2019b).
To achieve their predatory and defensive functions, snake venom arsenals comprise mixtures of dozens to hundreds of bioactive compounds that belong to a limited number (2 < n < 20) of protein families (Fry et al., 2009; Calvete, 2013; Junqueira-de-Azevedo et al., 2016; Tasoulis and Isbister, 2017; Barua and Mikheyev, 2019), suggesting that relatively few protein folds are amenable for evolving a toxic role (Reeks et al., 2015). As in other venomous lineages, each of these protein toxin families represent molecular phenotypic innovations or novelties that emerged through functional recruitment of single-copy gene products, gene duplication massive structural and functional diversification, or both (Zancolli and Casewell, 2020). Interestingly, venom proteomics evidence gathered from 236+ snake species, mainly within the families Elapidae (Fig. 1) and Viperidae (Fig. 2), has revealed that variation in venom composition -within- and between-species, in-space (geographic) and in-time (ontogenetic)- is a common feature at all taxonomic levels. Much of this variation is due to differences and changes in prey composition, highlighting the influence that trophic ecology has on the evolution of snake venom. Importantly, this influence of ecological factors also extend to other venom-associated traits and characters—the venom system— including both morphological and behavioural traits (Kazandjian et al., 2021; Westeen et al., 2020). Extant venomous snakes thus represent fascinating models for studying the emergence of novel functional traits within clear adaptive contexts and across multiple levels of biological complexity that span from molecule to morphology to behavior (Casewell et al., 2013; Schendel et al., 2019; Zancolli and Casewell, 2020; Post et al., 2020; Jackson and Koludarov, 2020).
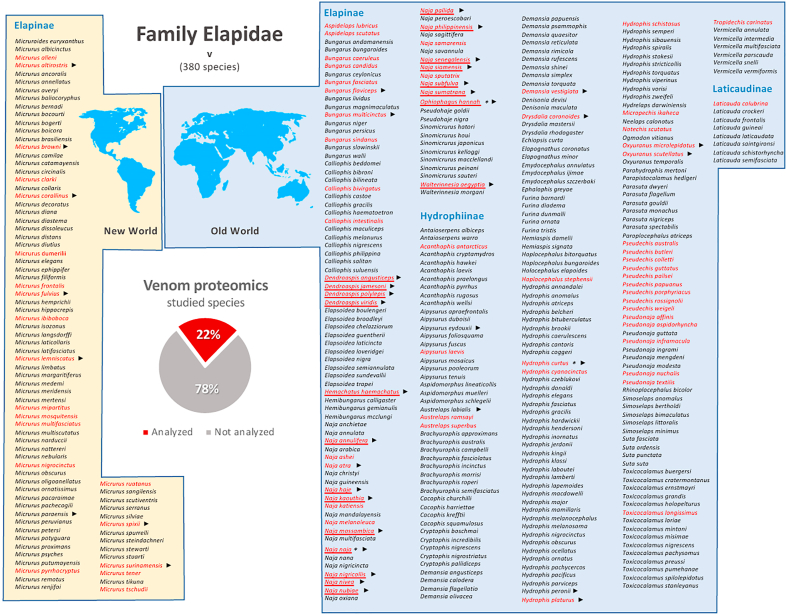
Fig. 1.
Snake species of the family Elapidae whose venoms have been studied by using proteomic tools (names indicated in red) among all snake species within this family (371; based on lists provided by www.reptile-database.org). As shown in the inset pie chart, studied species cover 24% of all elapids. ► and * denote, respectively, the availability of specific venom gland transcriptome and genome sequence. Species whose venom proteome was gathered using top-down MS are underlined. Last update: April 26, 2021.
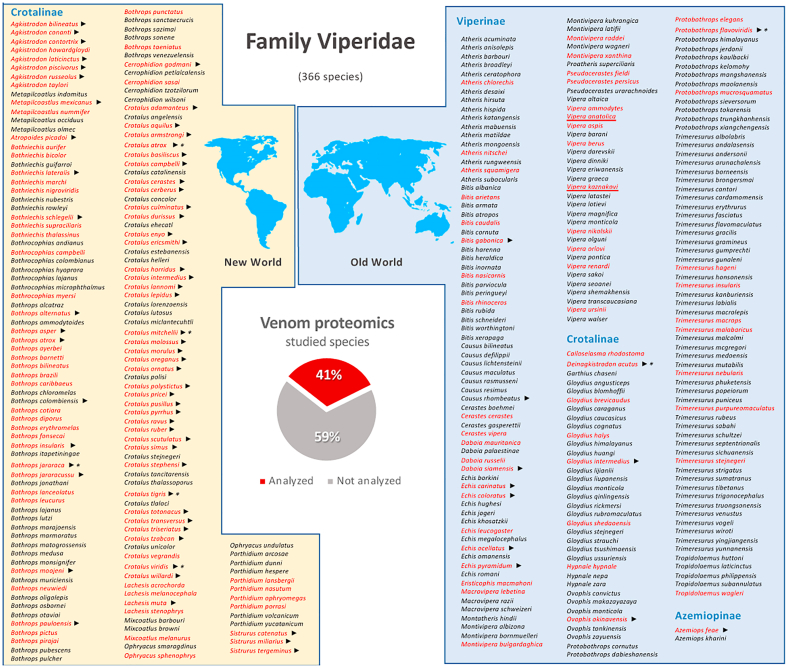
Fig. 2.
Snake species of the family Viperidae whose venoms have been studied by using proteomic tools (names indicated in red) among all snake species within this family (355; based on lists provided by www.reptile-database.org). As shown in the inset pie chart, studied species cover 35% of all viperids. ► and * denote, respectively, the availability of specific venom gland transcriptome and genome sequence. Species whose venom proteome was gathered using top-down MS are underlined. Last update: April 26, 2021.
The genome contains traces of sterile recombination events as well as those that have contributed to the functional genome of the extant species. However, snake genomics is in its infancy. The first genome draft of a venomous snake, the king cobra (Ophiophagus hannah), was published in 2013 (Vonk et al., 2013). Since then, the sequences of only a handful of venomous snake genomes have been compiled, including the speckled rattlesnake (Crotalus mitchellii) (Gilbert et al., 2014); the five-pacer viper (Deinagkistrodon acutus) (Yin et al., 2016); the habu snake (Protobothrops flavoviridis) (Shibata et al., 2018); the prairie rattlesnake (Crotalus viridis) (Schield et al., 2019); two hydrophiins (Hydrophis melanocephalus and Emydocephalus ijimae) and two laticaudins (Laticauda laticaudata and L. colubrina) (Kishida et al., 2019); the Shaw's sea snake (Hydrophis curtus) (Peng et al., 2020); the western diamondback rattlesnake (Crotalus atrox) (Giorgianni et al., 2020); the Indian cobra (Naja naja) (Suryamohan et al., 2020); the tiger rattlesnake (Crotalus tigris) (Margres et al., 2021); the olive sea snake (Aiysurus laevis) (Ludington and Sanders, 2021), and the jararaca (Bothrops jararaca) (Almeida et al., 2021). The foreseeable exponential growth of comparative snake genomics studies in the coming years (Kerkkamp et al., 2016; Drukewitz and von Reumont, 2019) will undoubtedly result in a major leap forward in our understanding of the deep history of venom evolution. In addition, the comprehensive catalog of venom-gland-specific toxin genes complemented with transcriptomics, venomics and toxinological information may pave the way for the generation a la carte of synthetic antivenoms.
3. Snakebite and the “ecological stance”
Snakebite envenoming is a biological problem and understanding a biological problem requires a biological lens. Biology is a science of complex functional systems with myriad sub-components of endless variety. Nonetheless, certain basic principles appear to hold across all biological systems. Acknowledgement of these basic principles should therefore be a core component of any attempt to understand biological systems and thereby solve (or mitigate) biological problems. Two of biology's fundamental principles are evolution – all biological systems evolve and have an evolutionary history – and ecology – all biological systems interact with other biological systems. These principles hold at all levels of analysis within biology, from the molecular to the organismal. Evolution and ecology themselves share a common principle – the transcendent role of context. In biology, what things are determined by their context – both their temporal and “genetic” context in an evolutionary lineage, and their extant context in an ecological assemblage. Again, this applies to molecules as much as it applies to organisms (Guttinger, 2018; Jackson and Koludarov, 2020). An appreciation of the role of context thus seems to be a core component of a minimal biological perspective – an evolutionary or ecological stance, akin to the “intentional stance” (Dennett, 1995) or “empirical stance” (van Fraassen, 2008). These “stances” represent heuristic strategies for understanding and getting to grips with phenomena. Here, we will discuss three ways in which the evolutionary and ecological stances, with their shared emphasis on context, may help us get to grips with the problem of snakebite envenoming.
3.1. Circumstances of bites
Snakebite itself, the result of an interaction between a snake and a human, is an uncontroversial ecological phenomenon. One of the most important factors in reducing the burden of snakebite envenoming is gaining detailed knowledge of the circumstances in which bites occur. Such knowledge may help prevent many snakebites from occurring, thus reducing the burden on healthcare services. What are the contexts in which snakebites occur? Answering this question requires knowledge of the ways in which snake ecology and human ecology become human-snake ecology. Currently we have much anecdotal evidence on the circumstances of bites, but very little research has been conducted in this space.
Naturally, the context of bites differs considerably from place to place, both regionally and locally. Thus, the circumstances in which bites occur in India may or may not exist in (for example) sub-Saharan Africa. Thus, ecological studies of human-snake interactions must be conducted in all snakebite-afflicted regions. Some examples of simple, testable hypotheses include:
-
•
Do snakebites typically occur when a snake is attempting to defend itself from a human it perceives as a potential threat?
-
•
Are bites to sleeping humans (e.g. by spitting cobras in Africa and kraits in India) a case of mistaken identity – when a confused snake mistakes the smell of a human for that of the mammalian (e.g. rodent) prey it seeks – or can be considered defensive (e.g. when a sleeping human accidently rolls over onto a snake that was seeking warmth)?
-
•
Does carrying a torch at night or wearing appropriate footwear reduce the incidence of snakebite?
-
•
Does clearing away rubbish or overgrown vegetation from around human dwellings reduce the number of human-snake interactions?
Questions like these can be combined to elaborate an educational program to instruct people about the behavior of snakes in their area, which may lead to a significant reduction in snakebites.
3.2. Molecular evolution and function of toxins
The composition and activity of snake venoms is strongly influenced by the feeding ecology of the snakes that deploy them, often resulting in compositional variation across biogeographical regions and developmental stages (Sunagar et al., 2016; Jackson et al., 2019). However, the mechanisms that generate this variation remain largely elusive, partly due to the paucity of both venomous and non-venomous snake genomic data (e.g., Aird et al., 2017). The evolution of this continuous spectrum of venom phenotypes is best understood through the application of quantitative genetic methods (Hansen and Houle, 2008; Walsh and Lynch, 2018), which have yet to be applied to snake venom systems. In addition, identifying the ecological correlates of particular venom mixtures from the current composition of venoms is not straightforward: We must take into account both phylogenetic constraints and the potentially confounding effect of having to use atypical prey species as models to measure venom potency and bioactivities (see below).
Contrasting our lack of knowledge on the mechanisms that underlie the population and developmental variational properties and dynamics of snake venoms, we have a much better knowledge about the processes that shape snake venoms on macroevolutionary scales (between species). For example, we can infer nodes where particularly influential shifts were made in genotype-phenotype space by mapping the venom phenotype profiles of a species clade across its phylogeny (Pla et al., 2013, 2017a; Gibbs et al., 2013; Lomonte et al., 2014; Calvete et al., 2017; Ainsworth et al., 2018; Calvete, 2019; Sanz et al., 2019a; Zaher et al., 2019; Kazandjian et al., 2021). Further, ancestral structure inference may allow the reconstruction of the evolutionary origin of key mutations across the history of individual toxins (Whittington et al., 2018) and functional snake venom clades (Blanchet et al., 2017; Ainsworth et al., 2018; Jackson and Koludarov, 2020; Holding et al., 2021).
3.3. The effectiveness or “ecological validity” of therapeutics
“Ecological validity” is a concept with a long history in psychology and refers to the generalizability of data from one context (e.g. the laboratory) to another (e.g. the world outside the laboratory) (Schmuckler, 2001). Whilst the term might not be commonly used in association with the design and development of snakebite therapeutics, it is a familiar concept within pharmacological studies – a drug may be effective in vitro without those promising results being translated to in vivo contexts. This may be described as the difference between “efficacy” and “effectiveness”, a distinction that has been previously highlighted in research on snake antivenoms (Isbister, 2010). Whether or not a snakebite therapeutic (e.g. an antivenom) is ecologically valid is partly determined by factors including whether or not an antibody that binds a toxin under laboratory circumstances will do so under physiological circumstances; whether binding a toxin translates to neutralisation (or elimination); whether the biodistribution of injected antibodies (or other therapeutics) is comparable to that of their targets (e.g. toxins); and a host of other factors. In addition to these molecular and physiological factors, the ecological validity of a snakebite therapeutic is influenced by the simple fact of whether the therapeutic can be delivered to a bite victim in a timely manner. Thus, as the World Health Organisation's Snakebite Envenoming Roadmap indicates (WHO, 2019), antivenom/drug distribution programs and other aspects of health infrastructure are crucial components in determining the ecological validity of a potential snakebite therapeutic. Much like any other functional trait—tools such as drugs can be considered part of an extended phenotype (Dawkins, 1982) of the human organism—the effectiveness of a snakebite therapeutic is contingent on its deployment in the appropriate context. Studying and engineering (e.g., through the creation of efficient distribution pipelines; the training of clinicians; etc.) this context is part of the landscape of snakebite research.
3.4. The evolutionary ecology of venoms and its impact on snakebite
Being primarily a trophic adaptation, the composition and potency of a snake venom is shaped by complex intrinsic (the genetic propensities of a lineage) and extrinsic (ecological interactions and environmental conditions) factors (Casewell et al., 2014, 2020; Sunagar et al., 2016). Hence, understanding the evolutionary ecology of venom becomes invaluable, not only from an academic perspective but also for designing efficient snakebite therapeutics. Ecological factors that are well-known to affect snake venoms include feeding ecology, ontogeny, predator pressure, inter- and intraspecific competition, and geographic distribution and population ecology.
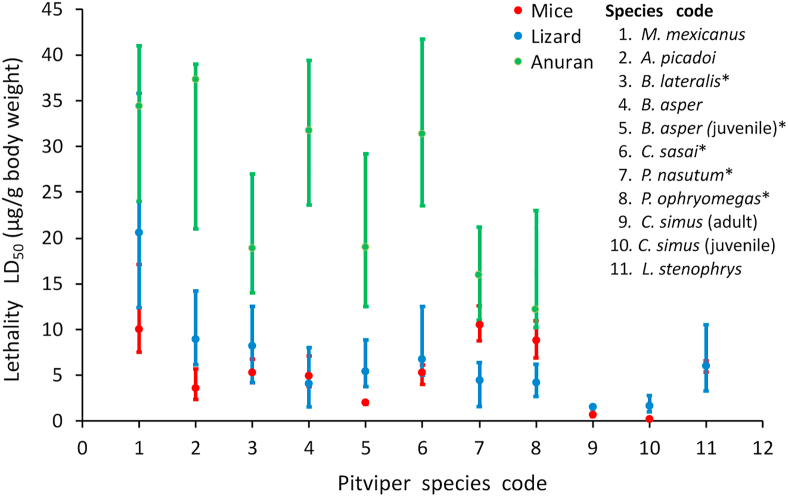
Variations in snake venoms are believed to be primarily driven by differences in the physiologies of the prey and/or predatory animals against which venom is deployed and are especially pronounced in snake species that are distributed across distinct biogeographical or agroclimatic zone. Dietary shifts, which may result from transitions in ecological niches, or from the differences in prey abundance from one region to another, have led to the extensive evolutionary fine-tuning of the snake venom cocktail (Daltry et al., 1996; Gibbs et al., 2011). The effect that dietary modifications and food specialisations have on the venom's composition has some restrictions since there appears to be a gradient of susceptibility to venom among different prey types. Thus, anurans appear to be much more resistant to snake toxins than lizards and rodents, sometimes by several orders of magnitude, even in species that feed on them regularly (Fig. 3).
Fig. 3.
Median Lethal Dose (LD50) estimates (95% CI, i.p. route) for several Neotropical pitvipers in three models of preys (mouse, Mus musculus; lizard, Hemidactylus frenatus; anuran, Engystomops pustulosus). Species that consume regularly lizards and frogs are highlighted from those that specialize in mammals by asterisks.
Asiatic kraits (Bungarus spp.) provide an instructive example of the effects of such dietary shifts and feeding specialisations on snake venoms. Kraits that chiefly feed on mammals secrete some of the most potent and medically important venoms to humans (Sunagar et al., 2021) whereas those that specialize in feeding on reptiles possess venoms with greatly reduced toxicity to mammals (Senji Laxme et al., 2019). Similar shifts in diet from mammals to arthropods (Barlow et al., 2009; Senji Laxme et al., 2019), amphibians (Hutchinson et al., 2007), birds (Mackessy et al., 2006), and eggs of various animals (Li et al., 2005) have significantly influenced the compositions and toxicities of snake venoms and, in some cases, their delivery systems.
As venoms play an essential role in prey incapacitation and, perhaps, predigestion, dietary shifts that are associated with developmental stage or ontogeny can alter venom compositions across the lifetime of an individual snake (Mackessy et al., 2003; Gibbs et al., 2011; Jackson et al., 2016). A shift in prey preference, from ectotherms as juveniles to endotherms as adults, is associated with changes in the venom profiles of various species of Crotalus snakes (Mackessy et al., 2003). Paedomorphosis (i.e., the retention of larval or juvenile traits into adulthood) has been noted in some species (e.g., Crotalus durissus (Calvete et al., 2010); Pseudonaja modesta (Jackson et al., 2016)). Dichotomous venom profiles are documented in others, in which the juvenile snakes possess a neurotoxin-rich venom as opposed to the proteolytic venoms of their adult counterparts (e.g. Crotalus simus (Durban et al., 2013), P. textilis (Jackson et al., 2016; Cipriani et al., 2017)). While the underlying molecular mechanisms remain in most cases uninvestigated, it has been suggested that microRNAs (miRNA) play a role in underpinning such stark transitions in the proteomic compositions of venoms across the ontogeny of the Central American rattlesnake (Crotalus simus simus) (Durban et al., 2013). Snake venoms and their molecular targets in prey and/or predators can exert reciprocal selection pressures. The clearest example of selection pressure towards venom resistance is in predators (e.g., opossums (Marsupialia: Didelphidae) and mongoose, Herpestes ichneumon) preying on venomous animals (Voss and Jansa, 2012; Drabeck et al., 2015; Holding et al., 2016). On the other hand, the evolution of adaptive resistance in ground squirrels (Spermophilus beecheyi) against the venom of the Northern Pacific rattlesnake (Crotalus viridis oreganus) represents another example of evolutionary chemical arms races between a venomous snake and its prey (Holding et al., 2016; Margres et al., 2017; Gibbs et al., 2020). While several molecular mechanisms of resistance are clearly understood, the precise impact of venom resistance on venom composition and potency remains elusive.
In addition to biotic interactions, environmental conditions are a primary influence on the abundance of predators and prey in a region and, in turn, influence snake venom cocktails. In addition, many extrinsic factors, including humidity, temperature and altitude can determine the rate of transcription and translation (i.e., RNA and protein synthesis, respectively). Hence, the environment may directly or indirectly influence venom profiles in a multitude of ways. It is unsurprising, therefore, that considerable variation in venom compositions and toxicities have been documented in snakes with large geographical distributions across distinct biogeographic conditions (Strickland et al., 2018; Senji Laxme et al., 2021). Considerable differences in snake venoms have also been described at finer geographical scales (Glenn et al., 1983). While the underlying causes are mostly unknown, this variability may stem from subtle differences in microhabitat, dietary preference, intraspecific competition, and venom resistance in local prey at all taxonomic levels.
As with evolutionary diversification more broadly, venom variation arises from the interaction between a plethora of intrinsic factors with those that are extrinsic (described in the preceding paragraphs). This interaction is an example of the transcendent role of context in evolution, in which an organism's environmental context (i.e., extrinsic factors) influence trait evolution in a manner that is both constrained and facilitated by the organism's phylogenetic context (i.e., factors intrinsic to an organismal lineage). Many genetic mechanisms including gene duplication, domain loss, neofunctionalisation, pseudogenisation, evolutionary tinkering of expression levels, alternative splicing, trans-splicing, and gene co-option have been identified as influencing the molecular diversification of snake venoms (Sunagar et al., 2016; Casewell et al., 2020). While positive Darwinian selection is implicated in rapidly introducing sequence variations in venom coding genes (Lynch, 2007; Juárez et al., 2008; Sunagar et al., 2013), mostly in an episodic or a ‘two-speed’ fashion (Sunagar and Moran, 2015), venom variation can also arise from the phylogenetic divergence of snakes over time. Cryptic genetic divergence in clinically important species can be detrimental to snakebite therapy, as antivenoms raised against the venom of one lineage may not be efficient in neutralising toxin variants in genetically distinct snakes. The existence of cryptic evolutionary lineages within a single species, therefore, can have tremendous implications for snakebite treatment, as can the existence of phenotypic crypsis in distinct species with overlapping range-distributions (Carbajal-Márquez et al., 2020; Sunagar et al., 2021). More infrequently, evolutionary convergence may result in cryptic lineages which are relatively inconsequential to snakebite therapy, given the convergence of venom composition along with the rest of the phenotype (Ukuwela et al., 2012).
4. What's in a venom?
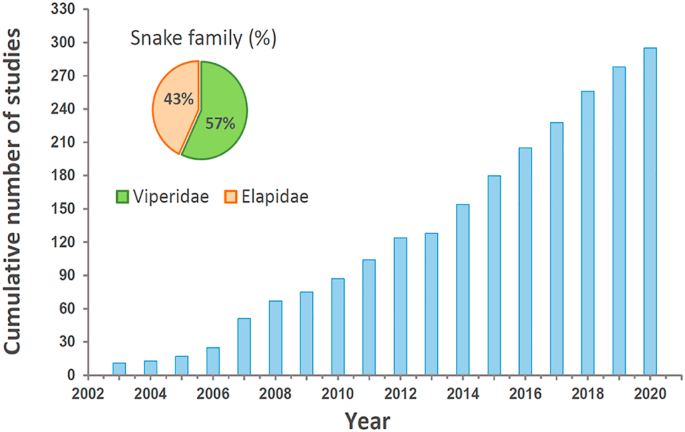
For more than a century, biochemists and pharmacologists have found snake venoms to be a rich and exciting ground for the discovery of novel molecules, mainly polypeptides, with a broad array of potent and specific bioactivities that explain their toxicity. Research on venoms has been continuously enhanced by technological advances, and a paradigm shift in the study of snake venoms came with the introduction of '-omic’ technologies at the turn of third millennium of our calendar (Fig. 4). However, proteomic strategies used to analyze venoms (“venomics") in different laboratories have been diverse and no general consensus has been adopted among molecular toxinologists that would facilitate comparison of results. For the most part, venomics studies have used peptide-centric ‘bottom-up’ approaches, with pre-mass spectrometry (MS) venom decomplexation, or applying direct ‘shotgun’ approaches (Fox and Serrano, 2008; Calvete, 2014; Zelanis and Tashima, 2014; Eichberg et al., 2015; Lomonte and Calvete, 2017).
Fig. 4.
Cumulative number of published studies dealing with the proteomic characterisation of snake venoms. Starting with pioneer works in 2003, up to mid-2020, a sustained increment in publications is evidenced (light blue bars). The inset pie chart illustrates the proportion of venomic studies that correspond to snake species from Elapidae (43%) (Fig. 1) and Viperidae (57%) (Fig. 2).
An important challenge in the characterisation of venoms (and other complex biological samples) is the inability of any single method to provide a complete view of its proteome. Proteome decomplexation and toxin identification and quantification represent major pillars of current state-of-the-art snake venom proteomics. Sample decomplexation protocols can be categorised as gel-based and gel-free methods (reviewed by Lomonte and Calvete, 2017). Among the multiple hyphenated approaches proposed by different research groups for integrating pre-MS venom proteome decomplexation and the identification and quantification of the individual toxins into a single workflow, arguably the most successful combination has turned out to be the one that takes advantage of venom fractionation to simultaneously quantitate the relative abundances of its different venom components. This strategy, which is usually referred to as ‘snake venomics’ (Juárez et al., 2004), relies on reverse-phase chromatographic decomplexation and eluate monitorisation at the absorbance wavelength of the peptide bond to estimate the relative abundances of the chromatographically separated fractions by peak area integration (Calvete, 2014, 2018). Among the gel-based approaches, two-dimensional electrophoresis (2-DE) generates an overview of a venom's protein complexity in a single image, including the approximate number of protein species and their apparent molecular masses and isoelectric points. In addition, comparison of non-reduced (IEF)/non-reduced (SDS-PAGE) and non-reduced/reduced 2-DE gels provides information on the quaternary structure of the venom proteins (Rabilloud, 2002; Eichberg et al., 2015). However, small (<3 kDa) peptides, which represent abundant components of some snake venoms, as well as proteins exhibiting extreme isoelectric points, may not be visualised in 2-DE venom reference maps, and there are a number of challenges in 2-DE proteome quantification: some spots tend to have severe tails in either dimension; contrast variations due to stain exposure, geometric distortions due to casting, polymerisation, and the running procedure may confound spot modeling, challenging reliable spot volume quantification.
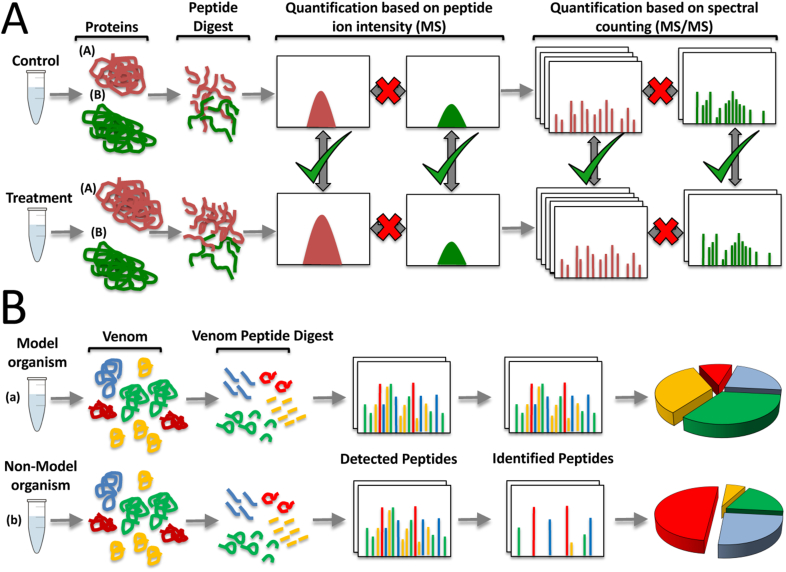
Gel-free LC-MS shotgun approaches often bypass the initial sample decomplexing step, and instead, the entire protein mixture is enzymatically digested into peptides that are subjected to mass spectrometric analysis (Wolters et al., 2001; Wu and MacCoss, 2002; Zhang et al., 2013). Conceptually, label-free quantification approaches are relatively straightforward, and are typically used to compare abundance changes of the same protein in different analyses. For example, comparing the abundance changes of protein A in control vs protein A in treated samples (Fig. 5A; Zhu et al., 2010; Neilson et al., 2011). In contrast, mass spectrometry is essentially a non-quantitative technique because proteolytic peptides exhibit a wide range of physicochemical properties that influence their ionisation efficiency, and hence, the quantitative estimations of parent protein abundances by label-free shotgun approaches is not appropriate for determining the abundances of different proteins in a single sample (Neilson et al., 2011; Lee et al., 2019). This point is discussed in more detail below. On the other hand, shotgun venomics, particularly when LC-MS data can be matched against a next generation species-specific venom gland transcriptome, may provide a deep catalogue of the protein/peptide components present in a venom (Rokyta et al., 2011; Aird et al., 2013; Calvete et al., 2018a). However, among the 236+ nominal snake species within families Elapidae and Viperidae that have their venom proteomes characterised through MS-based venomics, only for 114 taxa has the venom gland transcriptome been reported, and venom gland transcriptomes are also available for another 6 species whose venom proteomes have not been analysed (Fig. 1, Fig. 2). In the absence of a reference transcriptomic database, the data gathered through shotgun the relationship of shotgun MS/MS-derived peptides to their intact parent molecules may be lost, or very difficult to reconstruct (e.g., in the case of highly similar isoforms). With the advancement of next-generation approaches to sequence and assemble shotgun omics data, the limitation to generate a transcriptomic database is not technical but has more to do with accessibility to the necessary biological material, e.g. venom gland tissue. Very stringent national wildlife protection laws, notably the Wildlife Protection Act, 1972 in India, a country home to more than 60 species of venomous snakes some of which are sufficiently common to cause severe human envenomings (Whitaker and Captain, 2004), severely hinders the collection of venom and snake tissues for scientific research.
Fig. 5.
Panel A, generalised representation of label-free shotgun proteomics. Both control and treatment samples are separately prepared and subjected to individual mass spectrometric analysis. In shotgun, or bottom-up proteomics, proteins are enzymatically digested into peptides, often with trypsin, prior to mass spectrometry. Label-free quantification (LFQ) is based on comparison of the areas of the peptide precursor ions of the same peptide, or by counting the number of fragment ion spectra of the same protein, in the different analyses. Appropriate comparisons are highlighted by the green ✓. Red X highlight that label-free strategies are not appropriate for determining the abundances of different proteins in a single sample: in this example, there is no change in abundance for protein B between control and treatment samples, whereas protein A is most abundant in the treated sample. Panel B, simulation of the different outcomes of label-free quantification of the same shotgun-generated tryptic venom peptidome when a species-specific genomic/transcriptomic reference database is available (a) or not (b) to match the tryptic peptide set.
Recent venomics studies have applied standalone ‘top-down’ (TD) MS or combined top-down and bottom-up approaches (Petras et al., 2015, 2016, 2019; Göçmen et al., 2015; Melani et al., 2016; Ainsworth et al., 2018; Pla et al., 2018; Hempel et al., 2020; Kazandjian et al., 2021). Compared to bottom-up peptide-centric methods, top-down venomics has the capability to resolve the venom proteome to the level of individual proteoforms or closely related isoforms. On the other hand, TD-MS requires higher resolution instrumental configurations with extended mass-to-charge range to isolate and fragment multiply charged monoisotopic ions and interpret the resulting complex fragmentation patterns of daughter ions. A major limitation of current snake venomics arise from the paucity of genomic and venom gland transcriptomic sequence information for venomous snakes in public domain databases (Brahma et al., 2015; Drukewitz and von Reumont, 2019). This limitation is more pressing in the case of TD venomics, where the upper mass limit for efficient fragmentation and the poor understanding of the mechanism of gas-phase dissociation, makes the automatic interpretation of the complex fragmentation spectra of native protein ions impractical without specific database support. Conversely, in bottom-up venomics of orphan organisms, algorithms for aiding in the automated de novo interpretation of fragmentation mass spectra are becoming powerful tools. In addition, though demanding a high level of user skill, manual de novo sequencing of high-quality fragmentation spectra followed by BLAST analysis of the deduced sequence remains another valid option for low-throughput identification of venom proteins from poorly studied venoms.
5. The evils of quantification
The abundance and specific toxicity of the individual toxins are important features for inferring composition-activity correlations of venoms at any level of organisation, whether isolated from single snakes, within and between populations, or across phylogenetic clades. Mass spectrometry-based proteomics has emerged as a powerful tool for the global characterisation and quantification of proteins from complex biological samples (Cravatt et al., 2007; Han et al., 2008). However, intrinsic differences in the ionisation efficiency and/or detectability of peptide ions limit the applicability of label-free inferences to directly estimate relative parent protein abundances. Precise quantification of proteins generally requires modification of standardised protocols, and is contingent upon the specific scientific question being addressed, the proteome being investigated, and the type of sample being analysed (Megger et al., 2013). Further, the experimental workflow is largely dependent on the availability of comprehensive protein sequence databases. Hence, while ongoing and completed genome sequencing projects have resulted in high-quality reference databases for many model organisms (Collins et al., 2003), the lack of comprehensive protein sequence databases represents a major challenge for proteomic investigations of non-model organisms (Carpentier et al., 2008; Armengaud et al., 2014; Heck and Neely, 2020), such as the majority of venomous snakes. However, this situation may rapidly improve. Thus, not counting subspecies or distinct conspecific populations, there are 114+ venom gland transcriptomes available in the NCBI TSA (Transcriptome Shotgun Assembly Sequence) and/or the SRA (Short Read Archive) (Fig. 1, Fig. 2), and if species clade transcriptomics becomes the trend (Kazandjian et al., 2021; Holding et al., 2021) the snake venom gland transcriptomics database will see significant growth in the immediate future.
The strategy of label-free quantification by spectral counting or ion intensity is based on the assumption that the likelihood of data-dependent precursor ion selection is higher for abundant precursor ions, and that the number of peptide identifications, normalised to account for the fact that larger proteins tend to contribute more peptide/spectra (Powell et al., 2004), represents a proxy of the abundance of the parent protein. A normalised spectral abundance factor (NSAF) is calculated as the number of spectral counts identifying a protein “i" (SpCi), divided by the protein's length (Li), divided by the sum of SpC/L for all proteins in the experiment. Despite label-free proteome quantification strategies being developed for model organisms for which comprehensive genomic or transcriptomic databases are available, and thus protein identifications (SpCi) does not represent a limiting factor (Old et al., 2005), there has been increased use of label-free shotgun proteomic techniques to characterize venom compositional patterns and estimate the relative abundance of individual proteins within the proteome of non-model snake venoms, particularly by very active research groups in India (Kalita et al., 2018; Patra et al., 2019; Patra and Mukherjee, 2020) and Malaysia (Zainal Abidin et al., 2016; Tan et al., 2016, 2018, 2019). The conceptual shortcoming of this approach results from the fact when a comprehensive reference database is missing, quantification is biased toward the successful peptide identifications (Liu et al., 2004; Tang et al., 2006; Bantscheff et al., 2007, 2012; Choi et al., 2008; Neilson et al., 2011) (Fig. 5B). Instead, deriving reliable quantitative information from peptide-centric MS data should be based on an unbiased procedure, e.g., where there is complete coverage of the venom toxin transcriptome.
The “snake venomics" proteome quantification strategy, which uses offline pre-MS two-separation step (reverse-phase HPLC and SDS-PAGE) to decomplex and quantify the venom proteome (Calvete, 2014; Eichberg et al., 2015; Calvete, 2017), fulfills this condition. It is worth mentioning that unlike “label-free" strategies that refer to unitless figures, the “snake venomics" proteome quantification strategy provides an estimate of the relative abundance of the venom's toxin arsenal in percentage (mg/100 mg) of total venom proteins (Calderón-Celis et al., 2016, 2017, 2019). Novel instrumental configurations have been proposed that combine elemental and molecular mass spectrometry platforms to achieve locus-resolved absolute quantification of the venom proteome (Calvete et al., 2017). Implementing top-down and absolute quantification approaches into next-generation mass spectrometry systems promises a quantitative leap, in proteomics in general, and for the study of venoms in particular. Although not discussed in this work, a thorough characterisation of venom peptidomes, protein glycosylations, and other post-translational modifications, will be a relevant and necessary task to complete our understanding of 'what's in a venom'.
6. The road to integrative snake venomics
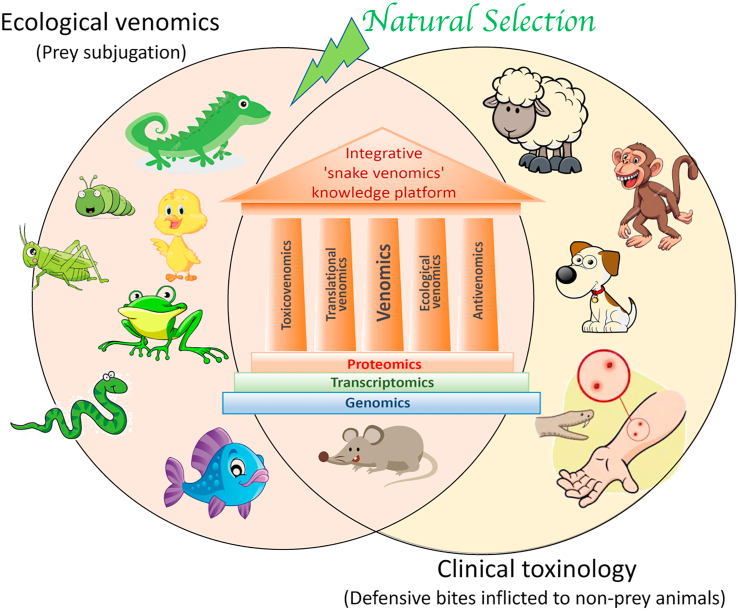
As addressed above, an increasing trend in venom analysis is “clade venomics", the identification of evolutionary trends across whole genera, taxonomic clades, and phylogenetic families. Clade venomics is contingent upon the specific scientific question to be addressed. This in turn implies that concepts, such as median lethal dose (LD50), must be interpreted differently depending on whether the question belongs to the ecology or clinical toxinology domain. Likewise, both ecological venomics and clinical venomics rely on complementary omic platforms that add new approaches to the study of venoms, contributing to the multidisciplinarity of the field. Venomics (sensu lato), “antivenomics" (Pla et al., 2017b; Calvete et al., 2018) and “toxicovenomics" (Lomonte, 2015; Lauridsen et al., 2017) are three complementary pillars of the molecular toxinologist's toolbox (Fig. 6). Strategic alliances between clade venomics, antivenomics and toxicovenomics highlight the versatility and adaptability of the original ‘venomics’ concept to the distinctive research of the different venomics domains.
Fig. 6.
Cartoon of the exclusive and overlapping domains of Ecological Venomics and Clinical Toxinology, emphasising the mutually enlightening relationship between evolutionary and translational venomics, guided by an integrative knowledge platform on snake venoms, supported by their compositional (venomics, aided by databases provided by transcriptomics and genomics), functional (toxicovenomics), and immunological (antivenomics) characteristics. Studies on adaptive variations that have evolved via natural selection and enhance the snake foraging success on preferred prey belongs exclusively to the field of ecological venomics, whereas the identification of the relevant toxins that should be neutralised to reverse the symptoms of snakebites inflicted to non-prey animals, such as pets, farm animals, or humans, falls into the exclusive scope of clinical toxinology. It is hoped that the expansion of integrative venomics may facilitate a deeper understanding of biological (ecological venomics) and medical (translational venomics) aspects of snake venoms, as well as contribute to confront the problem of snakebite envenomings by guiding improvements in the design and production of improved antivenoms.
6.1. Antivenomics provides key information to guide antivenom optimisation
Antivenoms are critical life-saving biologicals consisting of immunoglobulins (or derived F (ab')2 and Fab fragments) elicited by hyperimmunisation of horses or sheep with venom(s) from the most medically relevant snakes in the region where the antivenom is intended to be deployed. A strategy coined 'antivenomics' has been developed to evaluate the preclinical immunorecognition profile of an antivenom towards homologous and heterologous venoms. Successive “generations" of the antivenomics protocol have refined and extended its analytical scope (reviewed by Calvete et al., 2018). First generation consisted of the in-solution immunoprecipitation of antigen-antibody complexes followed by the chromatographic quantification of the uncomplexed venom proteins present in the supernatant (Lomonte et al., 2008). Affinity chromatography-based second generation (2G) antivenomics (Pla et al., 2012) yielded smoother and better resolved chromatographic profiles of both the non-binding and the bound venom proteins. The antivenom's toxin-resolved immunocapture ability can be estimated as the relative ratio of the chromatographic areas of the same protein recovered in the non-retained (NRi) and retained (Ri) affinity chromatography fractions using the equation: %NRi = 100 - [(Ri/(Ri + NRi)) x 100]. The inclusion of (R) in the equation compensates for possible losses during sample handling and chromatographic analysis, resulting in a more accurate quantification of the antivenom's immunorecognition profile than with the immunoprecipitation method. Polyclonal antivenoms exhibit distinct affinity concentration-dependent patterns of maximal binding toward different toxins of the venom. Optimising the immunoaffinity conditions for all venom components seems an impossible mission. However, the maximal immunocapturing capacity of an antivenom can be determined by the parallel running of identical antivenom affinity columns incubated with increasing amounts of venom until saturation is reached. This modified antivenomics protocol has been dubbed third generation (3G) antivenomics (Pla et al., 2017b).
Antivenomics informs on the maximal binding capacity of an antivenom's antibodies towards each of the venom toxins. However, the ability to bind toxins is an essential but not necessarily sufficient molecular property to endow an antivenom with clinical utility. A more relevant preclinical feature, the percentage of lethality neutralising antivenom antibodies (%AVneutr) can be calculated from the combined outcome of antivenomics and in vivo neutralisation assay as the ratio [P]/Maxbind (Calvete et al., 2018; Sanz et al., 2018; Al-Shekhadat et al., 2019; Sanz et al., 2020). [P] is the antivenom's potency (P [mg venom neutralised per mass unit (mL or mg) that rescue 100% of the test population] = [(n-1)/ED50] x LD50), where “n" is the number of median lethal doses (LD50s) used as challenge dose to determine the median effective dose (ED50) of the antivenom in the murine model) (Araujo et al., 2008; Morais et al., 2010). Maxbind stands for the antivenom's maximum toxin binding capacity.
6.2. Toxicovenomics in ecological venomics: LD50, and the relevance of contextualising toxinological studies
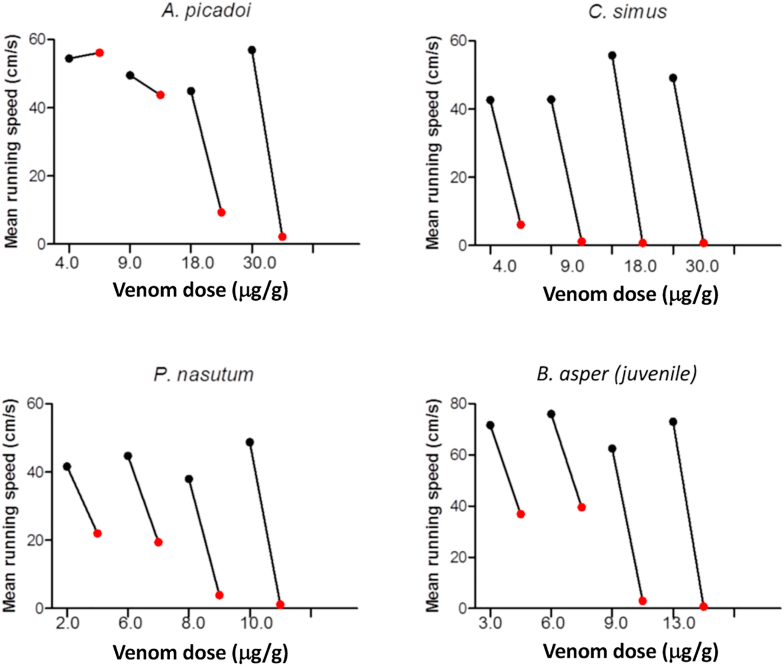
Antivenomics reveals information on the relative immunogenicity of the different venom components and quantifies the relative amounts of anti-toxin antibodies of an antivenom. However, venom toxins exhibit variable toxicity and it is the combination of abundance and potency that defines the ecological or clinical relevance of a toxin in the context of the venom injected into the prey or victim. Toxicovenomics aims to rank the different toxins present in a venom according to their contribution to the venom LD50. To this end a Toxicity Score (TS) is calculated by dividing the toxin (TXi) abundance (in moles/g of total venom proteins) by its median lethal dose (LD50) (in moles TXi/g animal body weight). The TS contributes to the identification of the most relevant toxins in a given venom (Calvete et al., 2010; Laustsen et al., 2015). The occurrence of taxon-preferent or taxon-specific toxins in certain venoms, particularly within the Colubridae family (Modahl et al., 2018; Modahl and Mackessy, 2019; Calvete et al., 2020), but also described in Micrurus (Elapidae) (Da Silva and Aird, 2001), underscores the importance of considering the adequate animal model for the purpose of the research aim (Jackson et al., 2019). Clearly, assessment of toxicity (median lethal dose, LD50) in the wild or in a controlled laboratory setting that mimics the natural ecosystem requires including natural prey in the study. Despite having stood the test of time, the LD50 is a somewhat coarse metric, which does not identify some important envenoming subtleties. The outcomes are reported at 24 or 48 h, which completely obscures events that occur during the first minutes of the envenoming. Under natural conditions, this period is crucial to the outcome of predation. The immobilisation or partial reduction of a prey's escape abilities or total paralysis are central aspects of a venom's efficiency (Fig. 7). For this reason, the knock-down time, the time to paralysis, could be a measure with greater biological significance when evaluating toxicity.
Fig. 7.
Changes in running speed after pitviper venom injection in the lizard Hemidactylus frenatus. Mean speed for four individuals before (black circle) and after (red circle) injection, tested on a 60 cm race track. Venoms from Atropoides picadoi, Crotalus simus, Porthidium nasutum, and Bothrops asper (juvenile) were tested. The first two species specialize in mammals; the last two include lizards in their diets. For any of the doses, lethal or sublethal, a significant decrease in escape velocity is observed few minutes after injection. In an ecological scenario, this reduction alone could represent an adaptive advantage for the snake.
The ecological limitations of the LD50 can be illustrated in the following example. The Duvernoy's gland secretory proteome of the Costa Rican false coral snake Rhinobothryum bovallii comprises a handful of toxins belonging to only three protein families (Calvete et al., 2020). Major monomeric (86.5 mol%) and minor dimeric (2.8 mol%) three-finger toxins (3FTxs) dominate its venom arsenal. The remaining 10.6% of the venom proteome comprises CRISP (8.2%) and PIII-SVMP (2.4%) molecules. Despite the large abundance of 3FTxs, a protein family whose elapid members are largely neurotoxic to mammals, in vivo lethality assays showed that R. bovallii venom is non-toxic to mice. Even intracerebroventricular injection of 100 μg of whole venom (containing 67.2 μg of 3FTxs) was harmless to mice. Intraperitonal administration of 50–650 μg of R. bovallii venom only killed one of two chickens at the maximum inoculated dose (14 μg/g), with death occurring around 18 h post-injection. Instead, a toxicovenomics analysis in R. bovallii's preferred prey, lizards e.g., baby green iguana, showed that both the major monomeric and minor dimeric 3FTxs at doses equivalent to 7.1–14.1 μg venom/g iguana body weight, 1 out of 3 animals was completely and irreversibly paralysed by 3 h post-injection and died overnight, <10 h later. At 450 μg/iguana (25.8–28.1 μg venom/g body weight) 100% (3/3) of the animals were immobilised entirely, but breathing, between 10 and 180 min post-injection. An identical effect was observed at the highest amount of venom injected (900 μg/iguana; 49–56 μg venom/g body weight), but at this dose all (3/3) iguanas collapsed between 10 and 35 min after venom administration, but only died hours later without showing during that time any signs of recovery. From these data, an “LD50″ of 315 μg/iguana was estimated, although this value was associated to broad 95% Confidence Intervals (CI95%, 62–569 μg/iguana). The fact that venom produces an irreversible immobilisation of the prey, which will then be swallowed alive, supports rapid prey immobilisation as the major role for the Costa Rican false coral snake venom, and questions the ecological meaning of LD50 in this, and likely other snake venoms. Perhaps it would be appropriate to define the irreversible immobilising activity of a venom as “the minimal dose of venom that induces irreversible immobilisation of the prey".
6.3. Toxicovenomics in preclinical research: allometric dose translation between model animals and human
In preclinical research, toxicovenomics complements antivenomics by uncovering the identity and lethal potential of the individual venom's toxins, thus informing which toxins should be primarily neutralised by an effective antivenom. In this sense, research on the likely toxic activities of venom from a snake species exhibiting a generalised or specialised diet towards endothermic prey, and how it would impact humans will involve the use of a laboratory animal of similar physiology to the victim, i.e. rodents. Making sense of the outcome of animal research in the context of human envenoming requires allometric dose translation between animal and human (West and Brown, 2005; Nair and Jacob, 2016). Allometry is used to describe how traits covary with body size, such as body surface area (BSA) and mass (White and Seymour, 2005). Allometric scaling is also an empirical approach where the exchange of drug dose is based on normalisation of dose to body surface area, and is used in research to predict an approximate dose on the basis of data existing in other species using the equation HED (human equivalent dose [mg/kg]) = animal dose [mg/kg] x [animal Km/human Km], where the correction factor (Km) is estimated by dividing the average body weight (kg) of the species to its body surface area (m2). For an average human body weight of 65 kg, and a body surface area of 1.75 m2 (calculated using the formula human body surface area (BSA) = [(weight [kg] x height [cm])/3600]1/2 (Mosteller, 1987; Verbraecken et al., 2006)), the Km factor for human is 65/1.75 = 37, whereas Km for a 20 g mouse is 3 (consult Table 1 in Nair and Jacob, 2016). Thus, allometric scaling of Egyptian cobra (Naja haje) i.p. LD50 for mice (20 g) of 1.8 μg/g mouse body weight translates into an LD50 for human of 0.15 mg/kg or 9.5 mg venom/65 kg person. For an estimated yield of 175–300 mg wet venom (52.5–100 mg dry proteins) (Branch, 1981; Mirtschin et al., 2006), a severe bite may deploy 6–11 median lethal doses into the victim. The amount of venom injected forms part of the “context” of a given snakebite treatment. Thus, for an antivenom dose to be effective in context – i.e. to possess ecological validity – it must be capable of neutralising at least 11 lethal doses of N. haje venom.
The venom of the Egyptian Cobra, N. haje, comprises a minor α-neurotoxin (αNTxs, 7.3 kDa, 9% of the total venom proteins) and two major cytotoxins (CTx-1, 32% (6.7 kDa) and CTx-2, 44% (6.8 kDa) of the venom proteome). Toxicovenomics analysis revealed disparity in LD50s of 0.4, 6.6, and 6 mg/kg mouse body weight, respectively, for the αNTx and cytotoxins CTx-1 and CTx-2 (JJC, data not published). These data correspond to allometrically calculated human LD50s [mg/human of 65 kg body weight] of 1.95 (αNTxs), 34.5 (CTx-1) and 31.5 (CTx-2). TS for these toxins are 45.6 (αNTxs), 2.38 (CTx-1), and 2.63 (CTx-2), clearly indicating that the major contributor to the venom's lethality are the α-NTXs. In an average 76 mg venom snakebite, the calculated number of LD50s deployed into a 65 kg human victim are unevenly distributed between the α-NTxs (3.5 LD50s) and the cytotoxins (CTx-1, 0.69 LD50s; CTx-2, 1.05 LD50s), highlighting the challenge for an effective antivenom to neutralize all three major 3FTxs present in the venom, but particularly the α-NTxs. According to allometric scaling predictions, only a mild snakebite, consisting of less than 20 mg of injected venom, would deploy sublethal amounts of any toxin into a victim, and in an envenoming where the amount of venom injected is in the range of 20–70 mg, only the α-NTXs would be life-threatening to the envenomed adult person. However, these calculations may vary if various toxins act synergistically.
6.4. Allometric scaling in ecological venomics
Continuing with the same species selected to illustrate the concept of allometry in preclinical venomics, in this section we highlight the relevance of allometric scaling in ecology in general, and ecological venomics in particular. The Egyptian cobra ranges across most of North Africa north of the Sahara, across the savannas of West Africa to the south of the Sahara, south to the Congo basin and east to Kenya and Tanzania. Knowledge on its dietary habits is based primarily on anecdotal data, suggesting they prey on a variety of small vertebrates, such as anurans, mammals, tortoises, birds and their eggs, lizards, fish, and occasionally snakes, including conspecifics (Schleich et al., 1996; Filippi and Petretto, 2013). Snake venom potency is generally prey-specific (Mebs, 2001; Healy et al., 2019). However, the potency of Egyptian cobra venom, and that of the vast majority of venomous snakes, has been studied only in the murine model, and venomics studies across its broad range and wide variety of habitats (steppes, dry to moist savannas, arid semi-desert regions with some water and vegetation) have not been reported. Adopting the “contextual stance” of evolution and ecology, this variation in diet and habitat is likely correlated with variation in venom composition and potency, including variations in potency towards distinct prey taxa.
Since the publication of Kleiber's influencial monograph, a great deal of effort has been invested in the investigation of both basal metabolic rate (BMR) scaling of drug doses from experimental animals to human, and the adaptive significance of BMR (Kleiber, 1932). During the last 35 years, investigation of BMR variation has gained prominence in ecology, and many studies have addressed the adaptive variation in BMR by correlating it with traits of interest (White and Seymour, 2005). Morphological and metabolic scaling result from adaptations evolved in the context of both physico-chemical and ecological constraints (reviewed by Glazier, 2005; but read also Magalhlaes Magalhaes Silva et al. (2018) regarding the interesting case of aquatic adaptations in a neotropical aquatic coral snake Micrurus surinamensis). Theoretical models of metabolic scaling have been proposed to explain interspecific relationships, including adaptive maximisation of foraging efficiency and maximisation of reproductive fitness. A ‘metabolic-level boundaries hypothesis’ focusing on two major constraints (surface-area limits on resource/waste exchange processes and mass/volume limits on power production), conceptually similar to the allometric dose translation between model animals and human discussed above, can explain much but not all the models of metabolic scaling with body mass (White et al., 2007).
A debate about the value of the allometric scaling exponent (b) relating metabolic rate (MR) to body mass (M) (MR = aMb) is still ongoing (Farrell-Gray and Gotelli, 2005; Glazier, 2005; White et al., 2007). A meta-analysis of 127 interspecific allometric exponents showed lack of support for a single exponent model (White et al., 2007). The analysis showed that the effect of size of mass on metabolic rate is significantly heterogeneous and that, on average, this effect is stronger for endotherms than for ectotherms. Significant differences between scaling exponents were also identified between ectotherms and endotherms, as well as between metabolic states (e.g., rest, field, and exercise). The absence of a universal metabolic allometry represents a significant challenge to any model between taxa or between metabolic states. A complete understanding of metabolic scaling will require the identification of both proximate (functional) and ultimate (evolutionary) causes.
6.5. Integrating biological reality into preclinical evaluation of antivenom efficacy
Preclinical studies of antivenom efficacy are typically based on elucidating the potency of antivenom against the potential lethal effects of venom in a model laboratory animal, generally mice. WHO considers the (murine) median effective dose (ED50) of antivenom to be a critical preclinical assessment bioassay that should be performed by manufacturers for all antivenoms, and enforced by the National Regulatory Authorities (NRAs) as part of the antivenom licensing procedure (WHO, 2017). The assay measures the dose of antivenom that prevents the death of 50% of test animals and may be reported in a number of ways (e.g.: mg venom/mL antivenom, μl antivenom/mg venom, mg venom/gram antivenom, etc.) (Ainsworth et al., 2020). The relevance to biological reality lies only in the premise that a product that prevents death in an animal model might also do the same in humans, albeit under different dosing, physiological and pharmacokinetic conditions. As we have discussed some venoms do not necessarily have death as primary endpoint; immobilisation, pre-digestion or pain (in the case of predator deterrence) may be the objective. Potency has been linked to prey-specificity and venom yields (the mass of venom the species produces in its venom glands), parameters that are likely associated with spatial environment, body size, metabolism (Healy et al., 2019; De Roodt et al., 2016), phenotypic, geographical and sexual factors (Mirtschin et al., 2002). How a venom is used also influences the type and quantity of venom produced. By relying on neutralisation of lethality in isolation the true biological context of antivenom design is often ignored, resulting in antivenoms that are ineffective in ‘field conditions’ where the ability to achieve clinical ‘cure’ (the objective of treatment) is crucial.
No minimum potency specifications currently exist for antivenoms marketed in sub-Saharan Africa (Gutiérrez et al., 2017b). Most products in the market have copied the specifications of FAV-Afrique and claim to neutralize a minimum of 200–250 murine (m) LD50 doses of each venom against which efficacy is claimed. The reality, however, is that neither yield of venom, nor mass of injected venom are equal across species. While 250 (m) LD50s/vial against Echis ocellatus venom from Ghana (LD50: 26.41 μg/19 g mouse) translates into 6602 μg venom, 250 LD50s/vial against Naja haje venom from Ugandan specimens (LD50: 1.90 μg/19 g mouse) is equal to 475 μg. When these figure are allometrically scaled to humans (LD50 E. ocellatus Ghana: 7.35 mg/65 kg human; LD50 N. haje Uganda: 0.52 mg/65 kg human), and the average venom yields are considered (E. ocellatus: 11.2 mg; Naja haje: 314.3 mg), it becomes clear that while 1.7 vials could potentially neutralize 50% of the lethality of E. ocellatus, the number needed for N. haje could be at least 604 vials (JJC, unpublished). However, current antivenom instructions for use recommend the same (typically small) doses be administered regardless of the species involved (DJW, unpublished). Venom yields obtained during laboratory-based venom extractions are typically much larger than the masses of venom released by snakes delivering defensive bites. For example, the mass of venom injected by the dreaded Dendroaspis polylepis during single defensive bites was 68.5% ± 12.5% less than the yield obtained from the same snakes during laboratory extractions (DJW, unpublished). Incorporating biologically relevant data such as mass of injected venom during defensive strikes and calculated human LD50 figures into the evaluation of preclinical antivenom efficacy could greatly improve formulation of products to ensure that they are likely to be effective against all of the species they are designed for use against. Similarly greater emphasis should be given to the specific activity of venom and the likely endpoints of envenoming. For example, antivenoms for species that are predominantly dermonecrotic or myotoxic should be assessed for their specific ability to abrogate these effects (WHO, 2017; Gutiérrez et al., 2017a) since lethality is often rare after bites by these species.
7. Concluding remarks and perspectives
Snakebite envenoming is a multifactorial health crisis afflicting the tropical developing world (WHO, 2019), which arises as a consequence of ecological interactions between humans and venomous snakes. The complexity of snakebite envenoming emerges from the reciprocal influence of the many factors involved – an interdependence that is characteristic of biological phenomena at all levels of analysis. Understanding such complex phenomena requires a combination of descriptive and explanatory knowledge. Descriptive knowledge consists of answers to “what?” questions – e.g., “what is there?” and “what is happening?“. Explanatory knowledge, on the other hand, consists of answers to “why?” questions – “why are things one way and not another?“. The achievement of explanatory understanding is contingent on the amassing of large quantities of descriptive knowledge – only when information about a large and diverse array of particulars is assembled can general patterns begin to be discerned.
In the present essay, we have detailed a number of approaches to the acquisition of highly resolved descriptive knowledge concerning the composition (venomics); activity and potency (toxicovenomics); and neutralisation (antivenomics and neutralisation assays) of snake venoms. In addition, we have provided perspectives, leveraging the contextual stance of ecology and evolutionary biology that provide a pathway towards explanatory knowledge. Mitigating the burden of snakebite envenoming will require strategies as multifactorial as the problem itself, including biotechnological approaches (e.g., knowledge-based refurbishing current antivenoms, creating recombinant antibodies, identifying small molecule inhibitors, etc. -Casewell et al., 2020) and health infrastructure engineering that have not been discussed in detail here. There is much work to be done. The specifics of venom variation and its impact in the clinical management of snakebite envenomings remain completely unknown, and thus our knowledge of the ecological validity of antivenoms against the venom of geographic variants of conspecific populations is extremely limited. Through the treatment of snakebite envenoming as the biological phenomenon that it is, by bringing to bear the resources of contemporary biological science in concert with those of biomedical science, we believe a pathway to deeper understanding is being forged. Understanding the problem will help us to solve it.
Author contributions
All the authors have contributed to the conceptualisation, original draft preparation, review and editing of the final manuscript version. All authors have read and agreed to the published version of the manuscript.
Ethical statement
Authors declare that international ethical guidelines for scientific papers were followed in the preparation of this manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
BOX.
-
-
Integration and contextualisation of ecological and clinical toxinology information can be mutually enlightening for both disciplines. In the present essay we have provided perspectives that should make the reader reflect on the appropriate use of concepts common to both disciplines, as well as on how to translate animal research outcome to humans.
-
-
Abundance and toxicity are conjugate variables in venom pharmacology. However, usage of this binomial is contingent on the appropriate toxinological context. The occurrence of taxon-preferent or taxon-specific venom toxins underscores the importance of considering the adequate animal model for the purpose of the research aim. The concept of venom median lethal dose (LD50) has also different meanings depending on animal species used and field of application, i.e., ecological vs. clinical toxinology.
-
-
Venomics, antivenomics and toxicovenomic are three complementary pillars of the molecular toxinologist's toolbox. The “ecological stance” should be also a vital part of the toxinologist's conceptual toolkit. The incorporation into preclinical antivenom evaluation of relevant snake's ecology data, such as mass of injected venom during defensive strikes of local medically relevant snakes and model animal LD50 figures allometrically scaled to human, could greatly impact the formulation of efficient snakebite therapeutics.
Acknowledgements
Studies by JJC's research group cited in this review were partially funded by grants from the Ministerio de Ciencia e Innovación, Madrid, Spain (BMC 2004-01432, BFU 2007-61563, BFU 2010-173730, BFU 2013-42833-P, and BFU 2017-89103-P). JJC wants to acknowledge and heartly thank all the researchers and collaborators of these projects who contributed laboratory work and numerous hours of scientific discussions. EABU was supported by a Norwegian Research Council FRIPRO-YRT Fellowship no. 287462. TNWJ was supported by National Health and Medical Research Grant 13/093/002 AVRU. KS was supported by DBT / Wellcome Trust India Alliance Fellowship (IA/I/19/2/504647). Support by Vicerrectoría de Investigación (University of Costa Rica) to work performed at Instituto Clodomiro Picado is also gratefully acknowledged.
Handling Editor: Glenn King
Contributor Information
Juan J. Calvete, Email: jcalvete@ibv.csic.es.
Bruno Lomonte, Email: Bruno.Lomonte@ucr.ac.cr.
Anthony J. Saviola, Email: anthony.saviola@cuanschutz.edu.
Fabián Bonilla, Email: fbonillamurillo@gmail.com.
Mahmood Sasa, Email: msasamarin@gmail.com.
David J. Williams, Email: dr_davidwilliams@outlook.com.
Eivind A.B. Undheim, Email: e.a.b.undheim@ibv.uio.no.
Kartik Sunagar, Email: ksunagar@iisc.ac.in.
Timothy N.W. Jackson, Email: timothy.jackson@unimelb.edu.au.
References
- Ainsworth S., Petras D., Engmark M., Süssmuth R.D., Whiteley G., Albulescu L.O., Kazandjian T.D., Wagstaff S.C., Rowley P., Wüster W., Dorrestein P.C., Arias A.S., Gutiérrez J.M., Harrison R.A., Casewell N.R., Calvete J.J. The medical threat of mamba envenoming in sub-Saharan Africa revealed by genus-wide analysis of venom composition, toxicity and antivenomics profiling of available antivenoms. J. Proteomics. 2018;172:173–189. doi: 10.1016/j.jprot.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Ainsworth S., Menzies S.K., Casewell N.R., Harrison R.A. An analysis of preclinical efficacy testing of antivenoms for sub-Saharan Africa: inadequate independent scrutiny and poor-quality reporting are barriers to improving snakebite treatment and management. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird S.D., Watanabe Y., Villar-Briones A., Roy M.C., Terada K., Mikheyev A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis) BMC Genom. 2013;14:790. doi: 10.1186/1471-2164-14-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird S.D., Arora J., Barua A., Qiu L., Terada K., Mikheyev A.S. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol. Evol. 2017;9:2640–2649. doi: 10.1093/gbe/evx199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida D.D., Viala V.L., Nachtigall P.G., Broe M., Gibbs H.L., Serrano S.M.T., Moura-da-Silva A.M., Ho P.L., Nishiyama M.Y., Jr., Junqueira-de-Azevedo I.L.M. Tracking the recruitment and evolution of snake toxins using the evolutionary context provided by the Bothrops jararaca genome. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2015159118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo H.P., Bourguignon S.C., Boller M.A., Dias A.A., Lucas E.P., Santos I.C., Delgado I.F. Potency evaluation of antivenoms in Brazil: the national control laboratory experience between 2000 and 2006. Toxicon. 2008;51:502–514. doi: 10.1016/j.toxicon.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Arbuckle K., Rodríguez de la Vega R.C., Casewell N.R. Coevolution takes the sting out of it: evolutionary biology and mechanisms of toxin resistance in animals. Toxicon. 2017;140:118–131. doi: 10.1016/j.toxicon.2017.10.026. [DOI] [PubMed] [Google Scholar]
- Armengaud J., Trapp J., Pible O., Geffard O., Chaumot A., Hartmann E.M. Non-model organisms, a species endangered by proteogenomics. J. Proteomics. 2014;105:5–18. doi: 10.1016/j.jprot.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 2007;389:1017–1131. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Bantscheff M., Lemeer S., Savitski M.M., Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- Barlow A., Pook C.E., Harrison R.A., Wüster W. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc. R. Soc. B. 2009;276:2443–2449. doi: 10.1098/rspb.2009.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua A., Mikheyev A.S. Many options, few solutions: over 60 My snakes converged on a few optimal venom formulations. Mol. Biol. Evol. 2019;36:1964–1974. doi: 10.1093/molbev/msz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet G., Alili D., Protte A., Upert G., Gilles N., Tepshi L., Stura E.A., Mourier G., Servent D. Ancestral protein resurrection and engineering opportunities of the mamba aminergic toxins. Sci. Rep. 2017;7:2701. doi: 10.1038/s41598-017-02953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J., Chesler A.T., Sharif-Naeini R., Medzihradszky K.F., Zhou S., King D., Sánchez E.E., Burlingame A.L., Basbaum A.I., Julius D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479:410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahma R.K., McCleary R.J., Kini R.M., Doley R. Venom gland transcriptomics for identifying, cataloging, and characterizing venom proteins in snakes. Toxicon. 2015;93:1–10. doi: 10.1016/j.toxicon.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Branch W.R. Venom of the south west African coral snake Aspidelaps lubricus infuscatus Mertens. J. Herpetol. Assoc. Afr. 1981;25:2–4. doi: 10.1080/04416651.1981.9650039. [DOI] [Google Scholar]
- Calderón-Celis F., Diez-Fernández S., Costa-Fernández J.M., Encinar J.R., Calvete J.J., Sanz-Medel A. Elemental mass spectrometry for absolute intact protein quantification without protein-specific standards: application to snake venomics. Anal. Chem. 2016;88:9699–9706. doi: 10.1021/acs.analchem.6b02585. [DOI] [PubMed] [Google Scholar]
- Calderón-Celis F., Cid-Barrio L., Encinar J.R., Sanz-Medel A., Calvete J.J. Absolute venomics: absolute quantification of intact venom proteins through elemental mass spectrometry. J. Proteomics. 2017;164:33–42. doi: 10.1016/j.jprot.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Calderón-Celis F., Sugiyama N., Yamanaka M., Sakai T., Diez-Fernández S., Calvete J.J., Sanz-Medel A., Encinar J.R. Enhanced universal quantification of biomolecules using element MS and generic standards: application to intact protein and phosphoprotein determination. Anal. Chem. 2019;91:1105–1112. doi: 10.1021/acs.analchem.8b04731. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Snake venomics: from the inventory of toxins to biology. Toxicon. 2013;75:44–62. doi: 10.1016/j.toxicon.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Next-generation snake venomics: protein-locus resolution through venom proteome decomplexation. Expert Rev. Proteomics. 2014;11:315–329. doi: 10.1586/14789450.2014.900447. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Venomics: integrative venom proteomics and beyond. Biochem. J. 2017;474:611–634. doi: 10.1042/BCJ20160577. [DOI] [PubMed] [Google Scholar]
- Calvete J.J. Snake venomics at the crossroads between ecological and clinical toxinology. Biochem (London) 2019;41:28–33. [Google Scholar]
- Calvete J.J., Sanz L., Cid P., de la Torre P., Flores-Díaz M., Dos Santos M.C., Borges A., Bremo A., Angulo Y., Lomonte B., Alape-Girón A., Gutiérrez J.M. Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res. 2010;9:528–544. doi: 10.1021/pr9008749. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Petras D., Calderón-Celis F., Lomonte B., Encinar J.R., Sanz-Medel A. Protein-species quantitative venomics: looking through a crystal ball. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017;23:27. doi: 10.1186/s40409-017-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete J.J., Rodríguez Y., Quesada-Bernat S., Pla D. Toxin-resolved antivenomics-guided assessment of the immunorecognition landscape of antivenoms. Toxicon. 2018;148:107–122. doi: 10.1016/j.toxicon.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Calvete J.J., Bonilla F., Granados-Martínez S., Sanz L., Lomonte B., Sasa M. Venomics of the Duvernoy's gland secretion of the false coral snake Rhinobothryum bovallii (Andersson, 1916) and assessment of venom lethality towards synapsid and diapsid animal models. J. Proteomics. 2020;225:103882. doi: 10.1016/j.jprot.2020.103882. [DOI] [PubMed] [Google Scholar]
- Carbajal-Márquez R.A., Cedeño-Vázquez J.R., Martínez-Arce A., Neri-Castro E., Machkour-M'rabet S.C. Accessing cryptic diversity in Neotropical rattlesnakes (Serpentes: Viperidae: Crotalus) with the description of two new species. Zootaxa. 2020;4729 doi: 10.11646/zootaxa.4729.4.1. zootaxa.4729.4.1. [DOI] [PubMed] [Google Scholar]
- Carpentier S.C., Panis B., Vertommen A., Swennen R., Sergeant K., Renaut J., Laukens K., Witters E., Samyn B., Devreese B. Proteome analysis of non‐model plants: a challenging but powerful approach. Mass Spectrom. Rev. 2008;27:354–377. doi: 10.1002/mas.20170. [DOI] [PubMed] [Google Scholar]
- Casewell N.R., Wüster W., Vonk F.J., Harrison R.A., Fry B.G. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol. Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Casewell N.R., Wagstaff S.C., Wüster W., Cook D.A., Bolton F.M., King S.I., Pla D., Sanz L., Calvete J.J., Harrison R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9205–9210. doi: 10.1073/pnas.1405484111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell N.R., Jackson T.N.W., Laustsen A.H., Sunagar K. Causes and consequences of snake venom variation. Trends Pharmacol. Sci. 2020;41:570–581. doi: 10.1016/j.tips.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Fermin D., Nesvizhskii A.I. Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani V., Debono J., Goldenberg J., Jackson T.N.W., Arbuckle K., Dobson J., Koludarov I., Li B., Hay C., Dunstan N., Allen L., Hendrikx I., Kwok H.F., Fry B.G. Correlation between ontogenetic dietary shifts and venom variation in Australian brown snakes (Pseudonaja) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2017;197:53–60. doi: 10.1016/j.cbpc.2017.04.007. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Green E.D., Guttmacher A.E., Guyer M.S. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Cravatt B.F., Simon G.M., Yates J.R., III The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- Da Silva N.J., Aird S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001;128:425–456. doi: 10.1016/s1532-0456(00)00215-5. [DOI] [PubMed] [Google Scholar]
- Daltry J.C., Wüster W., Thorpe R.S. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- Davies E.L., Arbuckle K. Coevolution of snake venom toxic activities and diet: evidence that ecological generalism favours toxicological diversity. Toxins. 2019;11:711. doi: 10.3390/toxins11120711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R. Oxford University Press; Oxford, UK: 1982. The Extended Phenotype: the Long Reach of the Gene. [Google Scholar]
- De Roodt A.R., Boyer L.V., Lanari L.C., Irazu L., Laskowicz R.D., Sabattini P.L., Damin C.F. Venom yield and its relationship with body size and fang separation of pit vipers from Argentina. Toxicon. 2016;121:22–29. doi: 10.1016/j.toxicon.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Dennett D. Simon & Schuster; New York, NY: 1995. Darwin's Dangerous Idea. [Google Scholar]
- Drabeck D.H., Dean A.M., Jansa S.A. Why the honey badger don't care: convergent evolution of venom-targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon. 2015;99:68–72. doi: 10.1016/j.toxicon.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Drukewitz S.H., von Reumont B.M. The significance of comparative genomics in modern evolutionary venomics. Front. Ecol. Evol. 2019;7:163. [Google Scholar]
- Durban J., Pérez A., Sanz L., Gómez A., Bonilla F., Rodríguez S., Chacón D., Sasa M., Angulo Y., Gutiérrez J.M., Calvete J.J. Integrated "omics" profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genom. 2013;14:234. doi: 10.1186/1471-2164-14-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichberg S., Sanz L., Calvete J.J., Pla D. Constructing comprehensive venom proteome reference maps for integrative venomics. Expert Rev. Proteomics. 2015;12:557–573. doi: 10.1586/14789450.2015.1073590. [DOI] [PubMed] [Google Scholar]
- Farrell-Gray C.C., Gotelli N.J. Allometric exponents support a 3/4-power scaling law. Ecology. 2005;86:2083–2087. [Google Scholar]
- Filippi E., Petretto M. Naja haje (Egyptian cobra) diet/ophiophagy. Herpetol. Rev. 2013;44:155–156. [Google Scholar]
- Fox J.W., Serrano S.M. Exploring snake venom proteomes: multifaceted analyses for complex toxin mixtures. Proteomics. 2008;8:909–920. doi: 10.1002/pmic.200700777. [DOI] [PubMed] [Google Scholar]
- Fry B.G., Vidal N., Norman J.A., Vonk F.J., Scheib H., Ramjan S.F., Kuruppu S., Fung K., Hedges S.B., Richardson M.K., Hodgson W.C., Ignjatovic V., Summerhayes R., Kochva E. Early evolution of the venom system in lizards and snakes. Nature. 2006;439:584–588. doi: 10.1038/nature04328. [DOI] [PubMed] [Google Scholar]
- Fry B.G., Roelants K., Champagne D.E., Scheib H., Tyndall J.D., King G.F., Nevalainen T.J., Norman J.A., Lewis R.J., Norton R.S., Renjifo C., Rodríguez de la Vega R.C. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genom. Hum. Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Fry B.G., Casewell N.R., Wüster W., Vidal N., Young B., Jackson T.N. The structural and functional diversification of the Toxicofera reptile venom system. Toxicon 2012. 2012;60:434–448. doi: 10.1016/j.toxicon.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Gibbs H.L., Sanz L., Chiucchi J.E., Farrell T.M., Calvete J.J. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri) J. Proteomics. 2011;74:2169–2179. doi: 10.1016/j.jprot.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Gibbs H.L., Sanz L., Sovic M.G., Calvete J.J. Phylogeny-based comparative analysis of venom proteome variation in a clade of rattlesnakes (Sistrurus sp.) PloS One. 2013;8 doi: 10.1371/journal.pone.0067220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs H.L., Sanz L., Pérez A., Ochoa A., Hassinger A.T.B., Holding M.L., Calvete J.J. The molecular basis of venom resistance in a rattlesnake-squirrel predator-prey system. Mol. Ecol. 2020;29:2871–2888. doi: 10.1111/mec.15529. [DOI] [PubMed] [Google Scholar]
- Gilbert C., Meik J.M., Dashevsky D., Card D.C., Castoe T.A., Schaack S. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc. Biol. Sci. 2014;281:20141122. doi: 10.1098/rspb.2014.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgianni M.W., Dowell N.L., Griffin S., Kassner V.A., Selegue J.E., Carroll S.B. The origin and diversification of a novel protein family in venomous snakes. Proc. Natl. Acad. Sci. U.S.A. 2020;117:10911–10920. doi: 10.1073/pnas.1920011117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier D.S. Beyond the ‘3/4-power law’: variation in the intra- and interspecific scaling of metabolic rate in animals. Biol. Rev. 2005;80:1–52. doi: 10.1017/S1464793105006834. [DOI] [PubMed] [Google Scholar]
- Glenn J.L., Straight R.C., Wolfe M.C., Hardy D.L. Geographical variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) venom properties. Toxicon. 1983;21:19–30. doi: 10.1016/0041-0101(83)90055-7. [DOI] [PubMed] [Google Scholar]
- Göçmen B., Heiss P., Petras D., Nalbantsoy A., Süssmuth R.D. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon. 2015;107:163–174. doi: 10.1016/j.toxicon.2015.09.013. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M. Snakebite envenoming from an Ecohealth perspective. Toxicon:X. 2020;7:100043. doi: 10.1016/j.toxcx.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nature Rev. Dis. Primers. 2017;3:17063. doi: 10.1038/nrdp.2017.63. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Solano G., Pla D., Herrera M., Segura Á., Vargas M., Villalta M., Sánchez A., Sanz L., Lomonte B., León G., Calvete J.J. Preclinical evaluation of the efficacy of antivenoms for snakebite envenoming: state-of-the-art and challenges ahead. Toxins. 2017;9:163. doi: 10.3390/toxins9050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S. A process ontology for macromolecular biology. In: Nicholson D.J., Dupré J., editors. Everything Flows: towards a Processual Philosophy of Biology. Oxford University Press; Oxford, UK: 2018. 198779631. [Google Scholar]
- Han X., Aslanian A., Yates J.R., III Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.F., Houle D. Measuring and comparing evolvability and constraint in multivariate characters. J. Evol. Biol. 2008;21:1201–1219. doi: 10.1111/j.1420-9101.2008.01573.x. [DOI] [PubMed] [Google Scholar]
- Harris R.J., Nekaris K.A.-I., Fry B.G. Coevolution between primates and venomous snakes revealed by α-neurotoxin susceptibility. bioRxiv preprint posted January. 2021;29:2021. doi: 10.1101/2021.01.28.428735. [DOI] [Google Scholar]
- Harrison R.A., Hargreaves A., Wagstaff S.C., Faragher B., Lalloo D.G. Snake envenoming: a disease of poverty. PLoS Neglected Trop. Dis. 2009;3:e569. doi: 10.1371/journal.pntd.0000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy K., Carbone C., Jackson A.L. Snake venom potency and yield are associated with prey-evolution, predator metabolism and habitat structure. Ecol. Lett. 2019;22:527–537. doi: 10.1111/ele.13216. [DOI] [PubMed] [Google Scholar]
- Heck M., Neely B.A. Proteomics in non-model organisms: a new analytical frontier. J. Proteome Res. 2020;19:3595–3606. doi: 10.1021/acs.jproteome.0c00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel B.F., Damm M., Mrinalini, Göçmen B., Karış M., Nalbantsoy A., Kini R.M., Süssmuth R.D. Extended snake venomics by top-down in-source decay: investigating the newly discovered anatolian meadow viper subspecies, Vipera anatolica senliki. J. Proteome Res. 2020;19:1731–1749. doi: 10.1021/acs.jproteome.9b00869. [DOI] [PubMed] [Google Scholar]
- Holding M.L., Drabeck D.H., Jansa S.A., Gibbs H.L. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integr. Comp. Biol. 2016;56:1032–1043. doi: 10.1093/icb/icw082. [DOI] [PubMed] [Google Scholar]
- Holding M.L., Strickland J.L., Rautsaw R.M., Hofmann E.P., Mason A.J., Hogan M.P., Nystrom G.S., Ellsworth S.A., Colston T.J., Borja M., Castañeda-Gaytán G., Grünwald C.I., Jones J.M., Freitas-de-Sousa L.A., Viala V.L., Margres M.J., Hingst-Zaher E., Junqueira-de-Azevedo I.L.M., Moura-da-Silva A.M., Grazziotin F.G., Gibbs H.L., Rokyta D.R., Parkinson C.L. Phylogenetically diverse diets favor more complex venoms in North American pitvipers. Proc. Natl. Acad. Sci. U.S.A. 2021;118 doi: 10.1073/pnas.2015579118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang A.Y., Field D.J., Webster T.H., Behlke A.D., Davis M.B., Racicot R.A., Gauthier J.A. The origin of snakes: revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol. Biol. 2015;15:87. doi: 10.1186/s12862-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson D.A., Mori A., Savitzky A.H., Burghardt G.M., Wu X., Meinwald J., Schroeder F.C. Dietary sequestration of defensive steroids in nuchal glands of the Asian snake Rhabdophis tigrinus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2265–2270. doi: 10.1073/pnas.0610785104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbister G.K. Antivenom efficacy or effectiveness: the Australian experience. Toxicology. 2010;268:148–154. doi: 10.1016/j.tox.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Jackson T.N.W., Koludarov I. How the toxin got its toxicity. Front. Pharmacol. 2020;11:574925. doi: 10.3389/fphar.2020.574925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T.N.W., Koludarov I., Ali S.A., Dobson J., Zdenek C.N., Dashevsky D., Op den Brouw B., Masci P.P., Nouwens A., Josh P., Goldenberg J., Cipriani V., Hay C., Hendrikx I., Dunstan N., Allen L., Fry B.G. Rapid radiations and the race to redundancy: an investigation of the evolution of Australian elapid snake venoms. Toxins. 2016;8:309. doi: 10.3390/toxins8110309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T.N.W., Jouanne H., Vidal N. Snake venom in context: neglected clades and concepts. Front. Ecol. Evol. 2019;7:332. [Google Scholar]
- Juárez P., Comas I., González-Candelas F., Calvete J.J. Evolution of snake venom disintegrins by positive Darwinian selection. Mol. Biol. Evol. 2008;25:2391–2407. doi: 10.1093/molbev/msn179. [DOI] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo I.L., Campos P.F., Ching A.T., Mackessy S.P. Colubrid venom composition: an -omics perspective. Toxins. 2016;8:230. doi: 10.3390/toxins8080230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita B., Mackessy S.P., Mukherjee A.K. Proteomic analysis reveals geographic variation in venom composition of Russell's Viper in the Indian subcontinent: implications for clinical manifestations post-envenomation and antivenom treatment. Expert Rev. Proteomics. 2018;15:837–849. doi: 10.1080/14789450.2018.1528150. [DOI] [PubMed] [Google Scholar]
- Kazandjian T.D., Petras D., Robinson S.D., van Thiel J., Greene H.W., Arbuckle K., Barlow A., Carter D.A., Wouters R.M., Whiteley G., Wagstaff S.C., Arias A.S., Albulescu L.O., Plettenberg Laing A., Hall C., Heap A., Penrhyn-Lowe S., McCabe C.V., Ainsworth S., da Silva R.R., Dorrestein P.C., Richardson M.K., Gutiérrez J.M., Calvete J.J., Harrison R.A., Vetter I., Undheim E.A.B., Wüster W., Casewell N.R. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science. 2021;371(6527):386–390. doi: 10.1126/science.abb9303. 2021 Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkkamp H.M., Kini R.M., Pospelov A.S., Vonk F.J., Henkel C.V., Richardson M.K. Snake genome sequencing: results and future prospects. Toxins. 2016;8:360. doi: 10.3390/toxins8120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida T., Go Y., Tatsumoto S., Tatsumi K., Kuraku S., Toda M. Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes. Proc. Biol. Sci. 2019;286:20191828. doi: 10.1098/rspb.2019.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber M. Body size and metabolism. Hilgardia. 1932;6:315–353. [Google Scholar]
- Lauridsen L.P., Laustsen A.H., Lomonte B., Gutiérrez J.M. Exploring the venom of the forest cobra snake: toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteomics. 2017;150:98–108. doi: 10.1016/j.jprot.2016.08.024. [DOI] [PubMed] [Google Scholar]
- Laustsen A.H., Lohse B., Lomonte B., Engmark M., Gutiérrez J.M. Selecting key toxins for focused development of elapid antivenoms guided by a Toxicity Score. Toxicon. 2015;104:43–45. doi: 10.1016/j.toxicon.2015.07.334. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Kim E.G., Jung H.R., Jung J.W., Kim H.B., Cho J.W., Kim K.M., Eugene C.Y. Refinements of LC-MS/MS spectral counting statistics improve quantification of low abundance proteins. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-49665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Fry B.G., Kini R.M. Eggs-only diet: its implications for the toxin profile changes and ecology of the marbled sea snake (Aipysurus eydouxii) J. Mol. Evol. 2005;60:81–89. doi: 10.1007/s00239-004-0138-0. [DOI] [PubMed] [Google Scholar]
- Liu H., Sadygov R.G., Yates J.R., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Calvete J.J. Strategies in 'snake venomics' aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J. Venom. Anim. Toxins Incl. Trop. Dis. 2017;23:26. doi: 10.1186/s40409-017-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte B., Escolano J., Fernández J., Sanz L., Angulo Y., Gutiérrez J.M., Calvete J.J. Snake venomics and antivenomics of the arboreal neotropical pitvipers Bothriechis lateralis and Bothriechis schlegelii. J. Proteome Res. 2008;7:2445–2457. doi: 10.1021/pr8000139. [DOI] [PubMed] [Google Scholar]
- Lomonte B., Tsai W.C., Ureña-Diaz J.M., Sanz L., Mora-Obando D., Sánchez E.E., Fry B.G., Gutiérrez J.M., Gibbs H.L., Sovic M.G., Calvete J.J. Venomics of New World pit vipers: genus-wide comparisons of venom proteomes across Agkistrodon. J. Proteomics. 2014;96:103–116. doi: 10.1016/j.jprot.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte B., Rey-Suárez P., Fernández J., Sasa M., Pla D., Vargas N., Bénard-Valle M., Sanz L., Corrêa-Netto C., Núñez V., Alape-Girón A., Alagón A., Gutiérrez J.M., Calvete J.J. Venoms of Micrurus coral snakes: evolutionary trends in compositional patterns emerging from proteomic analyses. Toxicon. 2016;122:7–25. doi: 10.1016/j.toxicon.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Longbottom J., Shearer F.M., Devine M., Alcoba G., Chappuis F., Weiss D.J., Ray S.E., Ray N., Warrell D.A., Ruiz de Castañeda R., Williams D.J., Hay S.I., Pigott D.M. Vulnerability to snakebite envenoming: a global mapping of hotspots. Lancet. 2018;392:673–684. doi: 10.1016/S0140-6736(18)31224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch V.J. Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes. BMC Evol. Biol. 2007;7:1–4. doi: 10.1186/1471-2148-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy S.P., Williams K., Ashton K.G. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: a case of venom paedomorphosis? Copeia. 2003. 2003:769–782. [Google Scholar]
- Mackessy S.P., Sixberry N.M., Heyborne W.H., Fritts T. Venom of the Brown Treesnake, Boiga irregularis: ontogenetic shifts and taxa-specific toxicity. Toxicon. 2006;47(5):537–548. doi: 10.1016/j.toxicon.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Magalhaes Silva F., da Costa Prudente A.L., Machado F.A., Santos M.M., Zaher H., Hingst-Zaher E. Aquatic adaptations in a Neotropical coral snake: a study of morphological convergence. J. Zool. Syst. Evol. Res. 2018;56:382–394. [Google Scholar]
- Margres M.J., Wray K.P., Hassinger A.T.B., Ward M.J., McGivern J.J., Moriarty Lemmon E., Lemmon A.R., Rokyta D.R. Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol. 2017;34:3099–3110. doi: 10.1093/molbev/msx231. [DOI] [PubMed] [Google Scholar]
- Margres M.J., Rautsaw R.M., Strickland J.L., Mason A.J., Schramer T.D., Hofmann E.P., Stiers E., Ellsworth S.A., Nystrom G.S., Hogan M.P., Bartlett D.A., Colston T.J., Gilbert D.M., Rokyta D.R., Parkinson C.L. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2014634118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebs D. Toxicity in animals. Trends in evolution? Toxicon. 2001;39:87–96. doi: 10.1016/s0041-0101(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Megger D.A., Bracht T., Meyer H.E., Sitek B. Label-free quantification in clinical proteomics. Biochim. Biophys. Acta Protein Proteonomics. 2013;1834:1581–1590. doi: 10.1016/j.bbapap.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Melani R.D., Skinner O.S., Fornelli L., Domont G.B., Compton P.D., Kelleher N.L. Mapping proteoforms and protein complexes from king cobra venom using both denaturing and native top-down proteomics. Mol. Cell. Proteomics. 2016;15:2423–2434. doi: 10.1074/mcp.M115.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirtschin P.J., Shine R., Nias T.J., Dunstan N.L., Hough B.J., Mirtschin M. Influences on venom yield in Australian tigersnakes (Notechis scutatus) and brownsnakes (Pseudonaja textilis: Elapidae, Serpentes) Toxicon. 2002;40:1581–1592. doi: 10.1016/s0041-0101(02)00175-7. [DOI] [PubMed] [Google Scholar]
- Mirtschin P.J., Dunstan N., Hough B., Hamilton E., Klein S., Lucas J., Millar D., Madaras F., Nias T. Venom yields from Australian and some other species of snakes. Ecotoxicology. 2006;15:531–538. doi: 10.1007/s10646-006-0089-x. [DOI] [PubMed] [Google Scholar]
- Modahl C.M., Mackessy S.P. Venoms of rear-fanged snakes: new proteins and novel activities. Front. Ecol. Evol. 2019;7:279. [Google Scholar]
- Modahl C.M., Mrinalini, Frietze S., Mackessy S.P. Adaptive evolution of distinct prey-specific toxin genes in rear-fanged snake venom. Proc. R. Soc. B. 2018;285:20181003. doi: 10.1098/rspb.2018.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais V., Ifran S., Berasain P., Massaldi H. Antivenoms: potency or median effective dose, which to use? J. Venom. Anim. Toxins Incl. Trop. Dis. 2010;16:191–193. [Google Scholar]
- Mosteller R.D. Simplified calculation of body surface area. N. Engl. J. Med. 1987;317:1098. doi: 10.1056/NEJM198710223171717. [DOI] [PubMed] [Google Scholar]
- Nair A.B., Jacob S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016;7:27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson K.A., Ali N.A., Muralidharan S., Mirzaei M., Mariani M., Assadourian G., Lee A., van Sluyter S.C., Haynes P.A. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- Old W.M., Meyer-Arendt K., Aveline-Wolf L., Pierce K.G., Mendoza A., Sevinsky J.R., Resing K.A., Ahn N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Patra A., Mukherjee A.K. Proteomic analysis of Sri Lanka Echis carinatus venom: immunological cross-reactivity and enzyme neutralization potency of Indian polyantivenom. J. Proteome Res. 2020;19:3022–3032. doi: 10.1021/acs.jproteome.0c00054. [DOI] [PubMed] [Google Scholar]
- Patra A., Chanda A., Mukherjee A.K. Quantitative proteomic analysis of venom from Southern India common krait (Bungarus caeruleus) and identification of poorly immunogenic toxins by immune-profiling against commercial antivenom. Expert Rev. Proteomics. 2019;16:457–469. doi: 10.1080/14789450.2019.1609945. [DOI] [PubMed] [Google Scholar]
- Peng C., Ren J.L., Deng C., Jiang D., Wang J., Qu J., Chang J., Yan C., Jiang K., Murphy R.W., Wu D.D., Li J.T. The genome of Shaw's sea Snake (Hydrophis curtus) reveals secondary adaptation to its marine environment. Mol. Biol. Evol. 2020;37:1744–1760. doi: 10.1093/molbev/msaa043. [DOI] [PubMed] [Google Scholar]
- Petras D., Heiss P., Süssmuth R.D., Calvete J.J. Venom proteomics of Indonesian king cobra, Ophiophagus hannah: integrating top-down and bottom-up approaches. J. Proteome Res. 2015;14:2539–2556. doi: 10.1021/acs.jproteome.5b00305. [DOI] [PubMed] [Google Scholar]
- Petras D., Heiss P., Harrison R.A., Süssmuth R.D., Calvete J.J. Top-down venomics of the East African green mamba, Dendroaspis angusticeps, and the black mamba, Dendroaspis polylepis, highlight the complexity of their toxin arsenals. J. Proteomics. 2016;146:148–164. doi: 10.1016/j.jprot.2016.06.018. [DOI] [PubMed] [Google Scholar]
- Petras D., Hempel B.F., Göçmen B., Karis M., Whiteley G., Wagstaff S.C., Heiss P., Casewell N.R., Nalbantsoy A., Süssmuth R.D. Intact protein mass spectrometry reveals intraspecies variations in venom composition of a local population of Vipera kaznakovi in Northeastern Turkey. J. Proteomics. 2019;199:31–50. doi: 10.1016/j.jprot.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla D., Gutiérrez J.M., Calvete J.J. Second generation snake antivenomics: comparing immunoaffinity and immunodepletion protocols. Toxicon. 2012;60:688–699. doi: 10.1016/j.toxicon.2012.04.342. [DOI] [PubMed] [Google Scholar]
- Pla D., Sanz L., Molina-Sánchez P., Zorita V., Madrigal M., Flores-Díaz M., Alape-Girón A., Núñez V., Andrés V., Gutiérrez J.M., Calvete J.J. Snake venomics of Lachesis muta rhombeata and genus-wide antivenomics assessment of the paraspecific immunoreactivity of two antivenoms evidence the high compositional and immunological conservation across Lachesis. J. Proteomics. 2013;89:112–123. doi: 10.1016/j.jprot.2013.05.028. [DOI] [PubMed] [Google Scholar]
- Pla D., Sanz L., Sasa M., Acevedo M.E., Dwyer Q., Durban J., Pérez A., Rodriguez Y., Lomonte B., Calvete J.J. Proteomic analysis of venom variability and ontogeny across the arboreal palm-pitvipers (genus Bothriechis) J. Proteomics. 2017;152:1–12. doi: 10.1016/j.jprot.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Pla D., Rodríguez Y., Calvete J.J. Third generation antivenomics: pushing the limits of the in vitro preclinical assessment of antivenoms. Toxins. 2017;9:E158. doi: 10.3390/toxins9050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pla D., Petras D., Saviola A.J., Modahl C.M., Sanz L., Pérez A., Juárez E., Frietze S., Dorrestein P.C., Mackessy S.P., Calvete J.J. Transcriptomics-guided bottom-up and top-down venomics of neonate and adult specimens of the arboreal rear-fanged Brown Treesnake, Boiga irregularis, from Guam. J. Proteomics. 2018;174:71–84. doi: 10.1016/j.jprot.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Post Y., Puschhof J., Beumer J., Kerkkamp H.M., de Bakker M.A.G., Slagboom J., de Barbanson B., Wevers N.R., Spijkers X.M., Olivier T., Kazandjian T.D., Ainsworth S., Iglesias C.L., van de Wetering W.J., Heinz M.C., van Ineveld R.L., van Kleef R.G.D.M., Begthel H., Korving J., Bar-Ephraim Y.E., Getreuer W., Rios A.C., Westerink R.H.S., Snippert H.J.G., van Oudenaarden A., Peters P.J., Vonk F.J., Kool J., Richardson M.K., Casewell N.R., Clevers H. Snake venom gland organoids. Cell. 2020;180:233–247. doi: 10.1016/j.cell.2019.11.038. [DOI] [PubMed] [Google Scholar]
- Powell D.W., Weaver C.M., Jennings J.L., McAfee K.J., He Y., Weil P.A., Link A.J. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- Reeks T.A., Fry B.G., Alewood P.F. Privileged frameworks from snake venom. Cell. Mol. Life Sci. 2015;72:1939–1958. doi: 10.1007/s00018-015-1844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D.R., Wray K.P., Lemmon A.R., Lemmon E.M., Caudle S.B. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Sanz L., Quesada-Bernat S., Chen P.Y., Lee C.D., Chiang J.R., Calvete J.J. Translational venomics: third-generation antivenomics of anti-siamese russell's viper, Daboia siamensis, antivenom manufactured in taiwan CDC's vaccine center. Trav. Med. Infect. Dis. 2018;3:66. doi: 10.3390/tropicalmed3020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L., Quesada-Bernat S., Ramos T., Casais-E-Silva L.L., Corrêa-Netto C., Silva-Haad J.J., Sasa M., Lomonte B., Calvete J.J. New insights into the phylogeographic distribution of the 3FTx/PLA2 venom dichotomy across genus Micrurus in South America. J. Proteomics. 2019;200:90–101. doi: 10.1016/j.jprot.2019.03.014. [DOI] [PubMed] [Google Scholar]
- Sanz L., de Freitas-Lima L.N., Quesada-Bernat S., Graça-de-Souza V.K., Soares A.M., Calderón L.A., Calvete J.J., Caldeira C.A.S. Comparative venomics of Brazilian coral snakes: Micrurus frontalis, Micrurus spixii spixii, and Micrurus surinamensis. Toxicon. 2019;166:39–45. doi: 10.1016/j.toxicon.2019.05.001. [DOI] [PubMed] [Google Scholar]
- Schendel V., Rash L.D., Jenner R.A., Undheim E. The diversity of venom: the importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins. 2019;11:666. doi: 10.3390/toxins11110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schield D.R., Card D.C., Hales N.R., Perry B.W., Pasquesi G.M., Blackmon H., Adams R.H., Corbin A.B., Smith C.F., Ramesh B., Demuth J.P., Betrán E., Tollis M., Meik J.M., Mackessy S.P., Castoe T.A. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019;29:590–601. doi: 10.1101/gr.240952.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich H.H., Kästle W., Kabisch K. Koeltz Scientific Books; 1996. Amphibians and Reptiles of North Africa: Biology, Systematics, Field Guide; p. 630. -13: 978-3874293778. [Google Scholar]
- Schmuckler M.A. What is ecological validity? A dimensional analysis. Infancy. 2001;2:419–436. doi: 10.1207/S15327078IN0204_02. [DOI] [PubMed] [Google Scholar]
- Senji Laxme R.R., Khochare S., de Souza H.F., Ahuja B., Suranse V., Martin G., Whitaker R., Sunagar K. Beyond the 'big four': venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senji Laxme R.R., Attarde S., Khochare S., Suranse V., Martin G., Casewell N.R., Whitaker R., Sunagar K. Biogeographical venom variation in the Indian spectacled cobra (Naja naja) underscores the pressing need for pan-India efficacious snakebite therapy. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H., Chijiwa T., Oda-Ueda N., Nakamura H., Yamaguchi K., Hattori S., Matsubara K., Matsuda Y., Yamashita A., Isomoto A., Mori K., Tashiro K., Kuhara S., Yamasaki S., Fujie M., Goto H., Koyanagi R., Takeuchi T., Fukumaki Y., Ohno M., Shoguchi E., Hisata K., Satoh N., Ogawa T. The habu genome reveals accelerated evolution of venom protein genes. Sci. Rep. 2018;8:11300. doi: 10.1038/s41598-018-28749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland J.L., Smith C.F., Mason A.J., Schield D.R., Borja M., Castañeda-Gaytán G., Spencer C.L., Smith L.L., Trápaga A., Bouzid N.M., Campillo-García G., Flores-Villela O.A., Antonio-Rangel D., Mackessy S.P., Castoe T.A., Rokyta D.R., Parkinson C.L. Evidence for divergent patterns of local selection driving venom variation in Mojave Rattlesnakes (Crotalus scutulatus) Sci. Rep. 2018;8:17622. doi: 10.1038/s41598-018-35810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Moran Y. The rise and fall of an evolutionary innovation: contrasting strategies of venom evolution in ancient and young animals. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Jackson T.N., Undheim E.A., Ali S.A., Antunes A., Fry B.G. Three-fingered RAVERs: rapid accumulation of variations in exposed residues of snake venom toxins. Toxins. 2013;5:2172–2208. doi: 10.3390/toxins5112172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K., Morgenstern D., Reitzel A.M., Moran Y. Ecological venomics: how genomics, transcriptomics and proteomics can shed new light on the ecology and evolution of venom. J. Proteomics. 2016;135:62–72. doi: 10.1016/j.jprot.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Sunagar K., Khochare S., Senji Laxme R.R., Attarde S., Dam P., Suranse V., Khaire A., Martin G., Captain A. A wolf in another wolf's clothing: post-genomic regulation dictates venom profiles of medically-important cryptic kraits in India. Toxins. 2021;13:69. doi: 10.3390/toxins13010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryamohan K., Krishnankutty S.P., Guillory J., Jevit M., Schröder M.S., Wu M., Kuriakose B., Mathew O.K., Perumal R.C., Koludarov I., Goldstein L.D., Senger K., Dixon M.D., Velayutham D., Vargas D., Chaudhuri S., Muraleedharan M., Goel R., Chen Y.J., Ratan A., Liu P., Faherty B., de la Rosa G., Shibata H., Baca M., Sagolla M., Ziai J., Wright G.A., Vucic D., Mohan S., Antony A., Stinson J., Kirkpatrick D.S., Hannoush R.N., Durinck S., Modrusan Z., Stawiski E.W., Wiley K., Raudsepp T., Kini R.M., Zachariah A., Seshagiri S. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020;52:106–117. doi: 10.1038/s41588-019-0559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.H., Tan K.Y., Tan N.H. Revisiting Notechis scutatus venom: on shotgun proteomics and neutralization by the “bivalent” Sea Snake Antivenom. J. Proteomics. 2016;144:33–38. doi: 10.1016/j.jprot.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Tan K.Y., Tan N.H., Tan C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell's viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-25955-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.H., Tan K.Y., Ng T.S., Quah E.S., Ismail A.K., Khomvilai S., Sitprija V., Tan N.H. Venomics of Trimeresurus (Popeia) nebularis, the Cameron Highlands pit viper from Malaysia: insights into venom proteome, toxicity and neutralization of antivenom. Toxins. 2019;11:95. doi: 10.3390/toxins11020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Arnold R.J., Alves P., Xun Z., Clemmer D.E., Novotny M.V., Reilly J.P., Radivojac P. A computational approach toward label-free protein quantification using predicted peptide detectability. Bioinformatics. 2006;22:e481–488. doi: 10.1093/bioinformatics/btl237. [DOI] [PubMed] [Google Scholar]
- Tasoulis T., Isbister G.K. A review and database of snake venom proteomes. Toxins. 2017;9:E290. doi: 10.3390/toxins9090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukuwela K., Sanders K., Fry B.G. Hydrophis donaldi (Elapidae, Hydrophiinae), a highly distinctive new species of sea snake from northern Australia. Zootaxa. 2012;3201:45–57. [Google Scholar]
- van Fraassen B.C. The Terry Lectures Series. Yale University Press; 2008. The empirical stance. [DOI] [Google Scholar]
- Verbraecken J., Van de Heyning P., De Backer W., Van Gaal L. Body surface area in normal-weight, overweight, and obese adults. A comparison study. Metabolism. 2006;55:515–524. doi: 10.1016/j.metabol.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J., Kerkkamp H.M., Vos R.A., Guerreiro I., Calvete J.J., Wüster W., Woods A.E., Logan J.M., Harrison R.A., Castoe T.A., de Koning A.P., Pollock D.D., Yandell M., Calderon D., Renjifo C., Currier R.B., Salgado D., Pla D., Sanz L., Hyder A.S., Ribeiro J.M., Arntzen J.W., van den Thillart G.E., Boetzer M., Pirovano W., Dirks R.P., Spaink H.P., Duboule D., McGlinn E., Kini R.M., Richardson M.K. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss R.S., Jansa S.A. Snake-venom resistance as a mammalian trophic adaptation: lessons from didelphid marsupials. Biol. Rev. Camb. Phil. Soc. 2012;87:822–837. doi: 10.1111/j.1469-185X.2012.00222.x. [DOI] [PubMed] [Google Scholar]
- Walsh B., Lynch M. Oxford University Press; Oxford, UK: 2018. Evolution and Selection of Quantitative Traits. -13: 978-0198830870. [Google Scholar]
- Ward-Smith H., Arbuckle K., Naude A., Wüster W. Fangs for the memories? A survey of pain in snakebite patients does not support a strong role for defense in the evolution of snake venom composition. Toxins. 2020;12:201. doi: 10.3390/toxins12030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G.B., Brown J.H. The origin of allometric scaling laws in biology from genomes to ecosystems: towards a quantitative unifying theory of biological structure and organization. J. Exp. Biol. 2005;208:1575–1592. doi: 10.1242/jeb.01589. [DOI] [PubMed] [Google Scholar]
- Westeen E.P., Durso A.M., Grundler M.C., Rabosky D.L., Davis Rabosky A.R. What makes a fang? Phylogenetic and ecological controls on tooth evolution in rear-fanged snakes. BMC Evol. Biol. 2020;20:80. doi: 10.1186/s12862-020-01645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R., Captain A. Draco Books; Chennai: 2004. Snakes of India: the Field Guide. -13: 978-8190187305. [Google Scholar]
- White C.R., Seymour R.S. Allometric scaling of mammalian metabolism. J. Exp. Biol. 2005;208:1611–1619. doi: 10.1242/jeb.01501. [DOI] [PubMed] [Google Scholar]
- White C.R., Cassey P., Blackburn T.M. Allometric exponents do not support a universal metabolic allometry. Ecology. 2007;88:315–323. doi: 10.1890/05-1883. [DOI] [PubMed] [Google Scholar]
- Whittington A.C., Mason A.J., Rokyta D.R. A single mutation unlocks cascading exaptations in the origin of a potent pitviper neurotoxin. Mol. Biol. Evol. 2018;35:887–898. doi: 10.1093/molbev/msx334. [DOI] [PubMed] [Google Scholar]
- WHO . WHO Document Production Services; Geneva, Switzerland: 2017. Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins. WHO Technical Report Series, No. 1004. available at: https://www.who.int/publications/i/item/9789241210133. [Google Scholar]
- WHO . WHO Document Production Services; Geneva, Switzerland: 2019. Snakebite Envenoming- A Strategy for Prevention and Control.https://www.who.int/snakebites/resources/9789241515641/en available at: 978-92-4-151564-1. [Google Scholar]
- Wolters D.A., Washburn M.P., Yates J.R. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- Yin W., Wang Z.J., Li Q.Y., Lian J.M., Zhou Y., Lu B.Z., Jin L.J., Qiu P.X., Zhang P., Zhu W.B., Wen B., Huang Y.J., Lin Z.L., Qiu B.T., Su X.W., Yang H.M., Zhang G.J., Yan G.M., Zhou Q. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 2016;7:13107. doi: 10.1038/ncomms13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H., Murphy R.W., Arredondo J.C., Graboski R., Machado-Filho P.R., Mahlow K., Montingelli G.G., Quadros A.B., Orlov N.L., Wilkinson M., Zhang Y.P., Grazziotin F.G. Large-scale molecular phylogeny, morphology, divergence-time estimation, and the fossil record of advanced caenophidian snakes (Squamata: serpentes) PloS One. 2019;14 doi: 10.1371/journal.pone.0216148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zainal Abidin S.A., Rajadurai P., Chowdhury M.E., Ahmad Rusmili M.R., Othman I., Naidu R. Proteomic characterization and comparison of Malaysian Tropidolaemus wagleri and Cryptelytrops purpureomaculatus venom using shotgun-proteomics. Toxins. 2016;8:299. doi: 10.3390/toxins8100299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancolli G., Casewell N.R. Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol. Biol. Evol. 2020;37:2777–2790. doi: 10.1093/molbev/msaa133. [DOI] [PubMed] [Google Scholar]
- Zelanis A., Tashima A.K. Unraveling snake venom complexity with ‘omics’ approaches: challenges and perspectives. Toxicon. 2014;87:131–134. doi: 10.1016/j.toxicon.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Zhu W., Smith J.W., Huang C.M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotech. 2010. 2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]