Abstract
Complex regional pain syndrome (CRPS) is a chronic pain condition characterized by inflammation and debilitating pain. CRPS patients with pain refractory to more conventional analgesics can be treated with subanesthetic doses of ketamine. Our previous studies found that poor responders to ketamine had a 22-fold downregulation of the miRNA hsa-miR-605 in blood prior to ketamine treatment. Hence, we sought to investigate the functional significance of miR-605 downregulation and its impact on target gene expression, as investigating target mRNAs of differentially expressed miRNAs can provide important insights on aberrant gene expression that may contribute to disease etiology. Using a bioinformatics prediction, we identified that miR-605 can target the proinflammatory chemokine CXCL5, which plays a role in leukocyte recruitment and activation. We hypothesized that downregulation of miR-605 in poor responders to ketamine could increase CXCL5 expression and thereby contribute to inflammation in these patients. We confirmed that miR-605 regulates CXCL5 by using a miRNA mimic and inhibitor in human primary endothelial cells. Inhibition of miR-605 increased CXCL5 secretion and migration of human monocytic cells, thereby demonstrating a functional impact of miR-605 on chemotaxis. Additionally, CXCL5 mRNA was upregulated in whole blood from poor responders to ketamine, and CXCL5 protein was increased in plasma from CRPS patients. Thus, our studies suggest that miR-605 regulation of CXCL5 can regulate inflammation.
Keywords: miR-605, CXCL5, complex regional pain syndrome, pain, inflammation, miRNA
Graphical abstract
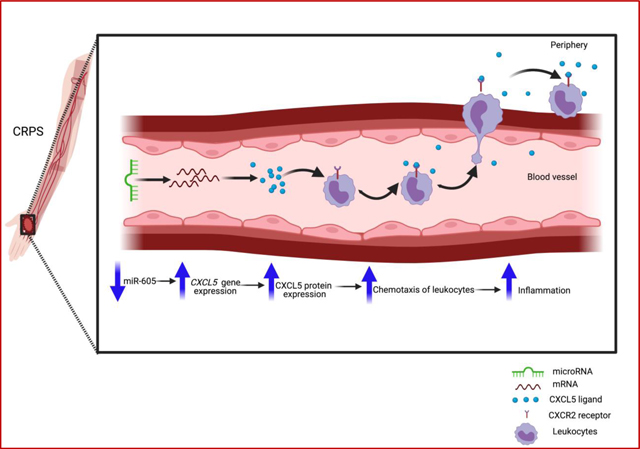
1. Introduction
Complex regional pain syndrome (CRPS) is a disabling disorder characterized by debilitating pain, inflammation, and sensory and motor abnormalities [1–3]. Recalcitrant CRPS can be treated with ketamine, an N-methyl-D-Aspartate (NMDA) receptor antagonist [4, 5]. Ketamine treatment is only considered for patients with moderate to severe CRPS symptoms meeting the Budapest criteria and for whom all other treatments have failed [6]. While continuous administration of subanesthetic doses of ketamine is usually effective, about 30% of patients fail to respond to the therapy [7, 8]. The etiology behind treatment failure is unknown.
There are beneficial prognostic and diagnostic biomarkers for CRPS, notably including circulating miRNAs [9]. miRNAs can negatively regulate gene expression by binding to the 3’ untranslated region (3’UTR) of a target mRNA, which leads to target mRNA degradation or translational inhibition, and thus, altered gene expression. We have shown that circulating miRNAs in whole blood from CRPS patients compared to healthy control donors can help stratify patient populations [10]. Additionally, miRNAs or their target mRNAs can also serve as therapeutic targets. We have observed that miR-939 targets several proinflammatory genes and is downregulated in CRPS patient circulation, suggesting that a single miRNA can regulate proinflammatory signaling networks [11]. Thus, investigating how differentially expressed miRNAs regulate target mRNAs can provide important insights on aberrant gene expression that contributes to disease pathology [9].
Our previous study in female patients with CRPS showed differential expression of circulating miRNAs in whole blood, both before and after ketamine treatment [12]. Poor ketamine responders, defined as patients with an increased average pain score or with a less than 50% decrease of pain score, had a 22-fold downregulation of miR-605 prior to ketamine treatment [12]. Therefore, we sought to assess the functional significance of miR-605 differential expression and how it may contribute to pain and inflammation in CRPS. Bioinformatics predictions show that miR-605 can potentially target the epithelial-derived neutrophil-activating peptide 78 (ENA78/CXCL5), a member of the CXC chemokine family. Chemokines are small proteins that induce migration of leukocytes and initiate a cascade of events leading to inflammation [13]. CXCL5 primarily binds to the chemokine receptor CXCR2 on immune cells to induce chemotaxis of leukocytes and thereby contribute to inflammation [14]. Indeed, CXCL5 expression was elevated in human and rodent skin after ultraviolet-B (UVB) radiation treatment [15–17], which induces a dose-dependent erythema and hypersensitivity to both thermal and mechanical stimulation that mimics inflammatory pain in rodents [18] and humans [19]. Autoinflammatory and autoimmune components are implicated in CRPS, and patients display complex neuroimmunological pathogenesis with dysfunction of both the innate and adaptive immune systems [20]. Although several studies have reported alterations of cytokines in CRPS patients [21–23], none of the studies so far has implicated CXCL5 in CRPS. We hypothesized that lower levels of miR-605 may lead to upregulation of CXCL5, which in turn may contribute to the inflammation and hyperalgesia observed in CRPS patients.
2. Materials and methods
2.1. Ethics statement
All subjects were enrolled after giving informed consent as approved by the Drexel University College of Medicine Institutional Review Board and met the clinical Budapest criteria for CRPS [24]. Standard protocol approvals, registrations, and patient consents, inclusion and exclusion criteria have been described in detail [10, 12] and banked RNA samples from these studies were used here.
2.2. Cell culture and maintenance
HEK293 cells (ATCC) were maintained in Dulbecco’s Modified Eagle Medium (DMEM), 10% heat-inactivated fetal bovine serum (FBS), and 1% penicillin-streptomycin. Human umbilical vein endothelial cells (HUVEC) (Sigma Aldrich, 200P-05n) were maintained in endothelial cell basal medium containing 10% FBS and 10% DMSO (Sigma Aldrich) at 37 °C in 5% CO2. Human monocytic leukemia THP-1 cells (TIB202, ATCC) were maintained in RPMI-1640 medium containing 10% FBS and 1% penicillin-streptomycin. SH-SY5Y cells were cultured in 1:1 mixture of ATCC-formulated Eagle’s Minimum Essential and F12 medium with 10% FBS.
2.3. Overexpression of miR-605 in HUVEC
Transfections were performed according to the manufacturer’s protocol for RNAiMax transfection reagent (Life Technologies) using either mimic control (CN-002000-01-05), inhibitor control (IP-004500-01-05), hsa-miR-605-5p inhibitor (Catalog number AM17000, 5nmol, Thermo Fisher Scientific), or hsa-miR-605–5p mimic (Catalog number 4464066, 5nmol, Thermo Fisher Scientific) with the following modifications. For each well of a 24-well plate, 3μl RNAiMax reagent was diluted in 50μl of Opti-MEM media (Thermo Fisher Scientific) and 10pmol of miR-605 mimic, miR-605 inhibitor, or their respective controls were diluted in 50 μl of Opti-MEM media individually, added to Lipofectamine RNAiMAX Reagent in a 1:1 ratio, and incubated for 5 min at room temperature. This transfection complex (100 μl) was added to 2 × 105 cells/well in 6 well plates and incubated for 24 hours at 37 °C, after which the cells were stimulated with 10 pmol of recombinant human tumor necrosis factor α (rhTNF-α)(210-TA-020, R&D Systems, Inc) and incubated for 24 h. Alternatively, for gene expression studies, the cells were stimulated with 1μg/ml bacterial lipopolysaccharide (LPS) (Sigma-Aldrich) for 6 h. The cell pellet was collected and stored in −80 °C until further use.
2.4. Luciferase Reporter Assay
The 3’ UTR clones for human CXCL5 (NM_002994.4) and the miR-605 (HmiR0158-MR04-B, precursor miRNA expression clone for hsa-mir-605) and precursor miRNA scrambled control for pEZX-MR04 were purchased from Genecopeia. Reporter assays were performed 48 h hours after co-transfection of HEK293 cells using Lipofectamine 2000 (Life Technologies). The Luc-Pair Duo-Luciferase assay (GeneCopoeia) was used to determine firefly and Renilla luciferase activity in accordance with the manufacturer’s protocol. Firefly luciferase measurements normalized to Renilla were used as a transfection control. Statistically significant differences from control were calculated using a Student’s t-test.
2.5. RNA isolation and qRT-PCR
For cells, total RNA was isolated using the mirVana RNA isolation kit (Ambion, Life technologies) according to manufacturer’s instructions. For patient samples, total RNA was isolated from whole blood as described previously [12]. RNA concentrations were determined using a NanoDrop ND1000 spectrophotometer (NanoDrop Technologies) and 200ng of total RNA was used to amplify cDNA using The Maxima cDNA synthesis kit (Thermo Fisher Scientific). Two μl of cDNA was used for the Taqman qPCR assay in a reaction volume of 20 μL comprised of 10 μL Taqman Fast Universal PCR master mix (2×), no AmpErase UNG (Life Technologies), 1 μL Taqman gene expression assay mix (20×), 2 μL cDNA, and 7 μL RNase-free water. 18S rRNA or GAPDH was used as the normalizer. Fold change of RNA was calculated using the raw cycle threshold (CT) values using the 2−ΔΔCT method [25]. The Assay ID for the Taqman primer probes were CXCL5 (Hs01099660-g1), 18S (4319413E-13U1058), and GAPDH (4326317E).
For miRNA qPCR, cDNA synthesis was performed using 40 ng total RNA, and qPCR for miR-605 was performed using TaqMan microRNA assay (Assay ID 001568 Applied Biosystems) as recommended by the vendor. U6 snRNA was used for normalization (Assay ID 001973, Applied Biosystems).
2.6. Enzyme-linked Immunosorbent Assay (ELISA)
Supernatants from HUVECs transfected with miR-605 mimic, inhibitor, or controls were used for ELISA. Briefly, 2×105 HUVECs were transfected with 10 pmol of mimic, inhibitor, and their respective controls. The transfection complex was incubated for 24 hours at 37 °C after which the cells were stimulated with 10 pmol of rhTNFα (R&D Systems) and incubated for 24 h. Twenty μL of media collected 48 h post transfection was used for ELISA, which was performed according to manufacturer’s protocol (Bio-legend) to determine CXCL5 concentration. For patient samples, aliquots of plasma stored in −80 °C were used. The numbers of samples from patients and controls used in these assays depended on plasma sample availability.
2.7. Leukocyte Transmigration Assay
Transmigration assay was performed according to manufacturer’s instructions (CBA-101, Cell Biolabs). Briefly, HUVECs were grown in endothelial media and plated to confluency (1×105cells/transwell insert). Cells were allowed to form a monolayer. After 72 hours, cells were transfected in Opti-MEM media as described previously and were treated with rhTNFα for 24 h. Supernatants from the transfected HUVECs were added to the bottom of the wells. THP-1 monocytes (1×105 cells) labelled with the LeukoTracker™ (1X) dye were placed in the upper chamber and allowed to migrate for 4 hours at 37 °C after which the media from the bottom well was collected and monocytes that had transmigrated were lysed and quantified with a fluorescent plate reader (Tecan Spark multimode plate reader) at 480nm/520 nm. Cells were also analyzed under an inverted fluorescence microscope and representative pictures were taken at the end of 4 h incubation.
2.8. Statistical analysis
Data shown as mean ± the standard error of the mean from three or more independent experiments. GraphPad Prism software was used for all statistical analysis. Student’s t-test was used for determining the statistical significance for comparisons between two groups. Pairwise comparisons between means were tested using one-way analysis of variance (ANOVA) for comparisons between two groups. A p-value less than 0.05 was considered significant.
3. Results
3.1. miR-605 was downregulated in CRPS patients who were poor responders to ketamine treatment.
We previously profiled circulating miRNAs in whole blood from female CRPS patients before and after treatment with subanesthetic doses of intravenous ketamine for five days and showed that a miRNA signature can help predict a treatment response to ketamine [12]. Of the 33 miRNAs that differed between responders and poor responders before therapy, we aimed to validate changes in miR-605 as it was highly downregulated in poor responders to ketamine and predicted to regulate inflammation-related gene targets. Our qPCR results confirmed that miR-605 expression was strongly downregulated in poor-responders prior to ketamine treatment (Fig 1A). With this validation, we moved to investigate the gene target(s) of miR-605.
Fig 1. Confirmation of hsa-miR-605 binding to the 3’UTR of CXCL5.
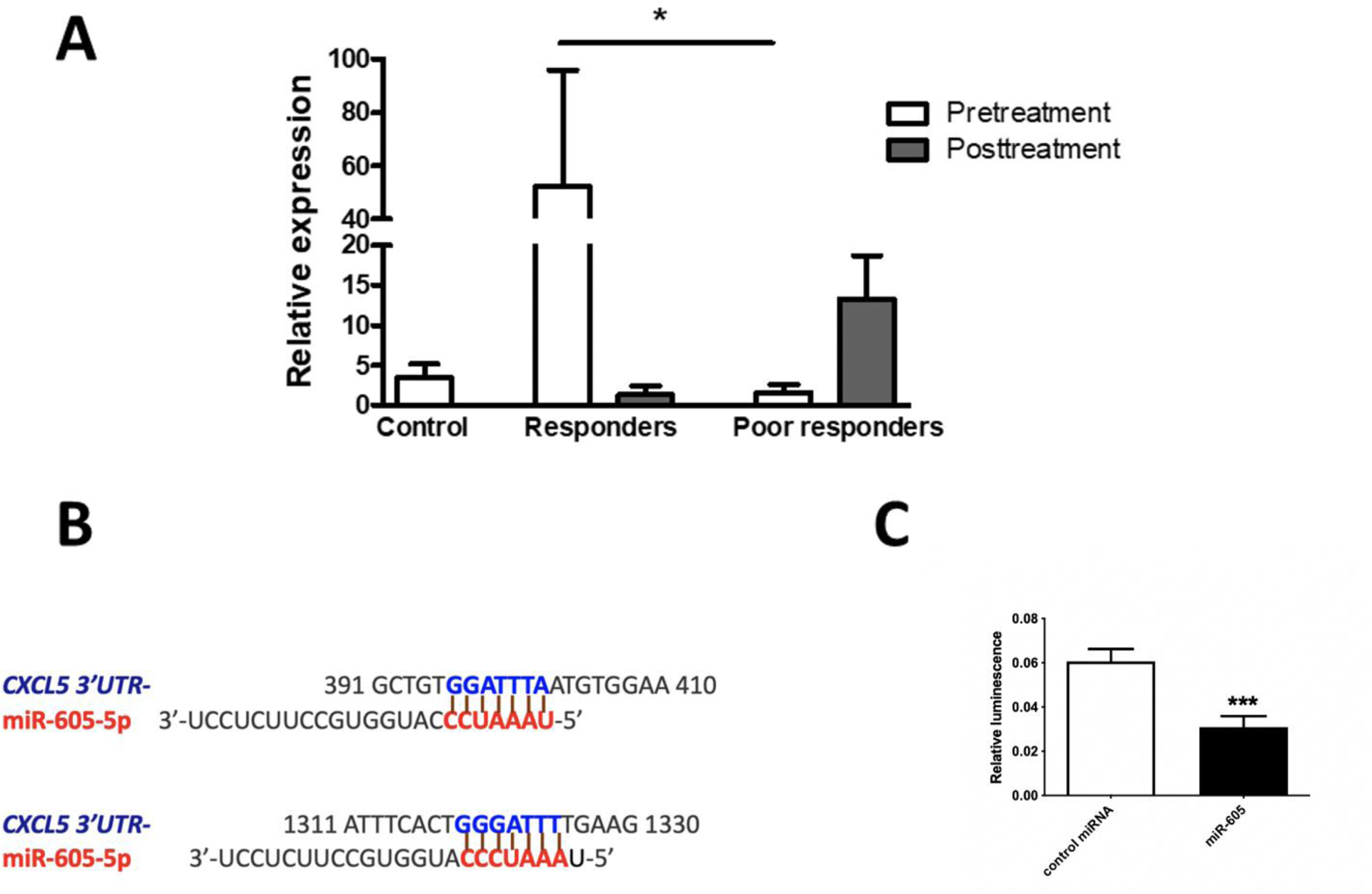
(A) Hsa-miR-605 expression in whole blood from healthy control donors and CRPS patient samples. Hsa-miR-605 in poor responders to ketamine therapy was lower relative to responders, prior to treatment. In responders, ketamine therapy reduced average pain scores by at least 50%. Relative expression of miR-605 was determined by qPCR. Controls n=19, responders n=8, poor responders n=7. miR-605 expression was normalized to U6 snRNA. Statistical significance was determined by one-way ANOVA, *p<0.05, data represent mean ± SEM. (B) The binding of seed sequence of hsa-miR-605-5p to 7mer-A1 and 7mer-m8 sites in 3’UTR of CXCL5 transcript at positions 396 and 1319 respectively. (C) To confirm miR-605 binding to 3’ UTR of CXCL5 mRNA, the 3’UTR of CXCL5 was cloned downstream of the luciferase open reading frame. Luciferase reporter assay was performed 48 h after co-transfection of HEK293 cells with CXCL5 3′UTR reporter plasmid and miR-605 or control miRNA. Firefly luciferase measurements normalized to Renilla luciferase was used as a transfection control. The average of three independent experiments is shown. Statistical significance was determined using unpaired Student t-test (two-tailed), data represent mean ± SEM ***p < 0.001.
3.2. Confirmation of miR-605 binding to the 3′UTR of CXCL5.
Bioinformatic analysis using miRDB, an online database for miRNA target prediction and functional annotations [26], predicted that the human CXCL5 3’UTR harbors two miR-605 binding sites (Fig 1B). Since CXCL5 is a proinflammatory chemokine that helps recruit and activate immune cells, we hypothesized that downregulation of miR-605 in poor responders leads to higher CXCL5 expression, which may contribute to inflammation and thus a potentially poor treatment response in these patients. First, we validated the binding of miR-605 to the CXCL5 3’UTR using a luciferase reporter assay where the 3’UTR of CXCL5 was cloned downstream of a luciferase open-reading frame. We transiently transfected HEK293 cells with plasmids encoding the 3’UTR construct of CXCL5 along with either miR-605 or a scrambled precursor miRNA control. As predicted, miR-605 could bind to the 3’UTR of CXCL5 mRNA, as demonstrated by a significant reduction in luciferase activity 48 h after transfection (Fig 1C).
3.3. miR-605 inhibits LPS and TNFα-stimulated CXCL5 mRNA expression in vitro
Next, we wanted to perform miR-605 transfection studies in a cell line that endogenously expresses CXCL5. We quantified CXCL5 mRNA expression levels in monocytic THP-1 cells, SH-SY5Y neuroblastoma cells, and human umbilical vein endothelial cells (HUVEC), and the highest CXCL5 expression was observed in HUVECs with a 29-fold upregulation upon LPS treatment (Supp Table 1). Prior studies have shown that CXCL5 is a potent chemokine involved in inflammation and known to have a role in neutrophil trafficking during LPS induced lung inflammation in mice [27]. CXCL5 expression can also be stimulated with TNFα in vitro [28].
LPS stimulation upregulated CXCL5 mRNA expression in SH-SY5Y cells, and transfection of these cells with miR-605 led to a small but significant decrease in CXCL5 mRNA levels (Fig 2A). We then transfected HUVECs with either a miR-605 mimic, miR-605 inhibitor, or their respective controls to evaluate the ability of miR-605 to modulate the levels of endogenous CXCL5 mRNA in vitro. Transfection with the miR-605 mimic significantly reduced the expression of CXCL5 transcripts, but the miR-605-5p inhibitor did not upregulate CXCL5 mRNA expression compared to the inhibitor control (Fig 2B). We then transfected HUVECs, and 24 h after transfection, stimulated the cells with LPS for 6 h. Cells transfected with the miR-605 inhibitor showed significantly increased expression of CXCL5 (Fig 2C), potentially by blocking binding of miR-605 to the 3’ UTR of CXCL5 mRNA.
Fig 2. Expression levels of CXCL5 mRNA in SH-SY5Y and HUVEC cells transfected with miR-605-5p.
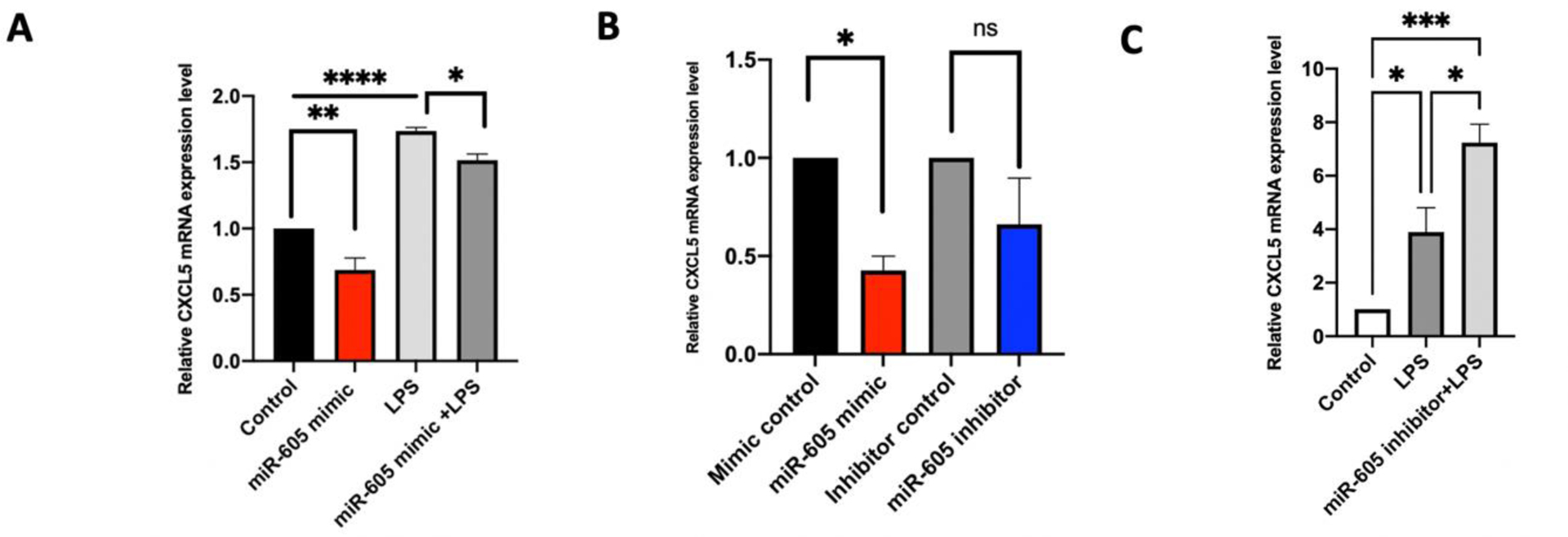
(A) Relative expression of CXCL5 was measured in miR-605 transfected SH-SY5Y cells. LPS induced upregulation of CXCL5 showed small but significant decrease upon miR-605 transfection compared to control (n=3). Statistical significance was determined using one-way ANOVA followed by Sidak’s multiple comparison tests, *p<.05, ***p < 0.001. (B) Taqman analysis of endogenous levels of CXCL5 in HUVECs transfected with miR-605 mimic, miR-605 inhibitor, mimic control and inhibitor control. Transfection with miR-605 mimic reduced CXCL5 transcripts compared to control miRNA transfection. miR-605 inhibitor did not significantly upregulate CXCL5 mRNA (n=3). Significance was determined by one-way ANOVA followed by Sidak’s multiple comparison tests, *p < 0.05. (C) Relative expression of CXCL5 mRNA in HUVEC cells transfected with miR-605 inhibitor. After 24 h, cells were stimulated with 1 μg/ml of LPS. Cells were harvested after 6 h of stimulation. Cell transfected with miR-605 inhibitor showed significant increase in CXCL5 expression (n=3). Statistical significance was determined using one-way ANOVA followed by Sidak’s multiple comparison tests, *p<.05, ***p < 0.001.
3.4. miR-605 reduces secretion of CXCL5 protein following TNFα stimulation in vitro
Since the miR-605 inhibitor was clearly functional in response to stimulation with LPS, we also tested its efficacy in cells stimulated with TNFα. We transfected HUVECs with either miR-605 mimic, miR-605 inhibitor, or their respective controls, then stimulated the cells with TNFα. At 24 h post TNFα treatment, we collected samples of culture media to assess changes in extracellular CXCL5 protein by ELISA. The miR-605 mimic did not change levels of secreted CXCL5, but as expected, the miR-605 inhibitor increased CXCL5 protein levels compared to control (Fig 3).
Fig 3. Changes in CXCL5 protein expression in HUVEC cells transfected with miR-605-5p.
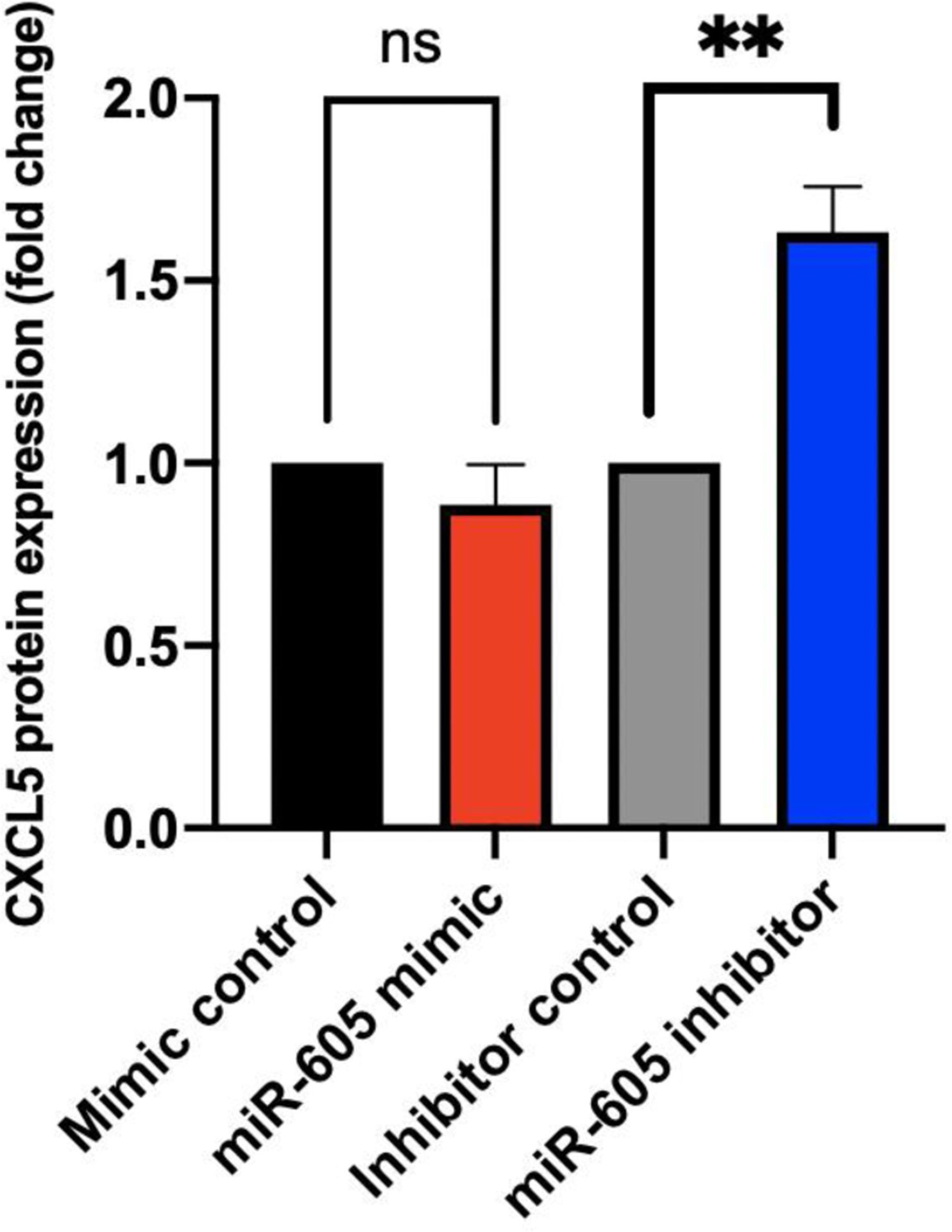
ELISA for CXCL5 secreted by HUVEC into culture media. HUVEC cells were transfected with miR-605 mimic, control mimic, miR-605 inhibitor, and control inhibitor followed by TNFα stimulation for 24 h. miR-605 inhibitor upregulated levels of secreted CXCL5 whereas transfection with miR-605 mimic did not significantly alter CXCL5 protein. Figure is derived from the average of three experiments. Statistical significance was determined using one-way ANOVA followed by Tukey’s multiple comparison test **p < 0.01.
3.5. miR-605 regulates leukocyte migration via CXCL5 secretion in vitro
CXCL5 promotes chemotaxis and acute inflammation by binding to the CXCR2 chemokine receptor on the surface of immune cells. As in vitro secretion of CXCL5 can be differentially regulated by transfection with a miR-605 mimic or inhibitor, we used this approach in a leukocyte transmigration assay to assess how altered CXCL5 secretion affects migration of human THP-1 monocytes. For this experiment, THP-1 cells were suspended in the upper chamber of a transwell, while the bottom chamber contained spent media from HUVECs transfected with either a miR-605 mimic, inhibitor, or their respective controls and treated with TNFα for 24 h (Fig 4A). We confirmed that inhibition of miR-605 in HUVECs increased secretion of CXCL5, which subsequently enhanced chemotaxis of human monocytic cells relative to control (Fig 4B, 4C). In contrast, migration of monocytes in response to supernatants from HUVECs transfected with miR-605 mimic was similar to the mimic control. Thus, miR-605-mediated downregulation of CXCL5 reduced chemotaxis of THP-1 monocytes.
Fig 4. miR-605 mediated regulation of CXCL5 impacts migration of human monocytes.
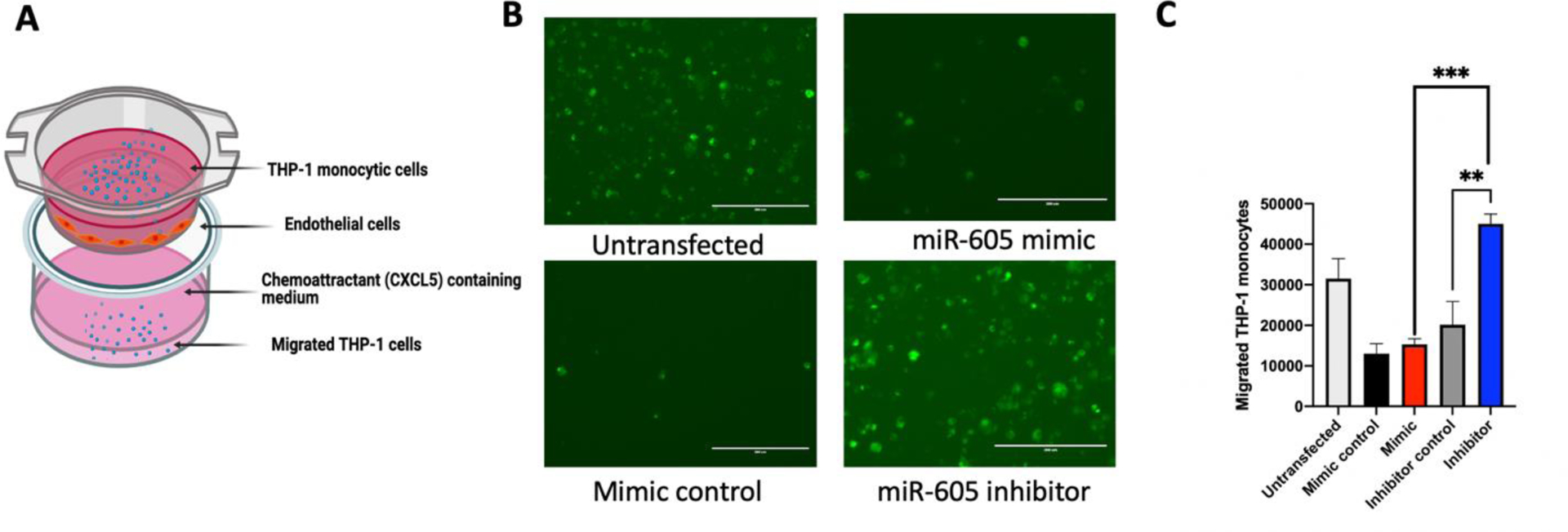
(A) Schematic of a transwell assay to determine if the regulation of CXCL5 levels by the miR-605/miR-605 inhibitor can functionally impact leukocyte migration. (B) Representative epifluorescence microscopic images of LeukoTracker labeled THP-1 cells from transwell assay. HUVEC cells were transfected with miR-605 mimic, mir-605 inhibitor, or control mimic. Supernatants from the transfected HUVECs were added to the bottom of the wells. Labelled THP-1 cells were added to the upper chamber and allowed to migrate for 4 hours. Transmigration of THP-1 monocytes is altered under the influence of miR-605 inhibitor. (C) An increased secretion of CXCL5 mediated by miR-605 inhibitor led to an upregulation of chemotaxis of THP1 cells, while miR-605 mimic did not affect secretion of CXCL5 or migration (n=3). Statistical significance was determined using one-way ANOVA followed by Sidak’s multiple comparison test, **p < 0.01, ***p<.001.
3.6. CXCL5 mRNA and protein expression in CRPS patients
To validate CXCL5 regulation in a disease-relevant context, we next investigated CXCL5 mRNA and protein levels in whole blood from CRPS patients that underwent ketamine therapy. Since poor responders had lower miR-605 levels prior to ketamine treatment, we hypothesized that these patients would also display upregulated CXCL5 levels. Using the same patient RNA samples, Taqman qPCR analysis showed a significantly higher expression of CXCL5 mRNA in poor responders compared to responders (Fig 5A). This suggests that there is an inverse correlation between the expression of miR-605 and its target CXCL5 mRNA in whole blood from poor responders to ketamine, prior to treatment. Since we did not have plasma samples for responders and poor responders, we further performed ELISA using banked plasma samples from CRPS patients. Indeed, CXCL5 protein expression was increased in plasma samples from CRPS patients (Fig 5B), suggesting elevation of this chemokine could have a role in inflammation, a hallmark of CRPS.
Fig 5. CXCL5 expression levels in CRPS patients.
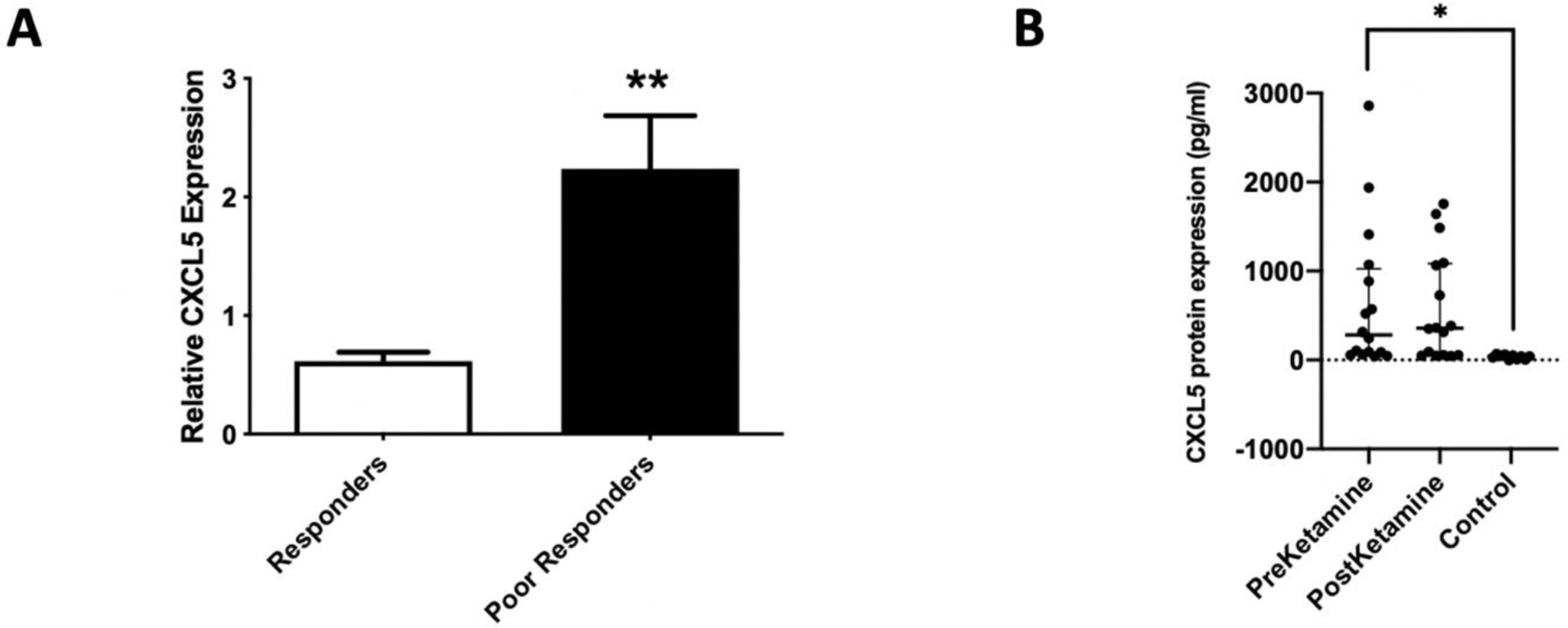
(A) Expression levels of CXCL5 mRNA in whole blood of CRPS patients prior to ketamine treatment. GAPDH was used as normalizer (n=8). Statistical significance was determined using t-test, **p < 0.01. (B) CXCL5 protein expression in plasma from CRPS patients. Plasma levels of CXCL5 protein measured by ELISA (preketamine n=16, postketamine n=16, control n=10). Statistical significance was determined using one way ANOVA, followed by Dunnett’s multiple comparison test, *p<0.05.
4. Discussion
In previous studies, circulating miRNAs have been identified as biomarkers to guide disease prognosis and treatment [29]. Therefore, we can better understand the pathophysiology of CRPS by linking changes in circulating miRNA expression to inflammation and pain. By extension, this approach may uncover new and much needed therapeutic targets. In this study, we investigated the function of miR-605 because it was significantly downregulated in CRPS patients who responded poorly to ketamine therapy. We validated that hsa-miR-605 regulates the pro-inflammatory chemokine CXCL5 and assessed potential implications related to the neuroimmunological pathogenesis of CRPS. We have previously linked some of the miRNAs identified by profiling blood samples from CRPS patients to target genes, which enhanced our understanding of the disease biology [11, 12, 30]. Examples of miRNA and target genes include miR-548d-5p and UDP-glucuronosyltransferase UGT1A1 mRNA [12], miR-34a and corticotrophin releasing hormone receptor 1 (CRHR1) [30], miR-939 and multiple proinflammatory genes including interleukin-6 (IL-6), vascular endothelial growth factor (VEGFA), tumor necrosis factor α (TNFα), nitric oxide synthase 2 (NOS2A or iNOS), and nuclear factor-κB2 (NFκB2) [11]. These examples suggest identifying miRNA and target interactions can be beneficial for diagnosis and potentially useful for discovering new therapeutic approaches for management of CRPS [31].
Initially, we identified putative target genes of mir-605 using the bioinformatic database miRDB [25]. From this query, mir-605 was predicted to bind to two potential sites in the 3’ UTR of CXCL5, a known proinflammatory chemokine. Inflammation is a crucial feature of CRPS, as several studies have identified elevations in inflammatory mediators in plasma, cerebrospinal fluid, and blisters from CRPS patients, as well as decreased levels of systemic anti-inflammatory cytokines [32]. To the best of our knowledge, CXCL5 has not been previously associated with CRPS. Thus, we hypothesized that the downregulation of miR-605 could increase expression of CXCL5 and exacerbate the chronic inflammatory states in CRPS that is unresponsive to ketamine therapy. Our luciferase reporter assay confirmed that miR-605 bound to the 3’UTR of CXCL5 and downregulated its expression.
The expression of chemokines is regulated at different levels, including mRNA expression [33]. Since endogenous expression of CXCL5 is low, we utilized LPS [27] and TNFα [28] to induce expression in cell lines of interest. With this approach, CXCL5 expression was reduced in cells transfected with miR-605 mimic and increased in cells transfected with a miR-605 inhibitor in the presence of LPS or TNFα. This further supports the hypothesis that CXCL5 expression is under miR-605 regulation. We also observed differences in CXCL5 upregulation in HUVECs transfected with a miR-605 inhibitor depending on the stimulating agent used. LPS stimulation upregulated CXCL5 mRNA whereas TNFα did not change mRNA expression but did increase CXCL5 protein levels and secretion. This could be due to differences in the signaling pathways of LPS and TNFα in addition to temporal differences in the effects of each stimulation. Several other factors could contribute to the differences between TNFα and LPS, including context-specific effects of other regulators. These can be transcription factors, RNA-binding proteins that modulate accessibility [34], or differential 3’ UTR isoform usage that influences either the inclusion of binding sites or their placement within contexts that may be more or less favorable [35].. The efficacy of the miR-605 inhibitor translated to higher CXCL5 protein expression in HUVECs. When delivered to a cell, miRNA inhibitors bind to endogenous mature miRNAs and this is thought to be irreversible. Thus, miRNA inhibitors sequester the endogenous miRNA, making it unavailable for normal function [36]. This could have contributed to the differences in functional efficacy from transfecting mimics and inhibitors.
Inflammation fundamentally requires the recruitment of leukocytes and other immune cells to target tissues [37]. Neutrophils are the first immune cells to be recruited to the site of inflammation [38]. These cells express CXCR2 and are attracted by CXCL5, which is produced in response to an inflammatory stimuli or infection by various cell types such as endothelial cells [39]. After recruitment to an inflammatory site, neutrophil adhesion molecules can bind to their respective receptors on activated endothelial cells under inflammation to enter inflamed tissue [40]. Other immune cells express the CXCL5 receptor CXCR2, including monocytes and endothelial cells, suggesting they also respond to secreted CXCL5. Our transwell assay confirmed that regulation of CXCL5 by miR-605 can significantly alter the number of migrating THP1 monocytic cells.
Since miR-605 expression was downregulated in blood from CRPS patients that poorly responded to ketamine therapy, we investigated if CXCL5 mRNA was higher in these patients due to the inverse relationship between the miRNA and its target gene. We observed a significantly higher expression of the CXCL5 gene in poor responders as compared to responders, prior to ketamine treatment. Since all the matching plasma samples for patients whose blood samples were used for miRNA profiling were not available, we used additional banked plasma samples from patients who received ketamine therapy to analyze CXCL5 protein levels. Though we were unable to match miRNA levels, we observed an increase in CXCL5 in CRPS patients compared to controls.
This leads to the question of how CXCL5 contributes to the pathogenesis of pain and inflammation in CRPS patients. The studies reported here aimed to only identify the target gene regulated by miR-605. We did not investigate the mechanisms leading to treatment-induced changes in miR-605 expression in these patients. Prior research has shown that CXCL5 mRNA was elevated in human and rodent skin punch biopsies after UV treatment, and that UV radiation induced CXCL5 protein in a dose-dependent manner in human skin [15–17]. Further, CXCL5 injection induced mechanical hypersensitivity that was associated with the infiltration of neutrophils and macrophages into the dermis and neutralizing the effects of CXCL5 attenuated pain hypersensitivity [15]. Since neutrophils and macrophages are both involved in the development of aberrant pain, the authors suggested that CXCL5 contributes to the sensory changes evoked by UVB irradiation through the recruitment of inflammatory cells and release of pro-algesic mediators [15]. They also reported an increase in mechanical hypersensitivity 30 min after CXCL5 application that was independent of leukocyte infiltration. This suggests that CXCL5 may act either directly on nociceptive terminals or via other resident cells expressing CXCR2 [15]. Elevated levels of CXCL5 were also reported in the synovial compartment of rheumatoid arthritis patients [41]. CXCL5 amplifies the release of NF-κB responsive proinflammatory cytokines in endothelial cells [14]. Mechanical allodynia after intrathecal injection of LPS in rats is generated by activating transcription factor STAT3 in spinal astrocytes [42]. STAT3 binds to the promoter regions inducing upregulation of three chemokines including CXCL5, which in turn could regulate the recruitment of peripheral immune cells [42]. Our observation that CXCL5 is upregulated in plasma samples from CRPS patients suggest this chemokine could exert a systemic effect. Additional studies in a rodent model of CRPS comparing WT and CXCL5 KO mice [43] will help elucidate the role of this chemokine in the injured limb as well as systemic effects in tissues that mediate pain.
5. Conclusions
A subset of CRPS patients that poorly responded to ketamine therapy had lower circulating levels of miR-605 and higher levels of CXCL5 mRNA. This miRNA-mRNA interaction was confirmed in vitro, and CXCL5 secreted in the absence of miR-605 enhanced migration of monocytes. This indicates that miR-605 negatively regulates CXCL5, and this interaction has functional consequences on immune cell chemotaxis. Plasma samples from CRPS patients showed an increase in CXCL5 protein, suggesting this chemokine could contribute to the inflammatory pain pathogenesis of CRPS.
Supplementary Material
Highlights.
CRPS patients responding poorly to ketamine has lower miR-605 in whole blood.
miR-605 is predicted to target proinflammatory chemokine CXCL5.
miR-605 binds to CXCL5 decreasing CXCL5 mRNA and protein expression in vitro.
miR-605 regulates leukocyte migration via CXCL5 impacting chemotaxis in vitro.
CRPS patients show increased CXCL5 mRNA and protein expression in whole blood.
Acknowledgement
We thank Dr. Bradley Nash for critical reading of the manuscript. The graphical abstract and Figure 4A was created with BioRender.com.
Funding
This study was funded by NIH NINDS R01NS102836 to Seena K. Ajit.
Abbreviations
- ANOVA
analysis of variance
- CRHR1
corticotrophin releasing hormone receptor 1
- CRPS
complex regional pain syndrome
- CXCL5
C-X-C motif chemokine 5
- CXCR2
chemokine receptor 2
- DMEM
Dulbecco’s modified eagle medium
- ELISA
enzyme-linked immunosorbent assay
- ENA78/CXCL5
epithelial-derived-neutrophil-activating-peptide78
- FBS
fetal bovine serum
- HEK293
human embryonic kidney 293 cells
- HUVEC
Human umbilical vein endothelial cells
- IL-6
interleukin-6
- KO
knockout
- LPS
Lipopolysaccharide
- miRNA
microRNA
- NFκB2
nuclear factor kappa B subunit 2
- NMDA
N-methyl-D-Aspartate
- NOS2A or iNOS
nitric oxide synthase 2
- rhTNFα
recombinant human tumor necrosis factor α
- STAT3
signal transducer and activator of transcription 3
- UDP
uridine diphosphate
- UGT1A1
UDP glucuronosyltransferase family 1 member A
- UVB
ultraviolet-B
- VEGFA
vascular endothelial growth factor
- 3’UTR
3’ untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- [1].Harden RN, Maihofner C, Abousaad E, Vatine JJ, Kirsling A, Perez R, Kuroda M, Brunner F, Stanton-Hicks M, Marinus J, van Hilten JJ, Mackey S, Birklein F, Schlereth T, Mailis-Gagnon A, Graciosa J, Connoly SB, Dayanim D, Massey M, Frank H, Livshitz A, Bruehl S, A prospective, multisite, international validation of the Complex Regional Pain Syndrome Severity Score, Pain 158(8) (2017) 1430–1436. [DOI] [PubMed] [Google Scholar]
- [2].Birklein F, O’Neill D, Schlereth T, Complex regional pain syndrome: An optimistic perspective, Neurology 84(1) (2015) 89–96. [DOI] [PubMed] [Google Scholar]
- [3].Shim H, Rose J, Halle S, Shekane P, Complex regional pain syndrome: a narrative review for the practising clinician, British journal of anaesthesia 123(2) (2019) e424–e433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zanos P, Moaddel R, Morris PJ, Riggs LM, Highland JN, Georgiou P, Pereira EFR, Albuquerque EX, Thomas CJ, Zarate CA Jr., Gould TD, Ketamine and Ketamine Metabolite Pharmacology: Insights into Therapeutic Mechanisms, Pharmacol Rev 70(3) (2018) 621–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schwartzman RJ, Alexander GM, Grothusen JR, The use of ketamine in complex regional pain syndrome: possible mechanisms, Expert Review of Neurotherapeutics 11(5) (2011) 719–734. [DOI] [PubMed] [Google Scholar]
- [6].Kiefer RT, Rohr P, Ploppa A, Dieterich HJ, Grothusen J, Koffler S, Altemeyer KH, Unertl K, Schwartzman RJ, Efficacy of ketamine in anesthetic dosage for the treatment of refractory complex regional pain syndrome: an open-label phase II study, Pain Med 9(8) (2008) 1173–201. [DOI] [PubMed] [Google Scholar]
- [7].Schwartzman RJ, Alexander GM, Grothusen JR, Paylor T, Reichenberger E, Perreault M, Outpatient intravenous ketamine for the treatment of complex regional pain syndrome: a double-blind placebo controlled study, Pain 147(1–3) (2009) 107–15. [DOI] [PubMed] [Google Scholar]
- [8].Zhao J, Wang Y, Wang D, The Effect of Ketamine Infusion in the Treatment of Complex Regional Pain Syndrome: a Systemic Review and Meta-analysis, Curr Pain Headache Rep 22(2) (2018) 12. [DOI] [PubMed] [Google Scholar]
- [9].Birklein F, Ajit SK, Goebel A, Perez R, & Sommer C (2018). Complex regional pain syndrome - phenotypic characteristics and potential biomarkers. Nature reviews. Neurology, 14(5), 272–284. 10.1038/nrneurol.2018.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK, MicroRNA modulation in complex regional pain syndrome, J Transl Med 9(1) (2011) 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McDonald MK, Ramanathan S, Touati A, Zhou Y, Thanawala RU, Alexander GM, Sacan A, Ajit SK, Regulation of proinflammatory genes by the circulating microRNA hsa-miR-939, Scientific reports 6 (2016) 30976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Douglas SR, Shenoda BB, Qureshi RA, Sacan A, Alexander GM, Perreault M, Barrett JE, Aradillas-Lopez E, Schwartzman RJ, Ajit SK, Analgesic Response to Intravenous Ketamine Is Linked to a Circulating microRNA Signature in Female Patients With Complex Regional Pain Syndrome, The Journal of Pain 16(9) (2015) 814–824. [DOI] [PubMed] [Google Scholar]
- [13].White FA, Bhangoo SK, & Miller RJ (2005). Chemokines: integrators of pain and inflammation. Nature reviews. Drug discovery, 4(10), 834–844. 10.1038/nrd1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chandrasekar B, Melby PC, Sarau HM, Raveendran M, Perla RP, Marelli-Berg FM, Dulin NO, Singh IS, Chemokine-cytokine cross-talk. The ELR+ CXC chemokine LIX (CXCL5) amplifies a proinflammatory cytokine response via a phosphatidylinositol 3-kinase-NF-kappa B pathway, The Journal of biological chemistry 278(7) (2003) 4675–86. [DOI] [PubMed] [Google Scholar]
- [15].Dawes JM, Calvo M, Perkins JR, Paterson KJ, Kiesewetter H, Hobbs C, Kaan TK, Orengo C, Bennett DL, McMahon SB, CXCL5 mediates UVB irradiation-induced pain, Sci Transl Med 3(90) (2011) 90ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dawes JM, Antunes-Martins A, Perkins JR, Paterson KJ, Sisignano M, Schmid R, Rust W, Hildebrandt T, Geisslinger G, Orengo C, Bennett DL, McMahon SB, Genome-wide transcriptional profiling of skin and dorsal root ganglia after ultraviolet-B-induced inflammation, PloS one 9(4) (2014) e93338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Reichert O, Kolbe L, Terstegen L, Staeb F, Wenck H, Schmelz M, Genth H, Kaever V, Roggenkamp D, Neufang G, UV radiation induces CXCL5 expression in human skin, Experimental dermatology 24(4) (2015) 309–12. [DOI] [PubMed] [Google Scholar]
- [18].Bishop T, Hewson DW, Yip PK, Fahey MS, Dawbarn D, Young AR, McMahon SB, Characterisation of ultraviolet-B-induced inflammation as a model of hyperalgesia in the rat, Pain 131(1–2) (2007) 70–82. [DOI] [PubMed] [Google Scholar]
- [19].Bishop T, Ballard A, Holmes H, Young AR, McMahon SB, Ultraviolet-B induced inflammation of human skin: characterisation and comparison with traditional models of hyperalgesia, Eur J Pain 13(5) (2009) 524–32. [DOI] [PubMed] [Google Scholar]
- [20].David Clark J, Tawfik VL, Tajerian M, Kingery WS, Autoinflammatory and autoimmune contributions to complex regional pain syndrome, Mol Pain 14 (2018) 1744806918799127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Üçeyler N, Eberle T, Rolke R, Birklein F, Sommer C, Differential expression patterns of cytokines in complex regional pain syndrome, Pain 132(1) (2007) 195–205. [DOI] [PubMed] [Google Scholar]
- [22].Alexander GM, Peterlin BL, Perreault MJ, Grothusen JR, Schwartzman RJ, Changes in plasma cytokines and their soluble receptors in complex regional pain syndrome, The journal of pain : official journal of the American Pain Society 13(1) (2012) 10–20. [DOI] [PubMed] [Google Scholar]
- [23].Birklein F, Drummond PD, Li W, Schlereth T, Albrecht N, Finch PM, Dawson LF, Clark JD, Kingery WS, Activation of Cutaneous Immune Responses in Complex Regional Pain Syndrome, The Journal of Pain 15(5) (2014) 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harden RN, Bruehl S, Perez RS, Birklein F, Marinus J, Maihofner C, Lubenow T, Buvanendran A, Mackey S, Graciosa J, Mogilevski M, Ramsden C, Chont M, Vatine JJ, Validation of proposed diagnostic criteria (the “Budapest Criteria”) for Complex Regional Pain Syndrome, Pain 150(2) (2010) 268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Schmittgen TD, Livak KJ, Analyzing real-time PCR data by the comparative C(T) method, Nature protocols 3(6) (2008) 1101–8. [DOI] [PubMed] [Google Scholar]
- [26].Chen Y, Wang X, miRDB: an online database for prediction of functional microRNA targets, Nucleic acids research 48(D1) (2019) D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jeyaseelan S, Manzer R, Young SK, Yamamoto M, Akira S, Mason RJ, Worthen GS, Induction of CXCL5 during inflammation in the rodent lung involves activation of alveolar epithelium, Am J Respir Cell Mol Biol 32(6) (2005) 531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu Y, Mei J, Gonzales L, Yang G, Dai N, Wang P, Zhang P, Favara M, Malcolm KC, Guttentag S, Worthen GS, IL-17A and TNF-α Exert Synergistic Effects on Expression of CXCL5 by Alveolar Type II Cells In Vivo and In Vitro, 186(5) (2011) 3197–3205. [DOI] [PubMed] [Google Scholar]
- [29].De Guire V, Robitaille R, Tetreault N, Guerin R, Menard C, Bambace N, Sapieha P, Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: promises and challenges, Clin Biochem 46(10–11) (2013) 846–60. [DOI] [PubMed] [Google Scholar]
- [30].Shenoda BB, Alexander GM, Ajit SK, Hsa-miR-34a mediated repression of corticotrophin releasing hormone receptor 1 regulates pro-opiomelanocortin expression in patients with complex regional pain syndrome, J Transl Med 14(1) (2016) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Birklein F, Ajit SK, Goebel A, Perez R, Sommer C, Complex regional pain syndrome - phenotypic characteristics and potential biomarkers, Nat Rev Neurol 14(5) (2018) 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Parkitny L, McAuley JH, Di Pietro F, Stanton TR, O’Connell NE, Marinus J, van Hilten JJ, Moseley GL, Inflammation in complex regional pain syndrome: A systematic review and meta-analysis, Neurology 80(1) (2013) 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ruddy MJ, Shen F, Smith JB, Sharma A and Gaffen SL (2004), Interleukin-17 regulates expression of the CXC chemokine LIX/CXCL5 in osteoblasts: implications for inflammation and neutrophil recruitment. Journal of Leukocyte Biology, 76: 135–144. doi: 10.1189/jlb.0204065. [DOI] [PubMed] [Google Scholar]
- [34].Hausser J, Zavolan M, Identification and consequences of miRNA-target interactions--beyond repression of gene expression, Nature reviews. Genetics 15(9) (2014) 599–612. [DOI] [PubMed] [Google Scholar]
- [35].Nam J-W, Rissland Olivia S., Koppstein D, Abreu-Goodger C, Jan Calvin H., Agarwal V, Yildirim Muhammed A., Rodriguez A, Bartel David P., Global Analyses of the Effect of Different Cellular Contexts on MicroRNA Targeting, Molecular cell 53(6) (2014) 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M, Silencing of microRNAs in vivo with ‘antagomirs’, Nature 438(7068) (2005) 685–9. [DOI] [PubMed] [Google Scholar]
- [37].A.V. Leick M, Newton G, Luscinskas FW. Leukocyte recruitment in inflammation: basic concepts and new mechanistic insights based on new models and microscopic imaging technologies. Cell Tissue Res. 2014;355(3):647–656. doi: 10.1007/s00441-014-1809-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tedgui A, Mallat Z. 2006. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol. Rev 86: 515–581. [DOI] [PubMed] [Google Scholar]
- [39].Goebeler M, Yoshimura T, Toksoy A, Ritter U, Brocker EB, Gillitzer R. 1997. The chemokine repertoire of human dermal microvascular endothelial cells and its regulation by inflammatory cytokines. J. Invest. Dermatol 108: 445–451. [DOI] [PubMed] [Google Scholar]
- [40].Luster A, Alon R & von Andrian U Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol 6, 1182–1190 (2005). 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- [41].Grespan R, Fukada SY, Lemos HP, Vieira SM, Napimoga MH, Teixeira MM, Fraser AR, Liew FY, McInnes IB, Cunha FQ, CXCR2-specific chemokines mediate leukotriene B4–dependent recruitment of neutrophils to inflamed joints in mice with antigen-induced arthritis, 58(7) (2008) 2030–2040. [DOI] [PubMed] [Google Scholar]
- [42].Liu X, Tian Y, Lu N, Gin T, Cheng CH, Chan MT, Stat3 inhibition attenuates mechanical allodynia through transcriptional regulation of chemokine expression in spinal astrocytes, PloS one 8(10) (2013) e75804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mei J, Liu Y, Dai N, Favara M, Greene T, Jeyaseelan S, Poncz M, Lee JS, Worthen GS, CXCL5 Regulates Chemokine Scavenging and Pulmonary Host Defense to Bacterial Infection, Immunity 33(1) (2010) 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


