Abstract
The corticotropin-releasing factor (CRF) system has been strongly associated with gastrointestinal pathophysiology, including colorectal cancer (CRC). We previously showed that altered expression of CRF receptors (CRFRs) in the colon critically affects CRC progression and aggressiveness through regulation of colonic inflammation. Here, we aimed to assess the potential of CRFR methylation levels as putative biomarkers in CRC. In silico methylation analysis of CRF receptor 1 (CRFR1) and CRF receptor 2 (CRFR2) was performed using methylome data derived by CRC and Crohn’s disease (CD) tissues and CRC-derived circulating cell-free DNAs (ccfDNAs). In total, 32 and 33 differentially methylated sites of CpGs (DMCs) emerged in CRFR1 and CRFR2, respectively, between healthy and diseased tissues. The methylation patterns were verified in patient-derived ccfDNA samples by qMSP and associated with clinicopathological characteristics. An automated machine learning (AutoML) technology was applied to ccfDNA samples for classification analysis. In silico analysis revealed increased methylation of both CRFRs in CRC tissue and ccfDNA-derived datasets. CRFR1 hypermethylation was also noticed in gene body DMCs of CD patients. CRFR1 hypermethylation was further validated in CRC adjuvant-derived ccfDNA samples, whereas CRFR1 hypomethylation, observed in metastasis-derived ccfDNAs, was correlated to disease aggressiveness and adverse prognostic characteristics. AutoML analysis based on CRFRs methylation status revealed a three-feature high-performing biosignature for CRC diagnosis with an estimated AUC of 0.929. Monitoring of CRFRs methylation-based signature in CRC tissues and ccfDNAs may be of high diagnostic and prognostic significance in CRC.
Keywords: CRF, colorectal cancer, methylation, bioinformatics, machine learning, liquid biopsy
1. Introduction
CRC is among the most common cancers worldwide, accounting for 10.2% of the newly diagnosed malignancies and 9.2% of cancer-related deaths in 2018 [1]. Although the five-year survival rates for early-stage CRC reach 90%, mortality is significantly high among patients with distant metastases [2]. Hence, tumor stage at the time of diagnosis is thought to be the most crucial prognostic factor. Early diagnosis is often delayed by lack of symptoms such as abdominal pain, rectal bleeding, weakness and weight loss which usually appear in late-stage CRC, accompanied by worse prognosis [3]. Therefore, preventive screening of asymptomatic individuals should be mandatory for reducing the incidence and mortality rates of CRC [4]. Currently, preventive colon cancer screening and early diagnosis are facilitated only by invasive methods such as colonoscopy and sigmoidoscopy. The success and accuracy of this method depend on several parameters, including the experience of the gastroenterologist and the correct implementation of the technical protocols, such as adequate bowel preparation and avoidance of bleeding complications [5,6]. In addition, randomized controlled trials have shown that sigmoidoscopy is limited in detecting cancers only in distal colon [7], while computed tomography (CT) colonography has shown lower sensitivity in detecting small polyps (<8 mm) [8]. Furthermore, radiation exposure and the need for additional colonoscopy in case of positive findings are also considerable disadvantages of the above methods [9]. The development of noninvasive approaches for early CRC screening has recently begun to gain ground. In this context, guaiac-based fecal occult blood test (gFOBT), a method that detects blood in feces, has demonstrated low sensitivity (51%) in detecting cancer cells [10], whereas DNA stool tests (i.e., Cologuard®) have high sensitivity for early CRC detection (92%) but lower performance in perceiving advanced pre-cancerous lesions (42.4%) [11]. Blood-based protein biomarkers have been suggested for the early detection of CRC [12]. Circulating cfDNA has been considered as a liquid biopsy material able to provide insights in cancer initiation and progression, meeting the need for a convenient, minimally invasive tool for precision medicine [13,14,15]. It is well-established that specific gene methylation changes that occur early in carcinogenesis and can be detected in ccfDNA could serve as valuable biomarkers for early cancer screening [16,17,18]. For example, a SEPT9 methylation test in ccfDNA has been introduced as a screening option for detecting CRC; however, it shows low rates of sensitivity (48%) and specificity (92%) [9]. Hence, the identification of novel biomarkers in liquid biopsy materials could be of great importance in early CRC screening.
Stress affects the function of the gastrointestinal tract in multiple ways and has been associated with its pathology [19,20]. The hypothalamic neuropeptide CRF and its homolog urocortin coordinate neuroendocrine pathways of the stress response via activation of two distinct receptors, CRFR1 and CRFR2. CRFRs are expressed throughout the gastrointestinal tract [19,20,21,22,23], while aberrant expression has been reported in CRC [24] and in Crohn’s disease [25,26,27,28] considered to be a high-risk premalignant condition. In CRC, expression of CRFR1 [29] and decreased expression of CRFR2 [30] are associated with tumorigenicity and progression, whereas the CRF system has been implicated in cancer development in multiple tissues [31].
Although the expression of CRFRs is known to be regulated by methylation in the central nervous system (CNS) [32,33], the methylation status of the CRFRs present in peripheral sites has not been thoroughly studied. We recently reported on an association of CRFR1 hypermethylation with the presence of steroid hormone receptors in breast cancer [34]. In addition, using genome-wide methylation approaches, Kobayashi et al. found that hypermethylation of CRFR2 is correlated with colitis-induced CRC [35]. This is in line with our previous reports indicating that diminished or lost CRFR2 expression in CRC promotes tumor development and aggressiveness, including tumor immunoescape, by regulating molecular circuits involved in sustaining inflammation in the gut [30,36]. Here, we explored the methylation status of CRFR1 and CRFR2 by performing bioinformatic analysis in DMCs identified in CRFR1 and CRFR2 genes obtained by publicly available high-throughput methylome data from healthy, CD and CRC colon tissue. We further examined the CRFRs methylation in patient-derived liquid biopsy samples (ccfDNA) and evaluated its diagnostic and prognostic value in CRC.
2. Materials and Methods
2.1. Dataset for the In Silico Analysis
Raw DNA methylation data from CRC, CD patients and normal colons as well as the corresponding clinicopathological parameters were obtained from the GEO (Gene Expression Omnibus) (https://www.ncbi.nlm.nih.gov/geo/, accessed on 1 September 2020) database [37]. CRC, CD and ccfDNA were used as keywords in the GEO query and ‘Methylation profiling by array’ as the study type. Thirty-four studies were found; of them, only those using Infinium Human Methylation 450K and EPIC BeadChips and providing adequate raw and clinical data were selected for further analysis. Four studies, namely GSE99788 [38], GSE149282 [39], GSE122126 [40] and GSE105798 [41], were recruited for our analysis. The description of study groups and correlations is presented in Table 1.
Table 1.
Methylome datasets used for the bioinformatic analysis of CRFR1 and CRFR2 methylation.
| Dataset | Platform | Correlated Groups | References |
|---|---|---|---|
| GSE149282 | EPIC | Twelve CRC vs. 12 adjacent colon tissues | [35] |
| GSE122126 | 450k | Four CRC ccfDNAs vs. four healthy ccfDNA | [36] |
| GSE105798 | 450k | Three CD vs. eight normal colon tissues | [37] |
| GSE99788 | EPIC | Thirteen CD vs. five normal colon fibroblasts | [34] |
CRC: colorectal cancer; CD: Crohn’s disease; NINF: noninflammatory; STEN: stenotic; INF: inflammatory.
2.2. Data Preprocessing and DNA Methylation Analysis
Raw DNA methylation data (IDAT files) and sample annotation files were processed in the Bioconductor R package RnBeads v2.0 (https://rnbeads.org/index.html, accessed on 18 September 2020) [42]. CpGs were chosen as the genomic region of interest. The MethyLumi-Noob method was used to normalize technical variation in the background fluorescence signal [43]. BMIQ was used as a normalization method to adjust beta values of type II design probes into a statistical distribution characteristic of type I probes [44]. Probes for SNPs or probes outside of the CpG context as well as probes on sex chromosomes were removed [45]. Probes/samples with the highest fraction of unreliable measurements were removed from further analysis using the Greedycut algorithm. Νormalized β-values for each CpG were generated, representing the methylated probe’s intensity divided by the overall intensity (sum of methylated and unmethylated probe intensities) plus an offset of 100 [46]. DNA methylation differences were analyzed using hierarchical linear models implemented in the limma package [47] provided in the RnBeads pipeline. Differentially methylated CpGs (DMCs) for CRFR1 and CRFR2 were identified based on the false discovery rate (FDR-adjusted p-value < 5.00 × 10−2).
2.3. In Silico Determination of Transcription Factor (TF) Binding
In order to examine if DMCs identified were correlated to CRFR1 and CRFR2 gene expression, we further analyzed promoter regions to locate TFs binding sites. First, the Methprimer (https://www.urogene.org/methprimer/, accessed on 5 November 2020) [48] tool was used for the identification of possible cytosine–guanine dinucleotides islands (CGIs) at these regions. CGIs are regions of transcription initiation with a high frequency of CpG sites. The criteria for CGIs prediction used were as follows: size region of at least 100 bp, GC percentage greater than 50% and an observed-to-expected CpG ratio greater than 60%. Then, the PROMO (http://alggen.lsi.upc.es/, accessed on 5 November 2020) [49] tool was used in order to define possible TFs binding in identified CGIs. Only human factors and human sites were considered for TFs search.
2.4. Clinical Samples
CRC patients who visited the Department of Medical Oncology of University General Hospital of Alexandroupolis between 2001 and 2016 were included in the study. Blood samples were collected following diagnosis from two patient groups: (a) 42 patients having undergone surgery for primary CRC within the previous month and before the initiation of adjuvant therapy (adjuvant group) and (b) 71 patients with metastatic disease following palliative surgery and before the initiation of first-line chemotherapy (metastatic group). The treatment modules used for the metastatic group were FOLFIRI (a combination of folinic acid, fluorouracil and irinotecan) or FOLFOX (a combination of folinic acid, fluorouracil and oxaliplatin) or the alternatives XELIRI (irinotecan and capecitabine) or XELOX (capecitabine and oxaliplatin) plus bevacizumab or panitumumab/cetuximab. The response of metastatic patients to the above regimens was correlated with CRFR1 and CRFR2 methylation. Clinicopathological and demographic characteristics are presented in Table 2. Follow-up data from 2001 to 2016 were also available. In parallel, blood samples from 20 healthy donors were collected from the blood donation unit of the same hospital and included in our study (11 males and nine females, mean age: 58.9 (± 9.0), median: 59.0 (range: 43.0–76.0), mean BMI: 26.0 (± 5.0), median: 24.5 (20.49–35.49)) (control group). Inclusion criteria of both patients and healthy individuals were age between 18 and 80 years old and the ability to give informed consent, for patients not to have initiated adjuvant or first-line treatment before sample collection and for healthy individuals to be free of cancer and cancer history. Peripheral blood was collected in an EDTA before treatment and processed immediately for plasma isolation within 2 h. The study was approved by the Scientific Board and the Ethics Committee of the University General Hospital of Alexandroupolis/Greece and was conducted according to the ethical principles of the Declaration of Helsinki. All the patients participated after signing a voluntary informed consent.
Table 2.
Demographic and clinicopathological characteristics of CRC patient and normal groups.
| Clinical Parameter | Total (n = 113) (%) | Adjuvant Group (n = 42) (%) | Metastatic Group (n = 71) (%) | Normal (n = 20) (%) |
|---|---|---|---|---|
| Age (years) | ||||
| Mean ± SD | 67.0 ± 9.7 | 69.5 ± 8.7 | 65.5 ± 10 | 58.9 (±9.0) |
| Median, range | 68, 44–87 | 70.5, 47–87 | 67, 44–85 | 59, 43–76 |
| Gender | ||||
| Male | 78 (69%) | 30 (71.4%) | 48 (67.6%) | 11 (55%) |
| Female | 35 (31%) | 12 (28.6%) | 23 (32.4%) | 9 (45%) |
| BMI | ||||
| <18.5 | 2 (1.8%) | 0 | 2 (2.8%) | 0 |
| 18.5–24.9 | 12 (10.6%) | 6 (14.3%) | 6 (8.5%) | 8 (40%) |
| 25–29.9 | 35 (31%) | 24 (57.1%) | 11 (15.5%) | 6 (30%) |
| ≥30 | 13 (11.5%) | 11 (26.2%) | 2 (2.8%) | 3 (15%) |
| Not available | 51 (45.1%) | 1 (2.4%) | 50 (70.4%) | 3 (15%) |
| Cancer location | ||||
| R | 19 (16.8%) | 13 (31%) | 6 (8.5%) | |
| A | 7 (6.2%) | 4 (9.5%) | 3 (4.2%) | |
| S | 28 (24.8%) | 18 (42.9%) | 10 (14.1%) | |
| T | 2 (1.8%) | 2 (4.8%) | 0 | |
| D | 3 (2.7%) | 3 (7.1%) | 0 | |
| C | 4 (3.5%) | 2 (4.8%) | 2 (2.8%) | |
| Νot available | 50 (44.2%) | 0 | 50 (70.4%) | |
| Dukes classification | ||||
| A | 14 (12.4%) | 14 (33.3%) | 0 | |
| B | 14 (12.4%) | 13 (31%) | 0 | |
| C | 13 (11.5%) | 13 (31%) | 0 | |
| D | 69 (61%) | 0 | 71 (100%) | |
| Not available | 3 (2.7%) | 2 (4.8%) | 0 | |
| Astler–Coller classification | ||||
| A | 2 (1.8%) | 2 (4.8%) | 0 | |
| B1 | 12 (10.6%) | 12 (28.6%) | 0 | |
| B2 | 13 (11.5%) | 13 (31%) | 0 | |
| B3 | 0 | 0 | 0 | |
| C1 | 1 (0.9%) | 1 (2.4%) | 0 | |
| C2 | 12 (10.6%) | 12 (28.6%) | 0 | |
| C3 | 0 | 0 | 0 | |
| D | 71 (62.8%) | 0 | 71(100%) | |
| Not available | 2 (1.8%) | 2 (4.8%) | 0 | |
| Stage | ||||
| Ι | 15 (13.3%) | 15 (35.7%) | 0 | |
| ΙΙ | 13 (11.5%) | 13 (31%) | 0 | |
| ΙΙΙ | 13 (11.5%) | 13 (31%) | 0 | |
| IV | 71 (62.8%) | 0 | 71(100%) | |
| Not available | 1 (0.9%) | 1 (2.4%) | 0 | |
| Grade | ||||
| 1 | 41 (36.3%) | 18 (42.9%) | 14 (19.7%) | |
| 2 | 50 (44.2%) | 18 (42.9%) | 41 (57.7%) | |
| 3 | 12 (10.6%) | 5 (11.9%) | 7 (9.9%) | |
| Not available | 10 (8.9%) | 1 (2.4%) | 9 (12.7%) | |
| Tumor size | ||||
| T1 | 3 (2.7%) | 3 (7.15%) | 0 | |
| T2 | 24 (21.2%) | 13 (30.95%) | 11 (15.5%) | |
| T3 | 66 (58.4%) | 21 (50%) | 45 (63.4%) | |
| T4 | 12 (10.6%) | 4 (9.5%) | 8 (11.3%) | |
| Not available | 8 (7.1%) | 1 (2.4%) | 7 (9.8%) | |
| LN status | ||||
| N0 | 52 (46%) | 27 (64.3%) | 0 | |
| N1 | 25 (22.1%) | 9 (21.4%) | 28 (39.4%) | |
| N2 | 26 (23%) | 4 (9.5%) | 35 (49.3%) | |
| Not available | 10 (8.9%) | 2 (4.8%) | 8 (11.3%) | |
| Metastatic site | ||||
| Lung | 21 (18.6%) | 0 | 21 (29.6%) | |
| Liver | 55 (48.7%) | 0 | 55 (77.5%) | |
| Pancreas | 1 (0.9%) | 0 | 1 (1.4%) | |
| Bone | 1 (0.9%) | 0 | 1 (1.4%) | |
| Peritoneum | 10 (8.9%) | 0 | 10 (14%) | |
| Brain | 2 (1.8%) | 0 | 2 (2.8%) | |
| Testis | 1 (0.9%) | 0 | 1 (1.4%) | |
| Uterus | 1 (0.9%) | 0 | 1 (1.4%) |
A: ascending; BMI: body mass index; C: cecum; D: descending; LN: lymph node; R: rectum; S: sigmoid; T = transverse.
2.5. Extraction of ccfDNA
Circulating cfDNA was extracted from plasma using a MagCore Nucleic Acid Extraction Kit (RBC BIOSCIENCE, New Taipei City, Taiwan) and a MagCore® Compact Automated Nucleic Acid Extractor (RBC BIOSCIENCE, New Taipei City, Taiwan) according to the manufacturer’s instructions. Briefly, ccfDNA was eluted from 1200 μL of plasma in 40 μL elution buffer and stored at −20 °C until further use.
2.6. Sodium Bisulfite Conversion of ccfDNA
Bisulfite conversion was performed by EZ DNA Methylation-Gold™ Kit (ZYMO Research Co., Orange, CA, USA) according to the manufacturer’s instructions and previous reports [50]. During conversion, all unmethylated but not the methylated cytosines of ccfDNA were converted to uracil. DNA was then eluted in 10 μL elution buffer and stored at −80 °C until use. In each experiment, CpGenome Human Methylated and Non-Methylated DNA standards (Merck Millipore, Germany) or H2O were included as positive and negative controls, respectively.
2.7. Quantitative Methylation-Specific PCR (qMSP)
Promoter methylation of CRFR1 and CRFR2 was analyzed by qMSP. A methylation-independent assay with non-CpG bearing sites for the β-actin gene (ACTB) was used in order to verify DNA quality and normalize results. Primer sequences and qMSP conditions are presented in detail in our previous work [34]. The samples were run in duplicates. The results were calculated using the Rotor-Gene 6000 Series Software 1.7 (Qiagen). The results were analyzed using the 2−ΔΔCT formula representing methylation levels, where ΔΔCT = ΔCTsample – ΔCTcalibrator [51]. Negative and positive control samples of 0% and 100% methylated converted DNA standards were included in each run.
2.8. Statistical Analysis
The Kolmogorov–Smirnov test was applied to check for normality in distribution and the chi-squared test for comparison between discrete variables. One-way ANOVA test followed by Bonferroni post-hoc or Kruskal–Wallis test were used for comparisons of continuous variables between three or more subgroups. In case of binary variables, t-test or Mann–Whitney test were also applied. Pearson or Spearman correlation was applied to compare two continuous variables. Metastatic patients who presented complete response (CR), partial response (PR) to treatment or stable disease (SD) at the first clinical examination after first-line treatment initiation according to response evaluation criteria in solid tumors (RESIST) version 1.1 [52] were considered as “responders,” whereas those who presented clinical progressive disease (PD) were considered as “non-responders.” Statistical significance was set at p-value < 0.05. Statistical analysis was performed using the IBM SPSS 19.0 statistical software (IBM Corp., 2010, IBM SPSS Statistics for Windows, version 19.0, Armonk, NY, USA).
2.9. Automated Machine Learning Analysis
Our data were further analyzed using an AutoML technology in order to construct signatures of diagnostic value, combining the liquid biopsy-based experimental parameters determined by our study and the clinicopathological features of the study groups. The Just Add Data Bio v1.1.118 (JADBio) (https://www.jadbio.com/, accessed on 25 January 2020) platform applicable to low-sample high-dimensional datasets [53] was employed and able to provide predictive models by employing standard, best-practice and state-of-the-art statistical and machine learning methods. JADBio works as follows: it first selects the appropriate algorithms to try for the task at hand depending on the outcome type, predictor type, user preferences (e.g., importance of quality of analysis vs. speed of analysis) using an artificial intelligence decision support system. The algorithms are selected to perform the following steps: data transformations, data preprocessing and imputation of missing values, feature selection, predictive modeling and data visualization. The AI system also selects which tuning hyperparameter values to try for each algorithm. All combinations of algorithms for each step and hyperparameter values (called configurations) are applied using a 10-fold cross-validation protocol. JADBio applies a bootstrap-based adjustment to the final reported performance [54] to remove this optimism and to return slightly conservative estimates of performance. JADBio performs biosignature discovery using SES (statistical equivalent signature) or LASSO (least absolute shrinkage and selection operator) algorithms for feature selection. A signature is defined as a minimal-size subset of predictors (features, molecular quantities, biomarkers, risk factors), which collectively (multivariately) lead to an optimal predictive model, neglecting all other features as irrelevant or redundant for prediction given the selected features. For classification modeling, JADBio employs an SVM (support-vector machine) [55] with full polynomial and Gaussian kernels, random forests [56], ridge logistic regression [57] and decision trees [58]. As most modern machine learning models are completely incomprehensible to a human, JADBio reports not only the best-out-of-all model, but also the best interpretable model (linear models or decision trees).
3. Results
3.1. In Silico Analysis of CRFR1 and CRFR2 Methylation in CRC and CD
In silico methylation analysis of CRFR1 and CRFR2 genes was performed using methylome data derived by CRC and CD tissues and CRC ccfDNAs. The results are described below.
3.1.1. Analysis in CRC and CD Tissue-Derived Datasets
For the methylation analysis of the indicated receptor genes in CRC and CD tissues, we used methylome data from four datasets (GSE149282, GSE122126, GSE105798, GSE99788) (Table 1). DMCs were identified among groups in all the studies (Supplementary Tables S1 and S2) except for GSE99788. In total, 32 and 33 DMCs were detected among healthy individuals and CD or CRC patients for CRFR1 and CRFR2, respectively. In general, lower methylation was noticed mostly in gene body intronic regions, while higher methylation was observed mainly in the first exon and the TSS200/1500 genome locations, known to be strongly associated with regulation of expression by methylation [59,60]. We therefore focused our analysis mostly in those regions. Among the DMCs identified in CRFR1, four CpGs (cg08473090, cg08929103, cg12577105 and cg18757974) were located at TSS1550 and two (cg11338426, cg13521908)—at the first exon (Table 3). Notably, methylation in all the CpGs located in the island region, known to have an important role in transcriptional regulation [61], was increased in CRC tissue in comparison to the adjacent normal tissue (GSE149282 dataset), whereas decreased methylation was noticed in the one found in the N shore, a region 0–2 kb upstream (5′) of the CpG island. Other CRFR1-related DMCs identified in CRC or CD tissue databases were located in the gene body or in the 5′UTR. A decrease of methylation was mainly noticed at these DMCs, while an increase was detected at DMCs located at islands or S shores of the gene body or the 5′UTR in cancer tissue in relation to the adjacent healthy colon (GSE149282 and GSE1222126). An increase of CRFR1 methylation was also noticed in gene-body DMCs of CD patients in comparison to normal tissues (GSE105798) (Supplementary Table S1). Finally, only one DMC (cg21773872) emerged from the GSE105798 dataset, showing decreased methylation in CD in relation to normal colon tissues.
Table 3.
DMCs at the first exon or close to the TSS identified by in silico analysis in CRFR1.
| STUDY | CpG ID |
Compared Study Groups |
Mean β-Value 1 * | Mean β-Value 2 * | Δ β-Value # | Methylation, CRC vs. Normal |
Gene Location |
Location Relative to CpG | FDR |
|---|---|---|---|---|---|---|---|---|---|
| GSE149 282 |
cg08473090 | Adjacent vs. CRC tissue | 0.089 | 0.330 | +0.241 | Up | TSS1500 | Island | 3.174 × 10−3 |
| GSE149 282 |
cg08929103 | Adjacent vs. CRC tissue | 0.563 | 0.239 | −0.323 | Down | TSS1500 | N shore | 4.453 × 10−5 |
| GSE149 282 |
cg11338426 | Adjacent vs. CRC tissue | 0.103 | 0.269 | +0.166 | Up | First Exon | Island | 1.786 × 10−2 |
| GSE149 282 |
cg12577105 | Adjacent vs. CRC tissue | 0.047 | 0.163 | +0.116 | Up | TSS1500 | Island | 8.456 × 10−3 |
| GSE149 282 |
cg13521908 | Adjacent vs. CRC tissue | 0.077 | 0.249 | +0.172 | Up | First Exon | Island | 3.136 × 10−3 |
| GSE149 282 |
cg18757974 | Adjacent vs. CRC tissue | 0.062 | 0.218 | +0.156 | Up | TSS1500 | Island | 7.654 × 10−3 |
|
GSE122 2126 |
cg08929103 | Healthy vs. CRC ccfDNA | 0.767 | 0.477 | −0.291 | Down | TSS1500 | N shore | 1.261 × 10−3 |
|
GSE122 2126 |
cg13521908 | Healthy vs. CRC ccfDNA | 0.117 | 0.213 | +0.096 | Up | First Exon | Island | 2.545 × 10−2 |
* Mean β-value 1 represents methylation in normal tissues and mean β-value 2—methylation in diseased tissues; # Δ β-value: mean β-value 2 − mean β-value 1; DMC: differentially methylated CpG; FDR: false discovery rate; ccfDNA: circulating cell-free DNA.
For CRFR2, nine CpGs located close to the TSS (cg01718447, cg02712145, cg04863452, cg07658503 cg13094036, cg14896516, cg15615793, cg18351440, cg21773872) and six CpGs located at the first exon (cg04922810, cg04923928, cg18266052, cg24214442, cg24610236, cg27430726) were identified as DMCs among the studied groups (Table 4). They were all found in the island except one found in the S shore (a region 0–2 kb downstream (3′) of the CpG island) and one in the N shelf (a region 2–4 kb upstream (5′) of the CpG island). In the GSE149282 dataset, all of the 11 CpGs (cg01718447, cg02712145, cg07658503, cg13094036, cg14896516, cg21773872, cg04922810, cg18266052, cg24214442, cg24610236, cg27430726) identified as DMCs were hypermethylated in cancer tissue in relation to the adjacent healthy colon.
Table 4.
DMCs at the first exon or close to the TSS identified by in silico analysis in CRFR2.
| STUDY | CpG ID |
Compared Study Groups |
Mean β-Value 1 * | Mean β-Value 2 * | Δ β-Value # | Methylation, Diseased vs. Normal |
Gene Location |
Location Relative to CpG | FDR |
|---|---|---|---|---|---|---|---|---|---|
| GSE149 282 |
cg01718447 | Adjacent vs. CRC tissue |
0.126 | 0.536 | +0.410 | Up | TSS200 | Island | 2.375 × 10−3 |
| GSE149 282 |
cg02712145 | Adjacent vs. CRC tissue |
0.166 | 0.471 | +0.305 | Up | TSS1500 | Island | 2.785 × 10−2 |
| GSE149 282 |
cg04922810 | Adjacent vs. CRC tissue | 0.077 | 0.435 | +0.358 | Up | First Exon | Island | 7.024 × 10−3 |
| GSE149 282 |
cg07658503 | Adjacent vs. CRC tissue | 0.051 | 0.313 | +0.262 | Up | TSS200 | Island | 3.165 × 10−2 |
| GSE149 282 |
cg13094036 | Adjacent vs. CRC tissue | 0.089 | 0.343 | +0.254 | Up | TSS1500 | Island | 3.663 × 10−3 |
| GSE149 282 |
cg14896516 | Adjacent vs. CRC tissue | 0.106 | 0.352 | +0.246 | Up | TSS1500 | Island | 1.861 × 10−4 |
| GSE149 282 |
cg18266052 | Adjacent vs. CRC tissue | 0.100 | 0.418 | +0.318 | Up | First Exon | Island | 4.872 × 10−2 |
| GSE149 282 |
cg21773872 | Adjacent vs. CRC tissue | 0.146 | 0.655 | +0.509 | Up | TSS200 | Island | 1.242 × 10−6 |
| GSE149 282 |
cg24214442 | Adjacent vs. CRC tissue | 0.123 | 0.463 | +0.340 | Up | First Exon | Island | 3.225 × 10−6 |
| GSE149 282 |
cg24610236 | Adjacent vs. CRC tissue | 0.074 | 0.451 | +0.378 | Up | First Exon | Island | 1.568 × 10−5 |
| GSE149 282 |
cg27430726 | Adjacent vs. CRC tissue | 0.133 | 0.457 | +0.325 | Up | First Exon | Island | 7.102 × 10−5 |
| GSE1222126 | cg04863452 | Healthy vs. CRC ccfDNA | 0.053 | 0.110 | +0.057 | Up | TSS200 | Island | 1.065 × 10−3 |
| GSE1222126 | cg04923928 | Healthy vs. CRC ccfDNA | 0.045 | 0.143 | +0.098 | Up | First Exon | Island | 1.665 × 10−4 |
| GSE1222126 | cg15615793 | Healthy vs. CRC ccfDNA | 0.486 | 0.629 | +0.142 | Up | TSS1500 | S_Shore | 9.913 × 10−3 |
| GSE1222126 | cg18351440 | Healthy vs. CRC ccfDNA | 0.885 | 0.806 | −0.079 | Down | TSS1500 | N_Shelf | 3.929 × 10−5 |
| GSE105799 | cg21773872 | Normal vs. CD | 0.089 | 0.041 | −0.049 | Down | TSS200 | Island | 2.386 × 10−4 |
* Mean β-value 1 represents methylation in normal tissues and mean β-value 2—methylation in diseased tissues; # Δ β-value: mean β-value 2 − mean β-value 1. DMCs: differentially methylated CpGs; FDR: false discovery rate; ccfDNA: circulating cell-free DNA.
3.1.2. Analysis of CRC-Derived ccfDNA Data
Two CpGs (cg08929103, cg13521908) identified in CRFR1 as DMCs in ccfDNA dataset GSE1222126 showed the same hypermethylation trend in patient-derived samples compared to normal counterparts, thus suggesting that ccfDNA may reflect the methylation status of tumor tissues. Methylation analysis of CRFR2 using the same dataset revealed that three CpGs (cg04863452, cg15615793, cg04923928) and one CpG (cg18351440, located in the N shelf) were hypermethylated and hypomethylated, respectively, in CRC ccfDNAs compared to their normal counterparts. It has to be noted that the DMCs found in CRC ccfDNA (GSE1222126 dataset) were not identified as DMCs in the CRC tissue (GSE149282).
3.2. In Silico Analysis of TFs Binding in CRFR1 and CRFR2 Promoters
DNA methylation regulates gene expression mainly by disturbing transcription factor (TF) and RNA polymerase binding to putative sites known to be necessary for initiation of transcription [62]. To address whether the identified DMCs may actually play a role in regulating CRFR1 and CRFR2 expression, we examined their presence within CGIs that are known to contain TFs binding that could initiate transcription.
For CRFR1, one CGI of 180 bp was found that contained DMCs between malignant and adjacent colon tissues. Further analysis by the PROMO tool predicted 31 putative TFs that could bind to this CGI (Figure 1A). For CRFR2, a CGI of 287 bp was also identified and 40 TFs were predicted (Figure 1B). Together, these findings show that CGIs containing the identified DMCs contain multiple sites for putative TFs binding and therefore altered methylation during carcinogenesis can affect CRFR1 and CRFR2 expression.
Figure 1.
Predicted TFs for binding to the CGIs of CRFR1 and CRFR2 promoters containing DMCs: (A) 31 TFs were predicted for CRFR1 and (B) 40 TFs were predicted for CRFR2 with a dissimilarity margin ≤ 15%. TFs: transcription factors; CGIs: CpG islands; CRFR1: human corticotropin-releasing factor receptor 1; CRFR2: human corticotropin-releasing factor receptor 2.
3.3. Methylation Analysis of CRFR1 and CRFR2 in CRC-Derived ccfDNA Clinical Samples
Following the in silico analysis, methylation of the CRF receptor genes was investigated in CRC patient-derived ccfDNAs and compared with their healthy counterparts. Quantitative MSP assays were performed in ccfDNAs isolated from 42 adjuvant CRC patients, 71 metastatic CRC patients and 20 healthy individuals (control). Primers of CRFR1 and CRFR2 were designed at the promoter region inside the studied CGI.
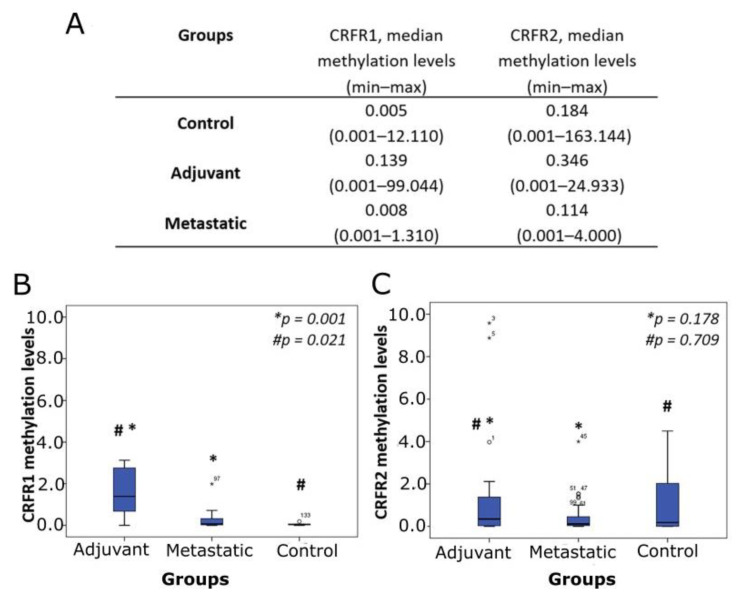
CRFR1 methylation was detected in 45.0, 35.7 and 36.6% of control, adjuvant and metastatic groups, respectively. For CRFR2, the respective numbers were 70.0, 64.3 and 67.6%. CRF receptor methylation levels are presented in Figure 2A. Significantly increased levels of CRFR1 methylation were found in the adjuvant group compared to the control group (p = 0.021) and the metastatic group (p = 0.001) (Figure 2B). For CRFR2, no statistically significant differences in methylation levels were observed between the studied groups (Figure 2C).
Figure 2.
Methylation levels of CRF receptor genes estimated by qMSP in ccfDNA from adjuvant and metastatic CRC patients and healthy individuals (control). (A) Median methylation levels of CRFR1 and CRFR2. Boxplots depict methylation levels in relation to study groups for (B) CRFR1 and (C) CRFR2; * p: adjuvant vs. metastatic; # p: adjuvant vs. control; ccfDNA: circulating cell-free DNA; qMSP: quantitative methylation-specific PCR; CRC: colorectal cancer.
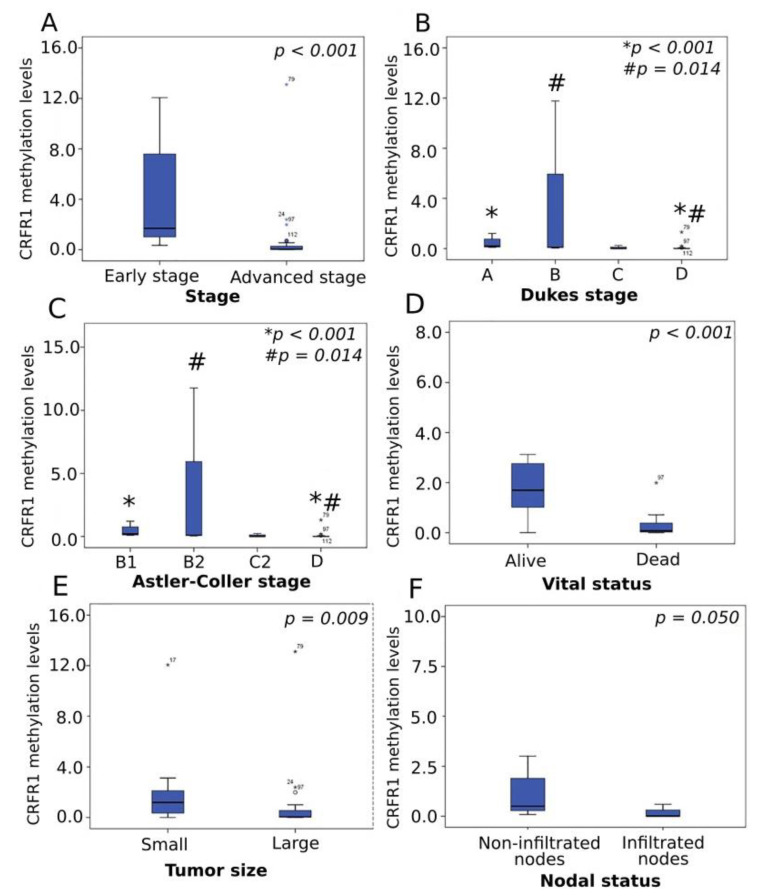
Interestingly, among cancer patients, lower methylation levels of CRFR1 were correlated with adverse clinicopathological characteristics and poor outcomes. Specifically, low methylation levels were significantly correlated with advanced disease stage (p < 0.001) (Figure 3A), D tumor stage (by Dukes) as compared to A (p < 0.001) and B (p = 0.014) stages (Figure 3B), while the same methylation trend was observed in the D stage when compared to B1 (p < 0.001) and B2 (p = 0.014) stages of the Astler–Coller classification system (Figure 3C).
Figure 3.
Correlation of CRFR1 methylation levels with clinical parameters. Boxplots depict CRFR1 methylation levels in relation to (A) stage, (B) Dukes stage, (C) Astler–Coller stage, (D) vital status, (E) tumor size and (F) nodal status; * p: A vs. D for Dukes stage and B1 vs. D for Astler–Coller stage; # p: B vs. D for Dukes stage and B2 vs. D for Astler–Coller stage.
Diminished CRFR1 methylation was also associated with the incidence of death (p < 0.001) (Figure 3D). Moreover, within the adjuvant group, larger tumors and those presenting lymph node infiltration showed decreased CRFR1 methylation levels in relation to small tumors (p = 0.009) (Figure 3E) and those without characteristics of LN infiltration (p = 0.050) (Figure 3F), respectively. No significant correlations were established between CRFR1 methylation levels and tumor location, grade, mutational status of the KRAS gene or demographic parameters. Similarly, CRFR1 methylation levels were not associated with the frequency of relapses in the adjuvant group or first-line treatment response in the metastatic group. CRFR2 methylation levels were not associated with any clinicopathological parameters or prognosis in a statistically significant manner. The methylation analysis of the ccfDNA study groups revealed that CRFR1 methylation was significantly increased in the adjuvant group of patients compared to that of healthy individuals and patients with metastatic disease (Figure 2B). Comparison of the CRFR1 methylation levels between the last two groups (control and metastatic) also showed a trend of hypermethylation in the metastatic group; however, this difference was not statistically significant. Within the patients’ groups (adjuvant and metastatic), the lower methylation levels were positively correlated with advanced tumor stage and size, infiltrated nodes and poor outcomes (Figure 3A–F). Furthermore, low methylation within the two malignant groups was significantly associated with advanced disease and adverse prognosis. The above findings demonstrate for the first time CRFR1 hypermethylation as a hallmark of early CRC status, which is remarkably diminished as the disease progresses to more aggressive phenotypes, reaching the almost normal levels.
3.4. Automated Machine Learning Analysis
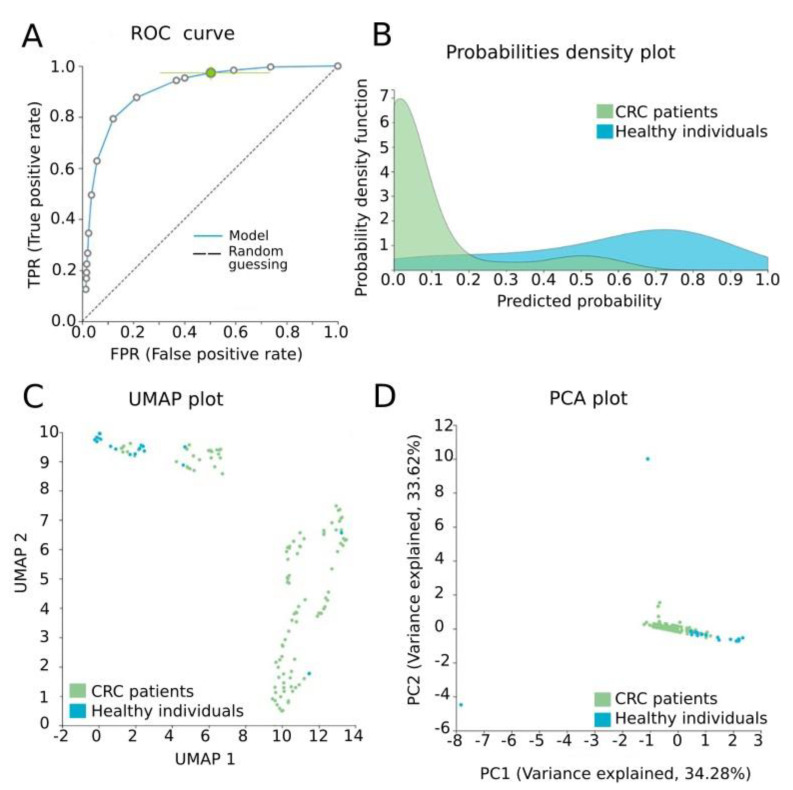
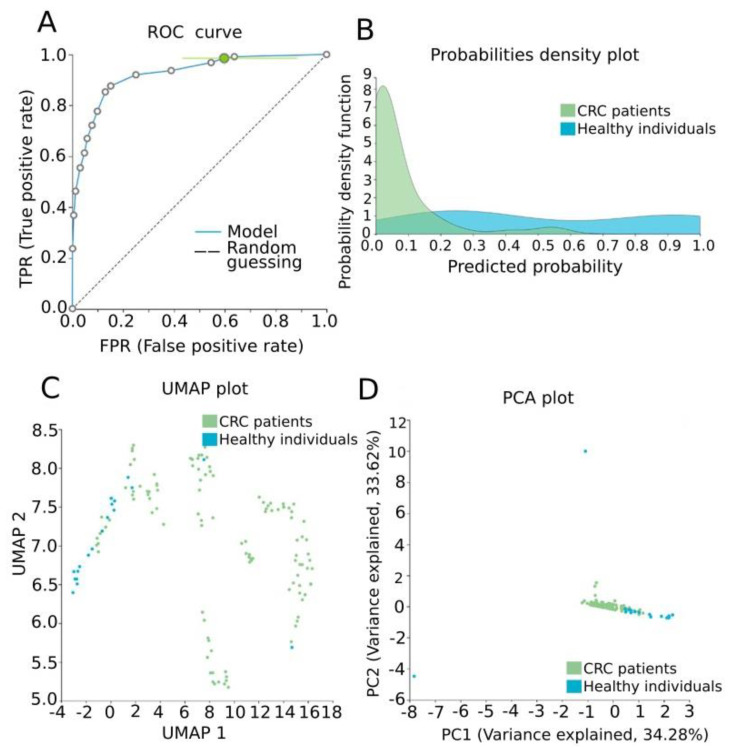
Our data were further analyzed by AutoML in order to construct a signature of diagnostic value, combining methylation measurements and established clinicopathological features of the study group. The task was a classification analysis in order to discriminate CRC patients from healthy individuals. Our best-performing model was a three-feature signature containing the methylation levels of CRFR1, the methylation levels of CRFR2 and age via the Classification Random Forests algorithm with an estimated area under the curve (AUC) of 0.929 (0.873, 0.972) and an average precision of 0.983 (0.965, 0.994) to discriminate healthy tissues from CRC. Model performance and model inspection are depicted in Figure 4. Furthermore, the best-interpretable model containing the same features via ridge logistic regression was produced reaching an AUC of 0.933 (0.887, 0.972) and an average precision of 0.983 (0.965, 0.994) (Figure 5).
Figure 4.
Model performance for the discrimination of CRC patients from healthy individuals. (A) ROC curve of the model, AUC: 0.929 (0.873, 0.972). (B) Probabilities density plot depicting distributions between normal (class 0, green) and CRC ccfDNA samples (class 1, blue). (C) UMAP plot showing sufficient discrimination between normal (class 0, blue) and CRC ccfDNA samples (class 1, green). (D) PCA plot presenting a good separation between normal (class 0, blue) and CRC ccfDNA samples (class 1, green). ROC: receiver operating characteristic; PCA: principal component analysis; UMAP: uniform manifold approximation and projection.
Figure 5.
Model performance for the discrimination of CRC patients from healthy individuals. (A) ROC curve of the model, 0.933 (0.887, 0.972). (B) Probabilities density plot depicting distributions between normal (class 0, green) and CRC ccfDNA samples (class 1, blue). (C) UMAP plot showing good discrimination between normal (class 0, blue) and CRC ccfDNA samples (class 1, green). (D) PCA plot presenting a good separation between normal (class 0, blue) and CRC ccfDNA samples (class 1, green).
4. Discussion
We and others have reported that CRF receptors play a critical role in regulating inflammation, carcinogenesis and disease progression in the colon [24]. Although the epigenetic regulation of CRF receptors has been thoroughly studied in stress-related CNS disorders, their methylation patterns and role in peripheral tissues have not been clearly elucidated. In this study, we first adopted a bioinformatic approach using publicly available datasets of CRC and CD to analyze methylation of CRF receptor genes. Although a large number of DMCs was identified in the entire genes (32 and 33 in CRFR1 and CRFR2, respectively) among healthy tissues and studied pathologies, we focused our analysis on the first exon and the TSS200/1500 region, which are genome locations known to be strongly associated with gene expression regulation by methylation [59,60].
Methylation of CRFR1 CpGs within the above regions and in particular those located in islands was significantly elevated in CRC compared to the adjacent healthy colon tissues, thus pointing to downregulation of CRFR1 expression. This pattern of CRFR1 hypermethylation was also observed in methylome datasets of ccfDNAs from CRC patients and clinical ccfDNA samples obtained in adjuvant CRC cases. Taken together, these findings suggest that assessing methylation in ccfDNA can dynamically reflect methylation events in the tumor’s lifespan. This notion is further supported by recent reports in pancreatic cancer, demonstrating that DNA methylation profiles of cancerous tissues and respective ccfDNAs significantly correlate with each other [38]. In concordance, our results for CRC ccfDNAs showed that methylation levels of CRFR1 were increased in the adjuvant group of patients in relation to healthy individuals. However, within the patients’ groups, lower methylation levels were correlated with advanced tumor stage and size, infiltrated nodes and poor outcomes. It can be postulated that CRFR1 methylation is a molecular event characterizing primary tumor formation and is later lost during transition to the metastatic phase. Our findings are the first demonstrating aberrant CRFR1 methylation in CRC. Given that decreased methylation may lead to increased CRFR1 expression, our findings strongly support an involvement of CRFR1 signaling in CRC aggressiveness, as previously shown in mouse models where CRFR1 showed a proinflammatory and a pro-tumorigenic effect in inflammation-related colon cancer [63]. In this context, a CRFR1-mediated CRC promoting pathway has also been described in CRC cells through modulation of IL-6/JAK2/STAT3 signaling and VEGF-induced angiogenesis [29], while CRFR1 hypermethylation in breast cancer models has been associated with the expression of steroid hormone receptors, a favorable prognostic factor [34]. Overall, our findings are quite novel, given that this is the first study reporting aberrant CRFR1 methylation in CRC, as per our knowledge.
Although we showed increased CRFR1 methylation levels in the adjuvant group compared to healthy individuals and metastatic group, we failed to establish any significant alterations in the methylation levels assessed in the control and metastatic study groups. At the same time, CRFR1 hypomethylation within the cancer groups (adjuvant and metastatic) revealed direct clinical relevance as it was positively correlated with adverse clinicopathological characteristics and poor outcomes. Based on the findings extrapolated by the two cancer groups, one could postulate that CRFR1 methylation might be a molecular event characterizing primary tumor onset which is significantly eliminated later, during tumor transition to a metastatic phase. Given the limitations of our in silico analysis in terms of the available patient study groups, we can further speculate that this trend of CRFR1 hypermethylation followed by hypomethylation as the disease progresses may be associated with the sample origin and thus be ccfDNA-specific. Our findings are novel, demonstrating that primary CRC is characterized by aberrant CRFR1 methylation which is progressively lost following disease aggressiveness and therefore the CRFR1 methylation levels may have a prognostic significance in CRC.
In silico analysis of CRFR2 methylation status revealed that all the emerged DMCs identified in transcription-related regions were hypermethylated in the CRC tissues compared to the healthy ones. This is in line with recent findings indicating positive correlation of hypermethylated CRFR2 (studied by an Infinium Human Methylation 450K array) with colitis-induced CRC [35]. Accordingly, the previously reported observations on diminished CRFR2 mRNA and protein expression in CRC cell lines and tissues [30] may be attributed to CRFR2 hypermethylation. We have further identified the CRFR2/Ucn2 signaling as a critical negative regulator in sustaining chronic inflammation and promoting cancer development and aggressiveness in the colon [24]. Based on the above findings we strongly suggest that CRFR2 methylation and expression levels in the colon tissues may be of prognostic significance in CRC management. In the CRC ccfDNA dataset, three more CpGs of CRFR2 were found hypermethylated. However, in our experimental part of the study, no statistically significant differences in CRFR2 methylation levels were observed between healthy and patient-derived ccfDNAs analyzed by qMSP, raising doubts about the clinical application in liquid biopsy.
Furthermore, methylation analysis in gene body intronic regions of CRFR1 and CRFR2 revealed lower methylation levels in both receptors in CRC compared to normal tissues. Gene body methylation is largely unexplored and very often connected to active transcription [64,65], which is known as the DNA methylation paradox [66]. Although gene body hypomethylation has been previously correlated to cancer [67], further targeted experimental analysis of gene body CRFR1 and CRFR2 methylation in relation to expression may be necessary in order to clarify their interconnection in our system.
Epigenetic modifications such as DNA methylation within CGIs are known to modulate the dynamic binding of transcription factors to regulatory elements, thus resulting in transcriptional repression [68]. In our study, we predicted 31 and 40 TFs that may regulate CRFR1 and CRFR2 transcription, respectively, through putative binding to DMC-containing CGIs. Therefore, any methylation events in these DMCs during malignant transformation may block TFs binding and transcriptional activities. Among the predicted TFs, NF kappa B, AP-2 and E2F have been previously shown to be sensitive to CpG methylation with consequent inhibition of their DNA binding activities [62]. In addition, p53 was also predicted, and CRF has been reported to inhibit cell proliferation and apoptosis in cell lines via the CRFR1-mediated p53 mechanism [69].
To further exploit our experimental observations and bring in a clinical perspective, we employed an autoML approach in order to build classifying models of diagnostic/prognostic performance. ML exploits a variety of algorithms to perform predictive analysis and its use in biomarker discovery in cancer is rapidly increasing [70,71]. Automated tools for ML promise to democratize data analysis for non-experts, increase productivity, improve replicability of the statistical analysis and shield against common methodological analysis errors such as overfitting [53]. We employed JADBio, an autoML platform designed for standard, best-practice and state-of-the-art statistical and machine learning methods. JADBio has previously been successfully used to produce signatures for clinical applications such as development of classifiers for metastatic BrCa based on novel ccfDNA methylation patterns [16], identification of risk of lung cancer in smokers [72] or suicide prediction among depressive patients [73]. Recently, by revisiting publicly available omics datasets via JADBio, we were able to deliver accurate highly-performing blood-based predictive biosignatures in Alzheimer’s disease [74] and in breast cancer [75].
In our study, JADBio analysis delivered a three-feature biosignature via the Classification Random Forests algorithm, including both CRF receptor methylation levels assessed in ccfDNA and age, with high-performance metrics in discriminating between CRC patients and healthy individuals. It has to be noted that although methylation of CRFR2 did not reach a statistically significant difference between groups when examined by standard univariate statistical analysis, multivariate analysis by JADBio selected it as a significant feature when combined with CRFR1 methylation and age. Previous ML-based studies performed in ccfDNA materials have also shown promising results in CRC. Wan et al. built an ML-based model for the early detection of CRC with an AUC of 0.92 (95% CI: 0.91–0.93) [76], while Luo et al. constructed a predictive model that accurately discriminated CRC patients from healthy individuals (AUC: 0.96) [77].
Overall, the identification of novel, noninvasive and high-performance biomarkers is of high importance for early, pre-asymptomatic CRC diagnosis. In this context, we strongly believe that our novel three-feature biosignature could potentially have a clinical application in early CRC screening. Upon perspective clinical validation, our model, built on qMSP rather than whole-genome deep-sequencing methodologies, may offer a feasible liquid biopsy-based solution for early CRC diagnosis that can be implemented in any standard molecular diagnostic laboratory. It is important to mention that for successful clinical implementation, a standard operational procedure for ccfDNA preanalytical preparation should be adopted between labs. However, limitations of our study, including the small number of participants in the available patient study groups, may be responsible for restricting putative correlations between CRFR1 and CRFR2 methylation levels and clinicopathological features such as tumor stage and metastasis. For future clinical implications, our preclinical model’s performance needs to be validated in a larger independent group of patients and confirmed in a prospective clinical study, which is the greatest challenge in translating such findings into clinical practice. Furthermore, studies employing in vitro models should clarify the exact timing of methylation events during the malignant transformation process and their results in receptor protein expression. Percentages of methylation detection in patient groups are a clear finding of our study without, however, some statistical significance. Based on our methodology, samples showing no methylation are unmethylated rather than unsuitable for methylation detection due to low sample quality or abundance. Given that ccfDNA is a valuable source for tracing molecular alterations of a tumor, detected changes in the methylation profile of important genes could reflect a dynamic tumor burden. Our results show that although the detection of CRFR1 and CRFR2 methylation in ccfDNA does not hold a diagnostic value, methylation levels considered in the context of an ML-built biosignature can help in differentiating healthy individuals from CRC patients.
5. Conclusions
CRC is one of the most lethal cancers due to a long asymptomatic phase and difficulties in diagnosis, prognosis and disease management. Our presented data introduce the methylation levels of both CRFR1 and CRFR2 as putative biomarkers in CRC based on our novel biosignature. The prognostic significance of the CRFR1 methylation status is further supported by the findings showing that decreased ccfDNA CRFR1 methylation levels among CRC patients are correlated with tumor aggressiveness and poor clinical outcomes. Along with our previously reported findings [20,26], we suggest here that the CRFR2 hypermethylation patterns in CRC determined by bioinformatic analysis may be associated with decreased receptor expression, which in turn contributes to CRC progression and metastatic potential in accordance with our previous findings. In addition, we demonstrate that the CRFR1 methylation levels assessed in liquid biopsy could offer a minimally invasive approach, overcoming current obstacles in CRC tissue methylation monitoring. To this end, a three-feature biosignature was built, which can help ensure accurate disease diagnosis upon prospective evaluation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jcm10122680/s1, Table S1: DMCs identified by in silico analysis in CRFR1 among the studied groups, Table S2: DMCs identified by in silico analysis in CRFR2 among the studied groups.
Author Contributions
Conceptualization, M.P., E.C. and S.B.; Methodology, M.P., A.C., M.K. and E.C.; Validation and formal analysis: M.P., A.C. and E.C.; Investigation, M.P., A.C., M.K., I.B., E.B. and E.C.; Data curation, M.P., I.B., E.B. and E.C.; Writing—review and editing, M.P., S.B., E.C., I.B., E.B., K.A., N.X. and S.K.; Visualization, M.P., E.C. and S.B.; Supervision: K.A., N.X., S.K., S.B. and E.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Special Account of Research Funds (ELKE) of the University of Crete (Grant No. KA 10419; PI: S.B.).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Democritus University of Thrace (protocol code 663/08-08-2016).
Informed Consent Statement
Informed consent was obtained from all the subjects involved in the study.
Data Availability Statement
Data are available upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Pellino G., Gallo G., Pallante P., Capasso R., De Stefano A., Maretto I., Malapelle U., Qiu S., Nikolaou S., Barina A., et al. Noninvasive Biomarkers of Colorectal Cancer: Role in Diagnosis and Personalised Treatment Perspectives. Gastroenterol. Res. Pract. 2018;2018:2397863. doi: 10.1155/2018/2397863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sammarco G., Gallo G., Vescio G., Picciariello A., De Paola G., Trompetto M., Currò G., Ammendola M. Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. J. Clin. Med. 2020;9:2852. doi: 10.3390/jcm9092852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garborg K., Holme Ø., Løberg M., Kalager M., Adami H.O., Bretthauer M. Current status of screening for colorectal cancer. Ann. Oncol. 2013;24:1963–1972. doi: 10.1093/annonc/mdt157. [DOI] [PubMed] [Google Scholar]
- 5.Kaminski M.F., Regula J., Kraszewska E., Polkowski M., Wojciechowska U., Didkowska J., Zwierko M., Rupinski M., Nowacki M.P., Butruk E. Quality indicators for colonoscopy and the risk of interval cancer. N. Engl. J. Med. 2010;362:1795–1803. doi: 10.1056/NEJMoa0907667. [DOI] [PubMed] [Google Scholar]
- 6.Rabeneck L., Paszat L.F., Saskin R. Endoscopist specialty is associated with incident colorectal cancer after a negative colonoscopy. Clin. Gastroenterol. Hepatol. 2010;8:275–279. doi: 10.1016/j.cgh.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Brenner H., Stock C., Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: Systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ Clin. Res. Ed. 2014;348:g2467. doi: 10.1136/bmj.g2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín-López J.E., Beltrán-Calvo C., Rodríguez-López R., Molina-López T. Comparison of the accuracy of CT colonography and colonoscopy in the diagnosis of colorectal cancer. Colorectal Dis. 2014;16:O82–O89. doi: 10.1111/codi.12506. [DOI] [PubMed] [Google Scholar]
- 9.Lin O.S.-T. Computed tomographic colonography: Hope or hype? World J. Gastroenterol. 2010;16:915–920. doi: 10.3748/wjg.v16.i8.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lansdorp-Vogelaar I., van Ballegooijen M., Boer R., Zauber A., Habbema J.D. A novel hypothesis on the sensitivity of the fecal occult blood test: Results of a joint analysis of 3 randomized controlled trials. Cancer. 2009;115:2410–2419. doi: 10.1002/cncr.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imperiale T.F., Ransohoff D.F., Itzkowitz S.H., Levin T.R., Lavin P., Lidgard G.P., Ahlquist D.A., Berger B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 12.Bhardwaj M., Terzer T., Schrotz-King P., Brenner H. Comparison of Proteomic Technologies for Blood-Based Detection of Colorectal Cancer. Int. J. Mol. Sci. 2021;22:1189. doi: 10.3390/ijms22031189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pantel K., Alix-Panabières C. Real-time liquid biopsy in cancer patients: Fact or fiction? Cancer Res. 2013;73:6384–6388. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 14.Alix-Panabières C., Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016;6:479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 15.Panagopoulou M., Esteller M., Chatzaki E. Circulating Cell-Free DNA in Breast Cancer: Searching for Hidden Information towards Precision Medicine. Cancers. 2021;13:728. doi: 10.3390/cancers13040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panagopoulou M., Karaglani M., Balgkouranidou I., Biziota E., Koukaki T., Karamitrousis E., Nena E., Tsamardinos I., Kolios G. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38:3387–3401. doi: 10.1038/s41388-018-0660-y. [DOI] [PubMed] [Google Scholar]
- 17.Panagopoulou M., Karaglani M., Balgkouranidou I., Pantazi C., Kolios G., Kakolyris S., Chatzaki E. Circulating cell-free DNA release in vitro: Kinetics, size profiling, and cancer-related gene methylation. Cell. Physiol. 2019;234:14079–14089. doi: 10.1002/jcp.28097. [DOI] [PubMed] [Google Scholar]
- 18.Roy D., Tiirikainen M. Diagnostic Power of DNA Methylation Classifiers for Early Detection of Cancer. Trends Cancer. 2020;6:78–81. doi: 10.1016/j.trecan.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatzaki E., Crowe P.D., Wang L., Million M., Taché Y., Grigoriadis D.E. CRF receptor type 1 and 2 expression and anatomical distribution in the rat colon. J. Neurochem. 2004;90:309–316. doi: 10.1111/j.1471-4159.2004.02490.x. [DOI] [PubMed] [Google Scholar]
- 20.Chatzaki E., Lambropoulou M., Constantinidis T.C., Papadopoulos N., Taché Y., Minopoulos G., Grigoriadis D.E. Corticotropin-releasing factor (CRF) receptor type 2 in the human stomach: Protective biological role by inhibition of apoptosis. J. Cell. Physiol. 2006;209:905–911. doi: 10.1002/jcp.20792. [DOI] [PubMed] [Google Scholar]
- 21.Larauche M., Kiank C., Tache Y. Corticotropin releasing factor signaling in colon and ileum: Regulation by stress and pathophysiological implications. J. Physiol. Pharmacol. 2009;60(Suppl. 7):33–46. [PMC free article] [PubMed] [Google Scholar]
- 22.Chatzaki E., Murphy B.J., Wang L., Million M., Ohning G.V., Crowe P.D., Petroski R., Taché Y., Grigoriadis D.E. Differential profile of CRF receptor distribution in the rat stomach and duodenum assessed by newly developed CRF receptor antibodies. J. Neurochem. 2004;88:1–11. doi: 10.1046/j.1471-4159.2003.02078.x. [DOI] [PubMed] [Google Scholar]
- 23.Chatzaki E., Charalampopoulos I., Leontidis C., Mouzas I.A., Tzardi M., Tsatsanis C., Margioris A.N., Gravanis A. Urocortin in human gastric mucosa: Relationship to inflammatory activity. J. Clin. Endocrinol. Metab. 2003;88:478–483. doi: 10.1210/jc.2002-020853. [DOI] [PubMed] [Google Scholar]
- 24.Baritaki S., de Bree E., Chatzaki E., Pothoulakis C. Chronic Stress, Inflammation, and Colon Cancer: A CRH System-Driven Molecular Crosstalk. J. Clin. Med. 2019;8:1669. doi: 10.3390/jcm8101669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paschos K.A., Kolios G., Chatzaki E. The corticotropin-releasing factor system in inflammatory bowel disease: Prospects for new therapeutic approaches. Drug Discov. Today. 2009;14:713–720. doi: 10.1016/j.drudis.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Yuan P.Q., Wu S.V., Elliott J., Anton P.A., Chatzaki E., Million M., Taché Y. Expression of corticotropin releasing factor receptor type 1 (CRF1) in the human gastrointestinal tract and upregulation in the colonic mucosa in patients with ulcerative colitis. Peptides. 2012;38:62–69. doi: 10.1016/j.peptides.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatzaki E., Anton P.A., Million M., Lambropoulou M., Constantinidis T., Kolios G., Taché Y., Grigoriadis D.E. Corticotropin-releasing factor receptor subtype 2 in human colonic mucosa: Down-regulation in ulcerative colitis. World J. Gastroenterol. 2013;19:1416–1423. doi: 10.3748/wjg.v19.i9.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatoo M., Li Y., Ma Z., Coote J., Du J., Chen X. Involvement of Corticotropin-Releasing Factor and Receptors in Immune Cells in Irritable Bowel Syndrome. Front. Endocrinol. 2018;9 doi: 10.3389/fendo.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang X., Hong Y., Dai L., Qian Y., Zhu C., Wu B., Li S. CRH promotes human colon cancer cell proliferation via IL-6/JAK2/STAT3 signaling pathway and VEGF-induced tumor angiogenesis. Mol. Carcinog. 2017;56:2434–2445. doi: 10.1002/mc.22691. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez J.A., Huerta-Yepez S., Law I.K., Baay-Guzman G.J., Tirado-Rodriguez B., Hoffman J.M., Iliopoulos D., Hommes D.W., Verspaget H.W., Chang L., et al. Diminished expression of CRHR2 in human colon cancer promotes tumor growth and EMT via persistent IL-6/Stat3 signaling. Cell. Mol. Gastroenterol. Hepatol. 2015;1:610–630. doi: 10.1016/j.jcmgh.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaprara A., Pazaitou-Panayiotou K., Kortsaris A., Chatzaki E. The corticotropin releasing factor system in cancer: Expression and pathophysiological implications. Cell. Mol. Life Sci. 2010;67:1293–1306. doi: 10.1007/s00018-010-0265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pape J.C., Carrillo-Roa T., Rothbaum B.O., Nemeroff C.B., Czamara D., Zannas A.S., Iosifescu D., Mathew S.J., Neylan T.C., Mayberg H.S., et al. DNA methylation levels are associated with CRF(1) receptor antagonist treatment outcome in women with post-traumatic stress disorder. Clin. Epigenetics. 2018;10:136. doi: 10.1186/s13148-018-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schartner C., Ziegler C., Schiele M.A., Kollert L., Weber H., Zwanzger P., Arolt V., Pauli P., Deckert J., Reif A., et al. CRHR1 promoter hypomethylation: An epigenetic readout of panic disorder? Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 2017;27:360–371. doi: 10.1016/j.euroneuro.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Koureta M., Karaglani M., Panagopoulou M., Balgkouranidou I., Papadaki-Anastasopoulou A., Zarouchlioti C., Dekavallas S., Kolios G., Lambropoulou M., Baritaki S., et al. Corticotropin Releasing Factor Receptors in breast cancer: Expression and activity in hormone-dependent growth in vitro. Peptides. 2020;129:170316. doi: 10.1016/j.peptides.2020.170316. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi M., Matsubara N., Nakachi Y., Okazaki Y., Uchino M., Ikeuchi H., Song J., Kimura K., Yasuhara M., Babaya A., et al. Hypermethylation of Corticotropin Releasing Hormone Receptor-2 Gene in Ulcerative Colitis Associated Colorectal Cancer. In Vivo. 2020;34:57–63. doi: 10.21873/invivo.11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pothoulakis C., Torre-Rojas M., Duran-Padilla M.A., Gevorkian J., Zoras O., Chrysos E., Chalkiadakis G., Baritaki S. CRHR2/Ucn2 signaling is a novel regulator of miR-7/YY1/Fas circuitry contributing to reversal of colorectal cancer cell resistance to Fas-mediated apoptosis. Int. J. Cancer. 2018;142:334–346. doi: 10.1002/ijc.31064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Yim A.Y.F., de Bruyn J.R., Duijvis N.W., Sharp C., Ferrero E., de Jonge W.J., Wildenberg M.E., Mannens M., Buskens C.J., D’Haens G.R., et al. A distinct epigenetic profile distinguishes stenotic from non-inflamed fibroblasts in the ileal mucosa of Crohn’s disease patients. PLoS ONE. 2018;13:e0209656. doi: 10.1371/journal.pone.0209656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishak M., Baharudin R., Rose I.M., Sagap I., Mazlan L., Azman Z.A.M., Abu N., Jamal R., Lee L.H., Mutalib N.S.A. Genome-Wide Open Chromatin Methylome Profiles in Colorectal Cancer. Biomolecules. 2020;10:719. doi: 10.3390/biom10050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moss J., Magenheim J., Neiman D., Zemmour H., Loyfer N., Korach A., Samet Y., Maoz M., Druid H., Arner P., et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018;9:5068. doi: 10.1038/s41467-018-07466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim T.O., Park D.I., Han Y.K., Kang K., Park S.G., Park H.R., Yi J.M. Genome-Wide Analysis of the DNA Methylation Profile Identifies the Fragile Histidine Triad (FHIT) Gene as a New Promising Biomarker of Crohn’s Disease. J. Clin. Med. 2020;9:1338. doi: 10.3390/jcm9051338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller F., Scherer M., Assenov Y., Lutsik P., Walter J., Lengauer T., Bock C. RnBeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biol. 2019;20:55. doi: 10.1186/s13059-019-1664-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Triche T.J., Jr., Weisenberger D.J., Van Den Berg D., Laird P.W., Siegmund K.D. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du P., Zhang X., Huang C.C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assenov Y., Müller F., Lutsik P., Walter J., Lengauer T., Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L.-C., Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 49.Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 50.Panagopoulou M., Lambropoulou M., Balgkouranidou I., Nena E., Karaglani M., Nicolaidou C., Asimaki A., Konstantinidis T., Constantinidis T.C., Kolios G., et al. Gene promoter methylation and protein expression of BRMS1 in uterine cervix in relation to high-risk human papilloma virus infection and cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017;39:1010428317697557. doi: 10.1177/1010428317697557. [DOI] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 53.Tsamardinos I., Charonyktakis P., Lakiotaki K., Borboudakis G., Zenklusen J.C., Juhl H., Chatzaki E., Lagani V. Just Add Data: Automated Predictive Modeling and BioSignature Discovery. bioRxiv. 2020 doi: 10.1101/2020.05.04.075747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsamardinos I., Greasidou E., Borboudakis G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018;107:1895–1922. doi: 10.1007/s10994-018-5714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boser B.E., Guyon I.M., Vapnik V.N. A training algorithm for optimal margin classifiers; Proceedings of the Fifth Annual Workshop on Computational Learning Theory; Pittsburgh, PA, USA. 27–29 July 1992; pp. 144–152. [Google Scholar]
- 56.Hoerl A.E., Kennard R.W. Ridge Regression: Biased Estimation for Nonorthogonal Problems. Technometrics. 1970;12:55–67. doi: 10.1080/00401706.1970.10488634. [DOI] [Google Scholar]
- 57.Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 58.Moore II D.H. In: Classification and Regression Trees. Breiman L., Friedman J.H., Olshen R.A., Stone C.J., editors. Volume 8. Brooks/Cole Publishing; Monterey, CA, USA: 1987. pp. 534–535. [DOI] [Google Scholar]
- 59.Brenet F., Moh M., Funk P., Feierstein E., Viale A.J., Socci N.D., Scandura J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner J.R., Busche S., Ge B., Kwan T., Pastinen T., Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tate P.H., Bird A.P. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437X(93)90027-M. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y., Fang X., Yuan J., Sun Z., Li C., Li R., Li L., Zhu C., Wan R., Guo R., et al. The role of corticotropin-releasing hormone receptor 1 in the development of colitis-associated cancer in mouse model. Endocr. Relat. Cancer. 2014;21:639–651. doi: 10.1530/ERC-14-0239. [DOI] [PubMed] [Google Scholar]
- 64.Jjingo D., Conley A.B., Yi S.V., Lunyak V.V., Jordan I.K. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3:462–474. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aran D., Toperoff G., Rosenberg M., Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011;20:670–680. doi: 10.1093/hmg/ddq513. [DOI] [PubMed] [Google Scholar]
- 66.Jones P.A. The DNA methylation paradox. Trends Genet. TIG. 1999;15:34–37. doi: 10.1016/S0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 67.Kulis M., Heath S., Bibikova M., Queirós A.C., Navarro A., Clot G., Martínez-Trillos A., Castellano G., Brun-Heath I., Pinyol M., et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 68.Héberlé É., Bardet A.F. Sensitivity of transcription factors to DNA methylation. Essays Biochem. 2019;63:727–741. doi: 10.1042/EBC20190033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Y., Wang L., Liu X., Wu Q., Zhang H., Hu F., Sun X. Human corticotrophin releasing factor inhibits cell proliferation and promotes apoptosis through upregulation of tumor protein p53 in human glioma. Oncol. Lett. 2018;15:8378–8386. doi: 10.3892/ol.2018.8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kourou K., Exarchos T.P., Exarchos K.P., Karamouzis M.V., Fotiadis D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chatzaki E., Tsamardinos I. Somatic copy number aberrations detected in circulating tumor DNA can hold diagnostic value for early detection of hepatocellular carcinoma. EBioMedicine. 2020;57:102851. doi: 10.1016/j.ebiom.2020.102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Markaki M., Tsamardinos I., Langhammer A., Lagani V., Hveem K., Røe O.D. A Validated Clinical Risk Prediction Model for Lung Cancer in Smokers of All Ages and Exposure Types: A HUNT Study. EBioMedicine. 2018;31:36–46. doi: 10.1016/j.ebiom.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adamou M., Antoniou G., Greasidou E., Lagani V., Charonyktakis P., Tsamardinos I., Doyle M. Toward Automatic Risk Assessment to Support Suicide Prevention. Crisis. 2019;40:249–256. doi: 10.1027/0227-5910/a000561. [DOI] [PubMed] [Google Scholar]
- 74.Karaglani M., Gourlia K., Tsamardinos I., Chatzaki E. Accurate Blood-Based Diagnostic Biosignatures for Alzheimer’s Disease via Automated Machine Learning. J. Clin. Med. 2020;9:3016. doi: 10.3390/jcm9093016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panagopoulou M., Karaglani M., Manolopoulos V.G., Iliopoulos I., Tsamardinos I., Chatzaki E. Deciphering the Methylation Landscape in Breast Cancer: Diagnostic and Prognostic Biosignatures through Automated Machine Learning. Cancers. 2021;13:1677. doi: 10.3390/cancers13071677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wan N., Weinberg D., Liu T.-Y., Niehaus K., Ariazi E.A., Delubac D., Kannan A., White B., Bailey M., Bertin M., et al. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. 2019;19:832. doi: 10.1186/s12885-019-6003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo H., Zhao Q., Wei W., Zheng L., Yi S., Li G., Wang W., Sheng H., Pu H., Mo H., et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. J. Sci. Transl. Med. 2020;12:eaax7533. doi: 10.1126/scitranslmed.aax7533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request.