Abstract
Moraxella catarrhalis (M. catarrhalis) is a Gram-negative bacterium that can cause serious respiratory tract infections and middle ear infections in children and adults. M. catarrhalis has demonstrated an increasing rate of antibiotic resistance in the last few years, thus development of an effective vaccine is a major health priority. We report here a novel designed multitope vaccine based on the mapped epitopes of the vaccine candidates filtered out of the whole proteome of M. catarrhalis. After analysis of 1615 proteins using a reverse vaccinology approach, only two proteins (outer membrane protein assembly factor BamA and LPS assembly protein LptD) were nominated as potential vaccine candidates. These proteins were found to be essential, outer membrane, virulent and non-human homologs with appropriate molecular weight and high antigenicity score. For each protein, cytotoxic T lymphocyte (CTL), helper T lymphocyte (HTL) and B cell lymphocyte (BCL) epitopes were predicted and confirmed to be highly antigenic and cover conserved regions of the proteins. The mapped epitopes constituted the base of the designed multitope vaccine where suitable linkers were added to conjugate them. Additionally, beta defensin adjuvant and pan-HLA DR-binding epitope (PADRE) peptide were also incorporated into the construct to improve the stimulated immune response. The constructed multitope vaccine was analyzed for its physicochemical, structural and immunological characteristics and it was found to be antigenic, soluble, stable, non-allergenic and have a high affinity to its target receptor. Although the in silico analysis of the current study revealed that the designed multitope vaccine has the ability to trigger a specific immune response against M. catarrhalis, additional translational research is required to confirm the effectiveness of the designed vaccine.
Keywords: Moraxella catarrhalis, vaccinomics, reverse vaccinology, immunoinformatics, epitope mapping, multitope vaccine
1. Introduction
Generally, M. catarrhalis is known to be a human-restricted commensal, but studies in recent decades have proved that it has become a major infectious agent in both the upper and lower respiratory tract and the causative agent of about 17% of acute otitis media infections in children [1]. Furthermore, M. catarrhalis was found to cause other types of infections, such as bacteremia, endocarditis and meningitis, especially in immunocompromised patients [2].
Usage of antibiotics without restrictions has contributed largely to the development of resistant bacterial strains which in turn shorten the list of available effective antimicrobial agents [3]. The resistance rates are rapidly increasing, particularly with regard to first-line antimicrobial agents. An alternative solution is the development of an effective vaccine; while several predicted vaccines against M. catarrhalis are in different stages of development, there is no FDA-approved one to date [4]. Adhesive proteins of M. catarrhalis were extensively targeted as potential vaccine candidates. They are outer membrane proteins that allow the microorganism to bind to its target receptor in the human respiratory tract, which is the first step in bacterial pathogenesis [5]. Application of this vaccine in animal models showed an enhanced lung clearance of bacteria [6].
Vaccine development through traditional approaches has the disadvantages of being expensive, time consuming and seldom giving successful outcomes [7]. Recent development in the fields of bioinformatics, immunoinformatics and structural vaccinomics has revolutionized the process of antigen screening. By merging these techniques, a new approach of vaccine development called reverse vaccinology was created [8] and applied for vaccine development against many bacterial and viral pathogens [9]. Recently, it was demonstrated that a multitope vaccine showed superior efficacy and protection against infectious agents, more so than single epitope-based vaccines [10].
In the current study, the reverse vaccinology approach has been applied to filter the complete proteome of M. catarrhalis and nominate potential vaccine candidates. This approach was chosen not only because of its advantages, but also because it selects the candidates from extracellular and outer membrane proteins which demonstrated promising results in animal models in previous trials. Additionally, T and B cell epitopes were mapped for the filtered proteins and the top-ranked epitopes were applied to construct a multitope vaccine which was analyzed according to its structural and immunological characteristics to support its nomination as a promising vaccine candidate against M. catarrhalis.
2. Materials and Methods
The flow of work started with the application of the reverse vaccinology approach to select which proteins would be our vaccine candidates, then B and T cell epitopes were mapped for these candidates and, following that, the multitope vaccine was constructed and, finally, it was analyzed for its physicochemical, structural and immunological characteristics.
2.1. Data Retrieval and Proteome Analysis
The whole proteome of M. catarrhalis BBH18 was retrieved (GenBank assembly accession no: GCA_000092265.1). This strain was selected as it represents the reference M. catarrhalis strain in NCBI. The reverse vaccinology approach was applied to perform protein prioritization on the whole proteome of M. catarrhalis. Firstly, essential proteins, without which M. catarrhalis cannot survive, were detected using the Geptop 2.0 webserver with an essentiality cutoff of 0.24. This server detects essential proteins through comparing the orthology and phylogeny of query proteins against the datasets defined experimentally in the database of essential genes (DEG) [11]. Following that, the PSORTb v3.0.2 online server [12] was used to analyze the subcellular localization of the essential proteins. The final outcome of this filtration step was the detection of exoproteome and secretome essential proteins. These proteins were applied in VICMpred [13] to estimate the virulence potential. Proteins filtered out from the previous step were searched using BLASTp from NCBI against the human proteome and proteins with ≥35% identity were excluded as they were considered human homologs. The presence of transmembrane helices in the proteins from the previous step was detected by TMHMM [14], while protein molecular weight was predicted on the Expasy webserver [15]. Proteins with a molecular weight of less than 110 kDa and with ≤1 transmembrane helix were moved to the final filtration step of antigenicity detection by Vaxijen V 2.0 [16]. After all the steps of proteome mining, top proteins that covered all the previous requirements and with the highest antigenicity score were nominated as the potential recombinant vaccine candidates of the study and their protein–protein interactions were assessed using the STRING webserver. Finally, they moved to the epitope mapping stage to construct the multitope vaccine.
2.2. T and B Cell Epitope Prediction
Before epitope prediction, the amino acid sequence of filtered proteins was submitted to the SignalP-5.0 Server to predict the presence and location of signal peptides. Following that, the obtained mature polypeptides were analyzed using the prediction tools in the Immune Epitope Database (IEBD). MHC-I binding was detected by applying NetMHCpan EL 4.0 as a prediction method and an HLA allele reference set which provides >97% in terms of population coverage [17], while IEBD recommended a 2.22 prediction method and a full HLA reference set that covers >99% in terms of population coverage [18] for analysis of MHC-II binding. Antibody epitope prediction was also performed with IEBD. Finally, the conservation profile of each selected epitope was analyzed through multiple sequence alignment available with Clustal Omega, while allergenicity assessment and toxicity analysis were performed with AllerTOP and ToxinPred servers, respectively.
2.3. Multitope Vaccine Construction
The designed vaccine started with a β defensin adjuvant, then top-ranked CTL, HTL and BCL of the nominated proteins were incorporated using GGGS, GPGPG and KK linkers, respectively. These linkers were added to make sure that successful separation in vivo was obtained [19]. The PADRE sequence was also added to our designed vaccine, and before moving to the next step, the constructed vaccine was confirmed to be antigenic and non-allergic using VaxiJen v2.0 and AllerTOP servers, respectively.
2.4. Physicochemical Characterization, Protein Solubility Assessment and Secondary Structure Prediction
ProtParam, a tool available on the Expasy server, was used for physicochemical characterization of the constructed vaccine. This tool detects many physicochemical characteristics of uploaded amino acids, such as molecular weight and isoelectric pH, in addition to several physicochemical properties. The propensity of the constructed vaccine upon overexpression in E. coli was assessed by SOLpro while the protein secondary structure was predicted on the PSIPRED server.
2.5. Tertiary Structure Prediction, Refinement and Validation
Protein tertiary structure prediction servers perform bending and twisting to generate a protein molecule with the lowest energy state and maximum stability. In the current study, 3Dpro [20] was employed for this purpose. 3Dpro performs its 3D modeling through analysis of the structural similarity between the query protein and the data available on PDB. The GalaxyRefine server [21] was used to refine the protein tertiary structure predicted by 3Dpro to enhance its accuracy, where the refined models were assessed by Ramachandran plot analysis [22] and ProSA [23].
2.6. Disulfide Engineering of the Designed Vaccine
Before moving to the next step and starting docking analysis, it is important to improve the designed model stability. Disulfide bonds improve protein geometric conformation and significantly increase their stability. Disulfide by Design 2.0 [24] was used to assign such bonds for the designed vaccine.
2.7. Docking of Designed Vaccine with TLR2
Molecular docking was applied to investigate the preferred orientation of the ligand to its corresponding receptor and analyze the binding affinity [25]. Inflammation triggered by M. catarrhalis mainly involves TLR2 [26]. Hence, TLR2 (PDB id: 2Z7X) was selected as a docking receptor while the refined model of vaccine tertiary structure was used as a ligand. Molecular docking was studied using the ClusPro 2.0 server [27].
2.8. Molecular Dynamics Simulation
Molecular dynamics is a computational approach that was applied in order to describe the molecules’ behavior as well as measure the stability of protein–protein complexes [28]. We employed the iMODS server to study and analyze the interaction between the designed vaccine and its receptor as it has the advantage of being fast and efficient [29]. This tool estimates the direction and range of the basic motions of the protein–ligand complex by measuring four main factors: deformability, eigenvalues, B-factors and covariance. Generally, deformation is much harder when the eigenvalue is high [30].
2.9. Reverse Translation and Codon Adaptation
E. coli k-12 was selected as the expression host of the recombinant vaccine and, to accelerate the rate of protein expression, codon adaptation was performed to adapt the codon to the chosen expression host. There are no similarities between the codon usage of humans and E. coli and, consequently, codon adaptation was performed to get a high expression rate of vaccine protein in the nominated host. The JCAT server [31] was employed for this process.
3. Results
3.1. In Silico Proteome Analysis
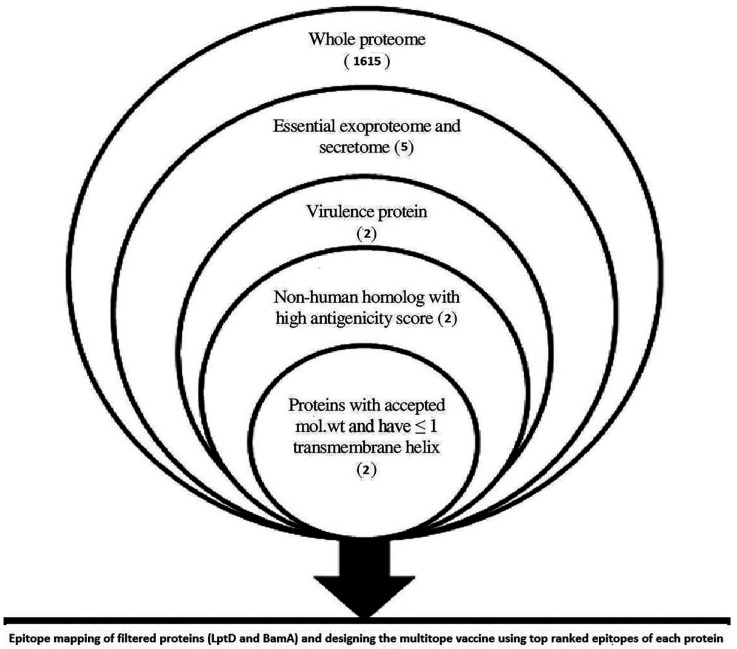
The total number of proteins of M. catarrhalis BBH18 was found to be 1615, and 395 of them were predicted as essential proteins. Subcellular localization of essential proteins demonstrated that two proteins, nucleoside-diphosphate kinase and tRNA (adenosine(37)-N6)-threonylcarbamoyltransferase complex transferase subunit TsaD, were located extracellularly while three proteins, LPS assembly protein LptD, outer membrane protein assembly factor BamA and outer membrane protein assembly factor BamD, were outer membrane proteins. Virulence assessment of these proteins revealed that only LPS assembly protein LptD and outer membrane protein assembly factor BamA were virulent, hence other protein filtration steps were performed only for these proteins (Figure 1) where analysis results (Table 1) confirmed their nomination as potential vaccine candidates of the current study.
Figure 1.
Overview of applied filtration steps for the nomination of potential vaccine candidates by applying reverse vaccinology technique.
Table 1.
Characteristics of potential vaccine candidates for M. catarrhalis BBH18.
| Protein Name | PSORb Result |
Essential | Virulent | Non-Human Homolog |
Molecular Weight (KDa) |
Transmembrane Helices |
Antigenicity Score |
|---|---|---|---|---|---|---|---|
| LptD | Outer membrane | √ | √ | √ | 104 | 1 | 0.59 |
| BamA | Outer membrane | √ | √ | √ | 90.9 | 0 | 0.52 |
3.2. Protein–Protein Interaction (PPI) Analysis
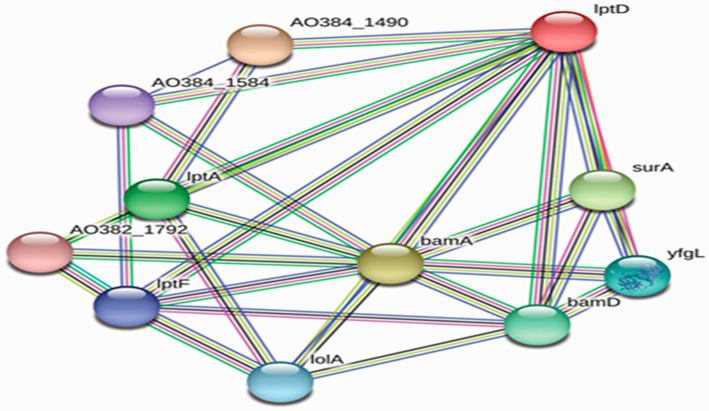
Interactions between protein candidates and other M. catarrhalis proteins were demonstrated using the STRING database (Figure 2). LptD and BamA were predicted to interact with a few other M. catarrhalis proteins and also each other. LptD was found to interact with other lipopolysaccharide assembly proteins such as LptF and LptA to form the lipopolysaccharide export system, while outer membrane protein assembly factor BamA interacts with BamD to constitute the outer membrane protein assembly complex. Both LptD and BamA were found to interact with other proteins (such as chaperone SurA and lipoprotein yfgL precursor) that are involved in the correct folding and assembly of outer membrane proteins.
Figure 2.
Protein–protein interaction network of vaccine candidates, estimated by STRING database.
3.3. T Cell Epitopes
For both MHC-I and MHC-II peptides, the top 1% predicted epitopes for each protein were analyzed according to antigenicity (estimated by VaxiJen v. 2.0), allergenicity (estimated by AllerTOP v. 2.0) and toxicity (estimated by ToxinPred), and the top 10 peptides that had the lowest percentile rank and antigenicity score >0.4 are listed in Table 2 (for MHC-I peptides) and Table 3 (for MHC-II peptides).
Table 2.
Top-ranked T cell epitopes (MHC-I peptides) of BamA and LptD proteins.
| BamA | LptD | ||||||
|---|---|---|---|---|---|---|---|
| Epitope | Antigenicity | Allergenicity | Toxicity | Epitope | Antigenicity | Allergenicity | Toxicity |
| YSAGVGATW | 1.16 | None | None | AELSGNVIM | 0.62 | Allergen | None |
| GEVVGGNAL | 1.42 | None | None | FEISTPYYL | 1.29 | Allergen | None |
| LTQDKQLRY | 0.73 | Allergen | None | RPYARLPQL | 0.41 | None | None |
| RYSAGVGATW | 1.31 | Allergen | None | RSRIQFDHTW | 0.49 | None | None |
| SETREVYSL | 0.78 | Allergen | None | SEYRLQHVM | 0.71 | Allergen | None |
| AEINFEGNRL | 0.95 | None | None | YEQLLNNNW | 0.48 | Allergen | None |
| VQFQIGSVF | 0.6 | None | None | SSRSSGLAW | 0.78 | None | None |
| AEEGFSQAM | 0.58 | None | None | SYEQLLNNNW | 0.41 | Allergen | None |
| IETELTNQY | 0.93 | Allergen | None | VFYLPYFNF | 2.99 | Allergen | None |
| DSYGGSLSY | 1.34 | None | None | FTASYPLLR | 0.93 | None | None |
Table 3.
Top-ranked T cell epitopes (MHC-II peptides) of BamA and LptD proteins.
| BamA | LptD | ||||||
|---|---|---|---|---|---|---|---|
| Epitope | Antigenicity | Allergenicity | Toxicity | Epitope | Antigenicity | Allergenicity | Toxicity |
| VDVEYYIDPVHPVYV | 0.63 | None | None | DTGRAIAKNTTLRIK | 0.5 | Allergen | None |
| DVEYYIDPVHPVYVR | 0.56 | Allergen | None | GRAIAKNTTLRIKKV | 0.4 | None | None |
| TVDVEYYIDPVHPVY | 0.44 | None | None | TGRAIAKNTTLRIKK | 0.43 | Allergen | None |
| VEYYIDPVHPVYVRR | 0.41 | None | None | STPYYLNLAPNYDAT | 0.91 | Allergen | None |
| TNPYFTVNGVSQSLS | 0.77 | Allergen | None | TPYYLNLAPNYDATI | 0.98 | Allergen | None |
| NPYFTVNGVSQSLSG | 1.02 | Allergen | None | RIKKVPVFYLPYFNF | 1.86 | Allergen | None |
| PYFTVNGVSQSLSGY | 0.92 | Allergen | None | VSYRYIDKMGRTRFE | 0.56 | Allergen | None |
| MTNPYFTVNGVSQSL | 0.75 | None | None | AVSYRYIDKMGRTRF | 0.45 | None | None |
| RPLLTQDKQLRYSAG | 0.6 | Allergen | None | DYNLDYVMDSLMGLN | 0.51 | None | None |
| ALATFGSELILPLPF | 0.99 | None | None | DANYLSDFNALGVES | 0.46 | None | None |
3.4. Construction of Multitope Vaccine with Physiochemical Property Assessment and Secondary Structure Prediction
The designed vaccine consisted of six CTL epitopes, six HTL epitopes and six BCL epitopes (three epitopes for each protein) linked in a sequential manner. Epitopes were selected from Table 2, Table 3 and Table 4 based on their characteristics, including epitopes that were non-allergic, non-toxic, conserved, covered different spots in the protein sequence and highly antigenic, were chosen for the multitope vaccine construction. In addition to that, the beta defensin adjuvant and PADRE peptide sequence were also added, resulting in a multiepitope vaccine that was 378 amino acids in length and its sequence was as follows.
Table 4.
Predicted B cell epitopes of BamA and LptD proteins.
| BamA | LptD | ||||||
|---|---|---|---|---|---|---|---|
| Epitope | Antigenicity | Allergenicity | Toxicity | Epitope | Antigenicity | Allergenicity | Toxicity |
| TGNFKTQDEV | 1.06 | Allergen | None | DGGASDHSAGI | 1.82 | Allergen | None |
| RREMRQLEGALASNQKIQ | 0.6 | None | None | KDQQYHDKD | 0.81 | None | None |
| RKTKYDNKNISNY | 0.7 | None | None | KKSIKDNSEPEKSG | 0.96 | Allergen | None |
| DLTVGFGDKT | 0.85 | None | None | SYDEDSLADQNIAKKNGR | 0.96 | None | None |
| LNKKQNDQT | 1.53 | None | None | APFGMHQDT | 0.42 | Allergen | None |
| HIRVGINDSESYSSRSS | 1.32 | None | None | ||||
| RKENRAFNQSAL | 0.43 | None | None | ||||
| YDYNLDYVMDSLM | 0.62 | None | None | ||||
| YRDAFNPHLSPD | 1.09 | None | None | ||||
“EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRCCRRKKEAAAKAKFVAAWTLKAAAGGGSYSAGVGATWGGGSAEINFEGNRLGGGSDSYGGSLSGGGSRPYARLPQLGGGSRSRIQFDHTWGGGSFTASYPLLRGPGPGVDVEYYIDPVHPVYVGPGPGMTNPYFTVNGVSQSLGPGPGALATFGSELILPLPFGPGPGAVSYRYIDKMGRTRFGPGPGDYNLDYVMDSLMGLNGPGPGDANYLSDFNALGVESKKRKTKYDNKNISNYKKDLTVGFGDKTKKLNKKQNDQTKKSYDEDSLADQNIAKKNGRKKHIRVGINDSESYSSRSSKKYRDAFNPHLSPDKKAKFVAAWTLKAAAGGGS”
The final construction was found to be non-allergenic, soluble with a SOLpro SVM score of 0.88 (SOLpro values > 0.5 are considered soluble) and antigenic with a Vaxijen antigenicity score of 1.07. The physiochemical properties of our construct were detected by using the ProtParam online tool (Table 5). Constructed vaccine secondary structure prediction demonstrated the presence of 18% he-lix, 19% strand and 63% coil structure (Figure 3).
Table 5.
Physicochemical characteristics of the designed multitope vaccine.
| Physicochemical Characteristic | Molecular Weight | Theoretical pI | Extinction Coefficient | GRAVY | Instability Index | Aliphatic Index |
|---|---|---|---|---|---|---|
| Score | 40.2 kDa | 9.71 | 53,290 M−1 cm−1 | −0.625 | 37.3 | 59.68 |
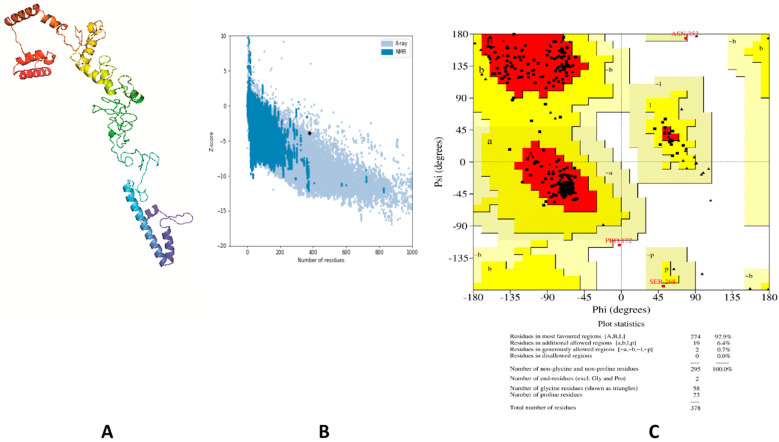
Figure 3.
Secondary structure prediction of designed multitope vaccine using PESIPRED server.
3.5. Tertiary Structure Prediction, Refinement and Validation
Structure validation of the primary structure, modeled by 3Dpro, showed that 87.5%, 12.2% and 0.3% of residues were located in favored, allowed and disallowed regions, respectively (estimated by Ramachandran plot analysis). In addition to that, the Z-score (estimated by ProSA webserver) of the same model was −3.2. These outcomes revealed that more refinement is required for this initial structure. Protein 3D structure refinement was performed using GalaxyRefine, where the best model (Figure 4A) showed the following enhancement: the Z-score improved from −3.2 to −3.88 (Figure 4B) and the Ramachandran plot analysis improved to show 92.9%, 7.1% and 0% of residues in favored, allowed and outlier regions, respectively (Figure 4C).
Figure 4.
Structural analysis of the designed vaccine. (A) The three-dimensional structure of the vaccine obtained after molecular refinements; (B) ProSA-web evaluation of the vaccine structure; (C) Ramachandran plot analysis of the protein structure after molecular refinements.
3.6. Vaccine Disulfide Engineering
Disulfide engineering was performed to stabilize the designed model of the vaccine. Regarding our vaccine, it was found that 29 pairs of amino acids had the capacity to make disulfide bonds by the DbD2 server but after considering other parameters, such as energy and chi3 value, only two pairs were recommended for mutation with cysteine. Hence, a total of four mutations were generated at the residue pairs, namely GLY36–ARG43 and PRO190–LEU202. The accepted values of energy and chi3 were less than 2.2 and −87: +97, respectively.
3.7. Molecular Docking of Vaccine with TLR2
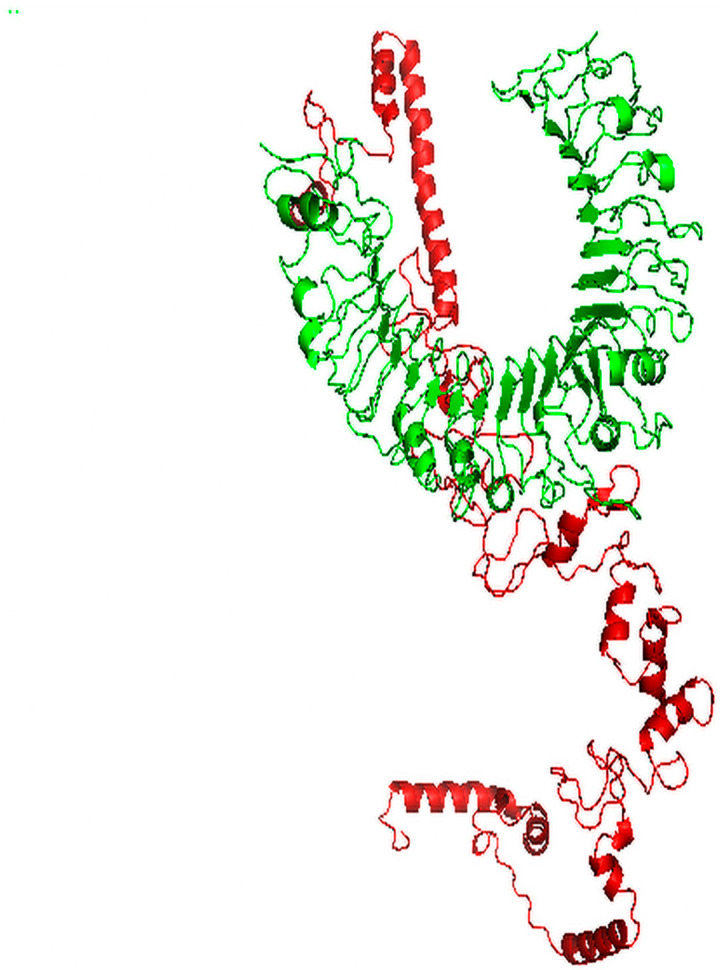
ClusPro 2.0 was employed to perform protein–protein molecular docking between the subunit vaccine and TLR2 receptor, where 30 models were created and, among them, we selected the model that occupied the receptor with the best orientation, resulting in high affinity and a low energy score. Model number 2 (Figure 5) fulfilled the selection criteria, where the measured energy score for the selected model was −1352.8, which was lower than the scores of all other predicted docked complexes, demonstrating a stronger affinity.
Figure 5.
Docked complex of vaccine construct (red) with human TLR2 (green).
3.8. Molecular Dynamics Simulation
The iMODS server was employed to perform normal mode analysis (NMA) to explore the stability of proteins. The deformability of the complex counts on the individual distortion of each residue, symbolized by hinges in the chain (Figure 6B). The estimated eigenvalue, which represents the motion stiffness of the complex, was 3.38e−07 (Figure 6D). Generally, an inverse relationship was found between the eigenvalue and the variance related to each normal mode (Figure 6C). The B-factor scores derived from NMA were equivalent to the RMS (Figure 6A). The covariance matrix indicated the coupling between pairs of residues, where different pairs demonstrated correlated, anti-correlated or uncorrelated motions, represented by red, blue and white, respectively (Figure 6E). An elastic network model was created (Figure 6F), and it revealed the pairs of atoms linked via springs according to the degree of stiffness between them, where stiffer strings appear as darker grays.
Figure 6.
Molecular dynamics simulation of multitope vaccine–TLR2 complex; stability of the protein–protein complex was investigated through B-factor values (A), deformability (B), variance (C), eigenvalue (D), covariance of residue index (E) and elastic network (F) analysis.
3.9. Vaccine Reverse Translation and Codon Optimization
The JCat server was employed to perform reverse translation and codon optimization on the amino acid sequence of the multitope vaccineAfter analysis, the measured GC content was 51.58%, which was accepted as it is within the accepted range (30:70%). Moreover, the codon adaptation index (CAI) was found to be 0.93, which demonstrates the high level of expression, as the value of CAI ranges from 0:1 and the accepted range is between 0.8 and 1.
4. Discussion
Conventional approaches for vaccine development, such as killed or live attenuated vaccines, are time consuming as it may take decades to develop a successful vaccine. Moreover, many microorganisms are difficult to cultivate or attenuate, resulting in adverse immune responses, which confirms our need for a revolution in the adopted approach for vaccine development [32]. Recently, the process of vaccine development has moved to a less time-consuming and more efficient genome pre-screening-dependent approach [22], and new studies have been based on a novel strategy through adopting immunoinformatics and reverse vaccinology approaches, where the generated subunit vaccine included the antigenic parts only and demonstrated a promising ability for fighting against pathogenic microorganisms [33].
In the current study, we have applied the reverse vaccinology approach on the proteome of M. catarrhalis BBH15 to select the most promising proteins as vaccine candidates. Only two proteins (lipopolysaccharide assembly protein LptD and outer membrane protein assembly factor BamA) out of 1615 (the number of all proteins of M. catarrhalis BBH15) were found to be essential, outer membrane localized, virulent, antigenic and non-human homologs, with appropriate molecular weight and an accepted number of transmembrane helices (fewer than two) and, therefore, they were selected to be our vaccine candidates. The same approach has been applied successfully to nominate vaccine candidates in bacterial models such as Staphylococcus aureus [34] and Shigella flexneri [35].
Bacterial outer membrane proteins are the first molecules to contact host cells, hence they were targeted to be promising vaccine candidates against the invading pathogen [36]. Additionally, they have a significant role in adhesion, nutrient acquisition and maintaining bacterial membrane integrity, therefore they are essential proteins for bacterial life and pathogenesis [37]. Lipopolysaccharide is a major structural component in most Gram-negative bacteria. It makes the outer membrane a barrier to the entry of many antibiotics [38] and, consequently, proteins that have a role in the assembly of lipopolysaccharide (vaccine candidates of the current study) have a high virulence potential which confirms their nomination to be vaccine candidates.
While pathogens grow quickly, protein extraction and testing for vaccine candidates on a large scale is a costly and time-consuming process, hence the role of immunoinformatics which is capable of decreasing the time needed and saving resources for the discovery of relevant vaccines through epitope mapping, where the non-antigenic regions of an antigen are neglected and only regions that can stimulate B cell and T cell immunity are selected [39]. This approach has been applied successfully and demonstrated promising results when these epitopes were tested on animal models. Actually, this approach has been applied to M. catarrhalis where 44 epitopes were predicted, and the top three epitopes were tested and showed superior activity in clearing M. catarrhalis from mouse lungs [40].
In the current study, B and T cell epitope prediction has been applied to filtered proteins (LptD and BamA). Th top epitopes have been selected based on percentile rank, antigenicity score, allergenicity, toxicity, conservation and reaction against a number of alleles to cover a high percentage in terms of population coverage. Top-ranked epitopes have been assembled together using appropriate linkers. In addition to that, the beta defensin adjuvant and PADRE peptide were also incorporated into the final construct of the multitope vaccine to strengthen the stimulated immune response and reduce the HLA polymorphism in the population [41].
Validation of the constructed vaccine has been assessed computationally. The constructed vaccine was analyzed according to its physicochemical, structural and immunological characteristics. It was found to be stable, hydrophilic, soluble, antigenic and non-allergenic and its structure validation using Ramachandran plots and ProSA proved that a high-quality 3D structure was modeled. Molecular docking of the designed vaccine with TLR2 showed a low energy score, which proves good binding and high affinity. Molecular dynamics simulation revealed that docked proteins have low deformability, which show the validity of our in silico-predicted vaccine.
5. Conclusions
Depending on modern in silico approaches for vaccine development, before biological experiments, is a successful technique that can guide experiments with a high probability of discovering an efficient vaccine with fewer trials, which is a great economic solution. In the current study, reverse vaccinology has been applied to the proteome of 1615 proteins of M. catarrhalis and generated two proteins (LptD and BamA) as potential vaccine candidates. Immunoinformatics along with structural vaccinology have been applied to these proteins to design a multitope vaccine, which was validated via online tools to be an efficient vaccine against M. catarrhalis. We recommend movement of the constructed vaccine to the biological validation phase using appropriate model organisms to validate our findings.
Author Contributions
Conceptualization, methodology, original draft preparation, M.A.S. and N.E.; writing—review and editing, M.A.E. and H.G.; supervision, project administration, A.A.A.-K., R.A.E. and E.B.E.; supervision, project administration, writing—review and editing, A.A.A.-K. All authors have read and agreed to the published version of the manuscript.
Funding
No funding was received for this study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bernhard S., Spaniol V., Aebi C. Molecular pathogenesis of infections caused by Moraxella catarrhalis in children. Swiss Med. Wkly. 2012;142:w13694. doi: 10.4414/smw.2012.13694. [DOI] [PubMed] [Google Scholar]
- 2.Singh B., Alvarado-Kristensson M., Johansson M., Hallgren O., Westergren-Thorsson G., Mörgelin M., Riesbeck K. The Respiratory Pathogen Moraxella catarrhalis Targets Collagen for Maximal Adherence to Host Tissues. mBio. 2016;7 doi: 10.1128/mBio.00066-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez A.C., Pang B., King L.B., Tan L., Murrah K.A., Reimche J.L., Wren J.T., Richardson S.H., Ghandi U., Swords W.E. Residence ofStreptococcus pneumoniaeandMoraxella catarrhaliswithin polymicrobial biofilm promotes antibiotic resistance and bacterial persistencein vivo. Pathog. Dis. 2014;70:280–288. doi: 10.1111/2049-632X.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren D., Pichichero M.E. Vaccine targets againstMoraxella catarrhalis. Expert Opin. Ther. Targets. 2016;20:19–33. doi: 10.1517/14728222.2015.1081686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertot G.M., Becker P.D., Guzmán C., Grinstein S. Intranasal Vaccination with Recombinant P6 Protein and Adamantylamide Dipeptide as Mucosal Adjuvant Confers Efficient Protection against Otitis Media and Lung Infection by Nontypeable Haemophilus influenzae. J. Infect. Dis. 2004;189:1304–1312. doi: 10.1086/382508. [DOI] [PubMed] [Google Scholar]
- 6.Verhaegh S.J., de Vogel C.P., Riesbeck K., Lafontaine E.R., Murphy T.F., Verbrugh H.A., Jaddoe V.W., Hofman A., Moll H.A., van Belkum A., et al. Temporal development of the humoral immune response to surface antigens of Moraxella catarrhalis in young infants. Vaccine. 2011;29:5603–5610. doi: 10.1016/j.vaccine.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Delfani S., Fooladi A.A.I., Mobarez A.M., Emaneini M., Amani J., Sedighian H. In silicoanalysis for identifying potential vaccine candidates againstStaphylococcus aureus. Clin. Exp. Vaccine Res. 2015;4:99–106. doi: 10.7774/cevr.2015.4.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegde N.R., Gauthami S., Sampath Kumar H.M., Bayry J. The use of databases, data mining and immunoinformatics in vaccinology: Where are we? Expert Opin. Drug Discov. 2018;13:117–130. doi: 10.1080/17460441.2018.1413088. [DOI] [PubMed] [Google Scholar]
- 9.Capelli R., Peri C., Villa R., Nithichanon A., Conchillo-Solé O., Yero D., Gagni P., Chiari M., Lertmemongkolchai G., Cretich M., et al. BPSL1626: Reverse and Structural Vaccinology Reveal a Novel Candidate for Vaccine Design Against Burkholderia pseudomallei. Antibodies. 2018;7:26. doi: 10.3390/antib7030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho T.F., Haddad J.P.A., Paixão T.A., Santos R.L. Meta-Analysis and Advancement of Brucellosis Vaccinology. PLoS ONE. 2016;11:e0166582. doi: 10.1371/journal.pone.0166582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen Q.-F., Liu S., Dong C., Guo H.-X., Gao Y.-Z., Guo F.-B. Geptop 2.0: An Updated, More Precise, and Faster Geptop Server for Identification of Prokaryotic Essential Genes. Front. Microbiol. 2019;10:1236. doi: 10.3389/fmicb.2019.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu N., Wagner J.R., Laird M., Melli G., Rey S., Lo R., Dao P., Sahinalp S.C., Ester M., Foster L.J., et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha S., Raghava G. VICMpred: An SVM-based method for the prediction of functional proteins of Gram-negative bacteria using amino acid patterns and composition. Genom. Proteom. Bioinforma. 2006;4:42–47. doi: 10.1016/S1672-0229(06)60015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 15.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; New York, NY, USA: 2005. pp. 571–607. [Google Scholar]
- 16.Doytchinova I.A., Flower D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A., Kolla R.V., De Silva A.D., de Silva A.M., et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2013;110:E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajighahramani N., Nezafat N., Eslami M., Negahdaripour M., Rahmatabadi S.S., Ghasemi Y. Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus. Infect. Genet. Evol. 2017;48:83–94. doi: 10.1016/j.meegid.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J., Randall A.Z., Sweredoski M.J., Baldi P. SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heo L., Park H., Seok C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41:W384–W388. doi: 10.1093/nar/gkt458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasan M., Ghosh P.P., Azim K.F., Mukta S., Abir R.A., Nahar J., Khan M.M.H. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb. Pathog. 2019;130:19–37. doi: 10.1016/j.micpath.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 23.Wiederstein M., Sippl M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig D.B., Dombkowski A.A. Disulfide by Design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinform. 2013;14:346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018;8:1–19. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira-Nascimento L., Massari P., Wetzler L.M. The role of TLR2 ininfection and immunity. Front. Immunol. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey R.K., Verma P., Sharma D., Bhatt T.K., Sundar S., Prajapati V.K. High-throughput virtual screening and quantum mechanics approach to develop imipramine analogues as leads against trypanothione reductase of leishmania. Biomed. Pharmacother. 2016;83:141–152. doi: 10.1016/j.biopha.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Awan F.M., Obaid A., Ikram A., Janjua H.A. Mutation-Structure-Function Relationship Based Integrated Strategy Reveals the Potential Impact of Deleterious Missense Mutations in Autophagy Related Proteins on Hepatocellular Carcinoma (HCC): A Comprehensive Informatics Approach. Int. J. Mol. Sci. 2017;18:139. doi: 10.3390/ijms18010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Blanco J.R., Aliaga J.I., Quintana-Ortí E.S., Chacón P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42:W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poland G.A., Ovsyannikova I.G., Jacobson R.M. Application of pharmacogenomics to vaccines. Pharmacogenomics. 2009;10:837–852. doi: 10.2217/pgs.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang P., Cai Y., Chen J., Ye X., Mao S., Zhu S., Xue X., Chen S., Zhang L. Evaluation of tandem Chlamydia trachomatis MOMP multi-epitopes vaccine in BALB/c mice model. Vaccine. 2017;35:3096–3103. doi: 10.1016/j.vaccine.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Soltan M.A., Magdy D., Solyman S.M., Hanora A. Design of Staphylococcus aureus New Vaccine Candidates with B and T Cell Epitope Mapping, Reverse Vaccinology, and Immunoinformatics. OMICS A J. Integr. Biol. 2020;24:195–204. doi: 10.1089/omi.2019.0183. [DOI] [PubMed] [Google Scholar]
- 35.Leow C.Y., Kazi A., Ismail C.M.K.H., Chuah C., Lim B.H., Singh K.K.B., Leow C.H. Reverse vaccinology approach for the identification and characterization of outer membrane proteins of Shigella flexneri as potential cellular- and antibody-dependent vaccine candidates. Clin. Exp. Vaccine Res. 2020;9:15–25. doi: 10.7774/cevr.2020.9.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizwan M., Naz A., Ahmad J., Naz K., Obaid A., Parveen T., Ahsan M., Ali A. VacSol: A high throughput in silico pipeline to predict potential therapeutic targets in prokaryotic pathogens using subtractive reverse vaccinology. BMC Bioinform. 2017;18:1–7. doi: 10.1186/s12859-017-1540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J., Huang S., Zhang Q. Outer membrane proteins: Key players for bacterial adaptation in host niches. Microbes Infect. 2002;4:325–331. doi: 10.1016/S1286-4579(02)01545-9. [DOI] [PubMed] [Google Scholar]
- 38.Okuda S., Sherman D.J., Silhavy T.J., Ruiz N., Kahne D. Lipopolysaccharide transport and assembly at the outer mem-brane: The PEZ model. Nat. Rev. Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majid M., Andleeb S. Designing a multi-epitopic vaccine against the enterotoxigenic Bacteroides fragilis based on immunoinformatics approach. Sci. Rep. 2019;9:1–15. doi: 10.1038/s41598-019-55613-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yassin G.M., Amin M.A., Attia A.S. Immunoinformatics Identifies a Lactoferrin Binding Protein A Peptide as a Promising Vaccine With a Global Protective Prospective AgainstMoraxella catarrhalis. J. Infect. Dis. 2016;213:1938–1945. doi: 10.1093/infdis/jiw062. [DOI] [PubMed] [Google Scholar]
- 41.Ghaffari-Nazari H., Tavakkol-Afshari J., Jaafari M.R., Tahaghoghi-Hajghorbani S., Masoumi E., Jalali S.A. Improving Multi-Epitope Long Peptide Vaccine Potency by Using a Strategy that Enhances CD4+ T Help in BALB/c Mice. PLoS ONE. 2015;10:e0142563. doi: 10.1371/journal.pone.0142563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.