Abstract
Simple Summary
Many molecular biology techniques have been widely used to study the pathogenesis of different diseases, particularly haematologic malignancies which are generally caused by abnormalities in the genome. TET2 gene is one of the commonly found mutated genes in BCR-ABL-negative myeloproliferative neoplasms. However, the prevalence of TET2 gene mutations in the disease remains unclear. Therefore, this study aims to estimate the prevalence of TET2 gene mutations in myeloproliferative neoplasms. The findings may be helpful for future research, diagnoses and the identification of better therapeutic strategies to manage the diseases.
Abstract
Multiple recurrent somatic mutations have recently been identified in association with myeloproliferative neoplasms (MPN). This meta-analysis aims to assess the pooled prevalence of TET2 gene mutations among patients with MPN. Six databases (PubMed, Scopus, ScienceDirect, Google Scholar, Web of Science and Embase) were searched for relevant studies from inception till September 2020, without language restrictions. The eligibility criteria included BCR-ABL-negative MPN adults with TET2 gene mutations. A random-effects model was used to estimate the pooled prevalence with 95% confidence intervals (CIs). Subgroup analyses explored results among different continents and countries, WHO diagnostic criteria, screening methods and types of MF. Quality assessment was undertaken using the Joanna Briggs Institute critical appraisal tool. The study was registered with PROSPERO (CRD42020212223). Thirty-five studies were included (n = 5121, 47.1% female). Overall, the pooled prevalence of TET2 gene mutations in MPN patients was 15.5% (95% CI: 12.1–19.0%, I2 = 94%). Regional differences explained a substantial amount of heterogeneity. The prevalence of TET2 gene mutations among the three subtypes PV, ET and MF were 16.8%, 9.8% and 15.7%, respectively. The quality of the included studies was determined to be moderate–high among 83% of the included studies. Among patients with BCR-ABL-negative MPN, the overall prevalence of TET2 gene mutations was 15.5%.
Keywords: essential thrombocythaemia, meta-analysis, myelofibrosis, myeloproliferative neoplasms, polycythaemia vera, TET2
1. Introduction
Myeloproliferative neoplasms (MPN) are a group of rare blood cancers characterised by the clonal expansion of a large number of abnormal haematopoietic stem cells. Classic Philadelphia-negative (BCR-ABL-negative) MPN can be divided into three categories: (i) polycythaemia vera (PV), (ii) essential thrombocythaemia (ET) and (iii) primary myelofibrosis (PMF). MPN can transform into acute myeloid leukaemia (AML) and may be associated with an elevated risk of thrombotic and haemorrhagic events [1,2]. Thrombosis and haemorrhage are the major causes of mortality and morbidity amongst patients with MPN and occur in about 34–39% of cases with PV, 10–29% with ET and 7.2–13.2% of patients with PMF [3].
Three main driver gene mutations, Janus kinase 2 (JAK2), Thrombopoietin receptor (MPL) and Calreticulin (CALR), have been identified in association with MPN and may have an important role in assisting the diagnosis of MPN [4]. In addition, epigenetic modification genes such as TET2, ASXL1, DNMT3A and EZH2 are also commonly mutated in cases of MPN with a frequency of 1–30% [5,6,7,8].
TET2 participates in one of the crucial steps in gene regulation, and mutations in this gene have been identified in 5–20% of people diagnosed with MPN [9]. Somatic missense mutations, somatic nonsense mutations and insertion–deletion mutations are detected in the TET2 gene among MPN patients. All of these mutations are loss-of-function mutations. Malfunction of TET2 protein may lead to the development of MPN and contributes to the disease progression [10,11]. However, some disagreement still exists about the relative significance of these TET2 gene mutations to MPN. Some researchers suggest that TET2 gene mutations are not important for MPN [12,13], whereas others have concluded that these mutations significantly contribute to their phenotype [14,15].
The prevalence of TET2 gene mutations among MPN has not yet been established. This meta-analysis aims to estimate the prevalence of TET2 gene mutations among all BCR-ABL-negative MPN and its three main subtypes.
2. Materials and Methods
PRISMA guidelines [16] were followed, and a study protocol was registered at the International Prospective Register of Systematic Reviews (PROSPERO), registration number CRD42020212223.
2.1. Data Sources and Searches
PubMed, Scopus, ScienceDirect, Google Scholar, Web of Science and Embase databases were searched from their inception till September 2020, without any language restrictions. Detailed search strategies are presented in Table S1. Any published studies or preprints with relevant data were included. Review articles, case reports and opinion articles were excluded. Data presented on websites or reported by press releases and news reports were not considered. Snowball searching was employed to review the references of included studies. Endnote X8 software was used to remove duplicate studies.
2.2. Study Selection
Study eligibility was determined by screening the title and abstract of the articles of interest. Two authors (Y.C.C. and M.A.I.) independently examined full-text reports of potentially relevant studies for inclusion. Any disagreements were resolved by consensus.
2.3. Extraction of Data
Data were independently extracted by two authors (Y.C.C. and M.A.I). The following data were obtained from each eligible study and inserted into a customised Excel spreadsheet: author surname, publication year, study design, study location, type of MPN, number of patients with MPN, demographic characteristics of patients including age and sex, clinical characteristics of the MPN patients including haemoglobin level, leucocyte and platelet counts, the total number of mutated ASXL1 and the screening method used to identify TET2 gene mutations and diagnostic criteria employed for MPN diagnoses.
2.4. Quality Assessment
A random-effects model was used to estimate the pooled prevalence of the TET2 gene mutations amongst patients with MPN, including 95% confidence intervals (Cis). Two authors (Y.C.C. and M.A.I.) independently assessed the quality of included studies using the Joanna Briggs Institute critical appraisal tools [13]. Study quality was categorised into three groups: low-quality or high risk of bias, moderate quality or moderate risk of bias, and high-quality or low risk of bias with overall scores of <50%, 50–69% and ≥70%, respectively [17].
2.5. Publication Bias
Funnel plots presenting estimates of prevalence plotted against standard error measures were used to assess the likelihood of publication bias. When a minimum of 10 studies were available, an Egger’s test was conducted to assess publication bias based on funnel plot asymmetry.
2.6. Data Synthesis and Sensitivity Analysis
The I2 statistic was used to gauge the heterogeneity between studies, with I2 > 75% indicating substantial heterogeneity. The statistical significance of study heterogeneity was also assessed using Cochran’s Q test; p < 0.05 was considered statistically heterogeneous. To help identify the outlier studies and the sources of heterogeneity, a Galbraith plot was constructed. Prevalence estimates were explored with sensitivity analyses. Three strategies were followed for these analyses: (i) studies with small sample sizes (<100) were excluded, (ii) low-quality studies were excluded and (iii) outlier studies were excluded. In each case, the results were then compared to the overall prevalence estimate. Metaprop codes in meta (version 4.15-1) and metaphor (version 2.4-0) packages of R (version 3.6.3) and RStudio (version 1.3.1093) were used for the analyses and graphs [18].
3. Results
3.1. Study Selection
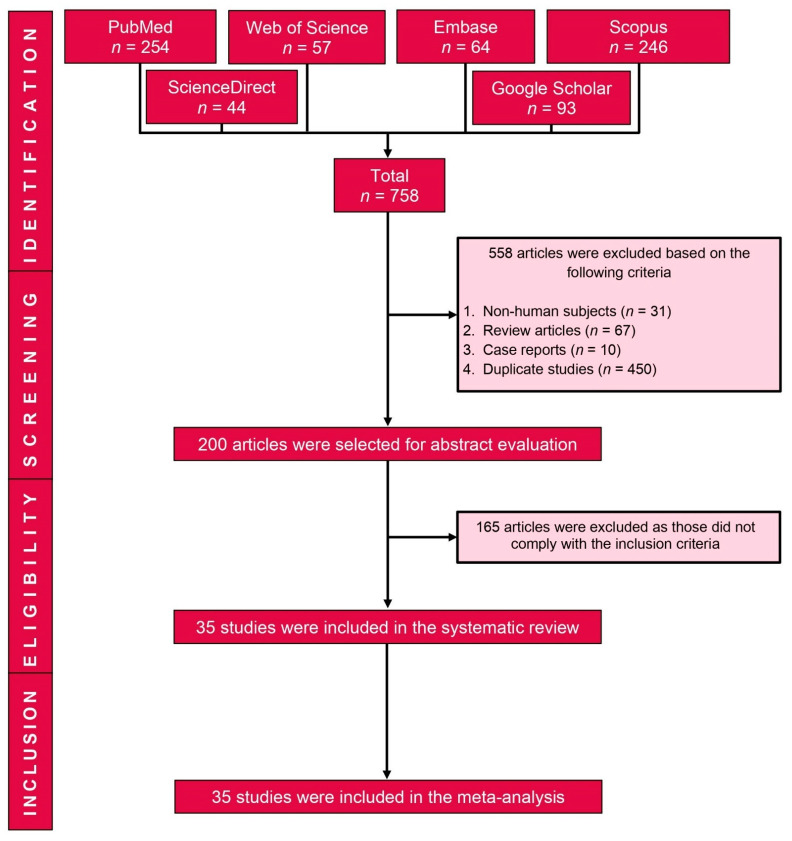
The search generated 758 potentially relevant studies. After excluding 558 studies (duplicates n = 450; review articles n = 67; non-human studies n = 31; and case reports, n = 10), 200 full-text studies were examined and 35 studies met the inclusion criteria and were included in the review (Figure 1).
Figure 1.
PRISMA flow diagram of study selection.
3.2. Characteristics of Included Studies
Table 1 presents the main characteristics of the 35 included studies. Overall, the meta-analysis includes data from 5121 patients with MPN (47.1% female). Study participants were located in four continents: Europe (n = 1758), Asia (n = 301), North America (n = 3019) and Australia (n = 43), and 12 countries (Australia, China, Denmark, France, Germany, Italy, Korea, Spain, Sweden, Switzerland, the United Kingdom and the United States of America). Most (27/35) studies used a version of the World Health Organization classification and diagnostic criteria (WHO 2016 7 studies, WHO 2008 17 studies and WHO 2001 3 studies) to determine MPN diagnoses. Many studies confirmed TET2 gene mutations with either next-generation sequencing (NGS) or Sanger sequencing, which have higher sensitivity in detecting mutations compared with other methods, such as high-resolution melting (HRM) analysis [19]. One study was published in Chinese Mandarin and was translated into English (Y.C.C.).
Table 1.
Major characteristics of the included studies.
| No | Study ID [References] |
Study Design |
Country | Type of MPN | Total Number of MPN Patients (Female) | Age (Years) [Mean ± SD/Median (IQR)/Range] | Haemoglobin (g/dL) [Mean ± SD/Median (IQR)/Range] |
Leucocyte Count (109/L) [Mean ± SD/Range/Median (IQR)] |
Platelet Count (109/L) [Mean ± SD/Range/Median (IQR)] |
Total Number of Mutated ASXL1 (%) | Screening Method for TET2 Gene Mutations | Diagnostic Criteria |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Andreasson 2020 [20] |
Cross-sectional | Sweden | PV | 85 (41) | 71.0 (37.0–94.0) |
NR | NR | NR | 8.2 | NGS | 2008 WHO |
| 2 | Barraco 2017 [21] |
Cross-sectional | USA | PV | 267 (125) | 64.0 (17.0–94.0) |
18.0 (14.8–24.3) |
11.5 (4.3–59.3) |
439.0 (37.0–2747.0) |
8.1 | NR | 2016 WHO |
| 3 | Bartels 2019 [22] |
Case–control | Germany | MF | 104 (53) | NR | NR | NR | NR | 9.6 | NGS | 2016 WHO |
| 4 | Brecqueville 2012 [23] |
Cross-sectional | France | PV, ET & MF | 127 (57) | NR (29.0–97.0) |
NR | NR | NR | 11.0 | SS | 2008 WHO |
| 5 | Brecqueville 2014 [24] |
Cross-sectional | France | MF | 68 (NR) | 69.0 (30.0–86.0) |
11.4 (5.8–17.8) |
8.9 (1.3–120.0) |
256.0 (5.0–1188.0) |
26.5 | SS | 2008 WHO |
| 6 | Carbuccia 2009 [25] |
Cross-sectional | France | PV, ET & MF | NR | NR | NR | NR | NR | 7.3 | SS | NR |
| 7 | Cerquozzi 2017 [26] |
Cross-sectional | USA | PV | 587 (302) | 60.0 (17.0–94.0) |
NR | NR | 476.0 (41.0–2747.0) |
10.5 | NGS | 2016 WHO, ELN |
| 8 | Delhommeau 2009 [11] |
Cross-sectional | France | PV, ET & MF | 203 (41) | NR | NR | NR | NR | NR | SS, SNP array, CGH | 2001 WHO |
| 9 | Delic 2016 [27] |
Cross-sectional | Germany | PV, ET & MF | 100 (NR) | 69.0 (28.0–87.0) |
NR | NR | NR | 21.0 | NGS | 2008 WHO |
| 10 | Gill 2018 [28] |
Cross-sectional | China | MF | 101 (39) | 60.0 (26.0–89.0) |
10.3 (3.0–18.5) |
12.1 (1.5–177.4) |
344.0 (19.0–1720.0) |
30.7 | NGS | 2016 WHO, IWG-MRT |
| 11 | Guglielmelli 2011 [29] |
Cross-sectional | Italy | MF | 518 (303) | NR | NR | NR | NR | 22.2 | HRM | 2008 WHO, IWG-MRT |
| 12 | Ha 2014 [14] |
Cross-sectional | Korea | PV, ET & MF | 99 (50) | 63.7 ± 13.0 | 13.7 ± 3.8 | 16.5 ± 15.4 | 825.4 ± 490.0 | NR | SS, SNP array, CGH | 2008 WHO |
| 13 | Huang 2020 [30] |
Cross-sectional | China | PV, ET & MF | 65 (32) | 62.0 (NR) | NR | NR | NR | 10.8 | NGS | 2016 WHO |
| 14 | Hussein 2010 [31] |
Cross-sectional | USA | PV, ET & MF | 199 (96) | 58.0 (19.0–93.0) |
NR | NR | NR | NR | NGS | 2001 WHO |
| 15 | Kröger 2017 [32] |
Cross-sectional | Germany | MF | 169 (73) | 58.0 (18.0–75.0) |
NR | NR | NR | 29.0 | SS | NR |
| 16 | Leibundgut 2020 [33] |
Cross-sectional | Switzerland | ET | 18 (10) | 59.5 (21.0–83.0) |
NR | 7.8 (3.0–14.6) |
788.0 (521.0–1359.0) |
11.1 | NGS | 2016 WHO |
| 17 | Magor 2016 [34] |
Cross-sectional | Australia | PV, ET & MF | 43 (16) | 61.0 (24.0–91.0) |
NR | NR | NR | 9.3 | Targeted exon resequencing | 2008 WHO |
| 18 | Martínez-Avilés 2012 [35] |
Cross-sectional | Spain | PV, ET & MF | 62 (43) | NR | NR | NR | NR | 4.8 | HRM, SS | 2008 WHO |
| 19 | Nielsen 2017 [36] |
Case–control | Denmark | MF | 16 (3) | 66.0 (52.0–80.0) |
10.3 (7.9–13.4) |
5.9 (2.3–64.4) |
155.5 (56.0–357.0) |
50.0 | PCR-DGGE | NR |
| 20 | Nischal 2013 [37] |
Cross-sectional | USA | PV, ET & MF | 25 (14) | 68.0 (54.0–72.0) |
NR | NR | NR | 24.0 | SS | NR |
| 21 | O’Sullivan 2019 [38] |
Cross-sectional | UK | ET | NR | NR | NR | NR | NR | NR | NGS | NR |
| 22 | Pardanani 2010 [39] |
Cross-sectional | USA | PV, ET & MF | 78 (34) | 64.0 (22.0–95.0) |
NR | NR | NR | NR | NGS | 2008 WHO |
| 23 | Patel 2015 [40] |
Cross-sectional | USA | MF | 95 (44) | 66.0 (40.0–84.0) |
10.7 (7.2–16.9) |
25.0 (2.5–159.0) |
339.0 (13.0–969.0) |
21.1 | NGS | IWG-MRT |
| 24 | Patriarca 2013 [41] |
Cross-sectional | Italy | PV, ET & MF | 97 (44) | NR | NR | NR | NR | NR | NGS | 2008 WHO |
| 25 | Saint-Martin 2009 [42] |
Cross-sectional | France | PV, ET & MF | NR | NR | NR | NR | NR | NR | SS | 2008 WHO |
| 26 | Schlenk 2016 [43] |
Cross-sectional | Germany | MF | 96 (33) | NR | NR | NR | NR | 30.2 | SS | 2008 WHO, IWG-MRT |
| 27 | Schnittger 2012 [44] |
Cross-sectional | Germany | ET & MF | NR | NR | NR | NR | NR | NR | SS, HRM | NR |
| 28 | Segura-Díaz 2020 [45] |
Cross-sectional | Spain | PV, ET & MF | 68 (40) | 68.0 (43.0–90.0) |
NR | NR | NR | 8.8 | NGS | 2016 WHO |
| 29 | Song 2017 [46] |
Cross-sectional | USA | PV, ET & MF | 135 (64) | NR | NR | NR | NR | 21.2 | NGS | 2008 WHO |
| 30 | Tefferi 2009 [47] |
Cross-sectional | USA | PV, ET & MF | 227 (111) | NR | NR | NR | NR | NR | NGS | 2001 WHO |
| 31 | Tefferi 2010 [48] |
Cross-sectional | USA | PV, ET & MF | 908 (487) | NR | NR | NR | NR | NR | NGS | 2008 WHO, IWG-MRT |
| 32 | Tefferi 2016 [49] |
Cross-sectional | USA | MF | 182 (64) | 63.0 (22.0–87.0) |
10.1 (5.8–16.0) |
10.5 (1.9–219.0) |
224.0 (11.0–1493.0) |
35.7 | NGS | 2008 WHO |
| 33 | Tefferi 2016a [50] |
Cross-sectional | USA | PV & ET | 316 (177) | NR | NR | NR | NR | 11.4 | NGS | 2008 WHO |
| 34 | Verger 2014 [51] |
Cross-sectional | France | PV, ET & MF | 27 (NR) | NR | NR | NR | NR | NR | SS | NR |
| 35 | Zhang 2015 [52] |
Cross-sectional | China | MF | 36 (15) | 65.0 (46.0–93.0) |
10.9 (3.0–16.0) |
22.3 (1.4–54.5) |
215.0 (3.0–1157.0) |
11.1 | WGS | 2008 WHO |
aCGH: array-comparative genomic hybridisation; ASXL1: Additional sex combs-like 1; CGH: comparative genomic hybridisation; ELN: European Leukemia Net; ET: essential thrombocythaemia; HRM: high-resolution melting analysis; IQR: interquartile range; IWG-MRT: International Working Group for Myelofibrosis Research and Treatment; MF: myelofibrosis; MPN: myeloproliferative neoplasms; SS: Sanger sequencing; NGS: next-generation sequencing; NR: not reported; PCR-DGGE: polymerase chain reaction-denaturing gradient gel electrophoresis; PV: polycythaemia vera; SD: standard deviation; SNP: single nucleotide polymorphism; TET2: Ten–eleven translocation 2; UK: United Kingdom; USA: United States of America; WGS: whole-genome sequencing; WHO: World Health Organization.
3.3. Meta-Analysis
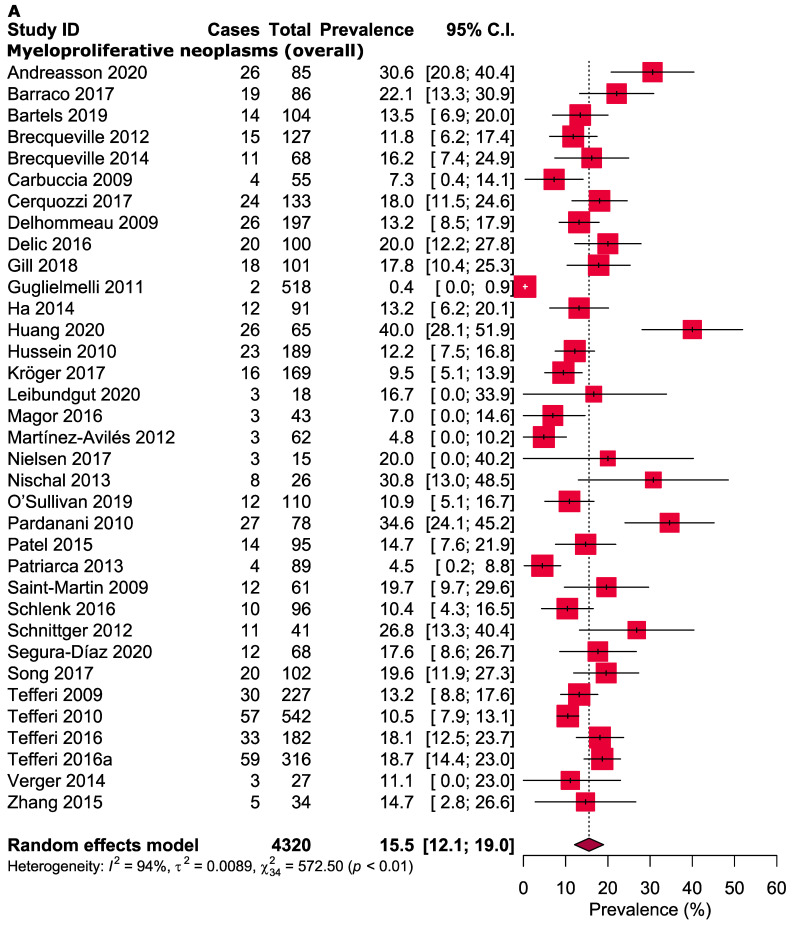
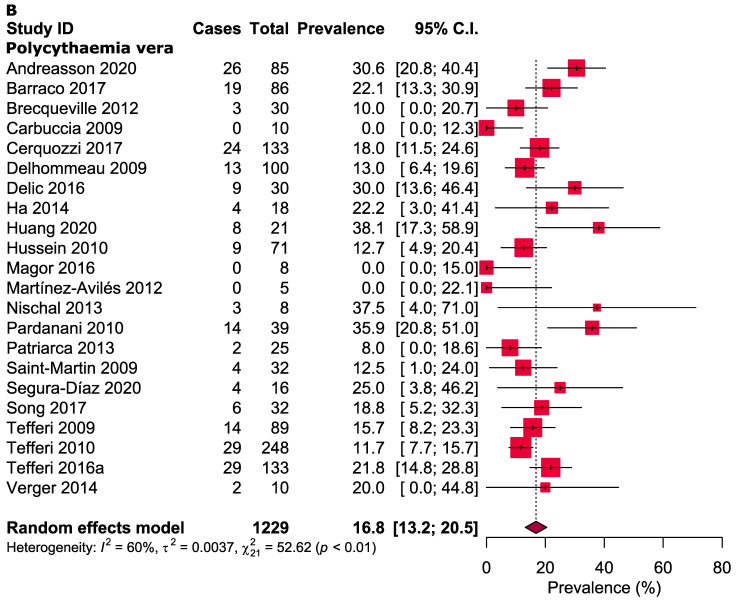
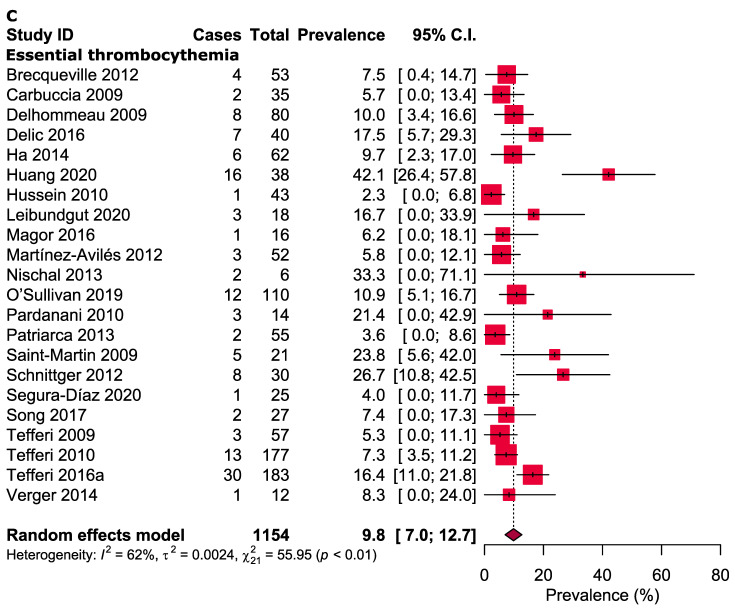
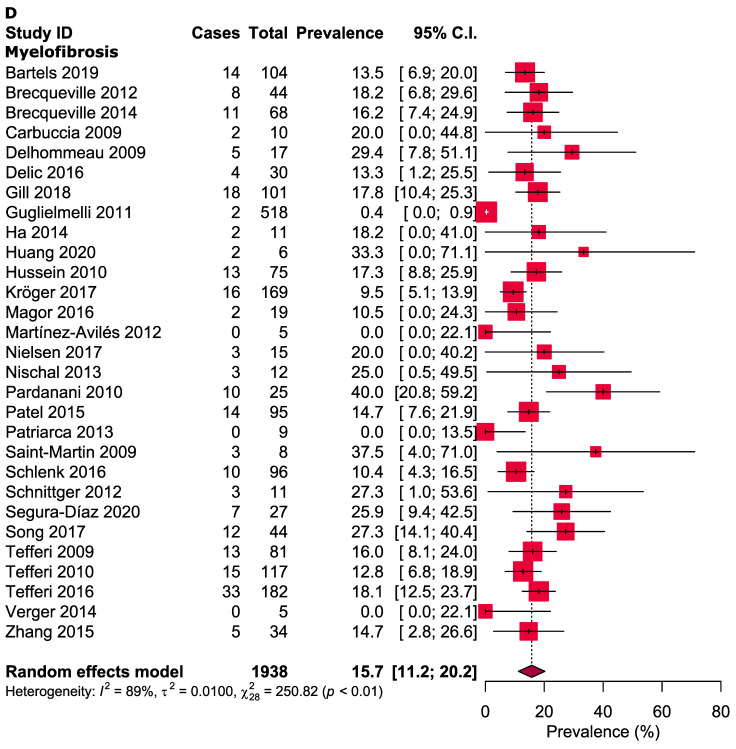
The overall pooled prevalence of TET2 gene mutations in patients with MPN was 15.5% (95% CI: 12.1–19.0%, I2 = 94%, Figure 2A). The prevalence of TET2 gene mutations in PV, ET and MF patients was 16.8% (95% CI: 13.2–20.5%, I2 = 60%, Figure 2B), 9.8% (95% CI: 7.0–12.7%, I2 = 62%, Figure 2C) and 15.7% (95% CI: 11.2–20.2%, I2 = 89%, Figure 2D), respectively. In other subgroup analyses, the pooled prevalence of TET2 gene mutations was compared between four continents: Europe (13.0%; 95% CI: 8.8–17.2%, I2 = 92%), North America (17.4%; 95% CI: 14.0–20.9%, I2 = 74%), Asia (20.8%; 95% CI: 10.5–31.1%, I2 = 80%) and Australia (7.0%; 95% CI: 0.0–14.6%, I2 = NA). The prevalence of TET2 gene mutations were further analysed based on countries: China (23.9%; 95% CI: 9.6–38.1%, I2 = 82%), France (13.6%; 95% CI: 10.6–16.7%, I2 = 0%), Germany (14.2%; 95% CI: 9.2–19.1%, I2 = 61%), Italy (1.9%; 95% CI: 0.0–5.7%, I2 = 71%), Spain (10.7%; 95% CI: 0.0–23.2%, I2 = 82%) and the United States (17.4%; 95% CI: 14.0–20.9%, I2 = 74%). Assessments of PV, ET and MF prevalence across the four continents (Figure S1) and in relation to different countries were also examined (Figure S2). Three forms of WHO criteria were used and the prevalence of TET2 gene mutations was highest in the 2016 version (WHO 2001 criteria 12.9%, 95% CI: 10.2–15.5%, I2 = 0%, WHO 2008 criteria 14.5%, 95% CI: 9.7–19.3%, I2 = 95% and WHO 2016 criteria 20.1%, 95% CI: 14.7–25.4%, I2 = 61%) (Figure S3). A higher prevalence of TET2 gene mutations were observed while using NGS (17.2%, 95% CI: 14.0–20.4%, I2 = 80%) and Sanger sequencing (12.7%, 95% CI: 9.6–15.9%, I2 = 52%), but not in HRM analysis (7.7%, 95% CI: 0.0–16.6%, I2 = 88%) (Figure S4). The MF subgroup was further divided into two subgroups (PMF and SMF), and the prevalence of TET2 gene mutations were studied in both and found to be similar (PMF 16.7%, 95% CI: 13.6–19.8%, I2 = 24% and SMF 14.8%, 95% CI: 9.3–20.2%, I2 = 0%) (Figure S5). Various levels of heterogeneity were observed in the main analyses (ranging from 60% to 94%) (Figure 2) and subgroup analyses (ranging from 0% to 93%) (Table 2, Figures S1–S5).
Figure 2.
(A) Prevalence of TET2 gene mutations in patients with MPN (overall). (B) Prevalence of TET2 gene mutations in patients with PV. (C) Prevalence of TET2 gene mutations in patients with ET. (D) Prevalence of TET2 gene mutations in patients with MF.
Table 2.
The pooled prevalence of TET2 gene mutations in different subgroups of MPN.
| Subgroups | Prevalence [95% CIs] (%) |
Number of Studies Analysed | Total Number of Patients | Heterogeneity | Publication Bias, Egger’s Test (p-Value) | |
|---|---|---|---|---|---|---|
| I 2 | p-Value | |||||
| Overall myeloproliferative neoplasms | ||||||
| Europe | 13.0 [8.8–17.2] | 19 | 2010 | 92% | <0.0001 | 0.004 |
| North America | 17.4 [14.0–20.9] | 11 | 1976 | 74% | <0.0001 | 0.0005 |
| Asia | 20.8 [10.5–31.1] | 4 | 291 | 80% | 0.001 | NA |
| Australia | 7.0 [0.0–14.6] | 1 | 43 | NA | NA | NA |
| China | 23.9 [9.6–38.1] | 3 | 200 | 82% | 0.003 | NA |
| France | 13.6 [10.6–16.7] | 5 | 480 | 0% | 0.67 | NA |
| Germany | 14.2 [9.2–19.1] | 5 | 510 | 61% | 0.03 | NA |
| Italy | 1.9 [0.0–5.7] | 2 | 607 | 71% | 0.06 | NA |
| Spain | 10.7 [0.0–23.2] | 2 | 130 | 82% | 0.01 | NA |
| USA | 17.4 [14.0–20.9] | 11 | 1976 | 74% | <0.0001 | 0.0005 |
| WHO criteria reported | 15.7 [11.8–19.7] | 27 | 3782 | 95% | <0.0001 | 0.0002 |
| WHO criteria not reported | 13.1 [8.9–17.3] | 8 | 538 | 47% | 0.06 | 0.005 |
| WHO 2001 criteria | 12.9 [10.2–15.5] | 3 | 613 | 0% | 0.93 | NA |
| WHO 2008 criteria | 14.5 [9.7–19.3] | 17 | 2594 | 95% | <0.0001 | 0.0004 |
| WHO 2016 criteria | 20.1 [14.7–25.4] | 7 | 575 | 61% | 0.01 | 0.40 |
| NGS method | 17.2 [14.0–20.4] | 18 | 2604 | 80% | <0.0001 | 0.0007 |
| SS method | 12.7 [9.6–15.9] | 11 | 965 | 52% | 0.02 | 0.001 |
| HRM method | 7.7 [0.0–16.6] | 3 | 621 | 88% | 0.0002 | NA |
| Polycythaemia vera | ||||||
| Europe | 14.6 [8.0–21.1] | 10 | 343 | 63% | 0.01 | 0.58 |
| North America | 18.2 [14.2–22.5] | 9 | 839 | 57% | 0.01 | NA |
| Asia | 29.6 [14.1–45.2] | 2 | 39 | 17% | 0.27 | NA |
| Australia | 0.0 [0.0–15.0] | 1 | 8 | NA | NA | NA |
| France | 12.5 [7.6–17.5] | 4 | 172 | 0% | 0.90 | NA |
| Spain | 12.7 [0.0–37.2] | 2 | 21 | 61% | 0.28 | NA |
| USA | 18.2 [14.0–22.5] | 9 | 839 | 57% | 0.01 | NA |
| WHO 2001 criteria | 13.7 [9.6–17.9] | 3 | 260 | 0% | 0.40 | NA |
| WHO 2008 criteria | 16.9 [11.3–22.6] | 12 | 685 | 69% | 0.0009 | 0.77 |
| WHO 2016 criteria | 21.4 [15.6–27.3] | 4 | 256 | 16% | 0.31 | NA |
| NGS method | 19.8 [15.1–24.6] | 12 | 922 | 67% | 0.0005 | 0.009 |
| SS method | 13.0 [8.4–17.7] | 7 | 203 | 0% | 0.71 | NA |
| HRM method | 0.0 [0.0–22.1] | 1 | 5 | NA | NA | NA |
| Essential thrombocythaemia | ||||||
| Europe | 8.8 [5.7–12.0] | 12 | 531 | 39% | 0.08 | 0.002 |
| North America | 8.7 [3.8–13.6] | 7 | 507 | 69% | 0.003 | NA |
| Asia | 25.1 [0.0–56.9] | 2 | 100 | 93% | 0.0002 | NA |
| Australia | 6.2 [0.0–18.1] | 1 | 16 | NA | NA | NA |
| France | 9.7 [5.3–14.2] | 4 | 166 | 0% | 0.44 | NA |
| Spain | 12.1 [0.0–21.2] | 2 | 46 | 74% | 0.04 | NA |
| USA | 8.7 [3.8–13.6] | 7 | 507 | 69% | 0.003 | NA |
| WHO 2001 criteria | 5.3 [1.1–9.6] | 3 | 180 | 44% | 0.16 | NA |
| WHO 2008 criteria | 9.4 [6.1–12.6] | 11 | 700 | 49% | 0.03 | 0.06 |
| WHO 2016 criteria | 20.3 [0.0–43.7] | 3 | 81 | 89% | <0.0001 | 0.41 |
| NGS method | 10.2 [6.1–14.4] | 12 | 787 | 75% | <0.0001 | 0.003 |
| SS method | 10.4 [6.2–14.6] | 8 | 316 | 31% | 0.18 | NA |
| HRM method | 14.9 [0.0–35.2] | 2 | 82 | 83% | 0.01 | NA |
| Myelofibrosis | ||||||
| Europe | 13.7 [7.9–19.5] | 15 | 1127 | 85% | <0.0001 | 0.008 |
| North America | 16.8 [12.3–23.7] | 9 | 640 | 52% | 0.09 | NA |
| Asia | 17.4 [11.4–23.5] | 4 | 152 | 0% | 0.82 | NA |
| Australia | 10.5 [0.0–24.3] | 1 | 19 | NA | NA | NA |
| China | 17.4 [11.2–23.6] | 3 | 141 | 0% | 0.63 | NA |
| France | 17.6 [9.9–25.3] | 5 | 142 | 20% | 0.51 | NA |
| Germany | 11.0 [8.0–14.0] | 5 | 410 | 0% | 0.61 | NA |
| Italy | 0.4 [0.0–0.9] | 2 | 527 | 0% | 0.50 | NA |
| Spain | 14.0 [0.0–39.4] | 2 | 32 | 70% | 0.21 | NA |
| USA | 17.7 [13.8–21.6] | 8 | 631 | 35% | 0.15 | NA |
| WHO 2001 criteria | 17.5 [11.9–23.3] | 3 | 173 | 0% | 0.52 | NA |
| WHO 2008 criteria | 14.4 [8.1–20.7] | 15 | 1210 | 90% | <0.0001 | 0.04 |
| WHO 2016 criteria | 16.5 [11.8–21.2] | 4 | 238 | 0% | 0.39 | 0.20 |
| NGS method | 16.5 [13.2–19.8] | 13 | 896 | 38% | 0.17 | 0.053 |
| SS method | 13.3 [9.1–17.5] | 11 | 446 | 24% | 0.35 | 0.01 |
| HRM method | 5.0 [0.0–18.2] | 3 | 534 | 50% | 0.10 | NA |
| Different types of myelofibrosis | ||||||
| PMF | 16.7 [13.6–19.8] | 20 | 853 | 24% | 0.41 | 0.06 |
| SMF | 14.8 [9.3–20.2] | 9 | 158 | 0% | 0.95 | NA |
CIs: confidence intervals; HRM: high-resolution melting analysis; NA: not applicable; NGS: next-generation sequencing; PMF: primary myelofibrosis; SMF: secondary myelofibrosis; SS: Sanger sequencing; WHO: World Health Organization.
3.4. Quality Assessment
Detailed quality assessments of the included studies are presented in Tables S2 and S3. Most studies were judged to be of high quality (68.6%), while the remainder were considered to be of either moderate (14.3%) or low quality (17.7%).
3.5. Publication Bias
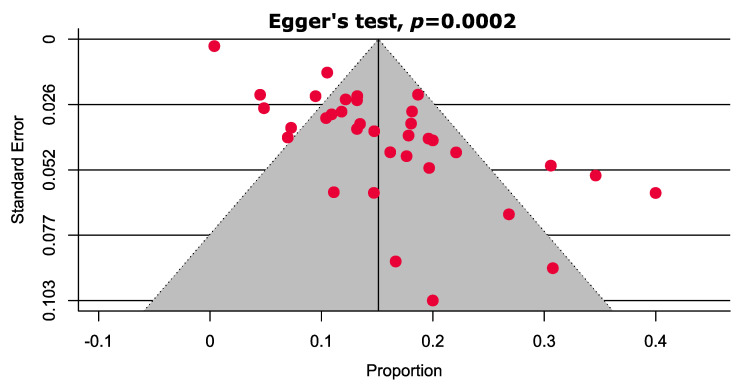
The results from the funnel plots and Egger’s tests suggest that there is only a small likelihood of publication bias (Figure 3 and Figure S6).
Figure 3.
Funnel plot estimating the prevalence of TET2 gene mutations in patients with MPN (overall).
3.6. Sensitivity Analyses
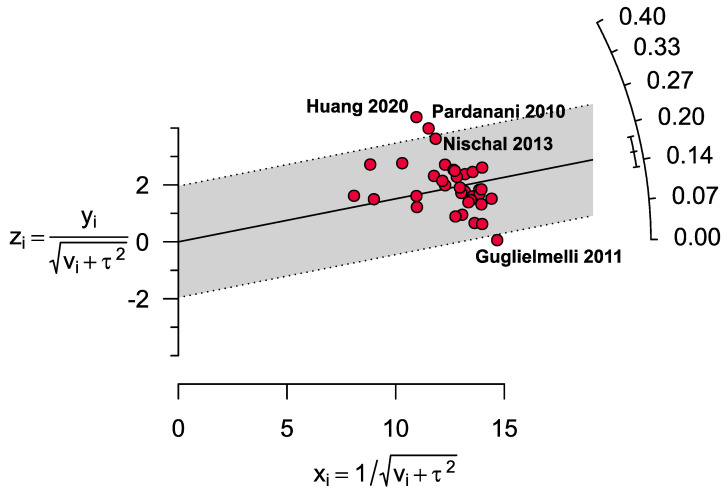
In the sensitivity analyses, only minor differences (ranging from 4.0% lower to 1.8% higher) were observed in the pooled prevalence estimates of TET2 gene mutations among cases of MPN compared to the main findings (Table 3 and Figure S7). A Galbraith plot was performed, and four outlier studies [29,30,37,39] were identified (Figure 4).
Table 3.
Sensitivity analyses.
| Strategies of Sensitivity Analyses | Prevalence [95% CIs] (%) |
Difference of Pooled Prevalence Compared to the Main Result | Number of Studies Analysed | Total Number of Subjects | Heterogeneity | |
|---|---|---|---|---|---|---|
| I 2 | p-Value | |||||
| Myeloproliferative neoplasms (overall) | ||||||
| Excluding small studies | 13.6 [8.8–18.4] | 1.9% lower | 15 | 3117 | 96% | <0.0001 |
| Excluding low- and moderate-quality studies | 15.4 [11.3–19.6] | 0.1% lower | 24 | 3485 | 95% | <0.0001 |
| Excluding outlier studies | 13.9 [12.0–15.9] | 1.6% lower | 31 | 3633 | 63% | <0.0001 |
| Polycythaemia vera | ||||||
| Excluding small studies | 15.6 [11.0–20.3] | 1.2% lower | 4 | 614 | 59% | 0.06 |
| Excluding low- and moderate-quality studies | 18.6 [14.4–22.7] | 1.8% higher | 13 | 946 | 59% | 0.003 |
| Excluding outlier studies | 15.4 [12.0–18.7] | 1.4% lower | 19 | 1161 | 54% | 0.01 |
| Essential thrombocythaemia | ||||||
| Excluding small studies | 11.3 [5.9–16.8] | 1.5% higher | 3 | 470 | 72% | 0.02 |
| Excluding low- and moderate-quality studies | 11.1 [7.1–15.0] | 1.3% higher | 12 | 839 | 72% | <0.0001 |
| Excluding outlier studies | 8.4 [6.0–10.8] | 4.4% lower | 19 | 1096 | 48% | 0.01 |
| Myelofibrosis | ||||||
| Excluding small studies | 11.7 [4.0–19.5] | 4.0% lower | 6 | 1191 | 95% | <0.0001 |
| Excluding low- and moderate-quality studies | 14.0 [8.9–19.1] | 1.7% lower | 18 | 1700 | 91% | <0.0001 |
| Excluding outlier studies | 14.5 [12.4–16.7] | 1.2% lower | 25 | 1377 | 18% | 0.46 |
CIs: Confidence intervals.
Figure 4.
Galbraith plot analysing MPN (overall) identified four outlier studies.
4. Discussion
The overall prevalence of TET2 gene mutations among BCR-ABL-negative MPN patients was estimated to be 15.5%. This estimate is similar to the occurrence of TET2 somatic mutations in patients with various myeloid cancers [11]. Compared with other myeloid malignancies, the prevalence of TET2 gene mutations among patients with BCR-ABL-negative MPN appears to be lower. Among patients with myelodysplastic syndromes (MDS), the prevalence of these mutations has been estimated to be 18–35% [11,53,54,55,56,57,58], 36–60% for those with chronic myelomonocytic leukaemia (CMML) [54,59,60,61], 24% in cases with AML, 22% in chronic myelogenous leukaemia (CML) [11], 20% in mastocytosis [62,63] and about 30% in patients with blastic plasmacytoid dendritic cell neoplasm [64,65].
The results appear to confirm the observation that epigenetic regulators like the TET2 gene have mutated more frequently among those patients with PV (p = 0.05), compared with those with either MF (p = 0.02) or ET (p = 0.023) [27]. According to a meta-analysis that analysed the frequency of three main genes (JAK2, MPL and CALR) in MPN from 2000 to 2018 [66], for the most common gene mutation JAK2 V617F, TET2 showed a lower prevalence as compared to JAK2 V617F in PV (46.7–100.0%), ET (31.3–72.1%) and MF (25.0–85.7%). For the MPL gene, our results displayed a higher proportion in PV (16.8% vs. 0%) but similar percentages in ET (9.8% vs. 0.9–12.5%) and MF (15.7% vs. 0–17.7%). Finally, in relation to the last common driver gene, CALR, TET2 manifested a higher prevalence in PV (16.8% vs. 0%), a lower prevalence in ET (9.8% vs. 12.6–50%) and a similar proportion in MF (15.7% vs. 10–100%). From these results, it appears that the TET2 gene mutations have distributed more evenly across MPN subcategories in contrast to the three main driver mutation genes [47].
This study has several notable strengths. To our knowledge, no meta-analysis has previously investigated the prevalence of TET2 gene mutations in patients with MPN. This meta-analysis included studies from six databases using robust search strategies without any language restrictions. All the sensitivity analyses produced similar results to the overall findings, suggesting that the main result is likely to be robust and credible.
Nevertheless, there are a few limitations. Several meta-analyses exhibited high heterogeneity, indicating considerable variability among the results from the included studies. After excluding the four outlier studies identified by the Galbraith plot, heterogeneity was reduced from 94% to 63% across all MPN studies, 60% to 54% for PV, 62% to 48% for ET and 89% to 18% for MF, suggesting that these four studies were an important source of heterogeneity. Several factors may further explain this heterogeneity. One of the outlier studies [29] recorded a very low prevalence of TET2 gene mutations (0.4%). This may be due to the use of a different method (HRM analysis) that may be less sensitive to identifying the mutations compared with most other studies. Notably, a similar result was also observed in one of the two other studies [35,44] that also employed the same analytical technique. Different etiological exposures might occur in different regions, resulting in differences in the prevalence estimates across the different studies [67]. In support of this hypothesis, a lower prevalence was recorded in Australia and Italy, whereas a higher result was identified in China and the United States. Variations in the use of different diagnostic guidelines may have also affected the estimates of prevalence and further contributed to the heterogeneity of results between studies. The discovery of the JAK2 gene mutations in 2005 and their subsequent inclusion in the diagnostic criteria [68] for MPN, PV and ET but not MF [2,69] may account for some of the differences observed among the studies. A stepwise increase in TET2 gene mutations in MPN was observed with subsequent versions of the WHO classification and diagnostic criteria among all cases of MPN (12.9% for WHO criteria 2001, 14.5% for WHO criteria 2008 and 20.1% for WHO criteria 2016), PV (13.7% for WHO criteria 2001, 16.9% for WHO criteria 2008 and 21.4% for WHO criteria 2016) and ET (5.3% for WHO criteria 2001, 9.4% for WHO criteria 2008 and 20.3% for WHO criteria 2016) but not in MF (17.5% for WHO criteria 2001, 14.4% for WHO criteria 2008 and 16.5% for WHO criteria 2016).
Another limitation of this meta-analysis is that the prevalence of MPN may be underestimated in some studies. Patients with MPN can be relatively symptom-free for many years so people, with little contact with health services, can remain undiagnosed for long periods [70]. Estimates of the prevalence of TET2 mutations in MPN may be underestimated, particularly in less-developed countries or among disadvantaged groups in well-developed countries.
Finally, the included studies largely focused on the allele frequencies of the main driver mutations (JAK2, MPL and CALR) and did not permit any analysis of the allelic frequencies of the TET2 mutant allele in MPN.
5. Conclusions
This meta-analysis provides the most comprehensive currently available estimate of the overall prevalence of TET2 gene mutations (15.5%) among patients with MPN. However, substantial heterogeneity was evident among the results included in this meta-analysis, likely related to factors such as regional differences in patients included in studies and variations in the diagnostic criteria employed by the studies. This heterogeneity suggests that caution should be employed with using the estimates of prevalence.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13123078/s1, Figure S1: prevalence of TET2 gene mutations based on continents, Figure S2: Prevalence of TET2 gene mutations based on countries, Figure S3: Prevalence of TET2 gene mutations based on the WHO classification and diagnostic criteria of MPN, Figure S4: Prevalence of TET2 gene mutations based on different methods used to detect TET2 gene mutations in MPN, Figure S5: Subgroup analysis. Prevalence of TET2 gene mutations in patients with (A) PMF and (B) SMF, Figure S6: Funnel plots estimating the prevalence of TET2 gene mutations in patients with (A) polycythaemia vera, (B) essential thrombocythaemia and (C) myelofibrosis, Figure S7: Sensitivity analyses, Table S1: Search strategies, Table S2: Quality assessment of the included cross-sectional studies, Table S3: Quality assessment of the included case–control studies.
Author Contributions
Conceptualisation, Y.C.C., M.A.I. and M.R.; methodology, Y.C.C., M.A.I. and P.H.; software, M.A.I.; validation, Y.C.C. and M.A.I.; formal analysis, Y.C.C. and M.A.I.; resources, Y.C.C. and M.A.I.; data curation, M.A.I., P.H., P.Y.W., M.F.J., R.H. and M.R.; writing—original draft preparation, Y.C.C.; writing—review and editing, M.A.I., P.H., P.Y.W., M.F.J., R.H. and M.R.; supervision, M.A.I., M.F.J., R.H. and M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Campregher P.V., de Souza Santos F.P., Perini G.F., Hamerschlak N. Molecular biology of Philadelphia-negative myeloproliferative neoplasms. Rev. Bras. Hematol. Hemoter. 2012;34:150–155. doi: 10.5581/1516-8484.20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tefferi A., Thiele J., Vardiman J.W. The 2008 World Health Organization classification system for myeloproliferative neoplasms: Order out of chaos. Cancer. 2009;115:3842–3847. doi: 10.1002/cncr.24440. [DOI] [PubMed] [Google Scholar]
- 3.Tefferi A., Elliott M. Seminars in Thrombosis and Hemostasis. Thieme Medical Publishers, Inc.; New York, NY, USA: 2007. Thrombosis in myeloproliferative disorders: Prevalence, prognostic factors, and the role of leukocytes and JAK2V617F; pp. 313–320. [DOI] [PubMed] [Google Scholar]
- 4.Nangalia J., Massie C.E., Baxter E.J., Nice F.L., Gundem G., Wedge D.C., Avezov E., Li J., Kollmann K., Kent D.G. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenedini E., Bernardis I., Artusi V., Artuso L., Roncaglia E., Guglielmelli P., Pieri L., Bogani C., Biamonte F., Rotunno G. Targeted cancer exome sequencing reveals recurrent mutations in myeloproliferative neoplasms. Leukemia. 2014;28:1052–1059. doi: 10.1038/leu.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinig E., Yang F., Traer E., Arora R., Brown S., Rattray R., Braziel R., Fan G., Press R., Dunlap J. Targeted next-generation sequencing in myelodysplastic syndrome and chronic myelomonocytic leukemia aids diagnosis in challenging cases and identifies frequent spliceosome mutations in transformed acute myeloid leukemia. Am. J. Clin. Pathol. 2016;145:497–506. doi: 10.1093/ajcp/aqw016. [DOI] [PubMed] [Google Scholar]
- 7.Vainchenker W., Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129:667–679. doi: 10.1182/blood-2016-10-695940. [DOI] [PubMed] [Google Scholar]
- 8.Chia Y.C., Islam M.A., Woon P.Y., Johan M.F., Hassan R., Ramli M. Molecular genetics of thrombotic myeloproliferative neoplasms: Implications in precision oncology. Genes Dis. 2021 doi: 10.1016/j.gendis.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott L.M., Rebel V.I. JAK2 and genomic instability in the myeloproliferative neoplasms: A case of the chicken or the egg? Am. J. Hematol. 2012;87:1028–1036. doi: 10.1002/ajh.23243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kameda T., Shide K., Yamaji T., Kamiunten A., Sekine M., Taniguchi Y., Hidaka T., Kubuki Y., Shimoda H., Marutsuka K., et al. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: Disease sustainer and disease accelerator. Blood. 2015;125:304–315. doi: 10.1182/blood-2014-04-555508. [DOI] [PubMed] [Google Scholar]
- 11.Delhommeau F., Dupont S., Della Valle V., James C., Trannoy S., Massé A., Kosmider O., Le Couedic J.P., Robert F., Alberdi A., et al. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 12.Swierczek S.I., Yoon D., Bellanné-Chantelot C., Kim S.J., Saint-Martin C., Delhommeau F., Najman A., Prchal J.T. Extent of hematopoietic involvement by TET2 mutations in JAK2V617F polycythemia vera. Haematologica. 2011;96:775–778. doi: 10.3324/haematol.2010.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moola S., Munn Z., Tufanaru C., Aromataris E., Sears K., Sfetcu R., Currie M., Lisy K., Qureshi R., Mattis P., et al. Chapter 7: Systematic reviews of etiology and risk. In: Aromataris E., Munn Z., editors. JBI Manual for Evidence Synthesis. JBI; Adelaide, Australia: 2020. [(accessed on 12 December 2020)]. Available online: https://synthesismanual.jbi.global. [Google Scholar]
- 14.Ha J.S., Jeon D.S., Kim J.R., Ryoo N.H., Suh J.S. Analysis of the ten-eleven translocation 2 (TET2) gene mutation in myeloproliferative neoplasms. Ann. Clin. Lab. Sci. 2014;44:173–179. [PubMed] [Google Scholar]
- 15.Abdel-Wahab O., Levine R.L. EZH2 mutations: Mutating the epigenetic machinery in myeloid malignancies. Cancer Cell. 2010;18:105–107. doi: 10.1016/j.ccr.2010.07.006. [DOI] [Google Scholar]
- 16.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Islam M.A., Alam S.S., Kundu S., Hossan T., Kamal M.A., Cavestro C. Prevalence of Headache in Patients with Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis of 14,275 Patients. Front. Neurol. 2020;11:562634. doi: 10.3389/fneur.2020.562634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 19.Ihle M.A., Fassunke J., König K., Grünewald I., Schlaak M., Kreuzberg N., Tietze L., Schildhaus H.-U., Büttner R., Merkelbach-Bruse S. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p. V600E and non-p. V600E BRAF mutations. BMC Cancer. 2014;14:13. doi: 10.1186/1471-2407-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andréasson B., Pettersson H., Wasslavik C., Johansson P., Palmqvist L., Asp J. ASXL1 mutations, Pevious vascular complications and age at diagnosis predict survival in 85 WHO-defined polycythaemia vera patients. Br. J. Haematol. 2020;189:913–919. doi: 10.1111/bjh.16450. [DOI] [PubMed] [Google Scholar]
- 21.Barraco D., Cerquozzi S., Gangat N., Patnaik M.M., Lasho T., Finke C., Hanson C.A., Ketterling R.P., Pardanani A., Tefferi A. Monocytosis in polycythemia vera: Clinical and molecular correlates. Am. J. Hematol. 2017;92:640–645. doi: 10.1002/ajh.24740. [DOI] [PubMed] [Google Scholar]
- 22.Bartels S., Faisal M., Büsche G., Schlue J., Hasemeier B., Schipper E., Vogtmann J., Westphal L., Lehmann U., Kreipe H. Mutations associated with age-related clonal hematopoiesis in PMF patients with rapid progression to myelofibrosis. Leukemia. 2019;34:1364–1372. doi: 10.1038/s41375-019-0668-5. [DOI] [PubMed] [Google Scholar]
- 23.Brecqueville M., Rey J., Bertucci F., Coppin E., Finetti P., Carbuccia N., Cervera N., Gelsi-Boyer V., Arnoulet C., Gisserot O., et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012;51:743–755. doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- 24.Brecqueville M., Rey J., Devillier R., Guille A., Gillet R., Adélaide J., Gelsi-Boyer V., Arnoulet C., Chaffanet M., Mozziconacci M.J., et al. Array comparative genomic hybridization and sequencing of 23 genes in 80 patients with myelofibrosis at chronic or acute phase. Haematologica. 2014;99:37–45. doi: 10.3324/haematol.2013.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbuccia N., Murati A., Trouplin V., Brecqueville M., Adelaide J., Rey J., Vainchenker W., Bernard O., Chaffanet M., Vey N. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–2186. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- 26.Cerquozzi S., Barraco D., Lasho T., Finke C., Hanson C.A., Ketterling R.P., Pardanani A., Gangat N., Tefferi A. Risk factors for arterial versus venous thrombosis in polycythemia vera: A single center experience in 587 patients. Blood Cancer J. 2017;7:662. doi: 10.1038/s41408-017-0035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delic S., Rose D., Kern W., Nadarajah N., Haferlach C., Haferlach T., Meggendorfer M. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and polycythaemia vera. Br. J. Haematol. 2016;175:419–426. doi: 10.1111/bjh.14269. [DOI] [PubMed] [Google Scholar]
- 28.Gill H., Ip H.W., Yim R., Tang W.F., Pang H.H., Lee P., Leung G.M.K., Li J., Tang K., So J.C.C., et al. Next-generation sequencing with a 54-gene panel identified unique mutational profile and prognostic markers in Chinese patients with myelofibrosis. Ann. Hematol. 2019;98:869–879. doi: 10.1007/s00277-018-3563-7. [DOI] [PubMed] [Google Scholar]
- 29.Guglielmelli P., Biamonte F., Score J., Hidalgo-Curtis C., Cervantes F., Maffioli M., Fanelli T., Ernst T., Winkelman N., Jones A.V., et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118:5227–5234. doi: 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 30.Huang X., Wu J., Deng X., Xu X., Zhang X., Ma W., Hu T., Yang J., Guan M., Tang G. Mutation profiles of classic myeloproliferative neoplasms detected by a customized next-generation sequencing-based 50-gene panel. J. Bio-X Res. 2020;3:13–20. doi: 10.1097/JBR.0000000000000061. [DOI] [Google Scholar]
- 31.Hussein K., Abdel-Wahab O., Lasho T.L., Van Dyke D.L., Levine R.L., Hanson C.A., Pardanani A., Tefferi A. Cytogenetic correlates of TET2 mutations in 199 patients with myeloproliferative neoplasms. Am. J. Hematol. 2010;85:81. doi: 10.1002/ajh.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kröger N., Panagiota V., Badbaran A., Zabelina T., Triviai I., Araujo Cruz M.M., Shahswar R., Ayuk F., Gehlhaar M., Wolschke C., et al. Impact of Molecular Genetics on Outcome in Myelofibrosis Patients after Allogeneic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2017;23:1095–1101. doi: 10.1016/j.bbmt.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 33.Oppliger Leibundgut E., Haubitz M., Burington B., Ottmann O.G., Spitzer G., Odenike O., McDevitt M.A., Röth A., Snyder D.S., Baerlocher G.M. Dynamics of mutations in patients with essential thrombocythemia treated with imetelstat. Haematologica. 2020 doi: 10.3324/haematol.2020.252817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magor G.W., Tallack M.R., Klose N.M., Taylor D., Korbie D., Mollee P., Trau M., Perkins A.C. Rapid Molecular Profiling of Myeloproliferative Neoplasms Using Targeted Exon Resequencing of 86 Genes Involved in JAK-STAT Signaling and Epigenetic Regulation. J. Mol. Diagn. 2016;18:707–718. doi: 10.1016/j.jmoldx.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Aviles L., Besses C., Alvarez-Larran A., Torres E., Serrano S., Bellosillo B. TET2, ASXL1, IDH1, IDH2, And c-CBL genes in JAK2-and MPL-negative myeloproliferative neoplasms. Ann. Hematol. 2012;91:533–541. doi: 10.1007/s00277-011-1330-0. [DOI] [PubMed] [Google Scholar]
- 36.Myrtue Nielsen H., Lykkegaard Andersen C., Westman M., Sommer Kristensen L., Asmar F., Arvid Kruse T., Thomassen M., Stauffer Larsen T., Skov V., Lotte Hansen L., et al. Epigenetic changes in myelofibrosis: Distinct methylation changes in the myeloid compartments and in cases with ASXL1 mutations. Sci. Rep. 2017;7:6774. doi: 10.1038/s41598-017-07057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nischal S., Bhattacharyya S., Christopeit M., Yu Y., Zhou L., Bhagat T.D., Sohal D., Will B., Mo Y., Suzuki M., et al. Methylome profiling reveals distinct alterations in phenotypic and mutational subgroups of myeloproliferative neoplasms. Cancer Res. 2013;73:1076–1085. doi: 10.1158/0008-5472.CAN-12-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Sullivan J.M., Hamblin A., Yap C., Fox S., Boucher R., Panchal A., Alimam S., Dreau H., Howard K., Ware P., et al. The poor outcome in high molecular risk, hydroxycarbamide-resistant/intolerant ET is not ameliorated by ruxolitinib. Blood. 2019;134:2107–2111. doi: 10.1182/blood.2019001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pardanani A., Lasho T., Finke C., Oh S.T., Gotlib J., Tefferi A. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia. 2010;24:1713–1718. doi: 10.1038/leu.2010.163. [DOI] [PubMed] [Google Scholar]
- 40.Patel K.P., Newberry K.J., Luthra R., Jabbour E., Pierce S., Cortes J., Singh R., Mehrotra M., Routbort M.J., Luthra M., et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood. 2015;126:790–797. doi: 10.1182/blood-2015-03-633404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patriarca A., Colaizzo D., Tiscia G., Spadano R., Di Zacomo S., Spadano A., Villanova I., Margaglione M., Grandone E., Dragani A. TET2 mutations in Ph-negative myeloproliferative neoplasms: Identification of three novel mutations and relationship with clinical and laboratory findings. BioMed Res. Int. 2013;2013:929840. doi: 10.1155/2013/929840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saint-Martin C., Leroy G., Delhommeau F., Panelatti G., Dupont S., James C., Plo I., Bordessoule D., Chomienne C., Delannoy A., et al. Analysis of the Ten-Eleven Translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood. 2009;114:1628–1632. doi: 10.1182/blood-2009-01-197525. [DOI] [PubMed] [Google Scholar]
- 43.Schlenk R.F., Stegelmann F., Reiter A., Jost E., Gattermann N., Hebart H., Waller C., Hochhaus A., Platzbecker U., Schafhausen P., et al. Pomalidomide in myeloproliferative neoplasm-associated myelofibrosis. Leukemia. 2017;31:889–895. doi: 10.1038/leu.2016.299. [DOI] [PubMed] [Google Scholar]
- 44.Schnittger S., Bacher U., Eder C., Dicker F., Alpermann T., Grossmann V., Kohlmann A., Kern W., Haferlach C., Haferlach T. Molecular analyses of 15,542 patients with suspected BCR-ABL1-negative myeloproliferative disorders allow to develop a stepwise diagnostic workflow. Haematologica. 2012;97:1582–1585. doi: 10.3324/haematol.2012.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segura-Díaz A., Stuckey R., Florido Y., González-Martín J.M., López-Rodríguez J.F., Sánchez-Sosa S., González-Pérez E., Perdomo M.N.S., del Mar Perera M., de la Iglesia S., et al. Thrombotic risk detection in patients with polycythemia vera: The predictive role of DNMT3A/TET2/ASXL1 mutations. Cancers. 2020;12:934. doi: 10.3390/cancers12040934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song J., Hussaini M., Zhang H., Shao H., Qin D., Zhang X., Ma Z., Hussnain Naqvi S.M., Zhang L., Moscinski L.C. Comparison of the Mutational Profiles of Primary Myelofibrosis, Polycythemia Vera, and Essential Thrombocytosis. Am. J. Clin. Pathol. 2017;147:444–452. doi: 10.1093/ajcp/aqw222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tefferi A., Pardanani A., Lim K.H., Abdel-Wahab O., Lasho T.L., Patel J., Gangat N., Finke C.M., Schwager S., Mullally A., et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tefferi A., Lasho T.L., Abdel-Wahab O., Guglielmelli P., Patel J., Caramazza D., Pieri L., Finke C.M., Kilpivaara O., Wadleigh M., et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tefferi A., Lasho T.L., Finke C.M., Elala Y., Hanson C.A., Ketterling R.P., Gangat N., Pardanani A. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;1:105–111. doi: 10.1182/bloodadvances.2016000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tefferi A., Lasho T.L., Guglielmelli P., Finke C.M., Rotunno G., Elala Y., Pacilli A., Hanson C.A., Pancrazzi A., Ketterling R.P., et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016;1:21–30. doi: 10.1182/bloodadvances.2016000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verger E., Andreoli A., Chomienne C., Kiladjian J.J., Cassinat B. TET2 gene sequencing may be helpful for myeloproliferative neoplasm diagnosis. Br. J. Haematol. 2014;165:416–419. doi: 10.1111/bjh.12738. [DOI] [PubMed] [Google Scholar]
- 52.Chunxia Z., Li G., Qianqian J., Zhenling L., Fanzhou H., Yayue G., Ming G., Shaohua X., Yin T., Yanrong C., et al. Symptom burden and its relationships with risk assessment and gene mutations in patients with primary myelofibrosis. J. Leuk. Lymphoma. 2015;24:453–456. [Google Scholar]
- 53.Tefferi A., Lim K.H., Abdel-Wahab O., Lasho T.L., Patel J., Patnaik M.M., Hanson C.A., Pardanani A., Gilliland D.G., Levine R.L. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 55.Guo Z., Zhang S.-K., Zou Z., Fan R.-H., Lyu X.-D. Prognostic significance of TET2 mutations in myelodysplastic syndromes: A meta-analysis. Leukemia Res. 2017;58:102–107. doi: 10.1016/j.leukres.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Bejar R., Stevenson K., Abdel-Wahab O., Galili N., Nilsson B., Garcia-Manero G., Kantarjian H., Raza A., Levine R.L., Neuberg D. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosmider O., Gelsi-Boyer V., Cheok M., Grabar S., Della-Valle V., Picard F., Viguié F., Quesnel B., Beyne-Rauzy O., Solary E. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 58.Itzykson R., Kosmider O., Cluzeau T., Mansat-De Mas V., Dreyfus F., Beyne-Rauzy O., Quesnel B., Vey N., Gelsi-Boyer V., Raynaud S. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia. 2011;25:1147–1152. doi: 10.1038/leu.2011.71. [DOI] [PubMed] [Google Scholar]
- 59.Kohlmann A., Grossmann V., Klein H.U., Schindela S., Weiss T., Kazak B., Dicker F., Schnittger S., Dugas M., Kern W., et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J. Clin. Oncol. 2010;28:3858–3865. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 60.Itzykson R., Kosmider O., Renneville A., Gelsi-Boyer V., Meggendorfer M., Morabito M., Berthon C., Adès L., Fenaux P., Beyne-Rauzy O. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J. Clin. Oncol. 2013;31:2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 61.Grossmann V., Kohlmann A., Eder C., Haferlach C., Kern W., Cross N., Haferlach T., Schnittger S. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 62.Tefferi A., Levine R.L., Lim K.H., Abdel-Wahab O., Lasho T.L., Patel J., Finke C.M., Mullally A., Li C.Y., Pardanani A., et al. Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23:900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soucie E., Hanssens K., Mercher T., Georgin-Lavialle S., Damaj G., Livideanu C., Chandesris M.O., Acin Y., Létard S., de Sepulveda P. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood. 2012;120:4846–4849. doi: 10.1182/blood-2011-12-397588. [DOI] [PubMed] [Google Scholar]
- 64.Jardin F., Ruminy P., Parmentier F., Troussard X., Vaida I., Stamatoullas A., Leprêtre S., Penther D., Duval A.B., Picquenot J.M. TET2 and TP53 mutations are frequently observed in blastic plasmacytoid dendritic cell neoplasm. Br. J. Haematol. 2011;153:413–416. doi: 10.1111/j.1365-2141.2010.08556.x. [DOI] [PubMed] [Google Scholar]
- 65.Menezes J., Acquadro F., Wiseman M., Gomez-Lopez G., Salgado R., Talavera-Casanas J., Buno I., Cervera J., Montes-Moreno S., Hernandez-Rivas J. Exome sequencing reveals novel and recurrent mutations with clinical impact in blastic plasmacytoid dendritic cell neoplasm. Leukemia. 2014;28:823–829. doi: 10.1038/leu.2013.283. [DOI] [PubMed] [Google Scholar]
- 66.Mejía-Ochoa M., Toro P.A.A., Cardona-Arias J.A. Systematization of analytical studies of polycythemia vera, essential thrombocythemia and primary myelofibrosis, and a meta-analysis of the frequency of JAK2, CALR and MPL mutations: 2000–2018. BMC Cancer. 2019;19:590. doi: 10.1186/s12885-019-5764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson L.A., Duncombe A.S., Hughes M., Mills M.E., Wilson J.C., McMullin M.F. Environmental, Lifestyle, And familial/ethnic factors associated with myeloproliferative neoplasms. Am. J. Hematol. 2012;87:175–182. doi: 10.1002/ajh.22212. [DOI] [PubMed] [Google Scholar]
- 68.Levine R.L., Wadleigh M., Cools J., Ebert B.L., Wernig G., Huntly B.J., Boggon T.J., Wlodarska I., Clark J.J., Moore S. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 69.Passamonti F., Maffioli M. Update from the latest WHO classification of MPNs: A user’s manual. Hematology. 2016;2016:534–542. doi: 10.1182/asheducation-2016.1.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tefferi A., Vardiman J.W. Classification and diagnosis of myeloproliferative neoplasms: The 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22:14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available within the article and supplementary materials.