Abstract
Pancreatic cancer has the lowest survival rate in all types of cancer. Pancreatic cancer patients are often diagnosed at advanced stages. A better therapeutic development for this devastating disease is urgently needed. Receptor for hyaluronan-mediated motility (RHAMM), not expressed in adult normal pancreas, has been suggested as a prognostic factor and a potential therapeutic target for pancreatic ductal adenocarcinoma (PDAC) and pancreatic neuroendocrine tumor (PNET). In this study, we initially sought to determine whether genetic deletion of RHAMM would slow down pancreatic cancer progression using Rhamm−/− mice. However, we found that Rhamm−/− mice expressed a truncated HMMRΔexon8-16 protein at higher abundance levels than wild-type RHAMM. While HMMRΔexon8-16 did not enable malignant progression of pancreatic intraepithelial neoplasia in p48-Cre; LSL-KRASG12D mice, it accelerated the formation of invasive PDAC and shortened the survival of p48-Cre; LSL-KRASG12D mice with heterozygous p53 knockout. KrasG12D PDAC mice with homozygous p53 knockout mice died around 10 weeks, and the effect of HMMRΔexon8-16 was not apparent in these mice with short life span. In addition, HMMRΔexon8-16 shortened the survival of PNET-bearing RIP-Tag mice, which had inactivated p53. In our analysis of TCGA dataset, pancreatic cancer patients with mutant TP53 or loss of one copy of TP53 had higher RHAMM expression, which, combined, predicted worse outcomes. Taken together, by collaborating with dysfunctional p53, high levels of HMMRΔexon8-16 that lacks the centrosome targeting domain and degrons for interaction with the Anaphase-Promoting Complex (APC) accelerated pancreatic cancer progression.
Keywords: RHAMM, pancreatic ductal adenocarcinoma, pancreatic neuroendocrine tumor, mouse models, p53
1. Introduction
Pancreatic cancer is the leading cause of cancer-related death. In contrast to the steady increase in survival observed for most cancer types, advances have been slow for pancreatic cancer, which is typically diagnosed at an advanced stage [1]. About 90% of all malignant pancreatic tissues are pancreatic ductal adenocarcinoma (PDAC) with a five-year survival rate of <10% [2]. KRAS, CDKN2A, TP53, and SMAD4 are the four frequently mutated genes that characterize PDAC [3]. Activating KRAS mutations are present in more than 99% of pancreatic intraepithelial neoplasia (PanIN) and over 90% of PDAC, and has been shown to be an initiating event of PDAC using genetically engineered mouse models [4]. Studies from mouse models of PDAC further demonstrated that loss of functional p53 cooperates with mutant KRAS to cause rapid PDAC progression and metastasis [5].
Pancreatic neuroendocrine tumor (PNET) is the second most common malignancy of the pancreas. The incidence of PNET is increasing and PNET presents with a wide variety of clinical manifestations and unfavorable survival rates [6]. Unlike PDAC, the major molecular driver for PNETs is not well -understood. Recent molecular characterization of PNETs reveals alterations in pathways/genes such as mTOR pathway, cyclin D1/Cdk4/retinoblastoma (Rb), TP53, MEN1, DAXX, ATRX, UCHL1, RHAMM, and microRNAs [7-9]. In the RIP-Tag mouse model of PNET, the rat insulin promoter (RIP) drives the expression of SV40 T antigen (Tag), providing the driving force for tumor initiation by inhibiting the activities of tumor suppressors, p53 and Rb [10]. Preclinical trials in the RIP-Tag mice have predicted that sunitinib and everolimus would be effective in treating human PNETs [11-13]. However, sunitinib and everolimus only extend the median patient survival by roughly 6 months, and all patients eventually develop resistance to both drugs [14, 15].
Receptor for hyaluronic acid-mediated motility (RHAMM or HMMR) was identified as a receptor of hyaluronan [16], and its expression peaks at G2/M [17, 18]. Studies have shown that RHAMM plays an important role in cell motility [19]. Human RHAMM encodes 18 exons and alternative splicing yields four isoforms [20]. Of the four isoforms (RHAMMv1-4), we have shown mRNA expression levels of RHAMMv3, also known as RHAMMB, to be the most prominent in human PDAC, PNET, and liver metastases of PNET [21]. Using the RIP-Tag; RIP-tva mouse model of PNET and cell line xenograft mouse models, we have discovered RHAMMB as the first protein that is able to promote PNET metastasis to the liver [21, 22], and we have also found RHAMMB expression to correlate with poor survival in PDAC patients [21]. Most normal human tissues, including pancreas, do not express RHAMM [17, 21], and the differential RHAMM expression in normal tissues and pancreatic cancer may thus provide novel therapeutic possibilities by targeting RHAMM. To investigate whether RHAMM is a potential therapeutic target in pancreatic cancer, we set out to determine the effects of RHAMM deletion on the progression and survival of PDAC and PNET in mouse models using Rhamm−/− mice [23]. However, after analyzing RHAMM mRNA and protein expression in Rhamm−/− mice, we unexpectedly discovered that a truncated HMMRΔexon8-16 protein was expressed in Rhamm−/− mice and was more abundant than the full-length protein in RHAMM wild-type (WT) mice. Based on these findings, we propose to rename Rhamm−/− mice as HMMRΔexon8-16/Δexon8-16 mice. In this study, we investigated the effects of the HMMRΔexon8-16 protein on tumor progression and survival of PDAC and PNET in mouse models.
2. Materials and Methods
2.1. Mouse Strains, Animal Husbandry, and Genotyping
Cohorts of A, B, and C groups were of mixed genetic background. p48-Cre; LSL-KRASG12D mice were bred with p53lox/lox mice and HMMRΔexon8-16/Δexon8-16 mice (previously known as Rhamm−/− [23]). Cohorts of PNETs were generated by crossing RIP-Tag mice (C57BL/6 genetic background) with HMMRΔexon8-16/Δexon8-16 mice. All procedures involving mice were approved by the Institutional Animal Care and Use Committee. There was no noticeable influence of sex on the results of this study. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All mice were housed in accordance with institutional guidelines.
Mouse genomic DNA was prepared for PCR genotyping through incubation of each earpiece in 200 μl 0.05 M NaOH for 20 minutes at 98°C followed by the addition of 20 μl 1 M Tris-HCl (pH 7.5) at room temperature. Primers used in the PCR genotyping are as follows: for HMMRΔexon8-16: 5’-TGCTCGAGATGTCATGAAGG-3’ and 5’-CGAGAGGTCCTTTTCACCAG-3’ (a 360-bp product); for HMMR wild-type: 5’-CCAGTGCCCGAGAGAATTTA-3’ and 5’-TCCACTTGATCAGATGCACA-3’ (a 338-bp product) or 5’-AGGGAGCAGTACAGAGGTGT-3’ and 5’-TCGACAGCGTGTTCGGATAG-3’ (a 535-bp product); for p48-cre: 5’-CTGATTTCGACCAGGTTCGT-3’ and 5’-ATTCTCCCACCGTCAGTACG-3’ (a 160-bp product); for β-actin: 5’-CAGCGTTTGCCTTTTATGGT-3’ and 5’-GCTTGCCACTCCCAAAGTAA-3’ (a 335-bp product); for LSL-KRASG12D: 5’-CGCAGACTGTAGAGCAGCG-3’ and 5’-CCATGGCTTGAGTAAGTCTGC-3’ (a 600-bp product; https://www.jax.org/); for p53 conditional knockout allele: 5’-GGTTAAACCCAGCTTGACCA-3’ and 5’-GGAGGCAGAGACAGTTGGAG-3’ (a 370-bp product for p53lox, 240-bp product for WT; https://www.jax.org/). RIP-Tag PCR genotyping was performed as described [24].
2.2. Sequencing of the HMMRΔexon8-16 Transcript and Western Blot Analysis
Mouse testes were removed and frozen on dry ice immediately after euthanasia. The testes were crushed into powder form with liquid nitrogen, and RNA was purified from the powder using the RNeasy Plus Mini Kit (QIAGEN, #74134). The purified RNA was used to synthesize cDNA using SuperScript™ IV VILO™ Master Mix with ezDNase™ Enzyme (Thermofisher, 11766050). An N-terminal HMMR fragment was amplified using AmpliTaq (Thermofisher, N8080153) or Q5 High-Fidelity DNA Polymerase (New England BioLabs, #M0491L) with the following primer pairs: (1) 5’-AACCAGAGCCAACGAGCTAC-3’ (on exon 5) and 5’-AGCCTTGGAAGGGTCAAAGT-3’ (on exon 17) (a 330 bp product to detect HMMRΔexon8-16); and (2) 5’-CTCCGGGTGCTTATGATGTT-3’ (on exon 2) and 5’-CCGTTTTTCCAGTGAAGCAT-3’ (on exon 5) (a 362-bp product for HMMRΔexon8-16 and WT to detect the missense mutation on exon 3). The resulting PCR products were purified using DNA Clean & Concentrator (Zymo, D4013) and were sent to PSOMAGEN, INC. along with the primer, 5’-CCGTTTTTCCAGTGAAGCAT-3’, for sequencing.
Frozen powders of mouse testes were lysed in NP-40 buffer (100 mM NaCl, 100 mM Tris pH 8.2, 0.5% NP-40) supplemented with a protease inhibitor mixture and PhosSTOP (Roche). Proteins were quantified by Bradford assay (Bio-Rad, Hercules, CA). Equal amounts of proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. To visualize equal protein loading, blots were stained with Ponceau S. Blots were incubated in 5% non-fat milk in TBST, probed with primary antibodies to RHAMM (clone EPR4054, Abcam, ab124729, 1:500) or Cyclin B2 (R&D, AF6204, 1 μg/mL), and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Protein bands were visualized by enhanced chemical luminescence (Pierce, Rockford, IL).
2.3. Immunohistochemistry
Mouse tissues were fixed in 10% buffered formalin overnight at room temperature and then transferred to 70% ethanol, paraffin-embedded, and sectioned. Slides were deparaffinized and rehydrated by passaging through a graded xylene/ethanol series before staining. The slides were then used for either hematoxylin and eosin (H&E) staining or immunohistochemical staining. Immunohistochemistry was performed using Vectastain ABC Peroxidase Kit (Peroxidase, Rabbit IgG, PK-6101) following the manufacturer’s instructions. The primary antibodies used are rabbit anti-RHAMM antibody (clone EPR4054, Abcam, ab124729, 1:500) and antibody against cytokeratin 17/19 (Cell Signaling, 3984, 1:500).
2.4. Statistical Analysis
Mice from this study were monitored daily for signs of disease and mortality up to 1 year. Kaplan-Meier method and log-rank test was used to test the survival differences between groups. Simulation method was used to adjust for multiple comparisons for log-rank test.
Publicly available gene expression (RNA-Seq version 2), harmonized mutations and gene level copy number datasets from The Cancer Genome Atlas (TCGA) were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov). Mann–Whitney U test and survival analysis were conducted for this dataset.
All analyses were performed in statistical software GraphPad Prism (version 7.0e) or SAS Version 9.4 (SAS Institute, Cary, NC). P < 0.05 was considered statistically significant.
3. Results
3.1. Detection of HMMRΔexon8-16 protein in Rhamm−/− mice
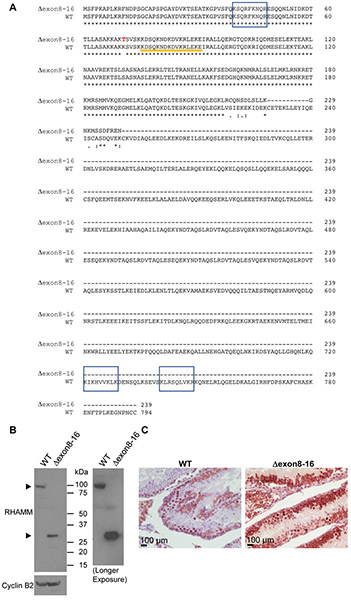
Mouse RHAMM/HMMR has 18 exons (NM_013552.2). The Rhamm−/− mouse strain was previously generated by deleting exons 8 ~16 of the HMMR gene through homologous recombination in embryonic stem (ES) cells [23]. It was reported that no HMMR protein was detected in the spleen lysate of Rhamm−/− mice using RHAMM polyclonal antibody in Western blot analysis [23]. However, because the polyclonal antibody was generated against recombinant RHAMM protein [23] and the specific immunogenic epitope(s) are unknown, we decided to investigate whether any truncated RHAMM protein was expressed in Rhamm−/− mice using a RHAMM-specific monoclonal antibody against the N-terminus (clone EPR4054). The immunogen for this clone EPR4054 was the first 50 amino acids of human RHAMM, and the antibody cross-reacted with mouse RHAMM. Full-length WT mouse RHAMM encodes 794 amino acids with a predicted molecular weight of ~92 kDa. We have previously reported that the testis has the most abundant RHAMM proteins among all adult human tissues, and most normal human tissues, including the pancreas, do not express RHAMM [17]. We predicted accordingly that RHAMM would also be highly expressed in mouse testis and it would be a better tissue than the spleen or the pancreas to evaluate whether a truncated RHAMM protein was expressed. To verify that exons 8 ~ 16 were deleted in the Rhamm−/− mice, we isolated mRNA from the testis of Rhamm−/− mice (n = 3), made cDNA, amplified RHAMM fragments by polymerase chain reaction (PCR), and performed DNA sequencing. Our analysis confirmed a fusion of exon 7 and exon 17, which resulted in a stop codon in exon 17 and would yield a truncated RHAMM of 239 amino acids with a predicted molecular weight of ~27 kDa (Figure 1A). In addition, we found an adenine to cytosine missense mutation at nucleotide position 290, which changed the amino acid from lysine to threonine (residue 71) (Figure 1A and Figure 2). Protein lysates and histological sections from testes of WT mice and Rhamm−/− mice were then evaluated for the expression of RHAMM proteins. Western blot analysis using EPR4054 antibody detected full-length RHAMM protein (~92 kDa) in WT mouse testes and truncated RHAMM protein (~27 kDa) in Rhamm−/− mouse testes (Figure 1B). Immunohistochemical staining using EPR4054 antibody also revealed RHAMM protein in Rhamm−/− mouse testes (Figure 1C). Both Western blot and immunohistochemical analyses showed that the truncated RHAMM protein (HMMRΔexon8-16) was highly expressed in Rhamm−/− mice, at levels more abundant than WT RHAMM protein in the control WT mice. Therefore, we renamed this mouse strain from Rhamm−/− to HMMRΔexon8-16/Δexon8-16.
Figure 1. Comparison of WT mouse RHAMM/HMMR protein and HMMRΔexon8-16.
(A) Deduced amino acid sequence of HMMRΔexon8-16 and WT mouse RHAMM/HMMR (translated from NM_013552.2). Potential hyaluronan-binding sites (BX7B motif) are shown in shaded boxes. Sequences from exon 4 are indicated by an orange line. A missense mutation from lysine (K) to threonine (T) at residue 71 is shown in red font. (B) Western blot analysis of mouse RHAMM in the testis of WT and HMMRΔexon8-16/Δexon8-16. The right panel was a longer exposure than the left panel for RHAMM. Cyclin B2 was used as a control. (C) Immunohistochemical staining showed RHAMM staining in the germ cells of testis from a HMMRΔexon8-16/Δexon8-16 mouse, at stronger intensity than that observed in the testis of a WT mouse.
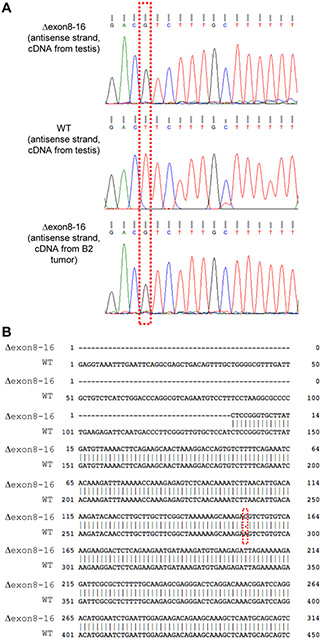
Figure 2. A missense mutation at nucleotide position 290 of the coding region of HMMRΔexon8-16.
(A) DNA sequencing showed a mutation from T to G (antisense strand) from HMMR transcripts in the testis of a HMMRΔexon8-16/Δexon8-16 mouse and in PDAC of B2: p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/Δexon8-16 mouse, as compared to transcripts derived from the testis of a WT mouse. (B) Sequence alignment between a partial fragment of HMMRΔexon8-16 and a partial coding sequence of NM_013552.2 showed an A to C missense mutation at nucleotide position 290.
3.2. HMMRΔexon8-16/Δexon8-16 mice have normal pancreas development
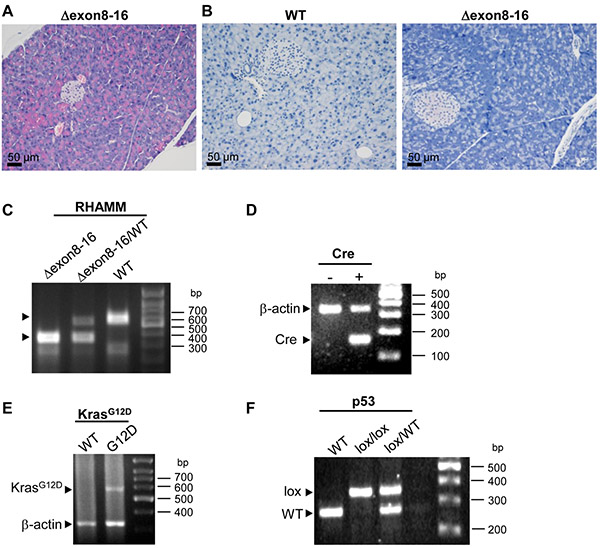
Consistent with the previous report [23], we found no embryonic lethality for HMMRΔexon8-16/Δexon8-16 mice. The appearance of HMMRΔexon8-16/Δexon8-16 mice was normal throughout their lifespan and live longer than 1 year without any spontaneous tumorigenesis (n = 10, data not shown). To determine whether the truncated RHAMM protein (HMMRΔexon8-16) causes any abnormality in pancreatic development, the pancreas from 5 adult HMMRΔexon8-16/Δexon8-16 was examined and no histologic abnormality was identified (Figure 3A). Similar to the lack of detectable RHAMM expression in human adult pancreas [17, 21], no immunohistochemical signal of RHAMM was found in the pancreas of WT mouse and Rhamm−/− mouse using the EPR4054 antibody (Figure 3B).
Figure 3. HMMRΔexon8-16/Δexon8-16 mice have normal pancreas.
(A) Hematoxylin and eosin (H&E) stain of representative pancreas from a 18-week-old HMMRΔexon8-16/Δexon8-16 mouse. (B) Immunohistochemical staining detected no RHAMM expression in the pancreas of a WT mouse and a HMMRΔexon8-16/Δexon8-16 mouse. (C-F) PCR genotyping results for HMMRΔexon8-16 (C), Cre (D), KrasG12D (E), and p53lox (F). β-actin was used as endogenous control.
3.3. HMMRΔexon8-16 did not alter the course of PanIN formation in p48-Cre; LSL-KRASG12D mice
It has been shown that the expression of mutant KRASG12D from the endogenous locus in the mouse pancreas leads to the development of premalignant ductal lesions, PanINs, in Pdx1-Cre; LSL-KRASG12D mice and p48-Cre; LSL-KRASG12D mice [4]. Subsequent studies revealed that p53 constrains the progression of PanIN to advanced PDAC [5, 25]. Pdx1-Cre; LSL-KRASG12D mice harboring heterozygous p53 loss or homozygous-null p53 developed PDAC with an average latency of 21.8 weeks or 6.2 weeks, respectively [5]. Because the expression of the Cre recombinase under p48 promoter is restricted to mouse pancreas compared to that under the Pdx1 promoter [3], we decided to use p48-Cre instead of Pdx1-Cre to target mutant KrasG12D expression in the pancreas in this study. To determine the effect of N-terminal RHAMM (HMMMRΔexon8-16) on the initiation and the progression of PanINs and pancreatic ductal adenocarcinomas (PDAC), we crossed HMMRΔexon8-16/Δexon8-16 to p48-Cre; LSL-KRASG12D mice with WT p53 (Group A), heterozygous p53 loss (p53lox/+, Group B), or homozygous-null p53 (p53lox/lox, Group C) (Table 1, and Figure 3C-3F for the PCR genotyping results). Within each group (A, B, and C), we evaluated the impact of WT HMMR (A0, B0, and C0), one copy of HMMRΔexon8-16 (HMMRΔexon8-16/WT: A1, B1, and C1), and two copies of HMMRΔexon8-16 (HMMRΔexon8-16/Δexon8-16: A2, B2, and C2) (Table 1).
Table 1.
Histological Phenotypes and Average Life Span of Mice
| Group | Genotype | Histology | Median survival (week) |
|---|---|---|---|
| A0 | p48-Cre; LSL-KRASG12D | PanIN | 46.79 |
| A1 | p48-Cre; LSL-KRASG12D; HMMRΔexon8-16/WT | PanIN | undefined |
| A2 | p48-Cre; LSL-KRASG12D; HMMRΔexon8-16/Δexon8-16 | PanIN | 48.86 |
| B0 | p48-Cre; LSL-KRASG12D; p53lox/+ | PDAC | 23.93 |
| B1 | p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/WT | PDAC | 17.50 |
| B2 | p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/Δexon8-16 | PDAC | 15.41 |
| C0 | p48-Cre; LSL-KRASG12D; p53lox/lox | PDAC | 9.14 |
| C1 | p48-Cre; LSL-KRASG12D; p53lox/lox; HMMRΔexon8-16/WT | PDAC | 9.86 |
| C2 | p48-Cre; LSL-KRASG12D; p53lox/lox; HMMRΔexon8-16/Δexon8-16 | PDAC | 9.57 |
| T0 | RIP-Tag | PNET | 17.57 |
| T1 | RIP-Tag; HMMRΔexon8-16/WT | PNET | 16.14 |
| T2 | RIP-Tag; HMMRΔexon8-16/Δexon8-16 | PNET | 15.50 |
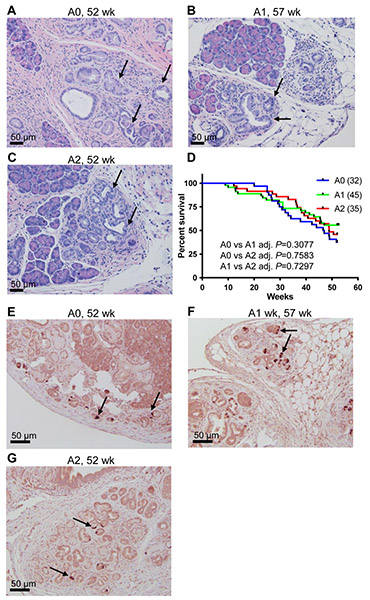
As expected, our cohort of p48-Cre; LSL-KRASG12D mice (A0) developed PanINs (Figure 4A). The majority of the A group mice lived longer than 40 weeks of age (Figure 4D). We found no significant survival difference among A0, A1, and A2 after following them up to 1 year (Figure 4D, Log-rank test, A0: 32 mice, A1: 45 mice, and A2: 35 mice). Similar results were obtained from Cox regression (data not shown). Mice in A0, A1, and A2 at the end point (1 year of age, , n > 3 for each subgroup) all developed PanINs, but no invasive PDAC (Figure 4A-4C). Immunohistochemical staining with RHAMM antibody showed expression of RHAMM in these PanIN lesions (Figure 4E-4G). This absence of invasive PDAC suggests that N-terminal RHAMM protein (HMMRΔexon8-16) cannot promote the progression of PanINs initiated by KRASG12D to PDAC.
Figure 4. HMMRΔexon8-16 does not significantly change the survival of p48-Cre; LSL-KRASG12D mice.
(A-C) Hematoxylin and eosin (H&E) stain of representative pancreas from (A) A0: p48-Cre; LSL-KRASG12D mouse, (B) A1: p48-Cre; LSL-KRASG12D; HMMRΔexon8-16/WT mouse, (C) A2: p48-Cre; LSL-KRASG12D; HMMRΔexon8-16/Δexon8-16 mouse. A0, A1 and A2 mice (1 year of age, n=3) all showed localized ductal proliferation with cytological dysplasia, consistent with PanINs (arrows), but no evidence of invasive PDAC. (D) Kaplan-Meier survival curve for A0: 32 mice, A1: 45 mice, and A2: 35 mice. Tukey adjusted P values from pairwise log-rank test were shown. (E-G) Immunohistochemical staining of these A0, A1 and A2 mice identified scattered RHAMM-positive cells in the PanIN lesions (arrows).
3.4. HMMRΔexon8-16 shortened the survival and accelerated PDAC formation in p48-Cre; LSL-KRASG12D; p53lox/+ mice
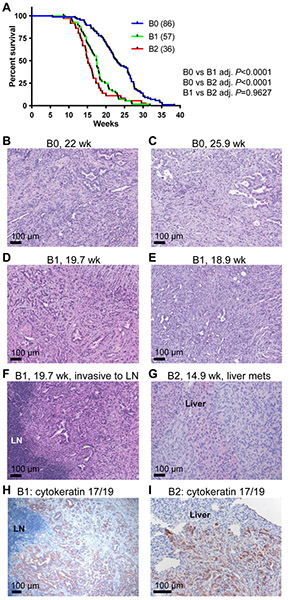
We established a cohort of B group, p48-Cre; LSL-KRASG12D mice with heterozygous p53 loss (p53lox/+) and different copy numbers of HMMRΔexon8-16 (B0, B1, and B2). Mice in the B group lived less than a year, and all mice in these three subgroups developed advanced PDAC at death or when they were sick (Figure 5 and data not shown). Having either one copy or two copies of HMMRΔexon8-16 significantly shortened the survival of p48-Cre; LSL-KRASG12D; p53lox/+ mice (Figure 5A, Log-rank test, B0: 86 mice, B1: 57 mice, and B2: 36 mice). The median survival ages for B0, B1, and B2 were 22.93 weeks, 17.5 weeks, and 15.14 weeks, respectively (Table 1). Cox regression analysis showed that B1 is 164% more likely to die at any time point than B0 (P < 0.0001) and B2 is 234% more likely to die than B0 (P < 0.0001). At the terminal stage of each subgroup, there was no significant difference in the PDAC histology found between p48-Cre; LSL-KRASG12D; p53lox/+ mice with either one copy or two copies of HMMRΔexon8-16 (Figure 5B-G, and data not shown). However, direct invasion of PDAC into peripancreatic lymph nodes and metastasis to liver were observed in mice with HMMRΔexon8-16 (Figure 5F and 5G). These foci of carcinoma in the lymph node and in liver were confirmed and highlighted by immunostaining for cytokeratin 17/19, a marker known to be expressed in PDAC (Figure 5H and 5I). We also confirmed the existence of the adenine to cytosine missense mutation at nucleotide position 290 of the coding region of HMMRΔexon8-16 in a PDAC tumor from a B2 mouse (Figure 2).
Figure 5. HMMRΔexon8-16 shortened the survival of p48-Cre; LSL-KRASG12D; p53lox/+ mice.
(A) Kaplan-Meier survival curve for B0: 86 mice, B1: 57 mice, and B2: 36 mice. Tukey adjusted P values from pairwise log-rank test were shown. (B-E) H&E stain of invasive PDAC from (B) a 22-week-old B0: p48-Cre; LSL-KRASG12D; p53lox/+ mouse, (C) a 25.9-week-old B0: p48-Cre; LSL-KRASG12D; p53lox/+ mouse, (D) a 19.7-week-old B1: p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/WT mouse, and (E) a 18.9-week-old B1: p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/WT mouse. (F) H&E stain of a peripancreatic lymph node (LN) invaded by PDAC from the same animal in (D). (G) H&E stain of liver metastasis of PDAC from a 14.9-week-old B2: p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/Δexon8-16 mouse. (H, I) Immunohistochemical staining of cytokeratin 17/19 of the lymph node (LN) invasion (H) and liver metastasis (I).
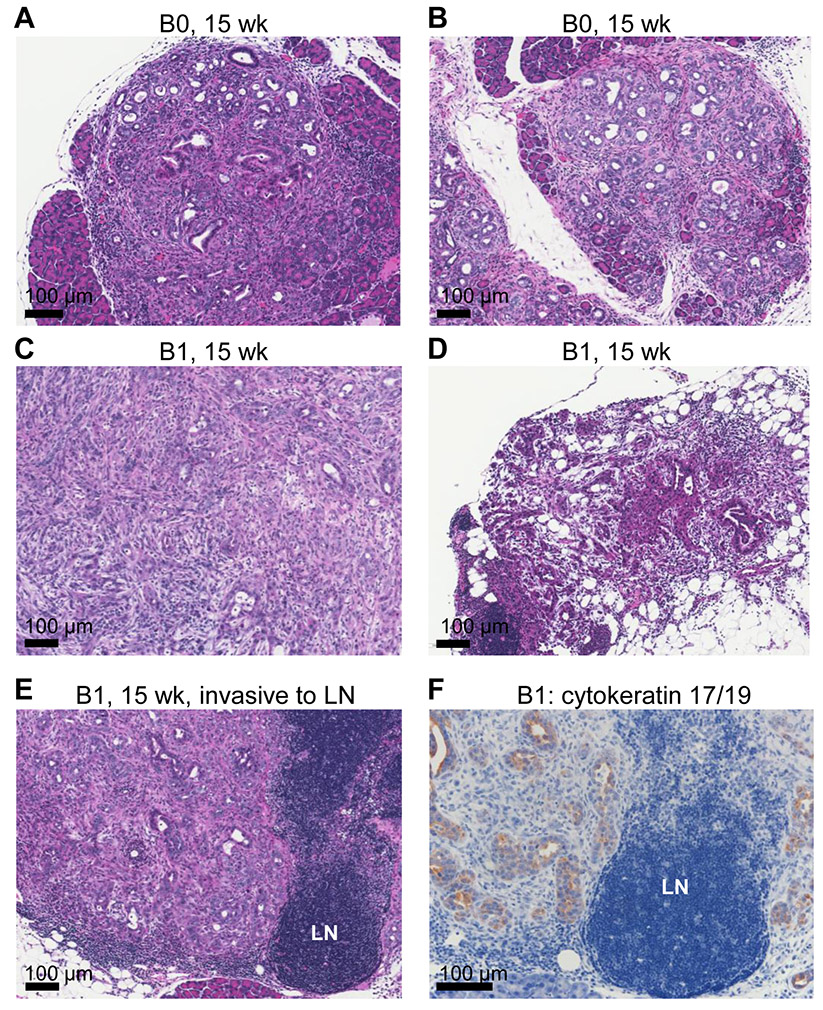
To investigate why p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/WT (B1) died earlier than p48-Cre; LSL-KRASG12D; p53lox/+ (B0), we sacrificed a cohort of B0 and B1 mice at the age of 15 weeks (n =3 per group). We found that whereas B0 mice showed only PanINs and not PDAC at 15 weeks (Figure 6A and 6B), B1 mice already developed invasive PDAC (Figure 6C, 6D), and direct invasion into a peripancreatic lymph node was also observed (Figure 6E), further illustrated by immunostaining for cytokeratin 17/19 (Figure 6F). Taken together, in the presence of heterozygous p53 loss, having one copy of HMMRΔexon8-16 was sufficient to accelerate progression of PanINs into invasive PDAC, leading to the shortened survival in the p48-Cre; LSL-KRASG12D; p53lox/+ mice.
Figure 6. HMMRΔexon8-16 accelerated PDAC formation in p48-Cre; LSL-KRASG12D; p53lox/+ mice.
H&E stain of representative PDAC from B group mice at 15 weeks of age. (A and B) two B0: p48-Cre; LSL-KRASG12D; p53lox/+ mice. (C and D) two B1: p48-Cre; LSL-KRASG12D; p53lox/+; HMMRΔexon8-16/WT mice, showing advanced PDAC (C) and early PDAC (D), respectively. (E) H&E stain of a peripancreatic lymph node (LN) invaded by PDAC from the same animal in (C). (F) Immunohistochemical staining of cytokeratin 17/19 of the LN invasion.
3.5. p48-Cre; LSL-KRASG12D; p53lox/lox mice had rapid PDAC progression irrespective of HMMRΔexon8-16 status
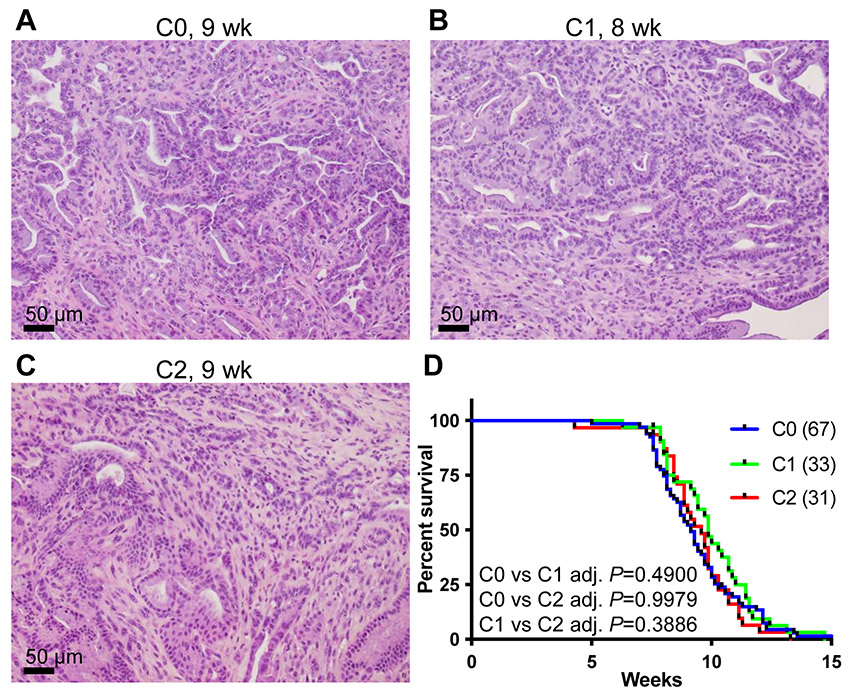
We established a cohort of group C mice of p48-Cre; LSL-KRASG12D; p53lox/lox. These mice were further divided into 3 subgroups (C0, C1, and C2) based on the copy numbers of HMMRΔexon8-16. At the terminal stage, all C0, C1, and C2 subgroups showed advanced invasive PDAC, and the histomorphology was similar and indistinguishable among subgroups. The C group mice developed advanced PDAC as the B group but at earlier ages (Figure 7A-7C and Table 1). The mice in the C group had very short life spans (Figure 7D and Table 1, Log-rank test, C0: 67 mice, C1: 33 mice, and C2: 31 mice). The median survival ages for C0, C1, and C2 were all between 9 and 10 weeks (Table 1). There was no significant difference in survival found by log-rank test.
Figure 7. The p48-Cre; LSL-KRASG12D; p53lox/lox mice had too rapid PDAC progression to potentiate the effect of HMMRΔexon8-16.
(A-C) H&E stain of representative PDAC from (A) a 9-week-old C0: p48-Cre; LSL-KRASG12D; p53lox/ lox mouse, (B) an 8-week-old C1: p48-Cre; LSL-KRASG12D; p53lox/ lox; HMMRΔexon8-16/WT mouse, and (C) a 9-week-old C2: p48-Cre; LSL-KRASG12D; p53lox/ lox; HMMRΔexon8-16/Δexon8-16 mouse. (D) Kaplan-Meier survival curve for C0: 67 mice, C1: 33 mice, and C2: 31 mice. Tukey adjusted P values from pairwise log-rank test were shown.
3.6. HMMRΔexon8-16 reduced the survival of RIP-Tag PNET mice
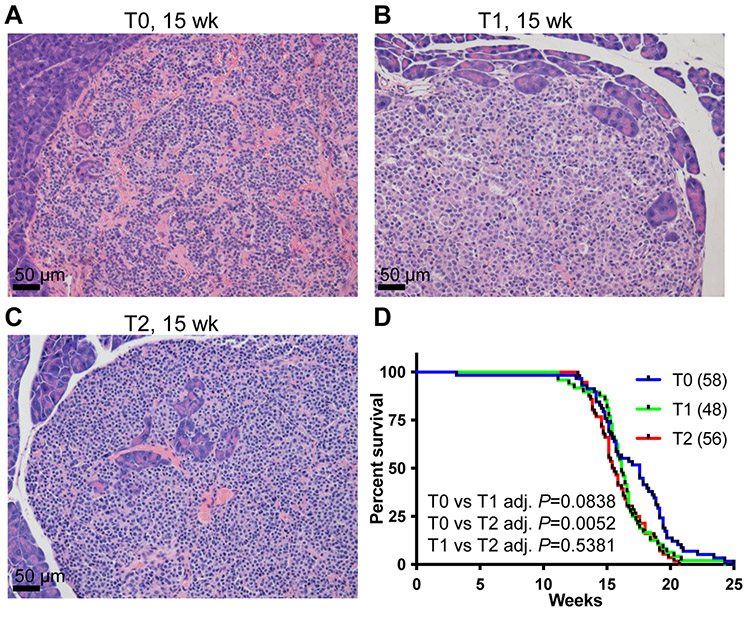
RIP-Tag mice developed insulinoma, one subtype of PNETs [10]. As a result, the RIP-Tag mice died before 18 weeks of age due to hypoglycemia. Because homozygous RIP-Tag mice develop PNET faster [24], only hemizygous RIP-Tag mice in C57BL/6 background were used here. We established a cohort of RIP-Tag mice (T group) with different copy numbers of HMMRΔexon8-16 (T0, T1, and T2). Healthy mice have non-fasting blood glucose levels around 200 mg/dL, and T group mice developed PNET with blood glucose levels lower than 50 mg/dL at the end point (data not shown). PNET histology at the terminal stage among the T subgroups were similar (Figure 8A-C). RIP-Tag; HMMRΔexon8-16/WT (T1) and RIP-Tag; HMMRΔexon8-16/Δexon8-16 (T2) died earlier than RIP-Tag (T0) (Figure 8D). Similar to the log-rank test results (Figure 8D), Cox regression analysis showed borderline significant difference between T1 vs T0: T1 is 46% more likely to die than T0 (P = 0.0573), and significant difference between T2 vs T0: T2 is 78% more likely to die than T0 (P = 0.0029). The median survival ages for T0, T1, and T2 mice were 17.57 weeks, 16.14 weeks, and 15.5 weeks, respectively (Table 1, T0: 58 mice, T1: 48 mice, and T2: 56 mice).
Figure 8. HMMRΔexon8-16 reduced the survival of RIP-Tag PNET mice.
H&E stain of representative PNETs from (A) a 15-week-old T0: RIP-Tag mouse, (B) a 15-week-old T1: RIP-Tag; HMMRΔexon8-16/WT mice, and (C) a 15-week-old T2: RIP-Tag; HMMRΔexon8-16/Δexon8-16 mouse. (D) Kaplan-Meier survival curve for T0: 58 mice, T1: 48 mice, and T2: 56 mice. Tukey adjusted P values from pairwise log-rank test were shown.
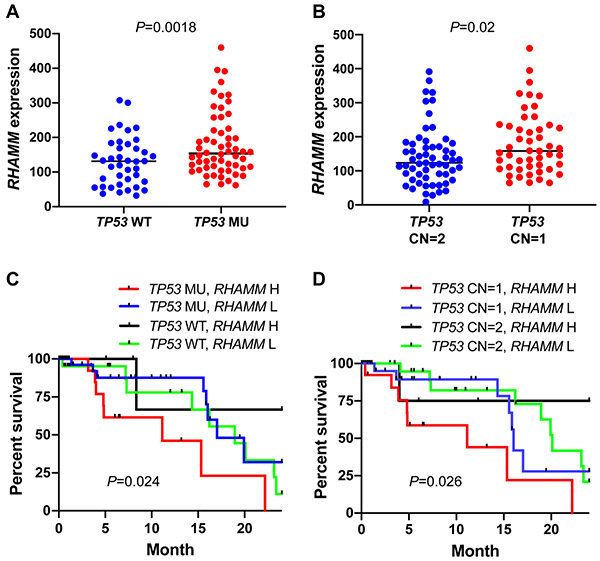
3.7. The association between RHAMM expression levels and TP53 mutations or copy number loss in TCGA pancreatic cancer cohort
It was previously reported that TP53 transcriptionally downregulates RHAMM in colon carcinoma cell lines [18]. We evaluated whether the RHAMM expression is associated with TP53 mutational status and copy number loss in pancreatic cancer patients using The Cancer Genome Atlas (TCGA) dataset that includes the data with gene expression (RNA-Seq), harmonized mutation, and gene level copy number. PDAC is the major pancreatic cancer type in the dataset. TP53 mutations in this dataset include missense mutation, nonsense mutation, frame shift deletion, frame shift insertion, in-frame insertion, and splice site. We found upregulation of RHAMM in the pancreatic cancer patients with TP53 mutations (Figure 9A, P < 0.05). Because TP53 can also be inactivated by copy number loss, we evaluated whether the RHAMM expression is associated with the copy number variation (CNV) of TP53. We observed that RHAMM levels were higher in patients with one TP53 copy number (CN = 1) than in patients with 2 copy numbers (Figure 9B, P < 0.05). These data suggested that WT TP53 protein possess a transcriptional repression activity for RHAMM/HMMR in vivo. Then, we conducted a survival analysis using RHAMM/HMMR expression and TP53 mutational status or its CNV. We found that patients have significantly different survival outcome among different TP53 status/RHAMM levels combination groups. Patients who have mutant TP53/high RHAMM had the shortest survival among all four groups (Figure 9C, overall log-rank P = 0.024). Similarly, patients who have TP53 CN = 1/high RHAMM had shortest survival among four groups (Figure 8D, overall log-rank P = 0.026).
Figure 9. The association between TP53 mutational status/copy number loss and RHAMM/HMMR expression levels in TCGA pancreatic cancer dataset.
(A) Upregulation of RHAMM/HMMR expression in the TCGA pancreatic cancer patients with TP53 mutations. WT: wild type (n = 53); MU: mutation (n = 66). (B) Upregulation of RHAMM/HMMR expression in the patients with copy number loss (CN = 1, n = 49) compared to autosomal copy number (CN = 2, n = 59). (C) Combination of TP53 mutations and high RHAMM/HMMR expression predicted worse outcome. (D) Combination of TP53 copy number loss (CN = 1) and high RHAMM/HMMR expression predicted worse outcome. High (H) and low (L) was based on the 75 quartile RHAMM/HMMR expression value. P values from log-rank test were shown in C and D.
4. Discussion
Pancreatic cancer is lethal and new treatment options for this devastating disease are urgently needed. We have reported the upregulation of RHAMM/HMMR in human PDAC, PNET, and many other cancer types, while RHAMM /HMMR protein is not expressed in most of the normal tissues including pancreas [17, 20, 21, 26, 27]. This property of RHAMM makes RHAMM blockage a very attractive therapeutic target in pancreatic cancer.
To test the above idea in pre-clinical mouse models of PDAC and PNET, we originally took advantage of an existing RHAMM deletion strain, Rhamm−/− mouse [23]. However, our molecular and histological characterization identified the expression of a truncated RHAMM (HMMRΔexon8-16) with a molecular weight of ~27 kDa in the Rhamm−/− mice. HMMRΔexon8-16 protein is more abundant than WT RHAMM/HMMR in WT mice. While HMMRΔexon8-16 by itself did not promote the progression of PanINs to PDAC (Group A, p48-Cre; LSL-KRASG12D mice), the combination of HMMRΔexon8-16 and heterozygous p53 loss promote earlier onset of invasive PDAC formation (Group B, p48-Cre; LSL-KRASG12D; p53lox/+ mice). The more copies of HMMRΔexon8-16, the more likely that mice would die early for Group B mice and Group T, RIP-Tag PNET mice. Consistent with our findings using mouse models, pancreatic cancer patients who have mutant TP53/high RHAMM or TP53 CN=1/high RHAMM had shortest survival among different TP53 status/RHAMM levels combination groups. These data together support that high levels of RHAMM/HMMR cooperated with dysfunctional p53 to accelerate the progression of pancreatic cancer in mice and in humans.
In addition to the inverse association between RHAMM expression levels and p53 mutational or deletion status found in human PDAC (Figure 8), we found the inverse association between RHAMM expression levels and p53 deletion in other genetically engineered mouse models of BrafV600E-driven thyroid carcinoma [28]. The analysis of the NCBI’s Gene Expression Ominibus (GEO) dataset from these mouse models (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS5645) showed that thyroid carcinomas with p53 deletions (TPO-CreER; BrafV600E; p53−/−) had higher expression levels of RHAMM compared to thyroid carcinomas without p53 deletions (TPO-CreER; BrafV600E) (P = 0.0043, t-test).
It has been shown that human RHAMM/HMMR is a G2/M protein [17, 18] and its C-terminus is recognized by the Anaphase-Promoting Complex (APC) for its degradation in the G1 phase [29]. Because this C-terminal homologous region is deleted in the mouse HMMRΔexon8-16 protein, the HMMRΔexon8-16 protein is likely not degradable by APC and this may contribute to the higher levels of the HMMRΔexon8-16 protein compared to the WT protein. Besides APC, two other E3 ubiquitin ligase activities, BRCA1/BARD1 and RFWD3, have been reported to regulate RHAMM/HMMR protein levels [30-32], but the consensus amino acid sequences for BRCA1/BARD1 and RFWD3 recognition are not known.
Exons 1 ~ 4 of RHAMM/HMMR encode a microtubule-binding region, exons 5 ~ 16 encode the central coiled-coil region, and exons 17 ~ 18 encode a centrosome targeting region. Full-length RHAMM has 3 putative hyaluronan-binding sites (BX7B motif) (Figure 1A). HMMRΔexon8-16 protein retains one of the 3 putative hyaluronan-binding sites and exon 4, but it loses the C-terminal centrosome targeting region because the fusion of exon 7 and exon 17 results in a stop codon in the middle of exon 17. Our study indicated that the N-terminal microtubule-binding region and/or part of the central coiled-coil region are important for its oncogenic function, but the C-terminal centrosome targeting region is not. Further investigation is required to determine whether the remaining putative hyaluronan-binding motif is essential for the oncogenic function of HMMRΔexon8-16 and whether the lysine to threonine mutation at residue 71 affects any function. Between two of the alternative splicing human RHAMM isoforms, we have previously demonstrated that the RHAMMB, which does not have exon 4, is crucial for in vivo liver metastatic capacity of PNET, but RHAMMA, which carries exon 4, cannot promote liver metastasis of PNET [21]. It remains to be investigated whether a mouse RHAMMB exists naturally or any mouse RHAMM isoform is upregulated in spontaneous mouse tumors. Because mouse HMMRΔexon8-16 protein still retains exon 4 and possesses an oncogenic function, we postulate that either loss of exon 4 or loss of the C-terminal centrosome targeting region confers the oncogenic function of RHAMM/HMMR.
In addition to HMMRΔexon8-16/Δexon8-16 (previously Rhamm−/−) mouse [23], HMMRm/m and HMMRtm1a/tm1a mice have been generated [33, 34]. A schematic of HMMR protein/gene in these mouse models can be found in the supplement figure 1 of [33]. The HMMRm/m mice expresses HMMRΔexon11-18 protein with a molecular weight of 65 kDa and HMMRΔexon11-18 protein is also more abundant than WT proteins in mouse testis [34]. The homozygous HMMRm/m mice are viable and normal in appearance [34]. However, HMMRΔexon11-18 causes hypofertility in older females (>24 weeks old) and in male mice independently of the male's ages [32, 34]. Because HMMRm (HMMRΔexon11-18) also loses C-terminal centrosome targeting region of RHAMM encoded by exons 17 ~ 18 and some of central coiled-coil region encoded by exons 5 ~ 16, HMMRm (HMMRΔexon11-18) might promote invasive PDAC progression and reduce the survival of mice with PDAC and PNET similar to this study with HMMRΔexon8-16. A side-by-side comparison of HMMRΔexon11-18 and HMMRΔexon8-16/Δexon8-16 for their oncogenic property would help to further dissect the functional domains of HMMR. In contrast, HMMRtm1a/tm1a mice only express the exon 1 and 2 of HMMR. HMMRtm1a/tm1a mice produced litters with decreased survival, along with decreased adult body size and morphological defects in the brain, and very few mice survive to adulthood [33]. Thus, HMMRtm1a/tm1a mice are not suitable for crossing to PDAC and PNET mouse models. A pancreas-specific knockout of HMMR would be needed to assess the effect of targeting RHAMM/HMMR in pancreatic cancer.
Highlights.
Rhamm−/− mice express a truncated HMMRΔexon8-16 protein.
HMMRΔexon8-16 accelerates invasive PDAC and shortens the survival of PDAC mice.
HMMRΔexon8-16 shortens the survival of PNET mice, in which p53 is inactivated.
Pancreatic cancer patients with dysfunctional TP53 have higher RHAMM levels.
Patients having higher RHAMM levels and dysfunctional TP53 have poor survival.
Acknowledgements
We thank Cornelia Tolg and Eva Ann Turley for providing Rhamm−/− mice. We thank Danny Huang, Annie Yang, Makheni Jean-Pierre, Lei Tan, Shuibing Chen, Bing He, Mai Ho, Leticia Dizon, Taotao Zhang, Vincent Sarno, and Ruben Diaz for assistance. Funding and project support were partially provided by NIH R01CA204916-01A1, DoD W81XWH-16-1-0619, STARR I12-0043, and the Center for Translational Pathology at the Department of Pathology and Laboratory Medicine, Weill Cornell Medicine.
Abbreviations
- APC
anaphase-promoting complex
- CN
copy number
- CNV
copy number variation
- ES
embryonic stem
- HMMRΔexon8-16
N-terminal RHAMM lacking exons 8 ~ 16
- MU
mutation
- OS
overall survival
- p53lox/+
conditional heterozygous p53 loss
- p53lox/lox
conditional homozygous-null p53
- PanIN
pancreatic intraepithelial neoplasia
- PDAC
pancreatic ductal adenocarcinoma
- PNET
pancreatic neuroendocrine tumor
- PCR
polymerase chain reaction
- RHAMM or HMMR
receptor for hyaluronic acid-mediated motility
- RHAMMB
receptor for hyaluronan-mediated motility isoform B
- Rb
retinoblastoma
- RIP
rat insulin promoter
- Tag
T antigen
- TCGA
The Cancer Genome Atlas
- WT
wild-type
Footnotes
Declaration of competing interest
There is no conflict of interest pertaining to this publication to be disclosed by any of the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018, CA Cancer J Clin, 68 (2018) 7–30. [DOI] [PubMed] [Google Scholar]
- [2].Pelosi E, Castelli G, Testa U, Pancreatic Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells, Biomedicines, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Westphalen CB, Olive KP, Genetically engineered mouse models of pancreatic cancer, Cancer J, 18 (2012) 502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA, Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse, Cancer Cell, 4 (2003) 437–450. [DOI] [PubMed] [Google Scholar]
- [5].Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, Depinho RA, Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse, Proc Natl Acad Sci U S A, 103 (2006) 5947–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, Shih T, Yao JC, Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States, JAMA Oncol, 3 (2017) 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Buicko JL, Finnerty BM, Zhang T, Kim BJ, Fahey TJ 3rd, Nancy Du YC, Insights into the biology and treatment strategies of pancreatic neuroendocrine tumors, Ann Pancreat Cancer, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang T, Choi S, Zhang T, Chen Z, Chi Y, Huang S, Xiang JZ, Du YN, miR-431 Promotes Metastasis of Pancreatic Neuroendocrine Tumors by Targeting DAB2 Interacting Protein, a Ras GTPase Activating Protein Tumor Suppressor, Am J Pathol, 190 (2020) 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Michael IP, Saghafinia S, Hanahan D, A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis, Proc Natl Acad Sci U S A, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hanahan D, Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes, Nature, 315 (1985) 115–122. [DOI] [PubMed] [Google Scholar]
- [11].Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D, Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors, J Clin Invest, 111 (2003) 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O, Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis, Cancer Cell, 15 (2009) 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pietras K, Hanahan D, A multitargeted, metronomic, and maximum-tolerated dose "chemo-switch" regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer, J Clin Oncol, 23 (2005) 939–952. [DOI] [PubMed] [Google Scholar]
- [14].Yao JC SM, Ito T, Bohas CL, Wolin EM, Van Cutsem E, Hobday TJ, Okusaka T, Capdevila J, de Vries EG, Tomassetti P, Pavel ME, Hoosen S, Haas T, Lincy J, Lebwohl D, Öberg K, Everolimus for advanced pancreatic neuroendocrine tumors, N Engl J Med, 364 (2011) 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Blumenthal GM CP, Zhang JJ, Tang S, Sridhara R, Murgo A, Justice R, Pazdur R, FDA approval summary: Sunitinib for the treatment of progressive well-differentiated locally advanced or metastatic pancreatic neuroendocrine tumors, Oncologist, 17 (2012) 1108–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Turley EA, Purification of a hyaluronate-binding protein fraction that modifies cell social behavior, Biochem Biophys Res Commun, 108 (1982) 1016–1024. [DOI] [PubMed] [Google Scholar]
- [17].Chen YT, Chen Z, Du YN, Immunohistochemical analysis of RHAMM expression in normal and neoplastic human tissues: a cell cycle protein with distinctive expression in mitotic cells and testicular germ cells, Oncotarget, 9 (2018) 20941–20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sohr S, Engeland K, RHAMM is differentially expressed in the cell cycle and downregulated by the tumor suppressor p53, Cell Cycle, 7 (2008) 3448–3460. [DOI] [PubMed] [Google Scholar]
- [19].Hardwick C, Hoare K, Owens R, Hohn HP, Hook M, Moore D, Cripps V, Austen L, Nance DM, Turley EA, Molecular cloning of a novel hyaluronan receptor that mediates tumor cell motility, J Cell Biol, 117 (1992) 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang D, Narula N, Azzopardi S, Smith RS, Nasar A, Altorki NK, Mittal V, Somwar R, Stiles BM, Du YN, Expression of the receptor for hyaluronic acid mediated motility (RHAMM) is associated with poor prognosis and metastasis in non-small cell lung carcinoma, Oncotarget, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Choi S, Wang D, Chen X, Tang LH, Verma A, Chen Z, Kim BJ, Selesner L, Robzyk K, Zhang G, Pang S, Han T, Chan CS, Fahey TJ 3rd, Elemento O, Du YN, Function and clinical relevance of RHAMM isoforms in pancreatic tumor progression, Mol Cancer, 18 (2019) 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Du YC, Chou CK, Klimstra DS, Varmus H, Receptor for hyaluronan-mediated motility isoform B promotes liver metastasis in a mouse model of multistep tumorigenesis and a tail vein assay for metastasis, Proceedings of the National Academy of Sciences of the United States of America, 108 (2011) 16753–16758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tolg C, Poon R, Fodde R, Turley EA, Alman BA, Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor), Oncogene, 22 (2003) 6873–6882. [DOI] [PubMed] [Google Scholar]
- [24].Zhang G, Chi Y, Du YN, Identification and Characterization of Metastatic Factors by Gene Transfer into the Novel RIP-Tag; RIP-tva Murine Model, J Vis Exp, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA, Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice, Cancer Cell, 7 (2005) 469–483. [DOI] [PubMed] [Google Scholar]
- [26].Maxwell CA, McCarthy J, Turley E, Cell-surface and mitotic-spindle RHAMM: moonlighting or dual oncogenic functions?, J Cell Sci, 121 (2008) 925–932. [DOI] [PubMed] [Google Scholar]
- [27].Schatz-Siemers N, Chen YT, Chen Z, Wang D, Ellenson LH, Du YN, Expression of the Receptor for Hyaluronic Acid-Mediated Motility (RHAMM) in Endometrial Cancer is Associated With Adverse Histologic Parameters and Tumor Progression, Appl Immunohistochem Mol Morphol, 28 (2020) 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].McFadden DG, Vernon A, Santiago PM, Martinez-McFaline R, Bhutkar A, Crowley DM, McMahon M, Sadow PM, Jacks T, p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer, Proc Natl Acad Sci U S A, 111 (2014) E1600–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Song L, Rape M, Regulated degradation of spindle assembly factors by the anaphase-promoting complex, Mol Cell, 38 (2010) 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM, The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly, Cell, 127 (2006) 539–552. [DOI] [PubMed] [Google Scholar]
- [31].Pujana MA, Han J-DJ, Starita LM, Stevens KN, Tewari M, Ahn JS, Rennert G, Moreno V, Kirchhoff T, Gold B, Assmann V, Elshamy WM, Rual J-F, Levine D, Rozek LS, Gelman RS, Gunsalus KC, Greenberg RA, Sobhian B, Bertin N, Venkatesan K, Ayivi-Guedehoussou N, Solé X, Hernández P, Lázaro C, Nathanson KL, Weber BL, Cusick ME, Hill DE, Offit K, Livingston DM, Gruber SB, Parvin JD, Vidal M, Network modeling links breast cancer susceptibility and centrosome dysfunction, Nat Genet, 39 (2007) 1338–1349. [DOI] [PubMed] [Google Scholar]
- [32].Li H, Frappart L, Moll J, Winkler A, Kroll T, Hamann J, Kufferath I, Groth M, Taudien S, Schutte M, Yaspo ML, Heuer H, Lange BM, Platzer M, Zatloukal K, Herrlich P, Ploubidou A, Impaired Planar Germ Cell Division in the Testis, Caused by Dissociation of RHAMM from the Spindle, Results in Hypofertility and Seminoma, Cancer Res, 76 (2016) 6382–6395. [DOI] [PubMed] [Google Scholar]
- [33].Connell M, Chen H, Jiang J, Kuan CW, Fotovati A, Chu TL, He Z, Lengyell TC, Li H, Kroll T, Li AM, Goldowitz D, Frappart L, Ploubidou A, Patel MS, Pilarski LM, Simpson EM, Lange PF, Allan DW, Maxwell CA, HMMR acts in the PLK1-dependent spindle positioning pathway and supports neural development, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li H, Moll J, Winkler A, Frappart L, Brunet S, Hamann J, Kroll T, Verlhac MH, Heuer H, Herrlich P, Ploubidou A, RHAMM deficiency disrupts folliculogenesis resulting in female hypofertility, Biol Open, 4 (2015) 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]