Abstract
Background
Recent studies proposed the whole-blood based IFN-γ-release assay to study the antigen-specific SARS-CoV-2 response. Since the early prediction of disease progression could help to assess the optimal treatment strategies, an integrated knowledge of T-cell and antibody response lays the foundation to develop biomarkers monitoring the COVID-19. Whole-blood-platform tests based on the immune response detection to SARS-CoV2 peptides is a new approach to discriminate COVID-19-patients from uninfected-individuals and to evaluate the immunogenicity of vaccine candidates, monitoring the immune response in vaccine trial and supporting the serological diagnostics results. Here, we aimed to identify in the whole-blood-platform the best immunogenic viral antigen and the best immune biomarker to identify COVID-19-patients.
Methods
Whole-blood was overnight-stimulated with SARS-CoV-2 peptide pools of nucleoprotein-(NP) Membrane-, ORF3a- and Spike-protein. We evaluated: IL-1β, IL-1Ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL- 15, IL-17A, eotaxin, FGF, G-CSF, GM-CSF, IFN-γ, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF, RANTES, TNF-α, VEGF. By a sparse partial least squares discriminant analysis we identified the most important soluble factors discriminating COVID-19- from NO-COVID-19-individuals.
Results
We identified a COVID-19 signature based on six immune factors: IFN-γ, IP-10 and IL-2 induced by Spike; RANTES and IP-10 induced by NP and IL-2 induced by ORF3a. We demonstrated that the test based on IP-10 induced by Spike had the highest AUC (0.85, p < 0.0001) and that the clinical characteristics of the COVID-19-patients did not affect IP-10 production. Finally, we validated the use of IP-10 as biomarker for SARS-CoV2 infection in two additional COVID-19-patients cohorts.
Conclusions
We set-up a whole-blood assay identifying the best antigen to induce a T-cell response and the best biomarkers for SARS-CoV-2 infection evaluating patients with acute COVID-19 and recovered patients. We focused on IP-10, already described as a potential biomarker for other infectious disease such as tuberculosis and HCV. An additional application of this test is the evaluation of immune response in SARS-CoV-2 vaccine trials: the IP-10 detection may define the immunogenicity of a Spike-based vaccine, whereas the immune response to the virus may be evaluated detecting other soluble factors induced by other viral-antigens.
Keywords: SARS-CoV-2, COVID-19, Biomarkers, T-cell, Immunity, IP-10, Whole-blood, Immune response, Spike, IFN-γ
Introduction
COronaVIrus Disease-2019 (COVID-19) pandemic is caused by the novel coronavirus designated as severe acute respiratory syndrome coronavirus (SARS-CoV)-2 [1] belonging to β-Coronovavirus genus. Its genome contains 14 open reading frames (ORFs) and encodes 27 different proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (NP) proteins [2]. The majority of people with COVID-19 develop mild (40%) or moderate (40%) symptoms, 15–20% develop a severe disease needing oxygen support and 5% have a critical disease with complications such as respiratory failure, acute respiratory distress syndrome (ARDS), sepsis and septic shock, thromboembolism, and/or multi-organ failure [3–5]. SARS-CoV-2 infection induces an immune response in the host characterized in severe COVID-19 cases by a decrease of lymphocytes number and a great increase of cytokines [6]. Currently, the mechanisms that lead to disease exacerbation remains largely undetermined. Thus, there is an urgent need to improve our understanding of the immunology of this disease to find correlate of protection or to monitor the course of the infection.
Several reports described different immune profiles of COVID-19-patients according to the diseases [7–15]. SARS-CoV-2 infection decreases the lymphocytes number and increases cytokines release in severe COVID-19-cases [14]. A significant increase of pro-inflammatory or anti-inflammatory cytokines, including T helper (Th) type-1 and type-2 cytokines and chemokines was described [10, 12, 16, 17], interleukin (IL)-1β, IL-6, IL-8, and Interferon (IFN)-γ-inducible protein (IP-10) were associated with severe or fatal course of disease [7–9]. Four immune signatures, constituted by growth factors, Th1-, Th2-, Th3-cytokines and chemokines, were correlated with distinct disease courses [9]. In acute and convalescent subjects, a coordinated immune response of T-cells and antibodies was associated with milder disease [13]. The importance of T-cell response against β-coronavirus infections has been underlined by a study on patients recovered from SARS, demonstrating the persistence of long-lasting memory T-cells reactive to SARS-CoV stimulation, years after the SARS-outbreak in 2003 [18, 19]. Recent studies highlighted the use of the whole-blood based IFN-γ released assay as a promising approach to study the antigen-specific SARS-CoV-2 response [10–12, 20, 21]. The use of a whole-blood-platform with SARS-CoV2 peptides to discriminate COVID-19-patients and uninfected-individuals [10, 20, 22], is a new potential approach to study the immunogenicity of vaccine candidate, to monitor the immune response in vaccine trial and to support the serological diagnostics.
In this study, we analyzed in a whole-blood-cytokine platform, the best approach to evaluate the SARS-CoV-2-T-cell response to the structural (N, S and M) [19] and accessory protein (ORF3a) [23, 24] of SARS-CoV-2. We aimed to identify (i) the best antigen to induce the SARS-CoV-2 specific T-cell response; (ii) the best subset of biomarkers to identify COVID-19-patients.
Results
Identification of plasma biomarkers for distinguishing COVID-19 from NO-COVID-19-individuals
Demographical and clinical information of the enrolled subjects are shown in Table 1. We stimulated the whole-blood of with SARS-CoV-2-specific peptide pools of NP (NP Pool1 and NP Pool2), Membrane, ORF3a, and Spike. Then, we evaluated by luminex the plasma level of 27 analytes. Among the different stimuli, the Spike and NP Pool1 peptides, belonging both to SARS-CoV-2 structural proteins, were the most recognized antigens by COVID-19-patients (Table 2). Spike peptide pool was the most immunogenic stimulus, modulating the highest number of cytokines, chemokines and growth factors (Table 2).
Table 1.
Demographical and clinical characteristics of the enrolled subjects
| COVID-19 | NO-COVID-19 | p value | |
|---|---|---|---|
| N (%) | 23 (56.1) | 18 (43.9) | – |
| Age median (IQR) | 45 (35–57) | 48.5 (33.25–59.5) | 0.73* |
| Male N (%) | 19 (82.6) | 11 (61.1) | 0.123§ |
| Origin N (%) | 0.014§ | ||
| West Europe | 11 (48) | 15 (83) | |
| East Europe | 0 (0) | 2 (11) | |
| Asia | 9 (39) | 0 (0) | |
| Africa | 1 (4.3) | 0 (0) | |
| South America | 2 (8.7) | 1 (6) | |
| Swab positive results N (%) | 23 (100) | 0 (0) | |
| Serology results IgM. N (%) | |||
| IgM + | 12 (52.2) | 0 (0) | |
| IgM − | 11 (47.8) | 18 (100) | |
| Serology results IgG. N (%) | |||
| IgG + | 13 (56.5) | 0 (0) | |
| IgG − | 8 (34.8) | 18 (100) | |
| IgG doubtful | 2 (8.7) | 0 (0) | |
| Severity N (%)a | |||
| Asymptomatic | 2 (8.7) | – | |
| Mild | 3 (13) | – | |
| Moderate | 11 (48) | – | |
| Severe | 5 (21.7) | – | |
| Critical | 2 (8.6) | – | |
| Cortisone N (%) | 6 (26) | – | |
| Severity of patients taking cortisone N (%) | |||
| Asymptomatic | 0 (0) | – | |
| Mild | 0 (0) | – | |
| Moderate | 2 (33) | – | |
| Severe | 3 (50) | – | |
| Critical | 1 (17) | – | |
COVID-19 Coronavirus Disease 19; N number
aWHO criteria (1)
*Mann Whitney test
§Chi-square test
Table 2.
Cytokines, chemokines and grow factors significantly modulated in COVID-19 and NO-COVID-19 individuals
| Main source | Analytes | NP Pool1 | NP Pool2 | Spike | ORF3a | Membrane | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID-19 | NO-COVID-19 | p* | COVID-19 | NO-COVID-19 | p* | COVID-19 | NO-COVID-19 | p* | COVID-19 | NO-COVID-19 | p* | COVID-19 | NO-COVID-19 | p* | |||
| Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | Median (IQR) | ||||||||
| Inflammatory cytokines/chemokines | Macrophages | IL-6 | – | – | – | – | – | – | 51.56 (22.2–98.48) | 8.98 (6.09–27.1) | 0.008 | – | – | – | – | – | – |
| Macrophages | TNF-α | 28.84 (15.72–40.99) | 4.5 (0.0–21.44) | 0.009 | 26.72 (7.04–55.16) | 4.84 (0.0–32.89) | 0.048 | 33.88 (16.32–70.8) | 14.08 (5.71–23.81) | 0.0067 | – | – | – | – | – | – | |
| Th1 | IFN-γ | – | – | – | – | – | – | 131.2 (101.2–243) | 48.2 (25.04–118.6) | 0.017 | – | – | – | – | – | – | |
| Th17 | IL-17 | – | – | – | – | – | – | 9.4 (4.44–15.44) | 2.88 (1.06–6.76) | 0.017 | – | – | – | – | – | – | |
| Monocytes | IP-10 | 546.2 (0.0–1751) | 26.78 (0.0–279.1) | 0.025 | 1349 (0.0–5328) | 34.56 (0.0–123.6) | 0.028 | 1108 (289.2–3145) | 19.36 (0.0–164.7) | < 0.0001 | – | – | – | – | – | – | |
| Fibroblasts | |||||||||||||||||
| Endothelial cells | |||||||||||||||||
| Monocytes | MCP-1 | 2548 (1526–3125) | 615.7 (138.4–1941) | 0.043 | – | – | – | 2679 (1350–4600) | 712.8 (220–1295) | 0.013 | 7754 (3901–10,540) | 11,377 (8489–17,050) | 0.01 | 7352 (2138–11,063) | 12,841 (7076–13,557) | 0.02 | |
| Macrophages | |||||||||||||||||
| Monocytes | MIP-1α | 16.84 (6.68–35.2) | 3.18 (1.57–11.18) | 0.0024 | – | – | – | 17.6 (10.48–56.12) | 7.2 (2.64–15.29) | 0.0137 | – | – | – | – | – | – | |
| Macrophages | |||||||||||||||||
| Monocytes | MIP-1β | 250.2 (128–335.9) | 51.66 (4.86–96.05) | 0.0012 | – | – | – | 382 (177.8–922) | 192.8 (65.69–323) | 0.014 | – | – | – | – | – | – | |
| Macrophages | |||||||||||||||||
| Platelets. macrophages | RANTES | 540.3 (259.8–1013) | 0 (0.0–229.6) | 0.001 | – | – | – | 590.8 (407.2–877) | 303.8 (0.0–690.9) | 0.049 | – | – | – | – | – | – | |
| Anti-inflammatory cytokines | Th2 | IL-4 | 0.92 (0.68–1.68) | 0.32 (0.12–0.94) | 0.0087 | – | – | – | 1.24 (0.64–2.64) | 0.38 (0.07–1.44) | 0.015 | – | – | – | – | – | – |
| Th2 | IL-10 | 1.84 (0.96–6.04) | 0.96 (0.0–2.13) | 0.046 | – | – | – | – | – | – | – | – | – | ||||
| Treg | |||||||||||||||||
| Th2 | IL-13 | – | – | – | – | – | – | 0.6 (0.28–2.44) | 0.08 (0.0–0.39) | 0.043 | – | – | – | – | – | – | |
| Growth factors | Th1 | IL-2 | 10.44 (2.88–32.84) | 1.78 (0.75–5.24) | 0.0023 | 21.72 (6.12–103) | 3.1 (0.3–10.89) | 0.0065 | 32.84 (9.44–100.5) | 2.34 (1.13–9.78) | 0.0018 | 59.28 (30.56–142) | 31.14 (20.52–62.02) | 0.039 | 64.16 (28.32–213.8) | 30.28 (12–62.14) | 0.03 |
| Th9 | IL-9 | – | – | – | – | – | – | 26.24 (0.76–32.2) | 6.02 (0.0–24.6) | 0.026 | – | – | – | – | – | – | |
| Stromal cells. Macrophages | FGF | – | – | – | – | – | – | 25.76 (13.88–40.24) | 10.62 (6.9–19.22) | 0.017 | – | – | – | – | – | – | |
COVID-19 CoronaVIrus Disease 19; N Number; IL interleukin; FGF: fibroblast growth factor, IFN: interferon, IP: IFN-γ-induced protein, MCP: monocyte chemoattractant protein, MIP: macrophage inflammatory protein, RANTES: regulated on activation, normal T cell expressed and secreted, TNF: tumour necrosis factor
*Mann Whitney test
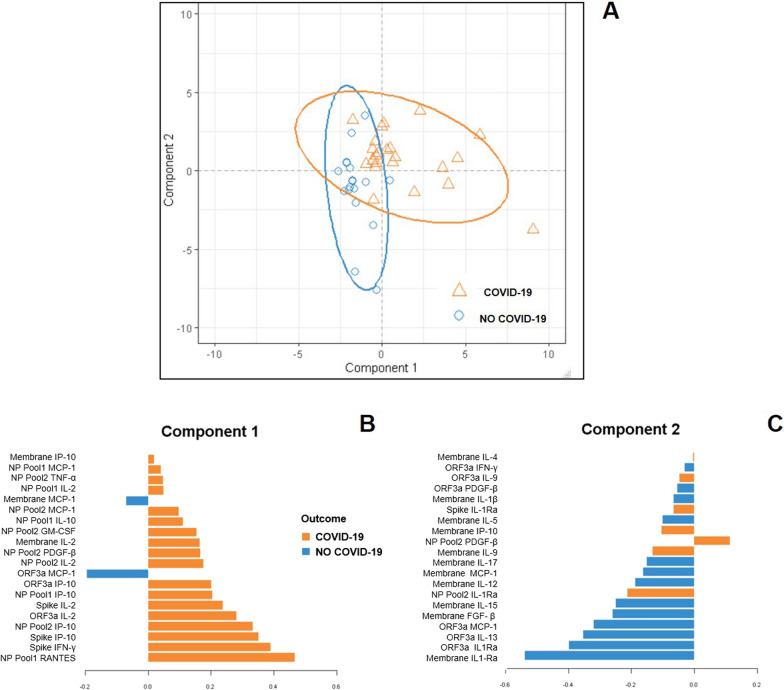
Applying a supervised sPLS-DA we aimed to identify the most important soluble factors, analyzing at the same time the luminex results and the different SARS-CoV-2-peptides pool stimulations (Fig. 1). Although the difference was not fully discriminative, the distribution of COVID-19 and NO-COVID-19-subjects in the space were quite separated (Fig. 1A). Evaluating the loading weights of each selected variable on each component, the mean level of production for the most important selected variables was maximal in COVID-19-patients within the component 1 (Fig. 1B), whereas the mean level of production was maximal in the NO-COVID-19 within the component 2 (Fig. 1C). Overall, the accuracy of the classification was high for both components (> 92%) (data not shown). Since the component 1 was represented mainly by factors upregulated in COVID-19-patients, we focused on this component. Then, we identified the six variables with the highest weight in the construction of component 1 (Fig. 1B–C): IL-2, IFN-γ and IP-10 induced by Spike, regulated on activation, normal T cell expressed and secreted (RANTES) induced by NP Pool1, IP-10 induced by NP Pool2, and IL-2 induced by ORF3a stimulation (hereafter referred as Spike IL-2, Spike IFN-γ, Spike IP-10, NP Pool1 RANTES, NP Pool2 IP-10, and ORF3a IL-2). Next, we evaluated, within the six variables signature associated to COVID-19, the proportion of response to each stimulus: IP-10 proportions induced by Spike and NP Pool2 were the most represented in COVID-19-patients (Fig. 2).
Fig. 1.
Sparse partial least squares discriminant analysis (sPLS-DA) on luminex data-set of COVID-19 and NO-COVID-19 subjects. The different soluble factors have been measured by luminex assay in plasma collected after stimulating whole-blood with different SARS-CoV-2 peptides (Spike, NP Pool1, NP Pool2, Membrane and ORF3a). A The samples are projected in the space spanned by the first two components with 95% confidence level ellipse plots. Colours and symbols indicate the class of each sample (orange triangle COVID-19 patients, blue circles NO-COVID-19 individuals). B, C Selected immune responses distinguish COVID-19 and NO-COVID-19 individuals over all the evaluated antigens stimulation and distinct immune response detected. The graphs represent the loading weights of the selected variables on each component (20 soluble factors for each component). The variables contribution ranked from the bottom, the most important, to the top. The colors indicate the class for which the selected variable has a maximal mean value: orange COVID-19 patients, blue NO-COVID-19
Fig. 2.
COVID-19 signature based on six selected immune factors. The graphs represent the proportion of immune factors secreted in response to SARS-CoV-2 peptides stimulation within the six variables immune signature associated to COVID-19: RANTES induced by NP Pool1; IFN-γ by Spike; IP-10 by Spike; IP-10 by NP Pool2; IL-2 by Spike, IL-2 by ORF3a. The different immune factors were measured by luminex assay in plasma collected after stimulating whole-blood with the different antigens. A Proportion of selected immune factors in COVID-19 patients. B Proportion of selected immune factors in NO-COVID-19 subjects. C Median proportion of selected immune factors in COVID-19 and NO-COVID-19 subjects. NP nucleoprotein, IL interleukin; IP interferon-γ inducible protein; IFN interferon; RANTES regulated on activation, normal T cell expressed and secreted
Comparison of AUC of the six immune factors
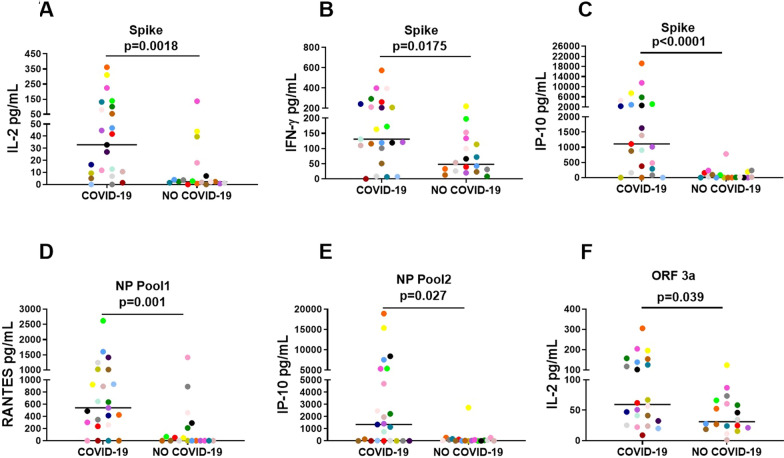
The selected six immune factors of component 1, as expected, had significant quantitative higher levels in COVID-19 compared to controls for: IL-2, IFN-γ, IP-10 induced by Spike (p = 0.0018; p = 0.0175; p < 0.0001; respectively), NP Pool1 RANTES (p = 0.001), NP Pool2 IP-10 (p = 0.027) and ORF3a IL-2 (p = 0.039) (Fig. 3; Table 2). ROC curve analysis of these factors showed that the highest AUC was related to IP-10 Spike (AUC 0.85; p < 0.0001; Fig. 4).
Fig. 3.
Increased antigen-specific response to selected SARS-CoV-2 antigens in whole-blood is associated with COVID-19. A IL-2 production induced by Spike stimulation. B IFN-γ production induced by Spike stimulation. C IP-10 production induced by Spike stimulation. D RANTES production induced by NP Pool1 stimulation. E IP-10 production induced by NP Pool2 stimulation. F IL-2 production induced by ORF3a stimulation. The different immune factors were measured by luminex assay in plasma collected after stimulating whole-blood with the different antigens. The horizontal lines represent the median; statistical analysis was performed using the Mann–Whitney test, and p value was considered significant when ≤ 0.05. NP nucleoprotein, IL interleukin; IP interferon-γ inducible protein; IFN interferon, RANTES regulated on activation, normal T cell expressed and secreted
Fig. 4.
Comparison of the AUC resulting from the SARS-CoV-2-specific responses. A The graph shows the AUC of seven different immune responses based on RANTES induced by NP Pool1; IFN-γ by Spike; IP-10 by Spike; IP-10 by NP Pool2; IL-2 by Spike, IL-2 by ORF3a, a combination of all above cited tests (combined test). Since one observation related to IL-2 induced by ORF3a is missing, the AUC comparison has been performed on 40 patients instead of 41. B Comparison of the single test AUCs with the combined test: *p values referred to correspondent ROC; **comparison of AUCs of NP Pool 1 RANTES, Spike IFN-γ, Spike IP-10, NP Pool 2 IP-10, Spike IL-2, ORF3a IL-2 with the AUC of combined test. C Comparison of the single test AUCs with IP-10 induced by Spike AUC: *p values referred to correspondent ROC; **comparison of AUCs of NP Pool 1 RANTES, Spike IFN-γ, NP Pool 2 IP-10, Spike IL-2, ORF3a IL-2, combined test, with the AUC of Spike IP-10. IL interleukin; IP interferon-γ inducible protein; IFN interferon; RANTES regulated on activation, normal T cell expressed and secreted, NP nucleoprotein, CI confidence interval; AUC area under the curve
Then, we generated a combined-test based on the six immune factors previously selected (Fig. 4). The combined-test showed a significantly further increase of AUC (AUC 0.94; p < 0.0001) compared to the AUCs of the other single tests except for IP-10 and IL-2 induced by Spike (Fig. 4B). Since IP-10 Spike test showed the highest AUC, we compared it with all the other AUCs and we did not find any significant differences among the different tests (Fig. 4C).
Impact of the clinical characteristics of patients on the COVID-19 signature
We investigated if any clinical characteristic of COVID-19-patients had an impact on the level of the six selected variables (Table 3). We found that age (p = 0.001), cortisone (p = 0.042) and severity of the disease (p = 0.015) had a significant impact on NP Pool1 RANTES. NP Pool2 IP-10 was modulated by symptoms (p = 0.036), IgM index (p = 0.003) and IgM score (p = 0.017). Finally, ORF3a IL-2 was modulated, by the number of days from the symptoms onset (p < 0.0001) and IgM index (p = 0.038). Similarly, Spike IL-2 was modulated by number of days from the symptoms onset (p = 0.001), IgM index (p = 0.028) and IgM score (p = 0.036).
Table 3.
Impact of the characteristics of COVID-19 patients on the six selected immune factors (IL-2. IFN-γ. IP-10 induced all by Spike; RANTES induced by NP Pool1; IP-10 induced by NP Pool2; IL-2 induced by ORF3a)
| Characteristics | NP Pool1 RANTES | Spike INF-γ | Spike IP-10 | NP Pool2 IP-10 | ORF3a IL-2 | Spike IL-2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median (IQR) | rs | p | Median (IQR) | rs | p | Median (IQR) | rs | p | Median (IQR) | rs | p | Median (IQR) | rs | p | Median (IQR) | rs | p | |
| Gender | ||||||||||||||||||
| Male | 540 (302–928) | Na | 0.935 | 164 (110–262) | Na | 0.194 | 1390 (375–4608) | Na | 0.33 | 1349 (0–5328) | Na | 0.513 | 59 (41–153) | Na | 0.67 | 33 (9–100) | Na | 0.935 |
| Female | 622 (0–1422) | 64 (7–166) | 385 (224–1677) | 2919 (580–6123) | 76 (24–132) | 29 (9–89) | ||||||||||||
| Age | Na | − 0.65 | 0.001 | Na | 0.03 | 0.893 | Na | − 0.14 | 0.513 | Na | − 0.06 | 0.792 | Na | − 0.15 | 0.519 | Na | 0 | 0.989 |
| Cortisone | ||||||||||||||||||
| No | 894 (302–1025) | Na | 0.042 | 131 (51–208) | Na | 0.484 | 1107 (375–3145) | Na | 0.779 | 1136 (0–2464) | Na | 0.34 | 53 (29–132) | Na | 0.376 | 42 (9–100) | Na | 0.916 |
| Yes | 380 (0–424) | 166 (116–243) | 1394 (81–2629) | 3025 (140–8414) | 109 (47–153) | 25 (12–54) | ||||||||||||
| Sympotms | ||||||||||||||||||
| No | 237 (0–1025) | Na | 0.391 | 208 (0–262) | Na | 0.92 | 375 (0–1107) | Na | 0.191 | 0 (0–0) | Na | 0.036 | 62 (9–67) | Na | 0.427 | 9 (1–42) | Na | 0.159 |
| Yes | 588 (302–928) | 147 (110–243) | 1510 (289–4608) | 1684 (140–5328) | 56 (32–153) | 46 (12–132) | ||||||||||||
| Days from onset sympotms | Na | − 0.21 | 0.405 | Na | 0.2 | 0.416 | Na | 0.35 | 0.15 | Na | 0.41 | 0.093 | Na | 0.8 | 0 | Na | 0.72 | 0.001 |
| Severity of disease | ||||||||||||||||||
| asy/mild/ | 770 (456–1135) | Na | 0.015 | 147 (114–235) | Na | 0.548 | 1510 (890–4488) | Na | 0.061 | 1684 (0–6464) | Na | 0.567 | 62 (32–156) | Na | 0.647 | 37 (10–120) | Na | 0.462 |
| mod | ||||||||||||||||||
| sev/crit | 260 (0–414) | 116 (7–243) | 289 (0–2307) | 1136 (18–2464) | 56 (22–125) | 16 (0–80) | ||||||||||||
| Lymphocytes (× 103) | ||||||||||||||||||
| Percentage | Na | 0.43 | 0.042 | Na | 0.46 | 0.026 | Na | 0.41 | 0.052 | Na | 0.06 | 0.772 | Na | 0.23 | 0.299 | Na | 0.22 | 0.309 |
| Number | Na | 0.28 | 0.204 | Na | 0.11 | 0.631 | Na | 0.1 | 0.653 | Na | − 0.05 | 0.83 | Na | 0.08 | 0.727 | Na | − 0.08 | 0.727 |
| Serology | ||||||||||||||||||
| IgG index | Na | − 0.12 | 0.594 | Na | − 0.12 | 0.596 | Na | − 0.15 | 0.499 | Na | 0.17 | 0.433 | Na | 0.19 | 0.385 | Na | 0.24 | 0.271 |
| IgG score | ||||||||||||||||||
| Negative | 491 (281–1153) | Na | 0.975 | 162 (106–345) | Na | 0.917 | 2251 (882–5219) | Na | 0.386 | 2091 (0–3896) | Na | 0.823 | 87 (28–147) | Na | 0.384 | 37 (6–90) | Na | 0.351 |
| Positive | 540 (237–1013) | 131 (116–208) | 902 (289–2629) | 1136 (26–5385) | 64 (41–139) | 42 (12–132) | ||||||||||||
| Doubtful | 671 (414–928) | 125 (7–243) | 1153 (0–2307) | 683 (18–1349) | 33 (20–47) | 8 (0–16) | ||||||||||||
| IgM index | Na | 0.13 | 0.563 | 0.21 | 0.333 | Na | 0.29 | 0.157 | Na | 0.59 | 0.003 | Na | 0.45 | 0.038 | Na | 0.46 | 0.028 | |
| IgM score | ||||||||||||||||||
| Negative | 414 (260–1013) | Na | 0.666 | 116 (51–243) | Na | 0.538 | 877 (0–2307) | Na | 0.157 | 18 (0–1960) | Na | 0.017 | 47 (24–117) | Na | 0.2 | 11 (1–54) | Na | 0.036 |
| Positive | 588 (331–1084) | 147 (119–237) | 1868 (692–4488) | 3462 (946–7979) | 102 (41–157) | 46 (23–135) | ||||||||||||
Bold values indicate p values<0.05
Mann-Whitney or Kruskall-Wallis Test for categorical variables; Spearman’s correlation for continuous variables
rs : Sperman’s correlation coefficient; Asy/mild/mod: asymptomatic/mild/moderate; Sev/crit: severe/critical; Na: not applicable
Differently, Spike IFN-γ and Spike IP-10 were not significantly modulated by any of the clinical characteristics considered.
Evaluation of IP-10 in different cohorts of COVID-19-patients
We demonstrated that Spike IP-10 had the highest AUC (0.85, p < 0.0001; Fig. 4) and that the clinical characteristics of the COVID-19-patients did not affect IP-10 production (Table 3). Based on these results, we further evaluated the production of IP-10 in a new study population of NO-COVID-19 and COVID-19-patients stratified according to the hospitalization status and symptoms onset (Table 4). To verify the consistency of our findings, we used a different experimental setting: IP-10 was detected using a routine approach as the enzyme-linked immunosorbent assay (ELISA) and Spike peptides were obtained from a commercial source (Miltenyi). IP-10 production significantly increased after Spike stimulation in the cohort A of “hospitalized COVID-19-patients enrolled between 1 and 14 days after symptoms onset” (p = 0.0014) and in the cohort B of “not hospitalized COVID-19-patients” (p = 0.0002), (Fig. 5A–B). ROC analysis demonstrated a high and significant AUC in cohort A and cohort B (AUC: 0.8167; p = 0.0020; AUC: 0.9056; p = 0.0005) (Fig. 5C–D). The specificity of the test to identify COVID-19 was 88.89% for both COVID-19-cohorts; the sensitivity was 66.67% for cohort A and 70% for cohort B (Fig. 5C–D).
Table 4.
Demographical and clinical characteristics of the enrolled subjects for the IP-10 study
| COVID-19 | COVID-19 | NO-COVID-19 | p value | |
|---|---|---|---|---|
| Cohort A | Cohort B | N = 18 | ||
| N = 15 | N = 10 | |||
| Hospitalized N (%) | 15 (100) | 0 (0) | 0 (0) | – |
| Enrolled “X” days after symptoms onset | 14-Jan | 35–100 | / | – |
| Age median (IQR) | 63 (52–70) | 55 (31–60) | 44 (38–53) | < 0.0001* |
| Male N (%) | 11 (73) | 0 (0) | 13 (68) | 0.0008§ |
| Origin N (%) | ||||
| West Europe | 15 (100) | 9 (90) | 18 (100) | 0.208§ |
| East Europe | 0 (0) | 1 (10) | 0 | |
| Asia | 0 (0) | 0 (0) | 0 | |
| Swab positive results N (%) | 14 (93) | 5 (50) | 0 (0) | 0.0036§§ |
| Serology results IgM N (%)a | ||||
| IgM + | 10 (66.7) | 2 (22.2) | 0 (0) | |
| IgM − | 4 (26.6) | 5 (55.6) | 0 (0) | 0.2999§§ |
| IgM doubtful | 1 (6.7) | 2 (22.2) | 0 (0) | |
| Serology results IgG N (%)a | ||||
| IgG + | 12 (80) | 8 (88.9) | 0 (0) | |
| IgG − | 2 (13.3) | 1 (11.1) | 0 (0) | 0.820§§ |
| IgG doubtful | 1 (6.7) | 0 (0) | 0 (0) | |
| Severity N (%)# | ||||
| Mild | 0 (0) | 9 (90) | / | |
| Moderate | 4 (26.7) | 0 (0) | / | < 0.0001§§ |
| Severe | 8 (53.3) | 1 (10) | / | |
| Critical | 3 (20) | 0 (0) | / | |
Bold values indicate p values<0.05,
COVID-19 COronaVIrus Disease 19; N number
aMissing information for one not hospitalized patients, the percentage has been calculated on 9 patients
*Mann Whitney test
§Chi-square test
§§Chi-square test performed only on COVID-19 cohorts A vs B
#WHO criteria [1]
Fig. 5.
IP-10 modulation in a second cohort of COVID-19 patients. IP-10 production was measured by ELISA in plasma collected after stimulating whole-blood with Spike peptides. A, B The horizontal lines represent the median of IP-10 production; statistical analysis was performed using the Mann–Whitney test, and p value was considered significant when ≤ 0.05. C, D The graphs represent the AUCs obtained by the ROC analysis comparing the NO-COVID-19 subjects with three cohorts of COVID-19 patients. A, C Hospitalized COVID-19 patients enrolled 1–14 days after symptoms onset. B, D Not-hospitalized COVID-19 patients enrolled 35–100 days after symptoms onset. IP interferon-γ inducible protein; CI confidence interval; AUC area under the curve
Discussion
In this study, by a multivariate exploratory analysis we found the best antigen and the best biomarker to distinguish COVID-19- and NO-COVID-19-individuals. To achieve our goal, we used a whole-blood-platform [10] with a luminex read-out. By the sPLS-DA, we identified a COVID-19 signature based on six immune factors. Our results showed that Spike IFN-γ, Spike IP-10, Spike IL-2; NP Pool1 RANTES; NP Pool2 IP-10 and ORF3a IL-2 are the most important in vitro conditions to distinguish COVID-19- from NO-COVID-19-subjects over all the antigen stimulations. Although we demonstrated that a combined test including all the immune factors reached the best AUC to identify COVID-19 and NO-COVID-19-individuals, we found that the single test based on Spike IP-10 could be a potential new biomarker assay of SARS-CoV-2 infection. Moreover, we validated the use of IP-10 as biomarker of SARS-CoV-2 infection in another cohort of COVID-19-patients with different clinical characteristics. In fact, to corroborate the reproducibility of our results, we performed a validation study testing Spike peptides from a commercial company and using a more feasible routine approach such the IP-10 ELISA. We demonstrated that IP-10 had a good accuracy to identify hospitalized COVID-19-patients in the first two weeks after symptoms onset and not-hospitalized-patients enrolled 35–100 days after symptoms onset.
IP-10 is a chemokine mainly secreted by monocytes, fibroblasts and endothelial cells in response to IFN-γ that attracts activated T-cells to foci of inflammation [25]; it has already been described as a potential biomarker for other infectious disease, such as tuberculosis and HCV [26–30] and may be easily measured in condition of immune-depression [30]. In acute COVID-19-patients, IP-10 production is a promising surrogate marker of impaired immune responses [13]. In our study IP-10 production induced by Spike stimulation was the only parameter not affected by any clinical characteristics. We reported that IP-10 identified SARS-CoV-2 infection in the acute phase of disease and in COVID-19-recovered subjects. This result has a double scientific implication. Firstly, it supports the specificity of the immune response to viral-peptides in different clinical conditions; secondly, it suggests a possible application of the “IP-10 and Spike whole-blood test” as a potential additional tool for diagnostic and immune response evaluation of COVID-19-patients during the acute phase of the disease. These findings are in agreement with other cytokine release-based tests applied for the diagnosis of several infectious diseases [31–34]. Moreover, an additional possible application of this whole-blood based cytokine assay is the evaluation of immune response in SARS-CoV-2 vaccine trials. In this context, the IP-10 detection may define the immunogenicity of a Spike-based vaccine, whereas the immune response to the virus infection may be evaluated detecting other factors as RANTES induced by NP.
Previous reports focused on the pre-existing immune response to SARS-CoV-2 in the general population, demonstrating that ORF1-specific T-cells were detected in SARS-CoV-2 unexposed donors [19, 35]. Differently, in recovered COVID-19-subjects, the T-cells mainly recognized the structural proteins [19]. In our study, we observed few modulations of immune factors among COVID-19 and NO-COVID-19 individuals in response to the peptides of accessory protein ORF3a; these data indicate that both groups have a similar immune response and suggest a minor contribution of ORF3a in the immune-specific response in acute-hospitalized COVID-19-patients. In line with previous evidence, the majority of immune modulations concerned to stimulations with structural proteins such as NP and Spike. As already reported [10, 16, 17] we observed a production of both inflammatory and anti-inflammatory cytokines and chemokines in response to the structural protein of SARS-CoV-2.
More than 90% of seroconverters COVID-19-individuals shows an immunological memory of T-cell compartment [36] and antibody response, for several months after infection [36, 37]. However, we need more longitudinal studies to understand exactly if the immune memory response remains stable over time. Considering that the early prediction of disease progression could be useful to assess the optimal treatment strategies, an integrated knowledge of the T-cell and antibody response lays the foundation to develop biomarkers to monitor the course of COVID-19 disease.
The limits of the present study are related to the low amount of patients evaluated. However, five different viral antigens and 27 markers were concomitantly evaluated and validated in different cohorts making the here generated evidence robust. Moreover, in the control group of NO-COVID-19 individuals, it would have been useful to include subjects with acute respiratory diseases, as Influenza. Indeed, it has been demonstrated that serum or plasma IP-10 is increased in several respiratory infections, as tuberculosis [26, 38] or influenza [39]. However, in 2020 and 2021 so far, in Europe the Influenza Virus positivity in sentinel specimens remained below the epidemic threshold due to the use of massive vaccination, masks and lockdown rules [40]. Further studies will help understanding if the coinfection of COVID-19 and other acute infectious diseases may have an impact of the SARS-CoV-2-specific IP-10 signature. Nevertheless, in a recent study [10] we showed that NO-COVID-19 patients with respiratory disease such as tuberculosis and bacterial pneumonia did not show IFN-γ-specific response to Spike stimulation. Similarly, in the present study, we did not find IP-10-specific response to Spike in NO-COVID-19 individuals. Interestingly, the NO-COVID-19 group included seven subjects with active tuberculosis under therapy and 5/7 in the acute phase of the disease as they were enrolled within 7 days of diagnosis and of starting the anti-TB specific therapy. These evidences support the specificity of our data even if generated with a low number of control patients.
In conclusion, we demonstrated the potential application of a whole-blood based platform that allowed the selection of the best antigen and best read out to evaluate the immune response to SARS-CoV-2 infection. We also identified IP-10 detection induced by Spike stimulation, as a good in vitro setting to distinguish COVID-19 from NO-COVID-19-individuals.
Materials and methods
Study design
This study was approved by the Ethical Committee of Lazzaro Spallanzani National Institute of Infectious Diseases (59/2020) and was conducted between July 15th and November 5th, 2020. Informed, written consent was required to prospectively enroll patients and controls by physicians. Demographic and clinical information were collected at enrollment (Table 1). The study complied with the principles of the Declaration of Helsinki. Inclusion criteria for COVID-19-patients: a diagnosis based on positive nasopharyngeal swab for SARS-CoV-2; a disease with specific clinical characteristics [41]. Exclusion criteria: HIV infection, inability to sign an informed consent and age younger than 18 years. To perform the multiplex analysis, we prospectively enrolled 23 COVID-19-patients and 18 individuals without COVID-19 (NO-COVID-19). COVID-19-patients were classified as asymptomatic (n = 2), mild (n = 3), moderate (n = 11), severe (n = 5), and critical (n = 2) (1). NO-COVID-19-individuals were healthy donors (n = 4), subjects with tuberculosis under therapy (n = 7) (5/7 were enrolled within 7 days of starting a specific anti-tuberculosis therapy), and subjects with latent tuberculosis infection (n = 7).
For the IP-10 study, we prospectively enrolled 18 NO-COVID-19-subjects and two cohorts of COVID-19-patients: cohort (A) 15 hospitalized-patients enrolled 1–14 days after symptoms onset; cohort (B) 10 not-hospitalized-patients (convalescent/recovered) enrolled 35–100 days after symptoms onset (Table 4).
Peptide pools and stimuli
For the exploratory study, SARS-CoV-2 peptide pools of 15-mers (55 peptides) at 2 µg/mL, covering the whole NP (Pool1 and Pool 2), M, ORF3a proteins and 40.5% of the Spike protein, were used as reported [42]. For the validation study, SARS‑CoV-2 PepTivator® Peptide Pool of the Spike protein at 0.1 µg/mL (Miltenyi, Biotec, Germany) were used. Stimulated whole-blood was overnight incubated at 37 °C, 5% CO2, plasma was collected and stored at − 80 °C until used.
SARS-CoV-2 serology
SARS-CoV-2 specific IgM and IgG levels were measured by ELISA according to manufacturer’s instructions (DIESSE Diagnostica Senese S.p.a., Monteriggioni, Italy). The ratio between the optical density (OD) of the sample and that one of the cut-off reagent (index) was calculated. The samples were scored positive (index > 1.1), doubtful (index between 1.1 and 0.9) and negative (index < 0.9).
Cytokines, chemokines and growth factors evaluation
Bio-Plex Pro Human Cytokine 27-plex Assay panel and the MagPix system (Bio-Rad, Hercules, CA, USA) were used to evaluate in harvested plasma: cytokines, chemokines and growth factors (IL-1β, IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL- 15, IL-17A, eotaxin, FGF, granulocyte-colony stimulating factor [G-CSF], granulocyte macrophage colony-stimulating factor [GM-CSF], IFN-γ, IP-10, monocyte chemoattractant protein-1 [MCP-1], macrophage inflammatory protein [MIP]-1α, MIP-1β, Platelet-derived growth factor [PDGF], RANTES, tumour necrosis factor-alpha [TNF-α], and vascular endothelial growth factor [VEGF]). Raw data were generated using the Bio-Plex manager software. Concentrations below the detection range were considered as zero. Concentrations above the detection range were converted to the highest value of the standard curve. Analyte levels were subtracted from the unstimulated control. Values generated from less than 50 beads reading were not calculated (one value was missing for: IL-2 ORF3a, IL-5 ORF3a; IL-12 ORF3a; IL-13 ORF3a; IL-15 ORF3a; IL-17 ORF3a; IL-17 NP Pool1; eotaxin Membrane; eotaxin Spike; GM-CSF NP Pool1; GM-CSF NP Pool2; GM-CSF ORF3a; GM-CSF Membrane; MIP-1 α ORF3a; TNF-a NP Pool1; VEGF NP Pool1. Two values were missing for TNF- α ORF3a; VEGF NP Pool 2; VEGF Spike. Three values were missing for VEGF-b ORF3a).
In the validation study, IP-10 was measured in plasma using Human CXCL10/IP-10 Quantikine ELISA (R&D Systems, Abingdon, UK) according to the manufacturer’s instructions. The samples were tested as diluted 1:50. The concentration range of detection was: 7.8–500 pg/mL.
Statistical analysis
Data were analysed using Graph Pad (GraphPad Prism 8 XML ProjecT), Stata (Stata 15, StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC) and R Project Software (version 3.6.1). Medians and interquartile ranges (IQRs) were calculated. Mann Whitney U test for comparisons among groups; Chi-squared test for categorical variables; receiver-operator characteristic (ROC) analysis for evaluating the area under the curve (AUC) and the diagnostic performance; Spearman Rank Correlation to measure the strength of association between two variables and the direction of the relationship (positive or negative).
We performed a multivariate exploratory analysis, sparse partial least squares discriminant analysis (sPLS-DA), to identify the most important soluble factors discriminating COVID-19- from NO-COVID-19-individuals. The sPLS-DA performed a variables reduction, generating latent components to synthetize the data information. For the sPLS-DA analysis, we considered in the model all the 135 analytes simultaneously (5 different stimuli, 27-factors each) limiting the components construction to the first 20 most important variables identified by the method. Data were analyzed with the R-package MixOmics. We performed a logistic regression analysis to evaluate the potential ability of a minimal subset of variables to classify COVID-19 from NO-COVID-19-patients; AUC and p values were reported.
Acknowledgements
The authors are grateful to all the patients and nurses who helped to conduct this study. The authors gratefully acknowledge the Collaborators Members of the National Institute for Infectious Diseases (INMI) COVID 19 study group: Maria Alessandra Abbonizio, Amina Abdeddaim, Chiara Agrati, Fabrizio Albarello, Gioia Amadei, Alessandra Amendola, Andrea Antinori, Mario Antonini, Tommaso Ascoli Bartoli, Francesco Baldini, Raffaella Barbaro, Barbara Bartolini, Rita Bellagamba, Martina Benigni, Nazario Bevilacqua, Gianlugi Biava, Michele Bibas, Licia Bordi, Veronica Bordoni, Evangelo Boumis, Marta Branca, Donatella Busso, Marta Camici, Paolo Campioni, Maria Rosaria Capobianchi, Ales sandro Capone, Cinzia Caporale, Emanuela Caraffa, Ilaria Caravella, Fabrizio Carletti, Concetta Castilletti, Adriana Cataldo, Stefano Cerilli, Carlotta Cerva, Roberta Chiappini, Pierangelo Chinello, Carmine Ciaralli, Stefania Cicalini, Francesca Colavita, Angela Corpolongo, Massimo Cristofaro, Salvatore Curiale, Alessandra D'Abramo, Cristina Dantimi, Alessia De Angelis, Giada De Angelis, Maria Grazia De Palo, Federico De Zottis, Virginia Di Bari, Rachele Di Lorenzo, Federica Di Stefano, Gianpiero D'Offizi, Davide Donno, Francesca Faraglia, Federica Ferraro, Lorena Fiorentini, Andrea Frustaci, Matteo Fusetti, Vincenzo Galati, Roberta Gagliardini, Paola Gallì, Gabriele Garotto, Saba Gebremeskel Tekle, Maria Letizia Giancola, Filippo Giansante, Emanuela Giombini, Guido Granata, Maria Cristina Greci, Elisabetta Grilli, Susanna Grisetti, Gina Gualano, Fabio Iacomi, Giuseppina Iannicelli, Giuseppe Ippolito, Eleonora Lalle, Simone Lanini, Daniele Lapa, Luciana Lepore, Raffaella Libertone, Raffaella Lionetti, Giuseppina Liuzzi, Laura Loiacono, Andrea Lucia, Franco Lufrani, Manuela Macchione, Gaetano Maffongelli, Alessandra Marani, Luisa Marchioni, Andrea Mariano, Maria Cristina Marini, Micaela Maritti, Alessandra Mastrobattista, Giulia Matusali, Valentina Mazzotta, Paola Mencarini, Silvia Meschi, Francesco Messina, Annalisa Mondi, Marzia Montalbano, Chiara Montaldo, Silvia Mosti, Silvia Murachelli, Maria Musso, Emanuele Nicastri, Pasquale Noto, Roberto Noto, Alessandra Oliva, Sandrine Ottou, Claudia Palazzolo, Emanuele Pallini, Fabrizio Palmieri, Carlo Pareo, Virgilio Passeri, Federico Pelliccioni, Antonella Petrecchia, Ada Petrone, Nicola Petrosillo, Elisa Pianura, Carmela Pinnetti, Maria Pisciotta, Silvia Pittalis, Agostina Pontarelli, Costanza Proietti, Vincenzo Puro, Paolo Migliorisi Ramazzini, Alessia Rianda, Gabriele Rinonapoli, Silvia Rosati, Martina Rueca, Alessandra Sacchi, Alessandro Sampaolesi, Francesco Sanasi, Carmen Santagata, Alessandra Scarabello, Silvana Scarcia, Vincenzo Schinina, Paola Scognamiglio, Laura Scorzolini, Giulia Stazi, Fabrizio Taglietti, Chiara Taibi, Roberto Tonnarini, Simone Topino, Francesco Vaia, Francesco Vairo, Maria Beatrice Valli, Alessandra Vergori, Laura Vincenzi, Ubaldo Visco Comandini, Serena Vita, Pietro Vittozzi, and Mauro Zaccarelli.Delia Goletti is a professor of Pathology of Unicamillus, International Medical University of Rome.
Authors’ contributions
EP Designed the data base, analysed and interpreted data and wrote the manuscript; SNF performed luminex assay, ELISA and serology evaluation, analysed and interpreted data; AN performed the statistical analysis and interpreted data; LiP participated in the interpretation of the data and revised the manuscript; VV processed blood samples; GC enrolled patients and collected clinical data; GG, LuP, EN, FP enrolled patients; AB provided peptide reagents and participated in the interpretation of data; GI participated in the interpretation of the data; DG designed and wrote the study, coordinated and supervised the project, contributed to the interpretation of the results, wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study has been funded by Line one-Ricerca Corrente “Infezioni Emergenti e Riemergenti” by Line four-Ricerca Corrente, by the projects COVID 2020-12371675 and COVID-2020-12371735, all funded by Italian Ministry of Health; by Società Numero Blu Servizi S.p.A., Fineco Bank S.p.A, Associazione magistrati della Corte dei conti, Società Mocerino Frutta Secca s.r.l., through a generous liberal donation funding on COVID-19 research (Issue No. 257; 14/04/2021).
Availability of data and materials
The data sets generated during and/or analyzed during the current study are available in our institutional repository after request at rawdata.inmi.it.
Declarations
Ethics approval and consent to participate
This study was approved by the Ethical Committee of Lazzaro Spallanzani National Institute of Infectious Diseases (59/2020). Informed, written consent was required to prospectively enroll patients and controls by physicians. The study complied with the principles of the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
AB submitted a patent to monitor SARS-Cov-2 specific T-cells and participated on advisory board for Oxford Immunotech and Quiagen; there are no conflicts of interest to declare for any of the other authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO . COVID-19 Clinical management: living guidance. Geneva: WHO; 2021. [Google Scholar]
- 2.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–8. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantini F, Goletti D, Petrone L, Najafi Fard S, Niccoli L, Foti R. Immune therapy, or antiviral therapy, or both for COVID-19: a systematic review. Drugs. 2020;80:1929–46. doi: 10.1007/s40265-020-01421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goletti D, Cantini F. Baricitinib therapy in covid-19 pneumonia—an unmet need fulfilled. N Engl J Med. 2021;384:867–9. doi: 10.1056/NEJMe2034982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laing AG, Lorenc A, Del Barrio IDM, Das A, Fish M, Monin L, Muñoz-Ruiz M, McKenzie DR, Hayday TS, Francos-Quijorna I, Kamdar S, Joseph M, Davies D, Davis R, Jennings A, Zlatareva I, Vantourout P, Wu Y, Sofra V, Cano F, Greco M, Theodoridis E, Freedman JD, Gee S, Chan JNE, Ryan S, Bugallo-Blanco E, Peterson P, Kisand K, Haljasmägi L, Chadli L, Moingeon P, Martinez L, Merrick B, Bisnauthsing K, Brooks K, Ibrahim MAA, Mason J, Lopez Gomez F, Babalola K, Abdul-Jawad S, Cason J, Mant C, Seow J, Graham C, Doores KJ, Di Rosa F, Edgeworth J, Shankar-Hari M, Hayday AC. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–35. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 7.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–45.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M, Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–43. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Dela Cruz C, Farhadian S, Ko AI, Omer SB, Iwasaki A, Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–9. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrone L, Petruccioli E, Vanini V, Cuzzi G, Najafi Fard S, Alonzi T, Castilletti C, Palmieri F, Gualano G, Vittozzi P, Nicastri E, Lepore L, Antinori A, Vergori A, Caccamo N, Cantini F, Girardi E, Ippolito G, Grifoni A, Goletti D. A whole blood test to measure SARS-CoV-2-specific response in COVID-19 patients. Clin Microbiol Infect. 2021;27:286.e7–286.e13. doi: 10.1016/j.cmi.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrone L, Petruccioli E, Vanini V, Cuzzi G, Gualano G, Vittozzi P, Nicastri E, Maffongelli G, Grifoni A, Sette A, Ippolito G, Migliori GB, Palmieri F, Goletti D. Coinfection of tuberculosis and COVID-19 limits the ability to in vitro respond to SARS-CoV-2. Int J Infect Dis. 2021 doi: 10.1016/j.ijid.2021.02.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrone L, Petruccioli E, Alonzi T, Vanini V, Cuzzi G, Najafi Fard S, Castilletti C, Palmieri F, Gualano G, Vittozzi P, Nicastri E, Lepore L, Grifoni A, Antinori A, Vergori A, Ippolito G, Cantini F, Goletti D. In-vitro evaluation of the immunomodulatory effects of Baricitinib: implication for COVID-19 therapy. J Infect. 2021;82:58–66. doi: 10.1016/j.jinf.2021.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, Crotty S. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ, Wang Q, Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020 doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, de Lafaille MAC, Mehandru S, Merad M, Samstein RM. Sinai immunology review project: immunology of COVID-19: current state of the science. Immunity. 2020;52:910–41. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, Haagmans BL, de Swart RL, Sette A, de Vries RD. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, Gozzi L, Iannone A, Lo Tartaro D, Mattioli M, Paolini A, Menozzi M, Milić J, Franceschi G, Fantini R, Tonelli R, Sita M, Sarti M, Trenti T, Brugioni L, Cicchetti L, Facchinetti F, Pietrangelo A, Clini E, Girardis M, Guaraldi G, Mussini C, Cossarizza A. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020 doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong CWM, Migliori GB, Raviglione M, MacGregor-Skinner G, Sotgiu G, Alffenaar JW, Tiberi S, Adlhoch C, Alonzi T, Archuleta S, Brusin S, Cambau E, Capobianchi MR, Castilletti C, Centis R, Cirillo DM, D'Ambrosio L, Delogu G, Esposito SMR, Figueroa J, Friedland JS, Ho BCH, Ippolito G, Jankovic M, Kim HY, Rosales Klintz S, Ködmön C, Lalle E, Leo YS, Leung CC, Märtson AG, Melazzini MG, Najafi Fard S, Penttinen P, Petrone L, Petruccioli E, Pontali E, Saderi L, Santin M, Spanevello A, van Crevel R, van der Werf MJ, Visca D, Viveiros M, Zellweger JP, Zumla A, Goletti D. Epidemic and pandemic viral infections: impact on tuberculosis and the lung: A consensus by the World Association for Infectious Diseases and Immunological Disorders (WAidid), Global Tuberculosis Network (GTN), and members of the European Society of Clinical Microbiology and Infectious Diseases Study Group for Mycobacterial Infections (ESGMYC) Eur Respir J. 2020;56:2001727. doi: 10.1183/13993003.01727-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, Chng MHY, Lin M, Tan N, Linster M, Chia WN, Chen MI, Wang LF, Ooi EE, Kalimuddin S, Tambyah PA, Low JG, Tan YJ, Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–62. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 20.Murugesan K, Jagannathan P, Pham TD, Pandey S, Bonilla HF, Jacobson K, Parsonnet J, Andrews JR, Weiskopf D, Sette A, Pinsky BA, Singh U, Banaei N. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echeverría G, Guevara Á, Coloma J, Ruiz AM, Vasquez MM, Tejera E, de Waard JH. Pre-existing T-cell immunity to SARS-CoV-2 in unexposed healthy controls in Ecuador, as detected with a COVID-19 interferon-gamma release assay. Int J Infect Dis. 2021;105:21–25. doi: 10.1016/j.ijid.2021.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aiello A, Fard SN, Petruccioli E, Petrone L, Vanini V, Farroni C, Cuzzi G, Navarra A, Gualano G, Mosti S, Pierelli L, Nicastri E, Goletti D. Spike is the most recognized antigen in the whole-blood platform in both acute and convalescent COVID-19 patients. Int J Infect Dis. 2021 doi: 10.1016/j.ijid.2021.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siu KL, Yuen KS, Castaño-Rodriguez C, Ye ZW, Yeung ML, Fung SY, Yuan S, Chan CP, Yuen KY, Enjuanes L, Jin DY. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–77. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–15. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrone L, Cannas A, Vanini V, Cuzzi G, Aloi F, Nsubuga M, Sserunkuma J, Nazziwa RA, Jugheli L, Lukindo T, Girardi E, Antinori A, Pucci L, Reither K, Goletti D. Blood and urine inducible protein 10 as potential markers of disease activity. Int J Tuberc Lung Dis. 2016;20:1554–61. doi: 10.5588/ijtld.16.0342. [DOI] [PubMed] [Google Scholar]
- 27.Petrone L, Chiacchio T, Vanini V, Petruccioli E, Cuzzi G, Di Giacomo C, Pucci L, Montalbano M, Lionetti R, Testa A, Lapa D, Navarra A, Visco-Comandini U, Goletti D. High urine IP-10 levels associate with chronic HCV infection. J Infect. 2014;68:591–600. doi: 10.1016/j.jinf.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Petrone L, Vanini V, Chiacchio T, Petruccioli E, Cuzzi G, Schininà V, Palmieri F, Ippolito G, Goletti D. Evaluation of IP-10 in Quantiferon-Plus as biomarker for the diagnosis of latent tuberculosis infection. Tuberculosis. 2018;111:147–153. doi: 10.1016/j.tube.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Santos VS, Goletti D, Kontogianni K, Adams ER, Molina-Moya B, Dominguez J, Crudu V, Martins-Filho PRS, Ruhwald M, Lawson L, Bimba JS, Garcia-Basteiro AL, Petrone L, Kabeer BS, Reither K, Cuevas LE. Acute phase proteins and IP-10 as triage tests for the diagnosis of tuberculosis: systematic review and meta-analysis. Clin Microbiol Infect. 2019;25:169–177. doi: 10.1016/j.cmi.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Vanini V, Petruccioli E, Gioia C, Cuzzi G, Orchi N, Rianda A, Alba L, Giancola ML, Conte A, Schininà V, Rizzi EB, Girardi E, Goletti D. IP-10 is an additional marker for tuberculosis (TB) detection in HIV-infected persons in a low-TB endemic country. J Infect. 2012;65:49–59. doi: 10.1016/j.jinf.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Petrone L, Albrich WC, Tamarozzi F, Frischknecht M, Gomez-Morales MA, Teggi A, Hoffmann M, Goletti D. Species specificity preliminary evaluation of an IL-4-based test for the differential diagnosis of human echinococcosis. Parasite Immunol. 2020;42:e12695. doi: 10.1111/pim.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrone L, Vanini V, Amicosante M, Corpolongo A, Gomez Morales MA, Ludovisi A, Ippolito G, Pozio E, Teggi A, Goletti D. A T-cell diagnostic test for cystic echinococcosis based on antigen B peptides. Parasite Immunol. 2017;39:e12499. doi: 10.1111/pim.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dammermann W, Bentzien F, Stiel EM, Kühne C, Ullrich S, Wiesch SZJ, Lüth S. Development of a novel IGRA assay to test T cell responsiveness to HBV antigens in whole blood of chronic hepatitis B patients. J Transl Med. 2015 doi: 10.1186/s12967-015-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goletti D, Lee MR, Wang JY, Walter N, Ottenhoff THM. Update on tuberculosis biomarkers: from correlates of risk, to correlates of active disease and of cure from disease. Respirology. 2018;23:455–66. doi: 10.1111/resp.13272. [DOI] [PubMed] [Google Scholar]
- 35.Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, Tan YJ. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–14. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, Nakao C, Rayaprolu V, Rawlings SA, Peters B, Krammer F, Simon V, Saphire EO, Smith DM, Weiskopf D, Sette A, Crotty S. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–30. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrone L, Vanini V, Chiacchio T, Petruccioli E, Cuzzi G, Schininà V, Palmieri F, Ippolito G, Goletti D. Evaluation of IP-10 in Quantiferon-Plus as biomarker for the diagnosis of latent tuberculosis infection. Tuberculosis. 2018;111:147–53. doi: 10.1016/j.tube.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Hayney MS, Henriquez KM, Barnet JH, Ewers T, Champion HM, Flannery S, Barrett B. Serum IFN-γ-induced protein 10 (IP-10) as a biomarker for severity of acute respiratory infection in healthy adults. J Clin Virol. 2017;90:32–7. doi: 10.1016/j.jcv.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ECDC/WHO Europe weekly influenza update: Flu News Europe 2020–2021 season overview. https://flunewseurope.org/SeasonOverview. Accessed 21 June 2021.
- 41.Nicastri E, Petrosillo N, Ascoli Bartoli T, Lepore L, Mondi A, Palmieri F, D’Offizi G, Marchioni L, Murachelli S, Ippolito G, Antinori A. National Institute for the Infectious Diseases “L. Spallanzani”, IRCCS. Recommendations for COVID-19 clinical management. Infect Dis Rep. 2020;12:8543. doi: 10.4081/idr.2020.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Bert N, Clapham HE, Tan AT, Chia WN, Tham CYL, Lim JM, Kunasegaran K, Tan LWL, Dutertre CA, Shankar N, Lim JME, Sun LJ, Zahari M, Tun ZM, Kumar V, Lim BL, Lim SH, Chia A, Tan YJ, Tambyah PA, Kalimuddin S, Lye D, Low JGH, Wang LF, Wan WY, Hsu LY, Bertoletti A, Tam CC. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J Exp Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during and/or analyzed during the current study are available in our institutional repository after request at rawdata.inmi.it.