Graphical abstract
Keywords: New coronavirus, Brain, Neurospheres, SARS-CoV-2, iPS
Abstract
Coronavirus disease 2019 (COVID-19) was initially described as a viral infection of the respiratory tract. It is now known, however, that several other organs are affected, including the brain. Neurological manifestations such as stroke, encephalitis, and psychiatric conditions have been reported in COVID-19 patients, but the neurotropic potential of the virus is still debated. Herein, we sought to investigate SARS-CoV-2 infection in human neural cells. We demonstrated that SARS-CoV-2 infection of neural tissue is non-permissive, however, it can elicit inflammatory response and cell damage. These findings add to the hypothesis that most of the neural damage caused by SARS-CoV-2 infection is due to a systemic inflammation leading to indirect harmful effects on the central nervous system despite the absence of local viral replication.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the 2019 coronavirus disease (COVID-19), an airborne infectious disease. Most affected patients have symptoms of respiratory infections, such as fever, dry cough, and dyspnea, and 5 to 10% of them evolve to severity/death. Respiratory failure is the main event that explains fatality in COVID-19. Some authors raised the possibility that at least part of the respiratory manifestations caused by COVID-19 could be due to direct viral damage to the respiratory center located in the brain stem (Gupta et al., 2020).
Apart from respiratory symptoms, anosmia and ageusia were identified as hallmarks of COVID-19. Indeed, evidence of neurological disease associated with COVID-19 is increasing; 30 to 60% of patients further present one or more neurological symptoms, including paresthesia, altered consciousness, and headache (De Felice et al., 2020, Wang et al., 2020). Besides, COVID-19 patients are at a 7-fold higher risk to suffer a stroke, and 2–6% progress to cerebrovascular disease (Fifi and Mocco, 2020). Less frequent neurological manifestations include encephalopathy, encephalitis and Guillain-Barré syndrome (Ellul et al., 2020, Mao et al., 2020). In addition, there is mounting evidence for neurological sequelae following recovery from COVID-19, such as cognitive decline and cortical hypofunction (Hosp et al., 2021).
Neuroinvasive capacity was shown for other highly pathogenic coronaviruses, including the Middle East Respiratory Syndrome coronavirus (MERS-CoV) (Li et al., 2016) and SARS-CoV (Glass et al., 2004, Xu et al., 2005). Similarly to SARS-CoV, SARS-CoV-2 uses the angiotensin‐converting enzyme 2 (ACE2) receptor as the main cellular entry (Yan et al., 2020). ACE2 is highly expressed in nasal epithelial cells, which underlies the initiation of respiratory tract infection (Sungnak et al., 2020), but is also present in multiple tissues (Hamming et al., 2004). Studies dissecting the putative mechanisms whereby SARS-CoV-2 enters the central nervous system (CNS) are still scarce. At first, it was speculated that the virus infects the olfactory neurons and reaches the brain by anterograde axonal transport. However, it was shown in rodents that SARS-CoV-2 infects non-neural cells in the olfactory epithelium rather than olfactory neurons, which could be attributed to the lack of expression of ACE2 in these cells (Brann et al., 2020, Bryche et al., 2020). In contrast, a recent study found viral particles throughout olfactory neuron projections and in the olfactory bulb of a few patients (Meinhardt et al., 2020) and other hypotheses have been proposed (Barrantes, 2021). Therefore, due to these conflicting findings, it is still debatable whether the mechanism of SARS-CoV-2 invasion in the CNS is via olfactory neurons.
Viral infections targeting the brain were successfully modeled in vitro using human neurospheres and brain organoids (Garcez et al., 2016, Qian et al., 2016). These models are comprised of neural cells in a tissular arrangement more like the CNS, which support more complex cellular interactions and consequently more reliable responses to viral infection. Because of that, these models became valuable for investigating SARS-CoV-2 infection in the brain tissue. While some researchers found that the virus infects neurons and glia (Jacob et al., 2020a, Mesci et al., 2020, Song et al., 2020), or a small subset of neurons (Dobrindt et al., 2021), others were unable to detect viral antigens in these cells (Pellegrini et al., 2020), suggesting SARS-CoV-2 neural infection was dependent on experimental conditions and/or viral isolates. Additionally, even fewer studies measured whether infection led to the production of new infective particles, or virions.
Here, we investigated SARS-CoV-2 infection in human neurospheres (NP) derived from induced pluripotent stem cells. Although viral infection was found to be non-permissive, it led to inflammatory response and cytotoxicity which was correlated to the amount of virus in the inoculum. We suggest that despite insufficient viral infection and replication in the brain parenchyma, there are consequences of the viral presence in the tissue, which may be enhanced by the entry of inflammatory molecules resulting from a generalized inflammatory response.
2. Materials and methods
2.1. Cell cultivation
Human induced pluripotent stem (iPS) cells were obtained from the Coriell Institute for Medical Research repository (GM23279A) or produced in house with CytoTune™-iPS 2.0 Sendai Reprogramming Kit (A16517- Invitrogen) from skin fibroblasts (Da Silveira Paulsen et al., 2012) or urine epithelial cells (Sochacki et al., 2016) (1 donor for each cell type) (Table 1 ). iPS cells were cultured in StemFlex media (A33494 - Thermo Fisher Scientific) on top of Matrigel (BD Biosciences, Franklin Lakes, NJ). Neural Stem Cells (NSCs) were generated from iPS cells following the published Life Technologies protocol for “Induction of Neural Stem Cells from Human Pluripotent Stem Cells” (Publication number: MAN0008031). NSCs were cultured in neural expansion medium (Advanced DMEM-F12 and Neurobasal medium 1:1 with 1% neural induction supplement) on a surface previously coated with Geltrex LDEV-Free (Thermo Fisher Scientific, A1413302) for expansion.
Table 1.
Cell lines used to generate human neurospheres.
| Cell line name | Primary cells | SC | Age at collection | Origin | References |
|---|---|---|---|---|---|
| GM23279A | Dermal Fibroblast | XX | 36 | Coriell biobank | (Takahashi et al., 2007, Casas et al., 2018) |
| CF1 | Dermal Fibroblast | XY | 37 | In house | (Casas et al., 2018, Ledur et al., 2020) |
| C19 | Urine | XX | 17 | In house | Unpublished |
SC - sex chromosomes
Neurospheres (NP) were prepared in suspension as follows: NSCs were split with Accutase (Merck Millipore, SCR005) and 3 × 106 cells (9 × 102 cells/NP) were plated in a 6 well-plate, being cultured on an orbital shaker at 90 rpm for 7 days in standard culture conditions. At day 3, the culture medium was changed to induce differentiation, containing Neurobasal medium (Thermo Fisher Scientific, 12348017)/DMEM-F12(Life Technologies, 11330–032) 1:1, supplemented with 1x N2 (Thermo Fisher Scientific, 17502001) and 1x B27 (Thermo Fisher Scientific, 17504001), and it was changed each 3–4 days (Casas et al., 2018). Alternatively, NP were prepared directly in 96 well plates. NSCs were split with Accutase (Merck Millipore, SCR005), centrifuged at 300×g for 5 min and resuspended in the same differentiation medium described for the 6 well-plate NP. Subsequently, 9 × 103 cells in 150 μL were plated per well of a round bottom Ultra-low attachment 96-well plate (Corning, 7007), followed by centrifugation at 300×g for 3 min to assure sedimentation. To minimize neurosphere damage, the medium of each well was changed every other day by only removing 70–100 μL and adding 150 μL of new medium. iPS cell-derived human cardiomyocytes (CM) were purchased from Pluricell (São Paulo, Brazil) (Cruvinel et al., 2020). CM were used between day 30 and day 40 of differentiation. Cell differentiation was determined by staining for cardiac specific troponin T protein and quantification by flow cytometry and found to be 86 ± 6.2%.
2.2. Total RNA isolation
NP were cultured in 6-well plates for 7 days, harvested and immediately frozen at −80 °C until further processing. Total RNA isolation was performed using ReliaPrep™ RNA Tissue Miniprep System (Promega Corporation) according to manufacturer’s instructions. For cardiomyocytes, total RNA was isolated using TRIzol™ reagent, according to manufacturer’s recommendations (Thermo Fisher Scientific). RNA concentration and quality were quantified on a NanoDrop™ 2000c Spectrophotometer (Thermo Fisher Scientific); and integrity and purity were evaluated by 1.8% agarose gel electrophoresis using a photo documentation device equipped with a UV lamp (L-PIX, Loccus Biotecnologia). Then, samples were digested with DNase I, Amplification Grade, following the manufacturer’s instructions (Invitrogen, Thermo Fisher Scientific). 2 μg of RNA from DNAse-treated samples were reverse transcribed using M−MLV for complementary DNA generation (cDNA) (Thermo Fisher Scientific).
2.3. Quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR)
For gene expression analysis of NP and cardiomyocytes, qRT-PCR reactions were conducted in three replicates with a final reaction volume of 10 µL in MicroAmp Fast Optical 96 Well Reaction Plates (Thermo Fisher Scientific) containing 1X GoTaq qPCR Master Mix (Promega Corporation), 300 nM CXR Reference Dye, a final concentration of 200 nM of each SYBR green designed primers [Angiotensin I Converting Enzyme 2 (ACE2; forward: 5′-CGAAGCCGAAGACCTGTTCTA-3′; reverse: 5′-GGGCAAGTGTGGACTGTTCC-3′) Thermo Fisher Scientific] and 10 ng of cDNA per reaction. Appropriate controls (no reverse transcriptase and template-negative controls) were incorporated into each run. Briefly, the reactions were performed on a StepOnePlusTM Real-Time PCR System thermocycler (Applied Biosystems). Thermal cycling program comprised of a denaturing step at 95 °C for 3 min, followed by 40 cycling stages at 95 °C for 15 sec, 57 °C for 15 sec, 72 °C for 15 sec and melt curve stage 95 °C, 15 sec; 60 °C, 1 min; 95 °C, 15 sec. The relative expression of the genes of interest was normalized by human reference genes: Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH; forward: 5′-GCCCTCAACGACCACTTTG-3′; reverse: 5′-CCACCACCCTGTTGCTGTAG-3′) and Hypoxanthine Phosphoribosyltransferase 1 (HPRT-1; forward 5′-CGTCGTGATTAGTGATGATGAACC-3′; reverse: 5′-AGAGGGCTACAATGTGATGGC-3′). qPCR data analysis was performed with the N0 method implemented in LinRegPCR v. 2020.0, which considers qPCR mean efficiencies estimated by the window-of-linearity method (Ramakers et al., 2003, Ruijter et al., 2009). Briefly, N0 values were calculated in LinRegPCR using default parameters. Then, the arithmetic mean of N0 values from gene of interest (GOI) was normalized by taking its ratio to the N0 geometric mean of the reference genes (REF: GAPDH and HRRT-1; N0GOI/N0REF).
2.4. SARS-CoV-2 propagation
SARS-CoV-2 obtained from a nasopharyngeal swab from a confirmed case in Rio de Janeiro, Brazil (GenBank accession no. MT710714, PANGO lineage B.1 (Rambaut et al., 2020), clade 20C (Hadfield et al., 2018) was expanded in African green monkey kidney cells (Vero, subtype E6). Virus isolation was performed after a single passage in cell culture in a 150 cm2 flask, previously infected with SARS-CoV-2 at multiplicity of infection (MOI) 0.01. All procedures related to virus culture were handled in a biosafety level 3 (BSL3) facility, according to WHO guidelines. Virus stocks were kept at −80 °C.
2.5. Cell infection
NP cultivated for 7 days were infected with SARS-CoV-2 at MOI 0.1 and 0.01. Following 1 h incubation, the inoculum was partially replaced by fresh medium and NP were cultivated for additional 2 and 5 days, with orbital shaking at 90 rpm in standard conditions (5% CO2 and 37 °C). The medium was not completely replaced to avoid excessive stress as explained above. NP exposed to uninfected Vero’s culture medium were used as controls of infection (Mock). NP (≈50 to 200) were used per experimental group for each analysis. Culture supernatants were collected at 2- and 5-days post-infection for virus titration by plaque forming units (PFU) assay and/or real time qRT-PCR. The assay was performed in a single experiment, consisting of three cell lines generated from 3 independent donors.
In lieu of the formerly described method, seven days-old NP were separately (one per well) infected for 1 h or 24 h with SARS-CoV-2 at different MOIs (0.1, 1 and 10). After incubation, the inoculum was partially replaced by fresh medium, for the reasons stated above, and NP were cultured in standard conditions (5% CO2 and 37 °C). Supernatants and NP were collected at 24 h, 48 h and 72 h post-infection. NP exposed to uninfected Vero’s culture medium were used as controls of infection (Mock). 5 to 24 NP were distributed by each experimental group.
Cardiomyocytes were infected with SARS-CoV-2 at MOI 0.1 for 1 h. Next, the inoculum was replaced by fresh medium and cultured in standard conditions for 48–72 h. After that, the monolayer was fixed with 4% paraformaldehyde (PFA) solution.
2.6. Immunofluorescence staining
NP were fixed in 4% paraformaldehyde solution (Sigma-Aldrich) for 1 h, followed by cryopreservation with 30% sucrose solution overnight. Then, samples were embedded in O.C.T. compound (Sakura Finetek, Netherland) and frozen at −80 °C. The O.C.T. blocks were sectioned in 20 μm slices with a Leica CM1860 cryostat. After washing with PBS, sections were incubated in permeabilization/blocking solution (0.3% Triton X-100/3% goat serum) for 2 h. The primary antibody was incubated overnight at 4 °C [anti-double-stranded RNA (dsRNA) monoclonal antibody (1:200, Scicons)]. Then, sections were incubated with secondary antibody goat anti-mouse Alexa Fluor 488; 1:400, A-11001 - Thermo Fisher Scientific) for 2 h. Nuclei were counterstained with 0.5 μg/mL 40–6-diamino- 2-phenylindole (DAPI) for 10 min and the slides were mounted with Aqua-Poly/Mount (Polysciences).
After fixation, cardiomyocytes were washed with PBS and then incubated with permeabilization/blocking solution (0.3% Triton X-100/3% bovine serum albumin) for 1 h. Then, cells were incubated with primary antibody anti-dsRNA (1:200) overnight at 4 °C. On the next day, cells were incubated with the secondary antibody goat anti-mouse Alexa Fluor 488 (1:400) for 1 h. Nuclei were counterstained with DAPI for 5 min and mounted with 50% PBS-Glycerol.
Images of NP and cardiomyocytes were acquired on a Leica TCS-SP8 confocal microscope with the 63× and 20× objective, respectively.
2.7. Flow cytometry
CM were plated on Geltrex™ (Thermo Scientific)-coated 6-well plates up to days 30–32 of differentiation. Then, cells were dissociated, fixed with 1% paraformaldehyde solution and permeabilized with 0.1% Triton (Sigma Aldrich) and 0.1% Saponin (Sigma Aldrich). CM were stained with anti-TNNT2 (1:2500, MA5-12960 - Thermo Scientific). The data of each batch of differentiation was acquired using a Canto BD Flow cytometer and analyzed using the FlowJo Software considering 1%–2% of false-positive events. iPS cells were used as negative control for the reaction for definitive CM. We used samples incubated only with secondary antibodies to draw the limits of positive events.
2.8. Identification and quantification of neural progenitor population
NP were fixed, cryopreserved, embedded in O.C.T. compound and frozen as described in the immunofluorescence staining section. After cryosectioning at 20 μm, NP were washed in PBS and incubated in permeabilization solution (0.3% Triton X-100 in PBS) for 20 min followed by incubation in blocking solution (3% bovine serum albumin/5% goat serum) for 2 h. The primary antibody was incubated overnight at 4 °C [human Pax6 polyclonal antibody (1:100, Thermo Fisher Scientific)]. Then, sections were incubated in blocking solution, as described previously; followed by secondary antibody goat anti-rabbit Alexa Fluor 488; 1:400, A-11008 - Thermo Fisher Scientific) for 2 h. Nuclei were counterstained with 0.5 μg/mL 40–6-diamino- 2-phenylindole (DAPI) for 10 min and slides were mounted with Aqua-Poly/Mount (Polysciences).
Images of NP were acquired on a Leica TCS-SP8 confocal microscope with the 63x objective and analyzed with Image J software. PAX6 + area was normalized over nuclei staining area. 10 NP per experimental group were analyzed for each cell line, totalling 20 NP from 2 different cell lines (CF1 and C19). Statistical analysis to compare differences among groups was performed using One way ANOVA with Tukey’s post-hoc test (significance set at p < 0.05).
2.9. Plaque forming unit assay
For virus titration, monolayers of Vero E6 cells (2 × 104 cell/well) in 96-well plates were infected with serial dilutions of supernatants containing SARS-CoV-2 for 1 h at 37 °C. Semi-solid high glucose DMEM medium containing 2% FBS and 2.4% carboxymethylcellulose was added and cultures were incubated for 3 days at 37 °C. Then, the cells were fixed with 10% formalin for 2 h at room temperature. The cell monolayer was stained with 0.04% solution of crystal violet in 20% ethanol for 1 h. Plaque numbers were scored in at least 3 replicates per dilution by independent readers blinded to the experimental group and the virus titers were determined by plaque-forming units (PFU) per milliliter.
2.10. Molecular detection of SARS-CoV-2 RNA
The total RNA from NP’s supernatant was extracted using QIAamp Viral RNA (Qiagen), according to manufacturer’s instructions. Quantitative RT-PCR was performed using QuantiTect Probe RT-PCR Kit (Quiagen®) in a StepOne™ Real-Time PCR System (Thermo Fisher Scientific). Amplifications were carried out in 25 µL reaction mixtures containing 2X reaction mix buffer, 50 µM of each primer, 10 µM of probe, and 5 µL of RNA template. Primers, probes, and cycling conditions recommended by the Centers for Disease Control and Prevention (CDC) protocol were used to detect the SARS-CoV-2. Alternatively, genomic (ORF1) (gRNA) and subgenomic (ORF(sgRNAE)) were detected, as previously described (Wölfel et al., 2020). In summary, for detection of gRNA, the following primers and probe targeting for sequences downstream of the start codons of the gene E were used (E_Sarbeco_F; ACAGGTACGTTAATAGTTAATAGCGT / E_Sarbeco_R; ATATTGCAGCAGTACGCACACA / E_Sarbeco_P1; FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ); and for sgRNA detection, a leader-specific forward primer (sgLeadSARSCoV2-F; CGATCTCTTGTAGATCTGTTCTC) was used with the reverse primer and probe for gene E above mentioned, and described in (Corman et al., 2020, Wölfel et al., 2020).
2.11. Western blot analysis
Two days post-infection (d.p.i), 100 µL of sample buffer without bromophenol blue (62.5 mM Tris-HCl, pH 6.8, containing 10% glycerol, 2% SDS and 5% 2-mercaptoethanol) was added to the NP and then, samples were frozen at −80 °C. Next, samples were gently broken down with a disposable pestle (BAF 199230001- Sigma, Bel-Art) and cell extracts were boiled at 95 °C for 10 min and centrifuged at 4 °C 16,000×g for 15 min to remove insoluble material. Protein content was estimated using the Bio-Rad Protein Assay (#5000006, Biorad). After addition of bromophenol blue (0.02%), extract samples (40 µg/lane for NP and 15 µg/lane for Vero cells) were separated by electrophoresis on a 10% SDS polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in 5% non-fat milk in Tris-Buffered Saline with 0.1% Tween-20 (TBS-T) for 1 h at room temperature. Membranes were then incubated overnight at 4 °C, in the presence of anti-SARS-CoV-2 SP (1:2000, #GTX632604 - GeneTex) and anti-actin (1:2000, MAB1501, Millipore) diluted in TBS-T with 5% non-fat milk. Then, membranes were incubated with peroxidase-conjugated antibody goat anti-Mouse IgG (H + L), HRP-conjugate (1:10,000, G21040 -Molecular Probes). The signals were developed using ECL Prime Western Blotting System (#GERPN2232, Sigma) for five minutes and chemiluminescence was detected with an Odyssey-FC System® (Imaging System - LI-COR Biosciences). Membranes were also stripped for re-staining by incubating for three cycles of 10 min in stripping buffer (pH 2.2, 200 mM glycine, SDS 0,1% and 1% Tween-20), the buffer was discarded, then the membranes were washed for 5 min with PBS (three times) and 5 min with 0.1% TBS-T (three times). Next, the membranes were blocked again and processed for immunolabeling as described above.
2.12. Cytokine multiplex assay and LDH cytotoxicity assay
A multiplex biometric immunoassay containing fluorescent dyed microbeads was used to measure cytokines in the cell culture supernatant (Bio-Rad Laboratories, Hercules, CA, USA). The following cytokines were quantified: IL-1β, IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12(p70), IL-13, IL-15, IL-17, basic FGF, Eotaxin, G-CSF, GM-CSF, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, and VEGF; and cytokine levels were calculated by Luminex technology (Bio-Plex Workstation; Bio-Rad Laboratories, USA). The analysis of data was performed using software provided by the manufacturer (Bio-Rad Laboratories, USA). A range of 0.51–8000 pg/mL recombinant cytokines was used to establish standard curves and the sensitivity of the assay. Cytotoxicity was determined according to the activity of lactate dehydrogenase (LDH) in the culture supernatants using a CytoTox® Kit (Promega, USA) according to the manufacturer’s instructions, and presented as OD (optical density) of infected cells minus OD of mock. Statistics were performed using GraphPad Prism software version 8. Numerical variables from NP experiments were tested regarding their distribution using the Shapiro-Wilk test. For those following a normal (parametric) distribution, One-way analysis of variance (ANOVA) with Dunnet’s post-hoc test was used to compare differences among groups; and for nonparametric data, Kruskal-Wallis test with Dunn’s post-hoc test was used to compare differences.
2.13. SARS-CoV-2 strains sequence analyses
Nucleotide sequences from our strain (GenBank accession No. MT710714), from strains used in others works (Song et al., 2020, Zhang et al., 2020), and from SARS-CoV-2 reference genome (Wuhan-Hu-1, GenBank accession No. NC_045512.2) were retrieved from NCBI database and from Multiple Sequence Alignment (MSA). Visualization of SARS-CoV-2 sequences was performed using ClustalW (Thompson et al., 1994) implemented in MEGA X program- version 10.1.8 (Kumar et al., 2018). Maximum Likelihood phylogenetic analyses were conducted using the JTT matrix-based model (Jones et al., 1992) with confidence assessed by bootstrap with 1000 replicates. For identification of amino acid mutations, the coding data was translated assuming a Standard genetic code and compared to the SARS-CoV-2 reference genome Wuhan-Hu-1.
3. Results
3.1. SARS-CoV-2 replication in human neural cells is non-permissive
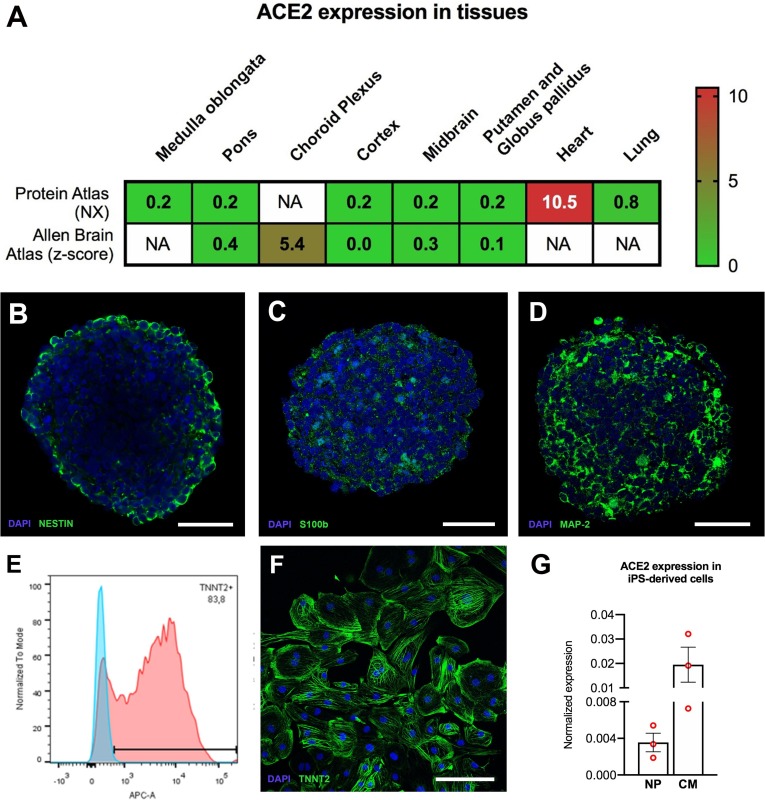
Histopathological analyses of several tissues from an infant deceased with COVID-19 fully described in a case report from our group (Gomes et al., 2020), showed viral presence by immunofluorescence (IF) with anti-SARS-CoV-2 spike protein (SP) in the choroid plexus (ChP), lateral ventricle (LV) lining, and in some locations of the frontal cerebral cortex (S1 Fig). SARS-CoV-2 infection was confirmed by quantitative Reverse Transcriptase–Polymerase Chain Reaction (qRT-PCR) (S1 Table). The heart and the lungs, which have been previously demonstrated to be permissive to SARS-CoV-2 infection (Gnecchi et al., 2020, Pérez-Bermejo et al., 2020, Schaller et al., 2020, Tabary et al., 2020), also showed presence of the virus (S1 Table and (Gomes et al., 2020)). We sought to verify whether the expression levels of the ACE2 receptor, which could be retrieved from Protein Atlas and Allen Brain Atlas databases (Hawrylycz et al., 2012, Uhlen et al., 2015) could correlate to the differences in viral infection in different brain areas. These databases contain RNAseq levels from donors between 0 and 96 years-old (Protein Atlas) and 24 and 57 years-old (Allen Brain Atlas). For comparison, we also looked at the expression levels in the heart and lungs, which are known to be infected by SARS-CoV-2. We found that ChP had higher ACE2 levels than other brain regions assessed (Fig. 1 ) and the heart and lungs expressed higher levels of ACE2 in comparison with most neural tissues (Fig. 1A).
Fig. 1.
ACE2 expression in adult human tissues and iPS-derived cells. (A) ACE2 expression in adult human tissues based on Protein Atlas and Allen Brain Atlas databases. Consensus normalized expression (NX) and z-score are used to represent the relative abundances within the respective databases (Citations in the main text). NA = not analyzed in the database. Characterization of NP and CM. NP were cultivated for 7 days and CM were differentiated for 35 days and characterized by IF (B-D and F) and flow cytometry (E). NP were immunostained for (B) Nestin (neural progenitors), (C) S100b, (astrocytes); and (D) MAP-2 (neurons). (E) Representative flow cytometry graph showing a quantification of 83.2% TNNT2 + cells from one of the cell batches used in this study; (F) CM immunostained for cardiac troponin T (TNNT2). Scale bars: (B-D) 40 µm and (F) 250 µm. (G) Relative mRNA expression levels of ACE2 in iPSC-derived cultures; NP cultivated for 7 days and CM differentiated for 35 days. (n = 3 cell culture batches for both), normalized to reference genes, GAPDH and HPRT-1. The data correspond to 3 independent cell lines for NP.
NP have been previously used to investigate ZIKV infection and in studies to address drug neurotoxicity and neural activity (Garcez et al., 2016, Garcez et al., 2017, Pampaloni et al., 2007, Sirenko et al., 2019, Woodruff et al., 2020). NP are obtained from neural stem cells (NSCs) cultured in suspension to form cellular aggregates in the presence of neural differentiation medium. This process triggers the differentiation of NSCs into neural cell types including neurons and astrocytes. Neuronal, astrocytic and neural progenitor proteins have been identified in a proteomic analysis of neurospheres recently published by our group (Goto-Silva et al., 2021). Here, a characterization of NP shows immunostaining for neurons (MAP-2), astrocytes (s100b) and neural progenitors (Nestin) (Fig. 1B-D). Human iPS cells-derived CM express the ACE2 receptor and were used as an in vitro model for cardiac SARS-CoV-2 infection (Marchiano et al., 2020, Pérez-Bermejo et al., 2020). The CM presently used were mostly TNNT2 positive (86 ± 6.2% positive cells at 30–32 days of differentiation) (Fig. 1E and F).
Then we checked whether human NP and CM would mirror ACE2 levels of brain and, consequently, serve the purpose of modeling SARS-CoV-2 infection. ACE2 expression in NP was, on average, almost one order of magnitude lower than in CM. (Fig. 1G). This shows that indeed NP reproduce the differences in tissue expression of ACE2 and validates the use of NP as a neural model of SARS-CoV-2 infection in vitro. Interestingly, NSCs express much less ACE2 and CD147, whereas AXL is unchanged, when compared to NP (S2B Fig), the former two well-established SARS-CoV-2 receptors. These differences suggest that neuroprogenitors may be less susceptible to SARS-CoV-2 infection than differentiated neural cells.
Having established the viability of these models, we then exposed NP and CM to the new coronavirus. SARS-CoV-2-infection in NP was addressed using multiplicity of infection (MOI) 0.01 and 0.1 and a 1 h exposure to viral particles, all conditions known to infect other virus-permissive cells, such as CM, Vero and Calu-3 cells (Han et al., 2020, Lamers et al., 2020, Marchiano et al., 2020, Pérez-Bermejo et al., 2020). Like other previous works, at first we evaluated SARS-CoV-2 infection using immunodetection of viral macromolecules (Dobrindt et al., 2021, Jacob et al., 2020b, Song et al., 2021, Zhang et al., 2020). At five days post infection (d.p.i.), SARS-CoV-2 could not be detected by immunostaining for double-stranded (ds)RNA (Fig. 2 A). Additionally, infected NP did not stain with anti-SARS-CoV-2 convalescent sera (CS) (S3A Fig). In contrast, NP infected with Zika virus (ZIKV) were immunoreactive for dsRNA (Fig. 2A and inset), and CM infected with SARS-CoV-2 were immunoreactive for dsRNA and CS (arrows, Fig. 2A and S3B Fig, correspondingly). The absence of immunostaining for SARS-CoV-2 was surprising because at 5 dpi it was expected that the virus would have replicated. At an earlier time point, 2 dpi, we also failed to detect viral proteins, using both anti-SARS-CoV-2 SP (Fig. 2B) and anti-SARS-CoV-2 CS (S3C Fig) with Western blotting (WB). On the other hand, infected Vero cells showed prominent bands corresponding to the molecular weights of SARS-CoV-2 spike protein and nucleoprotein, as detected by anti-SARS-CoV-2 SP (Fig. 2B) and anti-SARS-CoV-2 CS (S3C Fig), respectively.
Fig. 2.
SARS-CoV-2 replication in NP is non-productive. (A) IF staining for SARS-CoV-2, with anti-dsRNA (green), in cryosections of 5 d.p.i. NP (MOI 0.1). Nuclei were counterstained with DAPI (blue). ZIKV-infected NP (MOI 0.5 for 2 h, 3 d.p.i) and SARS-CoV-2-infected CM (MOI 0.1 for 1 h, 2 d.p.i) were used as positive controls for dsRNA labeling and SARS-CoV-2 infectivity, respectively. Scale bar: 50 µm (B) Detection of SARS-CoV-2 SP by WB. Protein extracts from Vero cells (MOI 0.1 for 24 h) were used as positive control. Gel loading was assessed by beta-actin staining. (C) Real-time qRT-PCR of genomic and subgenomic RNA levels of SARS-CoV-2 in the supernatants of NP 5 d.p.i. SARS-CoV-2-infected Vero cells were used for comparison. (D) Plaque forming units’ assay from the supernatants of the NP at 2 and 5 d.p.i (MOI 0.1). Data was plotted as average plus standard error. (E) Multiplex luminex assay for IL-7 and TNF-α from the supernatant of NP collected 5 d.p.i. (*p < 0.05, **p < 0.01). Collected data corresponds to an exposure of NP from 3 different cell lines (Table 1) to SARS-CoV-2 (PANGO lineage B.1, Nextstrain clade 20C) in one experimental infection. Each donor data point corresponds to the average of a duplicate technical measurement and is connected within the different experimental groups (cell lines). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite the absence of SARS-CoV-2 detection, the supernatant of NP harvested 5 d.p.i. showed approximately 104 and 105 copies of genomic and subgenomic viral RNA, respectively, which were at least 4 orders of magnitude lower than the observed in infected Vero cells (Fig. 2C). The detection of sgRNA indicates that there had been cell entry and initiation of negative-strand synthesis of the viral genome (Wölfel et al., 2020). The plaque forming unit (PFU) assay showed no infective viral progenies at 2 or 5 d.p.i in the supernatant of NP (MOI 0.1) (Fig. 2D), whereas for CM at 2 d.p.i (MOI 0.1) there was on average 2.8 × 106 PFU/mL (data not shown). The analysis of the virus inoculum in NP at day 0 contained on average 3 × 103 PFU/mL, confirming that they were exposed to infectious virus particles (Fig. 2D). The aforementioned detection of viral genomic RNA contrasts with the absence of infectious virus progeny and may indicate that neural cells were a dead-end for SARS-CoV-2 infection, or that the viral RNA detected in the supernatant was a residual of the inoculum.
3.2. SARS-CoV-2 infection of neural cells triggers an increase in pro inflammatory cytokines
SARS-CoV-2 infection has been associated with cytokine storm as a poor prognosis of disease progression (Tay et al., 2020). Misbalance of cytokine levels in the CNS microenvironment is associated with several pathologies (Guzman-Martinez et al., 2019, Lima et al., 2019, Mutso et al., 2020, Sochocka et al., 2017), hence the need to uncover the brain inflammation associated with SARS-CoV-2 infection. To investigate if SARS-CoV-2 generates a direct inflammatory response in neural tissues, the supernatant of infected NP was collected at 5 d.p.i. and analyzed by Multiplex Luminex assay to measure over 20 cytokines and chemokines (Fig. 2E and S4 Fig). The levels of IL-7 and TNF-α in the infected NP were higher compared with mock condition (Fig. 2E). The levels of other cytokines and chemokines were not affected by SARS-CoV-2 infection in these conditions (S4 Fig).
3.3. Prolonged exposure to higher inocula of SARS-CoV-2 does not elicit permissive infection of NP
The exposure time of NP to the virus inoculum and the low MOIs are sufficient to detect the virus in permissive cells, including Vero and CM. The same conditions, in contrast, did not prompt the detection of viral particles by neither IF nor WB in NP nor even infective particles in the culture supernatant. Arguably, this evidence of non-permissiveness could be explained by insufficient exposure time of the cells during viral inoculation. A previous report showed infection of NP after 24 h inoculation time at higher MOIs (Zhang et al., 2020). Additionally, it is speculated that the initial amount of virus in the inoculum is crucial in determining the severity of COVID-19 symptoms (Imai et al., 2020). To clarify these questions, NP were then exposed to a longer infection time and to higher MOIs. We emulated the protocols for neurospheres generation and infection with SARS-CoV-2 described by others in order to better compare our results with previously published data (Zhang et al., 2020).
We compared the capacity of SARS-CoV-2 to generate infectious viral particles in NP using 1 h and 24 h incubation times and MOIs of 0.1, 1 and 10. Despite longer exposure and higher viral titers, these conditions also did not support the production of novel infective particles, as measured by plaque forming units assay of the respective supernatants (Fig. 3 A). However, SARS-CoV-2 SP was detectable by IF in NP incubated for 24 h with the virus and analyzed 24 h post infection (Fig. 3B). On the other hand, after 48 h and 72 h of the same 24 h-infection, the NP had no detectable SP immunostaining, suggesting that viral proteins are only present transiently and then cleared (Fig. 3B and S3D Fig). Remarkably, at a time point in which no virus could be detected in NP, LDH measurements of culture supernatants revealed increased cytotoxicity at MOI 1 and 10, irrespective of incubation time with the virus inoculum (Fig. 3C). These data indicate that exposure of NP to SARS-CoV-2 at high MOIs is neurotoxic (Fig. 3). Also, the incubation time with the inoculum does not seem to affect the outcome of infection, since prolonged exposure time did not increase the production of viral particles or augment the extent of cell damage.
Fig. 3.
Prolonged exposure to higher inocula of SARS-CoV-2 does not elicit permissive infection of NP but higher MOIs are cytotoxic. NP were inoculated with SARS-CoV-2 for 1 h or 24 h at indicated MOIs. (A) At 72 h post-infection, culture supernatants were harvested to quantify infectious virus titers in Vero cells by plaque forming units’ assay. (B) NP inoculated for 24 h were immunostained with anti-SARS-CoV-2 SP (green) and counterstained with DAPI (blue) at 24 h and 48 h post-infection. Scale bars: 40 µm. (C) Lactate Dehydrogenase (LDH) levels were measured by colorimetric assay in NP exposed to viral particles for 1 h or 24 h and analyzed at 24 h, 48 h and 72 h post infection. Collected data correspond to the infection of 5 NP from 1 cell line, produced in house from urine epithelial cells, in one experimental infection. Each data point corresponds to measurements from single NP from one experimental infection. (D) Analysis of neural progenitor (PAX6+) population 5 d.p.i. at M.O.I. 0.01 and 0.1 from 2 cell lines (CF1 and C19), one experimental infection. Cryosections of NP were immunostained for PAX6 and the fraction of PAX6 + cells was quantified. Each data point represents 1 NP, and a total of 20 NP from 2 different cell lines were analyzed per group. **** p < 0.0001. (E) Representative IF staining for neural progenitors (PAX6+, green), in cryosections of NP (MOI 0.01 and 0.1, 5 d.p.i.). Nuclei were counterstained with DAPI (blue). Scale bar: 50 µm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Neural stem cells in the brain have an important role in multi-potency and self-renewal. In a study done on ocular donor tissues and iPScell-derived eye organoid model, it was observed that the limbus, an eye region containing stem cells, was particularly susceptible to infection, suggesting stemness was to blame (Makovoz et al., 2020). In order to investigate whether SARS-CoV-2 infection could affect the progenitor pool, we isolated NP nuclei by isotropic fractionation, a method previously reported to be effective to quantify cell populations in the brain (Herculano-Houzel and Lent, 2005). Using this method, the neural progenitor population in NP was identified as PAX6 + cells. We observed that SARS-CoV-2 infection did not change the proportion of neural progenitors in NP 5 days after infection but presented a trend towards progenitor pool reduction (S2A Fig). Since isotropic fractionation was done with a pool of NP which might have different degrees of infection individually, we decided to quantify the density of progenitors in single NP by immunostaining PAX6 + cells in cryosections. After quantifying about 20 NP per group from 2 different cell lines, we observed that PAX6 was significantly reduced at MOI 0.01 and 0.1 (p < 0.0001), with no significant difference between the different MOIs (Fig. 3D and E). These data suggest that infection impacts the amounts of progenitors by interfering with cell proliferation or inducing neural progenitor’s cell death. Considering that the expression of ACE2 is much lower in progenitors than in NP (S2B Fig), the mechanisms regulating reduction in progenitors during SARS-CoV-2 infection should be further explored.
3.4. Strain differences in SARS-CoV-2 RNA sequence could explain distinct infectivity of neural cells
Genetic variation of SARS-CoV-2 isolates has been described and could help to explain differences in the experimental infectivity of neural tissues previously reported (Korber et al., 2020). To investigate whether possible genetic components could account for the differences among SARS-CoV-2 infections reported in literature and in the current work, we compared the phylogeny of the nucleotide sequences of our strain with the strains used in these works, by means of the Wuhan strain genome as reference. The analysis showed <1% variation among the genetic sequences of our strain, Zhang and collaborators, and Song and collaborators (Table 2 ) (Song et al., 2020, Zhang et al., 2020). These small variations correlate with mutations in the receptor-binding domain of Spike protein and two non-structural proteins (nsp) (Table 3 ).
Table 2.
Estimates of evolutionary divergence between SARS-CoV-2 sequences.
| Virus Strain | NC_045512.2 Wuhan-01 | MT710714.1 This study | MT230904.1 Zhang et al | MT020880.1 Song et al | MT246667.1 Song et al |
|---|---|---|---|---|---|
| MT710714.1 -This study | 0.06 | – | – | – | – |
| MT230904.1 - Zhang et al | 0.03 | 0.09 | – | – | – |
| MT020880.1 - Song et al | 0.01 | 0.07 | 0.04 | – | – |
| MT246667.1 - Song et al | 0.01 | 0.07 | 0.04 | 0.00 | |
| MN985325.1 - Song et al | 0.01 | 0.07 | 0.04 | 0.00 | 0.00 |
The values correspond to the divergence in sequences of SARS-CoV-2 when comparing the strain in rows with the strain in columns. Sequences were compared with maximum likelihood phylogenetic analysis as described in the Material and Methods section.
Table 3.
Specific proteins mutations among SARS-CoV-2 strains.
| Virus Strain | NSP2 | NSP7 | NSP14 | SPIKE (RBD) | SPIKE | M | ORF8 |
|---|---|---|---|---|---|---|---|
| MT710714.1 - This study | T265I | S3884L | D614G | ||||
| MT230904.1 - Zhang et al | V367F | C207R | R183P | ||||
| MT020880.1 - Song et al | S5932F | ||||||
| MT246667.1 - Song et al | S5932F | ||||||
| MN985325.1 - Song et al | S5932F |
Each amino acid mutation was accessed through sequence pairing comparison to the SARS-CoV-2 reference genome (Wuhan-Hu-1).
Our viral isolate presented three missense mutations compared to the Wuhan sequence: D614G on spike protein, S3884L on nsp7 and T265I on nsp2. The D614G spike protein mutation has been linked to more infectious behavior of the strain (Korber et al., 2020, p. 61). Nsp 7 participates in SARS-CoV-2 replication, being a co-factor for RNA polymerase. The nsp2 role is unknown and may be related to the disruption of intracellular host signaling and mitochondrial biogenesis (Cornillez-Ty et al., 2009).
4. Discussion
Neurological manifestations of COVID-19 fueled research on the putative viral infection of the brain. Absence or low detection of SARS-CoV-2 was described in the cerebrospinal fluid (CSF), and so far many reports have described the presence of SARS-CoV-2 in the brain parenchyma (Crunfli et al., 2020, p.; Espíndola et al., 2020, Schaller et al., 2020, Song et al., 2020), while others suggested quick viral clearance in the brain due to swift innate immune responses (Schurink et al., 2020).
Here, we bring the attention to SARS-CoV-2 infection in the ChP in its LV lining, and in the frontal cortex of an infant’s brain (S1A Fig), which was confirmed by qRT-PCR for viral RNA (S1 Table) (pathological analysis of this case was described in detail in (Gomes et al., 2020)). The detection of SARS-CoV-2 was more pronounced in the ChP than in the cortex (S1A Fig). This matches expression data for the receptor ACE2, which is enriched in ChP both in vivo and in vitro, in comparison to other brain areas, including cortex (Jacob et al., 2020a, Pellegrini et al., 2020) (Fig. 1A). Our protocol for differentiating NP generates cortical brain-like tissue (Fig. 1B-D) (Garcez et al., 2017, Goto-Silva et al., 2021), which recapitulates the low expression levels of ACE2 mRNA observed in the human cortical tissue (Fig. 1G). ACE2 levels are increased in patients with comorbidities, suggesting a relationship between ACE2 expression and susceptibility to severe COVID-19 (Pinto et al., 2020). Strategies to regulate the expression of ACE2 have been proposed as therapies to fight viral infection (Annweiler et al., 2020, Liu et al., 2020). Also, it remains unaddressed if neuropsychiatric conditions and neurodegenerative diseases impact the expression of ACE2 in the brain (Rocha et al., 2018) and how they could interfere with the susceptibility of CNS infection. Indeed, there is an increased risk for mental illness in COVID-19 patients, especially those who had previously psychiatric episodes (Mazza et al., 2020). Moreover, there is a raised concern that in utero infection might cause future neurodevelopmental disorders in the offspring (López-Díaz et al., 2021). In this sense, the current work contributes to the understanding of brain developmental consequences of SARS-CoV-2 by demonstrating that progenitors express relatively lower levels of ACE2 and CD147 and are not damaged by exposure to low MOIs, most likely to occur during a COVID pregnancy. In addition, human neurospheres and brain organoids could be a model for in vitro drug testing, including monitoring changes in ACE2 expression and correlation with susceptibility to SARS-CoV-2 infection.
One experimental aspect that drew our attention was the amount of virus used to infect cells in other studies. Previous reports have demonstrated SARS-CoV-2 antigens in NP and brain organoids (Mesci et al., 2020, Song et al., 2020, Zhang et al., 2020) using MOIs at least 100 times higher than the lowest MOI used in our investigation. Although productive infection in neural cells was claimed at MOI 10 (Zhang et al., 2020), we did not obtain infectious viral particles at similar experimental conditions (Fig. 3A). Others observed viral proteins in a subset of neurons and SARS-CoV-2 sgRNA in the supernatant of these neuronal cultures, however, they did not gauged the production of virions (Dobrindt et al., 2021). This has been reviewed by Giani and Chen (2021) to show that SARS-CoV-2 is capable of entering neural cells (Giani and Chen, 2021). However, it remains possible that the viral variant and host cell genetics are determinant to permissiveness, i.e., presence of viral progeny, of SARS-CoV-2 infection in the brain.
The genetic comparison between our strain and the strains of other works showed little variations (Table 2) that, nevertheless, caused potentially significant alterations in the amino acid sequence of viral proteins (Table 3). Indeed, most of these variations might reflect the genetic diversity of the SARS-CoV-2 since the original outbreak (Phan, 2020). The strain used in the current work has a D614G mutation that renders the virus highly infectious by increasing ACE2 interaction (Korber et al., 2020). This D614G spike mutation has also been associated with increased binding of neutralizing antibodies (Weissman et al., 2020). Our strain has also a mutation in a cofactor of SARS-CoV-2 RNA-dependent RNA polymerase, nsp7, that did not lead to impairments in viral replication in permissive cells such as Vero (Fig. 2C), however, this mutation could not be excluded as responsible for the differences observed between the studies. Finally, the T265I nsp2 mutation falls within the extracellular N-terminus of the protein (Angeletti et al., 2020) and is thought to cause structural changes, and which can now be found in all geographical locations (Rehman et al., 2020). Similarly to our work, Zhang and collaborators have also used a strain with increased sensitivity to neutralizing antibodies (Li et al., 2020). On the other hand, Song and collaborators used a viral strain that is not world spread (Alouane et al., 2020, Koyama et al., 2020, Laamarti et al., 2020) and it has been reduced in frequency over time since February 2020 (Alouane et al., 2020). In addition, the S5932F mutation on nsp14 (ExoN), could lead to altered proofreading exonuclease activity via weaker interaction with nsp10, facilitating the emergence of diverse viral sequences with potential selective advantage (Ogando et al., 2019).
Cells from lungs, intestine, heart and kidney, which have already been demonstrated to be sites of infection (Bradley et al., 2020, Mallapaty, 2020, Remmelink et al., 2020, Tabary et al., 2020), produce infectious viral progeny in vitro with MOIs ranging from 0.01 to 1 (Han et al., 2020, Lamers et al., 2020). Therefore, it seems unlikely that even in severe neuro-COVID cases such as the one cited here, neural cells would be exposed to quantities equal to or more than one plaque forming virus per cell. Our observation that ChP is infected to a much greater extent than other regions of the brain, together with the lack of infectious virus particles in vitro with MOI < 1, argue against the clinical relevance of using high titers of SARS-CoV-2 for modeling the infection in vitro. A study using viral titers of MOI 0.5 showed that SARS-CoV-2 exclusively infects ChP cells in brain organoids, but no other cell types. In the same report, even at higher viral titers (MOI 10), infection of neurons was minor (Pellegrini et al., 2020). Our observation that SARS-CoV-2 infects the ChP without significantly spreading to other brain areas (S1 Fig) and the absence of a productive infection in human NP with SARS-CoV-2 at MOI of 0.1, 1 and 10 even at increased inoculation time (Fig. 3), corroborate these findings.
Despite non-permissiveness to SARS-CoV-2 infection of the brain parenchyma, the data presented here also points to direct pro-inflammatory and cytotoxic response of this tissue. This direct effect is probably related to the number of viral particles able to first get in contact with the CNS. It was observed in NP an increase in IL-7 and TNFα levels following infection with low MOIs, but not of other inflammatory cytokines such as IL-6. IL-7 has antiviral activity (Pellegrini et al., 2011) and neurotrophic effects in neurons (Araujo and Cotman, 1993, Michaelson et al., 1996) and its increase at MOIs 0.01 and 0.1 may correlate with a protective neuroimmune response dependent on virus titer amount. TNF-α is secreted by astrocytes and microglia in response to injury, including viral infection, inducing neuronal damage (Figueiredo et al., 2019). It also regulates the permeability of the blood–brain barrier and neuronal plasticity (McCoy and Tansey, 2008, Probert, 2015). Both cytokines are increased in ICU-committed COVID-19 patients when compared to non-ICU patients, suggesting they are indeed correlated with disease severity (Huang et al., 2020). Certainly, these results are interesting with respect to a direct inflammatory response from neural cells that had been in contact with SARS-CoV-2 and deserve further investigation in neural tissue culture models that present well stablished immune response. A pro-inflammatory response in NP was suggested as a consequence of the upregulation of innate immune response protein TLR4 by ZIKA infection (Garcez et al., 2017). However, the immune response in NP still needs to be further comprehended in terms of how it parallels developing and mature human brains’ responses. Others have observed enrolment of pro-inflammatory pathways using different iPS cell-derived models in contact with the new coronavirus (Makovoz et al., 2020, Giani and Chen, 2021). Nevertheless, the fact that proinflammatory cytokines were upregulated, however immature or unknown the NP immune response is, certainly merits mention and more studies. It is expected that in the complex scenario of the reaction to viral infection, multiple cytokines are regulated promoting cell protection and also limiting infection by inducing cell death. Therefore, the extent of neural damage should correspond to a balance between these responses.
We hypothesize that virus particles reaching the brain parenchyma could trigger inflammatory response leading to tissue damage. Notably, neurological symptoms are more frequent in severe patients with exacerbated inflammation, compared to mild or moderate ones, suggesting that an overall increase in the inflammatory response may set place for CNS damage (Mao et al., 2020). Low type I interferon response in infants’ brains may correlate with higher susceptibility for virus entry in CNS, but also patients with previous neurological and psychiatric disorders which may have weakened immune response or enhanced inflammation should be closely monitored (Mazza et al., 2020, Singh et al., 2020).
Acknowledgments
Acknowledgments
The authors would like to thank Claudia Figueiredo and Claudio Ferrari for helpful discussions during the elaboration of this manuscript; and Fernando Colonna Rosman and Leila Maria Cardão Chimelli for their support in the tissue’s analysis.
Funding: This work was supported by Foundation for Research Support in the State of Rio de Janeiro (FAPERJ) (grant number: E-26/210.182/2020); Brazilian Innovation Agency (FINEP) in partnership with Novageia Biotecnologia, in addition to scholarship grants by the National Council of Scientific and Technological Development (CNPq) and Coordination for the Improvement of Higher Education Personnel (CAPES) and intramural grants from the D’Or Institute for Research and Education.
Competing interests: The authors declare no competing interests.
References
- Alouane T., Laamarti M., Essabbar A., Hakmi M., Bouricha E.M., Chemao-Elfihri M.W., Kartti S., Boumajdi N., Bendani H., Laamarti R., Ghrifi F., Allam L., Aanniz T., Ouadghiri M., El Hafidi N., El Jaoudi R., Benrahma H., Attar J.E., Mentag R., Sbabou L., Nejjari C., Amzazi S., Belyamani L., Ibrahimi A. Genomic Diversity and Hotspot Mutations in 30,983 SARS-CoV-2 Genomes: Moving Toward a Universal Vaccine for the “Confined Virus”? Pathogens. 2020;9:829. doi: 10.3390/pathogens9100829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92(6):584–588. doi: 10.1002/jmv.v92.610.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annweiler C., Cao Z., Wu Y., Faucon E., Mouhat S., Kovacic H., Sabatier J.-M. Counter-regulatory “Renin-Angiotensin” System-based Candidate Drugs to Treat COVID-19 Diseases in SARS-CoV-2-infected patients. Infect. Disord. Drug Targets. 2020;20(4):407–408. doi: 10.2174/1871526520666200518073329. [DOI] [PubMed] [Google Scholar]
- Araujo D.M., Cotman C.W. Trophic effects of interleukin-4, -7 and -8 on hippocampal neuronal cultures: potential involvement of glial-derived factors. Brain Res. 1993;600(1):49–55. doi: 10.1016/0006-8993(93)90400-H. [DOI] [PubMed] [Google Scholar]
- Barrantes F.J. The unfolding palette of COVID-19 multisystemic syndrome and its neurological manifestations. Brain, Behav. Immunity - Health. 2021;14:100251. doi: 10.1016/j.bbih.2021.100251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley B.T., Maioli H., Johnston R., Chaudhry I., Fink S.L., Xu H., Najafian B., Deutsch G., Lacy J.M., Williams T., Yarid N., Marshall D.A. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396(10247):320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann, D.H., Tsukahara, T., Weinreb, C., Lipovsek, M., Berge, K.V. den, Gong, B., Chance, R., Macaulay, I.C., Chou, H.-J., Fletcher, R.B., Das, D., Street, K., Bezieux, H.R. de, Choi, Y.-G., Risso, D., Dudoit, S., Purdom, E., Mill, J., Hachem, R.A., Matsunami, H., Logan, D.W., Goldstein, B.J., Grubb, M.S., Ngai, J., Datta, S.R., 2020. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Science Advances 6, eabc5801. DOI:10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed]
- Bryche, B., Albin, A.S., Murri, S., Lacôte, S., Pulido, C., Gouilh, M.A., Lesellier, S., Servat, A., Wasniewski, M., Picard-Meyer, E., Monchatre-Leroy, E., Volmer, R., Rampin, O., Goffic, R.L., Marianneau, P., Meunier, N., 2020. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. bioRxiv 2020.06.16.151704. DOI:10.1101/2020.06.16.151704. [DOI] [PMC free article] [PubMed]
- Casas B.S., Vitória G., do Costa M.N., Madeiro da Costa R., Trindade P., Maciel R., Navarrete N., Rehen S.K., Palma V. hiPSC-derived neural stem cells from patients with schizophrenia induce an impaired angiogenesis. Transl. Psychiatry. 2018;8(1) doi: 10.1038/s41398-018-0095-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C.T., Liao L., Yates J.R., Kuhn P., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009;83(19):10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunfli, F., Carregari, V.C., Veras, F.P., Vendramini, P.H., Valenca, A.G.F., Antunes, A.S.L.M., Brandao-Teles, C., Zuccoli, G. da S., Reis-de-Oliveira, G., Silva-Costa, L.C., Saia-Cereda, V.M., Codo, A.C., Parise, P.L., Toledo-Teixeira, D.A., Souza, G.F. de, Muraro, S.P., Melo, B.M.S., Almeida, G.M., Firmino, E.M.S., Ludwig, R.G., Ruiz, G.P., Knittel, T.L., Davanzo, G.G., Gerhardt, J.A., Rodrigues, P.B., Forato, J., Amorim, M.R., Silva, N.B., Martini, M.C., Benatti, M.N., Batah, S., Siyuan, L., Silva, R.E.M.P., João, R.B., Silva, L.S., Nogueira, M.H., Aventurato, ítalo K., Brito, M.R. de, Alvim, M.K.M., Junior, J.R. da S., Damião, L.L., Stefano, M.E. de P.C., Sousa, I.M.P. de, Rocha, E.D. da, Gonçalves, S.M., Silva, L.H.L. da, Bettini, V., Campos, B.M. de, Ludwig, G., Viana, R.M.M., Martins, R., Vieira, A.S., Alves-Filho, J.C., Neto, E. de A., Sebollela, A., Cendes, F., Cunha, F.Q., Damásio, A., Vinolo, M.A.R., Munhoz, C.D., Rehen, S.K., Mauad, T., Duarte-Neto, A.N., Silva, L.F.F. da, Dolhnikoff, M., Saldiva, P., Fabro, A.T., Farias, A.S., Moraes-Vieira, P.M.M., Módena, J.L.P., Yasuda, C.L., Mori, M.A., Cunha, T.M., Martins-de-Souza, D., 2020. SARS-CoV-2 infects brain astrocytes of COVID-19 patients and impairs neuronal viability. medRxiv 2020.10.09.20207464. DOI:10.1101/2020.10.09.20207464.
- Cruvinel, E., Ogusuku, I., Cerioni, R., Rodrigues, S., Gonçalves, J., Góes, M.E., Alvim, J.M., Silva, A.C., Lino, V. de S., Boccardo, E., Goulart, E., Pereira, A., Dariolli, R., Valadares, M., Biagi, D., 2020. Long-term single-cell passaging of human iPSC fully supports pluripotency and high-efficient trilineage differentiation capacity. SAGE Open Med 8, 2050312120966456. DOI:10.1177/2050312120966456. [DOI] [PMC free article] [PubMed]
- Da Silveira Paulsen B., De Moraes Maciel R., Galina A., Da Silveira M.S., Souza C.D.S., Drummond H., Pozzatto E.N., Junior H.S., Chicaybam L., Massuda R., Setti-Perdigão P., Bonamino M., Belmonte-De-Abreu P.S., Castro N.G., Brentani H., Rehen S.K. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2012;21(7):1547–1559. doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- De Felice F.G., Tovar-Moll F., Moll J., Munoz D.P., Ferreira S.T. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the central nervous system. Trends Neurosci. 2020;43(6):355–357. doi: 10.1016/j.tins.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt K., Hoagland D.A., Seah C., Kassim B., O’Shea C.P., Murphy A., Iskhakova M., Fernando M.B., Powell S.K., Deans P.J.M., Javidfar B., Peter C., Møller R., Uhl S.A., Garcia M.F., Kimura M., Iwasawa K., Crary J.F., Kotton D.N., Takebe T., Huckins L.M., tenOever B.R., Akbarian S., Brennand K.J. Common Genetic Variation in Humans Impacts In Vitro Susceptibility to SARS-CoV-2 Infection. Stem Cell Rep. 2021;16:505–518. doi: 10.1016/j.stemcr.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellul M.A., Benjamin L., Singh B., Lant S., Michael B.D., Easton A., Kneen R., Defres S., Sejvar J., Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19(9):767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espíndola O.d.M., Siqueira M., Soares C.N., Lima M.A.S.D.d., Leite A.C.C.B., Araujo A.Q.C., Brandão C.O., Silva M.T.T. Patients with COVID-19 and neurological manifestations show undetectable SARS-CoV-2 RNA levels in the cerebrospinal fluid. Int. J. Infect. Dis. 2020;96:567–569. doi: 10.1016/j.ijid.2020.05.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fifi J.T., Mocco J. COVID-19 related stroke in young individuals. Lancet Neurol. 2020;19(9):713–715. doi: 10.1016/S1474-4422(20)30272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo C.P., Barros-Aragão F.G.Q., Neris R.L.S., Frost P.S., Soares C., Souza I.N.O., Zeidler J.D., Zamberlan D.C., de Sousa V.L., Souza A.S., Guimarães A.L.A., Bellio M., Marcondes de Souza J., Alves-Leon S.V., Neves G.A., Paula-Neto H.A., Castro N.G., De Felice F.G., Assunção-Miranda I., Clarke J.R., Da Poian A.T., Ferreira S.T. Zika virus replicates in adult human brain tissue and impairs synapses and memory in mice. Nat. Commun. 2019;10:3890. doi: 10.1038/s41467-019-11866-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez P.P., Loiola E.C., Madeiro da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., Rehen S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. doi: 10.1126/science:aaf6116. [DOI] [PubMed] [Google Scholar]
- Garcez P.P., Nascimento J.M., de Vasconcelos J.M., Madeiro da Costa R., Delvecchio R., Trindade P., Loiola E.C., Higa L.M., Cassoli J.S., Vitória G., Sequeira P.C., Sochacki J., Aguiar R.S., Fuzii H.T., de Filippis A.M.B., da Silva Gonçalves Vianez Júnior J.L., Tanuri A., Martins-de-Souza D., Rehen S.K. Zika virus disrupts molecular fingerprinting of human neurospheres. Sci. Rep. 2017;7(1) doi: 10.1038/srep40780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani A.M., Chen S. Human pluripotent stem cell-based organoids and cell platforms for modelling SARS-CoV-2 infection and drug discovery. Stem Cell Res. 2021;53:102207. doi: 10.1016/j.scr.2021.102207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173(6):4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- Gnecchi M., Moretti F., Bassi E.M., Leonardi S., Totaro R., Perotti L., Zuccaro V., Perlini S., Preda L., Baldanti F., Bruno R., Visconti L.O. Myocarditis in a 16-year-old boy positive for SARS-CoV-2. Lancet. 2020;395(10242):e116. doi: 10.1016/S0140-6736(20)31307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, I.C., Karmirian, K., Oliveira, J., Pedrosa, C., Rosman, F.C., Chimelli, L., Rehen, S., 2020. SARS-CoV-2 Infection in the Central Nervous System of a 1-Year-Old Infant Submitted to Complete Autopsy. DOI:10.20944/preprints202009.0297.v1. [DOI] [PMC free article] [PubMed]
- Goto-Silva, L., Martins, M., Murillo, J.R., Souza, L.R.Q., Vitória, G., Oliveira, J.T., Nascimento, J.M., Loiola, E.C., Nogueira, F.C.S., Domont, G.B., Guimarães, M.Z.P., Tovar-Moll, F., Rehen, S.K., Junqueira, M., 2021. Quantitative profiling of axonal guidance proteins during the differentiation of human neurospheres. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1869, 140656. DOI:10.1016/j.bbapap.2021.140656. [DOI] [PubMed]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y., Freedberg D.E., Kirtane A.J., Parikh S.A., Maurer M.S., Nordvig A.S., Accili D., Bathon J.M., Mohan S., Bauer K.A., Leon M.B., Krumholz H.M., Uriel N., Mehra M.R., Elkind M.S.V., Stone G.W., Schwartz A., Ho D.D., Bilezikian J.P., Landry D.W. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Martinez L., Maccioni R.B., Andrade V., Navarrete L.P., Pastor M.G., Ramos-Escobar N. Neuroinflammation as a Common Feature of Neurodegenerative Disorders. Front. Pharmacol. 2019;10:1008. doi: 10.3389/fphar.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield, J., Megill, C., Bell, S.M., Huddleston, J., Potter, B., Callender, C., Sagulenko, P., Bedford, T., Neher, R.A., 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34, 4121–4123. DOI:10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed]
- Hamming I., Timens W., Bulthuis M.L.C, Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/(ISSN)1096-989610.1002/path.v203:210.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y., Yang, L., Duan, X., Duan, F., Nilsson-Payant, B.E., Yaron, T.M., Wang, P., Tang, X., Zhang, T., Zhao, Z., Bram, Y., Redmond, D., Houghton, S., Nguyen, D., Xu, D., Wang, X., Uhl, S., Huang, Y., Johnson, J.L., Xiang, J., Wang, H., Pan, F.C., Cantley, L.C., tenOever, B.R., Ho, D.D., Evans, T., Schwartz, R.E., Chen, H.J., Chen, S., 2020. Identification of Candidate COVID-19 Therapeutics using hPSC-derived Lung Organoids. bioRxiv 2020.05.05.079095. DOI:10.1101/2020.05.05.079095.
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., Abajian C., Beckmann C.F., Bernard A., Bertagnolli D., Boe A.F., Cartagena P.M., Chakravarty M.M., Chapin M., Chong J., Dalley R.A., Daly B.D., Dang C., Datta S., Dee N., Dolbeare T.A., Faber V., Feng D., Fowler D.R., Goldy J., Gregor B.W., Haradon Z., Haynor D.R., Hohmann J.G., Horvath S., Howard R.E., Jeromin A., Jochim J.M., Kinnunen M., Lau C., Lazarz E.T., Lee C., Lemon T.A., Li L., Li Y., Morris J.A., Overly C.C., Parker P.D., Parry S.E., Reding M., Royall J.J., Schulkin J., Sequeira P.A., Slaughterbeck C.R., Smith S.C., Sodt A.J., Sunkin S.M., Swanson B.E., Vawter M.P., Williams D., Wohnoutka P., Zielke H.R., Geschwind D.H., Hof P.R., Smith S.M., Koch C., Grant S.G.N., Jones A.R. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489(7416):391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S., Lent R. Isotropic fractionator: a simple, rapid method for the quantification of total cell and neuron numbers in the brain. J. Neurosci. 2005;25:2518–2521. doi: 10.1523/JNEUROSCI.4526-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosp, J.A., Dressing, A., Blazhenets, G., Bormann, T., Rau, A., Schwabenland, M., Thurow, J., Wagner, D., Waller, C., Niesen, W.D., Frings, L., Urbach, H., Prinz, M., Weiller, C., Schroeter, N., Meyer, P.T., 2021. Cognitive impairment and altered cerebral glucose metabolism in the subacute stage of COVID-19. Brain. DOI:10.1093/brain/awab009. [DOI] [PMC free article] [PubMed]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., Yamada S., Fan S., Chiba S., Kuroda M., Guan L., Takada K., Armbrust T., Balogh A., Furusawa Y., Okuda M., Ueki H., Yasuhara A., Sakai-Tagawa Y., Lopes T.J.S., Kiso M., Yamayoshi S., Kinoshita N., Ohmagari N., Hattori S.-I., Takeda M., Mitsuya H., Krammer F., Suzuki T., Kawaoka Y. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U.S.A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, F., Pather, S.R., Huang, W.-K., Wong, S.Z.H., Zhou, H., Zhang, F., Cubitt, B., Chen, C.Z., Xu, M., Pradhan, M., Zhang, D.Y., Zheng, W., Bang, A.G., Song, H., Torre, J.C. de a, Ming, G., 2020a. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism. bioRxiv 2020.07.28.225151. DOI:10.1101/2020.07.28.225151. [DOI] [PMC free article] [PubMed]
- Jacob F., Pather S.R., Huang W.-K., Zhang F., Wong S.Z.H., Zhou H., Cubitt B., Fan W., Chen C.Z., Xu M., Pradhan M., Zhang D.Y., Zheng W., Bang A.G., Song H., Carlos de la Torre J., Ming G.-l. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27(6):937–950.e9. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K., 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35, 1547–1549. DOI:10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed]
- Laamarti, Meriem, Alouane, T., Kartti, S., Chemao-Elfihri, M.W., Hakmi, M., Essabbar, A., Laamarti, Mohamed, Hlali, H., Allam, L., Hafidi, N.E., Jaoudi, R.E., Allali, I., Marchoudi, N., Fekkak, J., Benrahma, H., Nejjari, C., Amzazi, S., Belyamani, L., Ibrahimi, A., 2020. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. bioRxiv 2020.05.03.074567. DOI:10.1101/2020.05.03.074567. [DOI] [PMC free article] [PubMed]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science:abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledur P.F., Karmirian K., Pedrosa C.d.S.G., Souza L.R.Q., Assis-de-Lemos G., Martins T.M., Ferreira J.d.C.C.G., de Azevedo Reis G.F., Silva E.S., Silva D., Salerno J.A., Ornelas I.M., Devalle S., Madeiro da Costa R.F., Goto-Silva L., Higa L.M., Melo A., Tanuri A., Chimelli L., Murata M.M., Garcez P.P., Filippi-Chiela E.C., Galina A., Borges H.L., Rehen S.K. Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-57914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C., Perlman S., Zhao J., Jewell A.K., Reznikov L.R., Gibson-Corley K.N., Meyerholz D.K., McCray P.B. Middle east respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J. Infect. Dis. 2016;213(5):712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L.i., Hao H., Liu S., Zhao C., Zhang Q.i., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima M.C., de Mendonça L.R., Rezende A.M., Carrera R.M., Aníbal-Silva C.E., Demers M., D’Aiuto L., Wood J., Chowdari K.V., Griffiths M., Lucena-Araujo A.R., Barral-Netto M., Azevedo E.A.N., Alves R.W., Farias P.C.S., Marques E.T.A., Castanha P.M.S., Donald C.L., Kohl A., Nimgaonkar V.L., Franca R.F.O. The Transcriptional and protein profile from human infected neuroprogenitor cells is strongly correlated to zika virus microcephaly cytokines phenotype evidencing a persistent inflammation in the CNS. Front. Immunol. 2019;10:1928. doi: 10.3389/fimmu.2019.01928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Wang T., Zhou Y., Zhao Y., Zhang Y., Li J. Potential Role of ACE2 in Coronavirus Disease 2019 (COVID-19) prevention and management. J. Transl. Int. Med. 2020;8:9–19. doi: 10.2478/jtim-2020-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Díaz Á., Ayesa-Arriola R., Crespo-Facorro B., Ruiz-Veguilla M. COVID-19 infection during pregnancy and risk of neurodevelopmental disorders in offspring: time for collaborative research. Biol. Psychiatry. 2021;89(5):e29–e30. doi: 10.1016/j.biopsych.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovoz, B., Moeller, R., Zebitz Eriksen, A., tenOever, B.R., Blenkinsop, T.A., 2020. SARS-CoV-2 Infection of Ocular Cells from Human Adult Donor Eyes and hESC-Derived Eye Organoids (SSRN Scholarly Paper No. ID 3650574). Social Science Research Network, Rochester, NY. DOI:10.2139/ssrn.3650574.
- Mallapaty S. Mini organs reveal how the coronavirus ravages the body. Nature. 2020;583(7814):15–16. doi: 10.1038/d41586-020-01864-x. [DOI] [PubMed] [Google Scholar]
- Mao, L., Wang, M., Chen, S., He, Q., Chang, J., Hong, C., Zhou, Y., Wang, D., Li, Y., Jin, H., Hu, B., 2020. Neurological Manifestations of Hospitalized Patients with COVID-19 in Wuhan, China: a retrospective case series study. medRxiv 2020.02.22.20026500. DOI:10.1101/2020.02.22.20026500.
- Marchiano, S., Hsiang, T.-Y., Higashi, T., Khanna, A., Reinecke, H., Yang, X., Pabon, L., Sniadecki, N.J., Bertero, A., Gale, M., Murry, C.E., 2020. SARS-CoV-2 infects human pluripotent stem cell-derived cardiomyocytes, impairing electrical and mechanical function. bioRxiv 2020.08.30.274464. DOI:10.1101/2020.08.30.274464. [DOI] [PMC free article] [PubMed]
- Mazza M.G., De Lorenzo R., Conte C., Poletti S., Vai B., Bollettini I., Melloni E.M.T., Furlan R., Ciceri F., Rovere-Querini P., Benedetti F. Anxiety and depression in COVID-19 survivors: Role of inflammatory and clinical predictors. Brain Behav. Immun. 2020;89:594–600. doi: 10.1016/j.bbi.2020.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy M.K., Tansey M.G. TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease. J. Neuroinflamm. 2008;5(1):45. doi: 10.1186/1742-2094-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R., Laue M., Schneider J., Brünink S., Greuel S., Lehmann M., Hassan O., Aschman T., Schumann E., Chua R.L., Conrad C., Eils R., Stenzel W., Windgassen M., Rößler L., Goebel H.-H., Gelderblom H.R., Martin H., Nitsche A., Schulz-Schaeffer W.J., Hakroush S., Winkler M.S., Tampe B., Scheibe F., Körtvélyessy P., Reinhold D., Siegmund B., Kühl A.A., Elezkurtaj S., Horst D., Oesterhelweg L., Tsokos M., Ingold-Heppner B., Stadelmann C., Drosten C., Corman V.M., Radbruch H., Heppner F.L. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2020;24(2):168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- Mesci, P., Macia, A., Saleh, A., Martin-Sancho, L., Yin, X., Snethlage, C., Avansini, S., Chanda, S.K., Muotri, A., 2020. Sofosbuvir protects human brain organoids against SARS-CoV-2. bioRxiv 2020.05.30.125856. DOI:10.1101/2020.05.30.125856.
- Michaelson M.D., Mehler M.F., Xu H., Gross R.E., Kessler J.A. Interleukin-7 is trophic for embryonic neurons and is expressed in developing brain. Dev. Biol. 1996;179(1):251–263. doi: 10.1006/dbio.1996.0255. [DOI] [PubMed] [Google Scholar]
- Mutso M., St John J.A., Ling Z.L., Burt F.J., Poo Y.S., Liu X., Žusinaite E., Grau G.E., Hueston L., Merits A., King N.J.C., Ekberg J.A.K., Mahalingam S. Basic insights into Zika virus infection of neuroglial and brain endothelial cells. J. General Virol. 2020;101:622–634. doi: 10.1099/jgv.0.001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The curious case of the nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampaloni F., Reynaud E.G., Stelzer E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Calzascia T., Toe J.G., Preston S.P., Lin A.E., Elford A.R., Shahinian A., Lang P.A., Lang K.S., Morre M., Assouline B., Lahl K., Sparwasser T., Tedder T.F., Paik J.-H., DePinho R.A., Basta S., Ohashi P.S., Mak T.W. IL-7 engages multiple mechanisms to overcome chronic viral infection and limit organ pathology. Cell. 2011;144(4):601–613. doi: 10.1016/j.cell.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Pellegrini, L., Albecka, A., Mallery, D.L., Kellner, M.J., Paul, D., Carter, A.P., James, L.C., Lancaster, M.A., 2020. SARS-CoV-2 infects brain choroid plexus and disrupts the blood-CSF-barrier. bioRxiv 2020.08.20.259937. DOI:10.1101/2020.08.20.259937. [DOI] [PMC free article] [PubMed]
- Pérez-Bermejo, J.A., Kang, S., Rockwood, S.J., Simoneau, C.R., Joy, D.A., Ramadoss, G.N., Silva, A.C., Flanigan, W.R., Li, H., Nakamura, K., Whitman, J.D., Ott, M., Conklin, B.R., McDevitt, T.C., 2020. SARS-CoV-2 infection of human iPSC-derived cardiac cells predicts novel cytopathic features in hearts of COVID-19 patients. bioRxiv 2020.08.25.265561. DOI:10.1101/2020.08.25.265561.
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet. Evol. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto B.G.G., Oliveira A.E.R., Singh Y., Jimenez L., Gonçalves A.N.A., Ogava R.L.T., Creighton R., Schatzmann Peron J.P., Nakaya H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020;222:556–563. doi: 10.1093/infdis/jiaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert L. TNF and its receptors in the CNS: The essential, the desirable and the deleterious effects. Neurosci., Inflamm. Nervous Syst. Disorders. 2015;302:2–22. doi: 10.1016/j.neuroscience.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H., Song M., Hadiono C., Ogden S., Hammack C., Yao B., Hamersky G., Jacob F., Zhong C., Yoon K.-J., Jeang W., Lin L.i., Li Y., Thakor J., Berg D., Zhang C.e., Kang E., Chickering M., Nauen D., Ho C.-Y., Wen Z., Christian K., Shi P.-Y., Maher B., Wu H., Jin P., Tang H., Song H., Ming G.-l. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J.M., Deprez R.H.L., Moorman A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339(1):62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman S., Mahmood T., Aziz E., Batool R. Identification of novel mutations in SARS-COV-2 isolates from Turkey. Arch. Virol. 2020;165(12):2937–2944. doi: 10.1007/s00705-020-04830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmelink M., De Mendonça R., D’Haene N., De Clercq S., Verocq C., Lebrun L., Lavis P., Racu M.-L., Trépant A.-L., Maris C., Rorive S., Goffard J.-C., De Witte O., Peluso L., Vincent J.-L., Decaestecker C., Taccone F.S., Salmon I. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Crit. Care. 2020;24(1) doi: 10.1186/s13054-020-03218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N.P., Simoes e Silva A.C., Prestes T.R.R., Feracin V., Machado C.A., Ferreira R.N., Teixeira A.L., de Miranda A.S. RAS in the central nervous system: potential role in neuropsychiatric disorders. Curr. Med. Chem. 2018;25(28):3333–3352. doi: 10.2174/0929867325666180226102358. [DOI] [PubMed] [Google Scholar]
- Ruijter, J.M., Ramakers, C., Hoogaars, W.M.H., Karlen, Y., Bakker, O., van den Hoff, M.J.B., Moorman, A.F.M., 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45. DOI:10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed]
- Schaller T., Hirschbühl K., Burkhardt K., Braun G., Trepel M., Märkl B., Claus R. Postmortem examination of patients with COVID-19. JAMA. 2020;323:2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurink B., Roos E., Radonic T., Barbe E., Bouman C.S.C., de Boer H.H., de Bree G.J., Bulle E.B., Aronica E.M., Florquin S., Fronczek J., Heunks L.M.A., de Jong M.D., Guo L., du Long R., Lutter R., Molenaar P.C.G., Neefjes-Borst E.A., Niessen H.W.M., van Noesel C.J.M., Roelofs J.J.T.H., Snijder E.J., Soer E.C., Verheij J., Vlaar A.P.J., Vos W., van der Wel N.N., van der Wal A.C., van der Valk P., Bugiani M. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1(7):e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Bhushan B., Maurya A., Mishra G., Singh S.K., Awasthi R. Novel coronavirus disease 2019 (COVID-19) and neurodegenerative disorders. Dermatol. Ther. 2020;33 doi: 10.1111/dth.13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O., Vargas-Hurlston S., Chandy G., Crittenden C., Gordon R., Guicherit O., Zanella F., Carromeu C. Functional evaluation of neurotoxic compounds using human iPSC-derived neuronal 3D cultures. J. Pharmacol. Toxicol. Methods. 2019;99:106595. doi: 10.1016/j.vascn.2019.05.191. [DOI] [Google Scholar]
- Sochacki J., Devalle S., Reis M., Mattos P., Rehen S. Generation of urine iPS cell lines from patients with Attention Deficit Hyperactivity Disorder (ADHD) using a non-integrative method. Stem Cell Res. 2016;17(1):102–106. doi: 10.1016/j.scr.2016.05.015. [DOI] [PubMed] [Google Scholar]
- Sochocka M., Diniz B.S., Leszek J. Inflammatory Response in the CNS: Friend or Foe? Mol. Neurobiol. 2017;54(10):8071–8089. doi: 10.1007/s12035-016-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, E., Zhang, C., Israelow, B., Lu, P., Weizman, O.-E., Liu, F., Dai, Y., Szigeti-Buck, K., Yasumoto, Y., Wang, G., Castaldi, C., Heltke, J., Ng, E., Wheeler, J., Alfajaro, M.M., Fontes, B., Ravindra, N.G., Dijk, D.V., Mane, S., Gunel, M., Ring, A., Wilen, C.B., Horvath, T.L., Louvi, A., Farhadian, S.F., Bilguvar, K., Iwasaki, A., 2020. Neuroinvasive potential of SARS-CoV-2 revealed in a human brain organoid model. bioRxiv 2020.06.25.169946. DOI:10.1101/2020.06.25.169946.
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.-E., Liu F., Dai Y., Szigeti-Buck K., Yasumoto Y., Wang G., Castaldi C., Heltke J., Ng E., Wheeler J., Alfajaro M.M., Levavasseur E., Fontes B., Ravindra N.G., Van Dijk D., Mane S., Gunel M., Ring A., Kazmi S.A.J., Zhang K., Wilen C.B., Horvath T.L., Plu I., Haik S., Thomas J.-L., Louvi A., Farhadian S.F., Huttner A., Seilhean D., Renier N., Bilguvar K., Iwasaki A. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N.i., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabary M., Khanmohammadi S., Araghi F., Dadkhahfar S., Tavangar S.M. Pathologic features of COVID-19: A concise review. Pathol. Res. Pract. 2020;216(9):153097. doi: 10.1016/j.prp.2020.153097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén, M., Fagerberg, L., Hallström, B.M., Lindskog, C., Oksvold, P., Mardinoglu, A., Sivertsson, Å., Kampf, C., Sjöstedt, E., Asplund, A., Olsson, I., Edlund, K., Lundberg, E., Navani, S., Szigyarto, C.A.-K., Odeberg, J., Djureinovic, D., Takanen, J.O., Hober, S., Alm, T., Edqvist, P.-H., Berling, H., Tegel, H., Mulder, J., Rockberg, J., Nilsson, P., Schwenk, J.M., Hamsten, M., Feilitzen, K. von, Forsberg, M., Persson, L., Johansson, F., Zwahlen, M., Heijne, G. von, Nielsen, J., Pontén, F., 2015. Tissue-based map of the human proteome. Science 347. DOI:10.1126/science.1260419. [DOI] [PubMed]
- Wang L., Shen Y., Li M., Chuang H., Ye Y., Zhao H., Wang H. Clinical manifestations and evidence of neurological involvement in 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J. Neurol. 2020;267(10):2777–2789. doi: 10.1007/s00415-020-09974-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman, D., Alameh, M.-G., Silva, T. de, Collini, P., Hornsby, H., Brown, R., LaBranche, C.C., Edwards, R.J., Sutherland, L., Santra, S., Mansouri, K., Gobeil, S., McDanal, C., Pardi, N., Hengartner, N., Lin, P.J.C., Tam, Y., Shaw, P.A., Lewis, M.G., Boesler, C., Sahin, U., Acharya, P., Haynes, B.F., Korber, B., Montefiori, D.C., 2020. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. medRxiv 2020.07.22.20159905. DOI:10.1101/2020.07.22.20159905.
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Woodruff G., Phillips N., Carromeu C., Guicherit O., White A., Johnson McCay, Zanella F., Anson B., Lovenberg T., Bonaventure P., Harrington A.W., Scemes E. Screening for modulators of neural network activity in 3D human iPSC-derived cortical spheroids. PLoS One. 2020;15:e0240991. doi: 10.1371/journal.pone.0240991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhong S., Liu J., Li L., Li Y., Wu X., Li Z., Deng P., Zhang J., Zhong N., Ding Y., Jiang Y. Detection of severe acute respiratory syndrome coronavirus in the brain: potential role of the chemokine mig in pathogenesis. Clin. Infect. Dis. 2005;41(8):1089–1096. doi: 10.1086/444461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L.u., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science:abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.-Z., Chu H., Han S., Shuai H., Deng J., Hu Y.-F., Gong H.-R., Lee A.-Y., Zou Z., Yau T., Wu W., Hung I.-N., Chan J.-W., Yuen K.-Y., Huang J.-D. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30(10):928–931. doi: 10.1038/s41422-020-0390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]