Abstract
Background
Drought is a common phenomenon worldwide. It is also one of the main abiotic factors that affect the growth and quality of strawberry. The dehydration-responsive element binding protein (DREB) members that belong to the APETALA2/ethylene-responsive element binding protein (AP2/EREBP) superfamily are unique transcription factors in plants that play important roles in the abiotic stress response.
Results
Here, a total of 119 AP2/EREBP genes were identified in Fragaria vesca, and the AP2/EREBP superfamily was divided into AP2, RAV, ERF, DREB, and soloist subfamilies, containing 18, 7, 61, 32, and one member(s), respectively. The DREB subfamily was further divided into six subgroups (A-1 to A-6) based on phylogenetic analysis. Gene structure, conserved motifs, chromosomal location, and synteny analysis were conducted to comprehensively investigate the characteristics of FvDREBs. Furthermore, transcriptome analysis revealed distinctive expression patterns among the FvDREB genes in strawberry plants exposed to drought stress. The expression of FvDREB6 of the A-2 subgroup was down-regulated in old leaves and up-regulated in young leaves in response to drought. Furthermore, qRT-PCR analysis found that FvDREB8 from the A-2 subgroup had the highest expression level under drought stress. Together, analyses with the expression pattern, phylogenetic relationship, motif, and promoter suggest that FvDREB18 may play a critical role in the regulation of FvDREB1 and FvDREB2 expression.
Conclusions
Our findings provide new insights into the characteristics and potential functions of FvDREBs. These FvDREB genes should be further studied as they appear to be excellent candidates for drought tolerance improvement of strawberry.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03095-2.
Keywords: DREB, Structural characteristics, Duplication, Drought stress, Fragaria vesca, Expression
Background
The cultivated strawberry (Fragaria × ananassa) is one of the most important and youngest crop species worldwide, originating approximately 300 years ago [11]. Furthermore, strawberry is popular and favored by consumers because of its disease-preventive and medicinal benefits as well as its wide array of aromas and flavors [8, 58]. Genomically, F. × ananassa is an allo-octoploid (2n = 8x = 56) plant derived from four different diploid ancestors. Considering the complex genome of F. × ananassa, the diploid woodland strawberry (F. vesca, 2n = 2x = 14), which has a small completely sequenced genome (240 Mb), is usually used as a model plant for studies on the functions of specific genes and molecular genomic analyses of Rosaceae [49]. A recent report on chromosome-scale assembly in the octoploid strawberry cultivar ‘Camarosa’ has identified a dominant subgenome that was derived from the F. vesca progenitor, which largely controls certain metabolic pathways [11]. Strawberry is very sensitive to osmotic stress caused by high-salt or drought stress [44]. Furthermore, strawberry has a large leaf area and a shallow root distribution, and upon drought stress, its growth and yield are greatly affected [15]. Therefore, drought is one of the main limiting factors that affect the growth and quality of strawberry. Understanding the regulatory mechanism of strawberry in response to drought stress can enhance the development and performance of strawberry when subjected to drought stress.
Drought, as a common phenomenon, is expected to intensify with global warming [3, 5]. In plants, a series of molecular, physiological, and biochemical changes caused by the reprogramming of stress-related genes occur in response and adaptation to drought stress [47]. In addition, numerous transcription factors (TFs) have been reported to regulate stress-responsive genes by binding to the promoter region of target genes [2, 6]. The dehydration-responsive element binding (DREB) TFs have been reported to play important roles in response to drought, low-temperature, or high-salt stress [56].
DREB is a subfamily of the APETALA2/ethylene-responsive element binding protein (AP2/EREBP) superfamily, which is categorized into six subgroups (A-1 to A-6) according to the genetic domain [37, 43]. The AP2/EREBPs are unique TFs in plants and characterized by at least one highly conserved AP2 domain. The AP2/EREBPs could be classified as AP2, Related to ABI3/VP1 (RAV), Ethylene Responsive Factor (ERF), DREB, and soloist in Arabidopsis and rice [46]. Generally, AP2 subfamily members contain two AP2 domains, RAV subfamily members contain one AP2 domain and an additional B3 domain, and ERF and DREB subfamily members both contain one AP2 domain [40]. The difference between DREB and ERF is based on the association of the AP2 domain with the 14th valine (V14) and 19th glutamic acid (E19) in DREB members, and the 14th alanine (A14) and 19th aspartic acid (D19) in ERF members [46].
With the release of whole-genome sequences for many plant species, more and more AP2/EREBP superfamilies, including DREB subfamilies, have been identified and studied at the genome-wide level [50]. Different DREB subgroups play different roles in different plants. For instance, the overexpression of AtDREB1A enhances drought and freezing tolerance in transgenic Lolium perenne plants but enhances heat stress tolerance in transgenic chrysanthemum [20, 30], whereas AtDREB1C (A-1) plays a central role in stress tolerance in Arabidopsis as a negative regulator [42]. Several studies have also reported that the expression of DREB2A and DREB2B, which belong to the A-2 subgroup, was induced in response to drought stress [31, 45].
Although a recent report had identified 91 FveERF genes, the authors had used an old version of the genome and mainly focused on tandem duplications for the expansion of the FveERF family [52]. Therefore, there is a lack of information on the expression of DREBs in response to drought. An updated annotated version, v4.0.a2, for the F. vesca genome has been recently published, adding 9,029 new genes and modifying 8,342 existing genes [32]. In the present study, we identified AP2/EREBP members in the F. vesca genome based on the latest version, v4.0.a2, and performed a comprehensive bioinformatics analysis of the DREB subfamily, including DREB classification and naming, gene structure and conserved motif analyses, and chromosomal localization and synteny analyses. Furthermore, the expression profiles of FvDREBs in response to drought stress were also obtained using transcriptome and qRT-PCR data. Our results will provide new insights into the biological roles of FvDREBs, which may improve drought tolerance in strawberry exposed to drought stress.
Results
Identification of AP2/EREBP in Fragaria vesca
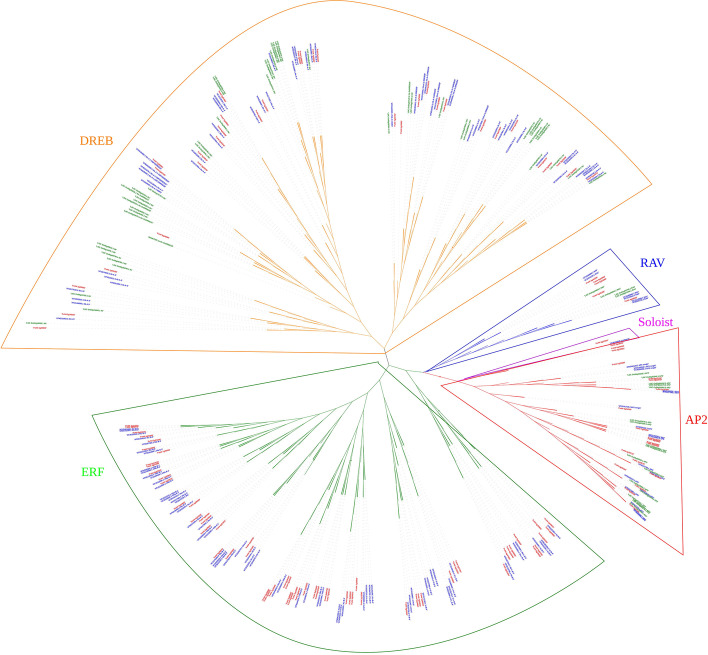
A total of 119 AP2/EREBP genes were identified through HMM searches, local BLAST analyses, and domain confirmations. These genes contained at least one AP2 domain (Supplementary information). A previous study has identified 115 AP2/EREBP genes in F. vesca [52]. The difference between that and this study is that the previous study used an older version of the genome, while our study used the latest version (v4.0.a2). The specific differences in numbers and gene IDs are shown in Table S1. According to the classification of AP2/EREBP in Arabidopsis and rice [40, 46], the 119 AP2/EREBP genes in F. vesca were divided into five groups. A phylogenetic tree was constructed based on the alignment of 337 AP2/EREBP proteins from Arabidopsis, rice, and F. vesca (Fig. 1; high-resolution Figure 1 in Supplementary material). The phylogenetic tree clearly classified the AP2/EREBP proteins from F. vesca into a soloist as well as four typical subfamilies, namely, the AP2, RAV, ERF, and DREB clades, which were comprised of 1, 18, 7, 61, and 32 proteins, respectively. Generally, the RAV subfamily has one AP2 domain and one B3 domain, and the FvRAV subfamily contains two members (FvH4_5g19881 and FvH4_6g29430), which had one AP2 domain and no B3 domain.
Fig. 1.
Phylogenetic tree of AP2/EREBP superfamily proteins from F. vesca, Arabidopsis, and rice. The color of Gene ID of F. vesca was red; the color of Gene ID of Arabidopsis was blue; the color of Gene ID of rice was green
Identification and phylogenetic analysis of FvDREBs
Based on the conserved 14th valine (V14) of the AP2 domain (Figure S1), 32 DREB genes were identified from AP2/ERF in F. vesca and named according to their chromosomal positions (Table 1). The identified FvDREBs proteins ranged from 150 to 579 amino acids in length, with theoretical isoelectric points (pI) ranging from 4.63 to 9.48 and molecular weights (MW) ranging from 16447.59 to 65304.13. Subcellular localization analysis predicted that most (26/32, 81.25%) FvDREBs were localized to the nucleus, whereas other (6/32, 18.75%) members were localized to the cytoplasm (Table 1).
Table 1.
Features of FvDREBs in F. vesca
| Gene Name | Gene ID(v1.1) | Gene ID (v4.02) | Chromosome location | Group | No. amino acids | pI | MW | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| FvDREB1 | mrna32378.1 | FvH4_5g01440 | Fvb5: 899521-900291 | A-1 | 256 | 5.20 | 28817.69 | Nucleus |
| FvDREB2 | mrna13329.1 | FvH4_7g28950 | Fvb7: 21384363-21385052 | A-1 | 229 | 5.13 | 24962.84 | Nucleus |
| FvDREB3 | mrna08479.1 | FvH4_2g38880 | Fvb2: 28047790-28050124 | A-2 | 226 | 9.48 | 24777.23 | Nucleus |
| FvDREB4 | mrna16710.1 | FvH4_6g01400 | Fvb6: 778696-779238 | A-2 | 180 | 6.14 | 19628.61 | Nucleus |
| FvDREB5 | mrna13783.1 | FvH4_6g10690 | Fvb6: 6419987-6422452 | A-2 | 579 | 4.63 | 65304.13 | Cytoplasm |
| FvDREB6 | mrna26463.1 | FvH4_6g23290 | Fvb6: 17276547-17277407 | A-2 | 286 | 5.78 | 32216.02 | Nucleus |
| FvDREB7 | mrna01985.1 | FvH4_6g43870 | Fvb6: 33904894-33907462 | A-2 | 192 | 9.40 | 20839.19 | Nucleus |
| FvDREB8 | mrna21047.1 | FvH4_7g25200 | Fvb7: 19265849-19267988 | A-2 | 378 | 4.89 | 41645.29 | Nucleus |
| FvDREB9 | mrna21003.1 | FvH4_7g24760 | Fvb7: 19064560-19066602 | A-3 | 322 | 6.30 | 35598.83 | Nucleus |
| FvDREB10 | mrna11882.1 | FvH4_1g05820 | Fvb1: 3086932-3088629 | A-4 | 254 | 4.95 | 27529.14 | Nucleus |
| FvDREB11 | mrna08838.1 | FvH4_2g35620 | Fvb2: 26279523-26280287 | A-4 | 254 | 5.01 | 27654.57 | Nucleus |
| FvDREB12 | mrna32380.1 | FvH4_5g01460 | Fvb5: 909587-910426 | A-4 | 279 | 4.71 | 29909.66 | Nucleus |
| FvDREB13 | mrna08876.1 | FvH4_5g19440 | Fvb5: 11268785-11269904 | A-4 | 225 | 5.44 | 25028.56 | Nucleus |
| FvDREB14 | mrna08873.1 | FvH4_5g19460 | Fvb5: 11295238-11295834 | A-4 | 187 | 5.05 | 20616.61 | Nucleus |
| FvDREB15 | mrna27021.1 | FvH4_5g33220 | Fvb5: 24034746-24036098 | A-4 | 196 | 5.26 | 21569.16 | Nucleus |
| FvDREB16 | mrna27017.1 | FvH4_5g33240 | Fvb5: 24050928-24051485 | A-4 | 185 | 4.97 | 20338.81 | Nucleus |
| FvDREB17 | mrna30159.1 | FvH4_6g18000 | Fvb6: 11817696-11821178 | A-4 | 239 | 6.43 | 25517.71 | Cytoplasm |
| FvDREB18 | mrna30226.1 | FvH4_6g18090 | Fvb6: 11874334-11874957 | A-4 | 207 | 5.02 | 22976.03 | Cytoplasm |
| FvDREB19 | mrna25758.1 | FvH4_6g32030 | Fvb6: 25127142-25128659 | A-4 | 258 | 5.03 | 27960.85 | Nucleus |
| FvDREB20 | mrna04810.1 | FvH4_7g09550 | Fvb7: 9175646-9176470 | A-4 | 274 | 5.24 | 29760.18 | Nucleus |
| FvDREB21 | mrna19141.1 | FvH4_7g16810 | Fvb7: 14364300-14365229 | A-4 | 198 | 5.43 | 21984.29 | Nucleus |
| FvDREB22 | mrna13327.1 | FvH4_7g28960 | Fvb7: 21396222-21397656 | A-4 | 235 | 4.92 | 24643.19 | Nucleus |
| FvDREB23 | mrna12919.1 | FvH4_1g09180 | Fvb1: 4892045-4893214 | A-5 | 216 | 4.69 | 24040.38 | Nucleus |
| FvDREB24 | mrna23873.1 | FvH4_1g16370 | Fvb1: 9445234-9446402 | A-5 | 150 | 9.45 | 16447.59 | Cytoplasm |
| FvDREB25 | mrna11145.1 | FvH4_2g26630 | Fvb2: 21353932-21356192 | A-5 | 234 | 5.37 | 25569.60 | Nucleus |
| FvDREB26 | mrna09137.1 | FvH4_2g34020 | Fvb2: 25362442-25364608 | A-5 | 159 | 9.18 | 18020.95 | Nucleus |
| FvDREB27 | mrna27062.1 | FvH4_5g33180 | Fvb5: 23967454-23968327 | A-5 | 165 | 5.50 | 17698.44 | Cytoplasm |
| FvDREB28 | mrna26530.1 | FvH4_5g34550 | Fvb5: 25211129-25211910 | A-5 | 209 | 7.62 | 23054.50 | Cytoplasm |
| FvDREB29 | mrna16350.1 | FvH4_1g21210 | Fvb1: 13220000-13221070 | A-6 | 356 | 6.00 | 39556.12 | Nucleus |
| FvDREB30 | mrna32084.1 | FvH4_5g04470 | Fvb5: 2609265-2610158 | A-6 | 297 | 8.55 | 32973.25 | Nucleus |
| FvDREB31 | mrna22114.1 | FvH4_5g37820 | Fvb5: 27851089-27852207 | A-6 | 372 | 5.85 | 40638.05 | Nucleus |
| FvDREB32 | mrna17698.1 | FvH4_6g26090 | Fvb6: 19953052-19954410 | A-6 | 452 | 5.76 | 51223.92 | Nucleus |
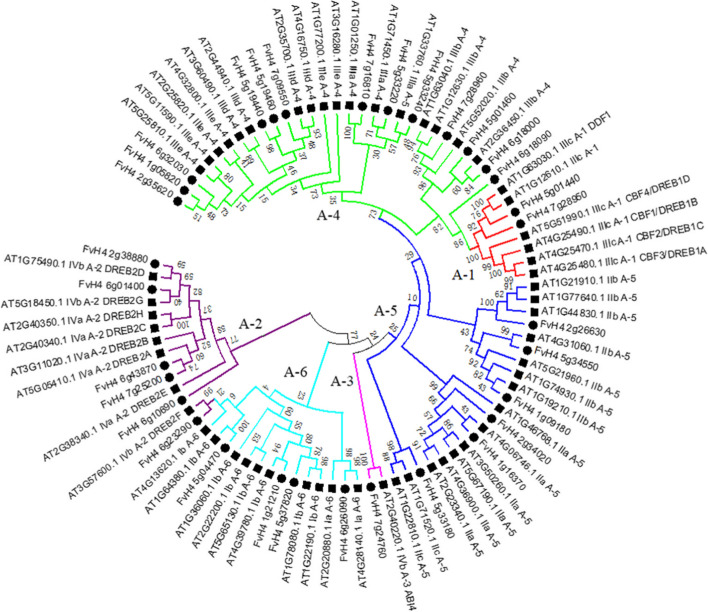
To investigate the phylogenetic relationships between DREBs in strawberry and other plants, a neighbor-joining phylogenetic tree was generated using the whole-protein sequences of the DREB subfamily between F. vesca and A. thaliana. As shown in Fig. 2, the phylogenetic tree was further divided into six subgroups (A-1 to A-6) as in Arabidopsis, in which the A-4 subgroup was the largest (13 members) and the A-3 subgroup was the smallest (one member). According to the similarities between AtDREB1/CBF and AtDREB2, the A-1 subgroup and A-2 subgroup included two and six members, respectively. Meanwhile, seven orthologous pairs were identified in F. vesca and A. thaliana, and one paralogous pair was identified in F. vesca based on a bootstrap value greater than 90 (Supplementary information).
Fig. 2.
Phylogenetic tree of DREB subfamily proteins from F. vesca and Arabidopsis. Different subgroups (A-1 to A-6) of DREB subfamily proteins are highlighted in different colors
Gene structure and conserved motif analysis of FvDREBs
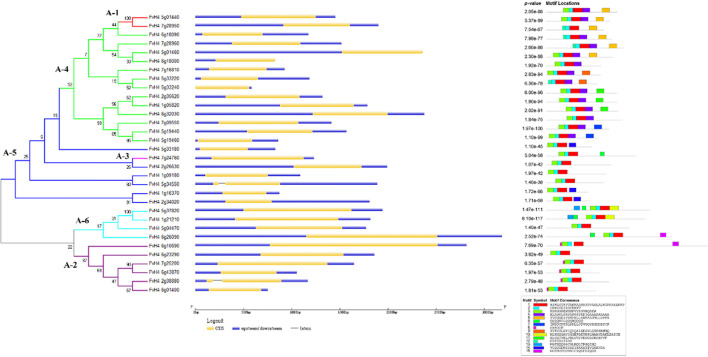
The exon-intron structures were analyzed to gain a better understanding of the structural characteristics of the FvDREBs genes. Almost all FvDREBs (30/32, 93.75%) were intronless, except for FvH4_2g38880 (FvDREB3) and FvH4_5g34550 (FvDREB28), which contained only one intron (Fig. 3).
Fig. 3.
Phylogenetic relationships, exon/intron structures, and conserved motifs of FvDREBs in F. vesca. Different subgroups (A-1 to A-6) are highlighted in different color. Different motifs are represented by blocks of different color and size
The conserved motifs of all FvDREBs were further examined using MEME. A total of 15 motifs were predicted and named as motifs 1 to 15. Motifs 1 and 2 were found in all FvDREB protein sequences and were related to the AP2 domain. The protein sequences of two members belonging to the A-1 subgroup both contained motifs 5 and 11. Motif 8 was only found in members of the A-2 subgroup, whereas motif 10 was only found in members of the A-6 subgroup. Some other motifs, such as motif 15, were distributed among various subgroups.
Chromosomal location and tandem duplication of FvDREBs
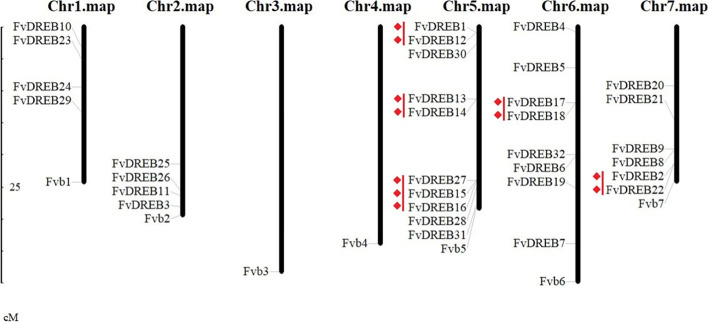
In order to explore the functional differentiation of FvDREB members, their positions on chromosomes were further investigated with the latest annotated genome (v4.0.a2). As shown in Fig. 4, 32 FvDREB members were distributed unevenly on five of the seven chromosomes, and there were no members on chromosomes 3 and 4. Chromosome 5 had the largest number (10, 31.25%) of FvDREB genes, containing one A-1 subgroup member, five A-4 subgroup members, two A-5 subgroup members, and two A-6 subgroup members. Chromosomes 6 and 7 had 25% (8/32) and 18.75% (6/32) FvDREB genes, respectively. The remaining 25% (8/32) members were evenly distributed on chromosomes 1 and 2. Moreover, five tandem duplication events involving eleven FvDREB genes were observed, namely, FvDREB1 and FvDREB12, FvDREB13 and FvDREB14, FvDREB17 and FvDREB18, FvDREB2 and FvDREB22, and FvDREB27, FvDREB15, and FvDREB16. Three of the five tandem duplication events were distributed on chromosome 5, including the three members with tandem duplications.
Fig. 4.
Chromosomal locations of FvDREBs in F. vesca. Red lines indicate tandem duplications
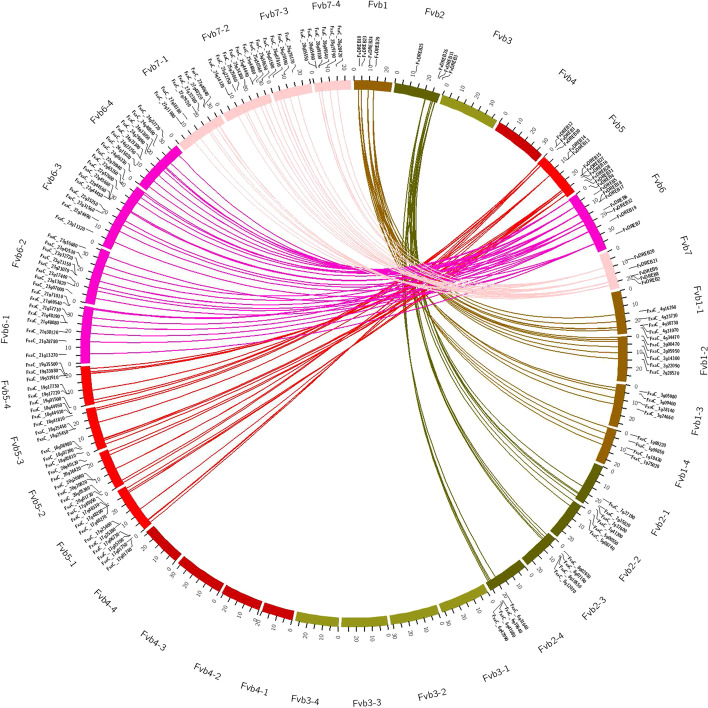
Synteny analysis of FvDREBs
There were 143 syntenic gene pairs that were syntenic between F. vesca and F. × ananassa, and high levels of collinearity were observed in all FvDREBs between F. vesca and their corresponding F. × ananassa, except FvDREB24 (Fig. 5). Moreover, each FvDREBs in F. vesca chromosomes corresponded to several syntenic genes in F. × ananassa chromosomes. For example, the FvDREB10 gene was on chromosome 1 in F. vesca and its syntenic corresponding genes were on chromosome Fvb1-1, Fvb1-2, Fvb1-3, and Fvb1-4, respectively, in F. × ananassa.
Fig. 5.
Syntenic relationships of DREB members from F. vesca and F. × ananassa
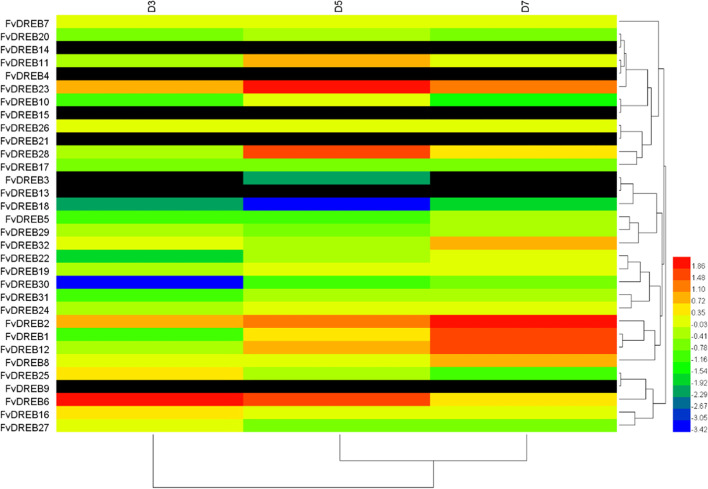
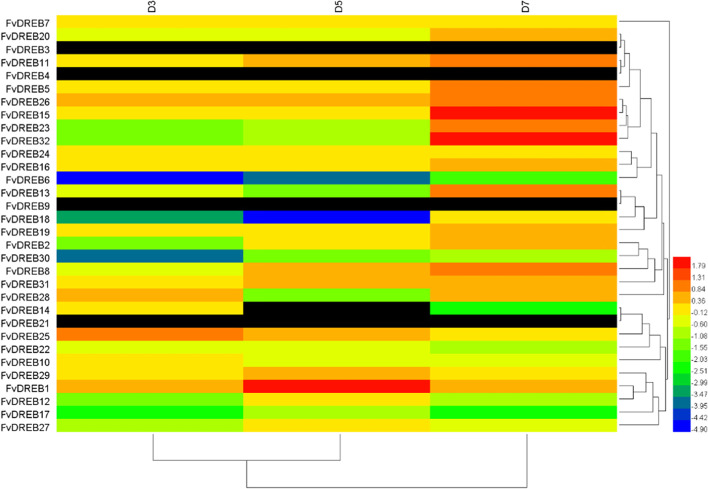
Expression profiles of FvDREBs in response to drought stress in different strawberry leaves
Transcriptome sequencing data from old and young leaves exposed to different drought stress conditions were previously generated by our group to investigate the expression profiles of FvDREBs. In general, different subgroups from different tissues showed different expression patterns (Figs. 6 and 7), suggesting functional divergence between different subgroups of FvDREB members. In old leaves, D5 and D7 showed a similar clustering relationship (Fig. 6), whereas D3 and D5 displayed a similar clustering relationship in young leaves (Fig. 7). Two genes (FvDREB1 and FvDREB2) from the A-1 subgroup were highly expressed in the later period of drought stress, whereas the expression of FvDREB6, which belonged to the A-2 subgroup, was significantly up-regulated at the initial stage of drought stress. The expression of FvDREB30 from the A-6 subgroup was lower in the early stages of drought stress and that of FvDREB18 from the A-4 subgroup was lower in the middle stages of drought stress. Their expression levels in old and young leaves were similar (Figs. 6 and 7), indicating that they are negative regulators in response to drought stress.
Fig. 6.
Heat map of differentially expressed FvDREBs in strawberry old leaves under drought stress. Red indicates up-regulation, blue and green indicate down-regulation, and black indicates data gaps
Fig. 7.
Heat map of differentially expressed FvDREBs in strawberry young leaves under drought stress. Red indicates up-regulation, blue and green indicate down-regulation, and black indicates data gaps
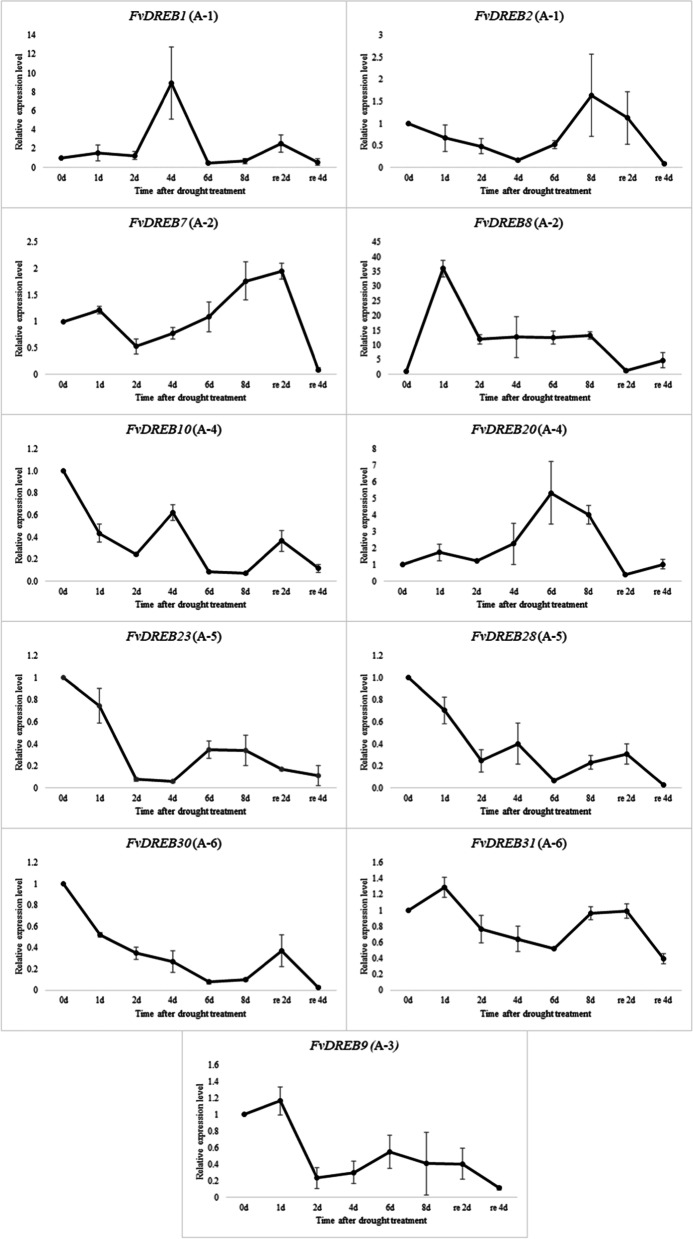
To further verify the expression of these identified FvDREB genes, two genes were randomly selected from each subgroup of the FvDREB gene family (the A-3 subgroup had only one member, so only one gene was selected) to detect their expression levels under different drought stress conditions by qRT-PCR analysis (Fig. 8). The results showed that the expression of FvDREB8 of the A-2 subgroup was significantly up-regulated, with the highest expression observed at 1d under drought stress (Fig. 8). The expression level gradually decreased, but the expression levels of all genes were ten times higher than those in the control group. The expression level of FvDREB1 from the A-1 subgroup reached the highest level when it was subjected to drought stress for 4 days, which was more than eight times that of the control group (Fig. 8). The expression level of FvDREB20 from the A-4 subgroup reached the highest level when subjected to drought stress for 6 days, which was more than five times that of the control group (Fig. 8). The longer the time of exposure to drought stress, the greater the down-regulation of FvDREB30 from the A-6 subgroup. The trend of the expression of FvDREB was consistent with the RNA-Seq data. It could be seen that the expression of FvDREB genes from different subgroups was variable and unstable under drought stress.
Fig. 8.
Expression profiles of the FvDREB genes in response to drought stress
Discussion
AP2/EREBP TFs are one of the largest families of plant-specific transcriptional regulators that modulate many plant processes such as plant growth, development, and response to various stresses. The AP2/EREBP superfamily has been widely identified and investigated in diverse plant species with the release of the whole-genome sequences of various plants. Furthermore, continuous improvements in high-throughput sequencing techniques and bioinformatic algorithms have allowed the accurate and accelerated delivery of information for researchers [1]. In this study, we identified the AP2/EREBP superfamily and comprehensively analyzed the DREB subfamily in the F. vesca genome based on the latest version, v4.0.a2. Moreover, we used RNA-Seq (transcriptome sequencing) and qRT-PCR to characterize differentially expressed FvDREB genes when plants were exposed to drought stress. Our study provides a better understanding of the FvDREB proteins, which may benefit strawberry biotechnological breeding and improve their adaptation and tolerance to drought.
The DREB TF family plays important roles in the response to abiotic stress. However, knowledge of FvDREB genes is limited. Gene structure analysis of FvDREBs revealed that 93.75% of FvDREBs were intronless, which was higher than that of rice, maize, and other plants [24, 27]. Motif analysis showed that all FvDREB protein sequences had motifs related to the AP2 domain and demonstrated that the AP2 domain was highly conserved in FvDREBs. The results also showed that different subgroups had specific motifs (Fig. 3). Generally, the DREB sequences were confirmed based on the presence of an AP2 domain with the 14th valine (V14) and 19th glutamic acid (E19) [46]. A comparison of the amino acid sequences of AP2 domains in FvDREBs revealed that although all of the amino acid sequences at position 14 were valine (V14), those at position 19 were not all glutamic acid (E19) (Figure S1). In addition, 81.25% of FvDREB proteins were predicted to be localized to the nucleus, suggesting the roles of these proteins in signal transduction and transcriptional regulation [48]. These results indicate that most domains or motifs of FvDREB proteins were highly conserved and functional divergence may occur by changing key amino acids. ERFs and DREBs exhibit distinctive features, the ERFs are bound to the GCC box (AGCCGCC), whereas the DREBs are bound to DRE (A/GCCGAC) [34].
Whole-genome duplications (WGD), tandem duplications, segmental duplications, or polyploidization events are widely believed to be the primary sources of plant evolution, they have also contributed to gene family expansion [4, 19, 25]. All FvDREB proteins were distributed unevenly on five of the seven chromosomes, which might have been the result of WGD. Tandem duplications and segmental duplications were also detected in FvDREBs, which also contributed to the expansion of the FvDREB gene family. Furthermore, we also performed an intergenomic synteny analysis between F. vesca and F. × ananassa to study the evolution of FvDREB in the Fragaria genome along with the species evolution and polyploidization. The high level of collinearity between FvDREBs of diploid and their corresponding octoploid homologs suggests a close relationship between genomes in Fragaria [11].
The expression patterns of FvDREBs reflect biological roles and gene functions in response to drought stress. Interestingly, we found that the expression of FvDREB1 from the A-1 subgroup correlated with the early and middle stages of drought stress, whereas the expression of FvDREB2 from the A-1 subgroup correlated with the middle and late stages of drought stress (Fig. 8). This suggests that the individual contribution and function of each DREB are different in response to the entire drought stress process. The A-1 of DREBs, also known as C-repeat binding factors (CBFs), were first identified as TFs in response to both low temperature and osmotic stress in Arabidopsis [56]. Furthermore, more and more studies have demonstrated that the overexpression of DREB1/CBF can improve the tolerance to environmental stresses such as freezing, drought, salt, and high temperature [16, 18, 23]. Our previous meta-analysis of the effect of CBF/DREB overexpression on drought stress response also confirms the premise that DREB overexpression can enhance drought stress tolerance in various crops and reflect the duration of the stress treatment (stress time). As a moderator, it had a clear effect on the response of the transgenic plants in relation to some parameters [10].
Equally important, the fact that FvDREB18 was down-regulated in response to whole drought stress, especially in the middle stage of drought stress of both old and young leaves, combined with the expression pattern of FvDREB1 and FvDREB2, was very similar to that of CBF2/DREB1C, a negative regulator of CBF1/DREB1B and CBF3/DREB1A in Arabidopsis [42]. The clade containing FvDREB18 was closest to A-1 subgroup clades in the phylogenetic tree with a bootstrap value of 86 (Fig. 2), and the number of conserved motifs in FvDREB18 was four, whereas the number of conserved motifs in FvDREB1 and FvDREB2 was five (Fig. 3). This may suggest that FvDREB18 plays a critical role in response to drought stress by precisely controlling the expression of FvDREB1 and FvDREB2, and, hence, that of the downstream genes. Moreover, we analyzed the promoters of these three genes and found that there were different cis-regulatory elements among them (Supplementary information and Figure S2). The cis-regulatory elements, such as ABRE (cis-acting element involved in the abscisic acid responsiveness), ARE (cis-acting regulatory element essential for the anaerobic induction), MBS (MYB binding site involved in drought-inducibility), MYB, and MYC, were present in the promoter regions of all three genes. By contrast, CAT-box, TGA-element, and HD-Zip 1 were only detected in the FvDREB2 promoter, which was related to meristem expression, auxin-responsive, and the differentiation of palisade mesophyll cells, respectively. There were three TCA-element elements (cis-acting element involved in salicylic acid responsiveness) in the FvDREB2 promoter and one in the FvDREB1 promoter, but none in the FvDREB18 promoter. The different types and numbers of cis-regulatory elements play essential roles in determining the stress-responsive or tissue-specific expression patterns of genes [13, 35], and those presenting in the promoter region of FvDREBs may indicate differential regulatory networks. However, the mechanisms by which FvDREB18 regulates FvDREB1 and FvDREB2 expression and the involvement of other regulators await further investigation.
In addition, FvDREB8 from the A-2 subgroup was significantly and positively regulated by drought stress and induced strongly and rapidly in the early stage. This suggests that FvDREB8 may be very sensitive to drought. The A-2 subgroup DREB members, which are involved in drought-responsive gene expression, were first referred to as DREB2 to distinguish them from DREB1, whereas DREB1 is thought to function in cold-responsive gene expression regulation [47]. DREB2A and DREB2B are induced under drought and salt stress conditions as two of the total eight DREB2-type genes in Arabidopsis, and only OsDREB2A and OsDREB2B were found to be induced by abiotic stress as two of all five DREB2-type genes in rice [41, 39, 46]. In this study, in addition to FvDREB8, FvDREB6 also showed drought stress-inducible gene expression among all six A-2 subgroup genes in F. vesca. The expression of FvDREB6 was down-regulated in old leaves and up-regulated in young leaves in response to drought and appeared to be tissue-specific (Figs. 6 and 7). Remarkably, the gene annotated as DREB2-2 was down-regulated in the leaves of dehydrated B. napus, whereas its expression was increased in roots [22, 33]. These findings provide new insights into the genetic control of drought tolerance in strawberry and offer some useful candidates for drought tolerance improvement.
Conclusions
We performed a genome-wide analysis on the AP2/EREBP family genes in F. vesca and identified 119 FvAP2/EREBP genes. A detailed investigation of the classification, phylogenetic evolution, structure, synteny, and expression profile of these FvDREBs in different tissues and in response to drought stress was carried out. Our results reveal that FvDREB8 from the A-2 subgroup play crucial roles in the early stage of drought stress response. FvDREB6 appeared to be tissue-specific and FvDREB18 may play a critical role in regulating the expression of FvDREB1 and FvDREB2. Overall, our findings provide new insights into the characteristics and potential functions of FvDREBs and offer a better understanding of their molecular basis in response to drought stress in strawberry.
Methods
Identification and classification of the DREB genes in strawberry
The most recent version of the F. vesca genome v4.0.a2 was downloaded from the Genome Database for Rosaceae (GDR) (https://www.rosaceae.org/species/fragaria_vesca/genome_v4.0.a2) to identify strawberry DREB TFs. Additionally, 53 DREB, 23 AP2, and six RAV genes from rice (Oryza sativa) were downloaded from the MSU Rice Genome Annotation Project Database (RGAP) (http://rice.plantbiology.msu.edu//), and 148 AP2/EREBP genes from Arabidopsis were downloaded from The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/index.jsp) database. The Hidden Markov Model (HMM) of the AP2 domain (PF00847) was downloaded from the Pfam protein analysis website (http://pfam.xfam.org/) and used to identify AP2/EREBPs with a defined e-value threshold < 1e−5. To search for all possible AP2/EREBPs, some AtDREB members were used as the query sequence in the local Basic Local Alignment Search Tool (BLAST). To validate the search results, all candidate sequences were examined and analyzed by a simple modular architecture research tool (SMART) (http://smart.embl.de/) [29] and the Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) [38].
All the AP2/EREBP candidate sequences encoding the conserved AP2 domain were constructed with multiple alignments by MUSCLE [12]. A neighbor-joining phylogenetic tree was generated using the 1000 bootstrap method and Poisson model with MEGA 6.06 [51]. All AP2/EREBP proteins were aligned to Arabidopsis and rice AP2/EREBP proteins to classify them into different groups. And the DREBs were identified based on the presence of only one AP2 domain with the 14th valine (V14) and 19th glutamic acid (E19) [40, 46]. The theoretical isoelectric point (pI) and molecular weight (MW) of the identified proteins were analyzed by the ProtParam Tool (https://web.expasy.org/protparam/) [14]. The subcellular localization of each protein was predicted with Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) [7].
Gene structure and conserved motif analysis of the FvDREB genes
The coding sequences (CDS) and full-length sequences of FvDREB genes were obtained from NCBI and graphically displayed with Gene Structure Display Server 2.0 (GSDS) (http://gsds.cbi.pku.edu.cn/) [21]. Conserved motifs in FvDREBs were predicted by the Multiple Em for Motif Elicitation Tool 5.1.1 (MEME) (http://meme-suite.org/tools/meme) using default parameters.
Chromosomal localization and synteny analyses
The chromosomal locations of the FvDREBs were retrieved from the annotated file of the F. vesca genome v4.0.a2 and graphically represented with MapInspect (http://www.softsea.com/review/MapInspect.htm). To investigate the effect of genome duplications on DREB evolution, we conducted an intergenomic synteny analysis on F. vesca and F. × ananassa. The syntenic information of FvDREBs was calculated with MCScanX [54], and the syntenic diagram was visualized with Circos (http://circos.ca/) [26]. Tandem duplications were characterized as multiple members of FvDREBs occurring in neighboring intergenic regions (distance < 100 kb) that were separated by ten or fewer non-homologous spacer genes [19]. Segmental duplications were identified from the Plant Genome Duplication Database (PGDD) (http://chibba.agtec.uga.edu/duplication/) [28].
Plant materials and stress treatment
The strawberry (F. vesca subspecies vesca) seeds were a kind gift from Dr. Janet Slovin (Fruit and Vegetable Lab at the USDA). They were grown on 1/2 MS medium after disinfecting with NaClO (20%, 20 mins) in a tissue culture room for 6 weeks with a photoperiod of 16-h light/8-h dark and a temperature of 24 ± 2°C. Light (~200 μmol m−2 s−1) was supplied by LED.
Drought stress was applied by transferring plants to soil medium and then withholding water. All leaves were collected from each 2-month-old plant which exposed to drought stress at 0 d, 1 d, 2 d, 4 d, 6 d and 8 d, as well as 2 d, 4 d post treatment. All samples were immediately placed into liquid nitrogen and stored at -80°C. Three biological replicates were analyzed for each treatment and control.
Total RNA was extracted from strawberry leaves using the CTAB method with minor modifications [17]. The RNase-free DNase Set with RNeasy/QIAamp® Columns (QIAGEN, USA) was used to eliminate contaminating genomic DNA. Total RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). All cDNAs were stored at -20°C.
Transcriptome analysis of strawberry under drought stress
The transcriptome data, which were unpublished, were provided by a member of our lab, Xiaolong Wang, and were only analyzed in his doctoral dissertation [53]. The strawberries were exposed to drought stress after 1 day of full water absorption. D3, D5, and D7 samples were collected on the third day (after 2 days of drought treatment), fifth day (after 4 days of drought treatment), and seventh day (after 6 days of drought treatment), respectively. Transcriptome data were obtained from a fixed amount of RNA collected from old (the stage of fully expanded mature leaves) and new leaves (the stage before fully expanded mature leaves). The log2 transformed Fragments per Kilobase per Million mapped reads (Log2FPKM) was used to calculate the expression levels of genes in control and treated plants at different times. The different expression patterns of FvDREB genes (Supplementary information) were clustered and visualized by HemI (http://hemi.biocuckoo.org/down.php) [9].
qRT-PCR and expression pattern analysis
The expression patterns of FvDREB genes were examined by quantitative real-time Polymerase Chain Reaction (qRT-PCR) using the QuantStudioTM Flex 96-Well PCR System (Applied Biosystems, USA) and SYBR® Green Reagents (Applied Biosystems, USA). The primer sequences used are listed in the Supplementary information. Some primers were designed with qPrimerDB (http://biodb.swu.edu.cn/qprimerdb), and others were designed with Beacon Designer 8.14. The total volume of each reaction mixture was 10 μL; it included 1 μL of cDNA as the template, 5 μL of PowerUpTM SYBR® Green Master Mix (Applied Biosystems, USA), 0.6 μL of each primer, and 3.4 μL of ddH2O. The PCR cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of amplification for 30 s at 95°C, annealing for 30 s at 58–60°C (depending on the primer’s annealing temperature), and extension for 20 s at 72 °C. The melting curve conditions were as follows: 72°C to 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. This was performed for each amplification immediately after the PCR. Four commonly used reference genes (actin, EF1, GAPDH, DBP) were amplified to test the stability of the expressed genes in strawberry. Finally, EF1 was used in this study because it was the most stable of the four reference genes.
The relative expression levels were calculated using the 2-ΔΔCT (ΔCT = CT target – CT reference; ΔΔCT = (CT target – CT reference) treatment – (CT target – CT reference) control) method [36]. The standard deviation (SD) were calculated by the three biological replicates [57].
Supplementary Information
Additional file 1. The details information of AP2/EPEBP and DREB contain gene name, gene ID, homologous, synteny, segmental duplication, promoter analysis, primer sequence and transcriptome data.
Additional file 2: Table S1. Comparison of group/subgroup size of AP2/EREBP superfamily between this study and the previous study. Figure S1. Comparison of amino acid sequences of the AP2 domains in the FvDREB subfamily. Figure S2. Characteristics of cis-regulatory elements in the promoter region of FvDREBs.
Acknowledgements
We thank Xiaolong Wang from Nanjing Agricultural University for sharing the unpublished transcriptome data.
Abbreviations
- AP2/EREBP
APETALA2/ethylene-responsive element binding protein
- BLAST
Basic Local Alignment Search Tool
- CBFs
C-repeat binding factors
- CDD
Conserved Domain Database
- DREB
Dehydration-responsive element binding protein
- ERF
Ethylene Responsive Factor
- F. × ananassa
Fragaria × ananassa
- F. vesca
Fragaria vesca
- GDR
Genome Database for Rosaceae
- GSDS
Gene Structure Display Server
- HMM
Hidden Markov Model
- MEME
Multiple Em for Motif Elicitation Tool
- MW
Molecular weight
- PGDD
Plant Genome Duplication Database
- pI
Isoelectric point
- qRT-PCR
Quantitative real-time Polymerase Chain Reaction
- RAV
Related to ABI3/VP1
- RGAP
MSU Rice Genome Annotation Project Database
- TAIR
The Arabidopsis Information Resource
- TFs
Transcription factors
- WGD
Whole-genome duplication
Authors’ contributions
CD and ZMC conceived the idea and designed the experiments; CD analyzed the data, conducted the experiments, and wrote the manuscript; YX helped with data analysis and offered constructive discussions; XC significantly contributed to data experiment preparation; ZMC contributed to the conception of the study and revised the manuscript. All authors have read and approved the final version of this manuscript.
Funding
This project was supported by the Jiangsu Province Graduate Student Training Innovation Project (KYCX18_0688), the Jiangsu Provincial Department of Education, China, and in part by the Tennessee Agricultural Experiment Station, University of Tennessee, USA. The authors gratefully acknowledge the financial support from the China Scholarship Council.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. The transcriptomic data supporting the conclusions of this article come from the doctoral dissertation of Xiaolong Wang [53], and the accession number of this transcriptomic data is PRJNA733854.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Afzal M, Alghamdi SS, Migdadi HH, Khan MA, Mirza SB, El-Harty E. Legume genomics and transcriptomics: From classic breeding to modern technologies. Saudi J Biol Sci. 2020;27(1):543–555. doi: 10.1016/j.sjbs.2019.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plant. 2010;54(2):201–212. doi: 10.1007/s10535-010-0038-7. [DOI] [Google Scholar]
- 3.Asadieh B, Krakauer NY. Global change in flood and drought intensities under climate change in the 21st century. Hydrol Earth Syst Sci Discuss. 2017;21:1–22. doi: 10.5194/hess-21-1-2017. [DOI] [Google Scholar]
- 4.Bennett MD. Perspectives on polyploidy in plants-ancient and neo. Biol J Linn Soc. 2004;82(4):411–423. doi: 10.1111/j.1095-8312.2004.00328.x. [DOI] [Google Scholar]
- 5.Burke EJ, Brown SJ, Christidis N. Modeling the recent evolution of global drought and projections for the twenty-first century with the Hadley Centre climate model. J Hydrometeorol. 2006;7(5):1113–1125. doi: 10.1175/JHM544.1. [DOI] [Google Scholar]
- 6.Chen WJ, Zhu T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004;9(12):591–596. doi: 10.1016/j.tplants.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3(2):153. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 8.Davik J, Aaby K, Buti M, Alsheikh M, Šurbanovski N, Martens S, Sargent DJ. Major-effect candidate genes identified in cultivated strawberry (Fragaria × ananassa Duch.) for ellagic acid deoxyhexoside and pelargonidin-3-O-malonylglucoside biosynthesis, key polyphenolic compounds. Hortic Res. 2020;7(1):1–17. doi: 10.1038/s41438-020-00347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS One. 2014;9(11):e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong C, Ma Y, Wisniewski M, Cheng ZM. Meta-analysis of the effect of overexpression of CBF/DREB family genes on drought stress response. Environ Exp Bot. 2017;142:1–14. doi: 10.1016/j.envexpbot.2017.07.014. [DOI] [Google Scholar]
- 11.Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, et al. Origin and evolution of the octoploid strawberry genome. Nat Genet. 2019;51(3):541–547. doi: 10.1038/s41588-019-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, You J, Xie K, Xie W, Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Gen Genomics. 2008;280(6):547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 14.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook. 2005. p. 571–607.
- 15.Ghaderi N, Siosemardeh A. Response to drought stress of two strawberry cultivars (cv. Kurdistan and Selva) Hortic Environ Biotechnol. 2011;52(1):6–12. doi: 10.1007/s13580-011-0019-6. [DOI] [Google Scholar]
- 16.Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124(4):1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Mendoza D, Moreno AQ, Zapata-Perez O. An improved method for the isolation of total RNA from Avicennia germinans leaves. Z Naturforsch C. 2008;63(1-2):124–126. doi: 10.1515/znc-2008-1-222. [DOI] [PubMed] [Google Scholar]
- 18.Haake V, Cook D, Riechmann J, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130(2):639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148(2):993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong B, Ma C, Yang Y, Wang T, Yamaguchi-Shinozaki K, Gao J. Over-expression of AtDREB1A in chrysanthemum enhances tolerance to heat stress. Plant Mol Biol. 2009;70(3):231–240. doi: 10.1007/s11103-009-9468-z. [DOI] [PubMed] [Google Scholar]
- 21.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janiak A, Kwaśniewski M, Szarejko I. Gene expression regulation in roots under drought. J Exp Bot. 2016;67(4):1003–1014. doi: 10.1093/jxb/erv512. [DOI] [PubMed] [Google Scholar]
- 23.Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280(5360):104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 24.Jin JH, Wang M, Zhang HX, Khan A, Wei AM, Luo DX, Gong ZH. Genome-wide identification of the AP2/ERF transcription factor family in pepper (Capsicum annuum L.) Genome. 2018;61(9):663–674. doi: 10.1139/gen-2018-0036. [DOI] [PubMed] [Google Scholar]
- 25.Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 26.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labbo AM, Mehmood M, Akhtar MN, Khan MJ, Tariq A, Sadiq I. Genome-wide identification of ap2/erf transcription factors in mungbean (vigna radiata) and expression profiling of the vrdreb subfamily under drought stress. Crop Pasture Sci. 2018;69(10):1009–1019. doi: 10.1071/CP18180. [DOI] [Google Scholar]
- 28.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2012;41(D1):D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021;49(D1):D458–D460. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li MR, Li Y, Li HQ, Wu GJ. Ectopic expression of FaDREB2 enhances osmotic tolerance in paper mulberry. J Integr Plant Biol. 2011;53(12):951–960. doi: 10.1111/j.1744-7909.2011.01087.x. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Cheng X, Liu J, Zeng H, Han L, Tang W. Heterologous expression of the Arabidopsis DREB1A/CBF3 gene enhances drought and freezing tolerance in transgenic Lolium perenne plants. Plant Biotechnol Rep. 2011;5(1):61–69. doi: 10.1007/s11816-010-0157-9. [DOI] [Google Scholar]
- 32.Li Y, Pi M, Gao Q, Liu Z, Kang C. Updated annotation of the wild strawberry Fragaria vesca V4 genome. Hortic Res. 2019;6(1):1–9. doi: 10.1038/s41438-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Zhang X, Zhang K, An H, Hu K, Wen J, et al. Comparative analysis of the Brassica napus root and leaf transcript profiling in response to drought stress. Int J Mol Sci. 2015;16(8):18752–18777. doi: 10.3390/ijms160818752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C, Zhang T. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genomics. 2017;18(1):1–16. doi: 10.1186/s12864-016-3406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Kong L, Zhang M, Lv Y, Liu Y, Zou M, et al. Genome-wide identification, phylogeny, evolution and expression patterns of AP2/ERF genes and cytokinin response factors in Brassica rapa ssp. pekinensis. PLoS One. 2013;8(12):e83444. doi: 10.1371/journal.pone.0083444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Magnani E, Sjölander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16(9):2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2010;39(suppl_1):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, et al. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Gen Genomics. 2010;283(2):185–196. doi: 10.1007/s00438-009-0506-y. [DOI] [PubMed] [Google Scholar]
- 40.Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140(2):411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K. Organization and expression of two arabidopsis dreb2 genes encoding dre-binding proteins involved in dehydration-and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42(4):657–65. [DOI] [PubMed]
- 42.Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci. 2004;101(11):3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7(2):173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perin EC, da Silva MR, Borowski JM, Crizel RL, Schott IB, Carvalho IR, et al. ABA-dependent salt and drought stress improve strawberry fruit quality. Food Chem. 2019;271:516–526. doi: 10.1016/j.foodchem.2018.07.213. [DOI] [PubMed] [Google Scholar]
- 45.Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LSP, et al. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007;50(1):54–69. doi: 10.1111/j.1365-313X.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- 46.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 47.Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18(5):1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer M, Paciorek T, Benková E, Friml J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc. 2006;1(1):98. doi: 10.1038/nprot.2006.15. [DOI] [PubMed] [Google Scholar]
- 49.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43(2):109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srivastava R, Kumar R. The expanding roles of APETALA2/Ethylene Responsive Factors and their potential applications in crop improvement. Briefings Funct Genomics. 2019;18(4):240–254. doi: 10.1093/bfgp/elz001. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Lin S, Liu D, McAvoy R, Ding J, Li Y. Characterization and expression analysis of ERF genes in Fragaria vesca suggest different divergences of tandem ERF duplicates. Front Genet. 2019;10:805. doi: 10.3389/fgene.2019.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X. Evolutionary and response analysis of bZIP genes in strawberry under drought stress. A Dissertation Presented to Nanjing Agricultural University in Partial Fulfillment of the Requirements for the Degree of Doctor of Agronomy in Pomology. 2017; 93-112
- 54.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wessler SR. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 2005;10(2):54–56. doi: 10.1016/j.tplants.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 56.Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6(2):251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xi Y, Liu J, Dong C, Cheng ZM. The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front Plant Sci. 2017;8:978. doi: 10.3389/fpls.2017.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan JW, Ban ZJ, Lu HY, Li D, Poverenov E, Luo ZS, et al. The aroma volatile repertoire in strawberry fruit: a review. J Sci Food Agric. 2018;98(12):4395–4402. doi: 10.1002/jsfa.9039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The details information of AP2/EPEBP and DREB contain gene name, gene ID, homologous, synteny, segmental duplication, promoter analysis, primer sequence and transcriptome data.
Additional file 2: Table S1. Comparison of group/subgroup size of AP2/EREBP superfamily between this study and the previous study. Figure S1. Comparison of amino acid sequences of the AP2 domains in the FvDREB subfamily. Figure S2. Characteristics of cis-regulatory elements in the promoter region of FvDREBs.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The transcriptomic data supporting the conclusions of this article come from the doctoral dissertation of Xiaolong Wang [53], and the accession number of this transcriptomic data is PRJNA733854.