SUMMARY
Understanding the structure and function of neural circuits underlying speech and language is a vital step toward better treatments for diseases of these systems. Songbirds, among the few animal orders that share with humans the ability to learn vocalizations from a conspecific, have provided many insights into the neural mechanisms of vocal development. However, research into vocal learning circuits has been hindered by a lack of tools for rapid genetic targeting of specific neuron populations to meet the quick pace of developmental learning. Here, we present a viral tool that enables fast and efficient retrograde access to projection neuron populations. In zebra finches, Bengalese finches, canaries, and mice, we demonstrate fast retrograde labeling of cortical or dopaminergic neurons. We further demonstrate the suitability of our construct for detailed morphological analysis, for in vivo imaging of calcium activity, and for multi-color brainbow labeling.
In Brief
Düring et al. describe a fast and efficient viral vector to dissect structure and function of neural circuits underlying learned vocalizations in songbirds. The AAV variant provides retrograde access to projection neuron circuits, including dopaminergic pathways in songbirds and additionally in mice, and allows for retrograde calcium imaging and multispectral brainbow labeling.
Graphical Abstract
INTRODUCTION
Speech and language disorders affect millions of children worldwide (Rosenbaum and Simon, 2016). These disorders are associated with a number of pathologies, including autism spectrum (Rapin and Dunn, 2003) and attention deficit hyperactivity disorders (Tirosh and Cohen, 1998), but they also occur in otherwise normally developing children. The causes of speech and language disorders are poorly understood, but abnormal brain structure likely plays a crucial role (Mayes et al., 2015). Interestingly, neither mice, the most common animal model for pre-clinical studies, nor non-human primates, our closest relatives, share the human ability to learn vocalizations by imitating a conspecific (Hauser et al., 2002). Songbirds, on the other hand, are gifted vocal learners and display many parallels with speech acquisition in humans (Bolhuis et al., 2010). Therefore, songbirds provide an excellent model system to examine the neural circuits associated with speech development. Songbird circuit research remains, however, limited by the current lack of transgenic animal lines and proper neurogenetic tools.
The virally mediated delivery of genetic cargo to a specific neuron population is a highly valuable approach for probing anatomy and circuit function. By entering at axon terminals, retrograde viral vectors can specifically transduce projection neuron populations and provide direct access to their wiring and function in the brain (Callaway, 2008; Larsen et al., 2008). Unfortunately, viral vectors are not easily transferable between model species. For example, rabies and canine adenovirus (CAV), the most commonly used retrograde vectors in rodents, have not been shown to successfully infect avian tissue. Other examples include adeno-associated viral (AAV) vectors that are commonly used in rodents and other mammals but suffer from major limitations in accessing songbird projection neurons (Chen et al., 2019; Daliparthi et al., 2019; Hisey et al., 2018; Roberts et al., 2012, 2017; Tanaka et al., 2018; Xiao et al., 2018).
The limitations of the AAV vectors so far applied for retrograde access to songbird projection neurons are 4-fold. First, these AAV vectors result in low transduction success rates, with from 18% to 43% of intracerebral injections resulting in successful transgene expression (Chen et al., 2019). Second, their retrograde transduction of projection neurons is sparse, resulting in transgene expression in small portions of the targeted populations (Chen et al., 2019; Hisey et al., 2018). This latter limitation makes these tools incompatible with studies requiring genetic manipulation of a majority of neurons among a given type. The third limitation is weak transgene expression (Chen et al., 2019; Roberts et al., 2017). For example, when used to induce fluorescent protein expression, the resulting limited dendritic and axonal labeling (Chen et al., 2019; Roberts et al., 2017) makes current retrograde tools unsuitable for morphological studies. Finally, the fourth limitation of the current AAV vectors is that they require long incubation periods of at least 4–6 weeks (and up to 6 months) post-injection until transgene expression in retrogradely transduced projection neurons becomes detectable (Daliparthi et al., 2019; Hisey et al., 2018; Roberts et al., 2017). Such long incubation periods typically hinder any vocal development studies in zebra finches (Taeniopygia guttata), the most common songbird model species in neuroscience, because zebra finches undergo short song learning phases during the critical period for vocal learning. This period ends 90 days after hatching and is accompanied by fast brain rewiring (Mooney, 2009). The rapid succession of vocal learning events in songbirds thus requires fast viral tools for circuit investigations. To date, no viral vector has proven to efficiently target songbird projection neurons resulting in robust transgene expression in a relevant time frame.
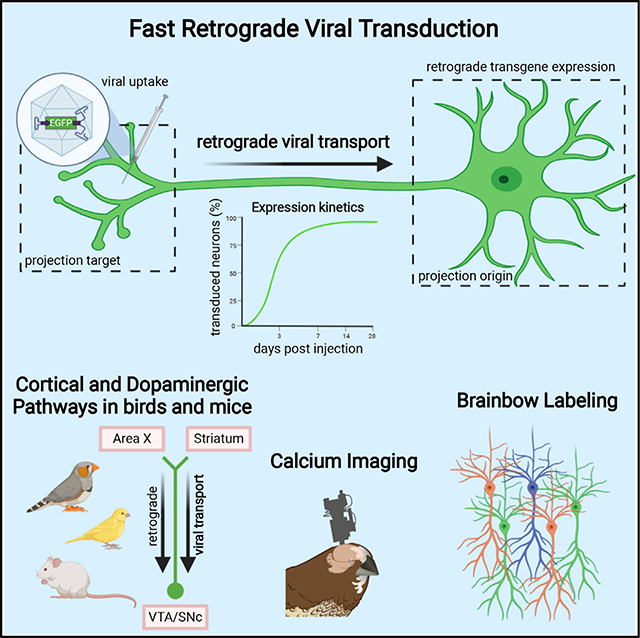
Here, we present a fast and reliable retrograde viral tool for the songbird brain. Briefly, we constructed self-complementary (sc) AAV-DJ/9 (Grimm et al., 2008) vectors according to scAAV-2-DJ/9-hCMV-I/E-chI-transgene-p(A) (Figures 1A and 1B; STAR Methods). In several songbird species and in mice, we found that our viral construct is efficient in retrogradely transducing projection neuron circuits within ~1 week after vector injection (Figures 1C–1O and 2). Furthermore, our construct is suitable for detailed morphological investigations, for multi-color brainbow imaging, and for in vivo recordings of neural activity using fluorescent calcium sensors.
Figure 1. AAV Capsid and Genome Structure for Efficient Retrograde Access to Songbird and Mouse Projection and Dopaminergic Neurons.
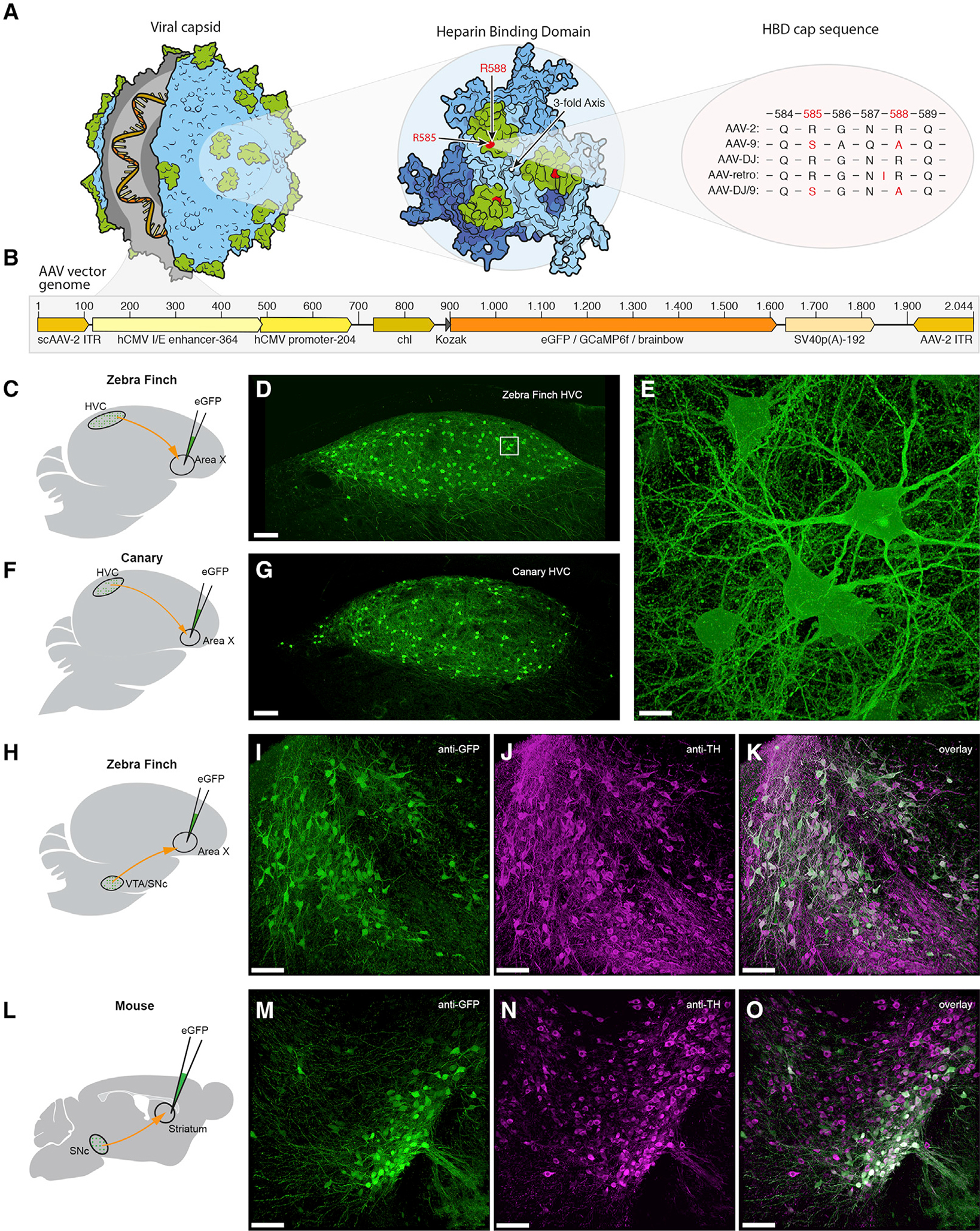
(A) Illustration of the AAV capsid, showing exposed surface proteins responsible for binding to the heparin sulfate proteoglycans (HSPG) receptor, thought to be important for cellular entry of the viral vector. Mutations of two crucial arginines in the heparin binding domain (HBD; highlighted in green) cap sequence at positions 585 and 588 (highlighted in red) reduce heparin binding affinity and potentially improve retrograde transport. Changes are shown for four serotypes that have been used for or possess potential for retrograde transduction.
(B) AAV vector genomes encoding diverse transgenes (orange region) including fluorescent proteins and the calcium indicator GCaMP6f. Numbers on top indicate the base pair length and relative position of encoded elements. Note that the actual size of the region encoding the transgene (orange) varies depending on the respective protein. Brainbow stands for three separate AAV vector genomes encoding either eGFP (enhanced green fluorescent protein), eCFP (enhanced cyan fluorescent protein), or mRuby3 (a red fluorescent protein).
(C, F, H, and L) Schematics of the virus injection sites (green) and the neuronal projections analyzed (red).
(D and G) Native fluorescence signal in retrogradely labeled HVCX neurons 7 days post-delivery of virus in zebra finch and in canary, respectively.(E, I–K, and M–O) Higher magnification z stack of the boxed region in (D). Dopaminergic projection neurons in zebra finch (I–K) and mouse (M–O) retrogradely labeled with our eGFP-construct, after immunostaining for anti-GFP (I and M) and anti-tyrosine hydroxylase (J and N). Scale bars: 100 μm in (D), (G), (I)–(K), and (M)–(O) and 10 μm in (E).
Figure 2. Retrograde Transduction Kinetics in Zebra Finch HVCX Neurons.
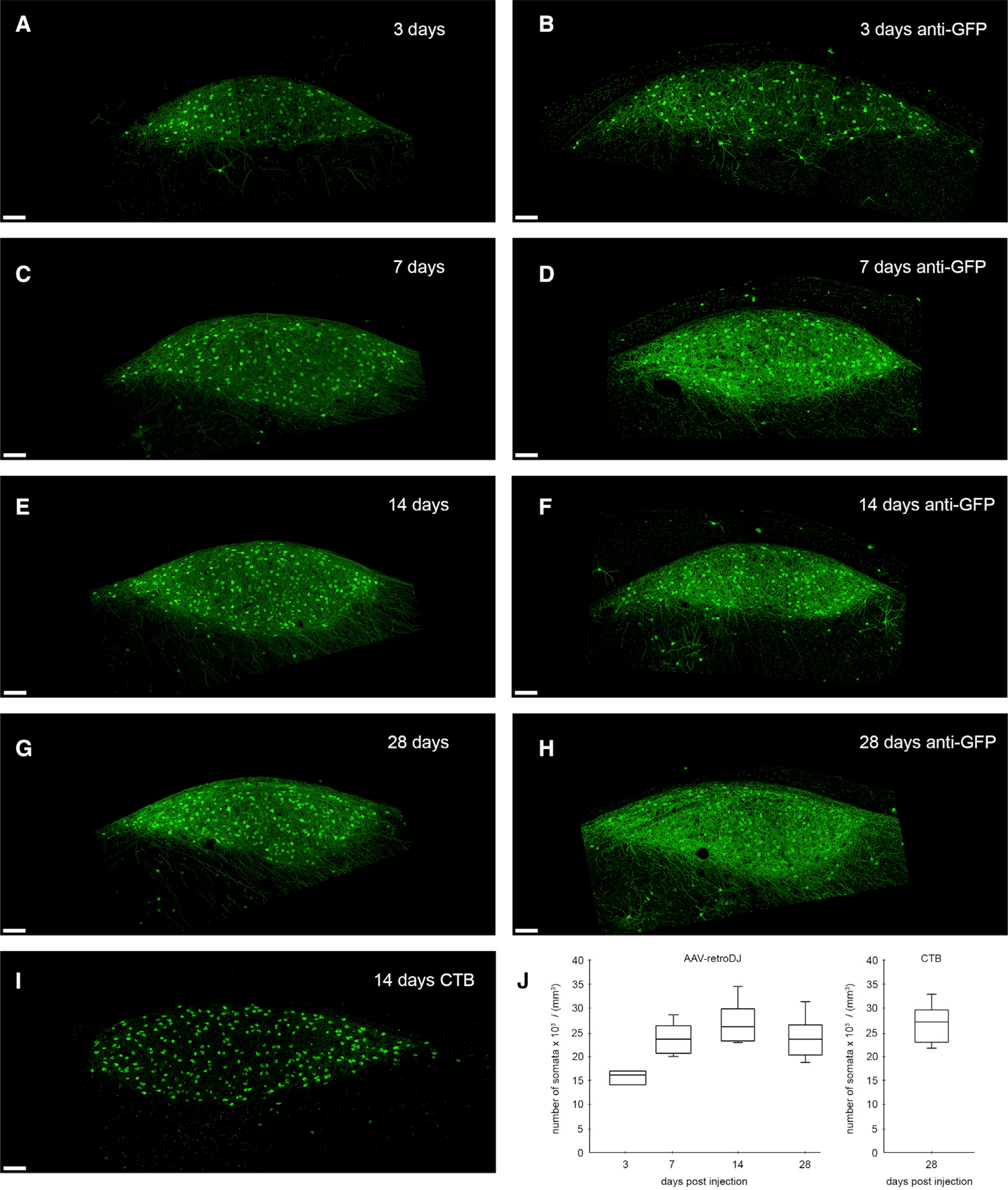
(A–H) We find strong retrograde expression of eGFP in area-X-projecting (HVCX) neurons as early as 3 days after injection of scAAV-2-DJ/9-hCMV-I/E-chI-eGFP-p(A) into area X. Confocal images show native eGFP signal post-delivery after (A) 3 days (C) 7 days, (E) 14 days, and (G) 28 days and confocal images (B, D, F, and H) of anti-GFP immunostainings of different sections in the same animal and hemisphere as in (A), (C), (E), and (G), respectively
(I) Comparative image of retrogradely labeled HVCX neurons 14 days after injections of cholera toxin B (CTB; a retrograde neural tracer) into area X. Note the strong eGFP signal in neurites of virally labeled cells as early as 3 days compared with the neural tracer.
(J) Number of retrogradely labeled HVCX somata at 3, 7, 14, and 28 days post-delivery into area X (based on native eGFP signal) compared with 14 days labeling with the retrograde tracer (boxplots indicating “minimum,” first quartile [Q1], median, third quartile [Q3], and “maximum” from N = 8 hemispheres per time point except for 3 days from N = 6 hemispheres). Scale bars, 100 μm.
RESULTS
Fast and Efficient Retrograde Transduction of Songbird Projection Neurons
First, we examined the transgene expression kinetics and stability of our viral construct. We assessed expression performance in the corticostriatal connection from nucleus HVC to area X. HVC, the songbird vocal premotor cortex analog, is a major hub during vocal learning, integrating information from auditory (Mooney, 2014), dopaminergic (Tanaka et al., 2018), and premotor (Roberts et al., 2008) afferents. HVC includes three major neuron populations: local interneurons; HVCRA neurons that project to the robust nucleus of the arcopallium (RA), which innervates vocal and respiratory motor neurons; and HVCX neurons that project to area X, the basal ganglia part of a corticobasal ganglia-thalamic loop involved in song learning (Dutar et al., 1998). HVCX neurons are analogous to mammalian corticostriatal projection neurons, which are involved in motor learning in mammals (Di Filippo et al., 2009).
We found robust native fluorescence of enhanced green fluorescent protein (eGFP) in HVCX neurons as early as 3 days post-injection of scAAV-2-DJ/9-hCMV-I/E-chI-eGFP-p(A) into area X (Figures 2A and 2B). Abundant eGFP-filled neurons could be seen within HVC throughout four different time points ranging from 3 to 28 days post-injection (Figures 2A–2H). Three days post-injection, we detected eGFP labeling on average among 59% (15.6 × 103 somata/mm3, N = 6 hemispheres) of HVCX somata compared with retrograde labeling with cholera toxin B (CTB; average 26.5 × 103 somata/mm3, N = 8 hemispheres; Figures 2I and 2J), a commonly used and highly efficient retrograde tracer in zebra finches (Mandelblat-Cerf et al., 2014). Already 7 days post-injection, the average eGFP labeling density (23.5 × 103 somata/mm3, N = 8 hemispheres; Figures 2C and 2J) was not significantly different from the average CTB labeling density (two-sided unpaired t test, p = 0.11). Longer eGFP expression times of 14 and 28 days did not further increase eGFP labeling densities (averages 25.3 × 103 and 23.7 × 103 somata/mm3, respectively, with N = 8 hemispheres per time point; Figures 2E, 2G, and 2J), which remained comparable to the average CTB labeling density (two-sided unpaired t test, p = 0.18). These results suggest that the peak of native fluorescence of eGFP expression is reached already 7 days post-delivery and that there is no obvious decay of fluorescence or neurotoxicity for at least 4 weeks post-injection. We further characterized the onset and the stability of eGFP fluorescence signal in HVC between 2 and 28 days post-injection by tracking the position and the fluorescence intensity of individual cells using in vivo two-photon microscopy, revealing no signs of elevated cell death in labeled cells (Figure S1).
Transduction Success Rate and Specificity of Retrograde Projection Neuron Labeling in Songbirds
We assessed the transduction success rate of our eGFP-construct and found consistent transduction of HVCX neurons in 24 of 24 single-injected hemispheres (N = 12 birds) after a minimum of 7 days after virus injection into area X. For comparison, previous studies applying AAV vectors for retrograde transduction in songbirds have reported overall successful transduction (i.e., presence of any retrograde signal in injected hemispheres) in from 18% (one brain pathway showing retrograde eGFP expression in 2 of 11 injected hemispheres) to 43% of virus injections (Chen et al., 2019). To date, we have not encountered a single injection (n > 100) of our eGFP-construct without successful retrograde transduction of projection neurons after 7 days.
We further assessed the specificity of retrograde labeling. HVC and area X are anatomically well separated, so given that no extensive spread from the injection site was apparent (Figure S2B), viral particles could not have passively diffused from area X to HVC. In addition, the fact that this connection is unidirectional makes it safe to assume that all labeled neurites in HVC belong to HVCX neurons. Nonetheless, some studies report the possibility of transsynaptic spread of AAV vectors (Zhao et al., 2017; Zingg et al., 2017). To rule out any potential mislabeling of RA-projecting neurons, we performed co-injections of CTB into RA and found no double-labeled cells (Figure S2). To assess whether all labeled neurons are indeed HVCX neurons and to further exclude any potential mislabeling, we examined the morphological characteristics of eGFP-labeled HVC neurons.
The three main HVC neuron classes are morphologically well described and can be separated based on dendritic spine densities (Kornfeld et al., 2017). HVC interneurons are mainly aspiny, HVCRA projection neurons are moderately spiny (0.21 ± 0.07 spines/μm), and HVCX neurons have the highest dendritic spine densities (0.70 ± 0.13 spines/μm). We analyzed 17 randomly chosen dendritic branches of various lengths (total branch length of 396 μm) across N = 4 adult male zebra finches (>7 days post-injection). A subset of histological brain sections was further subjected to anti-GFP immunolabeling to capture any potentially low-transgene-expressing cells. Analyzed dendritic branch fragments had dendritic spine densities ranging from 0.47 to 1.75 spines/μm (average 1.03 ± 0.35 spines/μm, which compares favorably with the previously reported 0.70 ± 0.13 spines/μm [Kornfeld et al., 2017]), further indicating that eGFP-expressing cells are most likely all HVCX neurons. Albeit some previously reported overlap in spine density between X- and RA-projecting cells, we did not encounter a single cell that was obviously either aspiny (interneuron) or had a very low spine density (HVCRA neurons). To also exclude the possibility of unspecific transduction of non-neuronal cell types such as astrocytes, we performed anti-neuronal protein HuC/HuD (Hu-antigen C, Hu-antigen D) counterstains, a common neuronal marker. We found that all examined eGFP-labeled cells were also labeled with HuC/HuD antibody (N = 4 sections of four different birds), confirming their neuronal identity (Figure 3).
Figure 3. Retrograde Expression Is Restricted to Neuronal Cell Types.

(A) Confocal microscopy image of zebra finch HVC with retrogradely labeled HVCX neurons.
(B) Immunostaining with anti-HuC/HuD, a commonly used antibody against neurons.
(C) Overlay of eGFP signal (green) and HuC/HuD signal (magenta), showing no HVCX neurons unlabeled for HuC/HuD.
(D–F) Higher magnification of the boxed regions in (A)–(C). Scale bars: 100 μm in (A)–(C) and 20 μm in (D)–(F).
Visualization of Morphological Details in Retrogradely Labeled Projection Neurons
The weak transgene expression produced by previous retrograde vectors for songbirds typically results in fluorescent protein labeling that is often limited to somata and to small extensions of proximal neurites (Chen et al., 2019; Roberts et al., 2017). Using our eGFP-construct thus opens exciting possibilities for highly detailed morphological analysis of songbird projection neurons. In fact, the high expression levels of eGFP in HVCX neurons 7 days post-delivery allowed a remarkable visualization of dendritic morphological details. Adjacent to imaged dendrites, we found clearly distinguishable dendritic spines throughout HVC (Figure 1E) at sufficient detail to distinguish and trace different spine types (Figure 4).
Figure 4. Strong eGFP Expression Permissive for Reconstructing Dendritic Spine Morphologies.
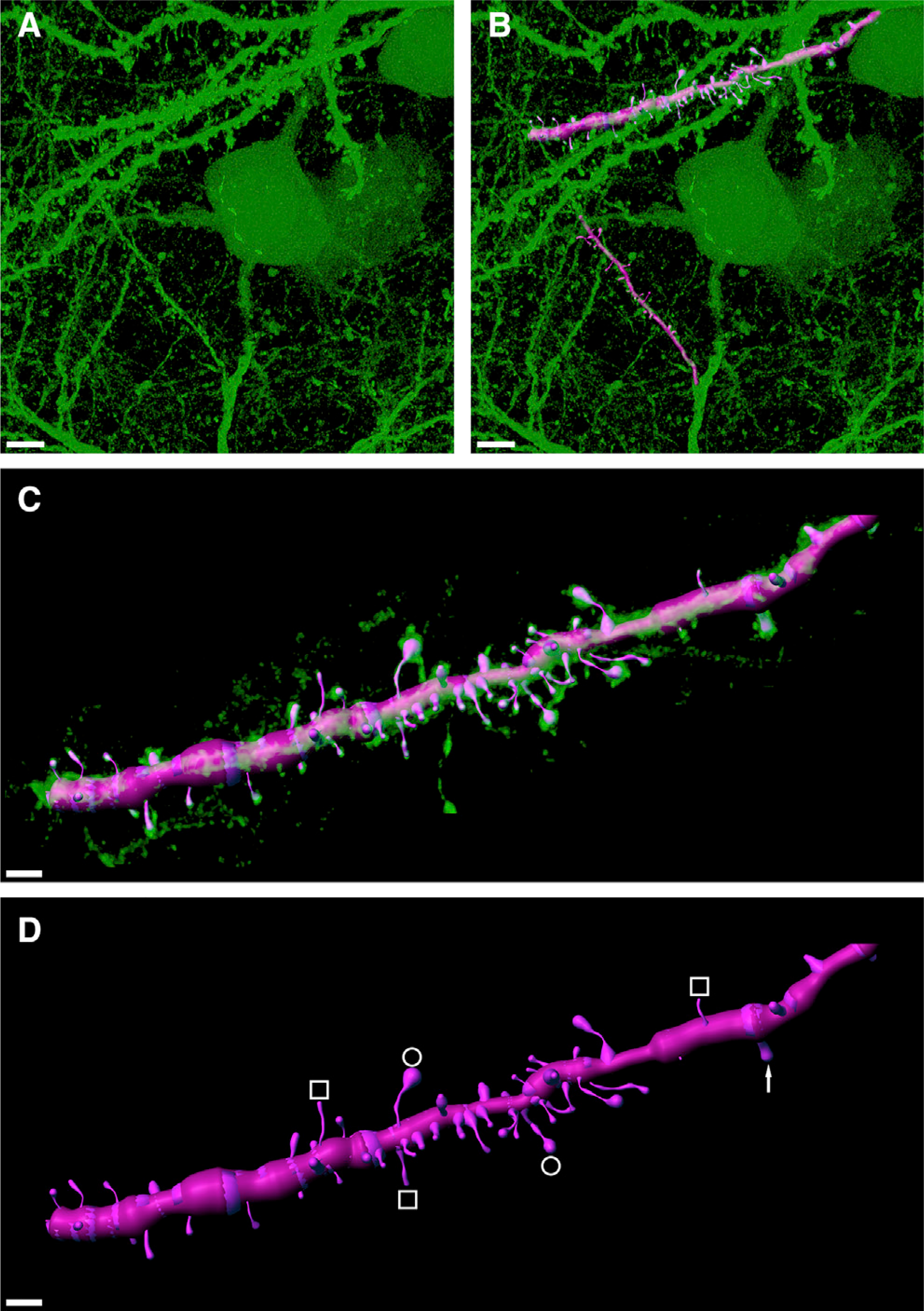
(A) High-magnification confocal image stack of retrogradely labeled HVCX neurons showing strong expression of eGFP in dendritic fragments.
(B) Digital 3D reconstruction (magenta overlay) of two exemplary dendritic fragments.
(C) Isolated and zoomed view of the upper dendritic fragment shown in (B), revealing high morphological detail.
(D) In the semi-automatically reconstructed dendritic fragment, various spine morphologies can be recognized including philopodia (white squares), mushroom heads (white circles), and stubby spines (white arrow). Scale bars: 5 μm in (A) and (B) and 2 μm in (C) and (D).
The extensive eGFP labeling makes our construct a promising candidate for in vivo imaging of morphological plasticity. To date, in vivo imaging of dendritic spine plasticity in songbirds has been achieved exclusively via lentivirus-mediated fluorescence labeling (Roberts et al., 2010, 2012), a tool that lacks retrograde transduction capabilities. Furthermore, the strong eGFP expression we demonstrate is highly beneficial for tissue processing techniques that require a strong initial fluorophore expression, including large-volume expansion (lattice) light-sheet microscopy (ExLSM [Düring et al., 2019], ExLLSM [Gao et al., 2019]) and whole-brain tissue clearing (Rocha et al., 2019).
Retrograde Expression in Canary Projection Neurons
The strong expression and efficient retrograde transport of our viral construct in both zebra finches and mice hint toward a broad species applicability of this vector. We therefore sought to test our construct on canaries (Serinus canaria). Canaries are seasonal breading songbirds that produce highly complex and flexible songs with extremely fast syllable repetitions, making this species a valuable model system for complex motor skill learning (Vellema et al., 2019). Additionally, canaries are a popular model for sex-hormone-induced neuroplasticity (Gahr, 2004). Seasonal changes in singing behavior in this species are correlated with sex hormone fluctuations and striking neural changes. To date, the application of viral vectors in the canary brain has been limited to the use of lentiviruses (Cohen et al., 2020), which do not retrogradely transduce projection neurons. Injections of our eGFP-construct into canary area X resulted in strong eGFP expression in HVCX neurons 7 days post-injection, confirming the rapid expression times observed in zebra finches (Figures 1F and 1G).
Retrograde Expression in Dopaminergic Projection Neurons
Similarly to the mammalian basal ganglia, area X is densely innervated by dopaminergic neurons of the ventral tegmental area-substantia nigra pars compacta complex (VTA/SNc), forming a continuous group of cells with diverse projection targets and innervation (Gale et al., 2008). In humans, the comparable dopaminergic system is crucially affected by neurodegenerative disorders such as Parkinson disease (Damier et al., 1999). Songbird VTA/SNcX neurons (traditionally referred to as VTAX neurons) play crucial roles in vocal learning, as they are responsible for encoding reward prediction errors associated with singing (Gadagkar et al., 2016).
In birds injected with our construct into area X, we observed retrograde eGFP labeling of VTA/SNcX neurons. To identify the labeled neuron types, we immunolabeled histological sections for tyrosine hydroxylase (TH), which is a good marker for dopamine and has been shown to label the majority of VTA/SNcX neurons (estimates ranging from 88% to 95% of VTA/SNcX neurons [Hisey et al., 2018; Person et al., 2008; Xiao et al., 2018]). We found that the majority of examined VTA/SNcX neurons were indeed positively labeled for TH (N = 4 birds; Figures 1H–1K), suggesting that these neurons are dopaminergic.
The strength of retrograde labeling of dopaminergic VTA/SNc projection neurons in zebra finches led us to investigate whether high labeling efficiency can also be observed in rodents. One of the most effective retrograde vectors in rodents, AAV-retro, only poorly transduces mouse dopaminergic projection neurons in the SNc (Tervo et al., 2016). Coincidently, we tested AAV-retro and found this construct to also be non-functional in zebra finches (N = 4 adult male zebra finches, single injections of the construct scAAV-2-retro-hCMV-I/E-chI-eGFP-p(A) into area X in both hemispheres), with post-delivery incubation times of 6 weeks. When we injected our eGFP-construct into the striatum of mice, a major projection target of the SNc, we found successful labeling of dopaminergic SNc projection neurons (confirmed with TH immunolabeling, N = 5 mice; Figures 1L–1O).
Brainbow Labeling of Projection Neuron Target Circuits
One common problem associated with studying the structure of local neural circuits is excessive labeling density, which can hinder the visual separation of adjacent neurons and their respective dendrites. One tool that can potentially overcome this problem is brainbow labeling (Lichtman et al., 2008). Brainbow labeling color-codes neurons via the relative expression ratios of diverse fluorescent proteins. One brainbow technique achieves diverse labeling colors via the selective uptake ratios of diverse viral particle types. This technique involves simply mixing and co-injecting three individual vector types encoding different chemical tags or fluorophores (Sakaguchi et al., 2018) (Figure 5A). Brainbow labeling in any of its variants has not yet been applied in songbirds.
Figure 5. Brainbow Labeling in Zebra Finch and Mouse.

(A) AAV-mediated brainbow labeling is achieved by injecting a mixture of three individual vectors encoding either eGFP, eCFP, or mRuby3. Each neuron ends up displaying a distinct color because of variable transduction of the three fluorescent proteins, largely depending on the number of vectors per cell per fluorophore.
(B) Brainbow labeling of locally transduced area X neurons allows for identification of individual neurons in a densely labeled z stack.
(C) High-magnification confocal images reveal consistent color labeling in dendritic fragments of area X neurons, including spines.
(D) Retrograde brainbow labeling in entorhinal cortex of neurons projecting to hippocampus in mouse. Scale bars: 100 μm in (B), 20 μm in (C), and 50 μm in (D).
In addition to the eGFP-expressing vector, we produced two additional vectors expressing spectrally distinct fluorescent proteins: enhanced cyan fluorescent protein (eCFP) and the red fluorescent protein mRuby3. Next, we injected the three aforementioned constructs in a ratio of 1:1:1 into area X of adult male zebra finches. Local expression within area X produced spectrally diverse neuron labeling (Figure 5B; Figure S3) with consistent and dense fluorophore expression patterns throughout neurons including morphological fine structures such as spines (Figure 5C). Although abundant brainbow labeling strategies exist for mice, we are not aware of any AAV-based retrograde labeling strategy that does not involve cre-recombination. We injected our eGFP-, eCFP-, and mRuby3-constructs in a ratio of 1:1:1 into the murine area CA1 of the dorsal hippocampus (N = 4). The injection of these three constructs produced spectrally diverse labeling of projection neurons in the mouse entorhinal cortex (Figure 5D). Unfortunately, retrograde brainbow labeling with our constructs did not achieve as satisfactory results in songbirds as in mice. Although all three fluorescent proteins are expressed retrogradely, the signal strength of each individual fluorophore is weakened when the three vectors are combined, thus failing to produce sufficient spectrally distinct labeling. Retrograde brainbow labeling with this technique can be potentially ameliorated by tinkering with the ratios and titers of the three vectors or by applying various modalities of signal amplification (Sakaguchi et al., 2018). This, however, would require extensive further testing, which is beyond the scope of this study.
Retrograde Expression of Calcium Sensors
In light of the obvious benefits of our construct for visualization of projection neuron circuits, we sought to investigate whether the expression of other genetic cargo could be mediated equally well. The packaging capacity of natural AAV serotypes is limited to a genome size of ~4.7 kb. Nevertheless, the absolute packaging limit of AAV vectors has been challenged, and one recent study found a brick-wall limit of 5.2 kb for AAV-8 (Wu et al., 2010). Because the maximum packaging size of an AAV likely also depends on the exact protein composition of the nucleocapsid, we decided to produce the same viral construct for GCaMP6f, with a total genome size of 5.286 kb, which is just over the reported maximum. Production was successful and yielded a physical viral titer of 5.9 × 1012 vg/mL (viral genomes per ml), which seemed promising to test for in vivo calcium imaging.
GCaMP is a genetically encoded calcium sensor (Nakai et al., 2001) consisting of a circular GFP, calmodulin (CaM), and a peptide chain (M13), which in its natural conformation shows only poor fluorescence. In the presence of calcium, CaM undergoes a structural change that entails a rapid increase in fluorescence. GCaMP6f was engineered for fast fluorescence dynamics and high calcium sensitivity, resulting in reliable single-spike detection at 50- to 75-ms inter-spike intervals (Chen et al., 2013).
We injected our GCaMP6f-construct into the RA of Bengalese finches (Lonchura striata var. domestica), another songbird species commonly used in birdsong research. We used a custom-built miniaturized fluorescence microscope (see STAR Methods) to image neuronal activity in vivo under isoflurane anesthesia. We imaged spontaneous activity in HVCRA neurons (Figure 6; Video S1), an HVC projection neuron population that generates precise temporal sequences (Hahnloser et al., 2002) during song production. In a field of view of (262.5 μm)2, we were able to detect 48 hypothetical spiking events in eight distinct HVCRA projection neurons over a time course of 100 s.
Figure 6. In Vivo Calcium Imaging in Retrogradely Labeled Bengalese Finch RA-Projecting (HVCRA) Neurons.

(A) Schematic of the scAAV-2-DJ/9-hCMV-I/E-chI-GCaMP6f-p(A) injection site and miniature microscope (finchscope) attachment in Bengalese finches.
(B) Fluorescence intensity traces (ΔF/F) for eight individual cells expressing GCaMP6f. All cells show clear spontaneous spike-like activity as indicated by a sudden increase in signal intensity.
(C) Single frame of in vivo calcium fluorescence movie under isoflurane anesthesia. Regions tracked over time are indicated in green. Bottom inset shows fluorescence signal of one cell over a period of 0.5 s (100 ms between snapshots). Left inset shows mean calcium transient (n = 43) in black and individual transients in gray (windows containing multiple events were excluded for visual clarity).
DISCUSSION
We present a viral construct for retrograde delivery of genetic cargo to projection neuron circuits in songbirds, with fast and robust transgene expression and high transduction efficiency. Our construct, scAAV-2-DJ/9-hCMV-I/E-chI-transgene-p(A), is suitable for studying the detailed connectivity and function of songbird corticostriatal and vocal motor pathways essential for vocal learning. Our construct also provides reliable access to probe the structure of cortical and of dopaminergic projection neuron circuits in both songbirds and mice. These findings illustrate the applicability of our vector in species and circuits that have been rather resistant to retrograde targeting with AAV vectors.
The current lack of reliable tools to target specific neuron populations in songbirds has been a possible contributor to the underrepresentation of songbirds as model species in medical and applied research. For targeted manipulations of projection neuron populations in the zebra finch, one serotype, AAV-9, has previously been used (Chen et al., 2019; Daliparthi et al., 2019; Hisey et al., 2018; Roberts et al., 2017). However, these vectors entail significant drawbacks, such as low transduction success rates, prolonged incubations times, and sparse and weak transgene expression, all of which limit their application. These limitations were not remedied by the use of the presumably stronger expressing sc variant of AAV-9 (Chen et al., 2019; Roberts et al., 2017), or by exploitation of the site-specific cre-recombinase system with the cre-recombinase vector injected at the projection target (Daliparthi et al., 2019; Hisey et al., 2018; Roberts et al., 2017). Unlike in songbirds, in mice, the cre system is very efficient and extremely low levels of cre are able to drive expression of otherwise silenced transcripts (Rothermel et al., 2013).
Despite previous limitations as retrograde tools for the songbird brain, AAV vectors still seemed worth exploration thanks to their extensive capsid variety, with more than 100 existing capsid variants. The inability of certain viral vectors to infect songbird tissue is likely owed to diverse factors. Such factors possibly include incompatibility of co-receptors for cellular access of the viral vector as well as differences in promoter sequences and intracellular physiology, both of which can impair transduction. A further potential limiting factor is a substantially different immune response in birds compared with mammals. Commonly, immune defense systems pose the first barrier when exploring viral tools. Accordingly, the low transduction success rate of AAV-9 vectors hints toward a possible strong immune response in zebra finches against this serotype. This hypothesis led us to explore serotypes focusing on low immune response, which is a trait of vectors engineered for human gene therapy. Human gene therapy vectors are required to have high transduction efficiency and low immunogenicity, both of which were highly desirable characteristics for the development of our songbird viral tool.
AAV serotypes have previously been selected in order to yield high efficiency and low immunogenicity (Grimm et al., 2008). In this study, the single prevailing capsid, the AAV-2, −8, and −9 chimera termed AAV-DJ, showed superior transduction characteristics, which motivated us to test this capsid. We found that the sc variant of AAV-DJ provides good local expression in songbird brain tissue, with good transduction characteristics, but unfortunately it shows no retrograde transduction. Although showing some potential for retrograde access to projection neurons (Castle et al., 2014; Hollis et al., 2008; Tervo et al., 2016), AAV vectors are traditionally not used for retrograde studies. One notable exception, the AAV-retro construct, has demonstrated a 10-fold higher retrograde transduction efficiency than AAV-DJ (Tervo et al., 2016). This motivated us to test AAV-retro, but we found this construct to be non-functional in zebra finches. We thus carefully examined the changes introduced in AAV-retro, which presumably led to increased efficacy of retrograde access. The critical changes appear to interact with the heparin sulfate proteoglycans (HSPG) binding domain (HBD) (DiMattia et al., 2012). The 10-mer insert between positions 587 and 588 (Figure 1A) probably both disrupts HBD’s functionality, as demonstrated by the reduced heparin binding affinity of AAV-retro, and creates a binding surface, which might improve vesicular trafficking or nuclear entry of viral particles (Tervo et al., 2016). Interestingly, the characteristics of the HBD were also carefully examined by the creators of the AAV-DJ construct (Grimm et al., 2008). One of their negative controls produced for comparison with AAV-DJ, AAV-DJ/9, included two point mutations in the HBD that disrupt heparin binding. These two point mutations seemed to reduce transduction efficiency of AAV-DJ/9 compared with AAV-DJ, but surprisingly also induced faster transduction kinetics. Faster transduction kinetics was a highly desirable characteristic for the songbird viral tool we were pursuing, which piqued our interest in the AAV-DJ/9 capsid variant. Coincidentally, the point mutations of the HBD in AAV-DJ/9 also rendered this serotype closer to its parental serotype AAV-9 (Grimm et al., 2008) (Figure 1A), which has been shown to have some potential for retrograde access in songbirds (Daliparthi et al., 2019; Hisey et al., 2018; Roberts et al., 2012; Xiao et al., 2018).
The exact composition of our construct combines many features that most likely all contribute constitutively to the highly efficient transgene expression and retrograde transport (Figures 1A and 1B). Clearly, the AAV-DJ/9 capsid structure of the HBD plays a major role for the viral access through axon terminals and the fast transduction kinetics. Nonetheless, this feature alone does not guarantee strong retrograde expression. The viral vector also needs to be transported by the transduced cells lysosomal system into the nucleus, where the genetic cargo has to be translated into messenger RNA (mRNA). The process of conversion into mRNA in the nucleus can be drastically accelerated by packaging two complementary copies of single-stranded DNA genomes in the scAAV vector instead of single-stranded DNA genomes. However, scAAV vectors have also been shown to induce a stronger immune response (Wu et al., 2012). When applying the scAAV-9-based vector (scAAV-9) for retrograde access in songbirds, the sc genome seems to neither have improved transduction success rate, transduction efficiency, nor expression kinetics (Chen et al., 2019). The disappointing results obtained with scAAV-9 might, however, be partially explained by the chosen promoter. Although a wide range of promoters have been applied to songbird brain tissue, we are not aware of a promoter that stands out in projection neurons. The scAAV-9 vector has been used in songbirds using a chicken beta actin hybrid (CBh) promoter (Chen et al., 2019). Nonetheless, CBh supposedly ensures strong expression through sc-vector-mediated transduction in neuron types that are also affected by human cytomegalovirus (hCMV) promoters (Gray et al., 2011). Given the poor transduction efficiency of scAAV-9-CBh, it could be that the hCMV immediate-early enhancer (hCMV-I/E) and/or the chimeric intron (chI) of our sc construct play crucial roles for retrograde transduction in projection neurons. The introduced intron might contribute to the improved transduction efficiency by regulating splicing of mRNA within the nucleus, which has been shown to improve nuclear export (Valencia et al., 2008). Further investigations would be necessary to fully elucidate the exact contributions of individual components to retrograde transport and transduction efficiency and kinetics.
In summary, in this work we present a viral construct for excellent access to specific projection neuron populations in songbirds. We demonstrated the suitability of our construct for in vivo imaging of calcium activity (Figure 6) and for detailed morphological analysis based on extensive axonal and dendritic labeling, using both single (Figure 4) and multi-color approaches (Figure 5). The high transduction success rates and great transduction efficiency hint at the potential for this tool to make a significant contribution to animal welfare by reducing the number of experimental animals required in future studies, as stated by the 3R (replace, reduce, refine) principles. Moreover, our tool will likely open exciting avenues of investigation into the structure and function of projection neuron populations and allow the study of the brain circuits underlying vocal learning in remarkable detail, including relevant dopaminergic inputs during song development.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Düring (dduering@ethz.ch).
Materials Availability
AAVs generated in this study are available from https://vvf.ethz.ch/.
Data and Code Availability
All datasets generated during and/or analyzed during the current study and all custom scripts and functions generated or used during the current study are available from the Lead Contact (dduering@ethz.ch) upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animals used in experiments had not previously been involved in other experiments or exposed to any drugs. Animal health was monitored daily and only healthy animals were used in experiments.
Zebra finches
Zebra finches (Taeniopygia guttata) used in this study were obtained from breeding colonies in Zurich, Switzerland, or Seewiesen, Germany, and kept on a 14h:10h light-dark cycle. All animals were male sex and ages ranged from 100 to 600 dph. Animal handling and all experimental procedures were conducted following the ethical principles and guidelines for animal experiments of Switzerland and Germany and approved by the respective authorities.
Bengalese finches
Bengalese finches (Lonchura striata var. domestica) used in this study were obtained from breeding colonies in Tokyo, Japan, and kept on a 14h:10h light-dark cycle. All animals were male sex and ages ranged from 100 to 300 dph. Animal handling and all experimental procedures were conducted following the ethical principles and guidelines for animal experiments of Japan and approved by the respective authorities.
Canaries
Canaries (Serinus canaria domestica) used in this study were obtained from breeding colonies in Seewiesen, Germany, and kept on a 14h:10h light-dark cycle. All animals were male sex and ages ranged from 300 to 600 dph. Animal handling and all experimental procedures were conducted following the ethical principles and guidelines for animal experiments of Germany and approved by the respective authorities.
Mice
Mice of the strain C57BL/6 were obtained from Charles River Germany and housed in the LASC animal husbandry at the University of Zurich on a 12h:12h light-dark cycle. All animals were male sex and ages ranged from 9 to 14 weeks. Animal handling and all experimental procedures were conducted following the ethical principles and guidelines for animal experiments of Switzerland and approved by the respective authorities.
METHOD DETAILS
Design of self-complementary adeno-associated virus vector plasmids
Self-complementary adeno-associated virus (AAV) vector plasmids (pscAAV) were constructed as previously described (McCarty et al., 2003; Wang et al., 2003). Briefly, the terminal resolution site (trs) and the packaging signal (D-sequence) from psub-2-CBA-WPRE (Paterna et al., 2000, 2004) were deleted by Bal I restriction digestion within the AAV serotype 2 (AAV-2) 5′ inverted terminal repeat (5′-ITR) resulting in the plasmid pscAAV-2-Δ3′-ITR. Subsequently, the multiple cloning site (MCS) of pBluescript II SK (+) (Stratagene) together with the AAV-2 3′-ITR and simian virus 40 late polyadenylation signal (SV40p(A)) containing fragment of psub-2-CMV-WPRE (Paterna et al., 2000) were inserted into pscAAV-2-Δ3′-ITR, resulting in the plasmid pscAAV-2-MCS-SV40p(A). In the AAV vector plasmids used here (Figure 1B), the human cytomegalovirus (hCMV) promoter/immediate-early enhancer (IE) of peGFP-N1 (Clontech) and the chimeric intron (chI) of pSI (Promega) were inserted into pscAAV-2-MCS-SV40p(A) resulting in pscAAV-2-hCMV-chI-SV40p(A).
The eGFP open reading frame (ORF) was amplified by PCR using pscAAV-2-hCMV-chI-floxedeGFP as the template DNA and primers 5′-ATACTAGTGCCACCATGGTGAGCAAGGGCG-3′ (forward) and 5′-TTGCGCGGCCGCTTACTTGTACAGCTCGTCCATG3′ (reverse). Amplicons were Spe I/Not I restriction digested and inserted into the Spe I/Not I restriction digested pscAAV-2-hCMV-chI-floxedeGFP to generate pscAAV-2-hCMV-chI-eGFP (eGFP vector plasmid). For construction of the mRuby3 vector plasmid (pscAAV-2-hCMV-chI-mRuby3-SV40p(A)), the mRuby3 ORF was amplified by PCR using Addgene #85146 as the template DNA and primers 5′-CATTACTAGTGTTTAAACACTCGAGGCTAGCGCCACCATGGTGTCTAAGG-3′ (forward) and 5′-TAGGCGCGCCTACGTACAATTGGGTACCTTACTTGTACAGCTCGTCCATG-3′ (reverse). The resulting PCR product was cut with SpeI and BsrGI and inserted into the Spe I and BsrG I sites of the eGFP vector plasmid.
For construction of the eCFP vector plasmid (pscAAV-2-hCMV-chI-eCFP-SV40p(A)), the eCFP ORF was isolated from plasmid pBV1 as Nhe I/Sac II fragment and inserted into the Sac II/Spe I opened eGFP vector plasmid.
For construction of the GCaMP6f vector plasmid (pscAAV-2-hCMV-chI-GCaMP6f-SV40p(A)), the GCaMP6f ORF was isolated by BglII/BstB I restriction digest from pssAAV-2-hSyn1-chI-GCaMP6f-WPRE-SV40p(A) (N-terminal part of GCaMP6f) and by BstB I/BssH II(blunt) restriction digest from pssAAV-2-hEF1a-dlox-GCaMP6f(rev)-dlox-WPRE-bGHp(A) (C-terminal part of GCaMP6f) and inserted into the BsrG I(blunt)/BglII restriction digested pscAAV-2-hCMV-chI-Lck_eGFP-SV40p(A) to generate pscAAV-2-hCMV-chI-GCaMP6f-SV40p(A) (GCaMP6f vector plasmid).
The identity of all constructs was confirmed by Sanger DNA sequencing and restriction endonuclease analyses.
Sequences of all viral vectors and their corresponding plasmids can be found in the repository of the Viral Vector Facility of the University of Zurich and ETH Zurich (https://vvf.ethz.ch/).
Production, purification, and quantification of self-complementary (sc) AAV vectors
Self-complementary (sc) AAV vectors were produced and purified as previously described (Paterna et al., 2004; Zolotukhin et al., 1999). Briefly, human embryonic kidney (HEK) 293 cells (Graham et al., 1977) expressing the simian virus (SV) large T-antigen (DuBridge et al., 1987) (293T) were transfected by polyethylenimine (PEI)-mediated cotransfection of AAV vector plasmids (providing the to-be packaged AAV vector genome, see above), the AAV helper plasmid pAAV-DJ/9; pAAV-DJ/9 providing the AAV serotype 2 rep proteins and the cap proteins of AAV-DJ/9) and adenovirus (AV) helper plasmids pBS-E2A-VA-E4 (Paterna et al., 2000) (providing the AV helper functions) in a 1:1:1 molar ratio.
At 120 to 168 hours post-transfection, HEK293T cells were collected and separated from their supernatant by low-speed centrifugation. AAV vectors released into the supernatant were PEG-precipitated over night at 4°C by adding a solution of polyethylenglycol 8000 (8% v/v in 0.5 M NaCl), and completed by low-speed centrifugation. Cleared supernatant was discarded and the pelleted AAV vectors resuspended in AAV resuspension buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.5). HEK293T cells were resuspended in AAV resuspension buffer and lysed by Bertin’s Precellys Evolution homogenizer in combination with 7 mL soft tissue homogenizing CK14 tubes (Bertin). The crude cell lysate was DENARASE (c-LEcta GmbH) treated (150 U/ml, 90 to 120 minutes at 37°C) and cleared by centrifugation (10 minutes at 17.000 g/4°C). The PEG-precipitated (1 hour at 3500 g/4°C) AAV vectors were combined with the cleared cell lysate and subjected to discontinuous density iodixanol (OptiPrep, Axis-Shield) gradient (isopycnic) ultracentrifugation (2 hours 15 minutes at 365’929 g/15°C). Subsequently, the iodixanol was removed from the AAV vector containing fraction by 3 rounds of diafiltration using Vivaspin 20 ultrafiltration devices (100’000 MWCO, PES membrane, Sartorius) and 1x phosphate buffered saline (PBS) supplemented with 1 mM MgCl2 and 2.5 mM KCl according to the manufacturer’s instructions. The AAV vectors were stored aliquoted at −80°C.
Encapsidated viral vector genomes (vg) were quantified using the Qubit 3.0 fluorometer in combination with the Qubit dsDNA HS Assay Kit (both Life Technologies). Briefly, 5 μl of undiluted (or 1:10 diluted) AAV vectors were prepared in duplicate. Untreated and heat-denaturated (5 minutes at 95°C) samples were quantified according to the manufacturer’s instructions. Intraviral (encapsidated) vector genome concentrations (vg/ml) were calculated by subtracting the extraviral (non-encapsidated; untreated sample) from the total intra- and extraviral (encapsidated and non-encapsidated; heat-denatured sample). All AAV vectors used in this study had vector genome concentrations between 3.8 × 1012 vg/ml and 7.4 × 1012 vg/ml.
Identity of encapsidated genomes were verified and confirmed by Sanger DNA-sequencing of amplicons produced from genomic AAV vector DNA templates (identity check).
Surgical procedure for viral vector delivery
Birds were treated with general analgesia 15 min prior to surgery by injecting Tramadol (5 mg/kg) in the pectoral muscle. After, birds were deeply anesthetized using an orally administered mixture of oxygen with 1%–3% isoflurane gas before being placed in a custom stereotaxic apparatus. After applying a topical anesthetic, a vertical incision was made in the skin over the skull, and a small craniotomy was performed at predetermined distances from the anatomical landmark lambda. For bi-lateral injections into Area X the following stereotaxic coordinates where used relative to lambda: anterior-posterior-axis (AP) +5.4 mm, medial-lateral-axis (ML) 1.6 mm, and dorsal-ventral-axis (DV) −3.2 mm, with an approximately 85 degree earbar-beak angle. For bi-lateral injections into RA of Bengalese finches we used the following coordinates relative to lambda: AP +5.1 mm, ML 2.0 mm, and DV −1.5 mm, with an approximately 180-degree earbar-beak angle. Following the craniotomy at the target locations, coordinates were confirmed by electrophysiological recordings using 1 MΩ tungsten sharp electrodes. Subsequently, 200 nL of undiluted viral vector were injected into each hemisphere using glass pipettes attached to either a Nanoject III (Drummond Scientific), with a constant flow rate of 1 nL per second, or a custom built pressure injector with a similar flow rate. To avoid backflow of viral vector and untargeted transduction, injection pipettes were raised about 200 μm relative to the injection site after content delivery and let sit for 5 minutes prior to ejection. Cranial holes were covered with quickcast, and the incision site in the skin was closed with tissue glue. At the end of surgery and the next day birds were treated with Meloxicam (1–2 mg/kg).
Mice were deeply anesthetized with Ketamine (90 mg/kg body weight)/Xylazine (8 mg/kg body weight), with buprenorphine (0.1 mg/Kg body weight, i.p.) given pre-emptive 20 minutes prior to anesthesia. Once pedal-reflex was absent, mice were mounted into a stereotaxic frame (Kopf), and 300 nL of undiluted viral vector were injected unillaterally into the dorsal striatum (DS) or into area CA1 of the dorsal hippocampus at a rate of 50 nl/min employing a 33G needle (nanofil 33 G, WPI) in a 10 μl syringe (nanofil, WPI) and a microinjector pump (WPI; UMP3 UltraMicroPump) directly mounted to the stereotaxic manipulator. The needle was left in place for an additional 5 minutes after the completed injection to avoid backflow of the viral vector. After the injection needle was removed, the incision was sutured. Animals received analgesic treatment for 3 days after surgery. The following stereotaxic coordinates where used relative to bregma: AP +1.0 mm, ML +1.8 mm, and DV −3.0 mm for DS; and AP −2.0 mm, ML +1.5 mm, and DV −1.6 mm for CA1.
Tissue preparation and immunohistochemistry
After isoflurane overdose, birds were transcardially perfused with PBS followed by 4% PFA (paraformaldehyde) in PBS (pH 7.4). Brains were extracted and post-fixed in 4% PFA overnight at 4°C, stored in PBS, and hemisected. Hemispheres were sectioned into either 30 or 60 μm-thick sagittal sections using a freezing sliding microtome (Leica Microsystems).
9 to 10 days after viral vector injection, mice were anesthetized with a lethal dose of Pentobarbital (200 mg/kg, i.p.), and transcardially perfused with saline followed by 4% PFA. Brains were extracted and post-fixed in 4% PFA for 24 hours. Sections were prepared with a Vibratome (Leica Microsystems) at 50 μm thickness.
Sections were either directly mounted onto glass slides with Vectashield antifade mounting medium (Vector Laboratories) and coverslipped for imaging of native fluorescence signal, or subjected to immunohistochemistry protocols, as described below.
Sections were washed with PBS, blocked in 10% normal goat serum/0.5% saponin/PBS for 90 minutes, and incubated overnight with primary antibodies in blocking solution at 4°C. Sections were then washed in 0.5% saponin/PBS, and incubated with secondary antibodies in blocking solution for 3 hours at room temperature. Finally, sections were washed in 0.5% saponin/PBS, mounted onto glass slides with Vectashield antifade mounting medium (Vector Laboratories), and coverslipped.
The following primary antibodies were used: chicken anti-GFP (1:1000, Aves, GFP-1020), mouse anti-HuC/HuD (1:20, Invitrogen, A21271), and rabbit anti-TH (1:500, Millipore, AB152). The following secondary antibodies were used: goat anti-chicken conjugated to Alexa Fluor 488 (1:500, Abcam, ab150169), goat anti-mouse conjugated to Alexa Fluor 555 (1:500, Invitrogen, A21422), and goat anti-rabbit conjugated to Alexa Fluor 555 (1:500, Invitrogen, A21428).
Image acquisition and analysis
Fluorescence images were acquired using a Leica SP8 confocal microscope (Leica Microsystems) using the 488 nm (eGFP/Alexa Fluor 488) and 561 nm (Alexa Fluor 555) laser lines for excitation. Spectral windows were set to 493 nm - 550 nm and 566 nm - 650 nm respectively and imaged using hybrid detectors (HyD). Low magnification images were acquired using a 20x/0.75 NA multi-immersion objective (HC PL APO CS2), and higher magnification images with a 63x/1.3 NA glycerol-immersion objective (HCX PL APO CS2). Pixel resolution and z section distances were set considering the nyquist criteria as defined at https://svi.nl/NyquistCalculator to ensure optimal post processing results.
When imaging the entire HVC multiple field of views where imaged with a 10% overlap and merged after acquisition using Leica’s imaging software (LASX). Prior to analysis of image stacks recorded with the higher magnification objective images were deconvolved using Huygens professional (SVI). Spine reconstructions and density measurements were performed with Imaris 9.2.1 (Bitplane) using the semi-automatic filament tracing function. Soma counts and densities were performed using Imaris’ spot tool with user defined thresholds. Sections for analysis were selected in a way that they roughly represent the same anatomical location (i.e., similar medio-lateral positions), and we performed soma counts in standardized volumes of 300 × 300 × 30 μm3 to accommodate for slight variations in HVC size or section thickness.
Calcium imaging
A custom-built, miniaturized fluorescence microscope was used to capture the temporal dynamics of calcium induced GFP signal from HVC projection neurons retrogradely transduced with our GCaMP6f-construct. The miniaturized microscope (named “finchscope”) was built in-lab according to the previously described manufacturing protocol (Liberti et al., 2017).
In order to image fluorescence signals, a cranial window was prepared in deeply anesthetized birds (by oral administration of a mixture of oxygen with 1%–3% isoflurane) as follows. The skull and dura over HVC were removed, and a ~3 mm hole on the skull just above HVC was made using a dental drill and scalpel. A round cover glass (3-mm diameter, CS-3R-0, Warner Instruments) was placed on the brain surface and fixed to the skull with tissue adhesive (Gluture, World Precision Instruments). The edge of glass window was secured to the skull with dental cement (PalaXpress, Kulzer). We then recorded spontaneous calcium activity by placing the finchscope just above the glass window with a stereotaxic manipulator.
Movies were recorded with the finchscope at 30 FPS and (0.75 μm)2 pixel size via a video capture device (GV-USB2, I-O DATA) and a recording software (AMcap), and were processed with in-house developed MATLAB (Mathworks) scripts as follows. First, we temporally smoothed each pixel using a 5-frame moving average. Next, we removed wide field intensity fluctuations by dividing each frame by a low-pass-filtered version of itself. For further analysis, we re-expressed each pixel in units of relative changes in fluorescence, given by ΔF(t)/F0 = (F(t) − F0)/F0, where F0 is the mean pixel value calculated by averaging over the entire movie. To obtain activity traces and spatial filters corresponding to the imaged cells, we applied an established automated signal extraction method to the preprocessed movie (Mukamel et al., 2009). Finally, we manually verified all extracted cells and discarded false positives.
In vivo two-photon imaging
In vivo two-photon imaging procedures were approved by the government of Upper Bavaria (ROB-55.2–2532.Vet_02–18-142). While experimental animals were under anesthesia using isoflurane inhalation, our GFP-construct was injected into Area X of adult male zebra finches, and a chronic cranial window with a headpiece was placed on the brain surface above HVC immediately after viral injection.
Imaging was carried out with a two-photon microscope (MOM, Sutter instruments) coupled to a pulsed Ti:Sapphire laser (Mai Tai DeepSee, Spectra-Physics) and controlled by ScanImage 5.4. A constant average power (56 mW) was applied on the brain tissue. Images were acquired using a 25x/0.95 NA water immersion objective lens (Leica) and photomultiplier tubes (Hamamatsu).
Time lapse images were acquired 2, 3, 7, 14, and 28 days post-injection (dpi). During imaging sessions, birds were anesthetized using isoflurane inhalation and head-fixed under the two-photon microscope objective. Images were acquired using ScanImage software at 1024×1024 pixels and analyzed using ImageJ. The cells on each dpi were manually identified and recorded with the ROI manager (ImageJ). Cell matching between different imaging sessions was done manually by visual inspection. Fluorescence intensity of cells was calculated by subtracting the background intensity from the mean pixel intensity within cell somata. Fluorescence intensity values were normalized to a range of values between 0 and 1. The pairwise distances between cells were calculated using the coordinates obtained by the ROI manager (ImageJ). Fluorescence intensities and the pairwise distances of matched cells were then compared between different dpi imaging sessions.
QUANTIFICATION AND STATISTICAL ANALYSIS
For quantifications and analyses we used MATLAB (Mathworks) unless stated otherwise. Quantifications are based on mean ± SD and information on sample size is indicated in each figure legend containing plots and/or in appropriate places in the text. Statistical significance analyses are performed using two-sided, unpaired t tests as indicated in the text. We did not perform any analysis to determine whether the data met assumptions of the statistical approach.
Supplementary Material
Supplementary Fig. 1 | In vivo two-photon imaging of retrogradely labeled HVCX neurons. Related to Figure 2. a, Time lapse in vivo two-photon images (top view) of eGFP-positive HVCX zebra finch neurons (maximum intensity projections of z-stacks) at 2, 3, 7, 14, and 28 days post-injection (dpi) with our GFP-construct. Middle row shows higher magnification views of the corresponding boxed regions in the upper row images. Bottom row shows scatter plots representing the 3D locations of HVCX neurons in the volumes corresponding to the regions of interest shown in the middle row. Neurons appearing on different time slices (dpi imaging sessions) are indicated by different colors. b, Time course of the appearance of eGFP positive cells in HVC across imaging sessions at different dpi. The minor fluctuations in cell numbers, can be attributed to imaging technicalities, where cells move into and out of the imaging region due to physiological movement of the tissue, or are covered by the growth of blood vessels and capillaries, and to the normal turnover of cells in songbird HVC. c, Changes in fluorescence intensity of eGFP-positive cells across diverse pairs of time slices (Mean +/− SD of all visible cells in one animal). The fluorescence intensity fluctuates over time, which is to be expected as it depends mainly on the activity of the promoter, but does seem stable as it does not drift in a particular direction. d, Changes in pairwise distances among eGFP-positive cells across diverse pairs of time slices, illustrating the spatio-temporal dynamics of neural tissue (Mean +/− SD of all visible cells in one animal). Scale bars: a, 100 μm (upper row); 50 μm (middle row). L, Lateral; C, Caudal.
Supplementary Fig. 2 | Retrograde expression of eGFP in HVCx neurons does not overlap with retrograde labelling of RA neurons. Related to Figure 3. We simultaneously injected birds with Cholera Toxin B linked to Alexa Fluor-555 (AF555) into RA and our eGFP-construct into Area X, and examined the brains 7 days post-injection for signal overlap in HVC. a, Schematic of the virus injection site (green) and the tracer injection site (red). b, Low magnification overview tile scan showing the eGFP signal at the virus injection site (Area X, green), the AF555 signal at the tracer injection site (RA, red), and both signals in HVC. c-e, Higher magnification z-stacks of the boxed region in b, showing maximum intensity projections of the AF555, eGFP, and the overlapping signals, respectively. Scale bars: 50 μm.
Supplementary Fig. 3 | Local brainbow expression in Area X produces spectrally diverse labelling. Related to Figure 5. a, We sampled 752 brainbow labelled neurons in Area X to quantify spectral diversity. b, Ternary plot showing the relative ratios of mRuby3 (red), eGFP (green) and eCFP (blue) for each neuron.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| chicken anti-GFP | Aves | Cat#GFP-1020, RRID:AB_10000240 |
| mouse anti-HuC/HuD | Invitrogen | Cat#A21271, RRID:AB_221448 |
| rabbit anti-TH | Millipore | Cat#AB152, RRID:AB_2201407 |
| goat anti-chicken conjugated to Alexa Fluor 488 | Abcam | Cat#ab150169, RRID:AB_2636803 |
| goat anti-mouse conjugated to Alexa Fluor 555 | Invitrogen | Cat#A21422, RRID:AB_141822 |
| goat anti-rabbit conjugated to Alexa Fluor 555 | Invitrogen | Cat#A21428, RRID:AB_141784 |
| Bacterial and Virus Strains | ||
| scAAV-2-DJ/9-hCMV-I/E-chI-eGFP-p(A) | this study, VVF | Cat#v65 |
| scAAV-2-DJ/9-hCMV-I/E-chI-GCaMP6f-p(A) | this study, VVF | Cat#v305 |
| scAAV-2-DJ/9-hCMV-I/E-chI-eCFP-p(A) | this study, VVF | Cat#v293 |
| scAAV-2-DJ/9-hCMV-I/E-chI-mRuby3-p(A) | this study, VVF | Cat#v286 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Vectashield antifade mounting medium | Vector Laboratories | Cat#H-1000–10 |
| PBS | FisherScientific | Cat#10010056 |
| Saponin | FisherScientific | Cat#ICN10285525 |
| PFA | CarlRoth | Cat#P087 |
| Isoflurane | Merck | Cat#Y0000858 |
| DENARASE | c-LEcta GmbH | Cat#20804 |
| Optiprep | Axis-Shield | Cat#1114542 |
| Buprenorphinum (Buapaq) | Streuli Pharma AG | Pharmacode 6563336 |
| Xylazin | Streuli Pharma AG | Pharmacode 6586900 |
| Ketanarkon | Streuli Pharma AG | Pharmacode 6586716 |
| CTB-488 | Thermo Fisher Scientific | Cat#C34775 |
| CTB-555 | Thermo Fisher Scientific | Cat#C22843 |
| Novaminsulfon | bela-pharm | Cat#799285 |
| Minocain | bela-pharm | Cat#799294 |
| Metacam | Boehringer Ingelheim Vetmedica | Cat#796617 |
| Critical Commercial Assays | ||
| Qubit dsDNA HS Assay Kit | Life Technologies/Thermo Fisher Scientific | cat#Q33230 |
| Experimental Models: Cell Lines | ||
| HEK293T | Clontech/Takara | 632273 |
| Experimental Models: Organisms/Strains | ||
| Mus musculus | Charles River | strain#C57BL/6-Crl1 |
| Software and Algorithms | ||
| MATLAB | mathworks | RRID:SCR_001622 |
| ImageJ | NIH | RRID:SCR_003070 |
| Huygens professional | SVI | RRID:SCR_014237 |
| Imaris | bitplane | RRID:SCR_007370 |
| AMcap | Genericom | N/A |
| ScanIamge | vidriotechnologies | RRID:SCR_014307 |
Highlights.
AAV/DJ variant for fast retrograde transduction in songbirds
Efficient retrograde expression of eGFP as early as 3 days post-injection
Retrograde access to dopaminergic pathways in mice and songbirds
Retrograde expression of GCaMP6f and brainbow labeling in songbirds
ACKNOWLEDGMENTS
This work was supported by European Union’s Horizon 2020 Marie Skłodowska-Curie grant 750055 (to D.N.D.); ETH grants (Switzerland) ETH-42 15-1 and ETH-20 19-01 (to R.H.R.H. and B.F.G.); NIH grant (US), BRAIN Initiative: Targeted BRAIN Circuits Projects, “Neural sequences for planning and production of learned Vocalizations” (to R.H.R.H. and M.D.R.); SNSF Sinergia grant (Switzerland) CRSII5-173721 (to B.F.G.); Swiss Data Science Center grant (Switzerland) C17–18 (to B.F.G.); and MEXT/JSPS, KAKENHI grants (Japan) 17H06380 and 17H01015 (to K.O.) and 19KT0023 (to R.O.T.). Part of the imaging was performed at the Center for Microscopy and Image Analysis, University of Zurich. Some figures were generated with the help of the Scientific Illustration and Visual Communication center of the University of Zurich. The AAV-DJ/9 helper plasmid was a kind gift from Mark A. Kay (Department of Pediatrics & Genetics, Stanford University). The plasmid pBV1 was a kind gift from Bernd Vogt (Institute of Virology, University of Zurich). We thank Veronika Bednarova and Benedikt Grothe (Max-Planck Institute of Neurobiology) for preliminary experiments in mice.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108364.
REFERENCES
- Bolhuis JJ, Okanoya K, and Scharff C (2010). Twitter evolution: converging mechanisms in birdsong and human speech. Nat. Rev. Neurosci. 11, 747–759. [DOI] [PubMed] [Google Scholar]
- Callaway EM (2008). Transneuronal circuit tracing with neurotropic viruses. Curr. Opin. Neurobiol. 18, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Gershenson ZT, Giles AR, Holzbaur ELFF, and Wolfe JH (2014). Adeno-associated virus serotypes 1, 8, and 9 share conserved mechanisms for anterograde and retrograde axonal transport. Hum. Gene Ther. 25, 705–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Puzerey PA, Roeser AC, Riccelli TE, Podury A, Maher K, Farhang AR, and Goldberg JH (2019). Songbird Ventral Pallidum Sends Diverse Performance Error Signals to Dopaminergic Midbrain. Neuron 103, 266–276.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Shen J, Semu D, Leman DP, Liberti WA III, Perkins LN, Liberti DC, Kotton DN, and Gardner TJ (2020). Hidden neural states underlie canary song syntax. Nature 582, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daliparthi VK, Tachibana RO, Cooper BG, Hahnloser RHR, Kojima S, Sober SJ, and Roberts TF (2019). Transitioning between preparatory and precisely sequenced neuronal activity in production of a skilled behavior. eLife 8, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damier P, Hirsch EC, Agid Y, and Graybiel AM (1999). The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain 122, 1437–1448. [DOI] [PubMed] [Google Scholar]
- Di Filippo M, Picconi B, Tantucci M, Ghiglieri V, Bagetta V, Sgobio C, Tozzi A, Parnetti L, and Calabresi P (2009). Short-term and long-term plasticity at corticostriatal synapses: implications for learning and memory. Behav. Brain Res. 199, 108–118. [DOI] [PubMed] [Google Scholar]
- DiMattia MA, Nam H-J, Van Vliet K, Mitchell M, Bennett A, Gurda BL, McKenna R, Olson NH, Sinkovits RS, Potter M, et al. (2012). Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 86, 6947–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, and Calos MP (1987). Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düring DN, Rocha MD, Dittrich F, Gahr M, and Hahnloser RHR (2019). Expansion Light Sheet Microscopy Resolves Subcellular Structures in Large Portions of the Songbird Brain. Front. Neuroanat. 13, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutar P, Vu HM, and Perkel DJ (1998). Multiple cell types distinguished by physiological, pharmacological, and anatomic properties in nucleus HVc of the adult zebra finch. J. Neurophysiol. 80, 1828–1838. [DOI] [PubMed] [Google Scholar]
- Gadagkar V, Puzerey PA, Chen R, Baird-Daniel E, Farhang AR, and Goldberg JH (2016). Dopamine neurons encode performance error in singing birds. Science 354, 1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahr M (2004). Hormone-dependent neural plasticity in the juvenile and adult song system: what makes a successful male? Ann. N Y Acad. Sci. 1016, 684–703. [DOI] [PubMed] [Google Scholar]
- Gale SD, Person AL, and Perkel DJ (2008). A novel basal ganglia pathway forms a loop linking a vocal learning circuit with its dopaminergic input. J. Comp. Neurol. 508, 824–839. [DOI] [PubMed] [Google Scholar]
- Gao R, Asano SM, Upadhyayula S, Pisarev I, Milkie DE, Liu T-L, Singh V, Graves A, Huynh GH, Zhao Y, et al. (2019). Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, and Nairn R (1977). Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36, 59–74. [DOI] [PubMed] [Google Scholar]
- Gray SJ, Foti SB, Schwartz JW, Bachaboina L, Taylor-Blake B, Coleman J, Ehlers MD, Zylka MJ, McCown TJ, and Samulski RJ (2011). Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 22, 1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, and Kay MA (2008). In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnloser RHR, Kozhevnikov AA, and Fee MS (2002). An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature 419, 65–70. [DOI] [PubMed] [Google Scholar]
- Hauser MD, Chomsky N, and Fitch WT (2002). The faculty of language: what is it, who has it, and how did it evolve? Science 298, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Hisey E, Kearney MG, and Mooney R (2018). A common neural circuit mechanism for internally guided and externally reinforced forms of motor learning. Nat. Neurosci. 21, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis ER II, Kadoya K, Hirsch M, Samulski RJ, and Tuszynski MH (2008). Efficient retrograde neuronal transduction utilizing self-complementary AAV1. Mol. Ther. 16, 296–301. [DOI] [PubMed] [Google Scholar]
- Kornfeld J, Benezra SE, Narayanan RT, Svara F, Egger R, Oberlaender M, Denk W, and Long MA (2017). EM connectomics reveals axonal target variation in a sequence-generating network. eLife 6, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DD, Wickersham IR, and Callaway EM (2008). Retrograde tracing with recombinant rabies virus reveals correlations between projection targets and dendritic architecture in layer 5 of mouse barrel cortex. Front. Neural Circuits 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti WA, Perkins LN, Leman DP, and Gardner TJ (2017). An open source, wireless capable miniature microscope system. J. Neural Eng. 14, 045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, and Sanes JR (2008). A technicolour approach to the connectome. Nat. Rev. Neurosci. 9, 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelblat-Cerf Y, Las L, Denisenko N, and Fee MS (2014). A role for descending auditory cortical projections in songbird vocal learning. eLife 3, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes AK, Reilly S, and Morgan AT (2015). Neural correlates of childhood language disorder: a systematic review. Dev. Med. Child Neurol. 57, 706–717. [DOI] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, and Samulski RJ (2003). Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 10, 2112–2118. [DOI] [PubMed] [Google Scholar]
- Mooney R (2009). Neural mechanisms for learned birdsong. Learn. Mem. 16, 655–669. [DOI] [PubMed] [Google Scholar]
- Mooney R (2014). Auditory–vocal mirroring in songbirds. Philos. Trans. R. Soc. B Biol. Sci. 369, 20130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, and Schnitzer MJ (2009). Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, and Imoto K (2001). A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141. [DOI] [PubMed] [Google Scholar]
- Paterna J-C, Moccetti T, Mura A, Feldon J, and Büeler H (2000). Influence of promoter and WHV post-transcriptional regulatory element on AAVmediated transgene expression in the rat brain. Gene Ther. 7, 1304–1311. [DOI] [PubMed] [Google Scholar]
- Paterna J-C, Feldon J, Büeler H, and Büeler H (2004). Transduction profiles of recombinant adeno-associated virus vectors derived from serotypes 2 and 5 in the nigrostriatal system of rats. J. Virol. 78, 6808–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person AL, Gale SD, Farries MA, and Perkel DJ (2008). Organization of the songbird basal ganglia, including area X. J. Comp. Neurol. 508, 840–866. [DOI] [PubMed] [Google Scholar]
- Rapin I, and Dunn M (2003). Update on the language disorders of individuals on the autistic spectrum. Brain Dev. 25, 166–172. [DOI] [PubMed] [Google Scholar]
- Roberts TF, Klein ME, Kubke MF, Wild JM, and Mooney R (2008). Telencephalic neurons monosynaptically link brainstem and forebrain premotor networks necessary for song. J. Neurosci. 28, 3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Tschida KA, Klein ME, and Mooney R (2010). Rapid spine stabilization and synaptic enhancement at the onset of behavioural learning. Nature 463, 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Gobes SMH, Murugan M, Ölveczky BP, and Mooney R (2012). Motor circuits are required to encode a sensory model for imitative learning. Nat. Neurosci. 15, 1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Hisey E, Tanaka M, Kearney MG, Chattree G, Yang CF, Shah NM, and Mooney R (2017). Identification of a motor-to-auditory pathway important for vocal learning. Nat. Neurosci. 20, 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha MD, Düring DN, Bethge P, Voigt FF, Hildebrand S, Helmchen F, Pfeifer A, Hahnloser RHR, and Gahr M (2019). Tissue Clearing and Light Sheet Microscopy: Imaging the Unsectioned Adult Zebra Finch Brain at Cellular Resolution. Front. Neuroanat. 13, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum S, and Simon P (2016). Speech and language disorders in children: Implications for the Social Security Administration’s Supplemental Security Income Program (The National Academies Press; ). [PubMed] [Google Scholar]
- Rothermel M, Brunert D, Zabawa C, Díaz-Quesada M, and Wachowiak M (2013). Transgene expression in target-defined neuron populations mediated by retrograde infection with adeno-associated viral vectors. J. Neurosci. 33, 15195–15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi R, Leiwe MN, and Imai T (2018). Bright multicolor labeling of neuronal circuits with fluorescent proteins and chemical tags. eLife 7, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sun F, Li Y, and Mooney R (2018). A mesocortical dopamine circuit enables the cultural transmission of vocal behaviour. Nature 563, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-YY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh E, and Cohen A (1998). Language deficit with attention-deficit disorder: a prevalent comorbidity. J. Child Neurol. 13, 493–497. [DOI] [PubMed] [Google Scholar]
- Valencia P, Dias AP, and Reed R (2008). Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA 105, 3386–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellema M, Diales Rocha M, Bascones S, Zsebők S, Dreier J, Leitner S, Van der Linden A, Brewer J, and Gahr M (2019). Accelerated redevelopment of vocal skills is preceded by lasting reorganization of the song motor circuitry. eLife 8, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma H-I, Li J, Sun L, Zhang J, and Xiao X (2003). Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 10, 2105–2111. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yang H, and Colosi P (2010). Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Töpfer K, Lin S-W, Li H, Bian A, Zhou XY, High KA, and Ertl HC (2012). Self-complementary AAVs induce more potent transgene product-specific immune responses compared to a single-stranded genome. Mol. Ther. 20, 572–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Chattree G, Oscos FG, Cao M, Wanat MJ, and Roberts TF (2018). A Basal Ganglia Circuit Sufficient to Guide Birdsong Learning. Neuron 98, 208–221.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Jiang H-F, Zeng W-B, Shu Y, Luo M-H, and Duan S (2017). Anterograde Trans-Synaptic Tagging Mediated by Adeno-Associated Virus. Neurosci. Bull. 33, 348–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, and Zhang LI (2017). AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron 93, 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S, Byrne BJ, Mason E, Zolotukhin I, Potter M, Chesnut K, Summerford C, Samulski RJ, and Muzyczka N (1999). Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6, 973–985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 | In vivo two-photon imaging of retrogradely labeled HVCX neurons. Related to Figure 2. a, Time lapse in vivo two-photon images (top view) of eGFP-positive HVCX zebra finch neurons (maximum intensity projections of z-stacks) at 2, 3, 7, 14, and 28 days post-injection (dpi) with our GFP-construct. Middle row shows higher magnification views of the corresponding boxed regions in the upper row images. Bottom row shows scatter plots representing the 3D locations of HVCX neurons in the volumes corresponding to the regions of interest shown in the middle row. Neurons appearing on different time slices (dpi imaging sessions) are indicated by different colors. b, Time course of the appearance of eGFP positive cells in HVC across imaging sessions at different dpi. The minor fluctuations in cell numbers, can be attributed to imaging technicalities, where cells move into and out of the imaging region due to physiological movement of the tissue, or are covered by the growth of blood vessels and capillaries, and to the normal turnover of cells in songbird HVC. c, Changes in fluorescence intensity of eGFP-positive cells across diverse pairs of time slices (Mean +/− SD of all visible cells in one animal). The fluorescence intensity fluctuates over time, which is to be expected as it depends mainly on the activity of the promoter, but does seem stable as it does not drift in a particular direction. d, Changes in pairwise distances among eGFP-positive cells across diverse pairs of time slices, illustrating the spatio-temporal dynamics of neural tissue (Mean +/− SD of all visible cells in one animal). Scale bars: a, 100 μm (upper row); 50 μm (middle row). L, Lateral; C, Caudal.
Supplementary Fig. 2 | Retrograde expression of eGFP in HVCx neurons does not overlap with retrograde labelling of RA neurons. Related to Figure 3. We simultaneously injected birds with Cholera Toxin B linked to Alexa Fluor-555 (AF555) into RA and our eGFP-construct into Area X, and examined the brains 7 days post-injection for signal overlap in HVC. a, Schematic of the virus injection site (green) and the tracer injection site (red). b, Low magnification overview tile scan showing the eGFP signal at the virus injection site (Area X, green), the AF555 signal at the tracer injection site (RA, red), and both signals in HVC. c-e, Higher magnification z-stacks of the boxed region in b, showing maximum intensity projections of the AF555, eGFP, and the overlapping signals, respectively. Scale bars: 50 μm.
Supplementary Fig. 3 | Local brainbow expression in Area X produces spectrally diverse labelling. Related to Figure 5. a, We sampled 752 brainbow labelled neurons in Area X to quantify spectral diversity. b, Ternary plot showing the relative ratios of mRuby3 (red), eGFP (green) and eCFP (blue) for each neuron.
Data Availability Statement
All datasets generated during and/or analyzed during the current study and all custom scripts and functions generated or used during the current study are available from the Lead Contact (dduering@ethz.ch) upon reasonable request.


