Abstract
Natural isolates of the soil‐dwelling bacterium Bacillus subtilis form robust biofilms under laboratory conditions and colonize plant roots. B. subtilis biofilm gene expression displays phenotypic heterogeneity that is influenced by a family of Rap‐Phr regulatory systems. Most Rap‐Phr systems in B. subtilis have been studied independently, in different genetic backgrounds and under distinct conditions, hampering true comparison of the Rap‐Phr systems’ impact on bacterial cell differentiation. Here, we investigated each of the 12 Rap‐Phr systems of B.subtilis NCIB 3610 for their effect on biofilm formation. By studying single ∆rap‐phr mutants, we show that despite redundancy between the cell–cell communication systems, deletion of each of the 12 Rap‐Phr systems influences matrix gene expression. These Rap‐Phr systems therefore enable fine‐tuning of the timing and level of matrix production in response to specific conditions. Furthermore, some of the ∆rap‐phr mutants demonstrated altered biofilm formation in vitro and colonization of Arabidopsis thaliana roots, but not necessarily similarly in both processes, indicating that the pathways regulating matrix gene expression and other factors important for biofilm formation may be differently regulated under these distinct conditions.
Keywords: Bacillus subtilis, biofilm formation, matrix gene expression, Rap‐Phr, root colonization
Bacillus subtilis NCIB3610 contains 12 cell–cell signaling Rap‐Phr systems that influence cell differentiation. Here, we systematically investigate how each of these systems impacts biofilm formation by studying WT and the 12 single ∆rap‐phr mutants in different phenotypic assays. Our approach revealed that while all 12 Rap‐Phr systems affect matrix gene expression, only certain Rap‐Phr systems influence the development of in vitro biofilms and root colonization, the latter representing an ecologically relevant environment for this bacterium.

1. INTRODUCTION
In nature, biofilms are the predominant lifestyle of bacteria and are known as surface‐associated microbial communities embedded in a self‐produced matrix (Flemming & Wingender, 2010; Hall‐Stoodley et al., 2004; López et al., 2010). Biofilms have been widely studied since they represent a fascinating example of microbial development in response to environmental cues (O'Toole et al., 2000). Furthermore, studying biofilms is of special interest due to their detrimental impact in clinical and industrial settings (Di Pippo et al., 2018; Stewart, 2002) as well as their promising potential within the biotechnology industry (Blake et al., 2021; Singh et al., 2006). Regarding the latter, the gram‐positive bacterium Bacillus subtilis has in the last two decades gained interest due to its promising potential as a biocontrol agent within agriculture (Kiesewalter et al., 2021; Ongena & Jacques, 2008). In its natural habitat, the soil‐dwelling bacterium colonizes plants by forming a biofilm on the root (Bais et al., 2004; Beauregard et al., 2013; Chen et al., 2012). After successfully colonizing the root, B.subtilis exerts its plant‐beneficial properties, including directly promoting plant growth and protecting the plant against diseases (Blake et al., 2021). Furthermore, B. subtilis forms spores that are highly resistant to extreme environments (Piggot & Hilbert, 2004) facilitating easy formulation (Ongena & Jacques, 2008).
Bacillus subtilis can easily be isolated from the rhizosphere of plants (Fall et al., 2004), and a study performed by Chen et al. (2013) showed that the majority of natural strains isolated from the rhizosphere formed architecturally complex biofilms under laboratory conditions, indicating that biofilm formation is an important trait for B. subtilis to thrive in its natural habitat. In the laboratory, B. subtilis has long been studied using different kinds of biofilm models including colonies at the air‐agar interface and floating biofilms formed at the air–liquid interface, termed pellicles (Arnaouteli et al., 2021).
A prevalent feature of B. subtilis biofilms is that they display complex phenotypic heterogeneity, where genetically identical cells differentiate into distinct cell types in response to external cues (López & Kolter, 2010; Lopez et al., 2009). The variation in environmental conditions throughout the biofilm (Costerton et al., 1994) thereby leads to a heterogeneous population with different cell types performing distinct tasks and occupying different micro‐niches. The extracellular signals triggering cell differentiation include quorum‐sensing molecules, natural products, and nutrient availability that activate a set of sensor kinases (Arnaouteli et al., 2021; Mhatre et al., 2014). Once activated, the sensor kinases phosphorylate their respective master transcriptional regulators, Spo0A, DegU, and ComA, each of which activates different sets of genes (López & Kolter, 2010). The Spo0A pathway governs differentiation into matrix‐producing cells and sporulating cells. In response to external cues, one or more of five histidine kinases, KinA‐E, are activated which results in phosphorylation of Spo0F. Spo0F~P then transfers its phosphoryl group to Spo0B, which in turn transfers the phosphoryl group to and thereby activates Spo0A (Fujita et al., 2005; Jiang, Shao, et al., 2000). At low Spo0A~P levels, the genes involved in the synthesis of matrix components, exopolysaccharide (EPS) and TasA protein fiber, are expressed (Cairns et al., 2014; Fujita et al., 2005). These two matrix components are well known to be required for biofilm formation in vitro and on the plant root (Beauregard et al., 2013; Branda et al., 2006). When high levels of Spo0A~P are reached, genes involves in sporulation are expressed (Fujita et al., 2005). The DegU response regulator is phosphorylated by its cognate histidine kinase DegS. Studies have indicated that inhibition of flagellar rotation, as may take place upon contact with a surface, acts as a mechanical trigger to activate the DegS‐DegU two‐component signaling pathway (Cairns et al., 2013). At very low levels of DegU~P, genes related to swarming motility are expressed, while elevated levels of DegU~P induce exoprotease production and at the same time represses motility genes (Belas, 2013; Verhamme et al., 2007). Finally, the pheromone ComX activates the histidine kinase ComP, which phosphorylates ComA, resulting in the expression of genes involved in competence development and surfactin production (Comella & Grossman, 2005).
This regulatory network governing cell differentiation in B. subtilis is further regulated by a family of response regulator aspartyl‐phosphate (Rap) phosphatases and their associated phosphatase regulator (Phr) peptides (Perego, 2013). In the B. subtilis group, 80 distinct putative rap‐phr alleles have been identified with a strain having on average 11 rap genes (Even‐Tov et al., 2016). The abundance of Rap and Phr peptides is transcriptionally controlled in response to different cellular signals (Auchtung et al., 2005; Jarmer et al., 2001; Jiang, Grau, et al., 2000; Lazazzera et al., 1999; McQuade et al., 2001; Mueller et al., 1992; Ogura et al., 2001; Perego et al., 1994). The genes encoding the Rap‐Phr pairs are found as gene cassettes with the phr gene immediately downstream of the rap gene and the expression of these being transcriptionally coupled, with some phr genes also being transcribed independently of their cognate rap genes from promoters controlled by σH (McQuade et al., 2001; Pottahil & Lazazzera, 2003; Reizer et al., 1997). Some exceptions to this exist, for example, the rapB gene is not followed by an active peptide encoding gene (Perego et al., 1996). Moreover, some Rap proteins are regulated by Phr peptides encoded in other cassettes, for example, RapB and J are both controlled by PhrC (Parashar, Jeffrey, et al., 2013). When expressed, the Rap phosphatases exert their effect within the cell by either dephosphorylating Spo0F~P (thus hindering Spo0A phosphorylation) or inhibiting the DNA‐binding activity of ComA or DegU (Perego, 2013). In contrast, the product of the phr gene is secreted out of the cell through the Sec‐dependent export pathway and processed into mature five to six amino acid signaling peptides. At high cell density, the Phr peptides reach threshold concentrations at which they are transported back into the cell by the oligopeptide permease (Opp) (Perego, 2013; Pottahil & Lazazzera, 2003). Once within the cell, the Phr peptides will inhibit their cognate Rap proteins, thereby relieving the inhibition of the master regulators resulting in altered gene expression (Perego, 2013; Pottahil & Lazazzera, 2003). The Rap‐Phr systems thereby act as cell–cell signaling systems in B.subtilis, allowing the bacteria to respond to environmental changes only at sufficient cell densities.
As expected by the diversity and abundance of multiple Rap‐Phr systems regulating the activity of these three master regulators, the Rap phosphatases show high redundancy in their regulatory function: RapA, B, E, H, I, J, and P have been shown to dephosphorylate Spo0F~P, RapC, D, F, H, K, and P regulate ComA, while RapG has been shown to regulate the activity of DegU (Auchtung et al., 2006; Ogura & Fujita, 2007; Perego, 2013; Omer Bendori et al., 2015) (Table A1). Furthermore, RapI is involved in the regulation of mobile genetic elements, as it activates the propagation of the mobile genetic element that encodes it (Auchtung et al., 2005). The regulation of the master regulators by multiple Rap phosphatases allows the integration of diverse signals to control cell differentiation in response to different conditions. However, this overall overview of the Rap‐Phr signaling network in B. subtilis is based on studies where most Rap‐Phr systems have been tested in different genetic backgrounds and under distinct cultivation conditions (Perego, 2013). Additionally, previous investigations in B. subtilis have directed their study toward certain targets of Rap‐Phr regulation, with RapA and B being mostly studied for their impact on sporulation (Perego & Hoch, 1996), while RapC and F are involved in competence development (Bongiorni et al., 2005; Core & Perego, 2003). So far, only RapP has been demonstrated to impact biofilm formation (Parashar, Konkol, et al., 2013; Omer Bendori et al., 2015). We previously studied all 12 Rap‐Phr systems of the undomesticated strain B. subtilis NCIB 3610 in the same genetic background by following the relative abundance of all possible single and double ∆rap‐phr mutants as well as the wild‐type WT (79 strains) in populations subjected to different selective conditions. This study highlighted that the variability in Rap‐Phr systems affected the ability to compete in diverse environments (Gallegos‐Monterrosa et al., 2021).
In this study, we systematically investigated the contribution of each of the 12 Rap‐Phr systems in B. subtilis 3610 to biofilm formation. We assessed wild type (WT) and the 12 single ∆rap‐phr mutants for matrix gene expression and biofilm formation under different conditions. We found that all 12 mutants showed altered matrix gene expression compared with the WT. Furthermore, we observed that the Rap‐Phr modules affect not only in vitro biofilm formation but also the colonization of plant roots that represents an ecologically relevant environment.
2. MATERIALS AND METHODS
2.1. Bacterial strains and cultivation methods
Strains used in this study are listed in Table 1. The B. subtilis DK1042 strain (a natural competent derivative of the undomesticated NCBI 3610) (Konkol et al., 2013) was used as WT. The 12 single ∆rap‐phr mutants were previously created and contain an antibiotic resistance cassette in place of the rap‐phr gene pair (except for the markerless ∆rapB mutant) (Gallegos‐Monterrosa et al., 2021). For flow cytometry, the ∆rap‐phr mutants were transformed with the plasmid phy_mKATE2 harboring the mKATE gene under the control of the hyper‐spank promoter (which is constitutive due to the absence of lacI) and a chloramphenicol (Chl) resistance gene within the flanking regions of the amyE gene (van Gestel et al., 2014). Transformants were identified by selecting for Chl resistance, and double crossovers were verified by the loss of amylase activity. The resulting mKATE‐labelled ∆rap‐phr mutants were transformed with genomic DNA from B. subtilis TB373, which harbors the PtapA‐gfp reporter construct with a kanamycin (Km) resistance gene integrated at the sacI locus. Successful transformants with the reporter construct inserted into the sacI locus were identified by selecting for Km resistance. The resulting reporter strains were verified for reporter activity under the fluorescence stereomicroscope. Importantly, due to the presence of a Km resistance cassette in place of the corresponding rap‐phr gene pair in ∆rapA, C, D, I, J, and K, markerless versions of those rap‐phr mutants were used to obtain the reporter strains for the tapA‐sipW‐tasA operon for the flow cytometry analysis. For all experiments, strains were grown overnight in Lysogeny broth (LB; Lennox, Carl Roth; 10 g·L−1 tryptone, 5 g·L−1 yeast extract and 5 g·L−1 NaCl) at 37°C while shaking (220 rpm). For transformation and stock preparation, antibiotics were used at the following working concentrations: Km: 5 µg ml−1 and Chl: 5 µg·ml−1. For analyzing biofilm formation and matrix gene expression, strains were grown in MSgg (5 mmol·l−1 potassium phosphate [pH 7], 0.1 mol·L−1 3‐(N‐morpholino)propanesulfonic acid (MOPS) [pH 7], 2 mmol·L−1 MgCl2, 700 μmol·L−1 CaCl2, 100 μmol·L−1 MnCl2, 50 μmol·L−1 FeCl3, 1 μmol·L−1 ZnCl2, 2 μmol·L−1 thiamine, 0.5 % glycerol, and 0.5 % K‐glutamate). For root colonization assay, strains were grown in MSNg (5 mmol·L−1 potassium phosphate buffer [pH 7], 0.1 mol·L−1 MOPS [pH 7], 2 mmol·L−1 MgCl2, 50 μmol·L−1 MnCl2, 1 µmol·L−1 ZnCl2, 2 μmol·L−1 thiamine, 700 μmol·L−1 CaCl2, 0.2 % NH4Cl2, and 0.05 % glycerol).
TABLE 1.
Strains used in this study
| Name | Description | Reference |
|---|---|---|
| DK1042 | NCIB 3610 coml Q12I | Konkol et al. (2013) |
| TB499 | DK1042 rapA‐phrA::KmR | Gallegos‐Monterrosa et al. (2021) |
| TB575 | DK1042 ∆rapB | |
| TB396 | DK1042 ∆rapC‐phrC:: KmR | |
| TB315 | DK1042 ∆rapD:: KmR | |
| TB339 | DK1042 ∆rapE‐phrE::SpecR | |
| TB341 | DK1042 ∆rapF‐phrF::SpecR | |
| TB404 | DK1042 ∆rapG‐phrG::SpecR | |
| TB405 | DK1042 ∆rapH‐phrH::SpecR | |
| TB272 | DK1042 ∆rapI‐phrI::KmR | |
| TB274 | DK1042 ∆rapJ::KmR | |
| TB557 | DK1042 ∆rapK‐phrK::KmR | |
| TB435 | DK1042 ∆rapP‐phrP::MlsR | |
| TB588 | DK1042 ΔrapA‐phrA | |
| TB410.1 | DK1042 ΔrapC‐phrC | |
| TB513 | DK1042 ΔrapD | |
| TB444 | DK1042 ΔrapI‐phrI | |
| TB411.2 | DK1042 ΔrapJ | |
| TB587 | DK1042 ΔrapK‐phrK | |
| DTUB189 | DK1042 ΔrapA‐phrA, amyE::Physpank‐mKATE ChlR | This study |
| DTUB165 | DK1042 ∆rapB, amyE::Physpank‐mKATE ChlR | This study |
| DTUB190 | DK1042 ΔrapC‐phrC, amyE::Physpank‐mKATE ChlR | This study |
| DTUB191 | DK1042 ΔrapD, amyE::Physpank‐mKATE ChlR | This study |
| DTUB159 | DK1042 ΔrapE‐phrE::SpecR, amyE::Physpank‐mKATE ChlR | This study |
| DTUB160 | DK1042 ΔrapF‐phrF::SpecR, amyE::Physpank‐mKATE ChlR | This study |
| DTUB166 | DK1042 ΔrapG‐phrG::SpecR, amyE::Physpank‐mKATE ChlR | This study |
| DTUB167 | DK1042 ΔrapH‐phrH::SpecR, amyE::Physpank‐mKATE ChlR | This study |
| DTUB192 | DK1042 ΔrapI‐phrI, amyE::Physpank‐mKATE ChlR | This study |
| DTUB193 | DK1042 ΔrapJ, amyE::Physpank‐mKATE ChlR | This study |
| DTUB194 | DK1042 ΔrapK‐phrK, amyE::Physpank‐mKATE ChlR | This study |
| DTUB173 | DK1042 ΔrapP‐phrP::MlsR, amyE::Physpank‐mKATE ChlR | This study |
| TB34 | DK1042 amyE::Physpank‐gfp ChlR | Mhatre et al. (2017) |
| TB35 | DK1042 amyE::Physpank‐mKATE ChlR | Hölscher et al. (2016) |
| TB865 | DK1042 amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | Dragoš et al. (2018) |
| DTUB284 | DK1042 ∆rapA‐phrA amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB285 | DK1042 ∆rapB, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB286 | DK1042 ∆rapC‐phrC, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB287 | DK1042 ∆rapD, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB288 | DK1042 ∆rapE‐phrE::SpecR, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB289 | DK1042 ∆rapF‐phrF::SpecR, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB290 | DK1042 ∆rapG‐phrG::SpecR, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB291 | DK1042 ∆rapH‐phrH::SpecR, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB292 | DK1042 ∆rapI‐phrI, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB293 | DK1042 ∆rapJ, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB294 | DK1042 ∆rapK‐phrK, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| DTUB295 | DK1042 ∆rapP‐phrP::MlsR, amyE::Physpank‐mKATE ChlR, sacA::PtapA‐gfp KmR | This study |
| TB373 | DK1042 sacA::PtapA‐gfp KmR | Gallegos‐Monterrosa et al. (2016) |
2.2. Microscopy imaging
All images were acquired with an Axio Zoom V16 stereomicroscope (Carl Zeiss, Germany) equipped with a Zeiss CL 9000 LED light source and an AxioCam MRm monochrome camera (Carl Zeiss, Germany).
2.3. Biofilm formation assay
For biofilm formation on a solid surface, 7 µl overnight cultures were spotted on MSgg supplemented with 1.5 % agar. For pellicle biofilm formation at the air–liquid interface, 15 µl inoculum of overnight cultures adjusted to OD600 of 5 was added to 1.5 ml MSgg medium in 24‐well plates, giving a starting OD600 of 0.05. Plates were incubated under static conditions at 30°C for 48 h, thereafter images of the arisen colonies and pellicles were obtained using the stereomicroscope.
2.4. Root colonization assay
Arabidopsis thaliana Col‐0 plants were used as a host for B. subtilis root colonization. A. thaliana seeds were sterilized in 2 % (v/v) sodium hypochlorite (NaOCl) for 10 min with an orbital shaker. Following this, NaOCl was removed and the seeds were washed five times in sterile water. Sterilized seeds were placed in MS agar plates (Murashige and Skoog basal salts, Sigma) (2.2 g·L−1) with approximately 20 seeds per petri dish. The Petri dishes were wrapped in parafilm and left for stratification at 4°C for 3 days to break seed dormancy and were then moved to the plant chamber (cycles of 16 h light at 24°C and 8 h dark at 20°C). After five to seven days, seedlings of 0.5–1.2 cm in size were placed in 48‐well plates containing 270 µl MSNg medium per well. To each well, 30 µl of overnight culture adjusted to OD600 of 0.2 was added resulting in a final starting OD600 of 0.02. The plates were sealed with parafilm and incubated in the plant chamber while shaking at 90 rpm for 16 h. Seedlings were then washed in minimal salts nitrogen glycerol (MSNg) to remove non‐attached cells from the root. The washed seedlings were placed in Eppendorf tubes containing 1 ml of NaCl (0.9 %) and subjected to standard sonication protocol to disperse the biofilm (Dragoš et al., 2018). The resulting bacterial cell suspension was diluted and plated on LB agar plates for CFU counting. To acquire the CFU per mm root, the obtained CFU was divided by the length of the corresponding root.
2.5. Growth profiling
To follow the growth of WT and mutants, two overnight cultures of each strain were independently inoculated into a 96‐well plate containing MSgg or LB broth, at a starting OD600 of 0.05. Growth was monitored in a plate reader (Infinite F200 PRO, TECAN, and BioTek Synergy HTX Multi‐Mode Microplate Reader) every 10 min for 48 h at 30°C under linear shaking conditions.
2.6. Matrix gene expression assays by flow cytometry
To measure expression from the matrix operon tapA‐sipW‐tasA, mKate‐labelled WT and ∆rap‐phr mutants harboring the reporter construct PtapA‐gfp were incubated in 10 ml MSgg in 100 ml bottles with a starting OD600 of 0.02. Lids were loosely on, so oxygen would not be depleted. Bottles were incubated at 37°C at 220 rpm for 6 h. After incubation, 1 ml of each sample was transferred to a 2 ml Eppendorf tube, and samples were run on the flow cytometer (MACSQuant® VYB, Miltenyibiotec). mKate‐positive cells were detected by yellow laser (561 nm) and filter Y3 (661/20 nm). Green fluorescent cells, representing the cells expressing the tapA‐sipW‐tasA operon, were detected by the blue laser (488 nm) and filter B1 (525/50 nm). Strain TB35 with constitutive mKate expression was used as a negative control for GFP expression, while strain TB34 with constitutive GFP expression was used as a positive control for GFP expression. In addition, TB34 and a medium control were used to identify the red background fluorescence noise due to autofluorescence, and cells above this background fluorescence were identified as the mKate‐positive cells representing B. subtilis cells producing the mKate protein. For each WT and mutant sample, single events were identified on the SSC‐H vs SSC‐A plot and gated into the mKate‐A vs SSC‐A plot, where mKate‐positive cells were identified. These gated cells were exported and read into Excel where the green fluorescence (GFP‐A) values were used for analysis. To get rid of negative values, 300 AU was added to all events in the samples. To obtain the distribution of GFP expression, data obtained from each replicate were subjected to binning with an identical bin size (of 50). Events with GFP expression from 0 to 10,000 were included as the majority of events were within this interval (>98%). Next, the number of events in each bin was divided by the total number of events in the given replicate, resulting in the normalized frequency. To obtain the mean distribution, a mean frequency for each bin was obtained by averaging the individual frequencies within this bin across the replicates, resulting in the mean distribution of single‐cell‐level expression. For statistics, the relative OFF (GFP values between 0 and 500) and ON (GFP values between 500–10,000) populations were calculated for each replicate, as was the mean fluorescence of the ON population.
2.7. Statistical analysis
Statistical analyses were performed in R Studio. For each root colonization assay, a one‐way analysis of variance (ANOVA) was performed on the log10‐transformed data. P‐values were adjusted using the Benjamini & Hochberg procedure. When adjusted P‐values were significant (p < 0.05), a Dunnett's multiple comparison test was performed. When data failed to meet parametric assumptions (normality and equal variance), a non‐parametric analysis (Kruskal–Wallis test) was used. If adjusted P‐values were significant (p < 0.05), treatment means were compared via a Dunn test. To test for significant differences in relative OFF/ON population and mean fluorescence of ON population between mutants and WT, an ANOVA followed by Dunnett's multiple comparison test was performed. The significance level for all tests was set at 5 %.
3. RESULTS
3.1. Single ∆rap‐phr mutants show altered matrix gene expression compared with the WT
To study the role of Rap‐Phr systems of B.subtilis in biofilm formation, we used the B.subtilis DK1042 strain (a natural competent derivative of the undomesticated NCBI 3610) (Konkol et al., 2013) as WT. In contrast to domesticated strains which may have acquired mutations to the Rap‐Phr systems or even lost some of them, NCIB 3610 and the derived DK1042 (hereafter referred to as 3610) contain all 12 rap‐phr modules (McLoon et al., 2011). Rap‐Phr systems of B. subtilis have been studied using both overexpression and deletion mutants of the Rap‐Phr systems as reviewed by Perego (2013). Interestingly, the absence of Rap modules in a natural isolate of B. subtilis was shown to influence sporulation timing (Serra et al., 2014). In this study, we employed single ∆rap‐phr mutants of B. subtilis created in our previous work (Gallegos‐Monterrosa et al., 2021) to study the impact of the Rap‐Phr modules on biofilm formation.
The diversity in Rap‐Phr regulatory systems and the reported role of several of the Rap phosphatases in regulating the activity of Spo0A (Perego, 2013; Omer Bendori et al., 2015), which controls matrix gene expression, prompted us to test WT and the 12 single ∆rap‐phr mutants for matrix gene expression. To quantify matrix gene expression during biofilm formation, we used a previously established approach (Kearns et al., 2005). This method utilizes the observation that expression from the promoter of the tapA‐sipW‐tasA operon, responsible for the production of the protein component of the extracellular matrix (Branda et al., 2006), is highly induced during the late exponential growth phase under shaking conditions in MSgg, a medium known to induce biofilm formation. After 6 h of growth in MSgg under shaking conditions, the fluorescence intensities of WT and mutants harboring a PtapA‐gfp reporter construct were measured using flow cytometry. Importantly, the side scatter (SSC‐A) vs forward scatter (FSC‐A) plots revealed no big changes in cell granularity or size of the mutants compared with the WT (Figure A1). Since RapA, B, E, H, I, J, and P have been reported to dephosphorylate Spo0F~P (Perego, 2013; Omer Bendori et al., 2015), we hypothesized that the mutants lacking one of these Rap phosphatases would have a larger relative ON population (i.e., cells in the high expression state), due to more cells committing to matrix production, and/or display a higher mean expression from PtapA in the ON population due to earlier matrix gene expression, as compared to the WT. Alternatively, if redundancy operates between some of the Rap‐Phr systems, the absence of one Rap‐Phr system may have only minor or insignificant effects due to the expression and function of another redundant system. However, ∆rapA showed a significantly smaller relative ON population (p = 0.022, n = 3–8) than the WT (Figure 1), while all other mutants showed a significantly higher mean expression of the ON population compared with the WT. The increased mean expression by the mutants could potentially be caused by enhanced growth in MSgg, as strains growing faster would reach the threshold density for biofilm formation earlier, and thus show increased matrix gene expression. However, when the optical density (OD) at 600 nm was measured after 6 h in minimal medium (MSgg) prior to flow cytometry analysis, the mutants showed either similar or reduced OD600 compared with WT, thus excluding this explanation. Interestingly, the increased matrix gene expression was not limited to the mutants lacking one of the Rap phosphatases reported to dephosphorylate Spo0F~P activity. These results show that all 12 Rap‐Phr regulatory systems influence matrix gene expression under these conditions.
FIGURE 1.
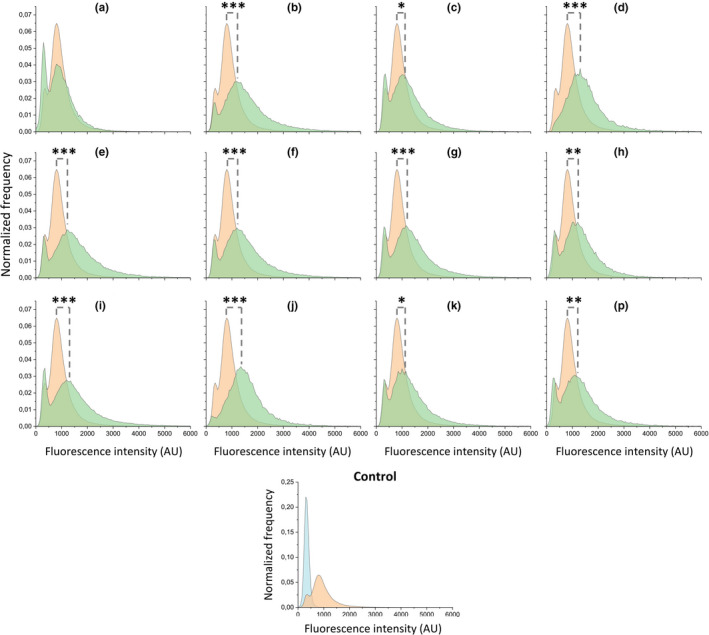
Expression of the tapA‐sipW‐tasA operon in Bacillus subtilis WT and ∆rap‐phr mutants after growth in MSgg under shaking conditions. Flow cytometry analysis showing the average distributions of GFP expression of ∆rap‐phr mutants and WT harboring the PtapA‐gfp construct (n = 3–8). The average WT distribution is shown in each graph for comparison. Orange = WT, green = mutant, blue = non‐labelled control strain. Letters denote the corresponding ∆rap‐phr mutants, that is, a = ∆rapA, b = ∆rapB and so forth. AU indicates arbitrary units. The significant difference in the mean fluorescence intensity of the ON population (GFP values between 500 and 10,000) between mutants and WT were tested by an ANOVA followed by Dunnett's multiple comparison test. *p < 0.05, **p < 0.01, ***p < 0.001
3.2. Absence of certain Rap‐Phr modules alters biofilm formation in vitro
Since matrix production is required for proper biofilm development (Branda et al., 2006; Dragoš et al., 2018), and most ∆rap‐phr mutants showed increased matrix gene expression under shaking conditions in MSgg, we hypothesized that this would manifest in more complex and robust biofilm formation. To investigate this, WT and mutants were tested for their ability to form biofilm on a solid surface (MSgg agar) and robust pellicle biofilms at the MSgg air‐liquid interface. A pellicle forms when oxygen in the medium is exhausted and B. subtilis moves toward higher oxygen concentrations, the air–liquid surface, where cells create a biofilm (Hölscher et al., 2015).
In accordance with previous work, B. subtilis WT produced a wrinkled colony biofilm, as well as a robust, wrinkled pellicle (Branda et al., 2001; Gallegos‐Monterrosa et al., 2016) (Figure 2). While ∆rapA, B, J, and K formed more wrinkled colonies, ∆rapI and P formed complex but very small colonies on MSgg agar compared with the WT. In contrast, ∆rapC formed a large, transparent colony with fewer wrinkles than WT. When testing for the development of pellicle biofilms, the ∆rapA, C, I, and P mutants formed thin and/or non‐homogenous pellicles, while the qualitative assessment revealed no mutants with more complex or robust pellicles than WT (Figure 2). Lastly, the ∆rapD, E, F, G, and H mutants formed comparable colony and pellicle biofilms to the WT. These results show that despite most ∆rap‐phr mutants showed increased matrix gene expression under shaking conditions compared with the WT, this did not necessarily manifest in the development of more complex biofilms on agar or at the air–liquid interface.
FIGURE 2.
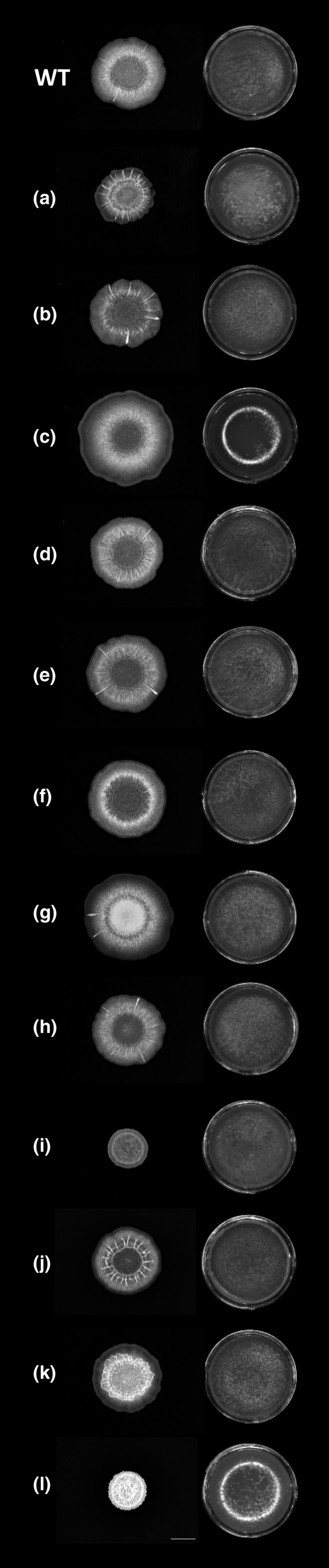
Biofilm formation of Bacillus subtilis WT and single ∆rap‐phr mutants. Overnight cultures of B. subtilis WT and ∆rap‐phr mutants were spotted on MSgg medium solidified with 1.5 % agar (left) or inoculated in MSgg with a starting OD600 = 0.05 (right). Images were taken from above after 48 h of incubation at 30°C using a stereomicroscope. Letters denote the corresponding ∆rap mutant, that is, a = ∆rapA, b = ∆rapB, and so forth, but l = ΔrapP. Bar denotes 5 mm for the biofilm colony images
3.3. Certain ∆rap‐phr mutants are affected in the colonization of Arabidopsis thaliana roots
To reveal how the Rap‐Phr modules affect biofilm formation in a more ecologically relevant environment, WT and mutants were tested for biofilm formation on the roots of the model plant organism A. thaliana. Similar to biofilm formation in vitro (Branda et al., 2006), previous studies have shown that matrix production is required for biofilm formation on A. thaliana roots (Beauregard et al., 2013; Dragoš et al., 2018). We, therefore, speculated that the increased matrix gene expression observed in most strains would enable enhanced biofilm formation on plant roots.
For this purpose, sterile A. thaliana seedlings with a root length of 0.5–1.2 cm were inoculated with one of the 12 ∆rap‐phr mutants or WT and incubated in a plant chamber at 24°C (90 rpm) for 16 h, after which root colonization was quantified as a colony‐forming unit (CFU) per mm root length. Of the 12 mutants tested, only the ∆rapD, J, and P mutants showed significantly increased root colonization compared with the WT (Figure 3). In contrast, the ∆rapI mutant was significantly reduced in root colonization. The observation that ∆rapI and P showed similar biofilm formation in vitro (Figure 2) but performed oppositely during root colonization (Figure 3) might indicate that the effect of each Rap phosphatase on biofilm formation depends on the environment. Furthermore, similarly to biofilm formation in vitro, these results show that increased matrix gene expression under shaking conditions did not necessarily manifest in increased root colonization.
FIGURE 3.
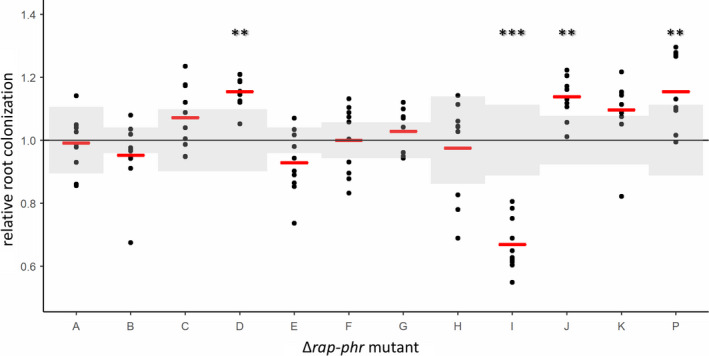
Arabidopsis thaliana root colonization by Bacillus subtilis WT and single ∆rap‐phr mutants. To estimate the impact of each of the Rap‐Phr modules on root colonization, WT and single ∆rap‐phr mutants were inoculated onto five‐day‐old seedlings of A. thaliana (n = 7–10). After 16 h, CFU per mm root length was quantified. The log10‐transformed value of CFU/mm root for each technical replicate was normalized to the mean of the WT from the same experiment. Each dot represents a root, while the mean value for each mutant is displayed as a red horizontal line. The black horizontal line represents the mean of the WT and the SD of the WT from each respective experiment is shown in shaded gray. For each assay, an ANOVA was performed on the log10‐transformed values of CFU/mm root length. When significant (p < 0.05), means were compared via Dunnett's multiple comparison test with WT as the control. When data failed to meet parametric assumptions, a Kruskal–Wallis test was performed followed by a Dunn's test. **p < 0.005, ***p < 0.001
3.4. Certain ∆rap‐phr mutants show altered growth compared with WT
The timing of biofilm initiation in vitro and on the root depends on the cell density, as only at sufficiently high cell density the Phr peptides will reach threshold concentrations allowing them to be imported into the cell, where they will inhibit their cognate Rap phosphatases. Consequently, a subset of the Rap phosphatases influences phosphorylation of Spo0A and the expression of biofilm genes. Biofilm formation is, therefore, affected by the growth rate, as strains growing faster will reach the threshold density for biofilm formation earlier. To test whether the observed changes in biofilm formation in vitro and on the root could be (partly) attributed to altered growth, WT and the 12 ∆rap‐phr mutants were tested for growth in MSgg under shaking conditions. For clear visualization, the growth curves of the 12 mutants were separated into four plots (Figure 4). Of the 12 mutants tested, only the ∆rapA, I, and P showed an altered growth profile compared with WT. ∆rapA and I showed a reduced growth rate, a delayed entry into the stationary phase, and reduced max OD590. In addition, ∆rapI had a prolonged stationary phase and a slower decline during the late stationary phase (possibly death phase). ∆rapP showed slower growth during the exponential phase, which continued for about 10 h longer than the WT, but displayed a higher max OD590 at the stationary phase. In LB medium, the same mutants were similarly or less affected in growth compared with WT (Figure A2). The impaired growth of ∆rapA and I is in accordance with these two mutants forming thin and/or non‐homogeneous pellicles (Figure 2), and ∆rapI displaying reduced root colonization (Figure 3). In contrast, the prolonged exponential growth of ∆rapP is inconsistent with the thin pellicle formed by this mutant, but correlating with the increased root colonization observed for this mutant. Finally, ∆rapC and ∆rapJ showed similar growth profiles as the WT but ∆rapC formed a thin pellicle, while ∆rapJ was increased in root colonization. The differential biofilm‐forming capacity in vitro and on the root observed for some of the mutants, interestingly, cannot be directly correlated with the growth rates and profiles of the mutants.
FIGURE 4.
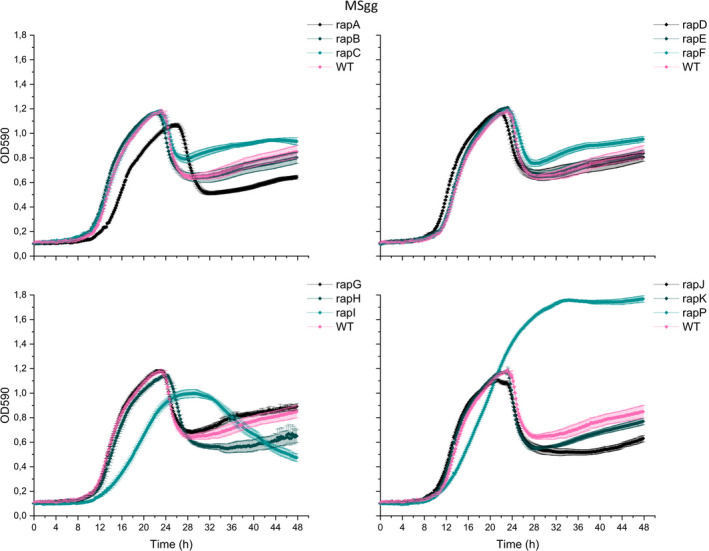
Growth of Bacillus subtilis WT and single ∆rap‐phr mutants in MSgg. WT and mutants were inoculated into 96‐well plates with a starting OD590 of 0.05. OD590 was measured every 10 min for 48 h at 30°C; each time point represents the mean of six technical replicates from two overnight cultures (N = 6). Error bars represent standard error (SE).
4. DISCUSSION
For decades, the Rap‐Phr regulatory systems which control the activity of the three master regulators governing cell differentiation in B. subtilis have been extensively studied (Perego, 2013; Pottahil & Lazazzera, 2003). However, since most studies have investigated the Rap‐Phr networks of B. subtilis independently from each other, in different genetic backgrounds, and under distinct conditions, it is difficult to compare the results from these studies. Here, we investigated all 12 Rap‐Phr systems found in B. subtilis 3610 for their effect on matrix gene expression and biofilm development in vitro. In addition, we examined the impact of Rap‐Phr systems in the colonization of plant roots for the first time.
Several of the Rap phosphatases have been reported to dephosphorylate Spo0F~P (Perego, 2013; Omer Bendori et al., 2015) which is expected to influence matrix production, but only RapP has previously been demonstrated to affect matrix gene expression (Parashar, Konkol, et al., 2013; Omer Bendori et al., 2015). We were, therefore, interested in testing the effect of each of the 12 Rap‐Phr systems on matrix gene expression. Inspired by a previously established method (Kearns et al., 2005), expression from the promoter of the tapA‐sipW‐tasA operon was measured for WT and the 12 ∆rap‐phr mutants in MSgg under shaking conditions. Only ∆rapA showed a significantly reduced relative ON population compared with the WT. Such reduced relative proportion of ∆rapA cells in the ON state may be due to more cells committing to activation of the sporulation pathway, and therefore, attenuating induction of matrix genes (Bischofs et al., 2009). Although only some of the Rap phosphatases have been reported to dephosphorylate Spo0F~P (Perego, 2013; Omer Bendori et al., 2015), we observed that all mutants except ∆rapA showed increased matrix gene expression. This indicates that despite the diversity of targets among Rap‐Phr systems, and the seeming redundancy of several Rap phosphatases regulating the same master regulator (Perego, 2013), each of the 12 Rap‐Phr systems has a regulatory role that affects matrix gene expression under the tested conditions. The involvement of 12 Rap‐Phr systems in influencing matrix gene expression suggests that the production of these costly public goods is under complex control, and allows the integration of multiple signals to fine‐tune the timing of matrix production in response to different conditions (Auchtung et al., 2006; Dragoš et al., 2018).
Matrix production is well known to be required for the formation of architecturally complex biofilms under laboratory conditions (Arnaouteli et al., 2021; Branda et al., 2006). Moreover, matrix production and localized cell death are responsible for the formation of wrinkles during biofilm development (Asally et al., 2012; Branda et al., 2006; Gallegos‐Monterrosa et al., 2017). We, therefore, speculated that the increased matrix gene expression observed for all mutants, except ∆rapA, would manifest in these 11 mutants forming more wrinkled colonies and more complex, robust pellicles compared with the WT. However, only some of the mutants were affected in biofilm formation. In accordance with increased matrix gene expression, the ∆rapB, J, and K mutants formed more wrinkled colonies, and ∆rapI and P formed complex, though very small colonies compared with the WT. Moreover, ∆rapC formed a larger, smoother, and more transparent colony. Furthermore, ∆rapA, with the similar mean expression of the tapA‐sipW‐tasA operon and a smaller relative ON population compared with WT, also formed a more wrinkled colony. Surprisingly, none of the mutants displayed increased pellicle robustness or complexity compared with the WT. In contrast, ∆rapA, C, I, and P formed thinner and/or non‐homogenous pellicles.
Next, we were interested in studying how Rap‐Phr systems affect biofilm formation of B. subtilis in a more ecologically relevant environment, that is, the plant root. Similar to biofilm formation in vitro, biofilm formation on the plant root by B. subtilis depends on matrix gene expression regulated by Spo0A (Beauregard et al., 2013; Chen et al., 2013). We, therefore, hypothesized that the increased matrix gene expression observed for most mutants would allow more bacterial cells to attach to and colonize the root. However, only ∆rapD, J, and P showed increased root colonization, while ∆rapI was reduced in root colonization. The biofilm and root colonization experiments of the ∆rap‐phr mutants thus show that the magnitude of matrix gene expression under shaking conditions does not directly correlate with the ability to develop complex biofilms in vitro (on agar and at the air–liquid interface) or to colonize the plant root (Table A1). A lack of positive correlation between matrix gene expression and biofilm formation was observed in a previous study which showed that the magnitude of expression of epsA‐O and tasA‐sipW‐tasA in B. subtilis 168 variants did not directly correlate with the formation of wrinkled biofilms (Gallegos‐Monterrosa et al., 2016). These experiments could thus support that biofilm formation in in vitro and on plant roots is influenced by additional factors than just matrix gene expression. For example, surfactin production, which is regulated by ComA—a target of several Rap phosphatases (Perego, 2013)—was shown to influence the colony structure of B. subtilis NCIB 3610 on MSgg, though this secondary metabolite was not essential for pellicle formation and root colonization (Thérien et al., 2020). However, it has to be noted that matrix gene expression was measured under heavily shaking conditions (220 rpm), while colonies and pellicles were developed under static conditions, and root colonization was assayed under mildly shaking conditions (90 rpm). An alternative explanation for the lack of correlation between matrix gene expression under shaking conditions and biofilm formation in vitro and on the root could be that the effect of the rap‐phr deletions on tapA operon expression may vary between these different conditions. Further work is needed to fully explain the discrepancies observed in this study between tapA operon expression and biofilm formation.
Interestingly, several studies have reported a correlation between the ability of strains to form robust biofilms in vitro and to colonize the root—both within and among strains (Chen et al., 2013; Gallegos‐Monterrosa et al., 2016). However, the ability of the ∆rap‐phr mutants to form biofilm in vitro did not necessarily reflect the ability to colonize the root (compare Figures 2 and 3 and the summary in Table A1). For example, ∆rapA and C formed thin and non‐homogenous pellicles but were able to colonize the root to similar levels as the WT. ∆rapD displayed comparable biofilm in vitro to the WT but was significantly better in root colonization. In addition, ∆rapJ formed a highly wrinkled colony, but a pellicle similar to the WT, and was increased in root colonization. Finally, both ∆rapI and P showed reduced colony size and thin and/or non‐homogenous pellicle formation, but while ∆rapI showed reduced root colonization, ∆rapP was increased in root colonization compared with the WT. These results indicate that the effect of the rap‐phr deletions on biofilm formation varies between in vitro and root conditions. This was similarly shown for a ∆tagE mutant (deficient in glycosylating wall teichoic acid), which was affected in root colonization but displayed similar biofilm formation on agar and at the air–liquid interface as the WT (Tzipilevich & Benfey, 2021).
In the study by Gallegos‐Monterrosa et al. (2016), showing that strains forming complex colonies and robust pellicles also efficiently colonize the root, the B. subtilis 168 stocks displayed genetic variation in distinct loci (e.g., epsC that encodes an enzyme that is directly involved in matrix production), resulting in large differences among the strains in their ability to form biofilm and colonize the root. In contrast, the ∆rap‐phr mutations studied here might only slightly modulate the regulatory pathways of B. subtilis; therefore, the ability of the mutants to form biofilm and colonize the root is less altered compared with WT. Nonetheless, the same study also demonstrated that biofilm development is influenced by medium composition (Gallegos‐Monterrosa et al., 2016). Besides static vs mild agitation and a temperature difference (30 vs. 24°C), the media used for testing colony and pellicle formation, and for testing root colonization also slightly differ. First, the MSgg medium used for colony and pellicle formation contains a 10 times higher concentration of glycerol (0.5 %) compared with the MSNg medium used for plant root colonization (0.05%). During plant root colonization, the bacteria thus depend on plant polysaccharides and root exudates as carbon sources. In addition, in vitro biofilm development depends on the availability of iron and manganese (Kolodkin‐Gal et al., 2013; Mhatre et al., 2016; Shemesh & Chai, 2013), while during plant root colonization in MSNg, biofilm formation is induced by plant polysaccharides and root exudates (Beauregard et al., 2013; Chen et al., 2013). The presence of plant polysaccharides may, therefore, allow ∆rap‐phr mutants that form weak pellicle biofilms in vitro to efficiently colonize the root (∆rapA, C and P, Figures 2 and 3). Thereby, the pathways regulating matrix gene expression and other factors important for biofilm formation may be differently regulated under distinct conditions.
Finally, the disparate results obtained in this study may be understood in light of the full set of 12 Rap‐Phr systems with redundant functions: the influence of a single rap‐phr deletion might be masked by the function of another redundant Rap‐Phr system. Furthermore, if such potential redundancy varies between the different conditions employed in this study (e.g., if the rap‐phr genes are differentially expressed under the distinct conditions tested), this could (partly) explain the observed discrepancy, for example, between biofilm formation in vitro and on the root.
To conclude, we here show that all 12 Rap‐Phr systems have an impact on matrix gene expression in liquid culture. Thereby, the diversity in Rap‐Phr systems in B. subtilis 3610 could function to integrate multiple signals to fine‐tune the timing and level of matrix gene expression in response to new ecological niches, such as those it will encounter in soil. Furthermore, we show that the ability to form biofilm in vitro not necessarily reflects the ability to colonize the root under the tested conditions. These findings thus support that the pathways involved in matrix gene expression and other components important for biofilm establishment could be differently influenced under distinct conditions.
CONFLICT OF INTERESTS
None declared.
AUTHOR CONTRIBUTIONS
Mathilde Nordgaard Christensen: Conceptualization (equal); Formal analysis (lead); Investigation (lead); Methodology (lead); Visualization (lead); Writing‐original draft (equal). Rasmus Møller Rosenbek Mortensen: Investigation (supporting); Writing‐review & editing (supporting). Nikolaj Kaae Kirk: Investigation (supporting); Writing‐review & editing (supporting). Ramses Gallegos‐Monterrosa: Methodology (supporting); Resources (supporting); Writing‐review & editing (equal). Ákos T. Kovács: Conceptualization (equal); Funding acquisition (lead); Methodology (supporting); Project administration (lead); Resources (equal); Supervision (lead); Writing‐original draft (equal).
ETHICS STATEMENT
None required.
ACKNOWLEDGEMENTS
We thank Mikael Lenz Strube for his suggestions on statistics. The work was supported by a DTU Bioengineering start‐up fund to ÁTK. Funding from Novo Nordisk Foundation (grant NNFOC0055625) for the infrastructure “Imaging microbial language in biocontrol (IMLiB)” is acknowledged.
APPENDIX 1.
FIGURE A1.
Single ∆rap‐phr mutants are not majorly affected in cell granularity or size. Flow cytometry analysis showing the side scatter (SSC‐A) vs forward scatter (FSC‐A) plots of ungated WT and ∆rap‐phr mutant cells harboring the PtapA‐gfp construct
FIGURE A2.
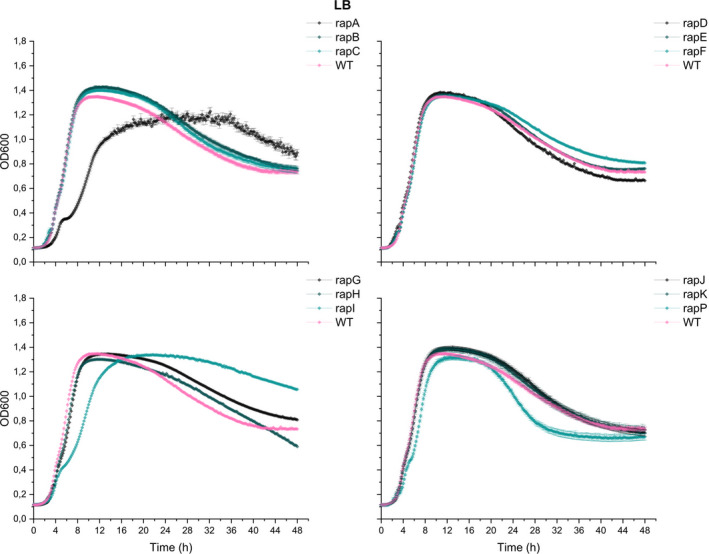
Growth of Bacillus subtilis WT and single ∆rap‐phr mutants in LB medium. WT and mutants were inoculated into 96‐well plates with a starting OD600 of 0.05. OD590 was measured every 10 min for 48 h at 30°C, and each time point represents the mean of six technical replicates from two overnight cultures (n = 6). Error bars represent standard error (SE)
TABLE A1.
Overview of phenotypes of the 12 single ∆rap‐phr mutants compared with WT
| Mutant | Known target(s) of respective Rap proteins | Matrix expr. | Colony | Pellicle | Root | Growth |
|---|---|---|---|---|---|---|
| rapA‐phrA | Spo0F | ↓ | ↑ | ↓ | – | ↓ |
| rapB | Spo0F | ↑ | ↑ | – | – | – |
| rapC‐phrC | ComA | ↑ | ↓ | ↓ | – | – |
| rapD | ComA | ↑ | – | – | ↑ | – |
| rapE‐phrE | Spo0F | ↑ | – | – | – | – |
| rapF‐phrF | ComA | ↑ | – | – | – | – |
| rapG‐phrG | DegU | ↑ | – | – | – | – |
| rapH‐phrH | Spo0F, ComA | ↑ | – | – | – | – |
| rapI‐phrI | Spo0F, regulation of mobile genetic elements | ↑ | ↑ (but small) | ↓ | ↓ | ↓ |
| rapJ | Spo0F | ↑ | ↑ | – | ↑ | – |
| rapK‐phrK | ComA | ↑ | ↑ | – | – | – |
| rapP‐phrP | Spo0F, ComA | ↑ | ↑ (but small) | ↓ | ↑ | ↑ |
Up arrows indicate increased while down arrows indicate decreased features compared with WT. For colony and pellicle formation, the direction of the arrow is related to wrinkles and complexity.
Nordgaard Christensen, M. , Mortensen, R. M. R. , Kirk, N. K. , Gallegos‐Monterrosa, R. , & Kovács, Á. T. (2021). Deletion of Rap‐Phr systems in Bacillus subtilis influences in vitro biofilm formation and plant root colonization. MicrobiologyOpen, 10, e1212. 10.1002/mbo3.1212
DATA AVAILABILITY STATEMENT
Data are provided in the results section and appendices of this article.
REFERENCES
- Arnaouteli, S. , Bamford, N. , Stanley‐Wall, N. , & Kovács, Á. T. (2021). Bacillus subtilis biofilm formation and social interactions. Nature Reviews Microbiology. 10.1038/s41579-021-00540-9 [DOI] [PubMed] [Google Scholar]
- Asally, M. , Kittisopikul, M. , Rue, P. , Du, Y. , Hu, Z. , Cagatay, T. , Robinson, A. B. , Lu, H. , Garcia‐Ojalvo, J. , & Suel, G. M. (2012). Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proceedings of the National Academy of Sciences of the United States of America, 109, 18891–18896. 10.1073/pnas.1212429109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung, J. M. , Lee, C. A. , & Grossman, A. D. (2006). Modulation of the ComA‐dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. Journal of Bacteriology, 188, 5273–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchtung, J. M. , Lee, C. A. , Monson, R. E. , Lehman, A. P. , & Grossman, A. D. (2005). Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proceedings of the National Academy of Sciences of the United States of America, 102, 12554–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais, H. P. , Fall, R. , & Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiology, 134, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard, P. B. , Chai, Y. , Vlamakis, H. , Losick, R. , & Kolter, R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proceedings of the National Academy of Sciences of the United States of America, 110, 1621–1630. 10.1073/pnas.1218984110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas, R. (2013). When the swimming gets tough, the tough form a biofilm. Molecular Microbiology, 90, 1–5. 10.1111/mmi.12354 [DOI] [PubMed] [Google Scholar]
- Bischofs, I. B. , Hug, J. A. , Liu, A. W. , Wolf, D. M. , & Arkin, A. P. (2009). Complexity in bacterial cell‐cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proceedings of the National Academy of Sciences of the United States of America, 106, 6459–6464. 10.1073/pnas.0810878106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, C. , Christensen, M. N. , & Kovács, Á. T. (2021). Molecular aspects of plant growth promotion and protection by Bacillus subtilis . Molecular Plant‐Microbe Interactions, 34, 15–25. [DOI] [PubMed] [Google Scholar]
- Bongiorni, C. , Ishikawa, S. , Stephenson, S. , Ogasawara, N. , & Perego, M. (2005). Synergistic regulation of competence development in Bacillus subtilis by two Rap‐Phr systems. Journal of Bacteriology, 187, 4353–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda, S. S. , Chu, F. , Kearns, D. B. , Losick, R. , & Kolter, R. (2006). A major protein component of the Bacillus subtilis biofilm matrix. Molecular Microbiology, 59, 1229–1238. 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- Branda, S. S. , González‐Pastor, J. E. , Ben‐Yehuda, S. , Losick, R. , & Kolter, R. (2001). Fruiting body formation by Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America, 98, 11621–11626. 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, L. S. , Hobley, L. , & Stanley‐Wall, N. R. (2014). Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Molecular Microbiology, 93, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, L. S. , Marlow, V. L. , Bissett, E. , Ostrowski, A. , & Stanley‐Wall, N. R. (2013). A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis . Molecular Microbiology, 90, 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Cao, S. , Chai, Y. , Clardy, J. , Kolter, R. , Guo, J. H. , & Losick, R. (2012). A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Molecular Microbiology, 85, 418–430. 10.1111/j.1365-2958.2012.08109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Yan, F. , Chai, Y. , Liu, H. , Kolter, R. , Losick, R. , & Guo, J. H. (2013). Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environmental Microbiology, 15, 848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comella, N. , & Grossman, A. D. (2005). Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum‐sensing transcription factor ComA in Bacillus subtilis . Molecular Microbiology, 57, 1159–1174. 10.1111/j.1365-2958.2005.04749.x [DOI] [PubMed] [Google Scholar]
- Core, L. , & Perego, M. (2003). TPR‐mediated interaction of RapC with ComA inhibits response regulator‐DNA binding for competence development in Bacillus subtilis . Molecular Microbiology, 49, 1509–1522. 10.1046/j.1365-2958.2003.03659.x [DOI] [PubMed] [Google Scholar]
- Costerton, J. W. , Lewandowski, Z. , DeBeer, D. , Caldwell, D. , Korber, D. , & James, G. (1994). Biofilms, the customized microniche. Journal of Bacteriology, 176, 2137–2142. 10.1128/jb.176.8.2137-2142.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pippo, F. , Di Gregorio, L. , Congestri, R. , Tandoi, V. , & Rossetti, S. (2018). Biofilm growth and control in cooling water industrial systems. FEMS Microbiology Ecology, 94, fiy044. [DOI] [PubMed] [Google Scholar]
- Dragoš, A. , Kiesewalter, H. , Martin, M. , Hsu, C.‐Y. , Hartmann, R. , Wechsler, T. , Eriksen, C. , Brix, S. , Drescher, K. , Stanley‐Wall, N. , Kümmerli, R. , & Kovács, Á. T. (2018). Division of labor during biofilm matrix production. Current Biology, 28, 1903–1913. 10.1016/j.cub.2018.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even‐Tov, E. , Omer Bendori, S. , Pollak, S. , & Eldar, A. (2016). Transient duplication‐dependent divergence and horizontal transfer underlie the evolutionary dynamics of bacterial cell‐cell signaling. PLoS Biology, 14, e2000330. 10.1371/journal.pbio.2000330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall, R. , Kinsinger, R. F. , & Wheeler, K. A. (2004). A simple method to isolate biofilm‐forming Bacillus subtilis and related species from plant roots. Systematic and Applied Microbiology, 27, 372–379. 10.1078/0723-2020-00267 [DOI] [PubMed] [Google Scholar]
- Flemming, H. C. , & Wingender, J. (2010). The biofilm matrix. Nature Reviews Microbiology, 8, 623–633. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- Fujita, M. , González‐Pastor, J. E. , & Losick, R. (2005). High‐ and low‐threshold genes in the Spo0A regulon of Bacillus subtilis . Journal of Bacteriology, 187, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos‐Monterrosa, R. , Christensen, M. N. , Barchewitz, T. , Koppenhöfer, S. , Priyadarshini, B. , Bálint, B. , Maróti, G. , Kempen, P. J. , Dragoš, A. , & Kovács, Á. T. (2021). Impact of Rap‐Phr system abundance on adaptation of Bacillus subtilis . Commun Biol, 4, 468. 10.1038/s42003-021-01983-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos‐Monterrosa, R. , Kankel, S. , Götze, S. , Barnett, R. , Stallforth, P. , & Kovács, Á. T. (2017). Lysinibacillus fusiformis M5 induces increased complexity in Bacillus subtilis 168 colony biofilms via hypoxanthine. Journal of Bacteriology, 199, e00204‐17. 10.1128/JB.00204-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos‐Monterrosa, R. , Mhatre, E. , & Kovács, Á. T. (2016). Specific Bacillus subtilis 168 variants form biofilms on nutrient‐rich medium. Microbiology, 162, 1922–1932. 10.1099/mic.0.000371 [DOI] [PubMed] [Google Scholar]
- Hall‐Stoodley, L. , Costerton, J. W. , & Stoodley, P. (2004). Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology, 2, 95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- Hölscher, T. , Bartels, B. , Lin, Y.‐C. , Gallegos‐Monterrosa, R. , Price‐Whelan, A. , Kolter, R. , Dietrich, L. E. P. , & Kovács, Á. T. (2015). Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. Journal of Molecular Biology, 427, 3695–3708. 10.1016/j.jmb.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher, T. , Dragoš, A. , Gallegos‐Monterrosa, R. , Martin, M. , Mhatre, E. , Richter, A. , & Kovács, Á. T. (2016). Monitoring spatial segregation in surface colonizing microbial populations. Journal of Visualized Experiments, 116, e54752. 10.3791/54752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmer, H. , Larsen, T. S. , Krogh, A. , Saxild, H. H. , Brunak, S. , & Knudsen, S. (2001). Sigma A recognition sites in the Bacillus subtilis genome. Microbiology, 147, 2417–2424. 10.1099/00221287-147-9-2417 [DOI] [PubMed] [Google Scholar]
- Jiang, M. , Grau, R. , & Perego, M. (2000). Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis . Journal of Bacteriology, 182, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, M. , Shao, W. , Perego, M. , & Hoch, J. A. (2000). Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis . Molecular Microbiology, 38, 535–542. 10.1046/j.1365-2958.2000.02148.x [DOI] [PubMed] [Google Scholar]
- Kearns, D. B. , Chu, F. , Branda, S. S. , Kolter, R. , & Losick, R. (2005). A master regulator for biofilm formation by Bacillus subtilis . Molecular Microbiology, 55, 739–749. 10.1111/j.1365-2958.2004.04440.x [DOI] [PubMed] [Google Scholar]
- Kiesewalter, H. T. , Lozano‐Andrade, C. N. , Wibowo, M. , Strube, M. L. , Maróti, G. , Snyder, D. , Jørgensen, T. S. , Larsen, T. O. , Cooper, V. S. , Weber, T. , & Kovács, Á. T. (2021). Genomic and chemical diversity of bacillus subtilis secondary metabolites against plant pathogenic fungi. mSystems 6, e00770‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin‐Gal, I. , Elsholz, A. K. W. , Muth, C. , Girguis, P. R. , Kolter, R. , & Losick, R. (2013). Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane‐embedded histidine kinase. Genes & Development, 27, 887–899. 10.1101/gad.215244.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkol, M. A. , Blair, K. M. , & Kearns, D. B. (2013). Plasmid‐encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis . Journal of Bacteriology, 195, 4085–4093. 10.1128/JB.00696-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera, B. A. , Kurtser, I. G. , McQuade, R. S. , & Grossman, A. D. (1999). An autoregulatory circuit affecting peptide signaling in Bacillus subtilis . Journal of Bacteriology, 181, 5193–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López, D. , & Kolter, R. (2010). Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis . FEMS Microbiology Reviews, 34, 134–149. [DOI] [PubMed] [Google Scholar]
- Lopez, D. , Vlamakis, H. , & Kolter, R. (2009). Generation of multiple cell types in Bacillus subtilis . FEMS Microbiology Reviews, 33, 152–163. [DOI] [PubMed] [Google Scholar]
- López, D. , Vlamakis, H. , & Kolter, R. (2010). Biofilms. Cold Spring Harbor Perspectives in Biology, 2, a000398. 10.1101/cshperspect.a000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoon, A. L. , Guttenplan, S. B. , Kearns, D. B. , Kolter, R. , & Losick, R. (2011). Tracing the domestication of a biofilm‐forming bacterium. Journal of Bacteriology, 193, 2027–2034. 10.1128/JB.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade, R. S. , Comella, N. , & Grossman, A. D. (2001). Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma‐H of Bacillus subtilis . Journal of Bacteriology, 183, 4905–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre, E. , Monterrosa, R. G. , & Kovács, Á. T. (2014). From environmental signals to regulators: modulation of biofilm development in Gram‐positive bacteria. Journal of Basic Microbiology, 54, 616–632. 10.1002/jobm.201400175 [DOI] [PubMed] [Google Scholar]
- Mhatre, E. , Sundaram, A. , Hölscher, T. , Mühlstädt, M. , Bossert, J. , & Kovács, Á. T. (2017). Presence of calcium lowers the expansion of Bacillus subtilis colony biofilms. Microorganisms, 5, 7. 10.3390/microorganisms5010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre, E. , Troszok, A. , Gallegos‐Monterrosa, R. , Hölscher, T. , Kuipers, O. P. , Lindst, S. , & Kovács, Á. T. (2016). The impact of manganese on biofilm development of Bacillus subtilis . Microbiology, 162, 1468–1478. 10.1099/mic.0.000320 [DOI] [PubMed] [Google Scholar]
- Mueller, J. P. , Bukusoglu, G. , & Sonenshein, A. L. (1992). Transcriptional regulation of Bacillus subtilis glucose starvation‐inducible genes: control of gsiA by the ComP‐ComA signal transduction system. Journal of Bacteriology, 174, 4361–4373. 10.1128/jb.174.13.4361-4373.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, M. , & Fujita, Y. (2007). Bacillus subtilis RapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiology Letters, 268, 73–80. [DOI] [PubMed] [Google Scholar]
- Ogura, M. , Yamaguchi, H. , Yoshida, K. , Fujita, Y. , & Tanaka, T. (2001). DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two‐component regulatory systems. Nucleic Acids Research, 29, 3804–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer Bendori, S. , Pollak, S. , Hizi, D. , & Eldar, A. (2015). The RapP‐PhrP quorum‐sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal‐insensitive allele of RapP. Journal of Bacteriology, 197, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena, M. , & Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends in Microbiology, 16, 115–125. 10.1016/j.tim.2007.12.009 [DOI] [PubMed] [Google Scholar]
- O'Toole, G. , Kaplan, H. B. , & Kolter, R. (2000). Biofilm formation as microbial development. Annual Review of Microbiology, 54, 49–79. 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- Parashar, V. , Jeffrey, P. D. , & Neiditch, M. B. (2013). Conformational change‐induced repeat domain expansion regulates Rap phosphatase quorum‐sensing signal receptors. PLoS Biology, 11, e1001512. 10.1371/journal.pbio.1001512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar, V. , Konkol, M. A. , Kearns, D. B. , & Neiditch, M. B. (2013). A plasmid‐encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. Journal of Bacteriology, 195, 2437–2448. 10.1128/JB.02030-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego, M. (2013). Forty years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biology, 11, e1001516. 10.1371/journal.pbio.1001516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego, M. , Glaser, P. , & Hoch, J. A. (1996). Aspartyl‐phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis . Molecular Microbiology, 19, 1151–1157. 10.1111/j.1365-2958.1996.tb02460.x [DOI] [PubMed] [Google Scholar]
- Perego, M. , Hanstein, C. , Welsh, K. M. , Djavakhishvili, T. , Glaser, P. , & Hoch, J. A. (1994). Multiple protein‐aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B.subtilis . Cell, 79, 1047–1055. 10.1016/0092-8674(94)90035-3 [DOI] [PubMed] [Google Scholar]
- Perego, M. , & Hoch, J. A. (1996). Cell‐cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America, 93, 1549–1553. 10.1073/pnas.93.4.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot, P. J. , & Hilbert, D. W. (2004). Sporulation of Bacillus subtilis . Current Opinion in Microbiology, 7, 579–586. 10.1016/j.mib.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Pottahil, M. , & Lazazzera, B. A. (2003). The extracellular Phr peptide‐Rap phosphatase signaling circuit of Bacillus subtilis . Frontiers in Bioscience, 8, 32–45. 10.2741/913 [DOI] [PubMed] [Google Scholar]
- Reizer, J. , Reizer, A. , Perego, M. , & Saier, M. H. (1997). Characterization of a family of bacterial response regulator aspartyl‐phosphate (RAP) phosphatases. Microbial and Comparative Genomics, 2, 103–111. 10.1089/omi.1.1997.2.103 [DOI] [PubMed] [Google Scholar]
- Serra, C. R. , Earl, A. M. , Barbosa, T. M. , Kolter, R. , & Henriques, A. O. (2014). Sporulation during growth in a gut isolate of Bacillus subtilis . Journal of Bacteriology, 196, 4184–4196. 10.1128/JB.01993-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh, M. , & Chai, Y. (2013). A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. Journal of Bacteriology, 195, 2747–2754. 10.1128/JB.00028-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. , Paul, D. , & Jain, R. K. (2006). Biofilms: implications in bioremediation. Trends in Microbiology, 14, 389–397. 10.1016/j.tim.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Stewart, P. S. (2002). Mechanisms of antibiotic resistance in bacterial biofilms. International Journal of Medical Microbiology, 292, 107–113. 10.1078/1438-4221-00196 [DOI] [PubMed] [Google Scholar]
- Thérien, M. , Kiesewalter, H. T. , Auria, E. , Charron‐Lamoureux, V. , Wibowo, M. , Maróti, G. , Kovács, Á. T. , & Beauregard, P. B. (2020). Surfactin production is not essential for pellicle and root‐associated biofilm development of Bacillus subtilis . Biofilm, 2, 100021. 10.1016/j.bioflm.2020.100021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipilevich, E. , & Benfey, P. N. (2021). Phage‐resistant bacteria reveal a role for potassium in root colonization. bioRxiv. 10.1101/2021.05.12.443821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gestel, J. , Weissing, F. J. , Kuipers, O. P. , & Kovács, A. T. (2014). Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME Journal, 8, 2069–2079. 10.1038/ismej.2014.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme, D. T. , Kiley, T. B. , & Stanley‐Wall, N. R. (2007). DegU co‐ordinates multicellular behaviour exhibited by Bacillus subtilis . Molecular Microbiology, 65, 554–568. 10.1111/j.1365-2958.2007.05810.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are provided in the results section and appendices of this article.


