Abstract
Purpose
Long non-coding RNA (lncRNA) is identified as an important regulator involved in oral squamous cell carcinoma (OSCC) tumorigenesis. This study aimed to investigate the functional role and underlying mechanism of LINC00662 in OSCC.
Materials and Methods
The expression levels of LINC00662, miR-144-3p, and enhancer of zeste homolog 2 (EZH2) mRNA were quantified with quantitative real-time polymerase chain reaction in OSCC tissues and cell lines. Western blot analysis was used to assay the expression levels of E-cadherin, Vimentin, and EZH2. Cell proliferation, migration, and invasion were monitored by cell counting kit-8 and Transwell assays. Dual-luciferase reporter and RNA immunoprecipitation assays were employed to verify the regulatory relationship between LINC00662 and miR-144-3p.
Results
The expression of LINC00662, positively associated with the increased TNM stage and lymph node metastasis of the patients, was up-regulated in OSCC tissues and cells. The overexpression of LINC00662 facilitated the proliferation, migration, and invasion of OSCC cells. MiR-144-3p could bind to LINC00662, and the promoting effect of LINC00662 overexpression was counteracted by miR-144-3p mimic. Moreover, EZH2 expression was negatively regulated by miR-144-3p and positively regulated by LINC00662. The silencing of EZH2 attenuated the promoting effects of overexpression of LINC00662 on cell proliferation, migration, invasion, and epithelial-mesenchymal transition.
Conclusion
LINC00662, as an oncogenic lncRNA of OSCC, accelerates OSCC progression by repressing miR-144-3p expression and increasing EZH2 expression.
Keywords: Oral squamous cell carcinoma, LINC00662, miR-144-3p, EZH2, cell growth, metastasis
INTRODUCTION
Oral squamous cell carcinoma (OSCC) is one of the common types of oral malignancies,1 and is characterized by invasive growth as well as inclination of lymph node metastasis.2 Currently, comprehensive treatment including surgery, radiotherapy, and chemotherapy remains the main therapeutic strategy for treating patients with OSCC. However, the life quality of patients with OSCC remains unimproved, and the prognosis is still unsatisfactory, with the 5-year survival rate merely about 40% to 50%.3,4 Towards this end, deciphering the molecular mechanism of occurrence, development, and metastasis of OSCC, as well as unearthing effective therapeutic targets for OSCC are necessary.
Long non-coding RNAs (lncRNAs), non-coding functional transcripts with lengths exceeding 200 nucleotides, are involved in regulating diverse biological events, including cellular differentiation, cell lineage choice, organogenesis, and tissue homeostasis.5,6 Importantly, the dysregulation of lncRNA is associated with various cancers.7,8 Reportedly, LINC00662 functions as a carcinogenic lncRNA in some cancers, including gastric cancer, prostate cancer, and OSCC.9,10,11 Notably, LINC00662 is capable of facilitating the occurrence and expediting the development of OSCC by regulating Wnt/β-catenin pathway, and it is associated with the detrimental prognosis of patients with OSCC.11 Notwithstanding the selected research referenced above, the relevant mechanism by which LINC00662 exerts a role in OSCC warrants further elucidation.
MicroRNA (miRNA), characterized as a small non-coding RNA composed of 19–23 nucleotides, possesses pivotal functions in biological and pathological processes.12 Abnormally expressed miRNA can serve as a potential biomarker for cancer diagnosis and prognosis evaluation. Additionally, miRNA figures prominently in tumors by regulating the expression levels of genes related to apoptosis, differentiation, metabolism, and migration. For instance, miR-133a-3p, whose expression is down-regulated in OSCC, impedes proliferation and migration of OSCC cells by directly targeting COL1A1.13 MiRNA-16 constrains OSCC growth by suppressing Wnt/β-catenin signaling pathway.14 Moreover, miR-144-3p expression is down-regulated in OSCC, and miR-144-3p suppresses the development of OSCC by down-regulating ERO1L expression.15 Nevertheless, the explicit molecular mechanism of miR-144-3p expression imbalance in OSCC is far from being elucidated.
Enhancer of zeste homolog 2 (EZH2), a member of the polycomb group protein family, mediates histone methylation and is considered as a candidate oncogene.16,17 For example, EZH2 accelerates the proliferation of gastric cancer cells by suppressing p21 expression.17 Likewise, small molecule inhibitor of EZH2 is capable of inducing the apoptosis of prostate cancer cells.18 Additionally, EZH2 represents high expression in OSCC, facilitating OSCC cell invasion and glycolysis by regulating STAT3 and FoxO1 signaling.19 Nonetheless, the underlying mechanism of EZH2 dysregulation in OSCC remains to be further clarified.
In the current study, LINC00662 was found to be highly expressed in OSCC tissues and cells. Moreover, LINC00662 accelerated the progression of OSCC by regulating miR-144-3p/EZH2 axis.
MATERIALS AND METHODS
Clinical samples
Fifty OSCC tissues and adjacent tissues (1.5 cm away from the tumor margins) were collected from the Affiliated Hospital of Shandong Medical College and cryopreserved in liquid nitrogen. All patients (29 males and 21 females, 31–78 years old, with an average age of 53 years old) were pathologically diagnosed as OSCC, and the patients did not receive chemotherapy or radiotherapy before surgery. Written informed consent was obtained. The ethical endorsement was granted by the Ethics Committee of Dongping Hospital Affiliated to Shandong First Medical University (Approval No. 2018-1203).
Cell culture
OSCC cell lines (SCC-25, ISG15, SCC-9, and CAL-27), normal oral keratinocyte (NOK) cell line and human gastric adenocarcinoma cell (AGS) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and the American Type Culture Collection (ATCC, Rockville, MD, USA). The cells were cultivated in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine serum (FBS, Biochrom AG, Berlin, Germany) and 1% penicillin/streptomycin (Invitrogen) at 37℃ with 5% CO2.
Cell transfection
LINC00662 overexpression plasmids (LINC00662; 50 ng), empty vector (vector; 50 ng), miR-144-3p mimics (miR-144-3p, 5′-UUCUCGAACGUGUCACGUUUU-3′; 50 nM), mimics negative control (miR-control, 5′-UCAUGUAGUAGAUAUGACAU-3′; 50 nM), siRNAs targeting EZH2 (si-EZH2, 5′-GAGGGAAAGUGUAUGAUAATT-3′; 100 nM), and siRNAs negative control (si-NC, 5′-GGGAAAGAGUAUAUAGUGATT-3′; 100 nM) were purchased from RiboBio Co., Ltd. (Guangzhou, China). SCC-9 and ISG15 cells were transfected with the oligonucleotide or plasmid employing Lipofectamine 2000 transfection reagent (Invitrogen) according to the protocols provided by the manufacturer.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from tissues/cells using the TRIzol reagent (Invitrogen). The RNA was reversely transcribed to cDNA with either the PrimeScript RT reagent Kit (Takara, Dalian, China) or miRNA reverse transcription PCR kit (RiboBio Co., Ltd.). Quantitative real-time polymerase chain reaction (qRT-PCR) assay was then conducted with SYBR Premix Ex Taq™ II kit (Takara), primers, and cDNA templates on the ABI Prism 7900HT Real-Time PCR System (Applied Biosystems Inc., Foster City, CA, USA). The 2−ΔΔCt method was employed for quantification, and GAPDH or U6 was used as the internal control. The primers were presented as follows: LINC00662 (Forward, 5′-ACTAACAAGCTGGGTGCAGA-3′; Reverse, 5′-CCTCCTGGTCTGCGAGAAAT-3′), miR-144-3p (Forward, 5′-GGCCGGCGTACAGTATAGATGA-3′; Reverse, 5′-GTGCAGGGTCCGAGGT-3′), EZH2 (Forward, 5′-GTGGAGAGATTATTCTCAAGATG-3′; Reverse, 5′-CCGACATACTTCAGGGCATCAGCC-3′), U6 (Forward, 5′-CTCGCTTCGGCAGCACA-3′; Reverse, 5′-AACGCTTCACGAATTTGCGT-3′), GAPDH (Forward, 5′-TCAAGGCTGAGAACGGGAAG-3′; Reverse, 5′-TGGACTCCACGACGTACTCA-3′).
Cell counting kit-8 assay
SCC-9 or ISG15 cells were seeded in a 96-well plate (2×103 cells/well). After 12, 24, 48, 72, and 96 h, cell counting kit-8 (CCK-8) reagent (Beyotime, Shanghai, China) was supplemented into each well (5 mg/mL), respectively, with which the cells were incubated in the dark at 37℃ for 2 h. Next, optical density value was determined at 450 nm wavelength.
Transwell assay
For assessing cell migration, Transwell chambers (8 mm pore size, Corning Costar, Cambridge, MA, USA) were placed in 24-well plates. SCC-9 and ISG15 cells (1×105) were transferred into the upper compartment of the Transwell system containing serum-free RPMI-1640 medium, and 0.6 mL of RPMI-1640 medium supplemented with 10% FBS was added to the lower compartment. The cells were cultured for another 24 h. After that, the cells on the upper surface of the filter were removed, while the cells below the surface of the filter were fixed with paraformaldehyde and stained with crystal violet. Subsequently, under the microscope (Olympus, Tokyo, Japan), five visual fields were randomly selected, and the cells were counted. For assessing cell invasion, diluted Matrigel (BD, San Jose, CA, USA) was used to cover the filter before the cells were seeded, and the other procedures were the same as the migration assay.
Luciferase reporter assay
The DNA fragment of wild type (WT) or mutant type (MUT) LINC00662 3′-UTR containing the predicted binding site for miR-144-3p was inserted into the psiCHECK2 vector (Promega, Madison, WI, USA) for constructing the LINC00662-WT reporter or LINC00662-MUT reporter. SCC-9 and ISG15 cells were then co-transfected with LINC00662-WT or LINC00662-MUT and miR-144-3p mimics or miR-control employing Lipofectamine 2000 according to the manufacturer's instructions. Next, the cells were cultured for 48 h. After that, luciferase activity of the cells was detected by the dual-luciferase reporter assay system (Promega). The luciferase activity of firefly was normalized to that of Renilla.
RNA immunoprecipitation
According to the manufacturer's instructions, the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) was used for RNA immunoprecipitation (RIP) experiments. Specifically, SCC-9 and ISG15 cells were lysed using RIP lysis buffer, and then the cell lysate was incubated with magnetic beads coupled with anti-argonaute2 antibody or immunoglobulin G (IgG) antibody. After that, the samples were incubated with proteinase K with shaking to remove the protein. Subsequently, the immunoprecipitated RNA was isolated by TRIzol reagent. Then, the enrichment of LINC00662 and miR-144-3p was tested by qRT-PCR.
Western blot
Cells were collected and lysed in RIPA buffer (Beyotime). Protein concentration was quantified using the BCA Protein Assay Kit (Invitrogen). The samples were separated with 10% SDSPAGE, and then electrophoretically transferred to polyvinylidene fluoride membranes (Millipore). After that, the membranes were blocked in 5% skimmed milk at room temperature for 1 h, followed by incubation with primary antibodies (anti-EZH2, ab186006, 1: 1000; anti-E-cadherin, ab1416, 1:1000; anti-Vimentin, ab92547, 1: 1000; anti-GAPDH, ab9485, 1:5000) for 2 h at 37℃. Subsequently, the membranes were rinsed with TBST three times, and then incubated with horseradish peroxidaseconjugated secondary antibody (Abcam, Cambridge, UK) for an additional 1 h at room temperature. At last, the protein blots were visualized employing an enhanced chemiluminescence kit (Promega). GAPDH was used as the internal reference.
Statistical analysis
All of the experiments were performed in triplicate. Statistical analysis was performed using the GraphPad Prism. Continuous variables are presented as mean±standard deviation, and their differences were analyzed by Student's t-test or one-way analysis of variance. p values<0.05 indicated statistical significance.
RESULTS
LINC00662 expression was up-regulated in OSCC and associated with unfavorable pathological characteristics
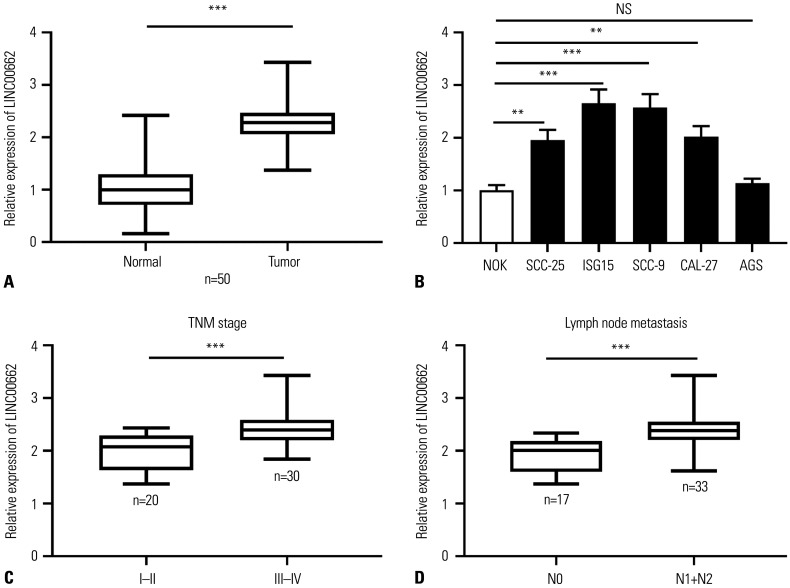
The expression of LINC00662 in 50 OSCC tissues and adjacent non-cancer tissues was quantified using qRT-PCR, and the results showed that it was up-regulated in OSCC tissues compared to adjacent non-cancerous tissues (Fig. 1A). Subsequently, the expression of LINC00662 was detected in OSCC cell lines, and the results showed that LINC00662 was remarkably highly expressed in all of the OSCC cell lines (SCC-25, ISG15, SCC9, and CAL-27) in contrast to NOK cell line and AGS (Fig. 1B), suggesting that the dysregulation of LINC00662 is a common biological event in OSCC cells during the pathogenesis of OSCC. Additionally, the elevated expression of LINC00662 was markedly correlated with the augmentation of TNM stage (Fig. 1C). The expression of LINC00662 in cancer tissues of OSCC patients with lymph node metastasis, compared with that of patients without lymph node metastasis, was also remarkably increased (Fig. 1D), suggesting that abnormally high expression of LINC00665 was probably associated with the progression of OSCC.
Fig. 1. LINC00662 expression was up-regulated in OSCC tissues and cells. (A) qRT-PCR was employed for detecting the expression of LINC00662 in OSCC tissues and adjacent normal tissues. (B) qRT-PCR was adopted for assaying the expression of LINC00662 in NOK cells, OSCC cells (SCC-25, ISG15, SCC-9, and CAL-27), and gastric cancer cell line AGS. (C) The expression level of LINC00662 in OSCC tissues with early stage (I–II) or advanced stage (III–IV) was measured. (D) The expression level of LINC00662 was detected in OSCC tissues with or without lymph node metastasis. **p<0.01, ***p<0.001, and NS: p>0.05. OSCC, oral squamous cell carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction; NOK, normal oral keratinocyte; AGS, human gastric adenocarcinoma cell.
LINC00662 served as a molecular sponge of miR-144-3p in OSCC cells
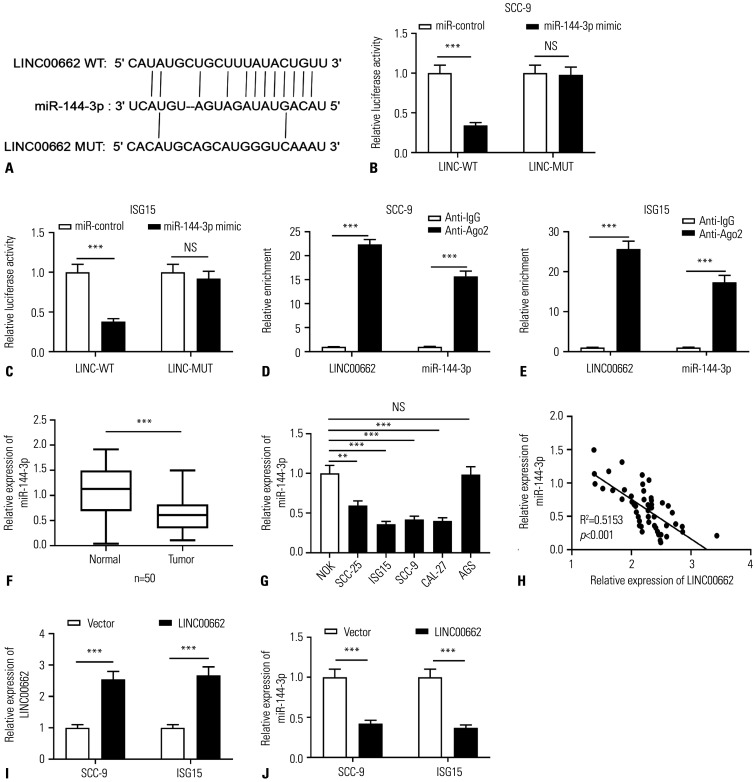
LncRNAs often play a role in cancer biology as the molecular sponge for miRNA.10 Based on this, by searching the StarBase database, target miRNAs of LINC00662 were predicted, and bioinformatics analysis predicted there was a binding sequence between miR-144-3p and LINC00662 (Fig. 2A). Subsequently, the interaction between LINC00662 and miR-144-3p was validated by dual-luciferase reporter assay, the results of which showed that miR-144-3p mimics suppressed the luciferase activity of LINC00662-WT reporter while no remarkable alteration was observed in the luciferase activity of LINC00662-MUT reporter (Fig. 2B and C). Additionally, RIP assay showed that LINC00662 and miR-144-3p were significantly enriched in the Ago2 group compared to the IgG group, suggesting that LINC00662 and miR-144-3p directly interacted in OSCC cells (Fig. 2D and E). Furthermore, qRT-PCR indicated that miR-144-3p expression in OSCC tissues was down-regulated compared to that in adjacent tissues (Fig. 2F), and miR-144-3p expression was down-regulated in all of the OSCC cell lines in contrast to that in NOK cells and AGS cells (Fig. 2G). In addition, the expression level of LINC00662 was shown to be negatively correlated with the expression level of miR-144-3p in OSCC tissues (Fig. 2H). Also, qRT-PCR showed that in SCC-9 and ISG-15 cells overexpressing LINC00662, the expression of miR-144-3p was reduced. Collectively, these data implied that LINC00662 was capable of directly targeting miR-144-3p to suppress its expression in OSCC cells (Fig. 2I and J).
Fig. 2. LINC00662 targeted miR-144-3p in OSCC cells. (A) Bioinformatics analysis was employed for predicting the binding sequence between miR-144-3p and LINC00662. Red-colored letters indicate complementary base pairing. (B and C) The luciferase activity in SCC-9 and ISG15 cells co-transfected with LINC00662-WT or LINC00662-MUT, and miR-control or miR-144-3p mimics was assayed using dual-luciferase reporter gene assay. (D and E) RIP assay was used to validate the direct binding relationship between LINC00662 and miR-144-3p in OSCC cells. (F) qRT-PCR was utilized for quantifying the expression of miR-144-3p in OSCC tissues and adjacent normal tissues. (G) qRT-PCR was used for assaying the expression of miR-144-3p in NOK cells, OSCC cells (SCC-25, ISG15, SCC-9, and CAL-27), and AGS cells. (H) Pearson correlation analysis was employed for evaluating the correlation between miR-144-3p and LINC00662 expression levels in OSCC tissue. (I and J). The expression of LINC00662 or miR-144-3p in SCC-9 and ISG15 cells with LINC00662 overexpression was detected employing qRT-PCR. **p<0.01, ***p<0.001, and NS: p>0.05. WT, wild type; MUT, mutant type; IgG, immunoglobulin G; NOK, normal oral keratinocyte; OSCC, oral squamous cell carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction; RIP, RNA immunoprecipitation; AGS, human gastric adenocarcinoma cell.
The up-regulation of LINC00662 expression expedited proliferation, migration, invasion, and epithelial-mesenchymal transition of OSCC cells by targeting miR-144-3p
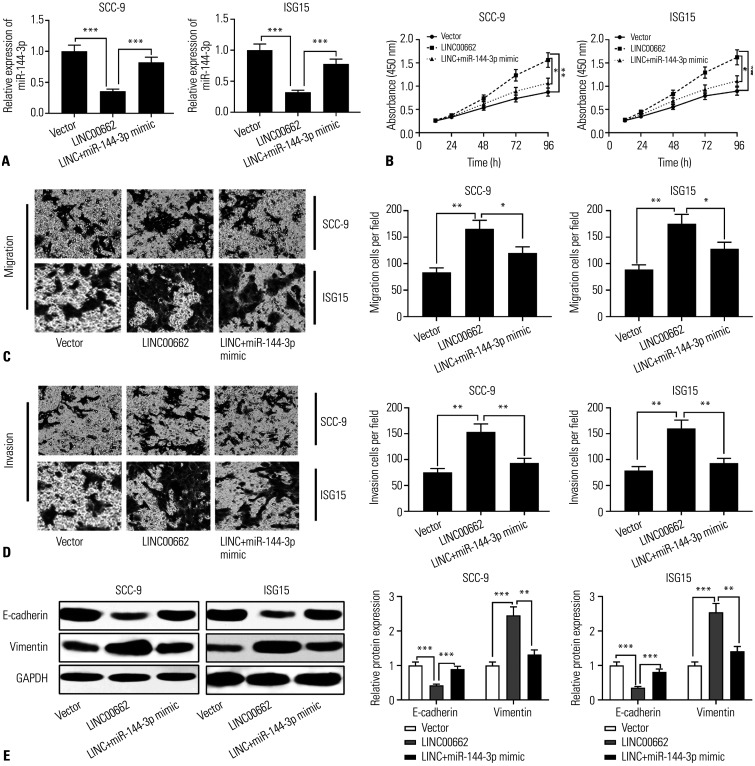
For unveiling the biological function and molecular mechanism of LINC00662 in OSCC, SCC-9 and ISG-15 cells were transfected with LINC00662 overexpression plasmid or co-transfected with LINC00662 overexpression plasmid and miR-144-3p mimics, respectively. qRT-PCR showed that LINC00662 overexpression repressed the expression of miR-144-3p, while miR-144-3p mimics partially restored the expression of miR-144-3p in SCC-9 and ISG-15 cells (Fig. 3A). Next, the proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) of OSCC cells were detected using the CCK-8 method, Transwell experiment, and Western blot, respectively. The results showed that LINC00662 overexpression accelerated OSCC cell proliferation, migration, invasion, and EMT; moreover, the promoting effects of LINC00662 overexpression on the abovementioned biological behaviors were partly counteracted by miR-144-3p mimics (Fig. 3B–E). These data implied that LINC00662 facilitated the proliferation, migration, invasion, and EMT of OSCC cells by repressing miR-144-3p.
Fig. 3. The up-regulation of LINC00662 expression facilitated proliferation, migration, invasion, and EMT of OSCC cells by inhibiting miR-144-3p expression. (A) qRT-PCR was used to detect the expression of miR-144-3p in OSCC cells transfected with LINC00662 overexpression plasmid or co-transfected with LINC00662 overexpression plasmid and miR-144-3p mimics, respectively. (B) The proliferation of transfected cells was detected by CCK-8 method. (C and D) Transwell experiment was used to detect the migration and invasion of transfected cells. (E) Protein expression levels of E-cadherin and Vimentin in transfected cells were detected by Western blot. *p<0.05, **p<0.01, ***p<0.001. EMT, epithelial-mesenchymal transition; qRT-PCR, quantitative real-time polymerase chain reaction; OSCC, oral squamous cell carcinoma; CCK-8, Cell counting kit-8.
The regulatory effects of LINC00662 and miR-144-3p on EZH2 expression
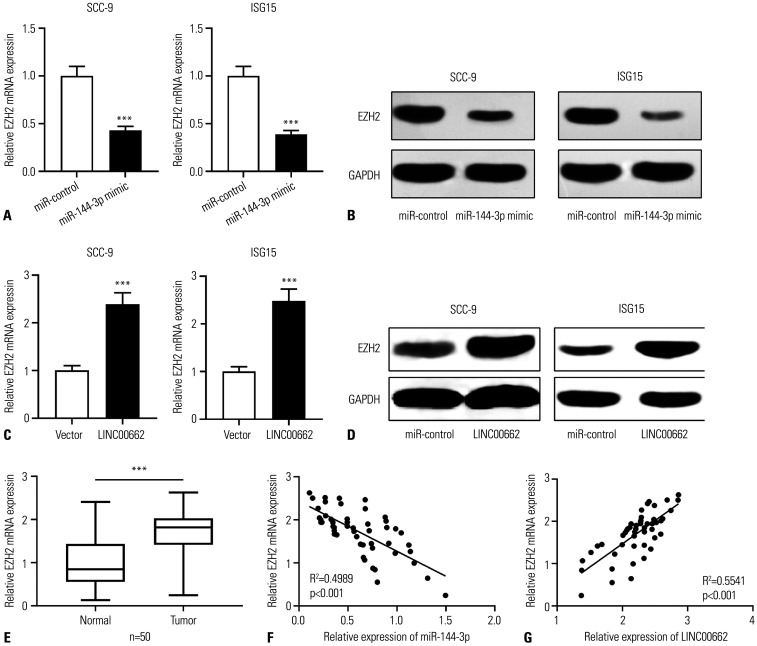
A previous study reported that miR-144-3p modulated the progression of lung adenocarcinoma by repressing EZH2.20 Moreover, EZH2, reportedly, is involved in the development of OSCC.19 Therefore, we investigated the regulatory effects of miR-144-3p and LINC00662 on EZH2 expression in OSCC cells. qRT-PCR and Western blot showed that, compared to the control group, the transfection of miR-144-3p mimics repressed the expression of EZH2 in OSCC cells, whereas LINC00662 overexpression worked oppositely (Fig. 4A–D). Furthermore, EZH2 expression was markedly up-regulated in OSCC tissues, in contrast to adjacent tissues (Fig. 4E). Also, EZH2 expression was negatively interrelated with miR-144-3p expression and positively associated with LINC00662 expression (Fig. 4F and G).
Fig. 4. The regulation of LINC00662/miR-144-3p on EZH2 expression. (A-D) qRT-PCR and Western blot were used to detect the expression level of EZH2 mRNA and protein in SCC-9 and ISG15 cells after the overexpression of miR-144-3p or LINC00662. (E) qRT-PCR was used to detect the expression of EZH2 mRNA in OSCC tissues and adjacent normal tissues. (F and G) Pearson correlation analysis was used to detect the correlation between EZH2 mRNA expression level and miR-144-3p expression level or LINC00662 expression level in OSCC tissues. ***p<0.001. EZH2, enhancer of zeste homolog 2; qRT-PCR, quantitative real-time polymerase chain reaction; OSCC, oral squamous cell carcinoma.
LINC00662 accelerated OSCC progression by regulating miR-144-3p/EZH2 axis
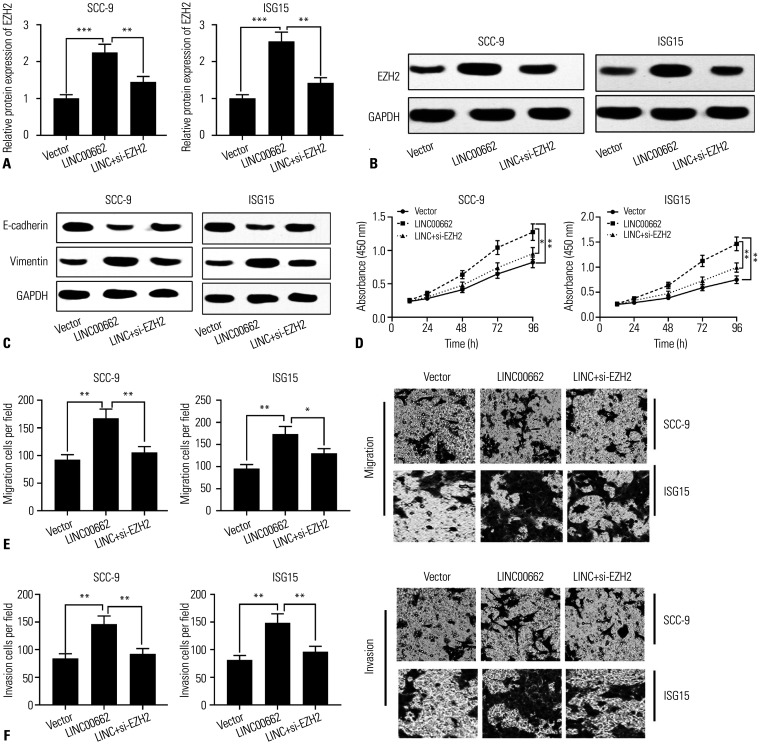
Subsequently, we investigated whether LINC00662 regulated the progression of OSCC by regulating EZH2. EZH2 siRNAs (si-EZH2) were transfected into SCC-9 and ISG-15 cells overexpressing LINC00662. Western blotting showed that the expression of EZH2 was augmented in OSCC cells with LINC00662 overexpression, whereas it was reduced after the transfection of si-EZH2 (Fig. 5A and B). As shown, the promoting effect of overexpression LINC00662 on EMT was partially weakened by si-EZH2 (Fig. 5C). CCK-8 and Transwell assays verified that the knockdown of EZH2 observably counteracted the promoting effects of overexpression of LINC00662 on OSCC cell proliferation, migration, and invasion (Fig. 5D–F). All in all, we concluded that LINC00662 promoted the proliferation, migration, invasion, and EMT of OSCC cells by regulating the miR-144-3p/EZH2 axis.
Fig. 5. Silencing EZH2 could reverse the cancer-promoting effects of LINC00662 in OSCC. (A and B) Western blot was used to detect the expression of EZH2 protein in SCC-9 and ISG15 cells transfected with LINC00662 overexpression plasmid or co-transfected with LINC00662 overexpression plasmid and si-EZH2, respectively. (C) Western blot was used to detect the protein expression levels of E-cadherin and Vimentin in transfected cells. (D and F) The proliferation, migration, and invasion of transfected cells were detected by CCK-8 method and Transwell experiment, respectively. *p<0.05, **p<0.01, ***p<0.001. EZH2, enhancer of zeste homolog 2; OSCC, oral squamous cell carcinoma; CCK-8, Cell counting kit-8.
DISCUSSION
LncRNA can regulate gene expression at the transcriptional level or post-transcriptional level.10 Previous research demonstrated that lncRNA plays a pivotal role in diverse biological processes, including tumorigenesis and cancer progression.7,21,22 Aberrantly expressed lncRNAs are observed in OSCC. For instance, lncRNA AC007271.3, highly expressed in OSCC, expedites cancer cell proliferation, migration and invasion, and represses apoptosis via regulating Wnt/β-catenin signaling;23 lncRNA CASC9, associated with tumor size and metastasis, works as a potential biomarker of detrimental prognosis of OSCC patients.24 Besides, LINC00662 has been reported to exert a carcinogenic role in diverse cancers, and facilitate the progression of liver cancer by altering genomic methylation status.25 The expression of LINC00662 in gastric cancer is markedly up-regulated, and its high expression indicates unfavorable prognosis; the knockdown of LINC00662 represses OSCC cell proliferation and sensitizes OSCC cells to chemotherapeutics.9 In the current study, LINC00662 expression was observed to be up-regulated in OSCC tissues and cell lines, and its high expression was associated with unfavorable pathological characteristics of the patients. Additionally, in vitro experiments showed that LINC00662 promoted the proliferation, migration, and invasion of OSCC cells, indicating that LINC00662 exerts a carcinogenic role in OSCC, which is consistent with the previous report.11
LncRNAs can exert their biological effects by serving as the molecular sponge for miRNAs, and this mechanism is involved in cancer biology. For example, lncRNA UCA1 promotes OSCC cell proliferation and chemoresistance by restraining miR-184 expression.26 LINC01234 enhances the aggressiveness of OSCC cells by regulating miR-637/NUPR1 axis.27 In the present work, LINC00662 was identified as a molecular sponge of miR-144-3p. Previous research demonstrated that miR-144-3p exhibits a tumor-suppressive role in various cancers, including renal clear cell carcinoma, glioblastoma, and thyroid papillary carcinoma.28,29,30 Specifically, miR-144-3p inhibits cancer cell proliferation and invasiveness by targeting TOP2A in HCMV-positive glioblastoma cells.29 Additionally, miR-144-3p can target PAX8 to repress the growth and metastasis of thyroid papillary cancer cells.30 Intriguingly, miR-144-3p can repress tumor cell proliferation, migration, and invasion by targeting the ERO1L/STAT3 signaling pathway in OSCC.15 In the present study, we demonstrated that LINC00662 could decoy miR-144-3p and suppress the expression of miR-144-3p in OSCC cells. Consistently, reduced expression of miR-144-3p was observed in OSCC tissues and cell lines. Moreover, miR-144-3p could counteract the biological effects of LINC00662 on OSCC cells. These results suggested that LINC00662 exerted its oncogenic functions by repressing miR-144-3p.
In this study, we also explored the downstream mechanism of miR-144-3p, and bioinformatics analysis predicted that EZH2, a catalytic subunit in polycomb repressive complex 2 with the function of methyltransferase, was one of the candidate targets of miR-144-3p.31 EZH2 contains an active SET catalytic domain, and it can alter the gene expression via the trimethylation of Lys-27 in histone 3 (H3K27me3); H3K27me3 is associated with reduced gene expression, and is considered an important epigenetic event during stem cell fate determination.32,33 EZH2 is highly expressed in multiple human malignancies and is associated with tumor progression and prognosis,19,34 regulating the proliferation, migration, invasion, apoptosis, and other malignant phenotypes of cancer cells.17,18,19 For example, EZH2 facilitates the recruitment of macrophages and changes the immune microenvironment in lung cancer by increasing the expression of CCL5.35 Moreover, EZH2-mediated up-regulation of ROS1 expedites OSCC cell metastasis.36 Notably, several previous studies reported that EZH2 is a target gene of miR-144-3p in endometrial cancer, osteosarcoma, and lung adenocarcinoma.20,37,38 Consistently, the results of the present study showed that EZH2 was negatively regulated by miR-144-3p. Additionally, EZH2 was observed to be positively regulated by LINC00662. Furthermore, the promoting effects of LINC00662 on the proliferation, migration, invasion, and EMT of OSCC cells were counteracted by EZH2 depletion. In light of these findings, we concluded that LINC00662 promoted the progression of OSCC partly by regulating the miR-144-3p/EZH2 axis.
In brief, we herein report that LINC00662 facilitates the proliferation, migration, invasion, and EMT by sponging miR-144-3p and up-regulating EZH2 expression in OSCC. This study partly explains the mechanism of the tumorigenesis of OSCC, and our data suggest that targeting LINC00662 may be a promising strategy for blocking OSCC progression.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Zhaohua Meng and Yongmei Yao.
- Data curation: Fengqin Jin.
- Formal analysis: Yang Liu.
- Funding acquisition: Zhaohua Meng.
- Investigation: Yongmei Yao.
- Methodology: Yongmei Yao and Yang Liu.
- Project administration: Zhaohua Meng and Fengqin Jin.
- Resources: Zhaohua Meng and Yongmei Yao.
- Software: Fengqin Jin, Yang Liu, and Yongmei Yao.
- Supervision: Zhaohua Meng.
- Validation: Yongmei Yao and Zhaohua Meng.
- Visualization: Fengqin Jin, Yang Liu, and Yongmei Yao.
- Writing—original draft: Yongmei Yao and Fengqin Jin.
- Writing—review & editing: Zhaohua Meng and Yongmei Yao.
- Approval of final manuscript: all authors.
References
- 1.Eckert AW, Wickenhauser C, Salins PC, Kappler M, Bukur J, Seliger B. Correction to: clinical relevance of the tumor microenvironment and immune escape of oral squamous cell carcinoma. J Transl Med. 2018;16:40. doi: 10.1186/s12967-018-1407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma--an update. CA Cancer J Clin. 2015;65:401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 3.Lee CF, Chiang NN, Lu YH, Huang YS, Yang JS, Tsai SC, et al. Benzyl isothiocyanate (BITC) triggers mitochondria-mediated apoptotic machinery in human cisplatin-resistant oral cancer CAR cells. Biomedicine (Taipei) 2018;8:15. doi: 10.1051/bmdcn/2018080315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie S, Xu H, Shan X, Liu B, Wang K, Cai Z. Clinicopathological and prognostic significance of survivin expression in patients with oral squamous cell carcinoma: evidence from a meta-analysis. PLoS One. 2015;10:e0116517. doi: 10.1371/journal.pone.0116517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Q, Li Q, Xie F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco Targets Ther. 2019;12:6297–6307. doi: 10.2147/OTT.S214689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Liu Y, Lu Q, Zhou X, Chen L, Liang W. The lncRNA TUG1 promotes epithelial ovarian cancer cell proliferation and invasion via the WNT/β-catenin pathway. Onco Targets Ther. 2018;11:6845–6851. doi: 10.2147/OTT.S167900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Liu Z, Yao Y, Huang S, Li L, Jiang B, Guo H, et al. LINC00662 promotes gastric cancer cell growth by modulating the Hippo-YAP1 pathway. Biochem Biophys Res Commun. 2018;505:843–849. doi: 10.1016/j.bbrc.2018.09.191. [DOI] [PubMed] [Google Scholar]
- 10.Li N, Zhang LY, Qiao YH, Song RJ. Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur Rev Med Pharmacol Sci. 2019;23:3688–3698. doi: 10.26355/eurrev_201905_17792. [DOI] [PubMed] [Google Scholar]
- 11.Xu D, Chen Y, Yuan C, Zhang S, Peng W. Long non-coding RNA LINC00662 promotes proliferation and migration in oral squamous cell carcinoma. Onco Targets Ther. 2019;12:647–656. doi: 10.2147/OTT.S188691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi GK, Deitz-McElyea S, Johnson M, Mali S, Korc M, Sardar R. Highly specific plasmonic biosensors for ultrasensitive microRNA detection in plasma from pancreatic cancer patients. Nano Lett. 2014;14:6955–6963. doi: 10.1021/nl503220s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B, Lin X, Tian F, Yu W, Qiao B. MiR-133a-3p inhibits oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J Cell Biochem. 2018;119:338–346. doi: 10.1002/jcb.26182. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Jiang H, Zhao J, Wen H. MiRNA-16 inhibited oral squamous carcinoma tumor growth in vitro and in vivo via suppressing Wnt/β-catenin signaling pathway. Onco Targets Ther. 2018;11:5111–5119. doi: 10.2147/OTT.S153888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Li Y, Jiang C, Chen L, Gan N. MicroRNA-144-3p inhibits tumorigenesis of oral squamous cell carcinoma by downregulating ERO1L. J Cancer. 2020;11:759–768. doi: 10.7150/jca.33267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae WK, Hennighausen L. Canonical and non-canonical roles of the histone methyltransferase EZH2 in mammary development and cancer. Mol Cell Endocrinol. 2014;382:593–597. doi: 10.1016/j.mce.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Wang Z, Lu W, Jiang H, Lu J, Qiu J, et al. EZH2 promotes gastric cancer cells proliferation by repressing p21 expression. Pathol Res Pract. 2019;215:152374. doi: 10.1016/j.prp.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Jin X, Yang J, Yang Y, He Y, Ding L, et al. Inhibition of EZH2 by chemo- and radiotherapy agents and small molecule inhibitors induces cell death in castration-resistant prostate cancer. Oncotarget. 2016;7:3440–3452. doi: 10.18632/oncotarget.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng M, Cao MX, Luo XJ, Li L, Wang K, Wang SS, et al. EZH2 promotes invasion and tumour glycolysis by regulating STAT3 and FoxO1 signalling in human OSCC cells. J Cell Mol Med. 2019;23:6942–6954. doi: 10.1111/jcmm.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Yang Z, Deng Z, Zhou Y, Gong Q, Zhao R, et al. Downregulated miR-144-3p contributes to progression of lung adenocarcinoma through elevating the expression of EZH2. Cancer Med. 2018;7:5554–5566. doi: 10.1002/cam4.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 22.Wang F, Ji X, Wang J, Ma X, Yang Y, Zuo J, et al. LncRNA PVT1 enhances proliferation and cisplatin resistance via regulating miR-194-5p/HIF1a axis in oral squamous cell carcinoma. Onco Targets Ther. 2020;13:243–252. doi: 10.2147/OTT.S232405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao TR, Zheng ZN, Chen YC, Wu QQ, Huang GZ, Li F, et al. LncRNA AC007271.3 promotes cell proliferation, invasion, migration and inhibits cell apoptosis of OSCC via the Wnt/β-catenin signaling pathway. Life Sci. 2019;239:117087. doi: 10.1016/j.lfs.2019.117087. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10:41. doi: 10.1038/s41419-018-1280-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo T, Gong C, Wu P, Battaglia-Hsu SF, Feng J, Liu P, et al. LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 2020;27:2191–2205. doi: 10.1038/s41418-020-0494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. 2017;6:2897–2908. doi: 10.1002/cam4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Cao J, Peng X. LINC01234 facilitates growth and invasiveness of oral squamous cell carcinoma through regulating the miR-637/NUPR1 axis. Biomed Pharmacother. 2019;120:109507. doi: 10.1016/j.biopha.2019.109507. [DOI] [PubMed] [Google Scholar]
- 28.Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C, et al. Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem. 2017;43:2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 29.Song J, Ma Q, Hu M, Qian D, Wang B, He N. The inhibition of miR-144-3p on cell proliferation and metastasis by targeting TOP2A in HCMV-positive glioblastoma cells. Molecules. 2018;23:3259. doi: 10.3390/molecules23123259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Su C, Chen Y, Li G. MiR-144-3p promotes the tumor growth and metastasis of papillary thyroid carcinoma by targeting paired box gene 8. Cancer Cell Int. 2018;18:54. doi: 10.1186/s12935-018-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150:3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li F, et al. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov. 2016;6:1006–1021. doi: 10.1158/2159-8290.CD-16-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia L, Zhu X, Zhang L, Xu Y, Chen G, Luo J. EZH2 enhances expression of CCL5 to promote recruitment of macrophages and invasion in lung cancer. Biotechnol Appl Biochem. 2020;67:1011–1019. doi: 10.1002/bab.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih CH, Chang YJ, Huang WC, Jang TH, Kung HJ, Wang WC, et al. EZH2-mediated upregulation of ROS1 oncogene promotes oral cancer metastasis. Oncogene. 2017;36:6542–6554. doi: 10.1038/onc.2017.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Ge L, Xu XJ, Yang T, Yuan Y, Ma XL, et al. LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the miR-144-3p/EZH2 axis. Radiol Oncol. 2019;53:434–442. doi: 10.2478/raon-2019-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao J, Han X, Qi X, Jin X, Li X. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol. 2017;51:1115–1123. doi: 10.3892/ijo.2017.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]