Graphical abstract
Keywords: COVID-19, Public health guidance, Taxonomy, Content analysis, Indexing, Knowledge management
Abstract
Background
Development and dissemination of public health (PH) guidance to healthcare organizations and the general public (e.g., businesses, schools, individuals) during emergencies like the COVID-19 pandemic is vital for policy, clinical, and public decision-making. Yet, the rapidly evolving nature of these events poses significant challenges for guidance development and dissemination strategies predicated on well-understood concepts and clearly defined access and distribution pathways. Taxonomies are an important but underutilized tool for guidance authoring, dissemination and updating in such dynamic scenarios.
Objective
To design a rapid, semi-automated method for sampling and developing a PH guidance taxonomy using widely available Web crawling tools and streamlined manual content analysis.
Methods
Iterative samples of guidance documents were taken from four state PH agency websites, the US Center for Disease Control and Prevention, and the World Health Organization. Documents were used to derive and refine a preliminary taxonomy of COVID-19 PH guidance via content analysis.
Results
Eight iterations of guidance document sampling and taxonomy revisions were performed, with a final corpus of 226 documents. The preliminary taxonomy contains 110 branches distributed between three major domains: stakeholders (24 branches), settings (25 branches) and topics (61 branches). Thematic saturation measures indicated rapid saturation (≤5% change) for the domains of “stakeholders” and “settings”, and “topic”-related branches for clinical decision-making. Branches related to business reopening and economic consequences remained dynamic throughout sampling iterations.
Conclusion
The PH guidance taxonomy can support public health agencies by aligning guidance development with curation and indexing strategies; supporting targeted dissemination; increasing the speed of updates; and enhancing public-facing guidance repositories and information retrieval tools. Taxonomies are essential to support knowledge management activities during rapidly evolving scenarios such as disease outbreaks and natural disasters.
1. Introduction and background
Pandemics create a challenging environment for public health (PH) officials to communicate guidance to the public at large and those making clinical and operational decisions in healthcare settings. PH organizations produce guidance pertaining to a wide range of topics, stakeholders and settings, e.g. protocols for collecting lab specimens, signage to be used in restaurants, and travel advisories for the general public. Moreover, state-of-the-art guidance required for decision-making is likely to rapidly change, which can be challenging for knowledge management (KM) strategies that rely on stable concepts to deliver the right guidance to the right stakeholder at the right time [1]. In the domain of PH, the ideal model of guidance dissemination is sometimes described as a knowledge “funnel” [2]. In this model, national- and international-level organizations synthesize best available evidence to inform PH control measures and then distribute guidance to organizations representing increasingly granular jurisdictions and stakeholders. Those organizations then curate available guidance, add further details to address local needs, and make relevant information accessible to their stakeholders, such as healthcare organizations, schools and individuals. In the United States, such a system typically involves guidance disseminated from the Center for Disease Control and Prevention (CDC) and the National Institutes of Health (NIH) to state or large city PH agencies, and then to more local (city or county) PH officials with greater familiarity with the needs of their communities.
In reality, implementing the knowledge “funnel” creates numerous challenges during large disease outbreaks. During the H1N1 pandemic, the “knowledge funnel” model created communication challenges and information overload [1], [3]. Similarly, during the present COVID-19 pandemic, PH organizations must rapidly produce, organize, and disseminate assets (guidelines and educational materials) to relevant stakeholders. Knowledge of the natural history of the disease, diagnosis and treatment changes rapidly, collapsing the typical years-long timeframe in which scientific research is transformed into guidelines [2]. Shorter and overlapping cycles create a significant challenge for typical KM asset management strategies. In addition, frequently updated guidance greatly increases the burden on healthcare systems, providers, and consumers to find and comprehend the most relevant and current information available. Unfortunately, pandemics tend to be accompanied by “infodemics” of misinformation that can spread rapidly via social media and other online platforms, and that PH officials strive to dispel [4], [5], [6], [7], [8], [9]. All of these factors contribute to a large volume of accurate and inaccurate information that PH officials must curate for their constituencies, and that stakeholders are faced with when seeking out guidance.
A customized taxonomy for classifying PH guidance is an important tool for addressing these problems and enabling informatics strategies that support the entire KM cycle, supporting expert activities like content development, curation, and indexing of assets designed by public health officials. A taxonomy of pH guidance can support guidance dissemination strategies by helping public health officials target the most relevant stakeholders and settings. Dissemination tools may include tools such as search engines, content feeds, recommender systems, and context–aware information retrieval (e.g. ‘Infobuttons’) [10]. A taxonomy also simplifies the systematic identification of gaps, redundancies, and conflicts in existing recommendations for public health seeking to produce new guidance. A well-designed taxonomy of public health guidance thus has the potential to help “harmonize” all aspects of knowledge authoring and dissemination, across organizations and settings.
KM strategies applied to PH emphasize the importance of coherent processes to manage the entire knowledge curation and dissemination life cycle, including the identification of relevant evidence, asset creation, updating, storage, evidence synthesis, intervention and evaluation [11]; and the shortcomings of approaches that fail to carefully target guidance [1]. Initial research on KM in PH has focused on the translation of scientific findings into evidence-based guidelines for healthcare systems [12], [13] and PH case reporting [14], [15]. In response to the COVID-19 pandemic, a number of studies have also sought to characterize knowledge search and dissemination patterns by the general public [6], [16], [17], [18]. Other important KM-related COVID-19 research initiatives include the COVID-19 Knowledge Accelerator [19] and the Text REtrieval Conference COVID project [20], both of which are primarily expert-facing projects to synthesize scientific evidence. Despite these important contributions to COVID-19 PH efforts, we identified a lack of means for systematically classifying COVID-19 or other guidance as it relates to diverse PH, healthcare and non-healthcare settings and stakeholders. Consequently, this study used the COVID-19 pandemic as a test case for developing a strategy to sample and taxonomically classify PH pandemic guidance to support dissemination efforts to individuals and organizations beyond public health agencies, including business, government and private organizations and members of the general public. The objectives of our project were thus to: i) design an efficient method for sampling guidance across multiple PH organizations; and ii) apply the sampling methodology to develop a taxonomy of COVID-19 PH guidance.
2. Methods
This study sought to develop a taxonomy of pH guidance related to the ongoing COVID-19 pandemic across multiple PH organizations in the US. Our unit of analysis was individual PH guidance “documents” in HTML, PDF or MS Word format. We defined “guidance” as a document containing recommended or required actions for a particular individual, population, or type of organization. This definition excluded documents intended to educate about aspects of COVID-19 without suggesting specific actions to be taken. Utah Department of Health’s “Travel Guidance” and “Overview of COVID-19 Surveillance” pages provide examples of documents that do and do not meet the study’s inclusion criteria, respectively [21], [22]. Streamlined content analysis of documents was used to assign documents to PH guidance categories and establish a taxonomy. We discuss methods in relation to three phases that were iterated over the course of the study: sample selection, content analysis, and sample strategy revision. Fig. 1 visualizes each of these phases. Steps in the diagram are referenced in the description of each phase.
Fig. 1.
Phases and steps within the iterated sampling and classification workflow. Circles indicate the point at which the method is iterated; text in the circles specifies what (if any) changes to sampling strategy are to be made in the next iteration. The workflow was terminated when stakeholders and settings reached saturation for two iterations [1]Channels included FL, IL, MA, UT and CDC [2]Purposive sampling relies on analyst judgment to select samples that maximize variability; random sampling selects a proportion of samples from a corpus stratified by a priori categories. [3]Interrater reliability is measured using kappa statistics. 0.75 is considered a desirable kappa. [4]Thematic saturation refers to the degree of exhaustiveness with which the taxonomy captures concepts in sampled documents. Less than or equial to 5% change between iterations is considered acceptable.
2.1. Sample selection
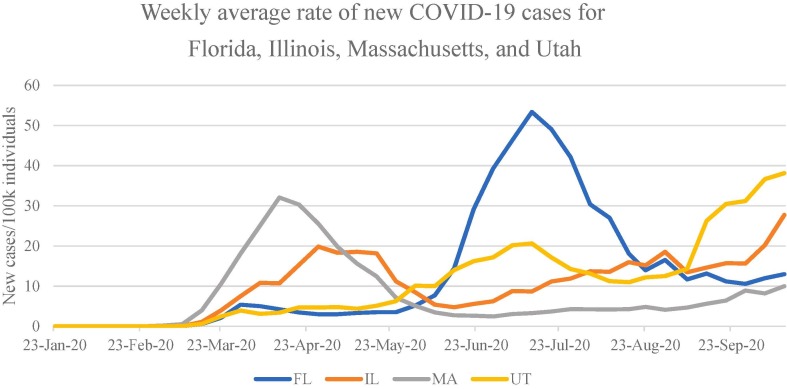
Step A1: Each iteration of sampling and taxonomic development began with the selection of states and externally linked organizations to target. Four U.S. state PH agencies were ultimately included in the study: Florida (FL), Illinois (IL), Massachusetts (MA), and Utah (UT). State agencies, rather than federal, were selected because we anticipated they would be responsive both to guidance changes at the federal level and shifting conditions in the state’s local communities. These four states were selected because they had differing patterns of COVID-19 infections which may impact the guidance PH is attempting to share; belong to different administrative regions of the U.S. Department of Health and Human Services (HHS); and are geographically dispersed. CDC, NIH and WHO websites were also sampled following procedures described below. Infection rate patterns in sampled states are shown in Fig. 2 . Sampling began with UT and MA because investigators were familiar with the respective state public health websites, and the issues that were at the forefront of public discourse in each state. This contextual knowledge helped to guide sampling. Sampling was later expanded to FL and IL. The iterative approach was intended to encourage rigor by ensuring that all coders were using codes the same way on a limited number of documents before expanding the corpus, a common strategy in content analysis.
Fig. 2.
Weekly average rate of new COVID-19 cases for Florida, Illinois, Massachusetts, and Utah. Y-axis is new cases/100 k individuals based on 2019 census data. Data drawn from JHU CSSE.[23].
Step A2: URLs for PH guidance were retrieved through an automated process followed by manual verification. PH agency websites were crawled using a commercial web-crawler tool (Versionista®; Portland, OR), generating an inventory of pandemic-related links. Two states (UT, FL) had web domains dedicated to COVID-19 guidance, which were exhaustively inventoried. For the two remaining states (IL, MA), filters were applied within the web-crawler to limit the search to COVID-19-related web pages. The script used for document retrieval can be found on Mendeley Data [24].
Step A3: To extract links to PH organizations external to the four state agencies, the inventories of links were processed using custom scripts developed in Apple Automator. First, URLs for HTML pages were scanned for external links. Second, static .pdf and .doc documents were downloaded and scanned for external links. Both sets of results were filtered to retain only links to CDC, NIH and WHO web pages. Duplicate links were removed. This semi-automated process produced an inventory of COVID-19 related URLs for each state and externally linked organizations. The scripts used to extract external links can be found on Mendeley Data [24].
Step A4: Each URL was manually checked by a team of four analysts to ascertain whether the link contained PH guidance, according to the study’s definition.
Step A5: If the linked document was determined to be guidance, it was assigned to one of three coarse stakeholder categories: general consumer, healthcare organization, or non-healthcare organization. “General consumer” guidance was defined as guidance related to individuals or the general population. “Healthcare organization” guidance was defined as guidance related to clinical care or the management of operations within healthcare organizations. “Non-healthcare organization” guidance was defined as guidance relating to any non-healthcare organization, including private businesses, government agencies and non-profit organizations. These categories offered a “first approximation” of the relevant stakeholders referenced by guidance documents to assist sampling. Stakeholder categories were refined via iterative classification, described further in Results.
Step A6: A determination was made in each iteration of sampling to use purposive or random sampling. Purposive sampling was used to maximize the concepts captured in small samples during early iterations, typically using 3–10 PH guidance documents. The final two iterations used random samples of 20% of all documents identified on the FL, IL, MA, UT and CDC websites (n = 234) to test the method at a larger scale and capture concepts missed by purposive sampling.
2.2. Content analysis
Step B1: Sampled PH guidance was classified via iterative content analysis [25]. Review of documents focused on capturing high level concepts or “gist”, rather than the details of guidance (e.g. capturing that a document indicated how to handle laboratory specimens, but not capturing what the recommended procedures were). All taxonomic classification was performed in NVivo 12. Documents were classified in terms of three broad domains: stakeholder, setting, and topic. “Stakeholder” refers to any individual, population or organization for which the guidance was explicitly related (e.g. business owner or manager; healthcare support staff). In some cases, the inference was made that guidance was intended for the general population when not explicitly stated. “Setting” refers to types of physical locations in which the guidance is expected to be relevant (e.g. freestanding clinics; schools). “Topic” refers to the general subjects that the guidance pertains to (e.g. COVID-19 lab testing procedures; physical distancing in private businesses). Documents could be assigned to multiple concepts within any of these domains. In iterations with purposive sampling, documents were independently reviewed and classified by four trained analysts simultaneously to ensure consistency in classification (PT, SP, ER, AL). The content analysis team included experts with backgrounds in anthropology, nursing, medicine, and global health, in addition to informatics.
Step B2: Interrater reliability was calculated to measure consistency of classification in the iteration. As is common in qualitative research and content analysis, kappa scores were used to quantify the degree of agreement between analysts and thus monitor the consistency of concept use and the study’s internal validity. Fleiss’ kappa is an interrater reliability metric used for three or more raters scaled between 0 and 1, interpreted as the likelihood that classification agreement is not by chance, with 0.75 or better deemed sufficient by convention [26]. Fleiss’ kappa was calculated in Stata via the kappaetc package.
Step B3: Concepts identified in guidance were discussed, concept names and definitions were designed by group consensus and concepts were merged into a master taxonomy. Definitions for clinical concepts drew on the taxonomy of generic clinical questions designed by Ely et al. [27]. In random sampling iterations, each document was assigned to one of four analysts, who then reported to the full group on changes that might be required in the taxonomy. Recommended changes were then discussed and made based on the consensus of all four analysts, resulting in an iteratively refined taxonomy.
Throughout the classification process, the taxonomy was periodically reviewed by a PH domain expert (CJS) to evaluate its coherence and relevance, as well as by the larger research team. This team-based approach conforms to best practices in qualitative coding and content analysis [28], [29], [30]. Classification at the level of the document (rather than paragraph or line) offered an unambiguous textual unit of analysis, another key component of interrater reliability and helped to streamline analysis [31].
Step B4: Thematic saturation was estimated for each iteration. Thematic saturation is a common means of conceptualizing sample adequacy in content analysis. It refers to the degree of exhaustiveness of a sample relative to the thematic content of a corpus of text. The conceptualization of thematic saturation varies [32], [33], although most approaches assume that the themes in a corpus are finite, such that iterated sampling will result in an increasingly exhaustive inventory of themes. This is not a realistic assumption in the context of a pandemic, in which new knowledge emerges rapidly in response to scientific research and real-world problems. However, thematic saturation metrics are still useful for quantifying the thematic “yield” of particular sampling strategies or phases (e.g. changes in the channels searched and topical focus), thus helping to guide ongoing sampling. This is the general purpose of the simple method of measuring thematic saturation proposed by Guest et al. [34], who recommend quantifying thematic saturation by measuring percent changes in the number of themes (or concepts) across iterations of a codebook (or taxonomy). Guest et al. suggest that sufficient saturation.has been reached when the number of identified concepts across sampling iterations change by 5% or less. This study assumed that stakeholder and setting concepts might stabilize sufficiently to achieve saturation, but that the topic domain would remain dynamic. For this reason, two consecutive iterations showing saturation in the stakeholder and setting domains was deemed an appropriate endpoint for analysis.
2.3. Sample strategy revision:
Step C1: Low kappa scores were taken as an indication that the sampling strategy of the previous iteration should be repeated to improve analysts’ shared understanding of existing categories through further use and discussion. If kappa scores were low (less than 0.75), the sampling strategy was repeated (skipping Step C2 and going to Step C3a). If kappa scores were high, adjustments to the sampling strategy were considered (continuing to Step C2).
Step C2: Percent change in the taxonomy was assessed in relation to a threshold of 10% change. The threshold was chosen because it indicated that change in the taxonomy was no longer as high as when channels or topics were first introduced, and that expanding the sampling strategy to new channels, settings or stakeholders would thus be more feasible. If percent change in taxonomy size was greater than 10 % analysts continued to Step C3b, repeating the previous sampling strategy without change. If percent change in taxonomy size was less than 10% , analysts continued to C3c, in which the sampling strategy was expanded to include more channels, settings or stakeholders.
Step C3a: Analysts reached this step if they returned low kappa scores. In this case, analysts repeated an iteration of sampling and taxonomy development with no modification to the targeted stakeholders, topics, channels, or sampling strategy.
Step C3b: Analysts reached this step if they measured high kappa scores and high percent change in taxonomy size (poor saturation). In this case, analysts considered expanding stakeholder or topic domains while retaining a purposive sampling strategy to efficiently capture variation.
Step C3c: Analysts reached this step if they measured high kappa scores and low percent change in taxonomy size (adequate saturation). In this case, analysts considered expanding stakeholder or topic domains, adding channels (e.g. additional states or linked documents from external organizations), and using random sampling to capture variation missed by purposive sampling in highly saturated domains. Sampling was terminated when setting and stakeholder categories remained at or below the 5% change threshold for two consecutive iterations.
3. Results
Study results are reported below for each of the two study aims.
3.1. Designing an efficient method for sampling guidance across multiple PH organizations
The four states inventoried varied drastically in the size and structure of their online PH resources. As noted in Methods, UT and FL had dedicated COVID-19-related web domains, while PH guidance for IL and MA was integrated into larger state government domains. After filtering URLs for relevance in IL and MA, the total number of URLs ranged from 54 (FL), 216 (IL), 229 (UT) to 581 (MA) (Table1 ). About half of the web pages for FL, IL and MA provided some form of guidance, while guidance pages for UT were only 32% of the state’s total inventory.
Table 1.
Number of URLs and proportion of randomly sampled guidance documents identified for each state public health agency. Links to NIH and WHO are omitted because of their infrequency.
| FL | IL | MA | UT | ||
|---|---|---|---|---|---|
| # URLs identified in state PH websites | 54 | 216 | 581 | 229 | |
| # (%) URLs containing PH guidance | 31 (57%) | 110 (51%) | 319 (55%) | 74 (32%) | |
| Proportion of guidance documents by stakeholder | % Healthcare | 42 | 41 | 35 | 23 |
| % Non-healthcare | 23 | 31 | 44 | 20 | |
| % General public | 35 | 28 | 22 | 57 | |
| # External links to CDC | 105 | 108 | 62 | 51 | |
| # (%) CDC links containing guidance | 78 (74%) | 67 (62%) | 37 (60%) | 38 (75%) | |
| Proportion of guidance documents by stakeholder | % Healthcare | 31 | 34 | 32 | 32 |
| % Non-healthcare | 8 | 24 | 24 | 16 | |
| % General public | 62 | 42 | 43 | 53 | |
| # External links to NIH or WHO | 0 | 4 | 5 | 0 | |
Across all states, the overwhelming majority of external links from state PH websites (n = 326; 97%) referred to guidance on the CDC website, ranging from 51 to 105 links. Links to NIH and WHO guidance were only present for IL and MA, and limited to only 4 and 5 links, respectively. Sixty to 75% of links from state PH agencies to the CDC led to guidance web pages vs. non-guidance. For all states, the general public was the stakeholder most frequently targeted by linked CDC guidance, followed by healthcare and non-healthcare stakeholders, respectively. A summary of URLs inventoried by this study is provided in Table 1.
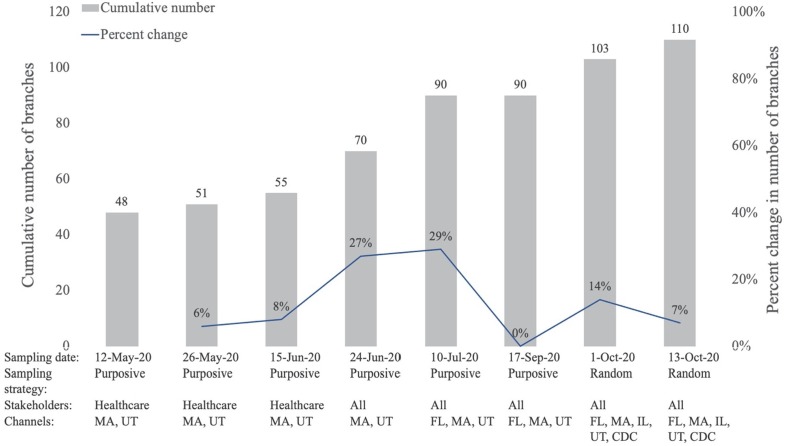
To illustrate thematic saturation, Fig. 3 describes the percent change in number of branches across major iterations of the taxonomy, which included purposive sampling between May and September 2020, followed by two rounds of random sampling in October 2020. Overall, this study achieved 5% or lower rates of change for numerous iterations in the stakeholder and setting domains of the taxonomy, as well as for topics related to clinical care. In early iterations, constraining the stakeholder (healthcare only), channel (UT, MA), and topic (clinical care) allowed the project to achieve saturation close to the ideal quickly (6–8% change in number of branches). Adding states and stakeholders resulted in greatly increased taxonomy size in June and July. After six rounds of purposive sampling and the addition of FL to the sample, one iteration achieved a 0% change. Notably, this was followed by two iterations that involved the analysis of a large volume of randomly sampled documents in October and resulted in a smaller increase in taxonomy size (14% and 7%, respectively) than the purposive sampling in June and July.
Fig. 3.
Cumulative and percent change in total taxonomy branches across major iterations of guidance sampling from FL, IL, MA, UT and/or CDC.
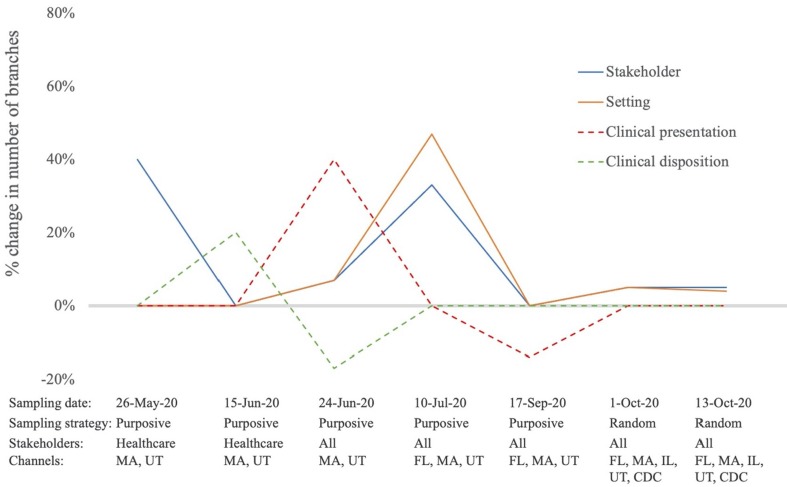
To better understand changes in the taxonomy over time, we also analyzed saturation in taxonomy subunits. We found that two iterations of purposive sampling were sufficient to bring the setting and stakeholder domains close to saturation, with the only spike in those domains related to the expansion of sampling from guidance targeted to healthcare stakeholders to guidance for all categories of stakeholders (healthcare, non-healthcare and general public). Similarly, we found that the concepts most directly related to clinical care, Topics/Clinical disposition and treatment and Topics/Clinical presentation and diagnosis, were highly stable over all phases of sampling. In contrast, many non-clinical topics tended to remain dynamic over many iterations of sampling. Topics/Economic dimensions, for example exhibited a 67% jump in taxonomy size with random sampling, and Topics/Reopening spiked by 25% with the first random sample. Fig. 3 shows the most stable taxonomy subunits we examined. Negative values indicate iterations where concepts were collapsed based on analyst consensus, resulting in fewer taxonomic branches than the previous iteration (see Fig. 4 ).
Fig. 4.
Percent change in subunits of the taxonomy across major iterations. (Note: negative values indicate iterations where the taxonomy was revised to collapse concepts.)
3.2. Testing the sampling methodology via rapid development of a taxonomy of COVID-19 PH guidance
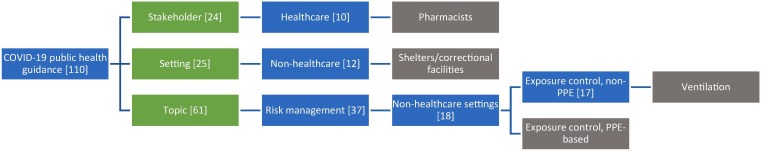
Our second, parallel study objective was to design a taxonomy to classify guidance sampled over the course of the COVID-19 pandemic. The preliminary taxonomy of COVID-19 public health guidance can be viewed in Appendix A. The taxonomy is also available on Mendeley Data in Excel and Simple Knowledge Organization System formats [24]. All branches of the preliminary taxonomy fall under three distinct “domains”: stakeholders, settings and topics. Branches in the stakeholder and setting domains terminated at the third level of the taxonomy; the topic domain reached up to six levels in depth.
The preliminary taxonomy contains a total of 110 distinct branches or possible “pathways” linking a terminal concept to one of the three domains at the top of the hierarchy. Some branches closely parallel each other in structure (e.g. Topics/Risk management strategies/Healthcare settings/Exposure control, non-PPE based and Topics/Risk management strategies/Non-healthcare settings/Exposure control, non-PPE based). However, these were kept independent from one another for the sake of clarity. The longest branches in the taxonomy belong to Topics/Risk management strategies/Healthcare settings/Exposure control, non-PPE based/Isolation strategies and Topics/Risk management strategies/Non-healthcare settings/Exposure control, non-PPE based/Isolation strategies. Example documents pertaining to major branches of the taxonomy can be found in Appendix B. To further illustrate the preliminary taxonomy, Fig. 5 visually depicts examples of branches in the three domains.
Fig. 5.
Example branches of the preliminary taxonomy of pH guidance. Green represents the taxonomy’s three domains: stakeholder, setting and topic. Blue indicates intermediate categories. Gray indicates the terminal concepts for the represented branches. The number in brackets indicates the total number of branches encompassed by a level in the taxonomy. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Random samples taken during the final two iterations of the project demonstrate the taxonomy’s utility in organizing a larger corpus of documents across numerous channels. We report the distribution of concepts applied to randomly sampled documents (n = 226) from all four states and the CDC (Table 2 ). For all channels, a large proportion of documents included the general public as a stakeholder (49–81%). Guidance for healthcare stakeholders was found in anywhere from 12 to 44% of documents, and for non-healthcare stakeholders in a range of 32–62% of documents. IL was the only state with a higher proportion of guidance oriented toward the healthcare setting, as opposed to non-healthcare settings. For all states, the topic concepts most frequently used concerned risk management which included distinct sub-levels relating to healthcare and non-healthcare settings. Of the two, the most frequently referenced was risk management for non-healthcare settings, which included basic precautions for the general public like wearing masks, physical distancing, washing hands and limiting group sizes in public. For topics related directly to clinical care, guidance most often referred to “Clinical presentation and diagnosis”, which included how, when and for whom to administer laboratory tests. Only five documents across all channels offered guidance on care coordination. MA had the highest proportion of guidance documents related to economic dimensions of the pandemic (26%), including unemployment benefits and paid leave, with UT second (19%). A list of all documents used in content analysis, along with corresponding URLs, is available in Appendix C.
Table 2.
Percentage of randomly sampled documents (n = 226) by stakeholder, setting and topic. (Note: a document may belong to multiple concepts within any taxonomic level; columns do not sum to 100%.)
| FL (n = 17) | IL (n = 34) | MA (n = 114) | UT (n = 21)_ | CDC (n = 40) | All (n = 226) | ||
|---|---|---|---|---|---|---|---|
| Domain | Subdomain | % | % | % | % | % | % |
| Settings | Healthcare | 12 | 44 | 35 | 29 | 28 | 33 |
| Non-healthcare | 47 | 32 | 56 | 62 | 35 | 49 | |
| Unassigned | 47 | 26 | 16 | 19 | 35 | 23 | |
| Stakeholders | General public | 76 | 65 | 49 | 81 | 65 | 59 |
| Healthcare | 29 | 38 | 33 | 38 | 40 | 35 | |
| Non-healthcare | 29 | 29 | 46 | 38 | 23 | 37 | |
| Topics | Risk management | 82 | 91 | 69 | 95 | 75 | 77 |
| Clinical presentation/diagnosis | 29 | 56 | 18 | 43 | 35 | 30 | |
| Reopening | 12 | 15 | 35 | 14 | 13 | 24 | |
| Economic dimensions | 6 | 9 | 26 | 19 | 8 | 18 | |
| Clinical disposition/treatment | 6 | 35 | 7 | 10 | 20 | 14 | |
| Resource management | 12 | 18 | 8 | 14 | 13 | 11 | |
| Mental health and wellness | 6 | 3 | 9 | 5 | 15 | 8 | |
| Discrimination/stigma/ethics | 0 | 12 | 1 | 0 | 8 | 4 | |
| Care coordination | 0 | 3 | 3 | 5 | 0 | 2 | |
4. Discussion
The number, diversity, and evolving nature of pH knowledge assets in the COVID-19 pandemic present major challenges to routine KM strategies. A system for classifying PH guidance can serve myriad functions, such as characterizing the breadth of extant knowledge; supporting tools to efficiently disseminate pertinent knowledge to stakeholders; identifying “bottlenecks” in dissemination pathways; and highlighting redundancies and conflicts in emerging or established guidance. Moreover, a PH guidance taxonomy may help to improve targeted communication strategies that can compete with social media and even the harmful impact of misinformation [8], [9].
Attempts to create taxonomies of either clinical or public health guidance are relatively rare. In a clinical context, Jones et al. classified possible applications of PPE through an analysis of tasks undertaken by healthcare workers [35]. Similarly, Nelson et al. have recently created an inductive classification of medical indications using automated methods [36]. Perhaps the most relevant example of a public health taxonomy is Gasser et al.’s categorization of COVID-19 digital heath tools to understand their ethical implications [37]. Despite the importance of the above contributions, to our knowledge ours is the first attempt to develop a systematic sampling method and taxonomy for dealing with the large scope of pH guidance documents that may be expected in a fast-moving crisis like the COVID-19 pandemic.
This study designed and implemented a rapid, semi-automated method for PH guidance sampling using widely available tools for guidance inventory. The study focused on developing a taxonomy that could be used in a “systems” paradigm, in the terms of the taxonomy of KM approaches developed by Earl (2001), given its orientation to facilitating the dissemination of pH guidance from dedicated online resources [38]. Our effort differs from many of the studies described by Earl, however, in its attempt to make knowledge available from specialized repositories to lay audiences. Additionally, unlike many case studies in health informatics KM, ours was not focused on a single, specific organization or electronic resource [39], [40]. Instead, the impetus for the study came from the dispersed nature of pH guidance. By making the taxonomy available on Github, we hope that it can be used in a wide variety of KM strategies, in whole or in part. And we expect that it will need to be adapted to local contexts and informal practices of KM to maximize its utility in guiding knowledge capture, sharing, application and creation [39], [41].
We successfully performed eight major iterations of sampling targeting four states (FL, IL, MA, and UT) and the CDC despite heterogeneity in the number of URLs related to PH guidance (54 for FL to 581 for MA) and the locations of COVID-19 guidance within the websites: guidance was located in dedicated domains in FL and UT but distributed in larger PH or state government domains for IL and MA, respectively. The CDC was overwhelmingly the most frequently linked external resource, with only 3 and 6 links to NIH and WHO resources, respectively. Some (25–40%) of the linked CDC URLs contained no actionable guidance (e.g., index pages that would require further navigation steps by a stakeholder seeking information). However, most of the externally linked URLs for all states focused on guidance for the general public, rather than healthcare or non-healthcare organizations. Extensive reliance on external resources indicates the ongoing prevalence of the “funnel” model of dissemination, and the challenges that stakeholders may confront in searching for relevant information from different sources. Similarly, the sheer volume of assets and differences in website structure highlight the organizational challenges for pandemic KM. A taxonomy such as the one proposed here can help to mitigate these issues.
The study’s method succeeded in developing a preliminary taxonomy of COVID-19 PH guidance that performed well relative to conventional measures of content analysis, including high interrater reliability and thematic saturation. The study began with a 48-branch taxonomy based on preliminary review of documents and expanded to 110 branches after eight iterations of sampling and content analysis. Sampling achieved saturation in a number of taxonomy subunits, including those most relevant to clinical decision-making, and the domains of “stakeholders” and “settings” that would be crucial to mobilizing the taxonomy. Thematic saturation for the entire taxonomy was 7% in the final round of sampling. A flexible method of pH guidance sampling and streamlined content analysis can capture a significant proportion of variation in PH guidance during an event such as the COVID-19 pandemic and can be continually updated as information that needs to be shared evolves over time.
The study found heterogeneity in the prevalence of topics across the sampled web domains. Only 2% of randomly sampled documents contained guidance for care coordination; the topic was absent from sampled CDC guidance. The distribution of guidance related to economic issues may reflect the economic impact of COVID-19 in the US over time and across different jurisdictions [42], [43], [44]. In MA where COVID-19 surged early in the pandemic, 26% of the documents were found to address an economic topic, whereas only 6% of the documents from FL addressed an economic topic. This topic and others were all found in guidance from the four states and the CDC with the exception of guidance related to ‘stigma, ethical considerations or discrimination’ or ‘care coordination’. These last two topics were not present or were infrequent (as shown in Table 2) in the documents we sampled and analyzed. Thus, it is important to aggregate concepts from guidance across sites to ensure critical concepts for organizing and retrieving documents are represented in a taxonomy, particularly concepts about ethical and equity concerns when managing a public health emergency such as the COVID-19 pandemic [45], [46], [47].
4.1. Future considerations and recommendations
The method described here can benefit from development in two major areas. First, natural language processing (NLP) could be used to automatically propose new concepts [48], [49], [50]. This approach would require mechanisms to safeguard against the excessive proliferation of concepts, as growth in the taxonomy increases the effort of applying and updating it, as well as the likelihood of inconsistent use. If implemented with care, NLP would relieve some burden from the process of content analysis. Second, the integration of stakeholder perspectives is a crucial addition to the process. Even the most flexible classification methods will not fulfill their function if their categories are not reflective of their target stakeholders’ search strategies, particularly how these audiences conceptualize problems and seek answers to common questions. Optimally, stakeholder feedback about information needs, search strategies and relevant topics should be integrated into future iterations of taxonomy development [51], [52], [53], [54].
Ultimately, the value of the study’s methods rests on whether they are capable of delivering the right PH guidance to the right stakeholder at the right time under rapidly evolving pandemic conditions [1], [3], [11], [13]. In this context, the rigor and terminological breadth of widely used indexing systems like Medical Subject Heading may be a liability if they result in long “refresh times” and struggle to keep pace with events. A key strength of our approach is that it can track and identify emerging categories of stakeholders, setting or topics. The resulting taxonomy should not be interpreted as a final product, but as a demonstration of the efficacy and efficiency of a process that can be deployed during periods of high-volume PH guidance production to help quickly develop and maintain a taxonomy. Indexing knowledge assets also increases the feasibility of utilizing them beyond the specific event of the COVID-19 pandemic. This taxonomy may be generalized, with modification, to future pandemics, as these are likely to exhibit similar features (e.g. overburdening of the healthcare system, restrictions on private businesses, detailed guidance regarding testing and social distancing measures). Beyond pandemics, our methods can be applied to other fast-moving scenarios that pose major KM challenges, such as environmental PH events [55] and natural disasters [3], [56].
4.2. Limitations
While the taxonomy presented here performed well in the final iteration of sampling, future work is needed to evaluate it against a separate corpus of guidance documents to assess its completeness and accuracy. Second, the focus on a taxonomy that could organize knowledge for those outside of public health meant that we omitted a number of important organizations that produce or disseminate public health guidance primarily for public health workers (e.g. Council of State and Territorial Epidemiologists; National Association of County and City Health Officials; Association of State and Territorial Health Officials). Some public-facing guidance may have been missed by omitting these organizations. An important limitation of the proposed approach is the labor-intensiveness of content analysis. This issue can be partially mitigated by carefully targeted sampling using thematic saturation measures as a guide to reduce effort in branches showing saturation when kappa scores indicate that classification has been performed with internal consistency. The methods required iterated interpretation of kappa and thematic saturation metrics, which are the subject of debate in content analysis and qualitative research [29], [30], [34]. In contrast with the use of these measures in many content analyses, here they figure primarily as a “steering mechanism”, rather than a retrospective evaluation of study rigor. Crucially, taxonomy development is only a preliminary step in the effort to classify extant PH guidance, which will require automated methods given the number and fast-paced production of relevant knowledge assets. Mechanisms must also be in place to update classified documents when changes are made to taxonomy structure. Finally, while we can measure internal consistency and scope of the sampling and taxonomy development processes, specifying which taxonomy categories “matter” is a question that demands input from a range of stakeholders. For example, the production and distribution of a vaccine will be a very important event, but related guidance could conceivably be captured in only a handful of concepts, constituting only a small percent change to overall taxonomy size. Stakeholder judgment is required to specify what the priorities of sampling and concept elaboration should be.
5. Conclusion
Knowledge taxonomies are an underemployed tool in the struggle to organize and disseminate PH knowledge to target audiences such as healthcare professionals and the general public. This study demonstrates a method to support efficient development of a taxonomy of pH guidance in the context of the COVID-19 pandemic. The method involves automated web crawling and document retrieval, as well as easily calculated interrater reliability and thematic saturation metrics. The taxonomy may be used by PH agencies to support the development and dissemination of pH guidance to target audiences such as healthcare professionals and the general public.
CRediT authorship contribution statement
Peter Taber: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Catherine J. Staes: Conceptualization, Investigation, Methodology, Project administration, Supervision, Visualization, Writing - review & editing. Saifon Phengphoo: Formal analysis, Investigation, Methodology, Validation. Elisa Rocha: Formal analysis, Investigation, Methodology, Validation. Adria Lam: Formal analysis, Investigation, Methodology, Validation. Guilherme Del Fiol: Conceptualization, Methodology, Project administration, Data curation, Resources, Supervision, Writing - review & editing. Saverio M. Maviglia: Conceptualization, Methodology, Data curation, Resources. Roberto A. Rocha: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Writing - review & editing, Software.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2021.103852.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Staes C.J., Wuthrich A., Gesteland P., et al. Public health communication with frontline clinicians during the first wave of the 2009 influenza pandemic. J. Publ. Health Manag. Pract. 2011;17(1):36–44. doi: 10.1097/PHH.0b013e3181ee9b29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green L.W., Ottoson J.M., García C., Hiatt R.A. Diffusion theory and knowledge dissemination, utilization, and integration in public health. Annu. Rev. Publ. Health. 2009;30:151–174. doi: 10.1146/annurev.publhealth.031308.100049. [DOI] [PubMed] [Google Scholar]
- 3.Lurie N., Manolio T., Patterson A.P., Collins F., Frieden T. Research as a part of public health emergency response. N. Engl. J. Med. 2013;368(13):1251–1255. doi: 10.1056/NEJMsb1209510. [DOI] [PubMed] [Google Scholar]
- 4.Eysenbach G. Infodemiology and infoveillance: framework for an emerging set of public health informatics methods to analyze search, communication and publication behavior on the Internet. J. Med. Int. Res. 2009;11(1):e11. doi: 10.2196/jmir.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naeem S.B., Bhatti R. The Covid-19 'infodemic': a new front for information professionals. Health Info. Libr. J. 2020;37(3):233–239. doi: 10.1111/hir.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rovetta A., Bhagavathula A.S. Global Infodemiology of COVID-19: Analysis of Google Web Searches and Instagram Hashtags. J. Med. Internet. Res. 2020;22(8):e20673. doi: 10.2196/20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tangcharoensathien V., Calleja N., Nguyen T., et al. Framework for managing the COVID-19 infodemic: methods and results of an online, crowdsourced WHO technical consultation. J. Med. Internet. Res. 2020;22(6):e19659. doi: 10.2196/19659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safarnejad L., Xu Q., Ge Y., Krishnan S., Bagarvathi A., Chen S. Contrasting misinformation and real-information dissemination network structures on social media during a health emergency. Am. J. Publ. Health. 2020;110(S3):S340–S347. doi: 10.2105/AJPH.2020.305854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylvia Chou W.Y., Gaysynsky A. A prologue to the special issue: health misinformation on social media. Am. J. Publ. Health. 2020;110(S3):270–272. doi: 10.2105/AJPH.2020.305943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Fiol G., Huser V., Strasberg H.R., Maviglia S.M., Curtis C., Cimino J.J. Implementations of the HL7 context-aware knowledge retrieval (“Infobutton”) standard: challenges, strengths, limitations, and uptake. J. Biomed. Inform. 2012;45(4):726–735. doi: 10.1016/j.jbi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balas E.A., Krishna S. From SARS to systems: developing advanced knowledge management for public health. Stud. Health Technol. Inform. 2004;100:149–156. [PubMed] [Google Scholar]
- 12.Staes C.J., Altamore R., Han E., Mottice S., Rajeev D., Bradshaw R. Evaluation of knowledge resources for public health reporting logic: implications for knowledge authoring and management. Online J. Publ. Health Inform. 2011;3(3) doi: 10.5210/ojphi.v3i3.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn E., Huckel-Schneider C., Campbell D., Seale H., Milat A.J. How can knowledge exchange portals assist in knowledge management for evidence-informed decision making in public health? BMC Publ. Health. 2014;14:443. doi: 10.1186/1471-2458-14-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobbins M., DeCorby K., Robeson P., Husson H., Tirilis D., Greco L. A knowledge management tool for public health: health-evidence.ca. BMC Publ. Health. 2010;10:496. doi: 10.1186/1471-2458-10-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooney M.A., Iademarco M.F., Huang M., MacKenzie W.R., Davidson A.J. The public health community platform, electronic case reporting, and the digital bridge. J. Publ. Health Manag. Pract. 2018;24(2):185–189. doi: 10.1097/PHH.0000000000000775. [DOI] [PubMed] [Google Scholar]
- 16.Li L., Zhang Q., Wang X., et al. Characterizing the propagation of situational information in social media during COVID-19 epidemic: a case study on Weibo. IEEE Trans. Comput. Social Syst. 2020;7(2):556–562. [Google Scholar]
- 17.Falcone R., Sapienza A. How COVID-19 changed the information needs of Italian Citizens. Int. J. Environ. Res. Publ. Health. 2020;17(19) doi: 10.3390/ijerph17196988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan S.L., Cui M., Qian J. Information resource orchestration during the COVID-19 pandemic: a study of community lockdowns in China. Int. J. Inf. Manage. 2020;54:102143. doi: 10.1016/j.ijinfomgt.2020.102143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.B. Alper, COVID-19 Knowledge Accelerator. GPS Health. https://www.gps.health/covid19_knowledge_accelerator.html. Published 2020. Accessed.
- 20.Roberts K., Alam T., Bedrick S., et al. TREC-COVID: rationale and structure of an information retrieval shared task for COVID-19. J. Am. Med. Inform. Assoc. 2020;27(9):1431–1436. doi: 10.1093/jamia/ocaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Health UDo. Travel Guidance. https://coronavirus.utah.gov/travel/. Published No date. (accessed May 9, 2021).
- 22.Health UDo. Overview of COVID-19 Surveillance. https://coronavirus.utah.gov/case-counts/. Published No date. (accessed May 9, 2021).
- 23.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taber, Peter., Staes, Catherine., Phengphoo, Saifon., Rocha, Elisa., Lam, Adria., Del Fiol, Guilherme., Maviglia, Saverio., Rocha, Roberto. (2021), “Preliminary Taxonomy of COVID-19 Public Health Guidance and Associated Files”, Mendeley Data, V1, doi: 10.17632/yjcv23t98b.1. [DOI] [PMC free article] [PubMed]
- 25.Graneheim U.H., Lindgren B.-M., Lundman B. Methodological challenges in qualitative content analysis: a discussion paper. Nurse Educ. Today. 2017;56:29–34. doi: 10.1016/j.nedt.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Klein D. Implementing a general framework for assessing interrater agreement in Stata. Stata Journal. 2018;18(4):871–901. [Google Scholar]
- 27.Ely J.W., Osheroff J.A., Gorman P.N., et al. A taxonomy of generic clinical questions: classification study. BMJ. 2000;321(7258):429–432. doi: 10.1136/bmj.321.7258.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascio M.A., Lee E., Vaudrin N., Freedman D.A. A team-based approach to open coding: considerations for creating intercoder consensus. Field Methods. 2019;31(2):116–130. [Google Scholar]
- 29.Bernard H.R., Wutich A., Ryan G.W. SAGE Publications; 2016. Analyzing Qualitative Data: Systematic Approaches. [Google Scholar]
- 30.Creswell J.W., Creswell J.D. Sage Publications; 2017. Research Design: Qualitative, Quantitative, and Mixed Methods Approaches. [Google Scholar]
- 31.Campbell J.L., Quincy C., Osserman J., Pedersen O.K. Coding in-depth semistructured interviews: problems of unitization and intercoder reliability and agreement. Sociol. Methods Res. 2013;42(3):294–320. [Google Scholar]
- 32.Lowe A., Norris A.C., Farris A.J., Babbage D.R. Quantifying thematic saturation in qualitative data analysis. Field Methods. 2018;30(3):191–207. [Google Scholar]
- 33.Varpio L., Ajjawi R., Monrouxe L.V., O'Brien B.C., Rees C.E. Shedding the cobra effect: problematising thematic emergence, triangulation, saturation and member checking. Med. Educ. 2017;51(1):40–50. doi: 10.1111/medu.13124. [DOI] [PubMed] [Google Scholar]
- 34.Guest G., Namey E., Chen M. A simple method to assess and report thematic saturation in qualitative research. PLoS ONE. 2020;15(5):e0232076. doi: 10.1371/journal.pone.0232076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones R.M., Bleasdale S.C., Maita D., Brosseau L.M., Program C.D.C.P.E. A systematic risk-based strategy to select personal protective equipment for infectious diseases. Am. J. Infect. Control. 2020;48(1):46–51. doi: 10.1016/j.ajic.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson S.J., Flynn A., Tuttle M.S. A bottom-up approach to creating an ontology for medication indications. J. Am. Med. Inform. Assoc. 2021 doi: 10.1093/jamia/ocaa331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasser U., Ienca M., Scheibner J., Sleigh J., Vayena E. Digital tools against COVID-19: taxonomy, ethical challenges, and navigation aid. Lancet Digit Health. 2020;2(8):e425–e434. doi: 10.1016/S2589-7500(20)30137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Earl M. Knowledge management strategies: toward a taxonomy. J. Manage. Inform. Syst. 2001;18(1):215–233. [Google Scholar]
- 39.Dixon B.E., McGowan J.J., Cravens G.D. Knowledge sharing using codification and collaboration technologies to improve health care: lessons from the public sector. Knowl. Manage. Res. Practice. 2009;7(3):249–259. [Google Scholar]
- 40.Sittig D.F., Wright A., Simonaitis L., et al. The state of the art in clinical knowledge management: an inventory of tools and techniques. Int. J. Med. Inform. 2010;79(1):44–57. doi: 10.1016/j.ijmedinf.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begoña Lloria M. A review of the main approaches to knowledge management. Knowl. Manage. Res. Practice. 2008;6(1):77–89. [Google Scholar]
- 42.V. Abedi, O. Olulana, V. Avula, et al., Racial, Economic and Health Inequality and COVID-19 Infection in the United States. medRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 43.Rollston R., Galea S. COVID-19 and the social determinants of health. Am. J. Health Promot. 2020;34(6):687–689. doi: 10.1177/0890117120930536b. [DOI] [PubMed] [Google Scholar]
- 44.L. Bauer, K.E. Broady, W. Edelberg, J. O’Donnell, Ten facts about COVID-19 and the U.S. economy. Brookings Institute. https://www.brookings.edu/research/ten-facts-about-covid-19-and-the-u-s-economy/. Published 2020. (accessed October 15, 2020).
- 45.Azar K.M.J., Shen Z., Romanelli R.J., et al. Disparities in outcomes among COVID-19 patients in a large health care system in California. Health Aff (Millwood). 2020;39(7):1253–1262. doi: 10.1377/hlthaff.2020.00598. [DOI] [PubMed] [Google Scholar]
- 46.McGuire A.L., Aulisio M.P., Davis F.D., et al. Ethical challenges arising in the COVID-19 pandemic: an overview from the association of bioethics program directors (ABPD) task force. Am. J. Bioeth. 2020;20(7):15–27. doi: 10.1080/15265161.2020.1764138. [DOI] [PubMed] [Google Scholar]
- 47.Rhodes R. Justice and guidance for the COVID-19 pandemic. Am. J. Bioeth. 2020;20(7):163–166. doi: 10.1080/15265161.2020.1777354. [DOI] [PubMed] [Google Scholar]
- 48.Friedman C., Borlawsky T., Shagina L., Xing H.R., Lussier Y.A. Bio-Ontology and text: bridging the modeling gap. Bioinformatics. 2006;22(19):2421–2429. doi: 10.1093/bioinformatics/btl405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu K., Hogan W.R., Crowley R.S. Natural Language Processing methods and systems for biomedical ontology learning. J. Biomed. Inform. 2011;44(1):163–179. doi: 10.1016/j.jbi.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreimeyer K., Foster M., Pandey A., et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J. Biomed. Inform. 2017;73:14–29. doi: 10.1016/j.jbi.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lobb R., Colditz G.A. Implementation science and its application to population health. Annu. Rev. Publ. Health. 2013;34:235–251. doi: 10.1146/annurev-publhealth-031912-114444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wensing M., Grol R. Knowledge translation in health: how implementation science could contribute more. BMC Med. 2019;17(1):88. doi: 10.1186/s12916-019-1322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodman M.S., Sanders Thompson V.L. The science of stakeholder engagement in research: classification, implementation, and evaluation. Transl Behav Med. 2017;7(3):486–491. doi: 10.1007/s13142-017-0495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Cathain A., Croot L., Duncan E., et al. Guidance on how to develop complex interventions to improve health and healthcare. BMJ Open. 2019;9(8):e029954. doi: 10.1136/bmjopen-2019-029954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Généreux M., Lafontaine M., Eykelbosh A. From science to policy and practice: a critical assessment of knowledge management before, during, and after environmental public health disasters. Int. J. Environ. Res. Public Health. 2019;16(4) doi: 10.3390/ijerph16040587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dorasamy M., Raman M., Kaliannan M. Knowledge management systems in support of disasters management: a two decade review. Technol Forecast Soc Change. 2013;80(9):1834–1853. doi: 10.1016/j.techfore.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.