Abstract
Over the last quarter century several genetic alterations have been implicated in hereditary breast cancer (HBC). Two papers recently published in the New England Journal of Medicine explored the mutation prevalence in breast cancer predisposition genes across a large population of affected and unaffected subjects. These analyses designated ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, RAD51C and RAD51D as the core set of genes associated with a significantly increased risk of developing breast cancer. A deeper understanding of the biological role of these genes unearths an intricate mechanism involving DNA repair and cell cycle regulation. Exploiting these inherited alterations for targeted treatments, as is currently the case with PARP inhibitors, may provide additional therapeutic opportunities for HBC patients.
Keywords: Breast cancer, Hereditary, Homologous recombination, Cell cycle, Targeted therapies
With the exception of BRCA1 and BRCA2, no definitive consensus is currently available on the candidate genes whose inherited mutations significantly increase the risk of developing breast cancer [1].
In a recent issue of the New England Journal of Medicine, two large epidemiological studies employed wide multigene panels to define the inherited genetic alterations predisposing to breast cancer [2,3]. Almost 180.000 women were included in the two analyses, divided between cases (i.e. subjects diagnosed with breast cancer) and controls (i.e. unaffected individuals). Results from both studies are concordant in delineating a core of 8 predisposition genes, namely ATM, BARD1, BRCA1, BRCA2, CHK2, PALB2, RAD51C and RAD51D, that can be considered hereditary breast cancer genes. A statistically significant correlation also emerged for CDH1 in one study and for TP53 in the other. While germline mutations of ATM, CDH1 and CHK2 are mainly associated with estrogen receptor (ER) positive breast cancer, BARD1, RAD51C and RAD51D predispose to ER negative and triple negative breast cancer [2,3].
While these data are more confirmatory than innovative, the large sample size and the consistency of the results conclusively defines a set of genes that should be investigated in all patients with hereditary breast cancer (HBC). However, in order to significantly impact clinical practice, these findings must improve breast cancer prevention and/or treatment. Indeed, in the era of personalized medicine, inherently acquired genomic alterations should be mechanistically understood and evaluated for their possible role as therapeutic targets.
A closer look at the molecular mechanisms emerging from the New England studies reveals alterations targeting DNA repair (via homologous recombination) and cell cycle regulation with the exception of the CDH1 gene that encodes for the E-cadherin cell adhesion protein.
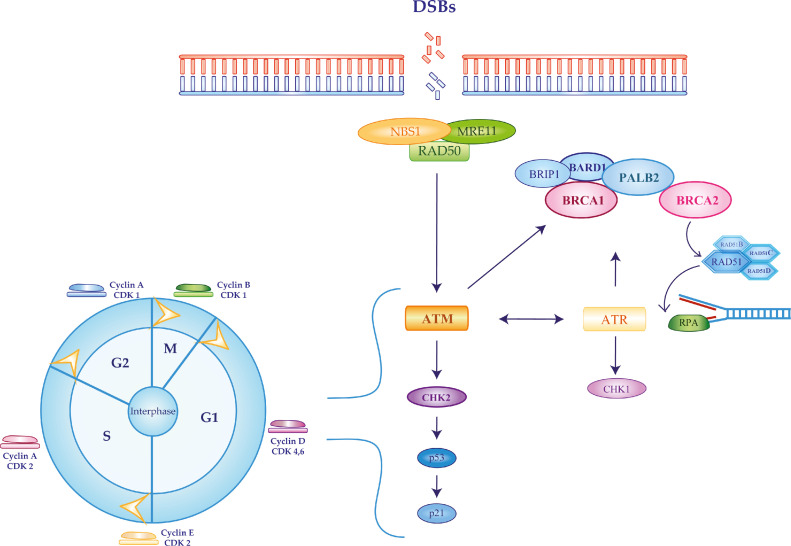
Normal cells use homologous recombination (HR) for the high-fidelity repair of DNA double-strand breaks (DSBs), employing the sister chromatid as a template during the S and G2 phases of the cell cycle. Hence, defects in HR increase genomic instability and mutation rates, contributing to cancer initiation. Mechanistically, HR repair is triggered by DSBs binding to the MRN (MRE11, RAD50 and NBS1) complex, leading to ATM and ATR recruitment. ATM and ATR phosphorylate BRCA1 that can then bind BRIP1, BARD1 and PALB2, the latter connecting BRCA1 and BRCA2. Activated BRCA2 and PALB2 mediate the incorporation of RAD51 and its paralogs (RAD51C and RAD51D) at the site of DSBs, enabling their assembly on the exposed single strand. Once RAD51-bound filaments are in place, the repair process can be finalized by the high fidelity copy of DNA from the sister chromatid (Figure 1) [4], [5], [6].
Fig. 1.
Homologous recombination pathway activated in response to double-strand DNA breaks. Homologous recombination repair (HRR) is activated in response to the generation of DNA double-strand breaks (DSB). A set of genes recently associated with significantly increased risk of developing breast cancer (ATM, BARD1, BRCA1, BRCA2, CHEK2, PALB2, RAD51C and RAD51D) is heavily implicated in modulating HRR and cell cycle progression. After DSB recognition by the MRE11-RAD50-NBS11 complex, activated ATM and ATR phosphorylate BRCA1 that in turn binds BRIP1, BARD1 and PALB2. The latter protein will then connect with BRCA2. The BRCA1-PALB2-BRCA2 complex recruits RAD51 and its paralogs RAD51C and RAD51D on the DSBs in order to finalize their repair.
A functional HR is also critical for genome replication and cell cycle progression [7]. CHK1 and CHK2 phosphorylation - by ATR and ATM, respectively - regulate G1/S, S and G2/M cell cycle checkpoints in order to provide the cell with the time required for DNA damage repair. BRCA1 and BARD1 also take part in cell cycle regulation activating G1/S, S and G2/M checkpoints. During the G1/S-checkpoint, p53 phosphorylation by ATM is required to activate the cyclin-dependent kinase (CDK) inhibitor p21 while, in S-phase, the BRCA1-BARD1-DNA topoisomerase 2-binding protein 1 complex stalls the replication forks in case of DSBs. Similarly, in the G2/M phase checkpoint, the BRCA1-abraxas-RAP80 complex determines late activation of CHK1 in response to DNA damage [6,7].
The detailed definition of these complex processes provides the basis for the therapeutic exploitation of the inherent vulnerability displayed by cells with a mutation in a hereditary breast cancer gene. Inhibiting compensatory DNA damage repair systems, such as base excision repair (BER), in HR-deficient cells eventually determines cell death, a concept known as synthetic lethality. Indeed, several classes of compounds have been developed following this assumption. Inhibitors of poly-adenosyl-ribose-polymerase (PARP), a key component of the BER pathway, have already entered clinical practice for the treatment of BRCA1/2-mutated breast cancer. A plethora of other molecules, targeting different components of the HR pathway and exploiting replicative stress for synthetic lethality are under development, including CHK1, ATM and ATR inhibitors [8]. Still, many questions remain. Indeed, if the role of PARP inhibitors in BRCA1/2 mutated breast cancer is well established, the activity of these drugs in patients harboring alterations on ATM, BARD1, CHK2, PALB2, RAD51C or RAD51D is still unknown. On the other hand, CHK1, ATM and ATR inhibitors are in the early phases of clinical development and their efficacy in patients with a germline mutation in hereditary breast cancer genes is yet to be determined.
Since HR is strictly intertwined with cell cycle progression and regulation, it is conceivable that CDK4/6 inhibitors may also exert a synthetic lethality effect in breast cancer patients with mutations in hereditary predisposition genes. By blocking tumor cells in the G1 phase, CDK4/6 inhibitors could impair the HR process, which requires cells to be in the S or G2 phase. When this phenomenon occurs in cells with defective HR (such as those harboring hereditary breast cancer mutations) the occurrence of fatal genomic instability may be increased [9]. As the association of a CDK4/6 inhibitor with endocrine therapy currently represents the mainstay for the treatment of patients with advanced hormone receptor-positive breast cancer, it would be of great interest to determine the magnitude of benefit of this approach in individuals with a germline mutation in a predisposition gene. Another unanswered question is whether CDK4/6 inhibitors may be exploited in hormone receptor-negative breast cancer presenting alterations in the same set of genes.
As the awareness of breast cancer predisposition genes will steadily increase so will the use of the genomic tests performed for their timely identification. Hence, the number of women diagnosed with HBC is destined to rise in the upcoming future. The development of tailored approaches for these patients will require detailed molecular knowledge and opportunistic thinking in order to transform an unfavorable genetic event in a therapeutic opportunity.
Author's contributions
Conceptualization: S.S., F.M. and P.V.; formal analysis: S.S.; data curation: S.S. and F.M.; writing-original draft preparation: S.S. and F.M..; writing-review and editing: S.S., F.M., L.M. and P.V.; supervision: L.M. and P.V.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Tung N, Lin NU, Kidd J. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J. Clin. Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu C, Hart SN, Gnanaolivu R. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breast Cancer Association Consortium. Dorling L, Carvalho S, Breast Cancer Association Consortium Breast Cancer Risk Genes - Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y, McCorvie TJ, Yates LA, Zhang X. Structural basis of homologous recombination. Cell. Mol. Life Sci. 2020;77:3–18. doi: 10.1007/s00018-019-03365-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonilla B, Hengel SR, Grundy MK, Bernstein KA. RAD51 Gene Family Structure and Function. Annu. Rev. Genet. 2020;54:25–46. doi: 10.1146/annurev-genet-021920-092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadeghi F, Asgari M, Matloubi M. Molecular contribution of BRCA1 and BRCA2 to genome instability in breast cancer patients: review of radiosensitivity assays. Biol Proced Online. 2020;22:23. doi: 10.1186/s12575-020-00133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pilie PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 2019;16:81–104. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Militello AM, Zielli T, Boggiani D. Mechanism of Action and Clinical Efficacy of CDK4/6 Inhibitors in BRCA-Mutated, Estrogen Receptor-Positive Breast Cancers: Case Report and Literature Review. Front. Oncol. 2019;9:759. doi: 10.3389/fonc.2019.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]