Abstract
Childhood lower respiratory tract infections (LRTI) are associated with dysbiosis of the nasopharyngeal microbiota, and persistent dysbiosis following the LRTI may in turn be related to recurrent or chronic respiratory problems.
Therefore, we aimed to investigate microbial and clinical predictors of early recurrence of respiratory symptoms as well as recovery of the microbial community following hospital admission for LRTI in children.
To this end, we collected clinical data and characterised the nasopharyngeal microbiota of 154 children (4 weeks–5 years old) hospitalised for a LRTI (bronchiolitis, pneumonia, wheezing illness or mixed infection) at admission and 4–8 weeks later. Data were compared to 307 age-, sex- and time-matched healthy controls.
During follow-up, 66% of cases experienced recurrence of (mild) respiratory symptoms. In cases with recurrence of symptoms during follow-up, we found distinct nasopharyngeal microbiota at hospital admission, with higher levels of Haemophilus influenzae/haemolyticus, Prevotella oris and other gram-negatives and lower levels of Corynebacterium pseudodiphtheriticum/propinquum and Dolosigranulum pigrum compared with healthy controls. Furthermore, in cases with recurrence of respiratory symptoms, recovery of the microbiota was also diminished. Especially in cases with wheezing illness, we observed a high rate of recurrence of respiratory symptoms, as well as diminished microbiota recovery at follow-up.
Together, our results suggest a link between the nasopharyngeal microbiota composition during LRTI and early recurrence of respiratory symptoms, as well as diminished microbiota recovery after 4–8 weeks. Future studies should investigate whether (speed of) ecological recovery following childhood LRTI is associated with long-term respiratory problems.
Short abstract
Composition of nasopharyngeal microbiota during LRTI in children is related to recurring respiratory symptoms in the following months, and to incomplete microbiota recovery. Future research may pinpoint host and microbial predictors of clinical outcomes. https://bit.ly/3aInAwN
Introduction
Lower respiratory tract infections (LRTIs) remain a leading cause of illness in early childhood, and are a risk factor for the development of recurrent and even chronic respiratory problems [1–3]. For instance, approximately half of infants with bronchiolitis will subsequently experience recurrent wheezing episodes [4], which may persist for years and might eventually develop into asthma [5]. Also, health-related quality of life may remain substantially decreased for months or even years following a LRTI in both children [6] and adults [7]. Causes of respiratory problems following a LRTI remain largely unknown, but emerging evidence suggests that the upper respiratory tract (URT) microbiota may play a role [8, 9].
As expected, during a LRTI, the microbial communities of the respiratory tract differ strongly from those of healthy, matched controls, with increased presence of potential pathogens (“pathobionts”) like Streptococcus pneumoniae and H. influenzae, and decreased presence of presumed beneficial bacteria like Corynebacterium spp. and Dolosigranulum spp. [10–13]. However, microbiota “recovery” following a LRTI is not extensively studied. One case–control study has shown that the abundance of pathobiont Haemophilus spp. decreased to normal levels within a month [11]. Conversely, a longitudinal study demonstrated persistent enrichment with Moraxella spp. up to 6 months after a LRTI [14]. Furthermore, in a prospective cohort of infants with bronchiolitis, increased nasal levels of Moraxella spp. and Streptococcus spp. in the weeks directly following hospitalisation were associated with recurrent wheeze at the age of 3 years, suggesting that persistent microbial “dysbiosis” may contribute to long-term respiratory outcomes following a LRTI [8].
Using a matched case–control design, we have previously demonstrated significant aberrations of the nasopharyngeal microbial community at time of hospital admission for childhood LRTI, when compared with asymptomatic age-, sex- and time-matched controls [13]. Currently, we extend our analysis to 4–8 weeks follow-up, and demonstrate that early recurrence of respiratory symptoms is related to the nasopharyngeal microbial community composition at times of LRTI as well as to impaired microbiota recovery following LRTI.
Methods
We refer to our previous publication on this cohort [13] for details on study design and microbiota analysis, and to the online data supplement for details on statistical analysis. Data are available from the NCBI Sequence Read Archive database (BioProject ID PRJNA428382).
Study design
We enrolled 154 cases aged 4 weeks to 5 years old, hospitalised for a LRTI, and 307 age-, time- and sex-matched, healthy controls from the community (1:2 ratio except for one case). Age, season and sex are known to influence microbiota composition [15–17], and were therefore used as matching criteria to avoid confounding. Nasopharyngeal swabs were obtained from cases at hospital admission and 4–8 weeks after discharge during a follow-up visit to the outpatient clinic or at home. Nasopharyngeal swabs from controls were obtained once during a home visit within 2 weeks from case admission (figure S1). Extensive information on medical history, lifestyle and environment was collected from all participants using questionnaires. From cases, we also collected clinical data during admission and at follow-up. Two expert paediatricians independently grouped cases into three major LRTI phenotypes (pneumonia, bronchiolitis and wheezing illness) based on medical records. Cases with an unclear or overlapping phenotype were classified as mixed infection. Recurrence of respiratory symptoms was defined as a parent-reported new episode of one or more respiratory symptoms (including rhinorrhoea, wheezing, earache, sore throat, coughing, hoarseness and “other” respiratory symptoms) between hospital discharge and the follow-up visit 4–8 weeks later. The study was approved by the Dutch National Ethics Committee (NL42019.094.12). Written informed parental consent was obtained from all participants.
Microbiota analysis
Bacterial DNA was isolated and quantified as previously described [17–19], with the following minor modifications: 16S real time PCR was conducted using TAMRA probe 16S-P1 (FAM-ATT AGA TAC CCT GGT AGT CCA-TAMRA) (Life Technologies, Carlsbad, CA, USA) and a PCR mixture containing 12.5 µL 2× Taqman universal master mix (Life Technologies), 1 µL of each primer (10 µM), 1 µL of the probe (5 µM), 6.5 µL HPLC graded water (Instruchemie, Delfzijl, the Netherlands) and 3 µL of template DNA, on the StepOnePlus System (Applied Biosystems, Foster City, CA, USA). Almost all samples (>99%) contained sufficient DNA for reliable analysis (figure S1). Following, amplicon libraries of the 16S-rRNA gene (V4 region) were generated, and sequencing was executed on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). Bioinformatic processing was performed as previously described [20, 21]. Contaminating sequences were identified using the decontam R-package, using their relation to bacterial biomass (frequency method) and their presence/absence in samples versus DNA isolation controls and PCR blanks (prevalence method) [22]. In total, 21 operational taxonomic units (OTUs) were identified as contaminants, and were removed prior to downstream analyses (table S1). Each OTU was assigned taxonomy and a rank number based on its abundance. Double annotations were assigned to OTUs that could be annotated to both species. Presence of S. pneumoniae, Staphylococcus aureus, H. influenzae and Moraxella catarrhalis was confirmed by quantitative PCR. Viral presence was detected by qualitative multiplex Real Time PCR (RespiFinder SMARTfast 22, Maastricht, the Netherlands).
Statistical analysis
Data analysis was performed in R version 3.4.3. Case–control comparisons accounted for matching. P-values or Benjamini–Hochberg adjusted q-values below 0.050 were considered statistically significant. Chi-squared and Wilcoxon tests were used to compare host characteristics between cases with and without recurrence of respiratory symptoms during follow-up. Independent relationships between antibiotic treatment, LRTI phenotype, age and recurrence of respiratory symptoms during follow-up were assessed using multivariable logistic regression, including pairwise interactions and correcting for follow-up time. Conditional logistic regression was used to compare viral presence between cases and controls.
Alpha diversity was assessed by the Chao1 index for microbial richness and the Shannon index for diversity (phyloseq [23]), and significance of differences between cases and controls was evaluated using linear mixed-effect models. Microbiota recovery was considered complete when the overall microbial composition was comparable between cases after 4–8 weeks follow-up and matched controls, which was evaluated by permutational analysis of variance (PERMANOVA) on the Bray–Curtis dissimilarity matrix (vegan [24]). We similarly analysed differences in microbial diversity at time of admission between cases with and without subsequent recurrence of respiratory symptoms, adjusting for age, sex and month of hospital admission.
Discriminant OTUs between cases and controls were identified by combining significant results from metagenomeSeq analysis [25] and cross-validated VSURF analysis [26], which were then filtered at a fold change of above 1.5 or below 0.5 (i.e. a 50% change).
Stratified analyses were performed of microbiota recovery in relation to recurrence of respiratory symptoms during follow-up, LRTI phenotype, antibiotic treatment and viral presence. In these stratified analyses, differential abundance testing was limited to the top 100 highest-ranked OTUs, because false positive results in low abundant OTUs are a known risk of metagenomeSeq analysis with smaller group sizes [27].
Results
Clinical and microbial factors during LRTI were associated with early recurrence of respiratory symptoms
Cohort characteristics were detailed previously [13]. Follow-up data were available for 149 (97%) cases, with a median (interquartile range) follow-up time of 39 days (35–46) (figure S1). In the 4–8 weeks following hospital discharge, 98 (66%) cases experienced recurrence of respiratory symptoms (from here on called recurrence of symptoms) (table S2). Of these cases, 57 (58%) specified at least two different respiratory symptoms, and 47 (48%) also reported fever (>38°C). Furthermore, 41 cases (42%) consulted a physician, and eight cases (8%) received antibiotics for these symptoms. Follow-up time was significantly longer in cases with recurrence of respiratory symptoms (p=0.015). Cases with and without recurrence of symptoms during follow-up were not significantly different in terms of baseline characteristics including age, lifestyle and environmental factors, medical history and clinical findings such as LRTI phenotype and antibiotic treatment (table 1). However, when age, LRTI phenotype and antibiotic treatment were included in a multivariable model as predictors of recurrence, we found a borderline significant, independent association between a diagnosis of wheezing illness and an increased rate of subsequent recurrence of respiratory symptoms (β 1.19 compared to pneumonia, 95% CI −0.097–2.54; p=0.074). Furthermore, an independent positive association was found between antibiotic treatment during admission and subsequent recurrence of respiratory symptoms (β 2.47, 95% CI 0.52–4.86; p=0.023), but this effect diminished with increasing age (interaction age and antibiotics: β −0.078, 95% CI −0.15– −0.013; p=0.028) (table S3). Viral presence, number of viruses and detection of respiratory syncytial virus (RSV) or human rhinovirus (HRV) was not significantly different between cases with and without recurrence (table 1).
TABLE 1.
Characteristics of cases with and without recurrence of respiratory symptoms during 4–8 weeks follow-up
| Recurrence | No recurrence | p-value | |
| Cases n | 98 | 51 | |
| Basics | |||
| Age (months) | 12.7 (5.5–21.6) | 16.1 (3.5–32.1) | 0.481 |
| Girl | 39 (39.8) | 21 (41.2) | 1.000 |
| Season of sampling | 0.679 | ||
| Spring | 13 (13.3) | 7 (13.7) | |
| Summer | 20 (20.4) | 14 (27.5) | |
| Autumn | 10 (10.2) | 3 (5.9) | |
| Winter | 55 (56.1) | 27 (52.9) | |
| Born at term | 90 (91.8) | 47 (92.2) | 1.000 |
| Mode of delivery | 0.190 | ||
| Vaginal | 76 (77.6) | 43 (84.3) | |
| Elective caesarean section | 9 (9.2) | 6 (11.8) | |
| Emergency caesarean section | 13 (13.3) | 2 (3.9) | |
| Lifestyle and environmental factors | |||
| Breastfeeding >3 months | 38 (38.8) | 18 (35.3) | 0.812 |
| Day care attendance | 65 (66.3) | 28 (54.9) | 0.235 |
| Tobacco smoke exposure | 19 (19.4) | 14 (27.5) | 0.359 |
| Number of siblings | 1.0 (0.2–2.0) | 1.0 (0.0–2.0) | 0.654 |
| Medical history | |||
| Previous LRTI | 29 (29.6) | 11 (21.6) | 0.393 |
| Previous hospitalisation for RTI | 29 (29.6) | 8 (15.7) | 0.096 |
| Prior wheezing | 25 (25.5) | 11 (21.6) | 0.740 |
| Clinical data | |||
| Main discharge diagnosis | 0.408 | ||
| Bronchiolitis | 37 (37.8) | 19 (37.3) | |
| Indeterminate | 18 (18.4) | 8 (15.7) | |
| Pneumonia | 18 (18.4) | 15 (29.4) | |
| Wheezing | 25 (25.5) | 9 (17.6) | |
| Antibiotic treatment during admission | 25 (25.5) | 14 (27.5) | 0.953 |
| Prednison during admission | 16 (16.3) | 9 (17.6) | 1.000 |
| Follow-up time (days after admission) | 42.0 (36.0–49.0) | 39.0 (34.5–44.0) | 0.015 |
| Viral detection at admission | |||
| Any virus (%) | 94 (98.9) | 47 (94.0) | 0.232 |
| Respiratory syncytial virus (%) | 42 (44.2) | 28 (56.0) | 0.240 |
| Human rhinovirus (%) | 51 (53.7) | 21 (42.0) | 0.245 |
| Number of viruses | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | 0.299 |
Data are presented as n (%) or median (interquartile range), unless otherwise stated. Data were acquired from parent questionnaires and medical records. Viral presence was detected by multiplex PCR in nasopharyngeal samples obtained at admission. p-values were calculated by Chi-squared tests or Wilcoxon rank-sum tests. LRTI: lower respiratory tract infection; RTI: respiratory tract infection.
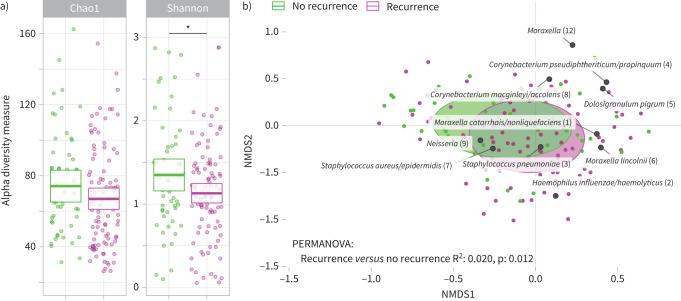
In this cohort, significant differences in the microbial community composition between cases at admission and controls were previously shown, with especially increased abundance of pathobionts H. influenzae/haemolyticus and S. pneumoniae, and decreased abundance of presumed beneficial bacteria like M. catarrhalis/nonliquefaciens, D. pigrum and C. pseudodiphtheriticum/propinquum [13]. Here, a modestly lower Shannon diversity at time of admission was related to a higher rate of recurrence of symptoms, even after adjusting for age, sex and month of hospital admission (p=0.049) (figure 1a). Furthermore, the overall microbial community composition at time of admission was significantly different between cases with versus cases without subsequent recurrence of symptoms, independent of age, sex and month of hospital admission (R2 0.020, p=0.012) (figure 1b), though not correlated with the severity of recurrence of symptoms (i.e. number of symptoms (1 or >1), presence of fever, or the parents’ decision to consult a physician during follow-up (data not shown)). Cases with recurrence of symptoms had higher abundances of gram-negatives like H. influenzae/haemolyticus, P. oris, Actinomyces spp. and Fusobacterium spp. and lower abundances of health-associated C. pseudodiphtheriticum/propinquum and D. pigrum at time of admission compared to controls. On the other hand, cases without recurrence had higher abundances of amongst others gram-positives S. aureus/epidermidis and S. pneumoniae at admission compared to controls (table 2 and figure S2).
FIGURE 1.
Nasopharyngeal microbiota during lower respiratory tract infection was associated with early recurrence of respiratory symptoms. a) Alpha diversity measures Chao1 index and Shannon diversity index estimated at time of admission for cases with compared to without recurrence of respiratory symptoms during follow-up. Boxes denote means with 95% confidence intervals. *: p<0.05, significance was tested by linear models adjusting for age, sex and month of admission. b) Nonmetric multidimensional scaling (NMDS) biplot based on Bray-Curtis dissimilarity depicts nasopharyngeal microbiota composition at time of admission for cases with compared to without recurrence of respiratory symptoms during follow-up, combined with the top 10 operational taxonomic units with highest relative abundance in the entire cohort. Ellipses represent the standard deviation of all points within a sub-cohort. Significance was tested using permutational analysis of variance (PERMANOVA), adjusting for age, sex and month of hospital admission.
TABLE 2.
Biomarker species during acute lower respiratory tract infection for subsequent recurrence of respiratory symptoms
| Differentially abundant at admission | OTU | Fold change | Significant within |
| In cases with subsequent recurrence of respiratory symptoms versus controls | Haemophilus influenzae/haemolyticus (2) | 5.97 | ms+V |
| Fusobacterium (83) | 2.57 | ms | |
| Prevotella oris (45) | 2.31 | ms | |
| Actinomyces graevenitzii (68) | 1.88 | ms+V | |
| Fusobacterium (74) | 1.76 | ms | |
| Actinomyces johnsonii (75) | 1.76 | ms+V | |
| Actinomyces odontolyticus (48) | 1.66 | ms+V | |
| Dolosigranulum pigrum (5) | 0.41 | ms | |
| Corynebacterium pseudodiphtheriticum/ propinquum (4) | 0.41 | ms | |
| In cases without subsequent recurrence of respiratory symptoms versus controls | Neisseria lactamica (19) | 2.97 | ms+V |
| Staphylococcus aureus/epidermidis (7) | 2.38 | ms+V | |
| Streptococcus pneumoniae (3) | 2.31 | V | |
| Atopobium (100) | 1.99 | ms | |
| Klebsiella (11) | 1.75 | ms+V | |
| Halomonas (14) | 1.57 | ms | |
| Prevotella melaninogenica (16) | 1.54 | V | |
| Prevotella nanceiensis (25) | 0.49 | ms | |
| Neisseria (9) | 0.43 | ms+V |
OTU: operational taxonomic unit; ms: metagenomeSeq analysis; V: VSURF analysis.
Nasopharyngeal microbiota and viral profiles after recovery from LRTI
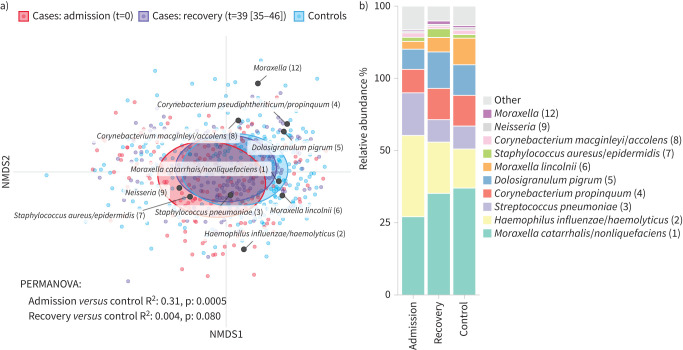
We also aimed to study remaining differences in microbial community diversity and composition between recovered cases and controls. In general, samples obtained 4–8 weeks after hospital discharge showed significantly higher microbial richness in (former) cases when compared to controls (p=0.042), while Shannon diversity and biomass were comparable (figure S3). Furthermore, despite the observed major differences in the overall microbial community composition at time of infection, after 4–8 weeks the microbiota of recovered cases had become more similar to controls, and only a small, non-significant difference remained (R2 0.004, p=0.080) (figure 2). On the OTU level, we observed 15 OTUs that were significantly differentially abundant between recovered cases and controls (table 3 and figure S4). Of these, Moraxella spp. and Helcococcus spp. were already under-represented at admission, and remained under-represented in recovered cases, whereas various gram-negative species including H. influenzae/haemolyticus, P. oris and Neisseria lactamica remained overrepresented in recovered cases compared to controls.
FIGURE 2.
Nasopharyngeal microbiota recovery following lower respiratory tract infection. a) Nonmetric multidimensional scaling (NMDS) biplot based on Bray-Curtis dissimilarity depicts nasopharyngeal microbiota composition for cases at admission, cases at follow-up (recovery), and controls, combined with the top 10 operational taxonomic units (OTUs) with highest relative abundance in the entire cohort. Time (t) in days between admission and the follow-up visit was reported as median (interquartile range). Ellipses represent the standard deviation of all points within a sub-cohort. Significance was tested using permutational analysis of variance (PERMANOVA). b) Mean relative abundances of the 10 OTUs with highest relative abundance.
TABLE 3.
Discriminant operational taxonomic unit (OTU) for cases during acute lower respiratory tract infection and after 4–8 weeks follow-up compared to controls
| Association | OTU | Cases at admission versus controls | Cases at recovery versus controls | ||
| FC | Significant within | FC | Significant within | ||
| Admission | Lactococcus lactis (67) | 2.06 | ms+V | ns | ns |
| Corynebacteriaceae (161) | 1.83 | ms+V | ns | ns | |
| Abiotrophia (111) | 1.66 | ms+V | ns | ns | |
| Corynebacterium (62) | 1.64 | ms | ns | ns | |
| Megasphaera (137) | 1.62 | ms | ns | ns | |
| Fusobacterium (74) | 1.61 | ms | ns | ns | |
| Actinomyces (48) | 1.56 | ms | ns | ns | |
| Fusobacterium (83) | 1.53 | ms | ns | ns | |
| Actinomyces graevenitzii (68) | 1.52 | ms+V | ns | ns | |
| Acinetobacter soli (125) | 1.51 | ms | ns | ns | |
| Actinomyces johnsonii (75) | 1.51 | ms | ns | ns | |
| Dolosigranulum pigrum (5) | 0.43 | ms | ns | ns | |
| Dolosigranulum (122) | 0.4 | ms+V | ns | ns | |
| Admission and recovery | Haemophilus influenzae/haemolyticus (2) | 3.85 | ms+V | 1.69 | V |
| Neisseria lactamica (19) | 2.22 | ms+V | 1.96 | ms+V | |
| Prevotella oris (45) | 2.03 | ms | 1.72 | ms+V | |
| Moraxella (54) | 0.32 | ms+V | 0.45 | ms+V | |
| Moraxella (58) | 0.19 | ms+V | 0.45 | ms+V | |
| Moraxella lincolnii (6) | 0.18 | ms+V | 0.3 | ms+V | |
| Moraxella (84) | 0.17 | ms+V | 0.34 | ms | |
| Moraxella (163) | 0.15 | ms+V | 0.39 | ms+V | |
| Moraxella (112) | 0.08 | ms | 1.68 | ms | |
| Helcococcus (43) | 0.07 | ms | 0.07 | ms+V | |
| Recovery | Neisseriaceae (15) | ns | ns | 2.26 | ms+V |
| Moraxella (12) | ns | ns | 1.83 | ms | |
| Porphyromonas (39) | ns | ns | 1.62 | ms | |
| Bradyrhizobium (17) | ns | ns | 1.54 | ms+V | |
| Janthinobacterium lividum (23) | ns | ns | 0.09 | ms+V | |
FC: fold change; ms: metagenomeSeq analysis; V: VSURF analysis; ns: not significantly different.
Assessment of microbiota recovery in relation to recurrence of respiratory symptoms, showed that when compared to matched controls, cases with recurrence of symptoms during follow-up had a significantly higher microbial richness at the end of follow-up (p=0.034) (figure S5a). This difference in microbial richness was not present between cases without recurrence of symptoms and their matched controls. Also, on microbial community composition level, the microbiota composition had failed to normalise in cases with recurrence of respiratory symptoms during follow-up (R2 0.008, p=0.028), while in cases without recurrence the microbiota were comparable to controls (R2 0.005, p=0.50). Especially the abundances of H. influenzae/haemolyticus, Neisseria spp., P. oris, and Porphyromonas spp. were persistently increased after recovery in cases with recurrence of symptoms during follow-up compared to controls, while in cases without recurrence, abundance of S. aureus/epidermidis and N. lactamica were increased after recovery compared to controls (figure S5b).
Viral presence at time of admission and after recovery was available for 70 cases and for 139 corresponding controls. As described previously [13], 97% of cases tested positive for any virus at time of admission versus 85% of controls (p=0.019). At follow-up, 91% of (former) cases were virus-positive, which was not significantly different from controls. RSV detection was higher in cases at time of admission compared to controls (40% versus 4%; p<0.001), but had normalised after recovery (3%). Interestingly, in recovered cases, HRV detection was more common (80%) than at time of admission (57%; p=0.006), though comparable to controls (70%) (figure S6). Detection of other respiratory viruses was low and not significantly different between cases and controls following recovery.
Microbiota recovery depends on LRTI phenotype, but not on antibiotic treatment or type of virus
Next, we used stratified analyses to investigate whether microbiota recovery was related to antibiotic treatment, LRTI phenotype or viral presence at time of admission. In total, 43 (28%) cases were treated with antibiotics (33 β-lactam, 10 macrolide) during admission. Interestingly, only cases not treated with antibiotics showed significantly higher microbial richness after follow-up compared to controls (p=0.001), while no differences in Shannon diversity were observed in either group compared to their respective controls (figure S7a). Microbiota composition at follow-up also showed little difference between both antibiotic-treated and not antibiotic-treated cases and their respective controls (treated cases versus controls R2 0.007, p=0.47; non-treated cases versus controls R2 0.005, p=0.17).
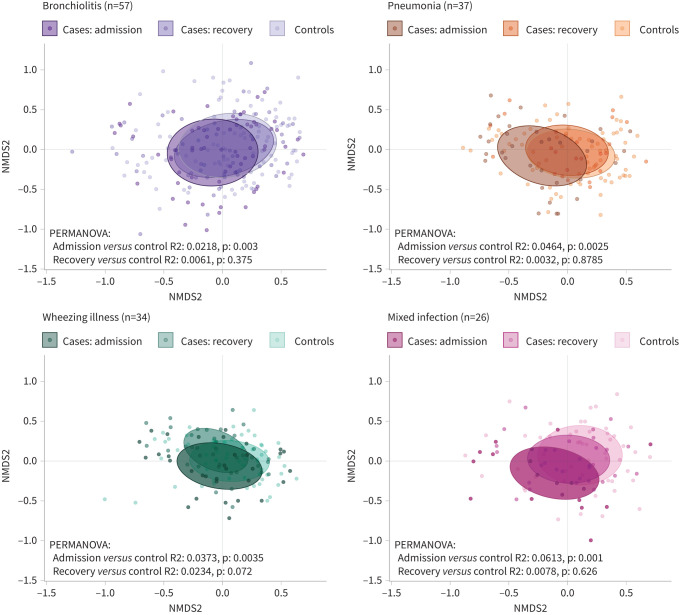
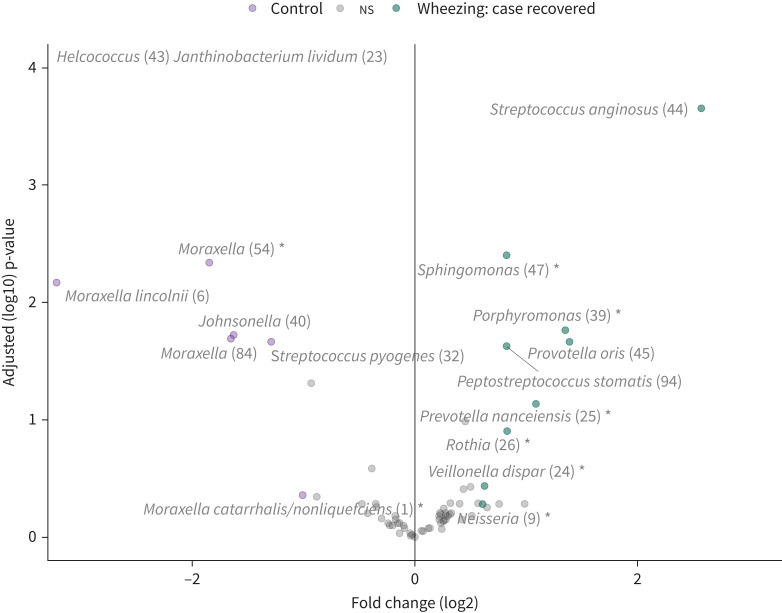
Regarding LRTI phenotype, 57 (37%) cases were classified as bronchiolitis, 37 (24%) as pneumonia, 34 (22%) as wheezing illness and 26 (17%) as mixed infection. Only (former) bronchiolitis cases had significantly increased microbial richness after follow-up compared with matched controls (p=0.013), though a similar trend was observed for former wheezing illness cases (p=0.088) (figure S7b). Furthermore, only (former) wheezing illness cases still showed a trend towards a different overall microbial community composition at follow-up compared with their respective controls (R2 0.023, p=0.072) (figure 3). As previously mentioned, cases with wheezing illness had the highest incidence of recurrence of respiratory symptoms, while cases with pneumonia had the lowest incidence (table 1). On OTU level, we observed that cases recovered from wheezing illness had increased levels of Streptococcus anginosus, and gram-negatives like P. oris, Porphyromonas spp. and Neisseria spp., which were all also associated with recurrence (figure 4). By contrast, cases recovered from pneumonia had increased levels of Klebsiella spp., Neisseriaceae family, and gram-positives S. aureus/epidermidis and Kocuria spp., possibly related to antibiotic selection (figure S8).
FIGURE 3.
Microbiota recovery depended on infection phenotype. Nonmetric multidimensional scaling (NMDS) plots based on Bray-Curtis dissimilarity depict nasopharyngeal microbiota composition for cases at admission, cases at follow-up (recovery), and controls, for each of the four phenotypes. Ellipses represent the standard deviation of all points within a sub-cohort. Significance was tested using permutational analysis of variance (PERMANOVA).
FIGURE 4.
Discriminant operational taxonomic units (OTUs) between cases recovered from wheezing illness and matched controls. Volcano plot of differentially abundant OTUs between cases recovered from wheezing illness and controls. Significance was assessed by metagenomeSeq analysis and cross-validated VSURF analysis limited to the top 100 most highly ranked OTUs, and combined results were filtered at a fold change of at least 1.5 or <0.5. OTUs marked by an asterisk were identified by cross-validated VSURF analysis. Results of data points falling beyond the limits of the plot: Helcococcus log2 fold change −5.93, adjusted p-value (log10) 13.48; Janthinobacterium lividum log2 fold change −3.67, adjusted p-value (log10) 10.01.
Finally, we stratified the analysis based on the most prevalent viruses at time of LRTI, i.e. RSV and HRV, which were detected in 72 (47%) and 73 (47%) cases, respectively, to rule out virus-mediated effects on microbiota recovery. Cases recovered from a RSV-associated LRTI had no significant differences in microbial richness, diversity or the overall microbial composition compared to their matched controls (R2 0.003, p=0.624). Cases recovered from a HRV-associated LRTI also showed no significant differences in overall microbial composition compared to controls (R2 0.008, p=0.165), though they had a slightly higher microbial richness upon recovery, which tended towards significance (p=0.070) (figure S7c).
Discussion
LRTI is strongly associated with dysbiosis of the nasopharyngeal microbiota [13]. Here, we found in children hospitalised for acute LRTI that lower microbial diversity and the overall microbial community composition in the nasopharynx were modestly associated with subsequent recurrence of even very mild respiratory symptoms within 1–2 months. Specifically, we identified gram-negatives like H. influenzae/haemolyticus, P. oris and Actinomyces spp. as potential biomarkers of an increased risk of recurrence of respiratory symptoms, and gram-positives like S. aureus/epidermidis and S. pneumoniae as potential biomarkers of a reduced risk.
These findings add to a small but growing body of literature suggesting that host-microbial interactions during and following acute (L)RTI may contribute to short- and long-term respiratory outcomes. Previously, Neumann et al. [28] found in noses of infants with their first RTI that lower microbial diversity and increased levels of bacterial families Moraxellaceae or Streptococcaceae were associated with persistent respiratory symptoms. Also in the nasal niche, Mansbach et al. [8] related persistently increased levels of Moraxella spp. and Streptococcus spp. in the weeks following hospitalisation for bronchiolitis in infancy to persistent wheeze at the age of 3 years. In case of RSV infection, increased nasopharyngeal levels of gram-negatives including H. influenzae have been associated with a pro-inflammatory systemic immune response with enhanced neutrophil recruitment and activation, and increased disease severity [29, 30]. Correlates with clinical outcomes during recovery remain unknown, but a severity-dependent relationship between RSV bronchiolitis and the risk of recurrent wheezing and asthma has been described [31]. Moreover, Haemophilus spp.-dominated nasopharyngeal microbiota during RSV infection has also been related to increased viral load [11] and delayed clearance [32], which might also contribute to persistent inflammation, slower recovery and more respiratory morbidity. Finally, in healthy infants, increased influx into the nasopharynx of gram-negatives typically found in the mouth, like Prevotella spp., Neisseria spp. and Fusobacterium spp., has also been associated with higher susceptibility to RTIs in general [20, 33]. Alternatively, gram-positive commensals might dampen inflammatory responses. For instance, S. epidermidis was shown to enhance mucosal innate immune responses in the nose and to thereby confer resistance to viral infection [34]. Moreover, dominance of Corynebacterium spp. and Dolosigranulum spp. in the infant nasopharyngeal microbiota has been related to decreased incidence of RTIs [35]. Future studies should therefore investigate whether antibiotic treatment targeted at gram-negatives and/or preservation or supplementation of gram-positive commensals may prevent recurrence of respiratory symptoms after LRTI and improve long-term respiratory outcomes.
In general, we observed that the nasopharyngeal microbiota had recovered 4–8 weeks after hospitalisation for LRTI, though subtle differences remained including a persistently lower abundance of Moraxella spp. than seen in healthy controls. Remarkably, this is opposite to observations by Teo et al. [14]. This study had an unmatched design, later timing of post-LRTI sampling and a different geographic location, but the discrepancy with our findings might also reflect biological differences between their cohort at high risk for atopy and our unselected cohort. In line with our findings, Kaul et al. [36] recently demonstrated that in some adult patients with acute influenza infection, microbial communities returned to a healthy state within 22 days from hospital admission. It thus appears that the URT microbiota are resilient, but the speed of recovery differs between individuals. In addition, we observed that children with early recurrence of respiratory symptoms following LRTI also had diminished microbiota recovery, despite a longer follow-up duration and more time for the microbiota to recover. The association between longer follow-up duration and recurrence of respiratory symptoms may be directly linked, though also explained by the fact that children had to be asymptomatic at the time of sampling, and as a consequence the follow-up visit was postponed when symptoms were present at that moment. Irrespectively, given the high incidence of symptom recurrence in our cohort, we theorise that while the microbiota gradually recover following LRTI, resistance to viral infection or bacterial pathobiont acquisition and overgrowth might remain diminished for some time, resulting in a (temporarily) elevated risk of new infections upon pathobiont exposure. Aberrant airway immune profiles in asymptomatic neonates and during RSV infection were previously associated with presence and abundance of gram-negatives colonising the respiratory tract [29, 37, 38], and might mediate this association, but this remains to be investigated. An alternative hypothesis is, however, that children may be genetically predisposed to both microbiota shifts and development of RTIs. For example, genetic variants were previously shown to increase susceptibility to otitis media by modifying the middle ear microbiome [39]. This would mean our findings are correlative in nature more than causally linked. Future studies should therefore take genetic factors into consideration to understand the potential mechanisms underpinning our findings.
Importantly, we observed early recurrence of respiratory symptoms followed by diminished microbiota recovery especially in children with wheezing illness, whereas microbiota changes during LRTI were previously shown to be phenotype-independent [13]. Together, our findings suggest that especially in children with inflammation-driven illness, the nasopharyngeal microbiota may have more limited property to recover to a state comparable to healthy, matched controls. This is in line with other observational studies where LRTIs accompanied by wheezing symptoms were particularly associated with aberrant respiratory microbiota development [14] and with later-life persistent wheeze and development of asthma [2, 3]. We speculate that wheezing illness patients, who were older than patients with a different LRTI phenotype, may have had a more elaborate medical history with LRTIs and atopic symptoms, which may have resulted in reduced resilience of the microbiota. Alternatively, wheezing illness patients often received treatment with inhaled corticosteroids upon hospital discharge, which might also have affected microbiota recovery [40, 41]. Unfortunately, limited power hampered us to study the effect of inhaled corticosteroids on the respiratory microbiota and its recovery. Antibiotic-treated patients also had an increased risk of subsequent recurrence, particularly the younger ones. Dutch physicians tend to reserve antibiotic treatment for the more severely ill patients. Therefore, we cannot rule out that this correlation between antibiotic treatment and recurrence might be influenced by more severe disease and accompanying inflammation. Following, the restorative capacity of the nasopharyngeal microbiota appeared in general not related to antibiotic treatment, though we had insufficient power to test if younger antibiotic-treated children may be more prone to prolonged microbiota disturbance, as has been suggested in previous reports [42, 43]. Finally, the type of virus detected at time of admission seemed unrelated to recurrence of respiratory symptoms and microbiota recovery in this cohort, despite known relationships between LRTIs associated with RSV or HRV and chronic respiratory morbidity [44].
Strengths of our study include the strictly matched case–control design that allowed us to preclude bias from age, sex and seasonality. Furthermore, inclusion of children up to 5 years old, hospitalised for all LRTI phenotypes allowed us to perform in-depth analyses. However, several limitations need to be acknowledged. We were unable to directly study the lung microbiota during recovery, but the high concordance between the microbiota in nasopharyngeal and endotracheal samples at time of LRTI in young children implies that the nasopharyngeal microbiota provides a valid proxy [13]. Furthermore, follow-up of cases entailed only one timepoint, and therefore, we could not distinguish at what pace microbial recovery occurred, and whether microbial recovery was still ongoing. Follow-up duration also varied and was different between cases with and without recurrence. However, since the microbiota of cases with recurrence had even more time to recover, this strengthens the likelihood that recurrence of respiratory symptoms following LRTI is associated with diminished microbiota recovery. Lastly, for our analyses of recurrence we relied on parental report of respiratory symptoms since hospital discharge, which may have introduced recall bias.
In conclusion, our results suggest that the composition of the nasopharyngeal microbiota during acute LRTI in children may increase susceptibility to new respiratory symptoms in the months following, while the microbiota gradually recover. Future prospective studies with higher resolution and longer follow-up duration, especially focussed on recovery from LRTI in high-risk groups of recurrent or chronic respiratory morbidity, are required to confirm and nuance our findings, and should strive to combine host and microbial factors into prediction models of (long-term) clinical outcomes. The current work may be a stepping stone to improved understanding of respiratory outcomes after childhood LRTI and potentially provide input for clinical studies on methods to alleviate recurrent respiratory problems.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00939-2020.SUPPLEMENT (1.4MB, pdf)
Acknowledgements
We thank the participating children and their families. We also thank all the members of the research team at Spaarne Gasthuis Academy, the paediatricians at Spaarne Gasthuis, the laboratory staff, the Streeklaboratorium Haarlem, the GGD Kennemerland and the JGZ Kennemerland for their support.
Footnotes
This article has supplementary material available from openres.ersjournals.com
Support statement: This work was supported in part by The Netherlands Organization for Scientific Research (NWO-VIDI grant 91715359) and CSO/NRS Scottish Senior Clinical Fellowship award (SCAF/16/03). The study was co-sponsored by the Spaarne Hospital Haarlem and the UMC Utrecht, The Netherlands. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of Interest: E.M. de Koff has nothing to disclose.
Conflict of interest: W.H. Man has nothing to disclose.
Conflict of interest: M.A. van Houten has nothing to disclose.
Conflict of interest: A.M. Vlieger has nothing to disclose.
Conflict of interest: M.L.J.N. Chu has nothing to disclose.
Conflict of interest: E.A.M. Sanders has nothing to disclose.
Conflict of interest: D. Bogaert has nothing to disclose.
References
- 1.Kusel MM, de Klerk NH, Kebadze T, et al. Early-life respiratory viral infections, atopic sensitisation, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol 2007; 119: 1105–1110. doi: 10.1016/j.jaci.2006.12.669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oddy WH, de Klerk NH, Sly PD, et al. The effects of respiratory infections, atopy, and breastfeeding on childhood asthma. Eur Respir J 2002; 19: 899–905. doi: 10.1183/09031936.02.00103602 [DOI] [PubMed] [Google Scholar]
- 3.Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med 2008; 178: 667–672. doi: 10.1164/rccm.200802-309OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bont L, van Aalderen WMC, Kimpen JLL. Long-term consequences of respiratory syncytial virus (RSV) bronchiolitis. Paediatr Respir Rev 2000; 1: 221–227. doi: 10.1053/prrv.2000.0052 [DOI] [PubMed] [Google Scholar]
- 5.Mansbach JM, Hasegawa K, Geller RJ, et al. Bronchiolitis severity is related to recurrent wheezing by age 3 years in a prospective, multicenter cohort. Pediatr Res 2020; 87: 428–430. doi: 10.1038/s41390-019-0589-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bont L, Steijn M, van Aalderen WMC, et al. Impact of wheezing after respiratory syncytial virus infection on health-related quality of life. Pediatr Infect Dis J 2004; 23: 414–417. doi: 10.1097/01.inf.0000122604.32137.29 [DOI] [PubMed] [Google Scholar]
- 7.el Moussaoui R, Opmeer BC, de Borgie CAJM, et al. Long-term symptom recovery and health-related quality of life in patients with mild-to-moderate-severe community-acquired pneumonia. Chest; 2006; 130: 1165–1172. doi: 10.1378/chest.130.4.1165 [DOI] [PubMed] [Google Scholar]
- 8.Mansbach JM, Luna PN, Shaw CA, et al. Increased Moraxella and Streptococcus species abundance after severe bronchiolitis is associated with recurrent wheezing. J Allergy Clin Immunol 2020; 145: 518–527.doi: 10.1016/j.jaci.2019.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015; 17: 704–715. doi: 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Steenhuijsen Piters WAA, Huijskens EGW, Wyllie AL, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J 2016; 10: 97–108. doi: 10.1038/ismej.2015.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ederveen THA, Ferwerda G, Ahout IM, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalised with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 2018; 6: 10. doi: 10.1186/s40168-017-0395-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakwinska O, Schmid VB, Berger B, et al. Nasopharyngeal microbiota in healthy children and pneumonia patients. J Clin Microbiol 2014; 52: 1590–1594. doi: 10.1128/JCM.03280-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Man WH, van Houten MA, Mérelle ME, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med 2019; 7: 417–426. doi: 10.1016/S2213-2600(18)30449-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teo SM, Tang HHF, Mok D, et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe 2018; 24: 341–352. doi: 10.1016/j.chom.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatsunenko T, Rey FE, Manary M, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486: 222–227. doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markle JGM, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013; 339: 1084–1088. doi: 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- 17.Bogaert D, Keijser B, Huse S, et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 2011; 6: 17035. doi: 10.1371/journal.pone.0017035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biesbroek G, Sanders EAM, Roeselers G, et al. Deep sequencing analyses of low density microbial communities: Working at the boundary of accurate microbiota detection. PLoS One 2012; 7: e32942. doi: 10.1371/journal.pone.0032942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyllie AL, Chu MLJN, Schellens MHB, et al. Streptococcus pneumoniae in saliva of Dutch primary school children. PLoS One 2014; 9: e102045. doi: 10.1371/journal.pone.0102045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A Prospective Cohort Study. Am J Respir Crit Care Med 2017; 196: 1582–1590. doi: 10.1164/rccm.201703-0554OC [DOI] [PubMed] [Google Scholar]
- 21.Bosch AATM, Levin E, van Houten MA, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMed 2016; 9: 336–345. doi: 10.1016/j.ebiom.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis NM, Proctor D, Holmes SP, et al. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018; 6: 226. doi: 10.1186/s40168-018-0605-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMurdie PJ, Holmes S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8: e61217. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oksanen J, Blanchet FG, Friendly M, et al. vegan: Community Ecology Package. 2016. https://CRAN.R-project.org/package=vegan [Google Scholar]
- 25.Paulson JN, Stine OC, Bravo HC, et al. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 2013; 10: 1200–1202. doi: 10.1038/nmeth.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genuer R, Poggi J, Tuleau-malot C. VSURF: An R package for variable selection using random forests. R J 2015; 7: 19–33. doi: 10.32614/RJ-2015-018 [DOI] [Google Scholar]
- 27.Thorsen J, Brejnrod A, Mortensen M, et al. Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome 2016; 4: 62. doi: 10.1186/s40168-016-0208-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neumann RP, Hilty M, Xu B, et al. Nasal microbiota and symptom persistence in acute respiratory tract infections in infants. ERJ Open Res 2018; 4: 00066–02018. doi: 10.1183/23120541.00066-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Steenhuijsen Piters WAA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med 2016; 194: 1104–1115. doi: 10.1164/rccm.201602-0220OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suárez-Arrabal MC, Mella C, Lopez SM, et al. Nasopharyngeal bacterial burden and antibiotics: influence on inflammatory markers and disease severity in infants with respiratory syncytial virus bronchiolitis. J Infect 2015; 71: 458–469. doi: 10.1016/j.jinf.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 31.Carroll KN, Wu P, Gebretsadik T, et al. The severity-dependent relationship of infant bronchiolitis on the risk and morbidity of early childhood asthma. J Allergy Clin Immunol 2009; 123: 1055–1061. doi: 10.1016/j.jaci.2009.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansbach JM, Hasegawa K, Piedra PA, et al. Haemophilus-dominant nasopharyngeal microbiota is associated with delayed clearance of respiratory syncytial virus in infants hospitalised for bronchiolitis. J Infect Dis 2019; 219: 1804–1812. doi: 10.1093/infdis/jiy741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Man WH, Clerc M, de Steenhuijsen Piters WAA, et al. Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am J Respir Crit Care Med 2019; 200:760–770. doi: 10.1164/rccm.201810-1993OC [DOI] [PubMed] [Google Scholar]
- 34.Kim HJ, Jo A, Jeon YJ, et al. Nasal commensal Staphylococcus epidermidis enhances interferon-λ-dependent immunity against influenza virus. Microbiome 2019; 7: 80. doi: 10.1186/s40168-019-0691-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biesbroek G, Tsivtsivadze E, Sanders EAM, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014; 190: 1283–1292. doi: 10.1164/rccm.201407-1240OC [DOI] [PubMed] [Google Scholar]
- 36.Kaul D, Rathnasinghe R, Ferres M, et al. Microbiome disturbance and resilience dynamics of the upper respiratory tract in response to influenza A virus infection in analog hosts. Nat Commun 2020; 11: 2537. doi: 10.1038/s41467-020-16429-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Følsgaard NV, Schjørring S, Chawes BL, et al. Pathogenic bacteria colonising the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am J Respir Crit Care Med 2013; 187: 589–595. doi: 10.1164/rccm.201207-1297OC [DOI] [PubMed] [Google Scholar]
- 38.Thorsen J, Rasmussen MA, Waage J, et al. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat Commun 2019; 10: 5001. doi: 10.1038/s41467-019-12989-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos-Cortez RLP, Chiong CM, Frank DN, et al. FUT2 variants confer susceptibility to familial otitis media. Am J Human Genet 2018; 103: 679–690. doi: 10.1016/j.ajhg.2018.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durack J, Lynch SV, Nariya S, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol 2017; 140: 63–75. doi: 10.1016/j.jaci.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singanayagam A, Glanville N, Cuthbertson L, et al. Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci Transl Med 2019; 11: 3879. doi: 10.1126/scitranslmed.aav3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasparrini AJ, Wang B, Sun X, et al. Persistent metagenomic signatures of early-life hospitalisation and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol; 4: 2285–2297. doi: 10.1038/s41564-019-0550-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bokulich NA, Chung J, Battaglia T, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 2016; 8: 343ra82. doi: 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo SM, Gooch KL, Korol EE, et al. A population-based study of childhood respiratory morbidity after severe lower respiratory tract infections in early childhood. J Pediatr 2014; 165: 123–128. doi: 10.1016/j.jpeds.2014.02.053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00939-2020.SUPPLEMENT (1.4MB, pdf)