Abstract
Background
The therapeutic efficacy of epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) in advanced EGFR-mutant lung squamous cell carcinoma (SCC) patients remains uncertain. Furthermore, the factors underlying the responsiveness have not been fully investigated. We therefore investigated the link between genomic profiles and EGFR-TKI efficacy.
Material and Methods
We consecutively enrolled stage IV, EGFR-mutant, and EGFR-TKI–treated patients with SCC. Patients with EGFR wild-type lung SCC and EGFR-mutant lung adenocarcinoma were consecutively enrolled as controls, and next-generation sequencing (NGS) was performed.
Results
In total, 28 EGFR-mutant lung SCC, 41 EGFR-mutant lung adenocarcinoma, and 40 EGFR wild-type lung SCC patients were included. Among the patients with EGFR mutations, shorter progression-free survival (PFS) was observed in SCC compared to adenocarcinoma (4.6 vs. 11.0 months, P<0.001). Comparison of the genomic profiles revealed that EGFR-mutant SCC patients had similar mutation characteristics to EGFR-mutant adenocarcinoma patients, but differed from those with EGFR wild-type SCC. Further exploration of EGFR-mutant SCC revealed that mutations in CREBBP (P = 0.005), ZNF217 (P = 0.016), and the Wnt (P = 0.027) pathway were negatively associated with PFS. Mutations in GRM8 (P = 0.025) were associated with improved PFS.
Conclusions
EGFR-mutant lung SCC has a worse prognosis than EGFR-mutant adenocarcinoma. Mutations in other genes, such as CREBBP, ZNF217, GRM8, or Wnt that had implications on PFS raise the possibility of understanding mechanisms of resistance to EGFR-TKI in lung SCC, which will aid identification of potential beneficial subgroups of patients with EGFR-mutant SCCs receiving EGFR-TKIs.
Keywords: lung squamous cell carcinoma, epidermal growth factor receptor, tyrosine kinase inhibitor, genomic profile, progression-free survival
Introduction
Epidermal growth factor receptor (EGFR) represents as the most frequently mutated driver gene in lung cancer. In comparison with lung adenocarcinoma, EGFR mutations are relatively rare lung squamous cell carcinoma (SCC), with a reported prevalence of 3% to 18% (1–10). In light of this, the therapeutic value of targeted therapy, EGFR tyrosine kinase inhibitors (EGFR-TKIs), in advanced lung SCC patients lacks in-depth multiple dimensional exploration with large cohorts. Pilot studies have reported moderate effectiveness of EGFR-TKIs in EGFR-mutant lung SCC, with an objective response rate (ORR) ranging from 25% to 49% (11–17). However, shortened progression-free survival (PFS) was shown, ranging from 1 to 5 months, in EGFR-mutant SCC patients (11–15, 17–21). Still, evidences derived from large-scale prospective cohorts are lacking.
Furthermore, the mechanism underlying the limited efficacy of EGFR-TKIs in EGFR-mutant lung SCC is poorly understood. Data from the Cancer Genome Atlas (TCGA) and Chinese cohorts have revealed that lung SCC exhibited a different genomic profile from that of lung adenocarcinoma (4, 22–24), providing insights into the study involving genome-based efficacy analysis. Regrettably, there are currently no studies exploring the genomic profile of EGFR-mutant lung SCC or analyzing the association between the genomic features and therapeutic efficacy.
In this study, we retrospectively recruited advanced lung SCC patients with EGFR mutations, and enrolled patients with EGFR-mutant adenocarcinoma and EGFR wild-type SCC. We aimed to characterize the genomic patterns of EGFR-mutant SCC, and analyze EGFR-TKI efficacy in EGFR-mutant SCC according to the genomic profiles of the patients.
Materials and Methods
Patients and Study Procedure
We enrolled 28 EGFR-mutant lung SCC, 41 EGFR-mutant lung adenocarcinoma, and 40 EGFR wild-type lung SCC patients from the Second Affiliated Hospital of Zhejiang University School of Medicine, the First Affiliated Hospital of Zhejiang University School of Medicine, and Sir Run Run Shaw Hospital of Zhejiang University School of Medicine from June 2015 to June 2019. Primary eligibility criteria included histologically confirmed SCC, stage IV non-small cell lung cancer (NSCLC) with EGFR mutations, and treatment with EGFR-TKIs. Other eligibility criteria included an Eastern Cooperative Oncology Group (ECOG) performance status of 0 to 2, at least one measurable lesion, and a life expectancy of 3 months or longer. EGFR-mutant adenocarcinoma patients and EGFR-wild-type SCC patients were consecutively enrolled at the same time.
All patients were diagnosed via percutaneous and transbronchial lung biopsy. Pathological diagnosis was confirmed by light microscopy and immunohistochemistry (IHC), and verified by staining for P40 (+) together with thyroid transcription factor (TTF)-1 (−) and napsin A (−). Formalin-fixed paraffin-embedded (FFPE) blocks or 10 to 15 PPFE slices with a thickness of 6 to 10 µm were obtained and the NGS testing was carried out in Nanjing Geneseeq Technology Inc (Nanjing, China) testing laboratory. Samples with tumor cell content above 20% were considered qualified.
The studies involving human patients were reviewed and approved by institutional review board of Second Affiliated Hospital of Zhejiang University School of Medicine.
Clinical Assessments and End Points
Tumor response was assessed using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. The treatment response was evaluated 1 month after the initiation of EGFR-TKI therapy and every 2 to 3 months thereafter, based on the patients’ computed tomography (CT) and magnetic resonance imaging (MRI) data.
The primary outcome was PFS, defined as the time from the start of treatment to disease progression, as confirmed by radiologic diagnosis or death from any cause. Patients who did not relapse or not die were censored at the last follow-up. Exploratory analyses included comparing the genomic profiles between EGFR-mutant adenocarcinoma and EGFR-mutant SCC patients, and EGFR-mutant SCC and EGFR wild-type SCC patients. The response to TKIs of EGFR-mutant SCC patients according to genomic profile was also analyzed.
DNA Extraction, Sequencing, and Bioinformatics Analysis
DNA extraction, sequencing library preparation, and targeted capture were carried out following previously described methods, with some modifications (25) (see Supplementary Methods ). Genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen), and libraries were prepared by KAPA Hyper Prep kit (KAPA Biosystems). Customized xGen lockdown probes panel (Integrated DNA Technologies) were used to selectively enrich for 425 predefined cancer-related genes (Geneseeq Prime panel) (see Table S1 ). Target-enriched libraries were sequenced on the HiSeq4000 platform (Illumina). Gene fusions were identified by FACTERA, copy number variations (CNVs) were analyzed with ADTEx, and allele-specific CNVs were analyzed by FACETS. Chromosome instability score (CIS) was defined as the proportion of the genome with aberrant (purity-adjusted segment-level copy number >=3 or <=1) segmented copy number. Tumor mutation burden (TMB) was defined as the number of somatic, coding, base substitution, and indel mutations per megabase of genome examined. Briefly, all base substitutions, including non-synonymous and synonymous alterations, and indels in the coding region of targeted genes were considered with the exception of known hotspot mutations in oncogenic driver genes and truncations in tumor suppressors. Synonymous mutations were counted in order to reduce sampling noise, and known driver mutations were excluded as they are over-represented in the panel. The summary of genomic aberrations among three cohorts is presented in Table S2 .
Statistical Analysis
Quantitative data are presented as the median (range) with percentages. Comparisons of proportions between groups were performed using Fisher’s exact test. For the survival analysis, Kaplan-Meier curves were generated, and p-values were determined with the log-rank test. Hazard ratios (HRs) were calculated by Cox proportional hazards model. A two-sided p-value of <0.05 was considered significant for all tests unless indicated otherwise. Univariable Cox regression was used to study the association between the different variables and PFS, and the results are presented as HRs with 95% confidence intervals (CIs). All analyses were performed with R 3.6.0 (R Development Core Team).
Results
Patient Characteristics
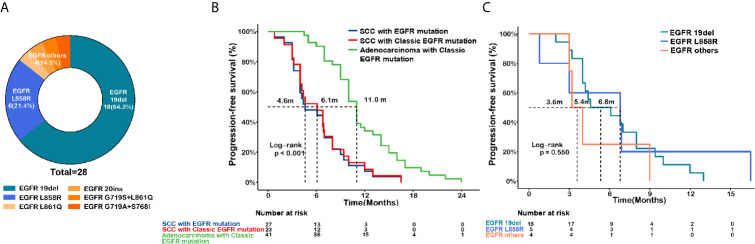
Baseline characteristics of the EGFR-mutant lung SCC (n = 28), EGFR-mutant lung adenocarcinoma (n = 41), and EGFR wild-type lung SCC (n = 40) groups were listed in Table 1 . Among lung SCC patients harboring the EGFR mutation, we identified 18 19Del (64.3%), 6 L858R (21.4%), 1 L861Q (3.6%), 1 20ins (3.6%), 1 G719S+L861Q (3.6%), and 1 G719S+S768I (3.6%) ( Figure 1A ). Among the EGFR-mutant adenocarcinoma patients, 19 (64.3%) harbored 19Del, and 22 (53.7%) harbored L858R changes. In patients with EGFR-mutant SCC, 27 (96.4%) were treated with first- or second-generation EGFR-TKIs, and only one (3.6%) with osimertinib. Similarly, the vast majority of adenocarcinoma patients (39, 95.1%) received first- or second-generation EGFR-TKIs, only two (4.9%) received osimertinib.
Table 1.
Baseline clinical characteristics of the study population.
| Characteristics | EGFR-mutant SCC(N=28) | EGFR-mutant adenocarcinoma(N=41) | EGFR wild-type SCC (N=40) |
|---|---|---|---|
| Age | 65 (46–83) | 64 (40–80) | 67 (50–87) |
| ≥65 | 13 (46.4%) | 21 (51.2%) | 35 (87.5%) |
| <65 | 15 (53.6%) | 20 (48.8%) | 15 (37.5%) |
| Gender | |||
| Male | 16 (57.1%) | 16 (39.0%) | 19 (47.5%) |
| Female | 12 (42.9%) | 25 (61.0%) | 21 (52.5%) |
| Smoking status | |||
| Ever-smokers | 10 (35.7%) | 19 (46.3%) | 28 (70.0%) |
| Never-smokers | 18 (64.3%) | 22 (53.7%) | 12 (30.0%) |
| EGFR Mutant | |||
| 19Del | 18 (64.3%) | 19 (46.3%) | – |
| L858R | 6 (21.4%) | 22 (53.7%) | – |
| Other mutation | 4 (14.3%) | 0 (0%) | – |
| EGFR-TKI | |||
| Icotinib | 14 (51.9%) | 18 (43.9%) | – |
| Gefitinib | 6 (22.2%) | 18 (43.9%) | – |
| Erlotinib | 3 (11.1%) | 3 (7.3%) | – |
| Afatinib | 3 (11.1%) | 0 (0%) | – |
| Osimertinib | 1 (3.7%) | 2 (4.9%) | – |
EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
Figure 1.
EGFR mutational subtypes and survival analysis in EGFR-mutated lung squamous cell carcinoma (SCC) and adenocarcinoma. (A) Mutation subtypes of EGFR-mutant lung SCC. (B) Kaplan-Meier curves of progression-free survival (PFS) comparing the SCC patients harboring EGFR mutation (n=27, blue) or classic EGFR mutation (19del or L858R, n=23, red), and in adenocarcinoma patients harboring classic EGFR mutation (19del or L858R, n=41, green). (C) Kaplan-Meier curves of PFS in SCC patients harboring 19del (n=18, green), L858R (n=5, blue), or other uncommon mutations (n=4, orange).
Efficacy of TKIs for EGFR-Mutant SCC and Adenocarcinoma
Among EGFR-mutant lung cancer patients receiving EGFR-TKIs, shorter PFS was observed in SCC compared to adenocarcinoma, (median PFS (mPFS): 4.6 vs. 11 months, P<0.001, Figure 1B ). Non-significant differences in PFS were found among the various EGFR mutation types in SCC (mPFS: 5.4 months for 19Del vs. 6.8 months for L858R vs. 3.6 months for other mutations, P = 0.550, Figures 1C ). Also, PFS was not affected by gender (P = 0.56) or smoking status (P = 0.14, Figure S1A, B ). These findings support the idea that, compared to EGFR-mutant adenocarcinoma, SCC with the EGFR mutation is less responsive to EGFR-TKIs.
Construction of Genomic Profiles and Molecular Features Correlated With Blunted Efficacy of EGFR-TKI in SCC
Comparison of Genomic Profiles Among EGFR-Mutant Adenocarcinoma, EGFR-Mutant SCC, and EGFR Wild-Type SCC
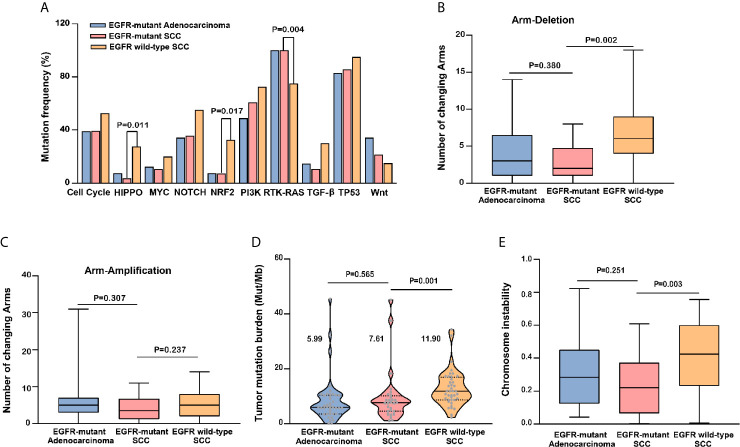
We compared genomic profiles between adenocarcinoma and SCC. Figure 2 shows the genomic changes detected in EGFR-mutant adenocarcinoma, EGFR-mutant SCC, and EGFR wild-type SCC, including genetic alterations, somatic copy-number alterations, and arm-level alterations.
Figure 2.
The landscape of genomic profiles in EGFR-mutant adenocarcinoma, EGFR-mutant SCC, and EGFR wild-type SCC. 32 top-ranking genetic alterations (top panel), 4 somatic copy number alterations (SCNAs, middle panel), and 22 arm level alteration (bottom panel) in three subgroups (EGFR-mutant adenocarcinoma, n = 28; EGFR-mutant SCC, n = 41; EGFR wild-type SCC, n = 40) were represented. Each column represented a sample.
Compared to EGFR-mutant adenocarcinoma, EGFR-mutant SCC exhibited a higher mutation ATR frequency [odds ratio (OR): 8.44, 95% CI: 0.87–420.30, P=0.037], BRCA1 (OR: 8.44, 95% CI: 0.87–420.30, P=0.037), and NF1 (OR: 8.44, 95% CI: 0.87–420.30, P=0.037), and more CNVs of EGFR (OR: 4.28, 95% CI: 1.03–21.60, P=0.028) ( Table S3 ). We also assessed responsiveness to EGFR-TKI according to the mutation profile. Either in EGFR-mutant SCC cohort or EGFR-mutant adenocarcinoma cohort, no difference was observed in PFS between mutant group and the wild-type group ( Table S4 , Figures S1C–E ). Notably, EGFR-mutant SCC and adenocarcinoma had a similar mutation frequency for TP53 (78.6% vs. 65.9%), where this mutation is known to reduce EGFR-TKI efficacy. However, compared to EGFR wild-type SCC, EGFR-mutant SCC showed a lower mutation frequency for FAT1 (OR: 0, 95% CI: 0–0.54, P = 0.004) and SMAD4 (OR: 0, 95% CI: 0–1.14, P=0.039), a higher CNV of EGFR (OR: 5.68, 95% CI: 1.23–36.46, P=0.020), and lower CNVs of SOX2 (OR: 0.06, 95CI: 0.001–0.47, P=0.001) and PIK3CA (OR: 0, 95% CI: 0–0.36, P<0.001) ( Table S3 ).
We also assessed signaling pathway involvement. In line with the above results, signaling pathway mutation frequency was identical between EGFR-mutant adenocarcinoma and SCC ( Figure 3A and Table S5 ). In contrast, the HIPPO pathway (OR: 0.10, 95% CI: 0.002–0.77, P=0.011), NRF2 pathway (OR: 0.16, 95% CI: 0.02–0.83, P=0.017), and RTK-RAS pathway (OR: infinity, 95% CI: 1.85–infinity, P=0.004) were significantly different between SCC with versus without the EGFR mutation.
Figure 3.
Comparison of genomic profiles among EGFR-mutant adenocarcinoma, EGFR-mutant squamous cell carcinoma (SCC), and EGFR wild-type SCC. Comparison of 10 signaling pathway mutation frequency (A), change number of arm deletion (B) and arm amplification (C), tumor mutation burden (TMB) (D), and chromosome instability (CIS) (E) among three subsets were represented.
Regarding mutation characteristics at the arm-level, the total number of arm deletions (P=0.380, Figure 3B ) and arm amplifications (P=0.307, Figure 3C ) was identical between EGFR-mutant adenocarcinoma and SCC. In contrast, a lower number of arm deletions (P=0.002, Figure 3B ) and similar number of arm amplifications (P=0.237, Figure 3C ) were detected in EGFR-mutant SCC compared to EGFR wild-type SCC.
Finally, we evaluated the TMB and CIS. TMB was comparable between EGFR-mutant SCC and adenocarcinoma patients (7.61 vs. 5.99 mutations/Mb, P=0.565), the median TMB (mTMB) was 7.61 (95% CI: 5.84–13.40) and 5.99 (95% CI: 5.77–11.13) mutations/Mb, respectively. However, it is interesting that the mTMB was significantly lower in EGFR-mutant SCC than EGFR wild-type SCC (7.61 vs. 11.9 mutations/Mb, P=0.001), the mTMB was 7.61 (95% CI: 5.84–13.40) and 11.90 (95% CI: 10.72–14.96) mutations/Mb, respectively ( Figure 3D ). The CIS was similar between EGFR-mutant SCC and adenocarcinoma (P = 0.251). In contrast, the CIS of EGFR-mutant SCC was significantly lower than that of EGFR wild-type SCC (P = 0.003; Figure 3E ).
Overall, EGFR-mutant SCC showed similar gene characteristics to EGFR-mutant adenocarcinoma, but different ones to EGFR wild-type SCC.
EGFR-TKI Efficacy in EGFR-Mutant SCC Patients According to Genetic Profile
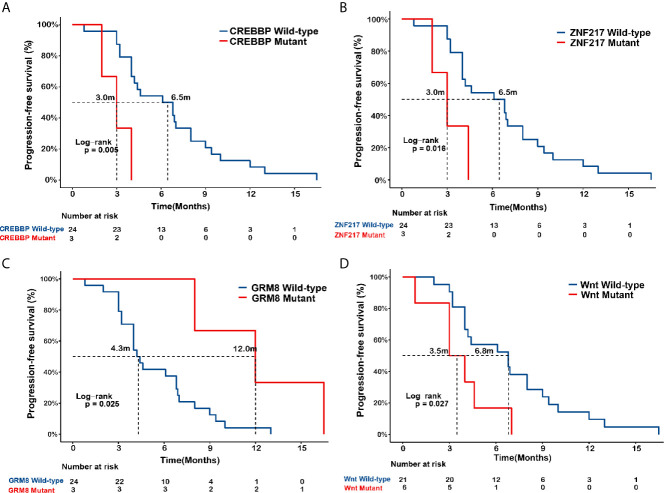
To identify the factors influencing EGFR-TKI efficacy in EGFR-mutant SCC, PFS was analyzed according to genomic profile within the EGFR-mutant SCC cohort.
As shown in Figure 4 , mPFS was poorer in the CREBBP-mutant group than the wild-type (3.0 vs. 6.5 months, P=0.005) and in the ZNF217-mutant SCC group than the wild-type (3.0 vs. 6.5 months, P=0.016). Conversely, mPFS was significantly improved in SCC patients with the GRM8 mutation than the wild-type (12.0 vs. 4.3 months, P=0.025). In addition, SCC patients with Wnt pathway mutations exhibited a shorter mPFS than the wild-type (3.5 vs. 6.8 months, P=0.027).
Figure 4.
Associations of CREBBP, ZNF217, GRM8, and Wnt pathway mutation with EGFR-TKI outcome in EGFR-mutant squamous cell carcinoma (SCC). Kaplan-Meier curves of progression-free survival (PFS) in EGFR-mutated lung squamous cell carcinoma (SCC) in terms of mutational status of CREBBP (A), ZNF217 (B), GRM8 (C), and Wnt pathway (D).
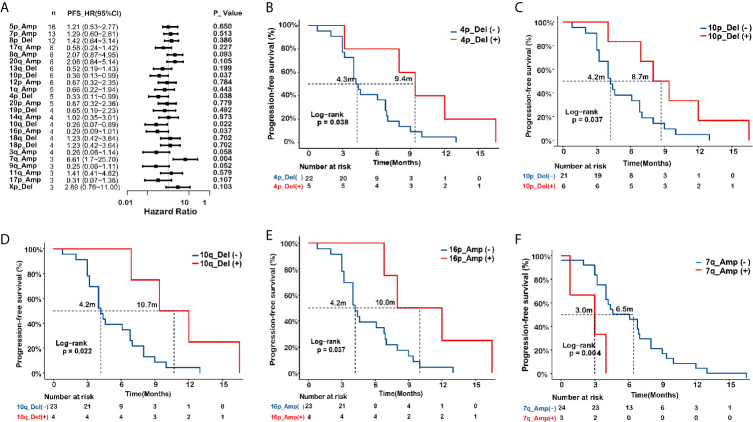
Next, we evaluated associations of arm-level changes with the responsiveness to EGFR-TKIs in the EGFR-mutant SCC patients. A forest plot is presented in Figure 5A . The mPFS was longer in SCC patients with chromosome arm changes on 4p_Del (9.4 vs. 4.3 months, P=0.038), 10p_Del (8.7 vs. 4.2 months, P=0.037), 10q_Del (10.7 vs. 4.2 months, P=0.022), and 16p_Amp (10 vs. 4.2 months, P=0.037) ( Figures 5B–E ), whereas mPFS was shorter in the SCC patients with 7q_Amp (3.0 vs 6.5 months, P=0.004) ( Figure 5F ).
Figure 5.
Associations of arm level changes with EGFR-TKI outcome in EGFR-mutant squamous cell carcinoma (SCC). (A) Forest plot presenting hazard ratios (HRs) of progression-free survival (PFS) comparing subgroups with and without specific arm-level changes in EGFR-mutated lung squamous cell carcinoma (SCC). (B–F) Kaplan-Meier curves of progression-free survival (PFS) in EGFR-mutated SCC in terms of arm-level changes on 4p_Del (B), 10p_Del (C), 10q_Del (D), and 16p_Amp (E), 7q_Amp (F). Del, deletion; Amp, amplification.
We also analyzed blunted reactivity to EGFR-TKI in SCC patients according to TMB and CIS. The mPFS did not differ significantly between patients with high and low TMB (8.2 vs. 4.2 months, P=0.095), or between patients with CIS ≥ 30% and < 30% (6.1 vs. 4.1 months, P=0.351, Figures S1F–G ). Since smokers were considered to bear higher TMB, this result was in line with the observation in different smoking status subset.
In summary, SCC patients harboring both the EGFR and CREBBP mutation, and those with the ZNF217 mutation, Wnt pathway mutation, and 7q_Amp were likely to have a shorter PFS, whereas those with the GRM8 mutation, or 4p_Del, 10p_Del, 10q_ Del, or 16p_Amp chromosome arm changes, tended to have prolonged PFS.
Discussion
To our knowledge, this is the first study to explore the therapeutic efficacy of EGFR-TKI in EGFR-mutant SCC patients with mutation profiles. In SCC patients harboring the EGFR mutation, identifying the factors that influence EGFR-TKI responsiveness, and the subgroups most likely to benefit from these agents, is of great importance. Our study provides evidence of a shorter PFS in EGFR-mutant SCC compared to EGFR-mutant adenocarcinoma patients. With discrepancy in TKI outcomes, EGFR-mutant SCC and adenocarcinoma, however, presented similar genomic patterns. We further observed that specific genomic features, such as CREBBP, ZNF217, or Wnt pathway mutation, may predict a shortened PFS in SCC patients.
Previous studies have shown that SCC patients with EGFR mutations conferred responsiveness to EGFR-TKIs, with a non-inferior ORR (25–49%) to that of lung adenocarcinoma patients (11–16). However, the survival benefit of EGFR-TKIs was not as pronounced in lung SCC patients. Our study confirmed an inferior mPFS (4.6 months) in the EGFR-mutant SCC cohort, in line with previous reports of a 1–5 month PFS (20, 26, 27). Previously, patients with lung non-adenocarcinoma, such as large cell lung carcinomas (LCLCs), harboring EGFR mutations and treated with EGFR-TKIs, also had a reduced mPFS (4.4 months) (20). In contrast, lung adenosquamous carcinoma patients showed a PFS of 8–14 months (20, 28).
The Cancer Genome Atlas (TCGA) study and data from Chinese lung cancer cohorts have confirmed differences in genomic profiles between lung SCC and adenocarcinoma patients (4, 22–24). In lung SCC, TP53, NFE2L2, CDKN2A, KEAP1, and PTEN were the most commonly mutated genes. Conversely, in adenocarcinoma, mutations were typically observed in the TP53, KRAS, EGFR, STK11, and RB1 genes. The higher mutation frequency of TP53 and lower mutation frequency of EGFR and KRAS were involved in lung SCC. Compared to the TCGA data, the Nanjing Lung Cancer Cohort (NJLCC) (4) and CHOICE cohort (24) of Chinese patients revealed higher rates of EGFR and RB1 mutations, and lower rates of KRAS, BRAF, and STK11 mutations, in adenocarcinoma patients. In SCC patients, there was a higher mutation frequency of TP53, RB1, and NFE2L2, and a lower mutation frequency of PIK3CA and CDKN2A. In EGFR wild-type SCC patients, TP53, NFE2L2, CDKN2A, KEAP1, PTEN, and RB1 were the most commonly mutated genes, consistent with previously reported genomic profiles.
We observed a higher mutation frequency of NF1, ATR, and BRCA1 in EGFR-mutant SCC compared to EGFR-mutant adenocarcinoma. NF1 is a tumor suppressor gene that negatively regulates RAS signaling (29). NF1‐mutant lung adenocarcinoma patients had inferior disease‐free survival (DFS), and overall survival (OS) compared to those with the EGFR-mutation (30), and downregulation of NF1 expression caused by truncating mutations was reported to confer resistance to EGFR-TKI in lung adenocarcinoma patients (31). Notably, in our study, the NF1 mutation was negatively, though not significantly, associated with PFS in lung SCC patients, which was possibly due to the relatively small size of the cohort. The ATR kinase encoded by ATR is implicated in the DNA damage response (DDR) (32), and is involved in lung cancer development (33). The ATR/CHK1 (the downstream effector kinases of ATR) axis was identified as a potential drug target for small-cell lung cancer (SCLC) patients (34), but no similar study in NSCLC has been reported. BRCA1 is a tumor suppressor gene that participates in DNA repair processes; mutations therein elevate the risk of developing breast, ovarian, and other cancers (35). Interestingly, EGFR-mutant NSCLC patients with a concurrent germline BRCA mutation showed a comparable PFS, and longer OS, in the context of EGFR-TKI treatment compared to patients with the wild-type germline BRCA (36).
We also explored the factors associated with EGFR-TKI efficacy in our EGFR-mutant SCC cohort. We identified the CREBBP mutation, ZNF217 mutation, and GRM8 mutation. CREBBP is a tumor suppressor gene that encodes a histone modifier, and the CREBBP mutation is considered a driver mutation in SCLC (37, 38). Pilot study indicated that CREBBP mutation may be involved in mutation profile that fitted Big Bang cancer evolution model (39). ZNF217 is an oncogene that has deleterious effects in various human cancers (40). Overexpression of the ZNF217 protein is associated with the development of spontaneous lung or node metastases in mice (41). In NSCLC, the positive expression rate of ZNF217 protein was higher in cancer tissues than that in paracancerous tissues, and increased with the increase of TMN stage. Poorer OS and PFS were noted in NSCLCs with positive ZNF217 ( 42). GRM8 is a member of the G-protein coupled receptors for the glutamate family. Mutations in GRM8 are reported in 8–16% of lung SCC patients (43, 44). GRM8 activation promotes lung SCC survival by inhibiting the cAMP pathway and activating the MAPK pathway (45); this agrees with our observations that patients with mutant GRM8 tended to show greater responsiveness to EGFR-TKI treatment. In the pathway analysis, poorer PFS was observed in the Wnt-mutant subgroup of the EGFR-mutant SCC cohort. The CTNNB1 gene, a key driver of Wnt signaling pathway activity, encodes the β-catenin protein, which regulates cellular proliferation (46). Mutations in CTNNB1 may lead to tumor proliferation and thus a poorer outcome. In our study, the presence of a Wnt pathway mutation was correlated with reduced responsiveness to EGFR-TKI treatment (47).
TMB, although with debates, has been employed to predict the beneficial population of ICIs. Recent meta-analysis compared the efficacy among first-line ICIs versus standard chemotherapy in TMB high and low patients. After analyzing eight different cohorts from five randomized controlled phase III studies (3848 patients), they found a proven benefit in OS in favor of IO agents in the TMB-high population (48). We did not identify significantly higher TMB in our EGFR-mutant SCC patients compared to those with adenocarcinoma, consistent with previous reports (24, 49). Some previous studies (50, 51) found that TMB in SCC was higher than that in adenocarcinoma, however, they did not distinguish between patients with EGFR mutations and those without EGFR mutations. Herein, the TMB of EGFR-mutant SCC seemed to be higher than that of adenocarcinoma, but no significant difference showed. This implied that as long as the patients harbored EGFR driver mutations, regardless of lung SCC or adenocarcinoma, the outcome for the mutation number was similar. On the contrary, the TMB was significantly higher in EGFR wild-type SCC than EGFR-mutant SCC in our study. Actually, the incidence of EGFR driver mutations in SCC is very low, which has little effect on the whole TMB value of all SCC patients. The higher TMB in EGFR-mutant SCC indicated that EGFR driver mutations had a great impact on mutation number of tumor cells, although they belonged to a same disease subtype in pathology. This phenomenon implied that the tumor evolutionary trajectories may had some difference between EGFR wild-type SCC and EGFR-mutant SCC. Of course, on the other hand, our sample size was limited. More studies with larger sample size are needed to further verify these findings. In addition, tissue specimen-based TMB analysis is frequently constrained by the inadequate tissue volume and tissue quality. Pilot study have confirmed the successful use of cytological samples for TMB analysis (52) and large-scale prospective studies are warranted.
With regard to our study, some limitations need to be acknowledged. First, our EGFR-mutated SCC cohort was relatively small, although multicenter recruitment has been undertaken to increase the cohort size. The prevalence of EGFR mutation is rather limited in SCC. Second, all of the patients in our study had advanced SCC or adenocarcinoma, diagnosed based on analysis of small samples rather than surgical resection. Third, although we have performed comprehensive genomic characterization of EGFR-mutant squamous cell lung cancer and try to figure out why EGFR-mutant SCC confers poor responsiveness to TKI in terms of mutation profiles, the underlying mechanisms have not been verified in vitro and in vivo. Between EGFR-mutant SCC and EGFR-mutant adenocarcinoma, the observation of similar genomic patterns presents, we are unable to adequately identify the unambiguous mechanism underlying the blunted responsiveness. However, we identified several aberrations as predictors for shortened PFS in SCC patients, and these associations are warranted to be validated in other cohort or via experimental approaches.
In conclusion, our study showed that EGFR-mutant lung SCC patients responded poorly to EGFR-TKI treatment compared to the EGFR-mutant adenocarcinoma patients. In lung SCC, EGFR mutation concomitant with a CREBBP, ZNF217, or Wnt pathway mutation was negatively associated with PFS. Conversely, a mutation in GRM8 was associated with improved PFS. These findings provide shed light on the mechanism underlying the blunted reactivity to EGFR-TKIs in SCC, and identify EGFR-mutant SCC patients as a subgroup likely to benefit from treatment with these agents.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the institutional review board of Second Affiliated Hospital of Zhejiang University School of Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HS, WL, and YX contributed to study conception and design. RJ, LP, JiaS, JW, YJ, FeiL, JZ, QL, BZ, FenL, and LX conducted patient recruitment and data collection. MW, XW, JY, and YS conducted DNA sequencing and bioinformatics analysis. RJ, QL, and YX drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation [LY20H010004] and the National Natural Science Foundation of China [81870022].
Conflict of Interest
MW, XW, JY and YS were employed by Geneseeq Technology Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.680804/full#supplementary-material
Associations of gender (A), smoking status (B), NF1 (C), ATR (D), BRCA1 mutation, tumor mutation (E) burden (TMB) (F), and chromosome instability (CIS) (G) with EGFR-TKI efficacy in EGFR-mutant squamous cell carcinoma (SCC).
References
- 1. Han B, Tjulandin S, Hagiwara K, Normanno N, Wulandari L, Konstantinovich LK, et al. Determining the Prevalence of EGFR Mutations in Asian and Russian Patients (Pts) With Advanced Non-Small-Cell Lung Cancer (ANSCLC) of Adenocarcinoma (Adc) and Non-Adc Histology: Ignite Study. Ann Oncol (2015) 26i29-30. 10.1093/annonc/mdv050.1 [DOI] [Google Scholar]
- 2. Zhang Q, Zhu L, Zhang J. Epidermal Growth Factor Receptor Gene Mutation Status in Pure Squamous-Cell Lung Cancer in Chinese Patients. BMC Cancer (2015) 15:88. 10.1186/s12885-015-1056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang R, Zhang Y, Pan Y, Li Y, Hu H, Cai D, et al. Comprehensive Investigation of Oncogenic Driver Mutations in Chinese Non-Small Cell Lung Cancer Patients. Oncotarget (2015) 6(33):34300–8. 10.18632/oncotarget.5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C, Yin R, Dai J, Gu Y, Cui S, Ma H, et al. Whole-Genome Sequencing Reveals Genomic Signatures Associated With the Inflammatory Microenvironments in Chinese NSCLC Patients. Nat Commun (2018) 9(1):2054. 10.1038/s41467-018-04492-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qian J, Chen R, Zhao R, Han Y, Yu Y. Comprehensive Molecular Characterizations of Chinese Patients With Different Subtypes of Lung Squamous Cell Carcinoma. Front Oncol (2020) 10:607130. 10.3389/fonc.2020.607130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang B, Zhang L, Yue D, Li C, Zhang H, Ye J, et al. Genomic Characteristics in Chinese Non-Small Cell Lung Cancer Patients and Its Value in Prediction of Postoperative Prognosis. Transl Lung Cancer Res (2020) 9(4):1187–201. 10.21037/tlcr-19-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Z, Chen Y, Kong W, Yu Z, He X. Genomic Features of Chinese Lung Squamous Cell Carcinoma Patients. Ann Oncol (2020) 31:S253. 10.1016/j.annonc.2020.08.181 [DOI] [Google Scholar]
- 8. Gao X, Zhu J, Chen L, Jiang Y, Zhou X, Shuai J, et al. Clinical And Imageological Features of Lung Squamous Cell Carcinoma With EGFR Mutations. Cancer Manag Res (2019) 11:9017–24. 10.2147/CMAR.S223021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lam VK, Tran HT, Banks KC, Lanman RB, Rinsurongkawong W, Peled N, et al. Targeted Tissue and Cell-Free Tumor DNA Sequencing of Advanced Lung Squamous-Cell Carcinoma Reveals Clinically Significant Prevalence of Actionable Alterations. Clin Lung Cancer (2019) 20(1):30–6.e3. 10.1016/j.cllc.2018.08.020 [DOI] [PubMed] [Google Scholar]
- 10. Joshi A, Mishra R, Desai S, Chandrani P, Kore H, Sunder R, et al. Molecular Characterization of Lung Squamous Cell Carcinoma Tumors Reveals Therapeutically Relevant Alterations. Oncotarget (2021) 12(6):578–88. 10.18632/oncotarget.27905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang W, Zhang J, Liang W, Huang Y, Yan Y, Wu X, et al. Efficacy of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors for Chinese Patients With Squamous Cell Carcinoma of Lung Harboring EGFR Mutation. J Thorac Dis (2013) 5(5):585–92. 10.3978/j.issn.2072-1439.2013.09.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hata A, Katakami N, Yoshioka H, Kunimasa K, Fujita S, Kaji R, et al. How Sensitive Are Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors for Squamous Cell Carcinoma of the Lung Harboring EGFR Gene-Sensitive Mutations? J Thorac Oncol (2013) 8(1):89–95. 10.1097/JTO.0b013e31827690b5 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y, Zhang Y, Zhang L, Liu B, Wang Y, Zhou X, et al. Efficacy of Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors for Lung Squamous Carcinomas Harboring EGFR Mutation: A Multicenter Study and Pooled Analysis of Published Reports. Oncotarget (2017) 8(30):49680–8. 10.18632/oncotarget.17915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu J, Chu T, Jin B, Dong X, Lou Y, Zhang X, et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Advanced Squamous Cell Lung Cancer. Clin Lung Cancer (2016) 17(4):309–14. 10.1016/j.cllc.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 15. Zhuang J, Yu Y, Li Z, Lu S. Efficacy of Epidermal Growth Factor Receptor (EGFR)-Tyrosine Kinase Inhibitors (TKIs) in Targeted Therapy of Lung Squamous Cell Carcinoma Patients With EGFR Mutation: A Pooled Analysis. Oncotarget (2017) 8(32):53675–83. 10.18632/oncotarget.15726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang S, Xu Y, Tan F, Ding L, Ma Y, Wang M. Efficacy of Icotinib in Advanced Lung Squamous Cell Carcinoma. Cancer Med (2018) 7(9):4456–66. 10.1002/cam4.1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou S, Wang H, Jiang W, Yu Q. Clinicopathological Characteristics and EGFR-TKIs Efficacies In Lung Squamous Cell Carcinoma Patients Harboring an EGFR Sensitizing Mutation. Onco Targets Ther (2019) 12:8863–71. 10.2147/OTT.S225760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fiala O, Pesek M, Finek J, Benesova L, Bortlicek Z, Minarik M. Gene Mutations in Squamous Cell NSCLC: Insignificance of EGFR, KRAS and PIK3CA Mutations in Prediction of EGFR-TKI Treatment Efficacy. Anticancer Res (2013) 33(4):1705–11. 10.1016/S0169-5002(13)70293-9 [DOI] [PubMed] [Google Scholar]
- 19. Hata A, Katakami N, Kunimasa K, Yoshioka H, Fujita S, Kaji R, et al. Erlotinib for Pretreated Squamous Cell Carcinoma of the Lung in Japanese Patients. Jpn J Clin Oncol (2011) 41(12):1366–72. 10.1093/jjco/hyr159 [DOI] [PubMed] [Google Scholar]
- 20. Xu J, Zhang Y, Jin B, Chu T, Dong X, Yang H, et al. Efficacy of EGFR Tyrosine Kinase Inhibitors for Non-Adenocarcinoma Lung Cancer Patients Harboring EGFR-Sensitizing Mutations in China. J Cancer Res Clin Oncol (2016) 142(6):1325–30. 10.1007/s00432-016-2133-4 [DOI] [PubMed] [Google Scholar]
- 21. Hu M, Zhang B, Xu J, Wang S, Zhao Y, Zhang L, et al. Clinical Outcomes of Different Generations of EGFR Tyrosine Kinase Inhibitors in Advanced Lung Adenosquamous Carcinoma. Mol Diagn Ther (2019) 23(6):773–9. 10.1007/s40291-019-00425-x [DOI] [PubMed] [Google Scholar]
- 22. Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature (2014) 511(7511):543–50. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, et al. Comprehensive Genomic Characterization of Squamous Cell Lung Cancers. Nature (2012) 489(7417):519–25. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, et al. Comprehensive Genomic and Immunological Characterization of Chinese Non-Small Cell Lung Cancer Patients. Nat Commun (2019) 10(1):1772. 10.1038/s41467-019-09762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tong L, Ding N, Tong XL, Li JM, Zhang Y, Wang XD, et al. Tumor-Derived DNA From Pleural Effusion Supernatant as a Promising Alternative to Tumor Tissue in Genomic Profiling of Advanced Lung Cancer. Theranostics (2019) 9(19):5532–41. 10.7150/thno.34070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song Z, Zhang Y. Efficacy of Gefitinib or Erlotinib in Patients With Squamous Cell Lung Cancer. Arch Med Sci (2015) 11(1):164–8. 10.5114/aoms.2013.39234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shukuya T, Takahashi T, Kaira R, Ono A, Nakamura Y, Tsuya A, et al. Efficacy of Gefitinib for Non-Adenocarcinoma Non-Small-Cell Lung Cancer Patients Harboring Epidermal Growth Factor Receptor Mutations: A Pooled Analysis of Published Reports. Cancer Sci (2011) 102(5):1032–7. 10.1111/j.1349-7006.2011.01887.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin G, Li C, Li PS, Fang WZ, Xu HP, Gong YH, et al. Genomic Origin and EGFR-TKI Treatments of Pulmonary Adenosquamous Carcinoma. Ann Oncol (2020) 31(4):517–24. 10.1016/j.annonc.2020.01.014 [DOI] [PubMed] [Google Scholar]
- 29. Le LQ, Parada LF. Tumor Microenvironment and Neurofibromatosis Type I: Connecting the GAPs. Oncogene (2007) 26(32):4609–16. 10.1038/sj.onc.1210261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pan Y, Yuan C, Cheng C, Zhang Y, Ma Y, Zheng D, et al. Frequency and Clinical Significance of NF1 Mutation in Lung Adenocarcinomas From East Asian Patients. Int J Cancer (2019) 144(2):290–6. 10.1002/ijc.31871 [DOI] [PubMed] [Google Scholar]
- 31. de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, et al. Reduced NF1 Expression Confers Resistance to EGFR Inhibition in Lung Cancer. Cancer Discov (2014) 4(5):606–19. 10.1158/2159-8290.CD-13-0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ciccia A, Elledge SJ. The DNA Damage Response: Making it Safe to Play With Knives. Mol Cell (2010) 40(2):179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El-Aarag SA, Mahmoud A, Hashem MH, Abd Elkader H, Hemeida AE, ElHefnawi M. In Silico Identification of Potential Key Regulatory Factors in Smoking-Induced Lung Cancer. BMC Med Genomics (2017) 10(1):40. 10.1186/s12920-017-0284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Doerr F, George J, Schmitt A, Beleggia F, Rehkamper T, Hermann S, et al. Targeting a Non-Oncogene Addiction to the ATR/CHK1 Axis for the Treatment of Small Cell Lung Cancer. Sci Rep (2017) 7(1):15511. 10.1038/s41598-017-15840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lord CJ, Ashworth A. PARP Inhibitors: Synthetic Lethality in the Clinic. Science (2017) 355(6330):1152–8. 10.1126/science.aam7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu XS, Yang DY, Li YL, Li L, Wang Y, Chen P, et al. Prevalence and Clinical Significance of Pathogenic Germline BRCA1/2 Mutations in Chinese Non-Small Cell Lung Cancer Patients. Cancer Biol Med (2019) 16(3):556–64. 10.20892/j.issn.2095-3941.2018.0506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative Genome Analyses Identify Key Somatic Driver Mutations of Small-Cell Lung Cancer. Nat Genet (2012) 44(10):1104–10. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, et al. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov (2018) 8(11):1422–37. 10.1158/2159-8290.CD-18-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kunimasa K, Hirotsu Y, Nakamura H, Tamiya M, Iijima Y, Ishida H, et al. Rapid Progressive Lung Cancers Harbouring Multiple Clonal Driver Mutations With Big Bang Evolution Model. Cancer Genet (2020) 241:51–6. 10.1016/j.cancergen.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 40. Cohen PA, Donini CF, Nguyen NT, Lincet H, Vendrell JA. The Dark Side of ZNF217, a Key Regulator of Tumorigenesis With Powerful Biomarker Value. Oncotarget (2015) 6(39):41566–81. 10.18632/oncotarget.5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vendrell JA, Thollet A, Nguyen NT, Ghayad SE, Vinot S, Bieche I, et al. ZNF217 Is a Marker of Poor Prognosis in Breast Cancer That Drives Epithelial-Mesenchymal Transition and Invasion. Cancer Res (2012) 72(14):3593–606. 10.1158/0008-5472.Can-11-3095 [DOI] [PubMed] [Google Scholar]
- 42. Chang S, Ran W, Luo X, Zhang B, Liu L. Expression of Zinc Finger Protein 217 in Non-Small Cell Lung Cancer and its Clinical Significance. Cancer Res Clin (2019) 31(5):310–4. 10.3760/cma.j.issn.1006-9801.2019.05.005 [DOI] [Google Scholar]
- 43. Choi M, Kadara H, Zhang J, Parra ER, Rodriguez-Canales J, Gaffney SG, et al. Mutation Profiles in Early-Stage Lung Squamous Cell Carcinoma With Clinical Follow-Up and Correlation With Markers of Immune Function. Ann Oncol (2017) 28(1):83–9. 10.1093/annonc/mdw437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse Somatic Mutation Patterns and Pathway Alterations in Human Cancers. Nature (2010) 466(7308):869–73. 10.1038/nature09208 [DOI] [PubMed] [Google Scholar]
- 45. Zhang PP, Kang B, Xie GY, Li SL, Gu Y, Shen Y, et al. Genomic Sequencing and Editing Revealed the GRM8 Signaling Pathway as Potential Therapeutic Targets of Squamous Cell Lung Cancer. Cancer Lett (2019) 442:53–67. 10.1016/j.canlet.2018.10.035 [DOI] [PubMed] [Google Scholar]
- 46. Gini B, Thomas N, Blakely CM. Impact of Concurrent Genomic Alterations in Epidermal Growth Factor Receptor (EGFR)-Mutated Lung Cancer. J Thorac Dis (2020) 12(5):2883–95. 10.21037/jtd.2020.03.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and Clinical Impact of Co-Occurring Genetic Alterations in Advanced-Stage EGFR-Mutant Lung Cancers. Nat Genet (2017) 49(12):1693–704. 10.1038/ng.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Galvano A, Gristina V, Malapelle U, Pisapia P, Pepe F, Barraco N, et al. The Prognostic Impact of Tumor Mutational Burden (TMB) in the First-Line Management of Advanced Non-Oncogene Addicted Non-Small-Cell Lung Cancer (NSCLC): A Systematic Review and Meta-Analysis of Randomized Controlled Trials. ESMO Open (2021) 6(3):100124. 10.1016/j.esmoop.2021.100124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin C, Shi X, Zhao J, He Q, Fan Y, Xu W, et al. Tumor Mutation Burden Correlates With Efficacy of Chemotherapy/Targeted Therapy in Advanced Non-Small Cell Lung Cancer. Front Oncol (2020) 10:480. 10.3389/fonc.2020.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 Human Cancer Genomes Reveals the Landscape of Tumor Mutational Burden. Genome Med (2017) 9(1):34. 10.1186/s13073-017-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational Heterogeneity in Cancer and the Search for New Cancer-Associated Genes. Nature (2013) 499(7457):214–8. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pepe F, Pisapia P, Gristina V, Rocco D, Micheli M, Micheli P, et al. Tumor Mutational Burden on Cytological Samples: A Pilot Study. Cancer Cytopathol (2021) 129(6)460-7. 10.1002/cncy.22400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Associations of gender (A), smoking status (B), NF1 (C), ATR (D), BRCA1 mutation, tumor mutation (E) burden (TMB) (F), and chromosome instability (CIS) (G) with EGFR-TKI efficacy in EGFR-mutant squamous cell carcinoma (SCC).
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.