Abstract
Treatment according to the dynamic changes of bacterial load in vivo is critical for preventing progression of bacterial infections. Here, we present a lead sulfide quantum dots (PbS QDs) based second near-infrared (NIR-II) fluorescence imaging strategy for bacteria detection and real-time in vivo monitoring. Four strains of bacteria were labeled with synthesized PbS QDs which showed high bacteria labeling efficiency in vitro. Then bacteria at different concentrations were injected subcutaneously on the back of male nude mice for in vivo imaging. A series of NIR-II images taken at a predetermined time manner demonstrated changing patterns of photoluminescence (PL) intensity of infected sites, dynamically imaging a changing bacterial load in real-time. A detection limit around 102–104 CFU/ml was also achieved in vivo. Furthermore, analysis of pathology of infected sites were performed, which showed high biocompatibility of PbS QDs. Therefore, under the guidance of our developed NIR-II imaging system, real-time detection and spatiotemporal monitoring of bacterial infection in vivo can be achieved, thus facilitating anti-infection treatment under the guidance of the dynamic imaging of bacterial load in future.
Keywords: bioimaging, in vivo imaging, NIR-II, bacterial infection, quantum dots
Introduction
Despite the rapid development of antibiotics, bacterial infection remains a major threat to human health, causing massive mortality and morbidity. Due to frequent failures to identify bacterial infections at an early stage in vivo with high sensitivity and accuracy, the infection can progress rapidly into a systemic disease along with irreversible damages, even leading to life-threatening conditions due to delayed treatment (Moore, 1993; Erasmus et al., 1999; Ning et al., 2011). Traditionally, diagnosis of bacterial infections mainly relies on in vitro approaches such as specimen culture, polymerase chain reaction (PCR), and enzyme linked immunosorbent assay (ELISA), which are not only time-consuming, but also lacking in sensitivity, not to say able to image the infection accurately in vivo (Gracias and McKillip, 2004; Heo and Hua, 2009; Fournier et al., 2013; Ahmed et al., 2014). Therefore, an imaging technology enabling early detection of bacterial infections in vivo is in urgent need.
For detecting bacterial infections in vivo, an ideal approach should firstly be non-invasive and biocompatible with the human body. Besides, an excellent signal-to-noise ratio (SNR) should be guaranteed to avoid interference from the complex dynamic environment inside the body. Furthermore, the overall imaging process should be cost-effective enough for general clinical applications in the future. Recently, fluorescence imaging based on the second near-infrared (NIR-II) window (1,000–1,700 nm) is gaining popularity for in vivo imaging due to minimized scattering and high SNR (Liu et al., 2009a; Liu et al., 2009b; Smith et al., 2009; Welsher et al., 2011). As a representative, lead sulfide quantum dots (PbS QDs) have been reported to be a promising semiconductor nanocrystal with excellent NIR-II fluorescence characteristics (Bruchez et al., 1998; Chan and Nie, 1998; Medintz et al., 2003; Wu et al., 2003; Michalet et al., 2005; Chen et al., 2016a), showing a huge potential for in vivo imaging with deep tissue penetration and low cytotoxicity (Chen et al., 2016a; Feng et al., 2016).
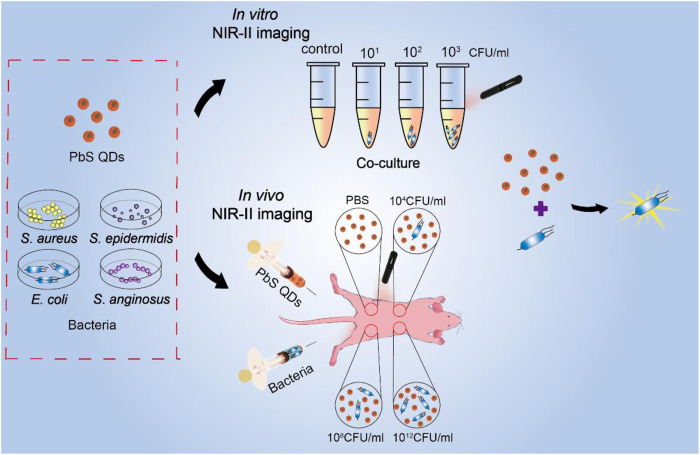
Here, we present a NIR-II imaging strategy based on fluorescence PbS QDs, facilitating dynamic detection of bacterial infection both in vitro and in vivo in a real-time manner (Scheme 1). Firstly, fluorescence properties of PbS QDs were validated. Then, the PbS QDs were co-cultured with three concentrations of four different bacteria strains including Staphylococcus aureus (S. aureus), Staphylococcus epidermidis (S. epidermidis), Escherichia coli (E. coli) and Streptococcus anginosus (S. anginosus) in vitro and highly efficient bacterial labeling was achieved. During in vivo observation, the PbS QDs displayed a quick detection of bacteria at the infected sites and allowed a real-time investigation on the change of bacterial load in a longitudinal manner. Besides, the biocompatible PbS QDs were eventually metabolized and cleared out of the body with no noticeable abnormality in major organs. Our work demonstrated a highly efficient probe for bacterial detection and a possible strategy to realize real-time observation and monitoring of bacterial infection in vivo, which provides novel thoughts into antibiotic treatment according to bacterial load.
SCHEME 1.
Schematic to represent the experimental process.
Materials and Methods
Reagents and Materials
All chemicals were used without further modification or purification procedure. Deionized water was utilized to arrange the working solutions. Lead acetate trihydrate [Pb(OAc)2·3H2O, ≥99.9%], sodium sulfide nonahydrate (Na2S·9H2O, ≥ 98.0%), bovine pancreatic ribonuclease A (MW:13.7 kDa, > 70 U/mg) and sodium hydroxide (NaOH, ≥ 98.0%) were purchased from Sigma-Aldrich. Aseptic phosphate buffer solutions (1 × PBS, pH = 7.4) was used in all experiments.
Bacteria
The bacteria were provided by the Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai, including S. aureus (ATCC 29213), E. coli (ATCC 25922), S. epidermidis (clinical isolates), and S. anginosus (clinical isolates).
Animals
Eight-week-old male nude mice with the weight of 20–24 g were purchased from Shanghai Jie Si Jie Laboratory Animal Co. Ltd. (Shanghai, China). All animal studies were done complying with guidelines of the Chinese Council for Animal Care.
Instrumentation
ELGA Purelab classic UVF system was used to generate deionized water. Microwave synthesis process was performed in a microwave reactor (Discover, CEM). The PbS QDs were neutralized and purified with Amicon ultracentrifugal filter tubes (MWCO: 10 kDa). The centrifugation was done with Thermo Scientific Heraeus Fresco 21 microcentrifuge and/or an Eppendorf 5810R centrifuge. NIR-II fluorescence spectra measurement was carried out with NS1 NanoSpectralyzer fluorimetric analyzer (Applied NanoFluorescence) and the fluorescence excitation wavelength is 785 nm. All NIR-II images of test tubes and mice were taken with a NIR-II imaging prototype connected with a 2D InGaAs CCD camera. 810 nm diode laser coupled with 850 nm and 1,000 nm short-pass filters was utilized as the excitation source. The emission light was filtered with an 1,100 nm long-pass filter before it was detected by the CCD sensor.
Synthesis of RNase-A@PbS QDs
500 μl of RNase-A (50 mg/ml) solution and 500 μl of Pb(OAc)2 (10 mM) solution was mixed to prepare RNase-A/Pb2+ precursor solution. 50 μl of NaOH solution (1 M) was added to adjust the pH to 9–11. Then 50 μl of freshly prepared Na2S (10 mM) solution was added to the mixture after the solution was stirred and mixed for 5 min. Subsequently a stir bar was put into the mixed solution and the test tube was inserted into the microwave reactor and heated at 70°C for 30 s with an input power of 30 W. The transparent dark brown mixtures can be seen when RNase-A@PbS QDs were formed. Then the PbS QDs were ultra-filtered against deionized water in order to adjust system pH to 7–8 and remove extra reagents (e.g. Pb2+, Na+, OAc−, etc.). After that, PBS was used to resuspend the solution to original volume. The final solution was stored at 4°C and kept away from light.
Bacteria culture
Four strains of bacteria including S. aureus, E. coli, S. epidermidis, and S. anginosus was used to investigate the labeling function of PbS QDs. Bacteria were cultured overnight in Luria-Bertani (LB) medium at 37°C under 5% CO2 in an incubator shaker with a set of 150 rpm. Then a single colony was collected and inoculated into a 6 ml of fresh LB medium and cultured to an optical density of 0.5–0.6 at 600 nm in a 10 ml flask. Suspensions of the bacteria were centrifuged at 2000 rpm for 15 min at 25°C. Then the bacterial sediments were collected after being washed by PBS twice and further resuspended in 5 ml of PBS. Finally, the suspensions of each strain of bacteria were diluted respectively into different concentrations for being inoculated to different culture media to calculate the colony-forming units (CFU) per milliliter.
Co-Culture of Bacteria and PbS QDs
A 50 μl portion of each bacteria strain at concentrations of 103, 102, 10 CFU/ml was added to a 1.5 mlcentrifuge tube and stirred with 50 μl of previously prepared PbS QDs respectively. Then the mixtures of PbS QDs and bacteria were incubated at 37°C under 5% CO2 for 1 h for co-culture. After that, the mixtures were centrifuged at 2,000 rpm for 10 min at 25°C. The transparent supernatants were separated from the sediments and removed to new 1.5 ml centrifuge tubes while the sediments were washed by PBS twice. Finally, NIR-II images were taken of the bacterial sediments in comparison with the original supernatants.
Preparation of Experimental Animal Models
Male nude mice which have body weight ranging from 20 to 24 g were included in all animal experiments. Four nude mice were randomly chosen to be injected with the four strains of bacteria. The nude mice were deeply anesthetized by intraperitoneal injection, then each of them was subcutaneously injected with 25 μl of one strain of bacteria on the back at three sites away from another at a bacterial concentration of 1012 CFU/ml, 108 CFU/ml and 104 CFU/ml. The injected sites were at the same positions across the four nude mice. Moreover, a fourth site was injected with the same amount of PBS as a control for each nude mouse.
In Vivo Observation
Right after the four nude mice were injected with bacteria, 50 μl of PbS QDs were injected subcutaneously at the same four sites on the back. Then the four nude mice were kept in cage under constant observation and the same circumstances. NIR-II images were collected from each nude mouse at 10 min, 1 h, 2 h, 1 day, 2 days, 1 w, 2 w and 3 w after the injection. The PL intensity and area of fluorescence of four injected sites of each nude mouse were measured by analyzing every NIR-II image using ImageJ. The data of 2 days post-infection were chosen to be further analyzed for detection limit of bacteria load in vivo with regression graphs applied to study the relevance between bacteria load and PL intensity using Graphpad Prism 9. To eliminate the potential background interference at each time point, PL intensity at the injected site/PL intensity of the background was calculated, marked as relative PL intensity.
Histological Analysis With Hematoxylin-Eosin Staining and Gram Staining
After 3 weeks, skin tissues of the sites with 1012 CFU/ml bacteria and the sites with PBS and major organs of the nude mice were collected and stored in 4% paraformaldehyde for at least 24 h to prepare pathological slides for histological analysis. Additionally, NIR-II images of collected major organs were taken and PL intensity was measured before preparing for histological analysis. After H&E and Gram staining were done, histological images were acquired on an optical microscope (Nikon Eclipse CI, Japan). Then immunohistochemical analysis including neutrophil count was done with immunohistochemical slides by calculating average number of neutrophils per square millimeter under three fields of 200× magnification using Image-pro plus 6.0 (Media Cybernetics, Inc., Rockville, MD, United States). Inflammatory factors including CD11b, IL-6, TNF-α, and monocyte chemoattractant protein-1 (MCP-1), which are highly associated with bacterial infection, were analyzed with immunohistochemical technique. Average Optical (AO) was calculated (AO = IOD/AREA, IOD: Integrate Optical Density).
Results and Discussion
Fluorescence Properties of PbS QDs
Firstly, PbS QDs were prepared according to the previous protocol (Kong et al., 2016). As shown in Figure 1A, a uniform light-brown liquid without sediments was formed. Then, NIR-II images of the freshly-prepared PbS QDs were achieved with a specialized system (Figures 1B1–B3). Under an increasing exposure time from 30 ms to 100 m, increasing fluorescence signals of the PbS QDs were detected, while the SNR kept low, which was in accordance with previous studies (Welsher et al., 2011; Li et al., 2014; Hong et al., 2015; Chen et al., 2016b; Wan et al., 2018). Furthermore, the photoluminescence (PL) spectrum of the PbS QDs was measured in Figure 1C, revealing an emission peak within the range of NIR-II window.
FIGURE 1.
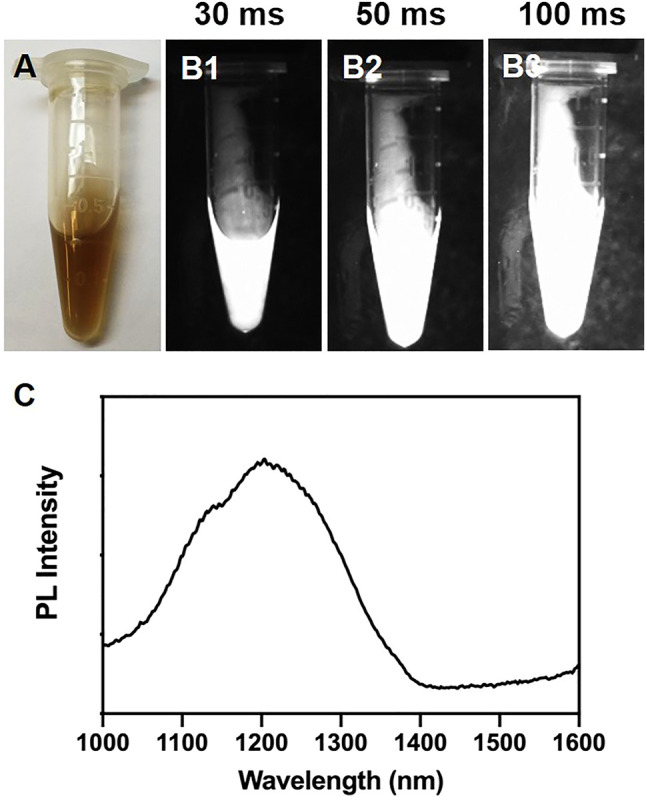
(A) Bright-field photograph of the prepared PbS QDs. (B1–B3) NIR-II fluorescence images of (A) under an exposure time of 30 ms, 50 ms and 100 ms. (C) PL intensity of the prepared PbS QDs.
In Vitro Labeling Efficiency of PbS QDs
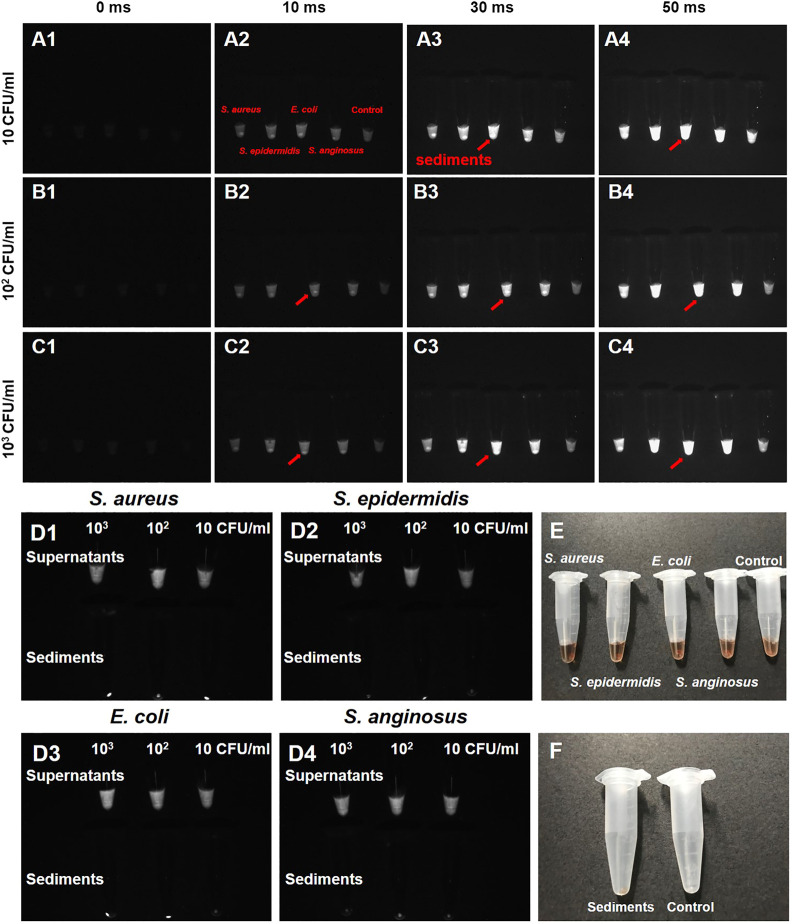
The labeling efficiency of PbS QDs was then investigated with S. aureus, S. epidermidis, E. coli and S. anginosus in vitro (Figure 2). Bacterial suspensions of the four typical bacterial strains were first diluted into different concentrations. Then, a 50 μl portion of each bacterial suspension at three different concentrations (10, 102 and, 103 CFU/ml) was mixed with 50 μl of PbS QDs and co-cultured at 37°C under 5% CO2 for 1 h respectively. After centrifugation, NIR-II fluorescence imaging was adopted for these mixtures under an exposure time of 0 ms, 10 ms, 30 ms and 50 ms in comparison with a control group (100 μl of pure PbS QDs). As shown in Figures 2A–C, fluorescence signals detected in each sample increased with exposure time. However, it was noted that a bright spot with high fluorescence signals was demonstrated at the bottom of the tube in samples with bacteria, compared with the control group which was a uniform liquid. Therefore, it was suggested that an aggregation of bacteria labeled with PbS QDs was formed after centrifugation.
FIGURE 2.
(A–C) NIR-II fluorescence images of four bacteria strains (S. aureus, S. epidermidis, E. coli and S. anginosus) co-cultured with PbS QDs under an exposure time of 0 ms, 10 ms, 30 ms and 50 ms at a bacterial concentration of 10 CFU/ml, 102 CFU/ml and 103 CFU/ml in comparison with a control group (red arrow: sediments presenting high fluorescence signal). (D1–D4) NIR-II fluorescence images of supernatants and bacterial sediments after co-culture. (E) Bright-field photograph of (A). (F) Bright-field photograph comparing the bacterial sediments and a control group after co-culture.
To further confirm the results, the original supernatants were separated, leaving the sediments rinsed by PBS till the supernatants became colorless. From the NIR-II images shown in Figure 2D, fluorescence signals could be detected in all supernatants of the four bacteria at the three concentrations, indicating relatively excessive PbS QDs against bacteria. On the other hand, comparatively high fluorescence signals were detected in the bacterial sediments. As pure PbS QDs did not form sediments after centrifugation at preparation, the current sediments were considered to be bacteria labeled with PbS QDs after co-culture. According to previous studies, the labeling capability of our PbS QDs could be explained by electrostatic interaction between the functional groups outside PbS QDs and the surface of the bacterial cell wall (Kloepfer et al., 2003; Hahn et al., 2005; Chen et al., 2015). Therefore, it was proved that our PbS QDs could label bacteria with a high efficiency in vitro, further presenting a promising imaging probe for bacteria detection in vivo.
In Vivo Real-Time NIR-II Fluorescence Imaging of Bacterial Infection
In vivo bacteria imaging is crucial for detecting bacterial infections, not only ensuring an early diagnosis but also facilitating monitoring the development of an infection over time. To test the in vivo imaging potential of our prepared PbS QDs, nude mice were subcutaneously infected with 25 μl of bacteria on the back at three sites away from another at a bacterial concentration of 1012 CFU/ml, 108 CFU/ml and 104 CFU/ml. Subsequently, the same amount of PBS was injected at a fourth site as a control group. Then, 50 μl of PbS QDs was injected subcutaneously at the same four sites. After that, NIR-II fluorescence images were recorded at 10 min, 1 h, 2 h, 1 day, 2 days, 1 week, 2 weeks and 3 weeks post-infection.
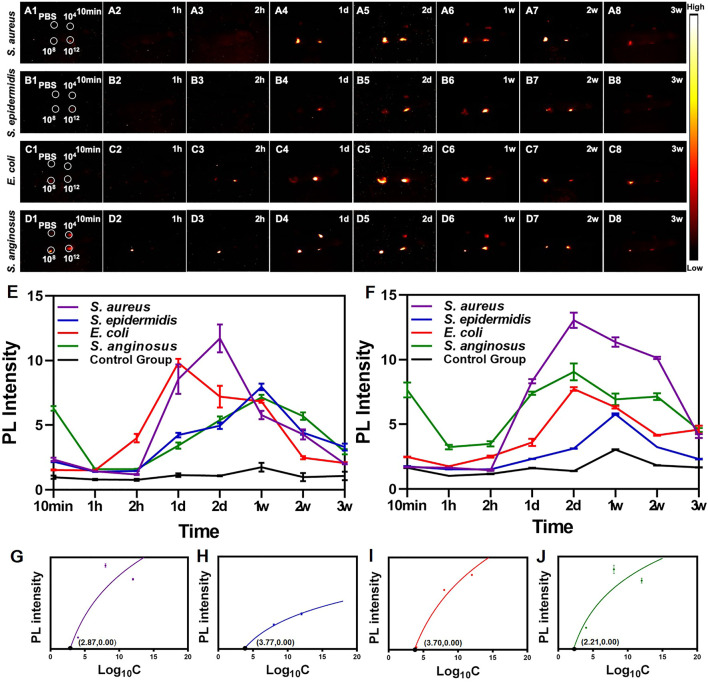
As shown in Figures 3A–D, after the injection of PbS QDs, generally higher fluorescence signals were detected at the bacteria-infected sites compared with the control group. It was suggested that PbS QDs rapidly labeled bacteria at the bacteria-infected sites, immediately showing high fluorescence signals. In contrast, the pure PbS QDs were diluted by PBS when mixed subcutaneously, resulting in a decrease in PL intensity with a faster rate of clearance. Furthermore, PL intensity at the site infected with 1012 CFU/ml (Figure 3E) and 108 CFU/ml (Figure 3F) of bacteria as well as the control group were measured. A wave pattern of PL intensity was shown at the bacteria-infected sites across the four bacteria strains, while a relatively flat line of PL intensity was displayed by the control group. The differences in the changing pattern of PL intensity further confirmed that the PbS QDs labeled bacteria in vivo.
FIGURE 3.
(A–D) NIR-II images of nude mice at 10 min, 1 h, 2 h, 1 day, 2 days, 1 weeks, 2 weeks and 3 weeks post-infection. (E) PL intensity of the site injected with 1012 CFU/ml of bacteria. (F) PL intensity of the site injected with 108 CFU/ml of bacteria. Graphs depicting the relevance between PL intensity and different concentration of four strains of bacteria including (G) S. aureus, (H) S. epidermidis, (I) S. anginosus and (J) E. coli.
Remarkably, the changing pattern of PL intensity of the PbS QDs labeled bacteria at the infected sites reflected a real-time bacterial load in vivo, which in turn demonstrated the process of development of bacterial infection. After the bacteria were injected and labeled with PbS QDs subcutaneously, there was firstly a decrease of PL intensity, then followed with an increase up to a peak, and finally the fluorescence signals became undetectable over time. As is known to all, short after the occurrence of a bacterial infection, the invading bacteria will be first attacked by the innate immune system (Albiger et al., 2007; Nizet, 2007), leading to a reduction of bacterial load. However, as the eradication of most bacteria mainly relies on the adaptive immune response which is stimulated in the later stage of bacterial infection, an interval for clonal expansion of lymphocytes will allow for bacterial propagation, resulting in a resurge of bacterial load (Albiger et al., 2007; Hall et al., 2014). For those with a robust immune system, the bacteria at the infected site will be gradually eliminated and eventually cleared out of the body, resulting in a drop of bacterial load. As the changing pattern of bacterial load was in consistent with the observed PL intensity of PbS QDs, it was suggested that PbS QDs not only successfully detected and labeled bacteria in vivo, but also could reflect and monitor the bacterial load in vivo.
Furthermore, the relationship between bacterial load and PL intensity in vivo were further analyzed. As shown in Figures 3G–J, the four strains of bacteria shared the same shape of logarithmic curves, showing increasing PL intensity with bacterial load under a certain bacterial concentration and gradually became flat. It was reported that high concentration of bacteria would inhibit the electrostatic interaction between the functional groups of PbS QDs and bacterial membrane due to steric effects, which might be due to the mechanism that the smaller interspace could make it difficult to realize further groups interactions in the labeling process (Albiger et al., 2007). Nevertheless, the detection limit of S. aureus, S. epidermidis, E. coli and S. anginosus in vivo was calculated as 102.87, 103.77, 103.70 and 102.21 CFU/ml respectively, indicating a relatively low limit achieved in vivo. Therefore, with longitudinal observation, in vivo monitoring of the development of bacterial infection could be further achieved, thus facilitating diagnosis and treatment under a real-time and long-time imaging guidance.
Analysis of Pathology and Inflammatory Factors
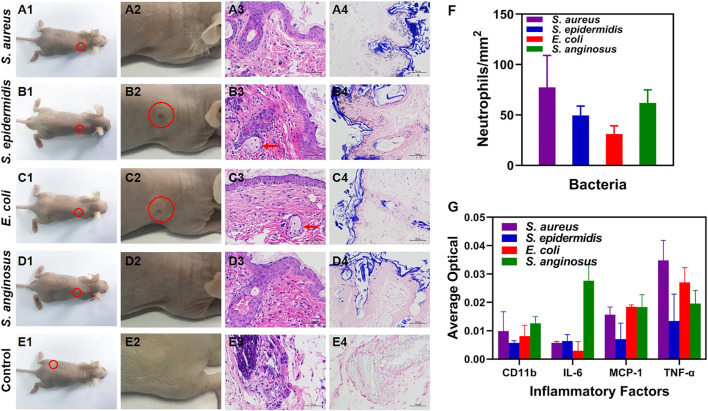
At 3 weeks post-infection, the nude mice were photographed right after the NIR-II imaging. An overall view of the whole nude mice was shown in Figures 4A1–E1 and photographs focusing on the site with 1012 CFU/ml of bacteria and the control group were shown in Figures 4A2–E2. As shown in Figures 4B2, C2, healed skin lesions were observed at the site infected with S. epidermidis (Figure 4B2) and E. coli (Figure 4C2), indicating a healing stage of bacterial infection. Furthermore, histological H&E staining was performed to study the histopathology of the infected tissues. The sites infected with 1012 CFU/ml of bacteria (Figures 4B3–D3) showed aggregation of neutrophils and appearance of foreign body giant cells, both of which were commonly seen in the late stage of infection. In comparison, the control group showed insignificant inflammatory infiltration (Figure 4E3). Additionally, Gram staining was carried out for bacteria analysis, with Gram-positive bacteria (S. aureus, S. epidermidis and S. anginosus) stained purple and Gram-negative bacteria (E. coli) stained pink (Figures 4A4–D4). No bacteria were found in the control group (Figure 4E4). Moreover, neutrophil count (Figure 4F) and analysis of several major inflammatory factors (CD11b, IL-6, MCP-1, TNF-α) (Figure 4G) were done with immunohistochemical slides to assess inflammatory response. There was no significant difference of neutrophil count among the four bacteria strains. As for inflammatory factors, the relatively high expression of TNF-α (mainly secreted by mononuclear macrophages) indicated the involvement of the innate immune system, while the others were at an insignificant level.
FIGURE 4.
(A1–E1) Bright-field photographs of the infected nude mice at 3 w post-infection (E1 is re-used image of A1 in which PBS injected site is circled). (A2–E2) Corresponding zoomed-in photographs of (A1–E1) at the site infected with 1012 CFU/ml of bacteria and the control group (red circle: healed skin rupture). (A3–E3) H&E staining micrographs of the skin tissues at the site infected with 1012 CFU/ml of bacteria (red arrow: foreign body giant cells; × 400 magnification). (A4–E4) Gram staining micrographs of the skin tissues at the site infected with 1012 CFU/ml of bacteria (×400 magnification). (F) Neutrophil count with the H&E staining micrographs. (G) Inflammatory factors (CD11b, IL-6, MCP-1 and TNF-α) measured with immunohistochemical micrographs.
Biosafety Validation of PbS QDs
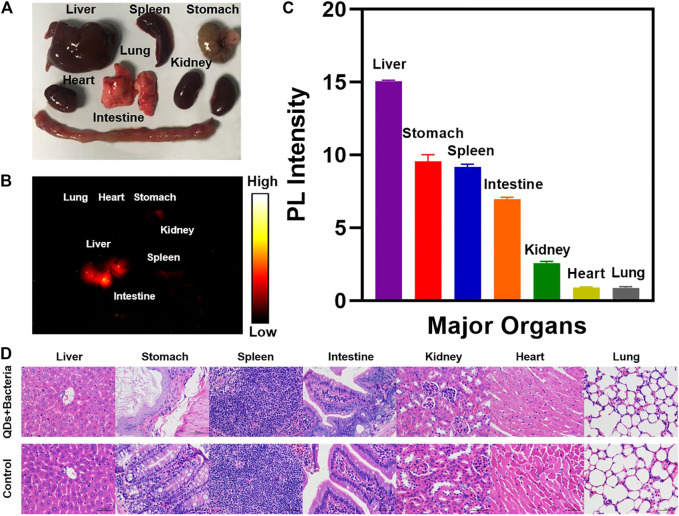
The distribution of PbS QDs in the major organs of the infected nude mice at 3 w post-infection was analyzed in Figure 5. There was no obvious abnormality in major organs from the bright-field photograph (Figure 5A). Among all the harvested organs, fluorescence signals were detected in the liver, spleen, stomach and parts of intestine from the NIR-II image (Figure 5B) and the PL intensity was measured in Figure 5C. It was in agreement with a previous study that major accumulation of PbS QDs was observed in the liver and the spleen after 24 h through intravenous injection (Chen et al., 2016a). As PbS QDs were injected subcutaneously in the current study, the metabolization rate was comparably slow, resulting in longer time for PbS QDs to accumulate in the organs and eventually to be cleared out of the body. From another perspective, the fluorescence signals detected in major organs indicated that the PbS QDs were metabolized by the body and then joined the circulation instead of simply quenched subcutaneously over time. Furthermore, H&E staining micrographs of major organ tissues at 3 w post-infection compared with a control group were shown in Figure 5D. No obvious signs of tissue injury or inflammation were noted.
FIGURE 5.
(A) Photograph of major organs harvested from the infected nude mouse. (B) NIR-II image of major organs harvested at 3 weeks post-infection. (C) PL intensity measured from the NIR-II image of the harvested major organs. (D) H&E staining micrographs of representative major organ tissues of the infected nude mice compared with a control group (magnification: ×400).
Conclusion
In conclusion, a novel imaging strategy based on highly fluorescence PbS QDs with NIR-II emission was developed for real-time in vivo detection of bacterial infection. After a high bacteria-labeling efficiency was proved in vitro, the PbS QDs enabled a rapid detection and imaging of bacteria in vivo, with a detection limit around 103–104 CFU/ml, allowing a real-time investigation on the bacterial load. Besides, the biocompatible PbS QDs were eventually metabolized and cleared out of the body with no noticeable abnormalities in major organs. Our work demonstrated a highly efficient imaging strategy for detection and monitoring of bacterial infection in vivo, facilitating the optimization of anti-infection treatment according to dynamically changing bacterial load under a real-time imaging guidance.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Department of Laboratory Animal Science of Fudan University.
Author Contributions
SF, HL, and CL contributed equally to this work. SF, HL, and CL performed the experiment and wrote the article. MC, HS, and MH helped perform the experiment. YL, JC assisted in the article polishment. JZ, YH and SC were in charge of the whole study.
Funding
This work was supported by National Key R&D Program of China (2016YFC1100300), National Natural Science Foundation of China (No. 81772339, 81972129, 81911530136, 81811530750, 82072521 and 8201101387), The Key Clinical Medicine Center of Shanghai (2017ZZ01006), Sanming Project of Medicine in Shenzhen (SZSM201612078), Shanghai Rising-Star Project (18QB1400500) and The Introduction Project of Clinical Medicine Expert Team for Suzhou (SZYJTD201714), Development Project of Shanghai Peak Disciplines-Integrative Medicine (20180101), Shanghai Talent Development Funding Scheme (2020080), Shanghai Committee of Science and Technology (19441901600 and 19441902000) and Shanghai Sailing Program (21YF1404100).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Ahmed A., Rushworth J. V., Hirst N. A., Millner P. A. (2014). Biosensors for Whole-Cell Bacterial Detection. Clin. Microbiol. Rev. 27 (3), 631–646. 10.1128/cmr.00120-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiger B., Dahlberg S., Henriques-Normark B., Normark S. (2007). Role of the Innate Immune System in Host Defence against Bacterial Infections: Focus on the Toll-like Receptors. J. Intern. Med. 261 (6), 511–528. 10.1111/j.1365-2796.2007.01821.x [DOI] [PubMed] [Google Scholar]
- Bruchez M., Moronne M., Gin P., Weiss S., Alivisatos A. P. (1998). Semiconductor Nanocrystals as Fluorescent Biological Labels. Science 281, 2013–2016. 10.1126/science.281.5385.2013 [DOI] [PubMed] [Google Scholar]
- Chan W. C., Nie S. M. (1998). Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science 281, 2016–2018. 10.1126/science.281.5385.2016 [DOI] [PubMed] [Google Scholar]
- Chen G., Roy I., Yang C., Prasad P. N. (2016). Nanochemistry and Nanomedicine for Nanoparticle-Based Diagnostics and Therapy. Chem. Rev. 116 (5), 2826–2885. 10.1021/acs.chemrev.5b00148 [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang M., Li B., Chen D., Dong X., Wang Y., et al. (2015). Versatile Antimicrobial Peptide-Based ZnO Quantum Dots for In Vivo Bacteria Diagnosis and Treatment with High Specificity. Biomaterials 53, 532–544. 10.1016/j.biomaterials.2015.02.105 [DOI] [PubMed] [Google Scholar]
- Chen J., Kong Y., Wang W., Fang H., Wo Y., Zhou D., et al. (2016). Direct Water-phase Synthesis of lead Sulfide Quantum Dots Encapsulated by β-lactoglobulin for In Vivo Second Near Infrared Window Imaging with Reduced Toxicity. Chem. Commun. 52 (21), 4025–4028. 10.1039/c6cc00099a [DOI] [PubMed] [Google Scholar]
- Erasmus J. J., McAdams H. P., Farrell M. A., Patz E. F. (1999). Pulmonary Nontuberculous Mycobacterial Infection: Radiologic Manifestations. Radiographics 19, 1487–1503. 10.1148/radiographics.19.6.g99no101487 [DOI] [PubMed] [Google Scholar]
- Feng S., Chen J., Wo Y., Li Y., Chen S., Zhang Y., et al. (2016). Real-time and Long-Time In Vivo Imaging in the Shortwave Infrared Window of Perforator Vessels for More Precise Evaluation of Flap Perfusion. Biomaterials 103, 256–264. 10.1016/j.biomaterials.2016.06.043 [DOI] [PubMed] [Google Scholar]
- Fournier P.-E., Drancourt M., Colson P., Rolain J.-M., Scola B. L., Raoult D. (2013). Modern Clinical Microbiology: New Challenges and Solutions. Nat. Rev. Microbiol. 11 (8), 574–585. 10.1038/nrmicro3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracias K. S., McKillip J. L. (2004). A Review of Conventional Detection and Enumeration Methods for Pathogenic Bacteria in Food. Can. J. Microbiol. 50 (11), 883–890. 10.1139/w04-080 [DOI] [PubMed] [Google Scholar]
- Hahn M. A., Tabb J. S., Krauss T. D. (2005). Detection of Single Bacterial Pathogens with Semiconductor Quantum Dots. Anal. Chem. 77 (15), 4861–4869. 10.1021/ac050641i [DOI] [PubMed] [Google Scholar]
- Hall B. G., Acar H., Nandipati A., Barlow M. (2014). Growth Rates Made Easy. Mol. Biol. Evol. 31 (1), 232–238. 10.1093/molbev/mst187 [DOI] [PubMed] [Google Scholar]
- Heo J., Hua S. Z. (2009). An Overview of Recent Strategies in Pathogen Sensing. Sensors 9 (6), 4483–4502. 10.3390/s90604483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong G., Diao S., Antaris A. L., Dai H. (2015). Carbon Nanomaterials for Biological Imaging and Nanomedicinal Therapy. Chem. Rev. 115 (19), 10816–10906. 10.1021/acs.chemrev.5b00008 [DOI] [PubMed] [Google Scholar]
- Kloepfer J. A., Mielke R. E., Wong M. S., Nealson K. H., Stucky G., Nadeau J. L. (2003). Quantum Dots as Strain- and Metabolism-specific Microbiological Labels. Aem 69 (7), 4205–4213. 10.1128/aem.69.7.4205-4213.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Chen J., Fang H., Heath G., Wo Y., Wang W., et al. (2016). Highly Fluorescent Ribonuclease-A-Encapsulated lead Sulfide Quantum Dots for Ultrasensitive Fluorescence In Vivo Imaging in the Second Near-Infrared Window. Chem. Mater. 28 (9), 3041–3050. 10.1021/acs.chemmater.6b00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhang Y., Wang M., Zhang Y., Chen G., Li L., et al. (2014). In Vivo real-time Visualization of Tissue Blood Flow and Angiogenesis Using Ag2S Quantum Dots in the NIR-II Window. Biomaterials 35 (1), 393–400. 10.1016/j.biomaterials.2013.10.010 [DOI] [PubMed] [Google Scholar]
- Liu Z., Fan A. C., Rakhra K., Sherlock S., Goodwin A., Chen X., et al. (2009). Supramolecular Stacking of Doxorubicin on Carbon Nanotubes for In Vivo Cancer Therapy. Angew. Chem. Int. Ed. 48 (41), 7668–7672. 10.1002/anie.200902612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Tabakman S., Welsher K., Dai H. (2009). Carbon Nanotubes in Biology and Medicine: In Vitro and In Vivo Detection, Imaging and Drug Delivery. Nano Res. 2 (2), 85–120. 10.1007/s12274-009-9009-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz I. L., Clapp A. R., Mattoussi H., Goldman E. R., Fisher B., Mauro J. M. (2003). Self-assembled Nanoscale Biosensors Based on Quantum Dot FRET Donors. Nat. Mater 2, 630–638. 10.1038/nmat961 [DOI] [PubMed] [Google Scholar]
- Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., et al. (2005). Quantum Dots for Live Cells, In Vivo Imaging, and Diagnostics. Science 307, 538–544. 10.1126/science.1104274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E. H. (1993). Atypical Mycobacterial Infection in the Lung: CT Appearance. Radiology 187, 777–782. 10.1148/radiology.187.3.8497629 [DOI] [PubMed] [Google Scholar]
- Ning X., Lee S., Wang Z., Kim D., Stubblefield B., Gilbert E., et al. (2011). Maltodextrin-based Imaging Probes Detect Bacteria In Vivo with High Sensitivity and Specificity. Nat. Mater 10 (8), 602–607. 10.1038/nmat3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V. (2007). Understanding How Leading Bacterial Pathogens Subvert Innate Immunity to Reveal Novel Therapeutic Targets. J. Allergy Clin. Immunol. 120 (1), 13–22. 10.1016/j.jaci.2007.06.005 [DOI] [PubMed] [Google Scholar]
- Smith A. M., Mancini M. C., Nie S. (2009). Second Window for In Vivo Imaging. Nat. Nanotech 4 (11), 710–711. 10.1038/nnano.2009.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H., Yue J., Zhu S., Uno T., Zhang X., Yang Q., et al. (2018). A Bright Organic NIR-II Nanofluorophore for Three-Dimensional Imaging into Biological Tissues. Nat. Commun. 9 (1), 1171. 10.1038/s41467-018-03505-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsher K., Sherlock S. P., Dai H. (2011). Deep-tissue Anatomical Imaging of Mice Using Carbon Nanotube Fluorophores in the Second Near-Infrared Window. Proc. Natl. Acad. Sci. 108 (22), 8943–8948. 10.1073/pnas.1014501108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu H., Liu J., Haley K. N., Treadway J. A., Larson J. P., et al. (2003). Immunofluorescent Labeling of Cancer Marker Her2 and Other Cellular Targets with Semiconductor Quantum Dots. Nat. Biotechnol. 21, 41–46. 10.1038/nbt764 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.