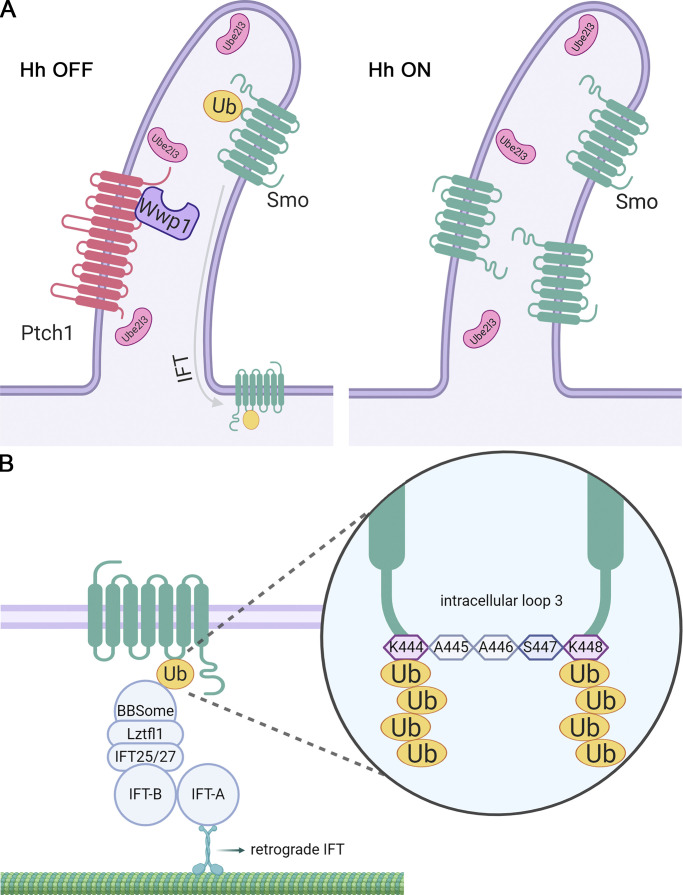
At the basal state, the ciliary levels of Smo are kept low through the actions of a ciliary localized E2 ubiquitin–conjugating enzyme, Ube2l3, and an E3 ubiquitin ligase, Wwp1. Pathway activation leads to removal of Wwp1 from cilia, allowing Smo to accumulate in cilia and become activated.
Abstract
The Hedgehog pathway, critical to vertebrate development, is organized in primary cilia. Activation of signaling causes the Hedgehog receptor Ptch1 to exit cilia, allowing a second receptor, Smo, to accumulate in cilia and activate the downstream steps of the pathway. Mechanisms regulating the dynamics of these receptors are unknown, but the ubiquitination of Smo regulates its interaction with the intraflagellar transport system to control ciliary levels. A focused screen of ubiquitin-related genes identified nine required for maintaining low ciliary Smo at the basal state. These included cytoplasmic E3s (Arih2, Mgrn1, and Maea), a ciliary localized E3 (Wwp1), a ciliary localized E2 (Ube2l3), a deubiquitinase (Bap1), and three adaptors (Kctd5, Skp1a, and Skp2). The ciliary E3, Wwp1, binds Ptch1 and localizes to cilia at the basal state. Activation of signaling removes both Ptch1 and Wwp1 from cilia, thus providing an elegant mechanism for Ptch1 to regulate ciliary Smo levels.
Introduction
Cilia are microtubule-based, hair-like organelles protruding from the surface of most eukaryotic cells. These organelles are evolutionarily conserved from unicellular eukaryotes to mammals and provide diverse motility and sensory functions to cells (Satir and Christensen, 2007). Ciliary dysfunction causes a variety of degenerative and developmental diseases collectively known as the ciliopathies (Reiter and Leroux, 2017). Cilia are complex structures made up of 1,000 or more unique proteins (Pazour et al., 2005). These organelles rely on proteins synthesized in the cell and transported into the cilium by intraflagellar transport (IFT; Bhogaraju et al., 2013; Kozminski et al., 1993; Rosenbaum and Witman, 2002; Scholey, 2003). The IFT system consists of trains built from IFT-A, IFT-B, and BBSome subcomplexes carried along cilia by kinesin-2 (Cole et al., 1998; Snow et al., 2004) and dynein-2 motors (Pazour et al., 1998). The ∼22 proteins of IFT-A and IFT-B are thought to be adaptors that connect the motors to the diverse cargos (Bhogaraju et al., 2013; Mukhopadhyay et al., 2010; Palenik et al., 2007). The eight subunits of the BBSome function primarily for the removal of ciliary proteins such as Smoothened (Smo) and Gpr161 (Lechtreck et al., 2009; Liew et al., 2014; Nozaki et al., 2019; Nozaki et al., 2018), although they may have roles in delivery as well (Berbari et al., 2008). Mutations in the BBSome-encoding genes do not block ciliary assembly but cause functional defects in sensory perception and signaling. In contrast, mutations in most IFT-A and IFT-B genes cause ciliary assembly defects. However, there are exceptions, such as Ift25 and Ift27. These two subunits of IFT-B are not required for ciliary assembly, but mutants show phenotypes indicative of Hedgehog signaling dysfunction (Eguether et al., 2018; Eguether et al., 2014; Keady et al., 2012).
The Hedgehog pathway (Bangs and Anderson, 2017; Huangfu et al., 2003) plays pivotal roles in embryonic patterning, tissue homeostasis, and tumorigenesis (Choudhry et al., 2014; Emechebe et al., 2016; Gupta et al., 2010; Petrova and Joyner, 2014). In vertebrates, this pathway is organized in cilia, with the receptors and effectors dynamically redistributing during signaling. At the basal state, the Hedgehog receptor Ptch1 localizes to cilia and prevents the ciliary accumulation and activation of Smo (Rohatgi et al., 2007). In addition, Gpr161 localizes to cilia at the basal state and directs the processing of Gli3 into Gli3R via cAMP signaling (Mukhopadhyay et al., 2013). Once bound by Hedgehog ligand, Ptch1 leaves cilia and no longer suppresses Smo. Smo becomes activated, accumulates in cilia, and promotes the removal of Gpr161 from cilia (Corbit et al., 2005; Mukhopadhyay et al., 2013). The Gli transcription factors then concentrate at the ciliary tip and become activated before translocating into the nucleus to induce Hedgehog target genes (Haycraft et al., 2005).
The Hedgehog pathway is extensively regulated by post-translational modifications that control the activity of the components and their localization to subcompartments in the cell. For example, Smo becomes highly phosphorylated upon activation (Jia et al., 2004). Ubiquitination also modulates numerous steps in the Hedgehog pathway, including regulating levels and localization of Ptch1 and the processing of the Gli transcription factors (Hsia et al., 2015). Adding to this, we recently found that the ubiquitination of Smo regulates its ciliary localization by modulating its interaction with the IFT system (Desai et al., 2020). Covalent conjugation of ubiquitin (Ub), a small protein of 76 residues (8.6 kD), regulates the function and fate of many cellular proteins. Typically, the carboxy terminus of Ub is conjugated to the ε-amino group of a lysine or to the N-terminal residue of a substrate (Breitschopf et al., 1998). Ub itself contains seven lysines and an N-terminal methionine that can be further ubiquitinated to produce poly-Ub chains. These poly-Ub chains are named by the Ub residue that is extended (K6, K11, K27, K29, K33, K48, K63, M1; Komander and Rape, 2012). The type of poly-Ub chain determines the fate of the modified protein. The best understood are K48, which triggers degradation by the proteasome, and K63, which regulates protein complex formation. Ubiquitination is catalyzed by a cascade of three enzymes. First, Ub is activated by an E1 in an ATP-dependent process (Schulman and Harper, 2009). The activated Ub is then transferred to the active site of an E2 (van Wijk and Timmers, 2010). Finally, the Ub is covalently attached to substrates with the help of E3 ligases (Pickart, 2001). Ub can be removed from substrates by deubiquitinases, which is important for Ub homeostasis (Kimura and Tanaka, 2010). Enormous complexity exists in the Ub system, with the human genome encoding 2 E1 Ub–activating enzymes, >30 E2 Ub–conjugating enzymes, >600 E3 Ub ligases (Jin et al., 2007), and >100 deubiquitinases (Nijman et al., 2005).
Our recent work showed that Smo is ubiquitinated at the basal state, and this causes Smo to be removed from cilia in an IFT-dependent process (Desai et al., 2020). To identify the enzymes regulating Smo ubiquitination and ciliary localization, we performed a focused screen of Ub-related components that previously had been connected to cilia or Hedgehog signaling. Starting from 147 Ub-related components with connections to cilia or Hedgehog signaling, we identified 40 genes whose loss altered Hedgehog signaling as measured by a Gli-GFP reporter. 9 of these 40 caused aberrant Smo accumulation in cilia at the basal state. The nine included an E2-conjugating enzyme (Ube2l3), four E3 ligases (Wwp1, Arih2, Mgrn1, and Maea), a deubiquitinating enzyme (DUB; Bap1), and three adaptors (Kctd5, Skp1a, and Skp2). The E3 ligases appear to function in discrete steps where Arih2 regulates cellular levels of Smo, Wwp1 regulates ciliary levels directly, and Mgrn1 and Maea function in the cytoplasm. Our findings shed light on the long-standing enigma of how the ciliary levels of Smo are regulated by Hedgehog signaling.
Results
Development of a Hedgehog reporter cell line
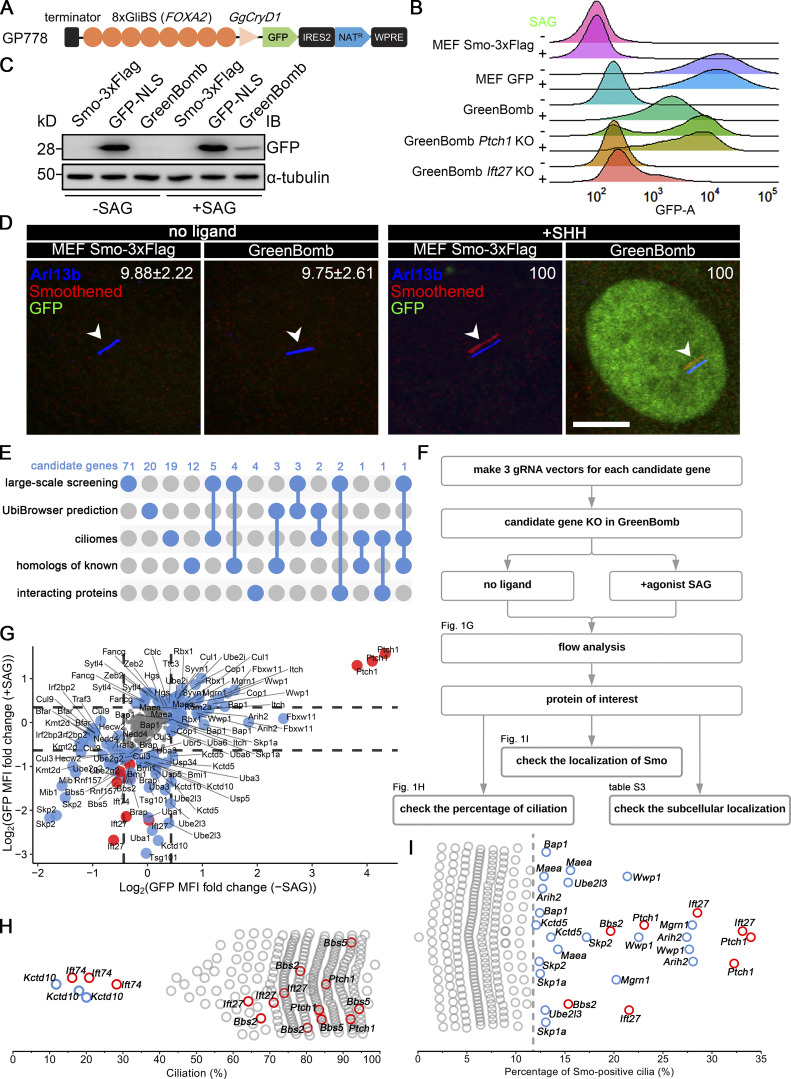
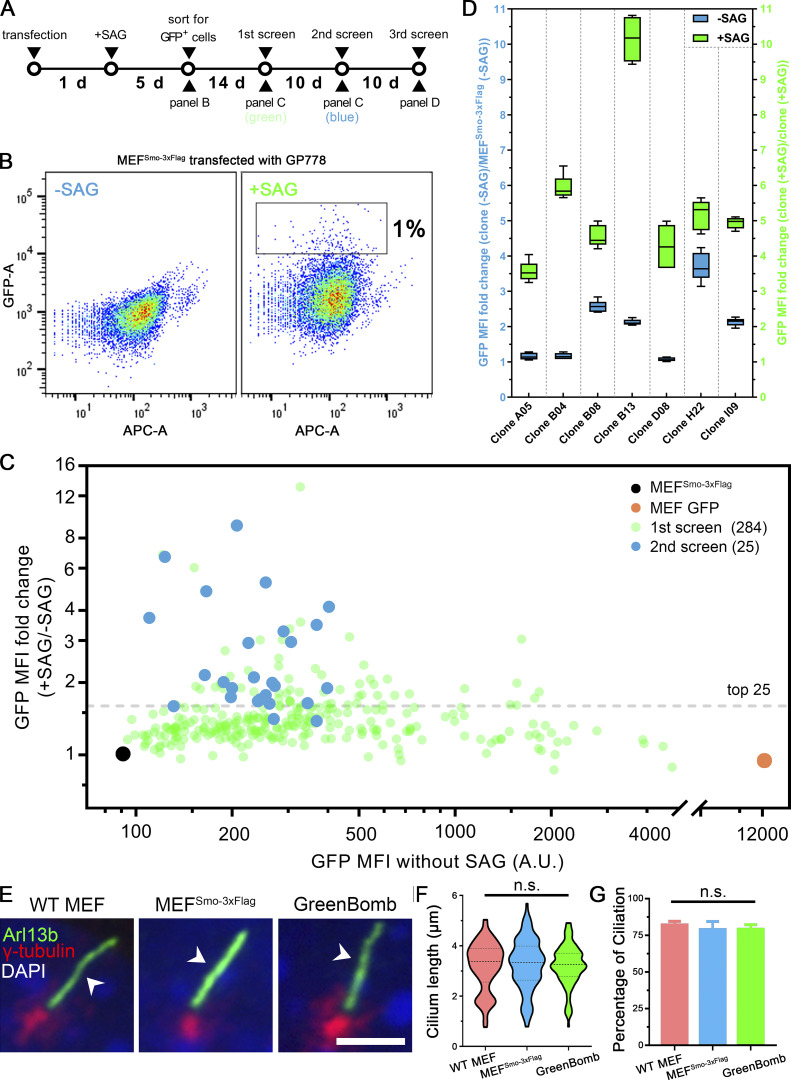
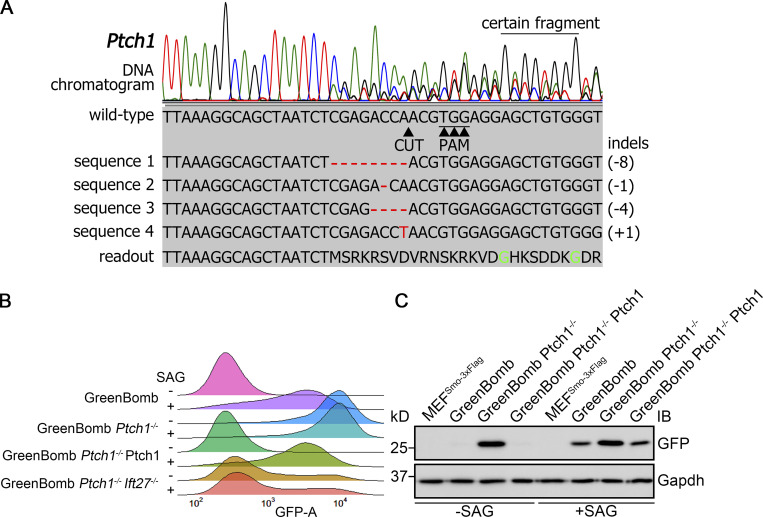
To identify genes involved in regulating the dynamics of Smo localization during Hedgehog signaling, we developed a cell line that monitors ciliary Smo and activation of Hedgehog-responsive genes. To do this, we first transfected immortalized mouse embryonic fibroblasts (MEFs) with a Smo-3xFlag construct (PD22) and cloned out a line (11479.6T PD22 clone 3, called “MEFSmo-3xFlag” hereafter) showing low ciliary Smo-3xFlag at the basal state and high ciliary Smo-3xFlag after pathway activation. This cell line was then transfected with a reporter construct (GP778) where nuclear localized GFP is driven from a promoter consisting of eight tandem Gli binding sites derived from the FOXA2 gene placed upstream of the chicken CryD1 minimal promoter (Fig. 1 A). Flow cytometry and single-cell cloning were used to identify a line, B13 (“GreenBomb” hereafter), where GFP fluorescence is enhanced ∼10-fold by addition of Smo agonist (SAG; Fig. 1, B–D; and Fig. S1, A–D). As predicted, knockout of Ift27 by CRISPR blocked induction of the GFP reporter by SAG, and knockout of Ptch1 caused GFP production without the need for pathway activation (Fig. 1 B). Characterization of GreenBomb cells indicated that they were similar to the parental MEF and MEFSmo-3xFlag cells with regard to percentage ciliation and cilia length (Fig. S1, E–G), and ciliary Smo-3xFlag was regulated normally (Fig. 1 D). These results indicate that GreenBomb is sensitive to perturbations of the Hedgehog pathway and can be used to characterize genes with unknown roles in ciliation and Hedgehog signaling.
Figure 1.
Identification of Ub-related genes regulating ciliary Smo levels. (A) Diagram of the GP778 Hedgehog reporter construct. GP778 contains a transcription terminator to stop read-through of upstream transcripts, eight copies of the Gli binding site (GliBS) derived from FOXA2, the chicken CryD1 minimal promoter, nuclear localized GFP, an IRES2 sequence followed by a Nat selectable marker, and the woodchuck post-transcriptional regulatory element (WPRE) RNA stabilization sequence. (B) Ridgeline plot of flow cytometry analysis of untransfected control cells (MEFSmo-3xFlag), cells transfected with a construct (BL17) expressing GFP from the CMV promoter (MEF GFP), GreenBomb, and GreenBomb transfected with Ptch1 or Ift27 gRNAs. Traces are shown for unstimulated cells (−SAG) and cells after pathway activation (+SAG). Each trace represents the fluorescence intensity of 10,000 individual cells. KO, knockout. (C) Western blot analysis of GFP expression before pathway activation (−SAG) and after activation (+SAG). α-Tubulin is included as a loading control. IB, immunoblot. (D) Immunofluorescence showing GFP (green), Smo (Flag, red), and cilia (Arl13b, blue, arrowheads) in MEFSmo-3xFlag or GreenBomb with or without SHH treatment. The Smo channel is shifted 10 pixels in both up and left directions. Scale bar, 5 microns. Numbers in the upper right corners are percentages of Smo-positive cilia. (E) UpSet diagram showing overlap of the five sources of candidate genes. Details are included in Table S1. (F) Workflow of the CRISPR-based screen. (G) GFP mean fluorescence intensity (MFI) fold change in GreenBomb with or without SAG treatment after candidate gene knockout. Each dot represents flow analysis of one gRNA transfected cell line. Red dots mark control cells. Data are plotted as the log2 ratio of GFP fluorescence of the experimental cells compared with control cells not transfected with a gRNA. Basal cells (−SAG) are plotted on the x axis, and stimulated cells (+SAG) are plotted on the y axis. Dashed lines indicate the mean ± 1 SD. Gray dots represent genes that were within this cutoff. Blue dots mark genes that fell outside of this cutoff and were further examined for effects on ciliation (H) and Smo localization (I). (H) Quantitation of the presence of cilia in cells with altered Hedgehog signaling. Each circle represents one gRNA transfected cell line. Red circles mark control cells. Blue circles mark genes causing reduced ciliation. (I) Quantitation of ciliary Smo localization in cells that had altered Hedgehog signaling. Each dot represents one gRNA transfected cell line. Red dots mark control cells. The dashed line marks the mean + 3 SD. Detailed data for G–I are included in Table S2.
Figure S1.
Development of a Hedgehog reporter line. (A) Flowchart for the development of the Hedgehog reporter cell line. Cells expressing Smo-3xFlag (MEF 11479.6T PD22 clone 3) were transfected with the reporter (GP778), grown in the presence of SAG and Nat for 5 d, and then the highest 1% of GFP expressers were sorted into 96-well plates. After expansion, these were put through three rounds of analysis to identify the best reporter line. (B) Sorting for GFP-positive cells. Transfected cells were selected for Nat resistance in the presence of SAG for 5 d and then analyzed by flow cytometry. Plot shows GFP fluorescence versus allophycocyanin (APC)-A, which reports autofluorescence. Box in +SAG marks the top 1% of cells that were collected for further analysis. −SAG are transfected cells that were not treated with SAG to determine the level of autofluorescence and unstimulated GFP expression. (C) The first and second screens for the development of the Hedgehog reporter line. After expansion, 284 individual clones that grew from the top 1% of cells were analyzed by flow cytometry with and without stimulation with SAG (green dots). The top 25 cell lines were repeated a second time (blue dots). The black dot marks untransfected control, and the orange dot marks a control transfected with CMV-GFP (BL17). (D) The third screen for the development of the Hedgehog reporter line. The top seven cell lines from the second screen were reanalyzed in quintuplicate. Data are presented as box-and-whisker plots that represent the range, spread, and median of GFP mean fluorescence intensity (MFI) fold change. Blue indicates unstimulated, and green represents after SAG stimulation. B13 showed a relatively low basal level of GFP expression and the highest level of induction by SAG, so it was chosen as the parental line for the screen and named GreenBomb. Error bars indicate SD. (E) Immunofluorescence showing cilia (Arl13b, green, arrowheads), centrosomes (γ-tubulin, red), and nuclei (DAPI, blue) in WT MEF, MEFSmo-3xFlag, and GreenBomb. Scale bar, 5 microns. (F) Ciliary length in WT MEF, MEFSmo-3xFlag, or GreenBomb. The lines in the violin plots mark the median and the quartiles. n = 110 cilia measured per condition. Nonsignificant values were determined by one-way ANOVA. (G) Percentage ciliation of WT MEF, MEFSmo-3xFlag, and GreenBomb. n = 3 repeats with >360 cells counted per experiment. Nonsignificant values were determined by one-way ANOVA. Error bars indicate SD.
Screening Ub-related genes for roles in Hedgehog signaling
To identify proteins regulating the dynamics of Hedgehog signaling, we identified Ub-related genes in published proteomic, CRISPR, and siRNA screens designed to identify genes related to Hedgehog signaling or cilia (a complete list of genes and sources can be found in Table S1; Breslow et al., 2018; Ishikawa et al., 2012; Mick et al., 2015; Pusapati et al., 2017). This list was supplemented with Ub-related genes suggested to play roles in Drosophila melanogaster Hedgehog signaling and ones predicted by UbiBrowser (http://ubibrowser.ncpsb.org.cn) to ubiquitinate Hedgehog components (Li et al., 2017). Finally, interacting proteins of the regulators we identified were also included. From these various sources, 147 genes were identified among 907 Ub-related genes (Fig. 1 E). Three gRNAs were synthesized for each gene and cloned in lentiCRISPR v2 or lentiCRISPR v2 PuroP93S. The clones were verified by sequence, packaged into lentiviral particles, and transduced into GreenBomb cells. After drug selection, the GreenBomb cells transfected with gRNAs were analyzed by flow cytometry with and without stimulation with SAG (Fig. 1, F and G). From this screen, we identified 18 genes whose loss reduced both basal Hedgehog signaling and signaling in response to SAG treatment (positive regulators); 20 genes whose loss activated the pathway without need for ligand (negative regulators); and two genes, Kctd5 and Ube2l3, that increased basal expression but reduced expression driven by SAG (Fig. 1 G and Table S2).
To determine the cellular localization of the Hedgehog regulators identified in this work, we cloned all except Kmtd2 and Usp34 and expressed epitope-tagged forms in fibroblasts. Kmtd2 and Usp34 had extremely large ORFs and resisted attempts to amplify either as whole cDNAs or in fragments. The collection of regulators has diverse distributions throughout the cell with Bfar, Cop1, Cul9, Fancg, Skp1a, Uba3, Ube2l3, and Wwp1 showing weak localization to cilia. In addition, Sytl4 localized to the ciliary pocket at the base of the cilium, and Fbxw11 localized to centriolar satellites (Table S3).
This collection of potential regulators was further screened to determine if their loss affected ciliation because this could have a major impact on Hedgehog signaling. As expected, knockout of Ift74, but not Bbs2, Bbs5, Ift27, or Ptch1, blocked ciliogenesis (Fig. 1 H). Importantly, none of the regulators affected ciliation except Kctd10 (Fig. 1 H), which is known to participate in CEP97 degradation (Nagai et al., 2018). This indicates that the reduced expression caused by the knockout of most positive regulators is not caused by the failure of ciliogenesis.
Screening the 40 regulators identified nine genes whose loss caused the accumulation of Smo in cilia when the Hedgehog pathway was at the basal state (Fig. 1 I). Of these nine genes, Ube2l3 encodes an E2 Ub–conjugating enzyme, and Arih2, Mgrn1, and Wwp1 encode E3 Ub ligases. Maea is a member of the RING family of E3 ligases but lacks a detectable RING-type zinc finger domain, raising questions of whether it retains E3 activity (Maitland et al., 2019). However, it is required for the ubiquitination of substrates as part of a complex (Lampert et al., 2018), so we refer to it as an E3 ligase even though it may function more like an adaptor. Bap1 is a deubiquitinase. The others, Kctd5, Skp1a, and Skp2, may act as substrate adaptors in E3 ligase complexes.
Arih2, Maea, Mgrn1, and Wwp1 regulate the ciliary localization of Smo
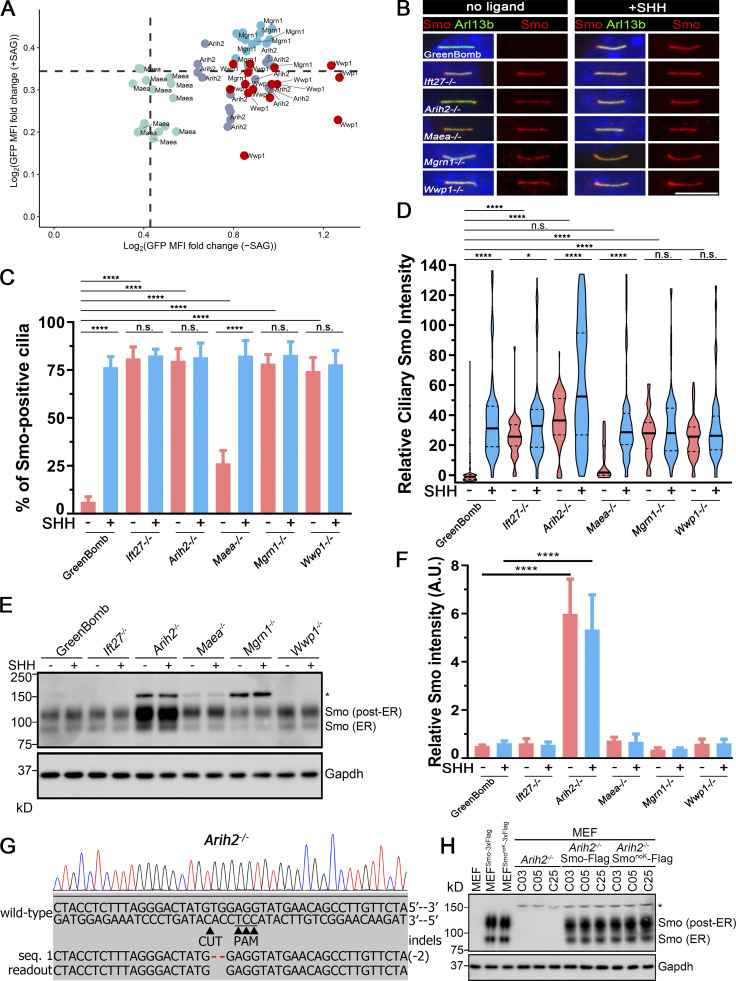
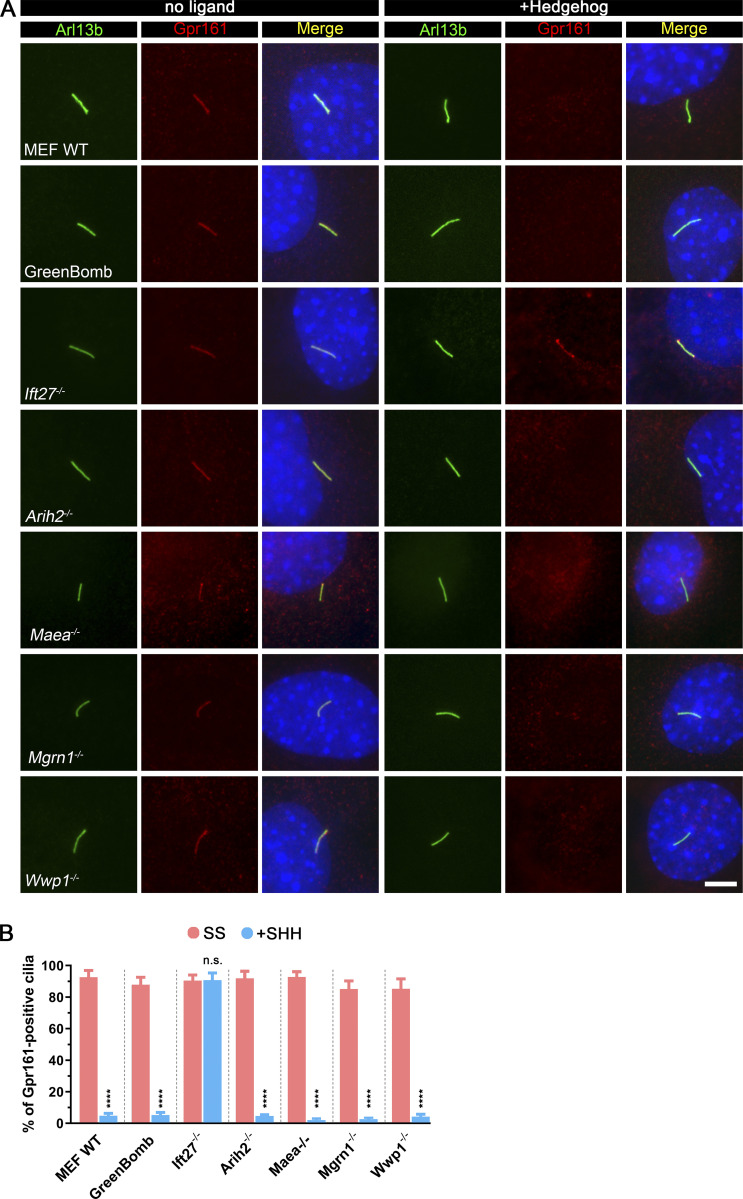
For further study of the E3 ligases that showed Smo accumulation in cilia, we isolated clones from each of the knockout populations. The clones behave like the uncloned populations as measured by flow cytometry of GFP expression from the reporter (Fig. 2 A) or by direct measures of Gli1 mRNA (Fig. S3 B). Similar to previous observations with Ift27−/− cells, clones of Arih2−/−, Mgrn1−/−, and Wwp1−/− had a high percentage of Smo-positive cilia in unstimulated cells, and no further increase was seen after treatment with sonic hedgehog (SHH)-conditioned medium (Fig. 2, B and C). Maea−/− cells had ∼26% Smo-positive cilia at the basal state, which increased to normal levels with SHH treatment (Fig. 2, B and C). Measurement of ciliary Smo intensity showed that SHH could further increase ciliary Smo levels in Arih2−/− and Maea−/− cells, but not Mgrn1−/− and Wwp1−/− cells (Fig. 2 D). Among the four E3 ligases, Arih2−/− cells had the highest ciliary Smo levels with or without SHH, whereas Maea−/− cells were the least affected (Fig. 2 D).
Figure 2.
E3 ligases regulating ciliary Smo levels.(A) GFP mean fluorescence intensity (MFI) fold change in Arih2−/−, Maea−/−, Mgrn1−/−, and Wwp1−/− knockout clones with or without SAG treatment. n = 5 repeats with 10,000 cells sorted per clone. Data are plotted as the log2 ratio of GFP fluorescence of the knockout cells compared with GreenBomb. Basal cells (−SAG) are plotted on the x axis, and stimulated cells (+SAG) are plotted on the y axis. Dashed lines indicate the mean + 1 SD as shown in Fig. 1 G. (B) Parental GreenBomb cells and subclones with Ift27, Arih2, Maea, Mgrn1, and Wwp1 knocked out were stained for Smo (Flag, red) and cilia (Arl13b, green) with and without SHH. Scale bar, 5 microns. (C) Quantification of the percentage of Smo-positive cilia from B. n = 6 repeats with at least 100 cilia counted per experiment. ****, P < 0.0001 by two-way ANOVA. Error bars indicate SD. (D) Quantification of the intensity of the ciliary Smo signal of cells from B. 100 cilia were measured for each condition. *, P < 0.05; ****, P < 0.0001 by two-way ANOVA. The solid lines in the violin plots mark the median and the dashed lines mark the quartiles. (E) Western blot of whole-cell extracts from parental GreenBomb cells and subclones with Ift27, Arih2, Maea, Mgrn1, or Wwp1 knocked out, with and without SHH. The asterisk marks a 150-kD band that binds nonspecifically to the Flag antibody. See G and H for details. GAPDH was used as a loading control. (F) Quantification of the Smo ER and post-ER signal intensity of the Western blot in D. n = 4 repeats. ****, P < 0.0001. Only Arih2−/− showed significant differences from control by two-way ANOVA. Error bars indicate SD. (G) Chromatogram showing one typical clone (gOC173 C19) of a MEF Arih2−/− cell with a deconvolved sequence. Red dashes mark deletions. (H) Western blot of whole-cell extracts from parental MEF cells and MEF cells expressing Smo-3xFlag or SmonoK-3xFlag (left three lanes). On the right, three clones of MEF Arih2−/− cells are shown untransfected and after transfection with Smo-3xFlag or SmonoK-3xFlag. Note that the band marked with an asterisk appears when Arih2 is knocked out and is present even when Smo-3xFlag is not expressed. GAPDH was used as a loading control.
Figure S3.
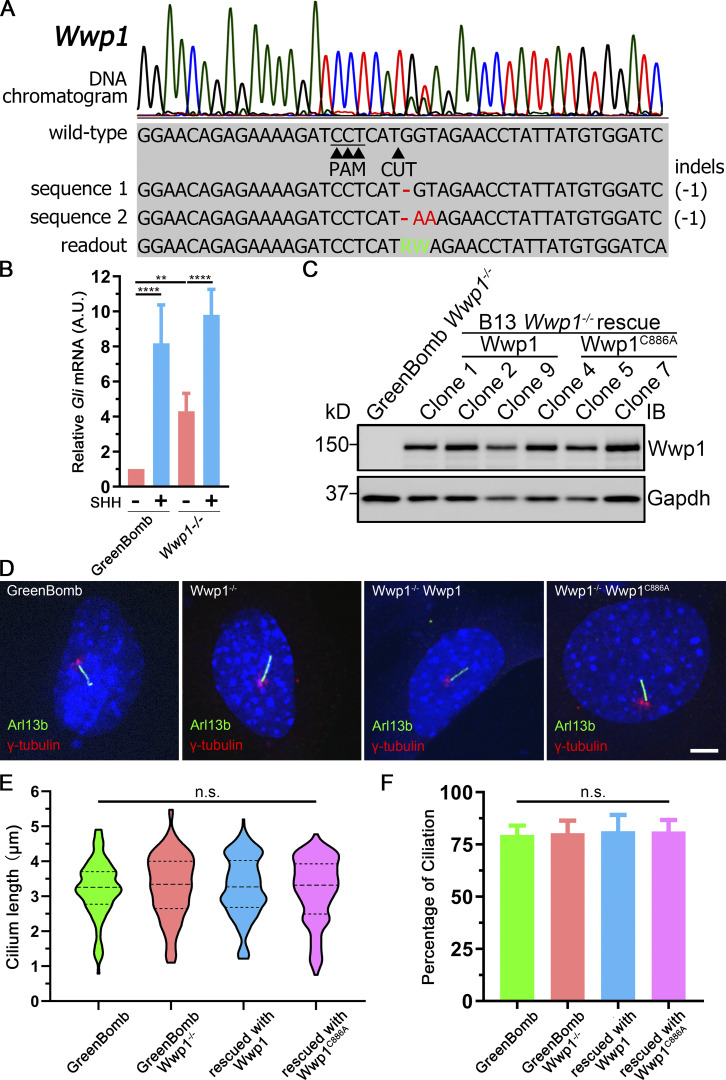
Wwp1 knockout does not affect ciliogenesis.(A) Chromatogram showing one typical clone (gBL79 C2) of GreenBomb Wwp1−/− cell with a deconvolved sequence. Red dashes and letters mark deletions and substitutions, respectively. Green letters in the sequence mean defined bases in the readout. (B) qRT-PCR showing the relative Gli mRNA in GreenBomb and GreenBomb Wwp1−/− cells with or without SHH treatment. **, P < 0.01; ****, P < 0.0001 by two-way ANOVA. Error bars indicate SD. (C) Western blot analysis of whole-cell extracts from GreenBomb Wwp1−/− and GreenBomb Wwp1−/− rescued with Wwp1 or the active site mutant Wwp1C886A. GAPDH was used as a loading control. (D) Immunofluorescence showing cilia (Arl13b, green) and centrosomes (γ-tubulin, red) in GreenBomb Wwp1−/− and GreenBomb Wwp1−/− rescued with Wwp1 or the active site mutant Wwp1C886A. Nuclei were stained with DAPI (blue). Scale bar, 5 microns. (E) Quantification of ciliary length of cells in C. At least 111 cilia were measured per condition. Nonsignificant values were derived by one-way ANOVA. The lines in the violin plots mark the median and the quartiles. (F) Percentage ciliation of cells in C. n = 7 repeats with >200 cells counted per experiment. Nonsignificant values were derived by one-way ANOVA. Error bars indicate SD.
To determine if these four E3 ligases affect Gpr161 dynamics in cilia, we stained cells for Gpr161 with and without SHH treatment. As previously reported, Gpr161 is present in cilia at the basal state but not after SHH treatment, and mutation of Ift27 prevents its removal after SHH treatment (Eguether et al., 2014; Mukhopadhyay et al., 2013). All four E3 mutants behaved like WT controls, indicating that they are not critical for Gpr161 dynamics (Fig. S2).
Figure S2.
Arih2, Maea, Mgrn1, or Wwp1 do not regulate the ciliary localization of Gpr161.(A) Immunofluorescence showing the cilia (Arl13b, green), Gpr161 (red), and nuclei (DAPI, blue) in WT MEF, GreenBomb, GreenBomb Ift27−/−, GreenBomb Arih2−/−, GreenBomb Maea2−/−, GreenBomb Mgrn1−/−, or GreenBomb Wwp1−/−. Scale bar, 5 microns. (B) Quantitation of Gpr161-positive cilia described in A. n = 6 repeats with at least 200 cilia counted per experiment. ****, P < 0.0001 as compared with serum-starved cells by two-way ANOVA. Error bars indicate SD.
As previously described (Marada et al., 2015), Western blot analysis of Smo in control cells shows a band of ∼90 kD that is likely to be immature Smo transiting the ER and an unfocused band at ∼120 kD that is thought to be a post-ER glycosylated form. The loss of Ift27, Maea, Mgrn1, or Wwp1 did not significantly alter the amount of these two forms of Smo (Fig. 2, E and F). However, the loss of Arih2 dramatically increased the levels of these forms (Fig. 2, E and F), suggesting that Arih2 is responsible for controlling the cellular levels of Smo by regulating its degradation. The 150-kD band (marked with an asterisk in the figures) is a protein that binds nonspecifically to the Flag antibody and appears to be up-regulated in the Arih2−/−, Maea−/−, and Mgrn1−/− cells (Fig. 2, E and H).
The localization of Wwp1 is regulated by Hedgehog signaling
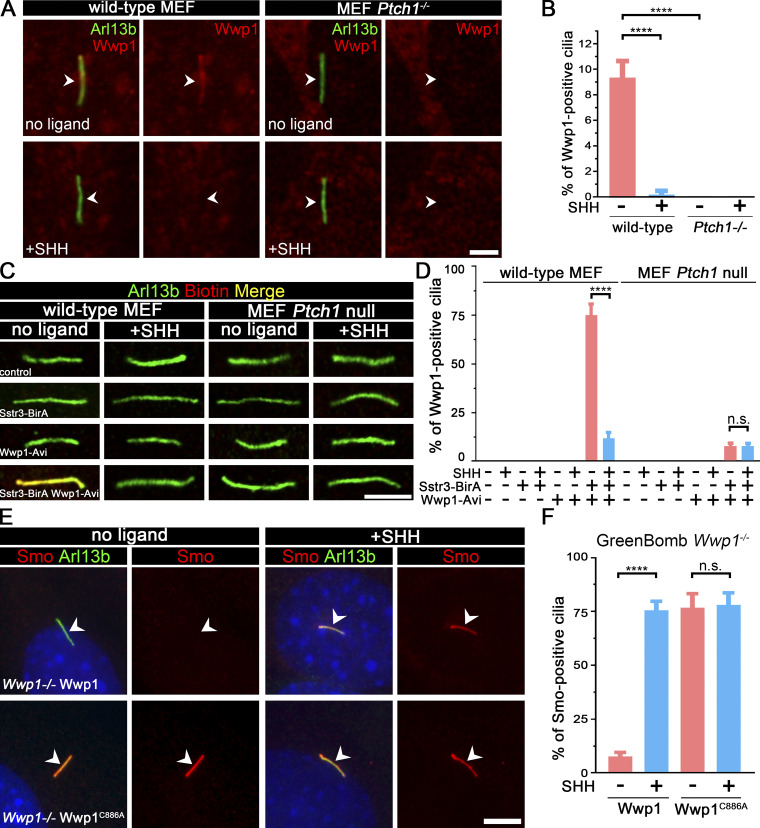
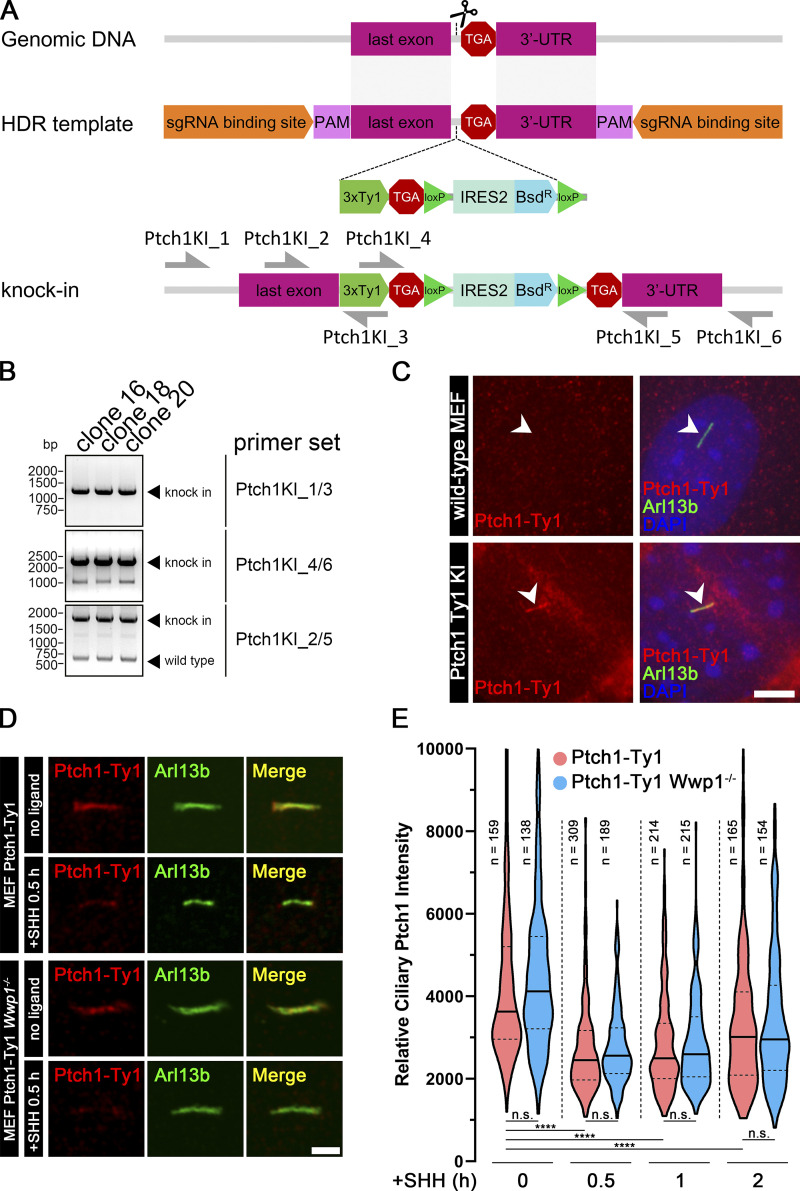
Of the four E3 ligases that are needed to maintain low ciliary Smo at the basal state, only Wwp1 localized to cilia (Fig. 3, A and B; and Table S3). Interestingly, the ciliary localization of Wwp1 is regulated by Hedgehog signaling, because Wwp1 is found in ∼9% of cilia at the basal state and nearly none after stimulation with SHH (Fig. 3, A and B). The localization of Wwp1 to cilia was lost in Ptch1-null cells (Fig. 3, A and B). The low percentage of positive cilia was unexpected because the loss of Wwp1 caused most cilia to accumulate Smo. It is possible that more cilia have Wwp1 present, but below our detection level. To increase the sensitivity of detection, we fused an Avi tag to the C-terminus of Wwp1 and coexpressed this with BirA targeted to cilia by fusion with Sstr3. The Avi tag is a short sequence that can be biotinylated by BirA (Beckett et al., 1999). By coexpressing this with cilia-targeted BirA, any Wwp1-Avi that enters the cilium will be biotinylated, which can be detected by fluorescent avidin. Using this compartmentalized labeling method, Wwp1 was found in ∼74% of cilia at the basal state and ∼11% after stimulation with SHH (Fig. 3, C and D). The localization of Wwp1 to cilia decreased to ∼7% in Ptch1-null cells (Fig. 3, C and D). The Smo accumulation phenotype in Wwp1−/− cells can be rescued by the reintroduction of Wwp1, but not by the Wwp1C886A active site mutant (Fig. 3, E and F), indicating that its E3 ligase activity is required for the ciliary removal of Smo. Moreover, Wwp1−/− cells are normal compared with the parental cells in terms of percentage ciliation and ciliary length (Fig. S3).
Figure 3.
Wwp1 localizes in cilia, and its E3 ligase activity is required for the ciliary removal of Smo.(A) WT MEF or MEF Ptch1-null cells expressing Wwp1-Flag were stained for Wwp1 (Flag, red) and cilia (Arl13b, green, arrowheads) with or without exposure to SHH-conditioned medium. Scale bar, 2 microns. (B) Quantification of the percentage of Wwp1-positive cilia from A. n = 6 replicates with at least 200 cilia counted each time. ****, P < 0.0001 by two-way ANOVA. Error bars indicate SD. (C) WT MEF or MEF Ptch1-null cells expressing Sstr3-BirA and Wwp1-Avi were stained for Wwp1 (biotin, red) and cilia (Arl13b, green) with and without SHH. Scale bar, 3 microns. (D) Quantification of the percentage of Wwp1-positive cilia from C. n = 6 repeats with 200 cilia counted per experiment. ****, P < 0.0001 by two-way ANOVA. Error bars indicate SD. (E) Wwp1 mutant cells transfected with Wwp1-Flag show rescue of ciliary Smo levels. Wwp1C886A is enzymatically dead, indicating that E3 activity is required for rescue. Arrowheads mark cilia. (F) Quantitation of Smo-positive cilia described in E. n = 6 repeats with 200 cilia counted per experiment. ****, P < 0.0001 as compared with serum-starved cells by two-way ANOVA. Error bars indicate SD.
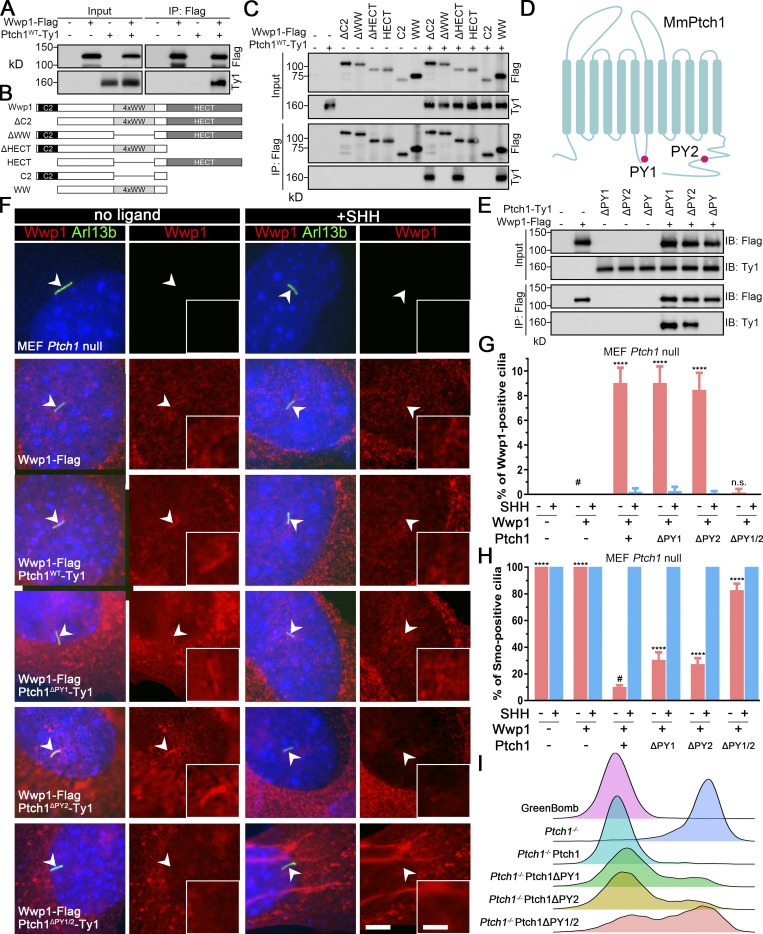
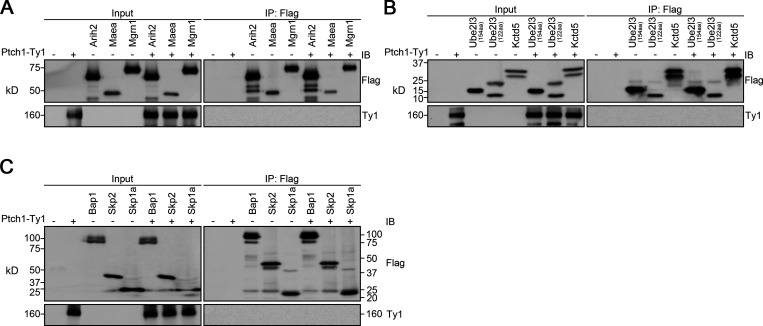
Wwp1 is one of the eight Nedd4 family HECT E3 ligases that bind to Ptch1 (Kim et al., 2015). Thus, Ptch1 may control Wwp1’s ciliary localization by providing a binding platform. Confirming published results, we found that Ptch1 binds to Wwp1 (Fig. 4 A). This binding is specific because Ptch1 did not bind Arih2, Maea, Mgrn1, Ube2l3, Kctd5, Bap1, Skp2, or Skp1a (Fig. S4). The WW domain of Wwp1 mediates the interaction with Ptch1 (Fig. 4, B and C). WW domains often interact with PY motifs, of which there are two in Ptch1 (Fig. 4 D). The deletion of either of the Ptch1 PY motifs does not affect the binding of Ptch1 to Wwp1, but the deletion of both PY motifs prevents its binding to Wwp1 (Fig. 4 E). Ptch1-knockout cells lost ciliary localization of Wwp1, but this could be rescued by reexpression of Ty1-tagged Ptch1 (Fig. 4, F and G; and Fig. S5). Consistent with the binding data, Wwp1 localized to cilia when either of the Ptch1 PY motifs was deleted individually, but the deletion of both Ptch1 PY motifs blocked the localization of Wwp1 to cilia (Fig. 4, F and G). The deletion of either PY motif caused a modest increase in Smo-positive cilia at the basal state, while the deletion of both motifs caused most cilia to be Smo positive (Fig. 4 H). Similar experiments by Kim et al. (2015) did not detect increased ciliary Smo levels at the basal state when the Ptch1 PY motifs were mutated, but the reason for the difference is not known. At the level of gene expression, the loss of either PY motif caused a small subset of cells to show increased basal signaling, while the loss of both PY motifs increased the number of cells with elevated basal signaling (Fig. 4 I). These data suggest that Wwp1 activity against Smo is regulated by controlling its ciliary localization through Ptch1 binding.
Figure 4.
Wwp1 activity against Smo is regulated by controlling its ciliary localization through Ptch1 binding.(A) Immunoprecipitation of Ptch1 and Wwp1. HEK 293T cells expressing Ptch1-Ty1 and Wwp1-Flag were immunoprecipitated with anti-Flag resin and probed for Flag and Ty1. Input is equivalent to 10% of the precipitate. (B) Domain structure of Wwp1 and diagram of deletion constructs used for coimmunoprecipitation. (C) Constructs described in B were coexpressed with Ptch1-Ty1 in HEK 293T cells and precipitated (IP) with anti-Flag resin. Input is equivalent to 10% of input. Note that deletion of the 4xWW domain of Wwp1 prevents binding to Ptch1. (D) Schematic diagram of Ptch1 showing the PY motif locations. (E) Immunoprecipitation of Wwp1 and Ptch1 PY motif mutants. Ptch1-Ty1 constructs lacking the first PY (ΔPY1), the second PY (ΔPY2), or both PY motifs (ΔPY1/2) were coexpressed in HEK 293T cells with Wwp1-Flag and immunoprecipitated with anti-Flag resin. Note that the loss of both PY motifs prevents binding of Wwp1 to Ptch1, but Ptch1 retaining either PY motif is still able to bind Wwp1. IB, immunoblotting. (F) Immunofluorescence showing Wwp1 (Flag, red) in cilia (Arl13b, green, arrowheads) with or without SHH treatment in Ptch1−/− cells expressing the Ptch1 mutants. Nuclei were stained with DAPI (blue). Scale bar, 5 microns. Insets are 2.5× enlargements. The scale bar in the inset represents 2 microns. (G) Quantitation of Wwp1-positive cilia in MEF Ptch1-null cells expressing Ptch1 or Ptch1 mutants. n = 5 repeats with >110 cilia counted per experiment. ****, P < 0.0001 compared with MEF Ptch1-null cells expressing Wwp1 (−SHH, labeled with #) by two-way ANOVA. Error bars indicate SD. (H) Quantitation of Smo-positive cilia in MEF Ptch1-null cells expressing Ptch1 or Ptch1 mutants. n = 4 repeats with 200 cilia counted per experiment. Smo was detected with an antibody against the endogenous protein. ****, P < 0.0001 compared with unstimulated MEF Ptch1-null cells expressing Ptch1 and Wwp1 (−SHH, labeled with #) by two-way ANOVA. Error bars indicate SD. (I) Ridgeline plot of flow cytometry analysis of GreenBomb, GreenBomb Ptch1-knockout cells, and GreenBomb Ptch1-knockout cells rescued with Ptch1, Ptch1ΔPY1, Ptch1ΔPY2, or Ptch1ΔPY1/2. Traces are shown for unstimulated cells. Each trace represents fluorescence intensity of 10,000 individual cells.
Figure S4.
Arih2, Maea, Mgrn1, Ube2l3, Kctd5, Bap1, Skp2, and Skp1a do not interact with Ptch1.(A) Immunoprecipitation of Ptch1 and Arih2, Maea, or Mgrn1. HEK 293T cells expressing Ptch1-Ty1 and Arih2-3xFlag, Maea-3xFlag, or Mgrn1-3xFlag were immunoprecipitated with anti-Flag resin and probed for Flag and Ty1. Input is equivalent to 10% of the precipitate. IB, immunoblotting. (B) Immunoprecipitation of Ptch1 and Ube2l3 or Kctd5. HEK 293T cells expressing Ptch1-Ty1 and Ube2l3-3xFlag or Kctd5-3xFlag were immunoprecipitated with anti-Flag resin and probed for Flag and Ty1. Input is equivalent to 5% of the precipitate. (C) Immunoprecipitation of Ptch1 and Bap1, Skp2, or Skp1a. HEK 293T cells expressing Ptch1-Ty1 and Bap1-3xFlag, Skp2-3xFlag, or Skp1a-3xFlag were immunoprecipitated with anti-Flag resin and probed for Flag and Ty1. Input is equivalent to 10% of the precipitate.
Figure S5.
Knockout of Ptch1 in GreenBomb activates the Hedgehog pathway.(A) Chromatogram showing one typical clone of a Ptch1−/− cell with a deconvolved sequence. Red dashes and letters mark deletions and substitutions, respectively. (B) Ridgeline plot of flow cytometry analysis of GreenBomb cells, Ptch1−/− cells, Ptch1−/− cells rescued with Ptch1-Ty1 and Ptch1−/−, and Ift27−/− double-knockout cells. Traces are shown for unstimulated cells (−SAG) and cells after pathway activation (+SAG). Each trace represents fluorescence intensity of 10,000 individual cells. (C) Western blot analysis of GFP expression before pathway activation (−SAG) and after activation (+SAG). α-Tubulin is included as a loading control. IB, immunoblot.
Wwp1 is not required for the removal of Ptch1 from cilia
Ptch1, the receptor for SHH, localizes in cilia at the basal state and is removed upon stimulation with SHH (Rohatgi et al., 2007). Because of Wwp1’s role in regulating Smo localization, it is important to determine if it is involved in regulating the dynamics of Ptch1. We were unable to obtain usable Ptch1 antibodies by injecting rabbits or from commercial sources, so we created a knock-in line where three copies of the Ty1 tag were inserted into the endogenous Ptch1 gene at its C terminus (Fig. 5, A–C). As expected, Ptch1-Ty1 localized to cilia at the basal state. Upon stimulation with SHH, Ptch1 levels dropped within 30 min and then started to increase after 2 h. Wwp1−/− cells responded like the controls, indicating that Wwp1 is not involved in regulating the dynamics of ciliary Ptch1 (Fig. 5, D and E).
Figure 5.
Wwp1 is not required for the removal of Ptch1 from cilia.(A) Diagram illustrating knocking three copies of the Ty1 epitope tag into Ptch1 at the C-terminal end just before the stop codon. HDR, homology-directed repair; PAM, protospacer adjacent motif. (B) PCR using primers shown in A was used to screen knock-in clones. Clones 16, 18, and 20 showed the correct pattern. Clone 20 was used for analysis. DNA size in base pairs (bp) is shown on the left. (C) Immunofluorescence showing cilia (Arl13b, green, arrowheads), Ptch1 (Ty1, red), and nuclei (DAPI, blue) in MEF Ty1 knock-in (KI) of Ptch1 clone 20. WT MEF cells were stained as control cells. Scale bar, 5 microns. (D) Immunofluorescence showing cilia (Arl13b, green) and Ptch1 (Ty1, red) in MEF Ty1 knock-in of Ptch1 or MEF Ty1 knock-in of Ptch1 with Wwp1−/−. Scale bar, 2 microns. (E) The loss of Wwp1 does not affect the removal of Ptch1 from cilia by pathway activation. Ptch1-Ty1 and Ptch1-Ty1,Wwp1−/− cells were fixed and stained for cilia and Ty1 before stimulation with SHH, and at 0.5, 1, and 2 h, control and Wwp1−/− cells behaved similarly. ****, P < 0.0001 as compared with serum-starved cells by repeated measures ANOVA. The solid lines in the violin plots mark the median and the dashed lines mark the quartiles.
Smo has K63-linked Ub chains
If the ciliary level of Smo is regulated by a specific type of poly-Ub linkage, DUBs that cleave the critical poly-Ub chain should increase basal ciliary Smo levels by interfering with Smo’s removal. To test this idea, we asked how the expression of Usp2, which is active against all types of poly-Ub linkages, affects ciliary Smo levels. Unfortunately, expression of full-length Usp2 or its catalytic domain (CD) tethered to Sstr3-Myc was toxic to cells. We overcame this by the use of a tetracycline (Tc)-inducible expression system to regulate expression of Usp2CD (Fig. 6 A). After induction of Sstr3-Usp2CD by Tc, Smo accumulated in Sstr3-Myc-Usp2CD–positive cilia, supporting the hypothesis that ubiquitinated Smo is removed from cilia at the basal state (Fig. 6 B).
Figure 6.
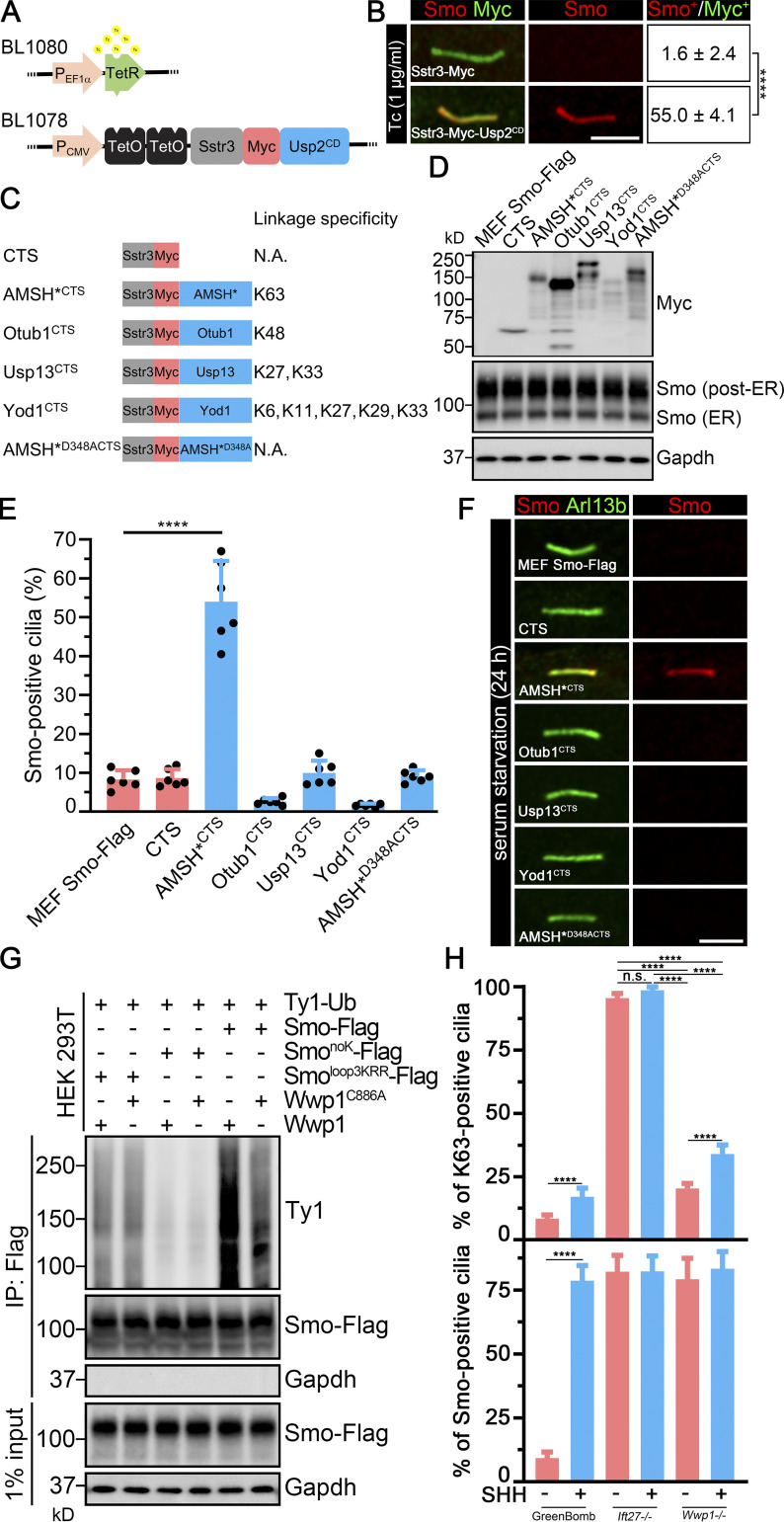
Cilia-targeted K63 DUB causes Smo accumulation in cilia.(A) Constructs used to regulate Usp2 expression. TetR forms a homodimer that binds to TetO in the absence of Tc and represses transcription. Tc inhibits the repression, allowing expression of Sstr3-Myc-Usp2CD. (B) MEFSmo-3xFlag cells expressing Sstr3-Myc or Sstr3-Myc-Usp2CD under the control of Tc-inducible promoter were treated with Tc and stained for Smo (Flag, red) and Myc (green). Scale bar is 3 microns. Numbers are the percentages of Smo-positive cilia among Myc-positive cilia. n = 3 repeats with 20 cilia counted per experiment. ****, P < 0.0001 as compared with serum-starved cells by independent samples t test. (C) Diagram of DUB constructs and specificity. Each of the DUBs was fused to the C terminus of the cilia-localized Sstr3 receptor with an Myc tag linking the two parts. These constructs are expressed from the CMV promoter and are not Tc regulated. CTS, ciliary targeting sequence; N.A., not applicable. (D) Western blot showing the expression of each of the DUB constructs and effects on total cellular Smo levels. (E) Quantitation of Smo-positive cilia described in C. n = 6 repeats with 200 cilia counted per experiment. ****, P < 0.0001 as compared with serum-starved cells by one-way ANOVA. Error bars indicate SD. (F) Immunofluorescence showing Smo (Flag, red) and cilia (Arl13b, green) in MEFSmo-3xFlag cells expressing the constructs shown in C. MEFSmo-3xFlag cells are the parental control cells. Scale bar is 3 microns. (G) Ligation of Ub onto Smo by Wwp1. HEK 293T cells transiently transfected with the constructs indicated on the top were lysed, and Smo was immunoprecipitated (IP) with Flag resin and examined by Western blot analysis. (H) Quantification of the percentage of Smo-positive cilia and K63-positive cilia in GreenBomb, GreenBomb Ift27−/−, and GreenBomb Ift27−/−. n = 6 repeats with 200 cilia counted per experiment. ****, P < 0.0001 by two-way ANOVA. Error bars indicate SD.
To understand the type of poly-Ub linkage involved in the ciliary removal of Smo, we targeted the DUBs AMSH* (Stam2-AMSH fusion), Otub1, Usp13, and Yod1 to cilia by fusing them to the C terminus of Sstr3. Each of these deubiquitinases recognizes a subset of poly-Ub chains and cleaves them selectively (Fig. 6 C; Mevissen et al., 2013; Sato et al., 2008; Tan et al., 2017). None of the tested DUBs had an appreciable effect on the total level of Smo in the cell (Fig. 6 D). Of the tested DUBs, only AMSH* caused Smo accumulation in cilia (Fig. 6, E and F). The accumulation was not seen when the active site (AMSH*D348A) was mutated (Fig. 6, E and F; McCullough et al., 2004), supporting the idea that the DUB activity was responsible for the Smo accumulation. AMSH* is thought to remove K63-linked Ub chains, suggesting that this is the form that is actively targeted for the removal of Smo. Furthermore, this is consistent with Wwp1 being the critical ciliary E3 because Wwp1 catalyzes K63 polymerization (French et al., 2017).
The ability of Wwp1 to ubiquitinate Smo was examined by coexpressing Ty1-tagged Ub and Wwp1 along with Smo in HEK 293T cells. Smo was then captured by immunoprecipitation, and the ligated Ub was detected with an anti-Ty1 antibody. As controls, Wwp1 was replaced with Wwp1C886A, which is enzymatically dead, and Smo was replaced with SmonoK or Smoloop3KRR, which should lack the expected sites of Wwp1 ubiquitination (Desai et al., 2020). A large amount of Ub was incorporated into Smo when coexpressed with Wwp1, and the amount was reduced in cells expressing enzymatically dead Wwp1. Likewise, replacement of Smo with SmonoK or Smoloop3KRR reduced the incorporation of Ub by Wwp1 (Fig. 6 G). These data support a model where Wwp1 is the E3 that catalyzes K63 polymerization of Ub onto Smo at K444 and K448. This was further supported by the observation that there are dramatically fewer K63-positive cilia in Wwp1−/− cells than K63-positive cilia in Ift27−/− cells, even though they all have equivalent levels of ciliary Smo accumulation (Fig. 6 H).
The E2 Ube2l3 regulates ciliary and cell body levels of Smo
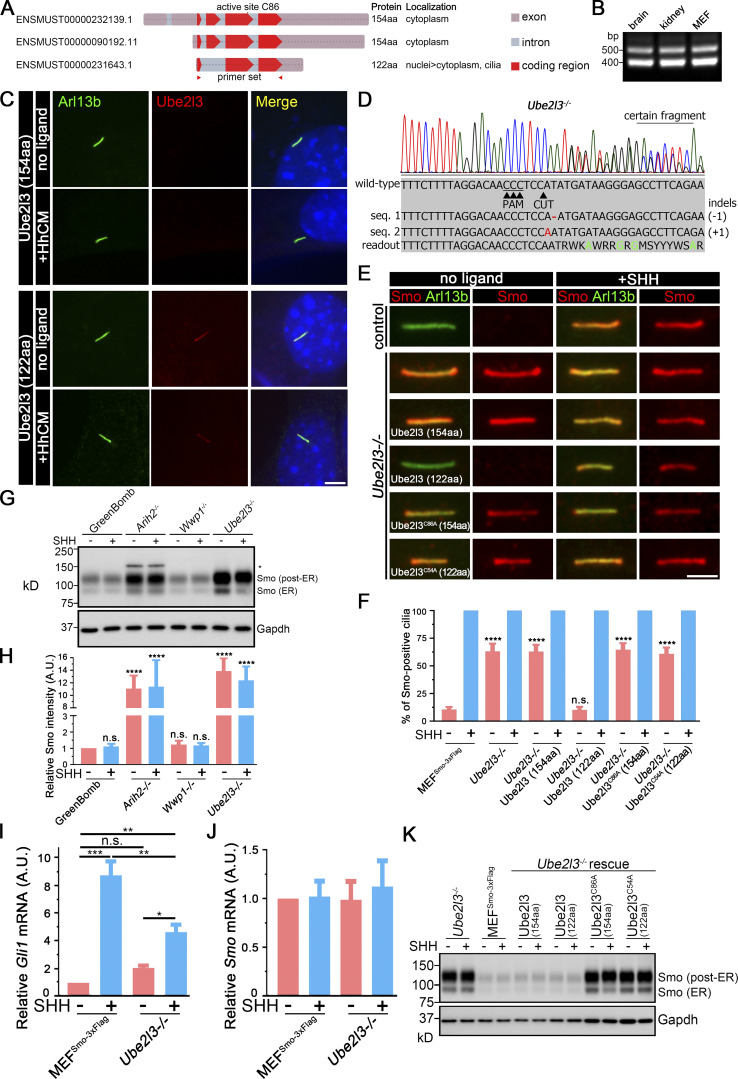
Our initial screen of Ub components with connections to cilia or Hedgehog contained 10 of the 37 putative E2-conjugating enzymes. Of these, knockout of Ube2g2 decreased SAG-induced expression, Ube2i increased basal expression, and Ube2l3 increased basal expression while decreasing SAG-induced expression (Table S2). Since we identified four E3 ligases from different families as critical to regulating ciliary Smo, we had concerns that an unexamined E2 might be important. Thus, we generated guides for the remaining 27 E2 Ub–conjugating enzymes (Table S4). Knockout of these E2s did not alter the output of GreenBomb (Table S2), indicating that one of the initial three is likely to be the critical E2. Of the initial three, only Ube2l3 knockout increased ciliary Smo at the basal level, so we focused on this enzyme. This E2 works with Arih2 and Wwp1 (French et al., 2017; Marteijn et al., 2009; Marteijn et al., 2005), consistent with a role in regulating ciliary Smo levels. Amplification of the ORF from fibroblast cDNA identified two splice variants (Fig. 7, A and B). The longer one encodes a 154-residue protein (NCBI accession no. NP_033482), while the shorter one encodes a 122-residue protein (RefSet accession no. XP_006522067). When epitope-tagged versions were expressed in fibroblasts, both isoforms localized to the cytoplasm but, interestingly, the short isoform also localized to cilia (Fig. 7 C). To understand the importance of the two splice variants to Smo localization, Ube2l3-knockout cells were generated (Fig. 7 D). Direct measurement of Gli1 mRNA showed that the loss of Ube2l3 increased basal expression and decreased SHH responses, similar to what was seen with the original reporter (Fig. 7 I). Mutant cells were rescued with the two isoforms as well as the enzyme-dead versions of each isoform. The parental Ube2l3 mutant cells accumulated Smo in cilia at the basal state (Fig. 7, E and F). This ciliary Smo accumulation could be rescued by the 122-residue isoform but not by the 154-residue isoform or by the enzyme-dead versions of either isoform (Fig. 7, E and F). Western blot analysis indicated that the loss of Ube2l3 increased the levels of both the ∼90 kD ER and the ∼120 kD post-ER forms of Smo (Fig. 7, G and H). The increased level of Smo was not due to increased transcription (Fig. 7, I and J). Reexpression of either the long or short form of Ube2l3, but not the enzyme-dead versions, restored the amount of Smo detected by Western blotting (Fig. 7 K). The observation that only the short form localizes to cilia and rescues the ciliary Smo accumulation phenotype while either isoform can rescue the accumulation of Smo in the cell body suggests that Ube2l3 needs to be localized to cilia to regulate ciliary Smo levels.
Figure 7.
The Ub-conjugating enzyme Ube2l3 regulates ciliary Smo levels. (A) Diagram of the three Ube2l3 transcripts from the mouse. The two longer ones encode the same 154-aa protein, while the shorter one lacks the second exon and encodes a 122-residue protein. Transcript identifiers from Ensembl are listed on the left. (B) RT-PCR indicates that transcripts representing both coding forms of Ube2l3 are expressed in mouse brain, kidney, and cultured fibroblasts. The primer set used here is labeled in A with red triangles. (C) Immunofluorescence showing the two isoforms of Ube2l3 (Flag, red) and cilia (Arl13b, green) with or without SHH treatment in WT MEFs. Nuclei were stained with DAPI (blue). Scale bar, 5 microns. (D) Chromatogram showing one typical clone of a Ube2l3−/− cell with a deconvolved sequence. Red dashes and letters mark deletions and substitutions, respectively. Green letters in the sequence mean defined bases in the readout. (E) Immunofluorescence showing Smo (Flag, red) and cilia (Arl13b, green) with or without SHH treatment in MEFSmo-3xFlag (control), Ube2l3-knockout cells, and Ube2l3-knockout cells rescued with Ube2l3 (154aa), Ube2l3 (122aa), Ube2l3C86A (154aa), or Ube2l3C54A (122aa). (F) Quantitation of Smo-positive cilia described in E. n = 6 repeats with 200 cilia counted per experiment. ****, P < 0.0001 as compared with MEFSmo-3xFlag cells (−SHH) by two-way ANOVA. Error bars indicate SD. (G) Western blot analysis of whole-cell extracts from GreenBomb and Arih2−/−, Wwp1−/−, Ube2l3−/− derivatives with and without SHH stimulation. The asterisk marks an unrelated band. GAPDH was used as a loading control. Numbers at the bottom are quantifications of the corresponding Smo signal intensity. n = 4 repeats. (H) Quantification of the whole Smo (all bands combined) signal intensity of the Western blot in G. n = 4 repeats. ****, P < 0.0001 as compared with GreenBomb (−SHH) by two-way ANOVA. Error bars indicate SD. (I) qRT-PCR showing the relative Gli mRNA in MEFSmo-3xFlag and MEFSmo-3xFlag Ube2l3−/− cells with or without SHH treatment. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-way ANOVA. Error bars indicate SD. (J) qRT-PCR showing the relative Smo mRNA in MEFSmo-3xFlag and Ube2l3−/− cells with or without SHH treatment. Nonsignificant values are as compared with MEFSmo-3xFlag without SHH by two-way ANOVA. Error bars indicate SD. (K) Western blot analysis of whole-cell extracts from Ube2l3−/− knockout cells, control cells (MEFSmo-3xFlag), and Ube2l3-knockout cells rescued with Ube2l3 (154aa), Ube2l3 (122aa), Ube2l3C86A (154aa), or Ube2l3C54A (122aa) with and without exposure to SHH-conditioned medium. GAPDH was used as a loading control.
Discussion
Our recent work demonstrated that ciliary Smo levels are regulated by ubiquitination, and we proposed a model where Smo becomes ubiquitinated if it diffuses into cilia when the pathway is inactive. This causes it to be removed from the cilium by the IFT/BBSome system, thus keeping ciliary Smo levels low at the basal state (Desai et al., 2020). In this work, we set out to identify the enzymes that control the ubiquitination state of Smo. We focused on Ub-related proteins that had been identified in large-scale ciliary screens and proteomic studies. From among 147 candidates, 40 genes were identified that altered Hedgehog signaling, 9 of which caused Smo to accumulate abnormally in cilia at the basal state. This collection included four E3 Ub ligases, one E2 Ub–activating enzyme, one DUB, and three adaptors. Previous work in flies indicated that cell surface Smo (which is analogous to the ciliary Smo levels in mammals) is regulated by ubiquitination. A variety of enzymes were implicated, including Cul4 (Li et al., 2018a), Herc4 (Jiang et al., 2019), Smurfs (Li et al., 2018b), Uchl5 (Zhou et al., 2018), and Usp8 (Li et al., 2012; Xia et al., 2012). Curiously, none of these enzymes showed a phenotype in our screen. This was unexpected because the core components of the pathway are conserved between Drosophila and vertebrates. However, ciliary localization is likely to require different regulatory mechanisms than bulk cell surface localization, and this probably explains the lack of conservation of regulatory proteins.
Of the E3 ligases, Wwp1 localized to cilia, suggesting that it is the key enzyme regulating ciliary Smo levels. In support of this idea, Wwp1 localization in cilia was Ptch1 dependent and regulated by Hedgehog ligand. The localization was facilitated by interactions between the PY motifs in Ptch1 and the WW domains in Wwp1. Wwp1 is not known for roles in cilia or in Hedgehog signaling but was found in a comparative proteomic study of cilia (Sigg et al., 2017) and was predicted by the UbiBrowser (Li et al., 2017) to ubiquitinate several Hedgehog components. Wwp1 is a HECT family ligase whose activity has been studied with regard to cancer, where its increased activity is correlated with aggressive tumors (Chen et al., 2007). Knockout mice have short tibias (Mouse Genome Informatics and the International Mouse Phenotyping Consortium) and are obese (Hoshino et al., 2020), two phenotypes that are commonly seen in cilia-defective mice. Wwp1 is reported to catalyze monoubiquitination and the extension of a poly-Ub chain through the rapid addition of two to four K63-linked subunits followed by slower addition of a variety of linkages onto the initial short K63 chain (French et al., 2017). The addition of K63 subunits is consistent with our finding that cilia-localized AMSH*, which catalyzes the removal of K63 poly-Ub, causes Smo to be retained in cilia in the absence of pathway activation. Recently, Shinde et al. (2020) also found that K63 Ub chains earmark Smo for BBSome-mediated removal from cilia.
The E3 ligase Arih2 appears to be critical in regulating the amount of Smo in the cell because its loss caused a dramatic increase in the overall level of Smo in addition to causing Smo accumulation in cilia. Loss of Arih2 in mice leads to embryonic lethality, while heterozygotes show a variety of cilia-related phenotypes, including polycystic kidney disease and bone morphology defects (Bult et al., 2019; Dickinson et al., 2016). Loss of Arih2 elevates total Smo levels suggests that Arih2 regulates the degradation of Smo, possibly through a quality control mechanism.
In addition to Arih2 and Wwp1, our studies also identified the E3 ligases Mgrn1 and Maea as important to regulation of ciliary Smo levels. Mgrn1 loss was previously shown to increase ciliary Smo levels (Pusapati et al., 2017) and has been proposed to regulate the internalization of Smo (Kong et al., 2020). Maea was identified as part of a 10-subunit complex called the glucose-induced degradation–deficient/C-terminal to LisH (GID–CTLH) complex. This complex, which includes Maea, Ypel5, Wdr26, Rmnd5a, Ranbp9, Ranbp10, Armc8, Mkln1, Gid8, and Ttyh2, plays roles in the degradation of metabolic enzymes and cell proliferation (Lampert et al., 2018). Within the GID/CTLH complex, E3 activity is thought to come from the Rmnd5, and Maea is thought to play an adaptor role because it lacks critical residues for enzymatic activity (Maitland et al., 2019). Whether Maea regulates ciliary Smo levels as part of the GID/CTLH complex or in an alternative complex will require more experimentation.
The transfer of Ub to substrates by E3 ligases requires the activity of E2-conjugating enzymes. Screening the entire family of E2-conjugating enzymes revealed that loss of Ube2g2 decreased SAG-induced Hedgehog expression, loss of Ube2i increased basal expression, and loss of Ube2l3 increased basal expression while decreasing SAG-induced expression. Of these, only Ube2l3 localized to cilia. Ube2l3 can work with both Arih2 and Wwp1 (French et al., 2017; Marteijn et al., 2009; Marteijn et al., 2005). The loss of Ube2l3 caused a large increase in Smo as detected by Western blot analysis and increased the amount of Smo localized to cilia at the basal state. We identified two isoforms of Ube2l3, and reexpression of both of these rescued the increase in total Smo levels, but only the short isoform rescued the ciliary accumulation of Smo. This suggests that Ube2l3 must be localized to cilia to function in regulating the ciliary levels of Smo.
Our previous work suggested a model where ciliary Smo levels are regulated by ubiquitination at K444 and K448 in its intracellular loop 3 (Desai et al., 2020). Our current work indicates that ciliary Smo levels are kept low at the basal state by the action of the E3 ligase Wwp1 working together with the E2-conjugating enzyme Ube2l3. These two enzymes are localized to cilia and are expected to ubiquitinate Smo because Smo diffuses into cilia when the pathway is inactive. Pathway activation promotes the removal of Wwp1 from cilia, thus allowing any Smo that enters the cilium to remain and become activated by other mechanisms (Fig. 8). The regulation of cellular and ciliary Smo levels is complex and involves other E3 ligases, including Arih2 and Mgrn1. Arih2 localizes in the cytoplasm and is likely involved in regulating Smo degradation. Mgrn1 is thought to ubiquitinate Smo for its internalization and degradation (Kong et al., 2020). Together, our work outlines a complex pathway where ciliary Smo dynamics are regulated by coordinating the cytoplasmic and ciliary Smo levels during signaling.
Figure 8.
Model for regulating ciliary Smo levels.(A) Ptch1-bound Wwp1 and the short isoform of Ube2l3 localize to cilia at the basal state and ubiquitinate any Smo that enters the cilium. Ubiquitinated Smo is the cargo for IFT, causing it to be removed from the cilium. Upon pathway activation, Ptch1 with associated Wwp1 exits the cilia, allowing Smo to accumulate in the cilia and activate the downstream steps of the pathway. Hh, Hedgehog. (B) Smo has K63-linked Ub chains at lysine 444 and lysine 448 in intracellular loop 3. The ubiquitinated Smo is recognized by the BBSome, which couples to IFT-B through Lztfl1/IFT25/IFT27 and removed out of cilia by the retrograde IFT.
Materials and methods
Plasmids
Plasmids were assembled by Gibson assembly (New England Biolabs) or the T5 exonuclease DNA assembly method (Xia et al., 2019) into the pHAGE lentiviral backbone (Wilson et al., 2008). All inserts are derived from the mouse unless otherwise stated. Inverse PCR, overlap extension PCR, and site-directed mutagenesis were performed as previously described (Lv et al., 2017). All inserts were fully sequenced and matched to the Ensembl reference sequence, National Center for Biotechnology Information reference sequence, or expected mutant forms. Plasmids are listed in Table S5, and SnapGene files will be provided upon request.
Cell culture
WT MEFs were derived from embryonic day 14 embryos and immortalized with simian virus 40 large T antigen. These cells were cultured in 95% DMEM (4.5 g/liter glucose), 5% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin (all from Gibco or Invitrogen). For the biotinylation of the Avi tag, 50 µM D-biotin was added for 24 h before fixing.
For SAG experiments, MEFs were plated at near-confluent densities and serum starved (same culture medium described above, but with 0.25% FBS) for 24 h before treatment to allow ciliation. SAG (Calbiochem) was used at 400 nM.
SHH-conditioned medium was generated from HEK 293T cells expressing Homo sapiens SHH (BL243), Xenopus tropicalis Scube2 (BL244), and Mus musculus Disp1 (BL323). Cells stably secreting SHH were grown to confluency in 90% DMEM (4.5 g/liter glucose), 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. The medium was then replaced with low-serum medium (0.25% FBS) and grown for 24 h. Medium was collected, filtered, sterilized with a 0.45-µm filter (EMD Millipore), and titered for the ability to relocate Smo to cilia. Dilutions similar in effect to 400 nM SAG were used for experiments.
For inducible protein expression from the Tc-inducible promoter, MEFs were plated at 70% confluent densities and then serum starved for 24 h. Medium was then renewed with low-serum medium (0.25% FBS) containing 1 µg/ml Tc for another 24 h.
Lentivirus production
Lentiviral vector–packaged pHAGE-derived plasmids (Wilson et al., 2008) were used for transfection. These vectors are packaged by a third-generation system comprising four distinct packaging vectors (Tat, Rev, Gag/Pol, and either vesicular stomatitis virus G or murine leukemia virus envelope) using HEK 293T cells as the host. DNA (plasmid of interest, 5 µg; Tat, 0.5 µg; Rev, 0.5 µg; Gag/Pol, 0.5 µg; vesicular stomatitis virus G or murine leukemia virus envelope, 1 µg) was delivered to the HEK cells as calcium phosphate precipitates. After 48 h, the supernatant was harvested, filtered through a 0.45-µm filter (EMD Millipore), and added to subconfluent cells. After 24 h, cells were selected with corresponding antibiotics: nourseothricin (Nat; 50 µg/ml), puromycin (Puro; 1 µg/ml), zeocin (Zeo; 500 µg/ml), or blasticidin (Bsd; 60 µg/ml for cytomegalovirus [CMV] promoter or 20 µg/ml for Gallus gallus CryD1 promoter).
Genome editing
gRNAs were selected from the Brie library (Doench et al., 2016) or designed using CHOPCHOP (Labun et al., 2019), E-CRISP (Heigwer et al., 2014), and the Dharmacon CRISPR Design Tool (https://dharmacon.horizondiscovery.com/gene-editing/crispr-cas9/crispr-design-tool/); corresponding oligonucleotides were cloned into lentiCRISPR v2 Puro (gift from Feng Zhang, Broad Institute, Cambridge, MA; Addgene plasmid 52961; Sanjana et al., 2014) or lentiCRISPR v2 PuroP93S (BL245) and screened by sequencing. lentiCRISPR v2 PuroP93S is similar to its parent except for a proline-to-serine mutation in the Puro N-acetyltransferase gene, which increases its resistance to Puro (https://www.addgene.org). The vectors were packaged into lentiviral particles and transfected into MEF cells. After selection, the pools were analyzed by flow cytometry. Individual cells were sorted into 96-well plates by flow cytometry or dilution cloning. Single mutant clones were identified with Sanger sequencing, GENEWIZ Amplicon-EZ sequencing, immunofluorescence, or immunoblotting. Sequencing results were analyzed with CRISPResso2, TIDE (Tracking of Indels by Decomposition), CRISP-ID, and SWS (Sanger sequencing and Microsoft Word wildcard searching) methods (Brinkman et al., 2014; Clement et al., 2019; Dehairs et al., 2016; Jie et al., 2017). Ty1 knock-in was achieved by using CRISPR-Cas9 genome editing. Cas9 and sgRNA were expressed from lentiCRISPR v2 Nat (OC166). The template for homology-directed repair was designed in Benchling (https://www.benchling.com/). Each homology arm was ∼800 bp. To enhance the editing efficiency, the target sequence of the gRNA and the protospacer adjacent motif sequences were added to both ends of the repair template (Zhang et al., 2017). Genome-edited cell lines were obtained and validated by Sanger sequencing and immunofluorescence. The following primers were used for detecting Ty1 knock-in of Ptch1: Ptch1KI_1, 5′-TGTGTTGCTCCGAGCTTTGT-3′; Ptch1KI_2, 5′-TCCATCCGGACTCCAGACAT-3′; Ptch1KI_3, 5′-GAACTTCAGCATCAAGCGGAT-3′; Ptch1KI_4, 5′-CATTTGCTTCTGGCCACACC-3′; Ptch1KI_5, 5′-AAAAGGTCCCCCAGGGTCTA-3′; Ptch1KI_6, 5′-ATGACCTCAGAAGCTGCCAC-3′. For the knock-in experiments, cells were transfected using a standard calcium phosphate method or Qiagen Effectene Transfection Reagent according to the manufacturer’s protocol.
Flow cytometry
For flow sorting, pelleted cells were resuspended in the corresponding media and sorted into tubes or 96-well plates with a BD FACS C-Aria II Cell Sorter (BSL-2+/BSC). For flow analysis, cells were seeded at nearly confluent densities in 12-well plates, serum starved for 24 h, and treated with or without SAG or SHH conditioned media for 48 h. Cells were trypsinized, pelleted, and then resuspended in 500 µl 1× PBS. Flow analysis was performed with a digital multiparameter flow cytometer analyzer (LSR II; BD Biosciences). Data were processed in FlowJo version 10. Ridgeline plots were drawn in RStudio with ggplot2 and ggridges (https://github.com/clauswilke/ggridges) in R 3.5.3.
Immunofluorescence
Cells were fixed with 2% PFA for 15 min, permeabilized with 0.1% Triton X-100 for 2 min, and stained as described elsewhere (Follit et al., 2006). In some cases, fixed cells were treated with 0.05% SDS for 5 min before prehybridization to retrieve antigens. The primary and secondary antibodies are described in Table S6.
Most confocal images were acquired at room temperature with an inverted microscope (TE-2000E2; Nikon) equipped with a Solamere Technology modified spinning disk confocal scan head (CSU10; Yokogawa). Three-image Z stacks were acquired at 0.2-micron intervals and converted to single planes by maximum projection with MetaMorph software (MDS Analytical Technologies). Confocal images were obtained at room temperature with an LSM910 microscope equipped with a 63× objective and converted to a maximum projection with ZEN 3.1 blue edition (Zeiss). Widefield fluorescence images were captured at room temperature using a forward-looking infrared camera on a Zeiss Axiovert 200M microscope equipped with a 100× Zeiss objective using μManager (Vale Lab, University of California, San Francisco). Images were processed in ZEN 3.1 blue edition (Zeiss) or Adobe Photoshop CS6 with brightness and contrast adjusted.
Protein and mRNA analysis
For Western blots, cells were pelleted and lysed directly with denaturing gel loading buffer (Tris-HCl 125 mM, pH 6.8, glycerol 20% vol/vol, SDS 4% vol/vol, β-mercaptoethanol 10% vol/vol, and bromophenol blue). The primary and secondary antibodies are described in Table S6. Western blots were developed by chemiluminescence (Pierce Super Signal West Dura; Thermo Fisher Scientific) and imaged using an Amersham Imager 600 imager (GE Healthcare Life Sciences). Bands were quantified with Gel-Pro Analyzer 4 (Meyer Instruments).
For immunoprecipitations, cells were serum starved for 48 h, and proteins were extracted with lysis buffer (20 mM Hepes, pH 7.5, 50 mM KCl, and 1 mM MgCl2) with 0.5% digitonin and protease inhibitor (cOmplete EDTA-Free; Roche). Insoluble components were removed by centrifugation at 20,000 g. Primary antibodies preadsorbed to protein G-sepharose beads (GE Healthcare) were added to the cell extract, and the mixture was incubated for 2 h at 4°C. After centrifugation, beads were washed with lysis buffer supplemented with 0.1% digitonin before elution in denaturing gel loading buffer for SDS-PAGE electrophoresis and Western blot analysis.
Isolation of mRNA and quantitative mRNA analysis was performed as previously described (Jonassen et al., 2008) using the primers tabulated in Table S7.
Smo ubiquitination assay
HEK 293T cells were plated at 60% confluent density onto a 10-cm plate. After 24 h, the cells were transfected with 5 µg of each vector using calcium phosphate transfection. 24 h after transfection, cells were treated with 1 µM MG132 for 4 h to block proteasomal degradation. Cells were lysed and Smo was captured as described in the immunoprecipitation method in Protein and mRNA analysis.
Statistical analysis
Statistical results were obtained from at least three independent experiments. Statistical differences between groups were tested by one-way ANOVA, two-way ANOVA, or repeated measures ANOVA in GraphPad Prism 7.04. Differences between groups were considered statistically significant if P < 0.05. Otherwise, nonsignificance was labeled on the figures. Statistical significance is denoted as *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars indicate SD.
Online supplemental material
Fig. S1 shows the development of a Hedgehog reporter line. Fig. S2 shows that Arih2, Maea, Mgrn1, or Wwp1 do not regulate the ciliary localization of Gpr161. Fig. S3 shows that Wwp1 knockout does not affect ciliogenesis. Fig. S4 shows that Arih2, Maea, Mgrn1, Ube2l3, Kctd5, Bap1, Skp2, and Skp1a do not interact with Ptch1. Fig. S5 shows that knockout of Ptch1 in GreenBomb activates the Hedgehog pathway. Table S1 provides a complete list of candidate genes and sources. Table S2 lists flow cytometry and other data from the screen. Table S3 describes subcellular localization of the Hedgehog regulators. Table S4 lists E2 ubiquitin-conjugating enzymes screened. Table S5 lists plasmids used in the study. Table S6 lists the primary and secondary antibodies and detection reagents used in the study. Table S7 lists RT-PCR and quantitative RT-PCR primers used in the study.
Supplementary Material
provides a complete list of candidate genes and sources.
lists the results of flow cytometry and other data from the screen.
describes subcellular localization of the Hedgehog regulators.
lists the E2 Ub–conjugating enzymes that were screened.
lists plasmids used in the study.
lists the primary and secondary antibodies and detection reagents used in the study.
lists RT-PCR and quantitative RT-PCR primers used in the study.
Acknowledgments
We thank Dr. Carol E. Schrader and the staff of the University of Massachusetts Medical School Flow Cytometry Core for assistance during this project. We thank Dr. Xiaoou Zhang for analyzing Amplicon-EZ next-generation sequencing data.
This work was supported by the National Institutes of Health (grant GM060992 to G.J. Pazour). Flow cytometry resources were supported by National Institutes of Health grant S10 1S10OD028576 to Carol Schrader, Ph.D., University of Massachusetts Medical School.
The authors declare no competing financial interests.
Author contributions: Conceptualization: B. Lv, O.A. Cabrera, and G.J. Pazour; methodology: B. Lv, O.A. Cabrera, M.W. Stuck, P.B. Desai, and G.J. Pazour; validation verification: B. Lv, M.W. Stuck, P.B. Desai, and G.J. Pazour; formal analysis: B. Lv, M.W. Stuck, P.B. Desai, and G.J. Pazour; investigation: B. Lv, O.A. Cabrera, M.W. Stuck, P.B. Desai, and G.J. Pazour; writing – original draft preparation: B. Lv and G.J. Pazour; writing – review and editing: B. Lv, O.A. Cabrera, M.W. Stuck, P.B. Desai, and G.J. Pazour; visualization preparation: B. Lv; supervision: G.J. Pazour; funding acquisition: G.J. Pazour.
Data availability
All data are available in the main text or the supplemental materials.
References
- Bangs, F., and Anderson K.V.. 2017. Primary cilia and mammalian Hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9:a028175. 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, D., Kovaleva E., and Schatz P.J.. 1999. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8:921–929. 10.1110/ps.8.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari, N.F., Lewis J.S., Bishop G.A., Askwith C.C., and Mykytyn K.. 2008. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA. 105:4242–4246. 10.1073/pnas.0711027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju, S., Cajanek L., Fort C., Blisnick T., Weber K., Taschner M., Mizuno N., Lamla S., Bastin P., Nigg E.A., and Lorentzen E.. 2013. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 341:1009–1012. 10.1126/science.1240985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschopf, K., Bengal E., Ziv T., Admon A., and Ciechanover A.. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17:5964–5973. 10.1093/emboj/17.20.5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow, D.K., Hoogendoorn S., Kopp A.R., Morgens D.W., Vu B.K., Kennedy M.C., Han K., Li A., Hess G.T., Bassik M.C., et al. 2018. A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat. Genet. 50:460–471. 10.1038/s41588-018-0054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman, E.K., Chen T., Amendola M., and van Steensel B.. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42:e168. 10.1093/nar/gku936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult, C.J., Blake J.A., Smith C.L., Kadin J.A., and Richardson J.E.. Mouse Genome Database Group . 2019. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47(D1):D801–D806. 10.1093/nar/gky1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Zhou Z., Ross J.S., Zhou W., and Dong J.T.. 2007. The amplified WWP1 gene is a potential molecular target in breast cancer. Int. J. Cancer. 121:80–87. 10.1002/ijc.22653 [DOI] [PubMed] [Google Scholar]
- Choudhry, Z., Rikani A.A., Choudhry A.M., Tariq S., Zakaria F., Asghar M.W., Sarfraz M.K., Haider K., Shafiq A.A., and Mobassarah N.J.. 2014. Sonic hedgehog signalling pathway: a complex network. Ann. Neurosci. 21:28–31. 10.5214/ans.0972.7531.210109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E., and Pinello L.. 2019. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37:224–226. 10.1038/s41587-019-0032-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., and Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit, K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Dehairs, J., Talebi A., Cherifi Y., and Swinnen J.V.. 2016. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep. 6:28973. 10.1038/srep28973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, P.B., Stuck M.W., Lv B., and Pazour G.J.. 2020. Ubiquitin links smoothened to intraflagellar transport to regulate Hedgehog signaling. J. Cell Biol. 219:e201912104. 10.1083/jcb.201912104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., et al. RIKEN BioResource Center . 2016. High-throughput discovery of novel developmental phenotypes. Nature. 537:508–514. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn, K.V., Hughes C.E., and Rohatgi R.. 2012. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell. 23:823–835. 10.1016/j.devcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether, T., San Agustin J.T., Keady B.T., Jonassen J.A., Liang Y., Francis R., Tobita K., Johnson C.A., Abdelhamed Z.A., Lo C.W., and Pazour G.J.. 2014. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell. 31:279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether, T., Cordelieres F.P., and Pazour G.J.. 2018. Intraflagellar transport is deeply integrated in hedgehog signaling. Mol. Biol. Cell. 29:1178–1189. 10.1091/mbc.E17-10-0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emechebe, U., Kumar P P., Rozenberg J.M., Moore B., Firment A., Mirshahi T., and Moon A.M.. 2016. T-box3 is a ciliary protein and regulates stability of the Gli3 transcription factor to control digit number. eLife. 5:e07897. 10.7554/eLife.07897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit, J.A., Tuft R.A., Fogarty K.E., and Pazour G.J.. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792. 10.1091/mbc.e06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, M.E., Klosowiak J.L., Aslanian A., Reed S.I., Yates J.R. III, and Hunter T.. 2017. Mechanism of ubiquitin chain synthesis employed by a HECT domain ubiquitin ligase. J. Biol. Chem. 292:10398–10413. 10.1074/jbc.M117.789479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Takebe N., and Lorusso P.. 2010. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2:237–250. 10.1177/1758834010366430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft, C.J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E.J., and Yoder B.K.. 2005. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1:e53. 10.1371/journal.pgen.0010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer, F., Kerr G., and Boutros M.. 2014. E-CRISP: fast CRISPR target site identification. Nat. Methods. 11:122–123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Hoshino, S., Kobayashi M., Tagawa R., Konno R., Abe T., Furuya K., Miura K., Wakasawa H., Okita N., Sudo Y., et al. 2020. WWP1 knockout in mice exacerbates obesity-related phenotypes in white adipose tissue but improves whole-body glucose metabolism. FEBS Open Bio. 10:306–315. 10.1002/2211-5463.12795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia, E.Y., Gui Y., and Zheng X.. 2015. Regulation of Hedgehog signaling by ubiquitination. Front. Biol. (Beijing). 10:203–220. 10.1007/s11515-015-1343-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., and Anderson K.V.. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 426:83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Ishikawa, H., Thompson J., Yates J.R. III, and Marshall W.F.. 2012. Proteomic analysis of mammalian primary cilia. Curr. Biol. 22:414–419. 10.1016/j.cub.2012.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, J., Tong C., Wang B., Luo L., and Jiang J.. 2004. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 432:1045–1050. 10.1038/nature03179 [DOI] [PubMed] [Google Scholar]
- Jiang, W., Yao X., Shan Z., Li W., Gao Y., and Zhang Q.. 2019. E3 ligase Herc4 regulates Hedgehog signalling through promoting Smoothened degradation. J. Mol. Cell Biol. 11:791–803. 10.1093/jmcb/mjz024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie, H., Li Z., Wang P., Zhao L., Zhang Q., Yao X., Song X., Zhao Y., and Yao S.. 2017. A simple method based on Sanger sequencing and MS Word wildcard searching to identify Cas9-induced frameshift mutations. Lab. Invest. 97:1500–1507. 10.1038/labinvest.2017.83 [DOI] [PubMed] [Google Scholar]
- Jin, J., Li X., Gygi S.P., and Harper J.W.. 2007. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 447:1135–1138. 10.1038/nature05902 [DOI] [PubMed] [Google Scholar]
- Jonassen, J.A., San Agustin J., Follit J.A., and Pazour G.J.. 2008. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 183:377–384. 10.1083/jcb.200808137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keady, B.T., Samtani R., Tobita K., Tsuchya M., San Agustin J.T., Follit J.A., Jonassen J.A., Subramanian R., Lo C.W., and Pazour G.J.. 2012. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell. 22:940–951. 10.1016/j.devcel.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Hsia E.Y.C., Brigui A., Plessis A., Beachy P.A., and Zheng X.. 2015. The role of ciliary trafficking in Hedgehog receptor signaling. Sci. Signal. 8:ra55. 10.1126/scisignal.aaa5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., and Tanaka K.. 2010. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147:793–798. 10.1093/jb/mvq044 [DOI] [PubMed] [Google Scholar]
- Komander, D., and Rape M.. 2012. The ubiquitin code. Annu. Rev. Biochem. 81:203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kong, J.H., Young C.B., Pusapati G.V., Patel C.B., Ho S., Krishnan A., Lin J.I., Devine W., Moreau de Bellaing A., Athni T.S., et al. 2020. A membrane-tethered ubiquitination pathway regulates Hedgehog signaling and heart development. Dev. Cell. 55:432–449.e12. 10.1016/j.devcel.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K.G., Johnson K.A., Forscher P., and Rosenbaum J.L.. 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 90:5519–5523. 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun, K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., and Valen E.. 2019. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47(W1):W171–W174. 10.1093/nar/gkz365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert, F., Stafa D., Goga A., Soste M.V., Gilberto S., Olieric N., Picotti P., Stoffel M., and Peter M.. 2018. The multi-subunit GID/CTLH E3 ubiquitin ligase promotes cell proliferation and targets the transcription factor Hbp1 for degradation. eLife. 7:e35528. 10.7554/eLife.35528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.-F., Johnson E.C., Sakai T., Cochran D., Ballif B.A., Rush J., Pazour G.J., Ikebe M., and Witman G.B.. 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187:1117–1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Chen Y., Shi Q., Yue T., Wang B., and Jiang J.. 2012. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol. 10:e1001239. 10.1371/journal.pbio.1001239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Xie P., Lu L., Wang J., Diao L., Liu Z., Guo F., He Y., Liu Y., Huang Q., et al. 2017. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat. Commun. 8:347. 10.1038/s41467-017-00299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Cho Y.S., Wang B., Li S., and Jiang J.. 2018a. Regulation of Smoothened ubiquitylation and cell surface expression through a Cul4-DDB1-Gβ E3 ubiquitin ligase complex. J. Cell Sci. 131:jcs218016. 10.1242/jcs.218016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Li S., Wang B., and Jiang J.. 2018b. Hedgehog reciprocally controls trafficking of Smo and Ptc through the Smurf family of E3 ubiquitin ligases. Sci. Signal. 11:eaan8660. 10.1126/scisignal.aan8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew, G.M., Ye F., Nager A.R., Murphy J.P., Lee J.S., Aguiar M., Breslow D.K., Gygi S.P., and Nachury M.V.. 2014. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell. 31:265–278. 10.1016/j.devcel.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, B., Wan L., Taschner M., Cheng X., Lorentzen E., and Huang K.. 2017. Intraflagellar transport protein IFT52 recruits IFT46 to the basal body and flagella. J. Cell Sci. 130:1662–1674. 10.1242/jcs.200758 [DOI] [PubMed] [Google Scholar]
- Maitland, M.E.R., Onea G., Chiasson C.A., Wang X., Ma J., Moor S.E., Barber K.R., Lajoie G.A., Shaw G.S., and Schild-Poulter C.. 2019. The mammalian CTLH complex is an E3 ubiquitin ligase that targets its subunit muskelin for degradation. Sci. Rep. 9:9864. 10.1038/s41598-019-46279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marada, S., Navarro G., Truong A., Stewart D.P., Arensdorf A.M., Nachtergaele S., Angelats E., Opferman J.T., Rohatgi R., McCormick P.J., and Ogden S.K.. 2015. Functional divergence in the role of N-linked glycosylation in Smoothened signaling. PLoS Genet. 11:e1005473. 10.1371/journal.pgen.1005473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn, J.A., van Emst L., Erpelinck-Verschueren C.A., Nikoloski G., Menke A., de Witte T., Löwenberg B., Jansen J.H., and van der Reijden B.A.. 2005. The E3 ubiquitin-protein ligase Triad1 inhibits clonogenic growth of primary myeloid progenitor cells. Blood. 106:4114–4123. 10.1182/blood-2005-04-1450 [DOI] [PubMed] [Google Scholar]
- Marteijn, J.A., van der Meer L.T., Smit J.J., Noordermeer S.M., Wissink W., Jansen P., Swarts H.G., Hibbert R.G., de Witte T., Sixma T.K., et al. 2009. The ubiquitin ligase Triad1 inhibits myelopoiesis through UbcH7 and Ubc13 interacting domains. Leukemia. 23:1480–1489. 10.1038/leu.2009.57 [DOI] [PubMed] [Google Scholar]
- McCullough, J., Clague M.J., and Urbé S.. 2004. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166:487–492. 10.1083/jcb.200401141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen, T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F., et al. 2013. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 154:169–184. 10.1016/j.cell.2013.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick, D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., and Nachury M.V.. 2015. Proteomics of primary cilia by proximity labeling. Dev. Cell. 35:497–512. 10.1016/j.devcel.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., and Jackson P.K.. 2010. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24:2180–2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Wen X., Ratti N., Loktev A., Rangell L., Scales S.J., and Jackson P.K.. 2013. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 152:210–223. 10.1016/j.cell.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Nagai, T., Mukoyama S., Kagiwada H., Goshima N., and Mizuno K.. 2018. Cullin-3-KCTD10-mediated CEP97 degradation promotes primary cilium formation. J. Cell Sci. 131:jcs219527. 10.1242/jcs.219527 [DOI] [PubMed] [Google Scholar]
- Nijman, S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., and Bernards R.. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell. 123:773–786. 10.1016/j.cell.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Nozaki, S., Katoh Y., Kobayashi T., and Nakayama K.. 2018. BBS1 is involved in retrograde trafficking of ciliary GPCRs in the context of the BBSome complex. PLoS One. 13:e0195005. 10.1371/journal.pone.0195005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki, S., Castro Araya R.F., Katoh Y., and Nakayama K.. 2019. Requirement of IFT-B-BBSome complex interaction in export of GPR161 from cilia. Biol. Open. 8:bio043786. 10.1242/bio.043786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik, B., Grimwood J., Aerts A., Rouzé P., Salamov A., Putnam N., Dupont C., Jorgensen R., Derelle E., Rombauts S., et al. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA. 104:7705–7710. 10.1073/pnas.0611046104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Wilkerson C.G., and Witman G.B.. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141:979–992. 10.1083/jcb.141.4.979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Agrin N., Leszyk J., and Witman G.B.. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, R., and Joyner A.L.. 2014. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development. 141:3445–3457. 10.1242/dev.083691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M.2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533. 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Pusapati, G.V., Kong J.H., Patel B.B., Krishnan A., Sagner A., Kinnebrew M., Briscoe J., Aravind L., and Rohatgi R.. 2017. CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen Sonic Hedgehog. Dev. Cell. 44:113–129.e8. 10.1016/j.devcel.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi, R., Milenkovic L., and Scott M.P.. 2007. Patched1 regulates hedgehog signaling at the primary cilium. Science. 317:372–376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Rosenbaum, J.L., and Witman G.B.. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Sanjana, N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir, P., and Christensen S.T.. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69:377–400. 10.1146/annurev.physiol.69.040705.141236 [DOI] [PubMed] [Google Scholar]
- Sato, Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., and Fukai S.. 2008. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 455:358–362. 10.1038/nature07254 [DOI] [PubMed] [Google Scholar]
- Scholey, J.M.2003. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 19:423–443. 10.1146/annurev.cellbio.19.111401.091318 [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., and Harper J.W.. 2009. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10:319–331. 10.1038/nrm2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde, S.R., Nager A.R., and Nachury M.V.. 2020. Ubiquitin chains earmark GPCRs for BBSome-mediated removal from cilia. J. Cell Biol. 219:e202003020. 10.1083/jcb.202003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg, M.A., Menchen T., Lee C., Johnson J., Jungnickel M.K., Choksi S.P., Garcia G. III, Busengdal H., Dougherty G.W., Pennekamp P., et al. 2017. Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways. Dev. Cell. 43:744–762.e11. 10.1016/j.devcel.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, J.J., Ou G., Gunnarson A.L., Walker M.R.S., Zhou H.M., Brust-Mascher I., and Scholey J.M.. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 6:1109–1113. 10.1038/ncb1186 [DOI] [PubMed] [Google Scholar]
- Tan, X.D., Pan M., Gao S., Zheng Y., Shi J., and Li Y.M.. 2017. A diubiquitin-based photoaffinity probe for profiling K27-linkage targeting deubiquitinases. Chem. Commun. (Camb.). 53:10208–10211. 10.1039/C7CC05504H [DOI] [PubMed] [Google Scholar]
- van Wijk, S.J., and Timmers H.T.. 2010. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24:981–993. 10.1096/fj.09-136259 [DOI] [PubMed] [Google Scholar]
- Wilson, A.A., Kwok L.W., Hovav A.H., Ohle S.J., Little F.F., Fine A., and Kotton D.N.. 2008. Sustained expression of alpha1-antitrypsin after transplantation of manipulated hematopoietic stem cells. Am. J. Respir. Cell Mol. Biol. 39:133–141. 10.1165/rcmb.2007-0133OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, R., Jia H., Fan J., Liu Y., and Jia J.. 2012. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 10:e1001238. 10.1371/journal.pbio.1001238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., Li K., Li J., Wang T., Gu L., and Xun L.. 2019. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 47:e15. 10.1093/nar/gky1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.-P., Li X.-L., Li G.-H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.-W., et al. 2017. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18:35. 10.1186/s13059-017-1164-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Yao X., Pang S., Chen P., Jiang W., Shan Z., and Zhang Q.. 2018. The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J. Mol. Cell Biol. 10:243–257. 10.1093/jmcb/mjx036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides a complete list of candidate genes and sources.
lists the results of flow cytometry and other data from the screen.
describes subcellular localization of the Hedgehog regulators.
lists the E2 Ub–conjugating enzymes that were screened.
lists plasmids used in the study.
lists the primary and secondary antibodies and detection reagents used in the study.
lists RT-PCR and quantitative RT-PCR primers used in the study.
Data Availability Statement
All data are available in the main text or the supplemental materials.