Abstract
Recent outbreak of 2019 novel Corona virus poses serious challenge for the global health system. In lieu of paucity of experimental data, tools and the very basic understanding of host immune responses against SARS-CoV-2, well thought effective measures are needed to control COVID-19 pandemic. We have identified specific overlapping antigenic peptide epitopes (OAPE) within the 4 structural proteins of SARS-CoV-2 predictive of triggering robust CD4 and CD8 T cell responses in host using bio-informatics tools (NetMHC4.0, IEDB, and Vaxijen2.0). We speculate an early release of pro-inflammatory cytokines for protection and later release of anti-inflammatory cytokines for prevention of immunopathology in designing a vaccine for Covid-19. Therefore, the selected immunogenic OAPE were subjected to in silico tools (IL-6-Pred, IFNepitope and PIP-EL) for analyzing their pro-inflammatory response. The OAPEs found to be pro-inflammatory in nature were further subjected to prediction servers (IL-4-Pred, IL-10-Pred, Pre-AIP) to characterize them as inducers of anti-inflammatory response as well. We finally filtered out 12 OAPE which had affinity for both CD4 and CD8 T cells as well as were inducers of pro-inflammatory and anti-inflammatory cytokines. On confirmation of OAPE binding affinity for respective T cell specific MHC allele using docking studies (pepATTRACT, Hex8.0 and Discovery studio) they were found to be have more immunogenic potential than the 3 negative control peptides (NCPs) included in the study. Additionally, we constructed CTxB-adjuvanated multi-epitopic vaccine inclusive of the 12 OAPEs which was non-toxic, non-allergenic and capable of inducing both pro-inflammatory and anti-inflammatory cytokines. A successful in silico cloning and docking of modeled subunit vaccine construct with toll like receptor-2 (TLR-2) confirmed the high efficacy of our multi-epitopic vaccine which can through a balanced interplay of cytokines help in creating a steady-state immune equilibrium. In silico immune simulation studies with the vaccine using C-ImmSim server also showed higher percentage of T cells along with production of pro-inflammatory as well as some anti-inflammatory cytokines. Experimental validation of this prediction based study on Peripheral Blood Mononuclear Cells (PBMCs) of un-infected individuals, patients and recovered individuals will facilitate production of high priority effective SARS -CoV-2 vaccine candidate.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40203-021-00098-7.
Keywords: SARS-CoV-2, COVID-19, Anti-inflammatory T cell epitopes, Peptide-based vaccine candidates, In silico cloning
Introduction
Pandemic or the worldwide spread of a new disease has been a crucial part in shaping the history of mankind since ages. The consequences of previous pandemics including the very recent 2009 H1N1 Swine flu has been very destructive but the havoc created by ongoing Corona virus disease 2019 (COVID-19) is certainly unprecedented. The causative agent of COVID-19 is severe acute respiratory syndrome corona virus 2 (SARS-CoV-2). The lipid envelope of these viruses are embedded with fringed projections which appears as crown and hence the name. CoVs possess a 5'capped single-strand positive-sense RNA with unusually large RNAgenome∼30 kb (Belouzard et al. 2012). CoVs encodes replicase proteins within 6 to 10 open reading frames (ORFs) and 4 main structural proteins namely spike protein, membrane protein, envelope protein, and nucleocapsid protein.The membrane and envelope proteins are involved in virus assembly and the spike protein is involved in the viral entry. The nucleocapsid is a distinct protein of CoVs, which favors the survival of virus by modulating various host mediated antiviral processes.
SARS-CoV-2 enters the host respiratory epithelial cells by adhering to angiotensin-converting enzyme 2 (ACE2) receptors on host cell (Zhou et al. 2020).The serine protease, trans-membrane protease serine 2 (TMPRSS2) present in the host cleaves viral spike protein into two subunits S1 and S2. The S2 subunit assist in fusion of viral and host cell membranes (Hoffmann et al. 2020). Following receptor binding, the virus enters the cell cytoplasm via endocytosis where it blocks various antiviral pathways which ultimately leads to defective type 1 interferon gene induction (Ou et al. 2019). This results in downstream enhanced NF-κB activation, pro-inflammatory cytokine production, and necroptosis (Siu et al. 2009). These alterations in signaling cascade leads to increased cell death, hyper-inflammation, and cytokine storm.
Once the virus is inside the host, viral antigens are processed into smaller peptide fragments and mounted on Major Histocompatibility Complex (MHC) molecules. MHC class II loaded with peptide epitope prime the CD4 helper T cells while MHC class I loaded peptide epitope prime the CD8 T cells. Studies have reported a higher frequency of SARS-CoV-2 specific CD4 T cells in patients with severe infection while a higher proportion of CD8 T cells in mild disease and patients who had recovered from COVID-19 (Grifoni et al. 2020; Sekine et al. 2020). Furthermore, CD8 T cell dominated over CD4 T cell in terms of frequency and magnitude of the responses. This observation suggests that while CD8 T cells imparts protective immunity during mild SARS-CoV-2 infection, excessive CD4 T cells stimulation causes immunopathology of COVID-19 during severe infection (Dong et al. 2020). Therefore, in case of SARS-CoV-2, inclusion of CD8 T cell epitope based vaccine becomes crucial in the absence of CD4 T cell mediated immunopathology. Moreover, CD8 T cell epitope based vaccine not only helps in identification and killing of viral infected host cells but also naturally supplements humoral immunity (Cosma and Eisenlohr 2018). Nevertheless, the role of CD4 T cell epitope based viral vaccine cannot be overlooked given their ability to enhance activation of CD8 T cell, inducing protective CD8 T cell memory and prolonging their antiviral function (Phares et al. 2012). Additionally, CD4 T cells also trigger antigen-specific B cells immunoglobulin class switching and affinity maturation (Campbell et al. 2020).Yet, an exaggerated and distinctive pro-inflammatory response is also reported with reference to severity of COVID-19 disease (Johnson and Laloraya 2020). Nonetheless, balance in the pro-inflammatory and anti-inflammatory immune responses determines the outcome of any bacterial or viral infection. We contemplate designing of CD4 and CD8 T cell specific peptide based SARS-CoV-2 vaccine candidates which are pro-inflammatory as well as anti-inflammatory in nature so as to counter the immunopathology of COVID-19 and elicit a protective immune response. The concept implicates creating a balance of pro-inflammatory and anti-inflammatory cytokine response for a favorable outcome along with killing of infected cells.
We found a huge repertoire of MHC restricted antigenic T cell epitopes within the 4 structural proteins of SARS-CoV-2. Shortlisting of OAPE having affinity for both classes I and class II MHC alleles makes our approach more stringent and inclusive. Almost all the selected OAPEs were characterized to be both pro-inflammatory as well as anti-inflammatory cytokine inducers. In comparison, the three negative control peptides included in the study were non-antigenic, have low binding affinity for MHC alleles and mostly pro-inflammatory in nature. Docking studies confirmed the strong interaction between the selected OAPEs and common MHC alleles in population. The in silicocloned and modelled vaccine construct was observed to bind in the major groove within the active sites of TLR-2, an important immune receptor. Furthermore in silico simulation studies with our multi-epitope vaccine resulted in Th1 type immune response along with production of certain anti-inflammatory cytokines. These results substantiate the efficacy of our vaccine candidate which can elicit strong protective immunity and preventing disease immunopathology.
Materials and methods
Sequence retrieval and identification of T cell epitopes within the SARS-CoV-2 proteins
The amino acid sequence for 4 putative proteins of SARS-CoV-2 was retrieved from NCBI database with following accession numbers: (envelope protein [E]—YP_009724392.1; spike protein [S] —YP_009724390.1; nucleocapsid phosphoprotein [N]—YP_009724397.2 and membrane glycoprotein [M]—YP_009724393.1).
For identifying MHC Class I restricted CD8 + T cell binding epitopes, 9mer peptides were selected from NetMHC4.0 software (Nielsen et al. 2003; Lundegaard et al. 2008). NetMHC4.0 is based on artificial neural networks and has 34 HLA-A, 33 HLA-B, and 10 HLA-C alleles. A default threshold value of 0.5% for strong binders and 2% for weak binders is recommended in NetMHC4.0. We found many epitopes with strong binding affinity exclusively for HLA-C and weak or no affinity for HLA-A and HLA-B. Because of higher population coverage, binding predictability with only HLA-A and HLA-B were included in this study. Extensively used immunological database IEDB was used for recognition of 15mer peptide epitopes having affinity for MHC Class II restricted CD4 + T cells (Fleri et al. 2017). SMM-align stabilization matrix algorithm was adopted in IEDB which predicts affinities for peptide: MHC complex (Nielsen et al. 2007). A total of 15 HLA DR alleles are covered under IEDB. Only strong binders with half maximal inhibitory concentration (IC) ≤ 250 were selected. Any cut off within the IC ≤ 500 is recommended as this range includes about 90% of immunogenic epitopes (Fleri et al. 2017).
Shortlisting of promiscuous and antigenic T cell epitopes
Promiscuity is defined as affinity of a given MHC molecule to bind various peptides and a promiscuous peptide epitope can be recognized by several different MHC molecules (Brusic et al. 1998). Peptides which bound to five or more alleles in both NetMHC4.0 and IEDB were considered promiscuous.Vaxijen2.0 server was employed for predicting the antigenic score for all the promiscuous peptide epitopes (Doytchinova and Flower 2007). The server recommends a default score of ≥ 0.4 for probable antigenic peptides. Top 5 promiscuous and antigenic peptides were shortlisted from both NetMHC4.0 and IEDB.
Shortlisting of OAPE (overlapping antigenic peptide T cell epitopes)
Each SARS CoV-2 protein was also scanned for the presence of any OAPE having affinity for both class I and class II MHC alleles. We have adopted the following criteria for selecting OAPE:
Promiscuous peptides in both NetMHC and IEDB.
Promiscuous in NetMHC and can bind 3 to 4 alleles in IEDB.
Promiscuous in IEDB and can bind 3 to 4 alleles in NetMHC.
Peptide epitopes which bind to 3 to 4 alleles in both NetMHC and IEDB.
All the selected OAPE were predicted antigens having vaxijen score of ≥ 0.4.
We included three peptides as negative control (NCPs) in our study to compare the immunogenicity of our shortlisted OAPEs:
NCP1-‘ARSVASQSI’ from Spike protein.
NCP2-‘KEELDKYFK’ from Spike protein.
NCP3-‘VSEETGTLI’ from Envelope protein.
All the selected NCPs were predicted non-antigens having vaxijen score of ≤ 0.4.
Prediction of OAPEs to be inducers of pro-inflammatory cytokines
We also subjected our shortlisted OAPE to other online servers like IL-6-Pred (Dhall et al. 2020), IFNepitope (Dhanda et al. 2013b) and PIP-EL (Manavalan et al. 2018). IL-6-Pred is designed on a wide range of machine learning techniques out of which Random Forest-based model achieves a maximum accuracy on dataset to see if they are predicted to be secreting IL-6. The default value of > 0.11 is recommended to consider a peptide being IL-6 inducer. IFNepitope uses SVM algorithm like IL-4-Pred server and a default value of more than 0.0 qualifies a peptide as IFN-γ inducer. PIP-EL server is designed as a cumulative datasets including ten independent models for amino acid sequence, di-peptide composition, distribution-transition and physiochemical properties. In PIP-EL, a peptide was considered as pro-inflammatory if it can induce pro-inflammatory cytokines like IL-8, IL-12, IL-18, IFN-γ and TNF-α. The output score is a probability where any value of more than 0.45 considers a peptide to be inducer of pro-inflammatory cytokine.
Prediction of OAEPs as anti-inflammatory T cell epitopes
The shortlisted overlapping antigenic peptides of four SARS CoV-2 proteins were evaluated in-silico for being anti-inflammatory in nature. These peptides were subjected to IL-4-Pred (Dhanda et al. 2013a) and IL-10-Pred (Nagpal et al. 2017) server which predicts the ability of these peptides to induce Interleukin-4 (IL-4) and Interleukin -10 (IL-10) respectively. These servers consist of experimentally validated epitopes from IEDB database. IL-4-Pred is based on SVM (Support Vector Machine based methods) and motif algorithm which incorporates the amino acid composition and propensity, di-peptide composition and physico-chemical properties in a peptide for prediction. IL-10-Pred is also based on composition based model using machine learning techniques like Random forest method which has maximum accuracy. In IL-4-Pred a default threshold value of > 0.2 is recommended while in IL-10-Pred a default threshold value of > 0.3 is recommended. Further, using Pre-AIP (Prediction of Anti-Inflammatory Peptides) server (Khatun et al. 2019), these peptides were characterized for being anti-inflammatory in nature. Pre-AIP systematically investigates different types of characters which includes primary sequence, evolutionary and structural information through a random forest classifier (Khatun et al. 2019). In Pre-AIP a peptide was considered as anti-inflammatory if it can induce anti-inflammatory cytokines like IL-10, IL-4, IL-13, IL-22 and TGF-β. In Pre-AIP; a score of ≥ 0.468 was labelled as high confidence AIP, 0.468 > score > 0.388 was labelled as medium confidence AIP and 0.388 > score > 0.342is labelled as low confidence AIP.
Peptide MHC docking by pepATTRACT and Hex
Docking studies were performed to define the binding of top scoring overlapping antigenic promiscuous peptides to their respective class I and class II alleles. MHC Class I allele-A2 supertype (PDB-1S9Y) and MHC Class II allele, HLA-DRB1*0101 (PDB ID-4MCZ) were included for docking studies because of their wide population coverage. Initially, the docking was carried out in pepATTRACT server (Schindler et al. 2015; De Vries et al. 2017) and the linear peptide and MHC molecule were separated from the docked structure. The linear peptide and MHC molecule were then docked again using Hex server (Macindoe et al. 2010) to obtain an energy score. The control peptide was a cytotoxic T lymphocyte (CTL) epitope (SLLMWITQS) belonging to NYESO-1 testicular cancer antigen for A2 supertype allele. For MHC class II, the control peptide was a rheumatoid arthritis epitope “GVYATRSSAVRLR”. A thorough analysis of binding energy (Kcal/mol) of the test and control peptide with the HLA allele was performed. The docked structures obtained from both the software were visualized using Discovery Studio Visualizer 4.1 (Accelyrs Inc., USA). The binding sites were also analyzed for the hydrogen bonds and hydrophobic interactions between the amino acid residues of peptide and the HLA molecule.
Designing of the T cell specific multi-epitopic adjuvanated vaccine against Covid-19
The proposed vaccine construct was designed by linking an adjuvant to the ten selected OAPEs inter-linked by specific linkers for proper demarcation of epitopes. The adjuvant used in this vaccine is Cholera Toxin Subunit B or CTxB. It is the B subunit of the cholera toxin and helps in translocation of Cholera Toxin Subunit A or CTxA. The use of CTxB as adjuvant provides proper stability to vaccine construct, increases the efficiency of its cellular uptake and antigen presentation (Walker et al. 2016). A stretch of ‘KPKPKP’ linker was used to join the adjuvant and OAPEs. Intra OAPEs were joined by AAY stretch.
Codon adaptation and in-silico cloning of vaccine construct
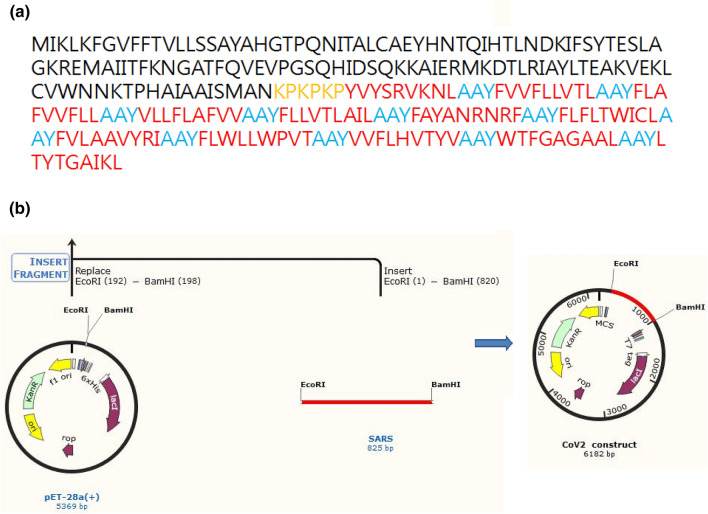
Java Codon Adaptation Tool (JCAT) (Grote et al. 2005) was employed for codon optimization and checking the expression of vaccine construct in E. coli strain K12. NEBcutter (Vincze et al. 2003) allowed us to choose the desired restriction enzyme cleavage sites in case of cloning performed in expression vector pET28a( +). In silico clone of the recombinant vaccine construct was generated using the SnapGene 1.1.3 restriction cloning tool.
Predicting physicochemical properties of designed vaccine construct
ProtParam tool of the ExPASy database server (Gasteiger et al. 2005) was used to evaluate the physiochemical characteristics of the vaccine construct.
Predicting toxicity and allergenicity of the vaccine construct
AllerTOP v2.0 (Dimitrov et al. 2014a) and AllergenFPv.1.0 (Dimitrov et al. 2014b) was used to evaluate whether the designed vaccine construct was allergenic. The toxicity of the vaccine construct was also analyzed using the ToxinPred server (Gupta et al. 2013). SVM based method with a default threshold value of 0.0 was chosen for predicting the toxicity and values greater than 0.0 was considered toxic.
Predicting pro-inflammatory and anti-inflammatory nature of the vaccine construct
The entire vaccine construct was subjected to Pre-AIP and PIP-EL servers to evaluating their anti-inflammatory and pro-inflammatory nature.
In silico structural modeling, validation, and docking of vaccine construct with host immune receptor
Secondary structure prediction and modeling of vaccine construct was done in I-TASSER (Roy et al. 2010). All the five models were subjected for structural validation using PROCHEK (Laskowski et al. 1996), ERRAT (Colovos and Yeates 1993) and Verify3D (Bowie et al. 1991) web servers available at UCLA Saves. The C score depicts the accuracy of model and is based on alignment and structure assembly simulations. Considering a value between −5 to 2; a higher C score signifies a higher confidence of model. Procheck evaluates the stereo-chemical quality of the structure via Ramachandran plot and Errat evaluates summative quality factor of the structure. Verify3D evaluates the protein structure based on comparisons with best models. Molecular docking studies were performed to confirm the binding affinity between the target vaccine construct and immune receptor Toll-like Receptor-2 (TLR-2) (PDB ID: 2Z80) using Hex 8.0.0. and Studio Visualizer 4.1 (Accelyrs Inc., USA).
In silico immune simulation studies in response to our vaccine
C-ImmSim server was used to predict the mammalian immune response on administration of our vaccine construct (Rapin et al. 2010). The time steps denote the number of times of simulation and each time step is approximately of 8 h. Time step value was selected to be 1000. Two injections of the target vaccine were administered at intervals of 4 weeks. Therefore, first dose was administered at time step 1 and the booster dose was administered at time step 84. The simulation volume and the steps were set at 50 µl with a random seed of 12,345 respectively. LPS was excluded from the injection.
Results
Identification and shortlisting of promiscuous, antigenic T cell epitopes
Among the four proteins of SARS Cov2, Surface glycoprotein (S) or Spike protein was the largest in size generating a total of 1265 nonamers with a step size of one amino acid residue. Envelope protein (E) was the smallest with a total of 67 overlapping nonamers. Number of binding epitopes was also highest in Spike (S) protein and lowest in Envelope protein (E). NetMHC4.0 predicts epitopes for total 70 MHC class I alleles including 36 alleles of HLA-A and 34 alleles of HLA-B using artificial neural network. Spike protein bound to all the alleles in NetMHC4.0 followed by Nucleocapsid phosphoprotein (N) which bound to 64 alleles. A total of 15 HLA-DRB alleles are documented in IEDB and the prediction is based on SMM align algorithm. Peptides of Spike protein (S), Membrane glycoprotein (M), and Nucleocapsid phosphoprotein (N) showed binding affinity for all 15 HLA-DRB alleles while peptides of Envelope protein (E) bound to 14 HLA-DRB alleles. The binding profile for all the selected proteins along with their evaluation parameters have been enlisted in Table 1. A detailed description of all the promiscuous peptides generated in both NetMHC and IEDB are enlisted in Table S2 and S3 respectively. Spike protein (S) generated maximum of 119 promiscuous epitopes (90 in NetMHC4.0 and 29 in IEDB) out of which 64 promiscuous epitopes were also antigenic. Despite the smallest size of Envelope protein (E); it generated 20 promiscuous antigenic epitopes (17 in NetMHC4.0 and 3 in IEDB) which was higher than 17 and 10 promiscuous epitopes generated by Membrane glycoprotein (M), and Nucleocapsid phosphoprotein (N) respectively (Table 1).
Table 1.
Prediction of CD4+ and CD8 + T cell binding epitopes of the selected SARS Cov2 proteins
| Software | Parameter | Envelope (E) protein | Membrane (M)protein | Spike (S) protein | Nucleocapsid (N) protein |
|---|---|---|---|---|---|
| NETMHC4.0 [Prediction of MHC Class I binders] | Overlapping nonamers | 67 | 214 | 1265 | 411 |
| Total number of binding epitopes | 33 | 103 | 411 | 101 | |
| Number of alleles bound | 62 | 61 | 70 | 64 | |
| Number of promiscuous epitopes | 23 | 26 | 90 | 20 | |
| Number of promiscuous antigenic epitopes | 17 | 13 | 50 | 9 | |
| IEDB [Prediction of MHC Class II binders] | Overlapping nonamers | 67 | 214 | 1265 | 411 |
| Total number of binding epitopes | 37 | 75 | 349 | 68 | |
| Number of alleles bound | 14 | 15 | 15 | 15 | |
| Number of promiscuous epitopes | 3 | 7 | 29 | 3 | |
| Number of promiscuous antigenic epitopes | 3 | 4 | 14 | 1 | |
| Vaxijen 2.0 [Prediction of whole protein antigenicity] | Antigenic score | 0.6025 | 0.5102 | 0.4646 | 0.5059 |
Spike protein (S) generated maximum number of overlapping nonamers and binding epitopes among all the four proteins. Because of its smallest size, Envelope (E) protein generated least overlapping nonamers and binding epitopes. Using NetMHC4.0: Spike protein (S) and Nucleocapsid (N) phosphoprotein bound to maximum number and Membrane (M) glycoprotein to least number of alleles. A total of 89 promiscuous antigenic epitopes were found in all the 4 proteins. Using IEDB: Surface glycoprotein or (S) Spike protein, Nucleocapsid (N) phosphoprotein and Membrane (M) glycoprotein bound to 15 class II alleles. A total of 22 promiscuous antigenic epitopes were found in all the 4 proteins. Vaxijen score of > 0.4 in all the 4 proteins shows their probable antigenic nature
For MHC class I and II, we have shortlisted top scoring promiscuous and antigenic peptides from both NetMHC4.0 and IEDB respectively. At least four top scoring promiscuous antigenic peptides were selected from all the four proteins in NetMHC4.0. Except for Nucleocapsid phosphoprotein (N) which included only 1 promiscuous antigenic epitope, remaining three proteins generated at least four top scoring promiscuous antigenic peptide epitopes in IEDB (Table 1).
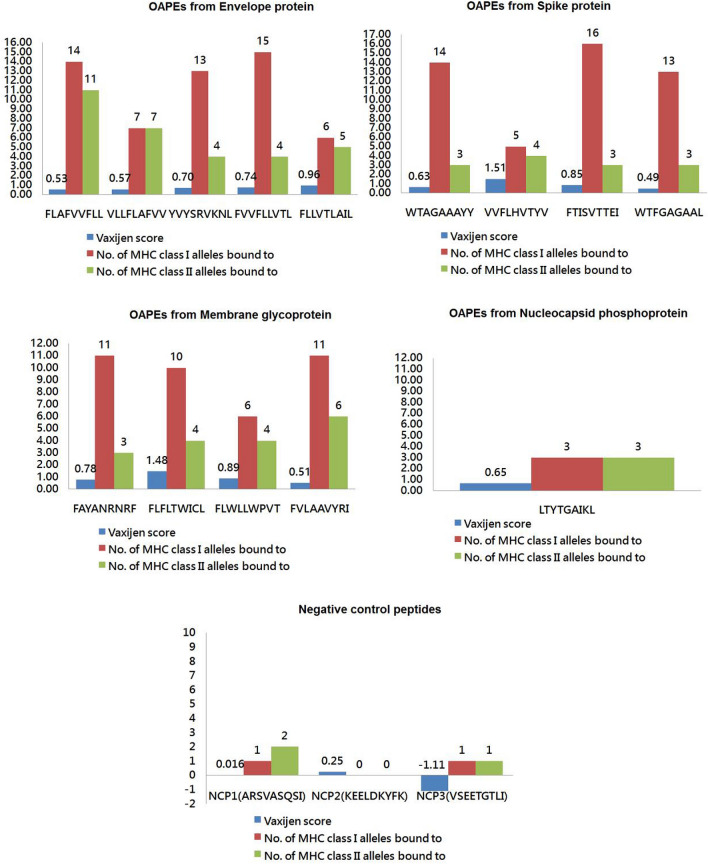
Additionally, all the four proteins were scanned for the presence of any OAPE having affinity for both class I and class II MHC alleles (Fig. 1 and Table S1). Two top scoring antigenic peptides in NetMHC4.0 and IEDB namely ‘FLAFVVFLL’ ‘VLLFLAFVV’ and peptide ‘FLLVTLAIL’ of Envelope protein (E) were promiscuous for both class I and II alleles and hence included as overlapping peptide epitopes. Two top scoring peptides in NetMHC4.0 and IEDB namely ‘FVLAAVYRI’ ‘FLFLTWICL’ of Membrane glycoprotein (M) were promiscuous for both class I and II alleles and were included as overlapping peptide epitopes. Except Nucleocapsid phosphoprotein (N), at least four overlapping peptide epitopes were shortlisted from all the SARS Cov2 proteins depending on the set criteria. For Nucleocapsid phosphoprotein (N), we considered peptide ‘LTYTGAIKL’ as overlapping epitope and it showed binding affinity for three class I and class II alleles each. A total of 14 OAPE were selected for further analysis. Out of the three selected NCPs, NCP2 (KEELDKYFK) of Spike protein had no binding affinity for class I and class II MHC alleles with a negative vaxijen score. NCP1 and NCP3 bound to 1 to 2 MHC class I and class II alleles and the vaxijen score was also not more than 0.4 (Fig. 1).
Fig. 1.
The shortlisted 14 OAPEs and the NCPs based on parameters of their vaxijen scores and binding affinity for class I and II MHC alleles. Depiction of all the 14 OAPEs selected from 4 different proteins of SARS CoV2 and the NCPs included in the study. Maximum of five epitopes belong to Envelope protein followed by four epitopes each from Spike protein and Membrane glycoprotein. Only one OAPE was selected from Nucleocapsid phosphoprotein. All the three NCPs are non-antigenic and their predicted affinity for class I and II alleles are almost negligible. The X axis in bar graph represents the number and the Y axis represents the OAPEs or NCPs
Evaluation of OAPEs for being pro-inflammatory epitopes
All our shortlisted 14 OAPEs were IL6-inducer (a biomarker cytokine for COVID-19) when subjected to IL-6-Pred (Table 2). Among them, ten OAPEs were also predicted to be inducers of pro-inflammatory cytokines in PIP-EL server. OAPEs ‘WTAGAAAYY’ and ‘WTFGAGAAL’ were not predicted to be pro-inflammatory cytokine inducers. In IFNepitope prediction, we found ten OAPEs were positive for being IFN-γ inducer. OAPEs ‘YVYSRVKNL’ ‘VVFLHVTYV’ ‘FTISVTTEI’ and ‘LTYTGAIKL’ were IFN-γ non-inducers (Table 2). The three NCPs included in the study were characterized to be pro-inflammatory in nature and non-inducers of IFN-γ as well as IL-6 (Table 2).
Table 2.
In-silico characterization of OAPEs of SARS- Cov-2 proteins as pro-inflammatory
| Peptide epitope | Random forest method score (Threshold 0.11) of peptide being IL-6 inducer/non-inducer4 | SVM method score (Threshold 0.0) of peptide being IFN-γ inducer/non-inducer5 | Probability of peptide being PIP/Non-PIP6 (Threshold 0.45) |
|||
|---|---|---|---|---|---|---|
| YVYSRVKNL (Envelope protein) | IL-6 inducer | 0.86 | IFN-γ non-inducer | − 0.39 | 0.632 | PIP |
| FVVFLLVTL (Envelope protein) | IL-6 inducer | 0.8 | IFN-γ inducer | 0.29 | 0.523 | PIP |
| FLAFVVFLL (Envelope protein) | IL-6 inducer | 0.89 | IFN-γ inducer | 1.24 | 0.512 | PIP |
| VLLFLAFVV (Envelope protein) | IL-6 inducer | 0.89 | IFN-γ inducer | 0.88 | 0.480 | PIP |
| FLLVTLAIL (Envelope protein) | IL-6 inducer | 0.76 | IFN-γ inducer | 0.24 | 0.522 | PIP |
| FAYANRNRF (Membrane Protein) | IL-6 inducer | 0.69 | IFN-γ inducer | 0.06 | 0.5828 | PIP |
| FLFLTWICL (Membrane Protein) | IL-6 inducer | 0.66 | IFN-γ inducer | 0.16 | 0.543 | PIP |
| FLWLLWPVT (Membrane Protein) | IL-6 inducer | 0.76 | IFN-γ inducer | 0.05 | 0.586 | PIP |
| FVLAAVYRI (Membrane Protein) | IL-6 inducer | 0.81 | IFN-γ inducer | 0.30 | 0.632 | PIP |
| WTAGAAAYY (Spike protein) | IL-6 inducer | 0.78 | IFN-γ inducer | 0.08 | 0.425 | Non-PIP |
| VVFLHVTYV (Spike protein) | IL-6 inducer | 0.85 | IFN-γ non-inducer | − 0.30 | 0.540 | PIP |
| FTISVTTEI (Spike protein) | IL-6 inducer | 0.82 | IFN-γ non-inducer | −0.29 | 0.5788 | PIP |
| WTFGAGAAL (Spike protein) | IL-6 inducer | 0.71 | IFN-γ inducer | 0.04 | 0.424 | Non-PIP |
| LTYTGAIKL Nucleocapsid phosphoprotein | IL-6 inducer | 0.88 | IFN-γ non-inducer | − 0.190 | 0.550 | PIP |
| NCP1 (ARSVASQSI) (Spike protein) | IL-6 non-inducer | < 0.11 | IFN-γ non-inducer | − 0.089 | 0.624 | PIP |
| NCP2 (KEELDKYFK) (Spike protein) | IL-6 non-inducer | < 0.11 | IFN-γ non-inducer | − 0.200 | 0.542 | PIP |
| NCP3 (VSEETGTLI) (Envelope protein) | IL-6 non-inducer | < 0.11 | IFN-γ non-inducer | − 0.732 | 0.561 | PIP |
All the shortlisted 14 OAPEs and the 3 NCPs were subjected to three different bio-informatics tools for characterizing them to be pro-inflammatory. Prediction of peptide for being Interleukin 6 (IL-6) inducer or non- inducer using IL-6-pred; Prediction of peptide being IFN-γ inducer or non- inducer using IFNepitope; Prediction of pro-inflammatory peptides (PIP) using PIP-EL server. We found all selected OAPEs were also predicted of inducing IL-6 in IL-6-Pred; IL-6 is a key biomarker for COVID-19 disease. Out of 14 OAPEs, 12 were predicted to be inducers of pro-inflammatory cytokines in PIP-EL server while 10 OAPEs were positive for inducing IFN-γ in IFNepitope server. All the 3 NCPs were predicted IL-6 non-inducer, IFN-γ non-inducer and pro-inflammatory cytokine inducer
Characterization of shortlisted overlapping epitopes as anti-inflammatory
The selected OAPEs were further subjected to three different bio-informatics tools for predicting their anti-inflammatory nature. All the five overlapping epitopes from Envelope (E) protein were IL-10 inducers and anti-inflammatory in nature. All the four overlapping epitopes of Membrane (M) protein were also inducers of IL-4, IL-10 and anti-inflammatory in medium to high confidence range. Among 4 overlapping epitopes from Spike (S) protein, two epitopes were anti-inflammatory in medium confidence range and capable of inducing either IL-4 or IL-10. Peptide ‘FTISVTTEI’ was predicted to induce IL-4 and IL-10 however, was low confidence anti-inflammatory epitope. Peptide ‘WTAGAAAYY’ was predicted to be non-inducers of IL-4, IL-10 and also was low confidence anti-inflammatory epitope. Peptide ‘LTYTGAIKL’ from Nucleocapsid (N) phosphoprotein was non-inducers of IL-4 and IL-10 but was high confidence anti-inflammatory epitope (Table 3). Therefore, out of total 14 OAPEs, we finally shortlisted 12 OAPEs characterized to produce a balance of pro-inflammatory and anti-inflammatory cytokines for further confirmatory studies. The NCPs were all non-inducers of IL-10 with a low confidence AIP scores for being anti-inflammatory in nature. However, all the three selected NCPs were IL-4 inducers.
Table 3.
In-silico characterization of antigenic promiscuous peptide epitopes of SARS- Cov-2 proteins as anti-inflammatory
| Peptide epitope | SVM + Motif based score (Threshold 0.2) of peptide being IL-4 inducer/non-inducera | Random forest method score (Threshold 0.3) of peptide being IL-10 inducer/non-inducerb | Combined RF score of peptide being Pre-AIP/Non- Pre-AIPc | |||
|---|---|---|---|---|---|---|
| YVYSRVKNL (Envelope protein) | IL-4 inducer | 0.38 | IL-10 inducer | 0.49 | 0.524 | High confidence AIP |
| FVVFLLVTL (Envelope protein) | IL-4 inducer | 0.26 | IL-10 inducer | 0.63 | 0.465 | Medium confidence AIP |
| FLAFVVFLL (Envelope protein) | IL-4 inducer | 0.28 | IL-10 inducer | 0.558 | 0.396 | Medium confidence AIP |
| VLLFLAFVV (Envelope protein) | IL-4 Non-inducer | – 0.72 | IL-10 inducer | 0.48 | 0.445 | Medium confidence AIP |
| FLLVTLAIL (Envelope protein) | IL-4 Non-inducer | – 0.94 | IL-10 inducer | 0.705 | 0.519 | High confidence AIP |
| FAYANRNRF (Membrane Protein) | IL-4-inducer | 0.33 | IL-10 inducer | 0.445 | 0.491 | High confidence AIP |
| FLFLTWICL (Membrane Protein) | IL-4-inducer | 0.28 | IL-10 inducer | 0.427 | 0.460 | Medium confidence AIP |
| FLWLLWPVT (Membrane Protein) | IL-4-inducer | 0.28 | IL-10 inducer | 0.778 | 0.586 | High confidence AIP |
| FVLAAVYRI (Membrane Protein) | IL-4-inducer | 0.28 | IL-10 inducer | 0.445 | 0.412 | Medium confidence AIP |
| WTAGAAAYY (Spike protein) | IL-4 Non-inducer | 0.16 | IL-10 non inducer | 0.095 | 0.384 | Low confidence AIP |
| VVFLHVTYV (Spike protein) | IL-4 Non-inducer | 0.14 | IL-10 inducer | 0.557 | 0.396 | Medium confidence AIP |
| FTISVTTEI (Spike protein) | IL-4-inducer | 0.42 | IL-10 inducer | 0.557 | 0.354 | Low confidence AIP |
| WTFGAGAAL (Spike protein) | IL-4-inducer | 0.25 | IL-10 non inducer | 0.128 | 0.448 | Medium confidence AIP |
| LTYTGAIKL Nucleocapsid phosphoprotein | IL-4 Non-inducer | 0.02 | IL-10 non inducer | − 0.097 | 0.482 | High confidence AIP |
| NCP1 (ARSVASQSI) (Spike protein) | IL-4-inducer | 0.45 | IL-10 non inducer | − 0.64 | 0.364 | Low confidence AIP |
| NCP2 (KEELDKYFK) (Spike protein) | IL-4-inducer | 0.29 | IL-10 non inducer | − 0.529 | 0.372 | Low confidence AIP |
| NCP3 (VSEETGTLI) (Envelope protein) | IL4-inducer | 0.45 | IL-10 non inducer | − 0.196 | 0.386 | Low confidence AIP |
All the shortlisted 14 OAPAs and the 3 NCPs were subjected to three different bio-informatics tools for characterizing them to be anti-inflammatory
aPrediction of peptide for being Interleukin 4 (IL-4) inducer or non- inducer using IL4-pred
bPrediction of peptide being Interleukin 10 (IL-10) inducer or non- inducer using IL-10pred
cPrediction of Anti-inflammatory peptides (Pre-AIP) using Pre-AIP software. Among the 14 selected OAPEs, 8 OAPEs were predicted of inducing IL-4 in IL-4-Pred while 11 OAPEs were also predicted of inducing IL-10 in IL-10-Pred. 12 OAPEs were predicted to be anti-inflammatory in nature with a medium to high confidence score in Pre-AIP. All the 3 NCPs were predicted IL-4 inducer but non-inducers of IL-10. The NCPs also had low confidence AIP score which characterize them to be non anti-inflammatory in nature
Docking of OAPE onto MHC alleles by pepATTRACT and Hex
All the 12 shortlisted overlapping peptide epitopes were docked in pepATTRACT with HLA-A*02 (PDB ID-1S9Y) and HLA-DRB1*01:01 (PDB ID- 4MCZ) and their energy scores were generated using Hex 8.0.0. The maximum energy score of −573.26 was observed for peptide ‘VLLFLAFVV’ of Envelope (E) protein docked to HLA-A*02 (Table 4). The maximum energy score of −564.92 was observed for peptide ‘FVLAAVYRI’ of Membrane (M) protein docked to HLA- DRB1*01:01 (Table 4). The energy score of control peptide was −560.06 for HLA-A*02 and −696.0 for HLA-DRB1*01:01. Docking of representative peptide from each protein with both HLA-A*02 and HLA-DRB1 has been shown in (Figs. 2, 3, 4, 5). These peptide epitopes had hydrophobic amino acid at second and ninth position for HLA-A*02 binding and at fourth and sixth position for HLA-DRB1 binding. The presence of hydrogen bond made these peptides tightly bound to the MHC groove.
Table 4.
Docking of OAPEs with a balanced cytokine response to HLA-A*02 and HLA-DRB1 MHC alleles
| Peptide epitopes | Hex energy score for HLA-A*02 binding (Kcal/mol) | Hex energy score for HLA-DRB1 binding (Kcal/mol) |
|---|---|---|
| YVYSRVKNL (Envelope protein) | − 462.20 | − 509.94 |
| FVVFLLVTL (Envelope protein) | − 508.04 | − 526.18 |
| FLAFVVFLL (Envelope protein) | − 535.93 | − 547.52 |
| VLLFLAFVV (Envelope protein) | − 573.26 | − 527.08 |
| FLLVTLAIL (Envelope protein) | − 538.32 | − 497.75 |
| FAYANRNRF (Membrane Protein) | − 478.24 | − 485.4 |
| FLFLTWICL (Membrane Protein) | − 505.28 | − 508.96 |
| FVLAAVYRI (Membrane Protein) | − 525.38 | − 564.92 |
| FLWLLWPVT (Membrane Protein) | − 485.5 | − 507.69 |
| VVFLHVTYV (Spike protein) | − 477.96 | − 499.42 |
| WTFGAGAAL (Spike protein) | − 464.22 | − 477.17 |
| LTYTGAIKL Nucleocapsid (N) phosphoprotein | − 512.19 | − 499.52 |
The straightened peptide and the MHC obtained from pepATTRACT server were docked on to Hex. These overlapping peptides were docked against HLA-A*02 allele (PDB ID-1S9Y) and HLA DR B1 allele (PDB ID- 4mcz). The energy score of control peptide was −560.06 for HLA-A*02 and −696.0 for HLA-DRB1*01:01. The maximum energy score of −573.26 and −564.92 was observed for peptide‘VLLFLAFVV’ of Envelope (E) protein docked to HLA-A*02 and peptide‘FVLAAVYRI’ of Membrane (M) protein docked to HLA-DRB1*01:01 respectively
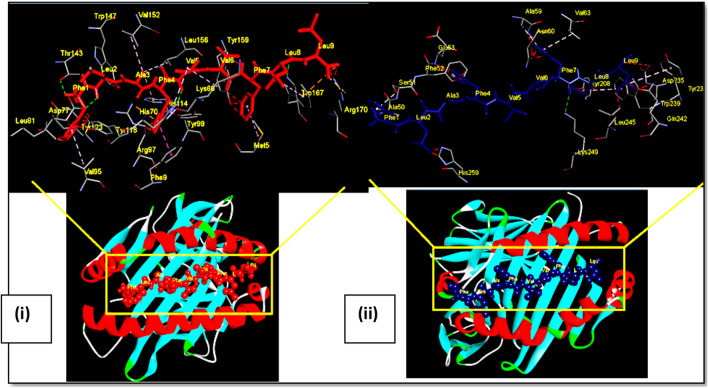
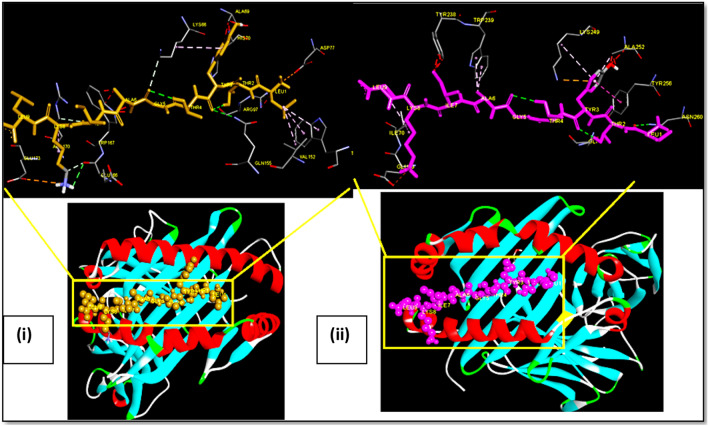
Fig. 2.
Docking of OAPE ‘FLAFVVFLL’ of Envelope protein with MHC class I HLA-A*02 and MHC class II HLA-DRB1. Ball and stick model of OAPE ‘FLAFVVFLL’ of Envelope protein docked onto the (i) HLA-A*02 allele (PDB ID-1S9Y) and (ii) HLA DR B1 allele (PDB ID- 4mcz). The inset shows the detailed interactions of epitope with MHC residue. Potential bonds formed between the two are shown as dotted lines
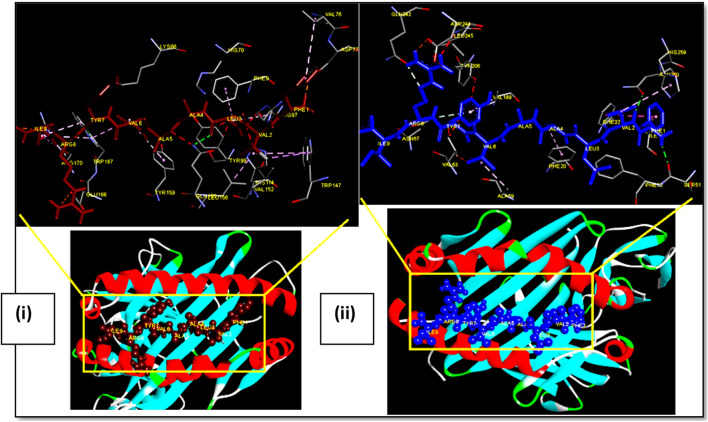
Fig. 3.
Docking of OAPE ‘FVLAAVYRI’ of Membrane protein with MHC class I HLA-A*02 and MHC class II HLA-DRB1. Ball and stick model of OAPE ‘FVLAAVYRI’ of Membrane protein docked onto the (i) HLA-A*02 allele (PDB ID-1S9Y) and (ii) HLA DR B1 allele (PDB ID- 4mcz). The inset shows the detailed interactions of epitope with MHC residue. Potential bonds formed between the two are shown as dotted lines
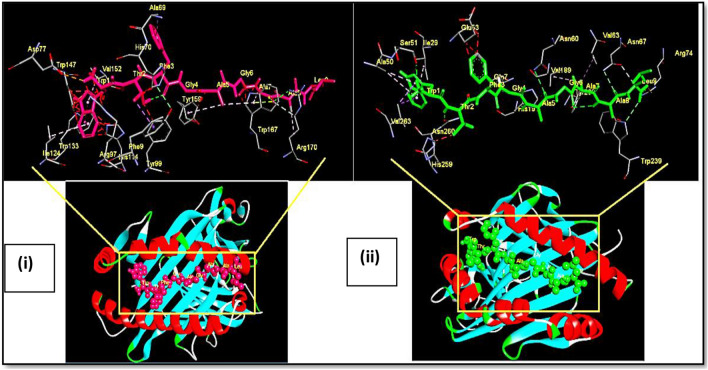
Fig. 4.
Docking of OAPE ‘WTFGAGAAL’ Spike protein with MHC class I HLA-A*02 and MHC class II HLA-DRB1. Ball and stick model of OAPE ‘WTFGAGAAL’ Spike protein docked onto the (i) HLA-A*02 allele (PDB ID-1S9Y) and (ii) HLA DR B1 allele (PDB ID- 4mcz). The inset shows the detailed interactions of epitope with MHC residue. Potential bonds formed between the two are shown as dotted lines
Fig. 5.
Docking of OAPE ‘LTYTGAIKL’ of Nucleocapsid protein with MHC class I HLA-A*02 and MHC class II HLA-DRB1. Ball and stick model of OAPE ‘LTYTGAIKL’ of Nucleocapsid protein docked onto the HLA-A*02 allele (PDB ID-1S9Y) and HLA DR B1 allele (PDB ID- 4mcz). The inset shows the detailed interactions of epitope with MHC residue. Potential bonds formed between the two are shown as dotted lines
Designing of the T cell specific multi-epitopic adjuvanated vaccine
Codon adaptation followed by in silico cloning of the designed vaccine construct with immunogenic CTxB as an adjuvant and the 10 selected OAPEs joined by specific linkers was performed (Fig. 6).
Fig. 6.
Design and in silico cloning of the vaccine construct. a The amino acid sequence of the vaccine construct. The CTxB adjuvant is highlighted in black, whereas linker KPKPKP joining adjuvant and rest of the construct is in yellow. The ten OAPEs are highlighted in red and the inter epitopic linker AAY is in blue respectively. b The methodology for in silico cloned construct in SnapGene 3.1.4
Physiochemical characterization of designed vaccine construct
The designed vaccine construct is composed of 271 amino acids with a molecular weight of approximately 30.46 kDa. The theoretical pI was 9.55, implicating the significant alkaline nature of vaccine. The half-life of the vaccine was estimated to be 30 h in mammalian reticulocytes (in vitro), > 20 h in yeast (in vivo), and > 10 h in E. coli (in vivo), suggesting that the construct is stable in vivo. The instability index was predicted to be 22.36, which clears the vaccine to be a stable protein. The estimated aliphatic index and Grand Average of Hydropathicity (GRAVY) score were found to be 113.54 and 0.710 respectively, implicating that the vaccine is thermo-stable and hydrophilic in nature.
Evaluating toxicity and allergenicity of the vaccine construct
The designed vaccine construct was predicted to be a non-allergen in both servers AllerTop v2.0 and AllergenFPv.1.0 and non-toxic in server ToxinPred.
Predicting pro-inflammatory and anti-inflammatory nature of the vaccine construct
The entire vaccine construct was predicted to be pro-inflammatory with a probability score of 0.724 in PIP-EL server and anti-inflammatory in nature with a high confidence score of 0.571 in Pre-AIP server.
Structure prediction and validation of designed vaccine construct
I-TASSER was employed for secondary structure prediction of the vaccine construct. The 5 models generated were evaluated using different structure validation servers. Model 4 having a C score of -3.94 in I-TASSER was selected based on all the analyzed parameters (Figure S1). Ramachandran plot of model 4 showed 84.6% residues were present in the favored region, 10.6% residues in the generously allowed region, and 4.7% residues in disallowed regions (Figure S1). Overall quality factor of 47.9 was generated in ERRAT for Model 4 and Verify3D score of the model was 92.25.
Molecular docking of vaccine construct with host immune receptor
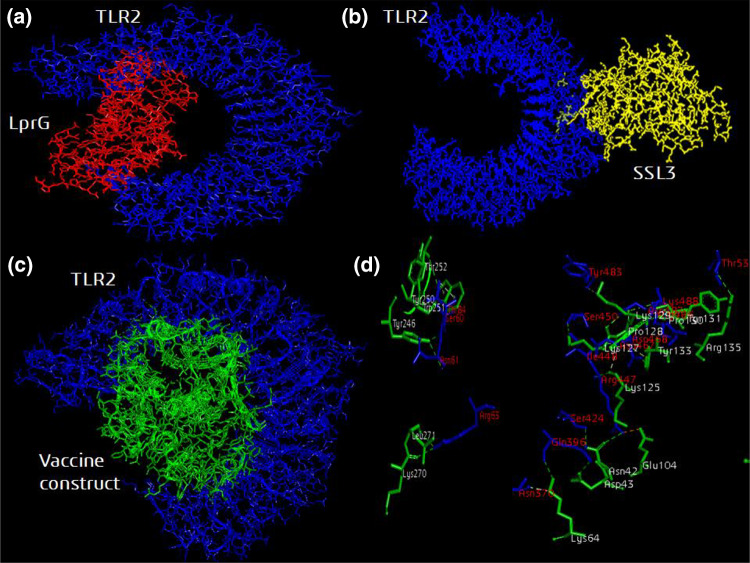
TLR2 is an important receptor for binding of pathogen an inducing a proinflammatory immune response required for clearance of pathogen. The structurally validated vaccine construct was then evaluated for immune receptor interaction through molecular docking. The positive control ligand protein LprG and the vaccine construct both were observed to bind within the active site groove of the TLR2 structure (Fig. 7). The vaccine construct forms 24 hydrogen bonds with Hex energy score of −860.60. LprG protein forms 27 hydrogen bonds with Hex energy score of −832.93. The negative control protein SSL3 did not bind to the groove of TLR2 molecule and generated a low Hex energy score of −556.01.
Fig. 7.
Molecular docking of the designed and validated vaccine construct with TLR2 immune receptor. a TLR2 agonist LprG protein (red color) and c modeled vaccine construct (green color) binds to the active site groove of TLR2 receptor (blue color). b The negative control SSL3 protein (yellow color) binds randomly to TLR2 receptor (blue color). d The hydrogen bonds formed between the active site residues of TLR2 molecule labeled in red with the vaccine construct residues labeled in white
In silico immune simulation studies in response to our vaccine
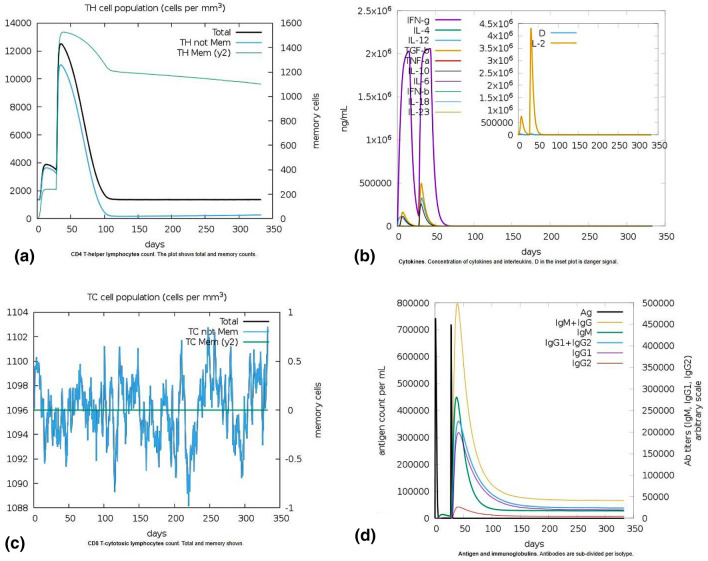
Our vaccine construct led to a significant simulation of mammalian immune response predicted through C-ImmSim server (Fig. 8). A booster administration of vaccine led to a gradual increase in immune responses. The generation of T- cells was quite significant with memory cells lasting for several months. One interesting observation was the profile of cytokines being produced after the injections. A sharp peak of pro-inflammatory IFN-γ, IL-12 and IL-2 was seen following repeated exposure to the vaccine. Additionally, we found peaks for some anti-inflammatory cytokines like TGF-β and IL-10 which increased with the booster dose. The results also indicated that our T cell specific multi-epitopic vaccine was able to generate good levels of B cells secreting antibodies; the more prominent ones being the IgM and IgG subclass.
Fig. 8.
Prediction of immune simulation response to the vaccine construct using C-ImmSim. In silico immune simulation study by C-ImmSim software in response to a double dose administration of the vaccine construct at intervals of 4 weeks. The population of activated a CD4 + T helper cells and b CD8 + cytotoxic T cells in terms of memory response generated after the injections. Plot c shows cytokines levels observed after the injections while d shows the levels of antibody titers generated after the injections. In plot c; the inset shows blue colored peak D which is defined as emergence of diversity of epitope specific T cell clones. Lower D value depicts lower diversity
Discussion
Testing traditional vaccines made of entire pathogen or pathogen proteins in animal models can be costly, allergenic, and time-consuming (Li et al. 2014; Lo et al. 2013). Only specific amino acid fragments within the pathogen acts as potent immunogens and are sufficient to mount a protective immune response within host (Li et al. 2014; Singh et al. 2019). This rationale leads to designing of ‘peptide based COVID-19 vaccine’ incorporating only specific immunogenic peptide epitopes. Prediction and identification of all the potential immunogenic epitopes within the viral genome is possible with immuno-informatics and reverse vaccinology approach (Singh et al. 2019; Rappuoli 2001). Most of the vaccines against SARS-CoV-2 are in development, under trials or in emergency usage are targeting the humoral immune response for making neutralizing antibodies (Amanat and Krammer 2020; Le et al. 2020). However, the concept of acquired, specific T cell immunity becomes crucial in context of vaccination to clear viral infections and enhance neutralizing antibody responses (Campbell et al. 2020). Many studies have reported a higher frequency of Spike protein specific CD4 T cell responses and heightened CD8 T cell responses specific to other internal proteins of SARS-CoV-2 in convalescent individuals (Grifoni et al. 2020; Sekine et al. 2020; Dong et al. 2020). Although the CD4 T cell response was maximal in acute and severe COVID-19 cases, the proportion of CD8 T cell response was higher in mild COVID-19 individuals. Thus, we have adopted an immuno-informatic approach focused on all the four structural proteins of SARS-CoV-2 for recognition of potential CD4 and CD8 T cell peptide epitopes which can trigger host immune response to the maximum. In silico analysis through software NetMHC4.0 and IEDB predicted an enormous reservoir of epitopes that could be identified in just four structural proteins of SARS-CoV-2. These four proteins were predicted to be highly immunogenic. Spike protein generated maximum promiscuous antigenic epitopes both in NetMHC4.0 and IEDB which could be attributed to the large size of this protein. The antigenicity and the binding affinity of the 14 shortlisted OAPEs to MHC alleles were much higher than the 3 included negative control peptides or NCPs in the study.
In case of viral infections, innate immune cells recognize pathogen-associated molecular pattern (PAMP) possessed by the virus which raises inflammatory alarm. Triggering of signaling cascade leads to secretion of several pro-inflammatory cytokines including IL-12, IFN-γ, TNF-a, IL-6 etc. In early phase of infection these cytokines act as signals to regulate the innate immune system via stimulation of CD4 T cell mediated protective immunity thus mounting an anti-viral immune response along with production of IgG-class antibodies. (Costela-Ruiz et al. 2020). SARS- CoV-2 disturbs the delicate balance of favorable inflammatory response making it detrimental systemic inflammation or cytokine storm by unrestrained secretion of pro-inflammatory markers, in particular cytokines TNF-α, IL-1β, IL-8, and IL-6 (Soy et al. 2020; Opal and DePalo 2000). Evidences support the role of anti-inflammatory cytokines such as IL-4, IL-10, and transforming growth factor β (TGF-β) in impeding the hyper-inflammatory cytokine storm (Asadullah et al. 2003). Therefore, our study encompasses the necessity of a balanced pro-inflammatory immune response to ensure killing of virus along with a counteracting anti-inflammatory response to avoid the hyperactive cytokine storm observed in severe COVID19 disease pathology. We shortlisted 12 OAPEs which were characterized as inducers of pro-inflammatory and anti-inflammatory cytokines. Given the exaggerated inflammatory cytokine storm in severe Covid19 cases; it was no surprise that all the 12 OAPEs were predicted to be IL6-inducers when subjected to IL-6-Pred server. However, at least 8 OAPEs were IL-4 inducer, 11 OAPEs were IL-10 inducer and 9 OAPEs were IFNγ inducer. In comparison, the NCPs included in our study were all pro-inflammatory in nature and non-inducers of IL-10, IL-6 and IFN-γ.
The affinity of these selected antigenic and anti-inflammatory peptide epitopes to their cognate MHC class I and II allele was confirmed by docking studies using pepATTRACT, HEX, Pymol, and Discovery Studio. A vaccine construct was designed inclusive of all the 12 OAPEs and Cholera Toxin subunit B (CTxB) as an adjuvant. The construct was non-toxix, non-allergenic and capable of inducing both anti-inflammatory and pro-inflammatory immune response. The vaccine construct was successfully cloned in silico followed by structural modelling to evaluate its interaction with immune receptor like Toll like Receptor 2 (TLR2). A step forward in this study was to perform the docking studies with the shortlisted 12 overlapping antigenic and anti-inflammatory peptides with globally present MHC class I and II alleles i.e. HLA-A*02 allele (PDB ID-1S9Y) and HLA-DR*B1 allele (PDB ID- 4mcz). The Hex energy scores for peptide:MHC alleles were comparable to the control peptide. The 6 pockets in MHC class I alleles are crucial in peptide MHC interactions (Webb et al. 2004). The second and 9th hydrophobic residues of the docked peptide epitopes engages in peptide’s interaction with 2nd and 6th pocket of HLA*A02. The docked peptide epitopes also have hydrophobic amino acid at 4th and 6th position which help in binding the peptide into the HLA-DR*B1 allele groove as reported (Werner and Freund 2017). The presence of hydrogen bonds makes these peptides tightly bound to the MHC groove. Hydrogen-bonding ensures the peptide’s N terminus and C-terminus independent of peptide sequence and also bridges the peptide and buried hydrophilic MHC residues (Killer et al. 1999). We further designed our vaccine construct followed by in silico cloning. CTxB was added as an adjuvant to heighten the immunogenicity of the subunit vaccine construct. CTxB has been used variedly as an adjuvant and also is reported for its efficiency in Influenza virus vaccine (Jafari et al. 2020). The construct was non-allergenic and non-toxic implicating its candidature for a good vaccine. Toll Like Receptors (TLRs) activate the innate immune system which subsequently direct production of effective adaptive immunity. To ensure that the vaccine will be effective in generation of immune response, we evaluated its in silico interaction with TLR-2 receptor and comparing the results with positive control TLR-2 agonist-LprG and a negative control-SSL3. The binding affinity of the vaccine construct in terms of energy score was more than LprG protein. Additionally, it was observed that the construct protein forms hydrogen bonds with TLR-2 receptor which involves interacting amino acids throughout the length of vaccine construct and not just the CTxB region. Immune simulation studies by C-ImmSim software in response to dual dose of vaccine exposure further substantiate efficacy of our subunit vaccine. A heightened Th1 type immune response was observed including an increased population of T cells with memory cells lasting for months. The observation that the vaccine resulted in generation of both pro-inflammatory (IFN-γ, IL-12 and IL-2) and anti-inflammatory cytokines (TGF-β and IL-10) in response to our vaccine construct further supported the hypothesis of this study. Although the proposed vaccine construct is specific for T cells; the server also predicted high levels of antibody secreting B cells with a prolonged memory response.
Conclusion
Immunogenicity, protectiveness and memory are the three crucial parameters to evaluate antigenic epitopes as peptide-based subunit vaccine candidates. Our study predicts SARS- CoV-2 peptide based vaccine candidate encompassing all the 3 parameters. Critically, both CD4 T and CD8 T cells along with immunological memory are essential to impart protective immunity against SARS-CoV-2 infection (Guo et al. 2020; Grifoni et al. 2020). However, uncontrolled inflammatory innate responses and deregulated T cell responses leads to immunopathology (Vabret et al. 2020; Cao 2019). Incorporation of CD4 and CD8 T cell specific OAPEs which could elicit both pro-inflammatory and anti-inflammatory immune response seems a possible solution to tackle the major concern of hyper-inflammatory cytokine storm in COVID-19 along with killing of virus infected host cells. The inclusiveness of peptide antigens from all the 4 structural proteins of SARS-CoV-2 in this study pinpoints the highly immunogenic targets within the virus. Nevertheless, the proposed peptide vaccine candidate needs to be validated experimentally ensuring its safety and immunogenic profile to help stop this global outbreak. We plan to evaluate these 12 immunogenic OAPE SARS-CoV-2 peptide epitopes based vaccine construct in PBMC from healthy individuals, active COVID-19 patients and recovered individuals. We also wish to study the poly-functionality of T cells in terms of cytokine production in these individuals. Given the current state of medical urgency and the lack of experimental data, this study is quite relevant and provides a novel basis for COVID-19 vaccine strategies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Medha and Priyanka are recipient of Senior and Junior Research Fellowship of Council of Scientific and Industrial Research (CSIR), Government of India, respectively.
Abbreviations
- COVID-19
Coronavirus disease 2019
- SARS-CoV-2
Severe acute respiratory syndrome corona virus 2
- OAPE
Overlapping antigenic peptide epitope
- NCP
Negative control peptide
- IL
Interleukin
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors have no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Medha, Email: medha1.singh@mirandahouse.ac.in.
Monika Sharma, Email: monika.sharma@mirandahouse.ac.in.
Sadhna Sharma, Email: sadhna.sharma@mirandahouse.ac.in.
References
- Amanat F, Krammer F. Perspective SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadullah K, Sterry W, Volk HD. Interleukin-10 therapy—review of a new approach. Pharmacol Rev. 2003;55:241–269. doi: 10.1124/pr.55.2.4. [DOI] [PubMed] [Google Scholar]
- Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie JU, Lüthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Brusic V, Rudy G, Honeyman G, Hammer J, Harrison L. Prediction of MHC class II-binding peptides using an evolutionary algorithm and artificial neural network. Bioinformatics. 1998;14:121–130. doi: 10.1093/bioinformatics/14.2.121. [DOI] [PubMed] [Google Scholar]
- Campbell VL, Nguyen L, Snoey E, McClurkan CL, Laing KJ, Dong L, Sette A, Lindestam Arlehamn CS, Altmann DM, Boyton RJ, Roby JA, Gale M, Stone M, Busch MP, Norris PJ, Koelle DM. Proteome-wide Zika virus CD4 T cell epitope and HLA restriction determination. ImmunoHorizons. 2020;4:444–453. doi: 10.4049/immunohorizons.2000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2019;2019:2019–2020. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma G, Eisenlohr L. CD8+ T-cell responses in vaccination: reconsidering targets and function in the context of chronic antigen stimulation [version 1; referees: 2approved] F1000 Res. 2018;7:1–8. doi: 10.12688/f1000research.14115.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries SJ, Rey J, Schindler CEM, Zacharias M, Tuffery P. The pepATTRACT web server for blind, large-scale peptide-protein docking. Nucleic Acids Res. 2017;45:W361–W364. doi: 10.1093/nar/gkx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhall A, Patiyal S, Sharma N, Usmani SS, Raghava GPS. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19 00. Brief Bioinform. 2020 doi: 10.1093/bib/bbaa259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Gupta S, Vir P, Raghava GP. Prediction of IL4 inducing peptides. Clin Dev Immunol. 2013;2013:263952. doi: 10.1155/2013/263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Vir P, Raghava GPS. Designing of interferon-gamma inducing MHC class-II binders. Biol Direct. 2013;8:1–15. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP vol 2–a server for in silico prediction of allergens. J Mol Model. 2014;20:2278. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- Dimitrov I, Naneva L, Doytchinova I, Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30:846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, López-camacho C, Slon-campos J, Zhao Y, Stuart DI, Paesen GC, Grimes JM, Antson AA, Bayfield OW, Hawkins DEDP, Ker D, Wang B, Turtle L, Subramaniam K, Thomson P, Zhang P, Dold C, Ratcliff J, Simmonds P, Silva TD, Sopp P, Wellington D, Rajapaksa U, Chen Y, Salio M, Napolitani G, Paes W, Borrow P, Kessler BM, Fry JW, Schwabe NF, Semple MG, Baillie JK, Moore SC, Openshaw PJM, Ansari MA, Dunachie S, Barnes E, Frater J, Kerr G, Goulder P. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Imunol. 2020 doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, Sette A. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:1–16. doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. The proteomics protocols handbook. Proteomics Protoc Handb. 2005 doi: 10.1385/1592598900. [DOI] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cao Q, Hong Z, Tan Y, Chen S, Jin H, Tan K, Wang D, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 ( COVID-19) outbreak—an update on the status. Millitary Med Res. 2020;7:1–10. doi: 10.1186/s40779-019-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Consortium OSDD, Raghava GPS In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013;8:73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-weber H, Schroeder S, Mu MA, Drosten C, Po S, Hoffmann M, Kleine-weber H, Schroeder S, Kru N. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven article SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cel. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari D, Malih S, Gomari MM, Safari M, Jafari R, Farajollahi MM. Designing a chimeric subunit vaccine for influenza virus, based on HA2, M2e and CTxB: a bioinformatics study. BMC Mol Cell Biol. 2020;21:1–13. doi: 10.1186/s12860-020-00334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Laloraya M. A cytokine super cyclone in COVID-19 patients with risk factors: the therapeutic potential of BCG immunization. Cytokine Growth Factor Rev. 2020;54:32–42. doi: 10.1016/j.cytogfr.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatun MS, Hasan MM, Kurata H. PreAIP: Computational prediction of anti-inflammatory peptides by integrating multiple complementary features. Front Genet. 2019 doi: 10.3389/fgene.2019.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killer KN, Fan BQR, Wiley DC. Structure of human histocompatibility leukocyte antigen. J Exp Med. 1999;190:113–123. doi: 10.1084/jem.190.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Le TT, Andreadakis Z, Kumar A, Román RG, Tollefsen S, Saville M, Mayhew S. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK. Peptide vaccine: progress and challenges. Vaccines. 2014;2:515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y, Pai T, Wu W, Chang H. Prediction of conformational epitopes with the use of a knowledge-based energy function and geometrically related neighboring residue characteristics. BMC Bioinform. 2013 doi: 10.1186/1471-2105-14-S4-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24:1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer: An FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan B, Shin TH, Kim MO, Lee G. PIP-EL: a new ensemble learning method for improved proinflammatory peptide predictions. Front Immunol. 2018;9:1783. doi: 10.3389/fimmu.2018.01783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal G, Usmani SS, Dhanda SK, Kaur H, Singh S, Sharma M, Raghava GPS. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep. 2017;7:1–10. doi: 10.1038/srep42851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform. 2007;8:1–12. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal SM, DePalo VA. Anti-inflammatory cytokines. Chest. 2000;117:1162–1172. doi: 10.1378/chest.117.4.1162. [DOI] [PubMed] [Google Scholar]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2019 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phares TW, Stohlman SA, Hwang M, Min B, Hinton DR, Bergmann CC. CD4 T cells promote CD8 T cell immunity at the priming and effector site during viral encephalitis. J Virol. 2012;86:2416–2427. doi: 10.1128/jvi.06797-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5:e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/S0264-410X(00)00554-5. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler CEM, de Vries SJ, Zacharias M. Fully blind peptide-protein docking with pepATTRACT. Structure. 2015;23:1507–1515. doi: 10.1016/j.str.2015.05.021. [DOI] [PubMed] [Google Scholar]
- Sekine T, Rivera-ballesteros O, Ljunggren H, Aleman S, Buggert M, Parrot T, Folkesson E, Covid- K. Article robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19 ll ll robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Bhatt P, Sharma M, Varma-Basil M, Chaudhry A, Sharma S. Immunogenicity of late stage specific peptide antigens of Mycobacterium tuberculosis. Infect Genet Evol. 2019 doi: 10.1016/j.meegid.2019.103930. [DOI] [PubMed] [Google Scholar]
- Siu K, Kok K, Ng MJ, Poon VKM, Yuen K, Zheng B, Jin D. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3∙TANK∙TBK1/IKK∈ complex. J Biol Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy M, Tabak F, Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Agrawal M, Aleynick M, Belabed M, Brown M, Casanova-Acebes M, Catalan J, Centa M, Charap A, Chan A, Chen ST, Chung J, Bozkus CC, Cody E, Cossarini F, Dalla E, Fernandez N, Grout J, Ruan DF, Hamon P, Humblin E, Jha D, Kodysh J, Leader A, Lin M, Lindblad K, Lozano-Ojalvo D, Lubitz G, Magen A, Mahmood Z, Martinez-Delgado G, Mateus-Tique J, Meritt E, Moon C, Noel J, O’Donnell T, Ota M, Plitt T, Pothula V, Redes J, Reyes Torres I, Roberto M, Sanchez-Paulete AR, Shang J, Schanoski AS, Suprun M, Tran M, Vaninov N, Wilk CM, Aguirre-Ghiso J, Bogunovic D, Cho J, Faith J, Grasset E, Heeger P, Kenigsberg E, Krammer F, Laserson U. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker WA, Tarannum M, Vivero-Escoto JL. Cellular endocytosis and trafficking of cholera toxin B-modified mesoporous silica nanoparticles. J Mater Chem B. 2016;4:1254–1262. doi: 10.1039/C5TB02079D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AI, Dunstone MA, Chen W, Aguilar MI, Chen Q, Jackson H, Chang L, Kjer-Nielsen L, Beddoe T, McCluskey J, Rossjohn J, Purcell AW. Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J Biol Chem. 2004;279:23438–23446. doi: 10.1074/jbc.M314066200. [DOI] [PubMed] [Google Scholar]
- Werner JM, Freund C. Major histocompatibility complex ( MHC ) Class i and MHC Class ii proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8:1–16. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.