Abstract
The increasing numbers of infected cases of coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) poses serious threats to public health and the global economy. Most SARS‐CoV‐2 neutralizing antibodies target the receptor binding domain (RBD) and some the N‐terminal domain (NTD) of the spike protein, which is the major antigen of SARS‐CoV‐2. While the antibody response to RBD has been extensively characterized, the antigenicity and immunogenicity of the NTD protein are less well studied. Using 227 plasma samples from COVID‐19 patients, we showed that SARS‐CoV‐2 NTD‐specific antibodies could be induced during infection. As compared to the results of SARS‐CoV‐2 RBD, the serological response of SARS‐CoV‐2 NTD is less cross‐reactive with SARS‐CoV, a pandemic strain that was identified in 2003. Furthermore, neutralizing antibodies are rarely elicited in a mice model when NTD is used as an immunogen. We subsequently demonstrate that NTD has an altered antigenicity when expressed alone. Overall, our results suggest that while NTD offers a supplementary strategy for serology testing, it may not be suitable as an immunogen for vaccine development.
Keywords: SARS‐CoV‐2, COVID‐19, N‐terminal domain, NTD, serology, immunogen
Receptor binding domain (RBD) of the SARS‐CoV‐2 spike protein has been extensively studied in functional antibody screening and COVID‐19 vaccine design. To explore the function of N‐terminal domain (NTD) of spike protein, we reveal that NTD is a supplementary antigen for serology testing, but not a suitable immunogen for vaccine design.
Introduction
The novel coronavirus SARS‐CoV‐2, which is the pathogen that has caused the COVID‐19 pandemic, has spread to over 216 countries [1]. COVID‐19 patients show varying disease severity ranging from asymptomatic to requiring intensive care [2]. Many studies have now shown that SARS‐CoV‐2‐specific IgG in COVID‐19 patients is a key signature of immune response upon the infection [3, 4, 5, 6, 7]. The spike glycoprotein is the immunodominant target for the neutralizing antibody response in COVID‐19 patients. Importantly, neutralizing antibodies to the spike are able to maintain at detectable levels for 5–8 months after infection [8, 9, 10]. The spike protein consists of S1 (head) and S2 (stem) subunits that are initially connected by a furin cleavage site [11]. The S1 contains two structurally well‐defined domains, namely the N‐terminal domain (NTD) and receptor binding domain (RBD).
SARS‐CoV‐2 initiates viral entry by engaging the host receptor angiotensin converting enzyme 2 (ACE2) through the RBD. Most known SARS‐CoV‐2 neutralizing antibodies to date are RBD‐specific [3, 4, 12, 13, 14, 15, 16]. Thus, detection of RBD‐specific antibodies is widely used in many serodiagnosis tests [17, 18]. RBD has also been a major focus in vaccine design [19, 20, 21]. In contrast, the immunogenicity and antigenicity of other domains on the spike is not very well characterized. An increasing number of neutralizing antibodies to the NTD have recently been identified from COVID‐19 patients [12, 22, 23, 24, 25, 26, 27]. Interestingly, tyrosine‐protein kinase receptor UFO (AXL) is suggested to be a co‐receptor for SARS‐CoV‐2 by interacting with the NTD [28]. Another recent finding shows that the NTD can interact with tetrapyrrole products that reduce the reactivity of the SARS‐CoV‐2 spike with human immune sera as a possible mechanism to evade antibody immunity [29]. It is thus believed that neutralizing antibodies to NTD antibodies may play an important role in protection against SARS‐CoV‐2. However, the NTD‐specific antibodies are so far mainly identified from clonal B cells of individuals. The serological response to the NTD in COVID‐19 patients, as well as the immunological properties of NTD are not yet well understood. In this study, we evaluated the human serological response to NTD protein from 227 plasma specimens collected from 141 COVID‐19 patients. The cross‐reactivity of NTD‐specific antibody response to different coronaviruses was also examined. We also explored the serological response by using NTD as an immunogen for immunization in mice.
Results
Serological responses to the NTD of SARS‐CoV‐2
We tested 227 plasma samples from 141 RT‐PCR confirmed COVID‐19 patients in Hong Kong and another 195 plasma samples from healthy blood donors that were collected prior to the emergence of SARS‐CoV‐2 as baseline controls. The samples were tested in parallel in ELISA assays for the IgG against NTD and RBD. For each assay, samples were defined as seropositive if the detection signal was three standard deviations above the mean of baseline controls. There was a progressive increase of seropositivity in the NTD ELISA after the first day of symptom onset, with 25% (12/48) being positive in the first 2 weeks and 84.9% (152 out of 179) after day 14 to day 141 (Table 1; Figure 1A). Consistent with our previous study [17], the positivity in the RBD assay also progressively increased with time after illness onset, with 58.3% (28/48) specimens positive in the first 2 weeks of illness onset and 98.3% (176 of 179) after day 14 to day 141 (Table 1, Figure 1B). Specimens that were found to be positive in the NTD ELISA (n = 164) were also positive in the RBD ELISA. In fact, there was a strong correlation between the serological response to NTD and RBD proteins after day 8 of symptom onset (Pearson correlation = 0.78; Figure 1C). A recent study has shown that neutralizing antibody titer to SARS‐CoV‐2 among severe patients were higher than mild and asymptomatic patient [10]. Moreover, another study has also shown that to RBD protein was significant higher in severe patients compared to those with mild condition [30]. We obtained the information of clinical severity from 20 severe, 38 moderate, and 51 mild/asymptomatic patients and compared the binding levels of RBD and NTD respectively from their plasma samples. Our data show that both RBD and NTD binding antibody level has significant differences among patients with different severities (Supporting information Figure 1A‐B).
Table 1.
RBD and NTD ELISA results from the plasma of COVID‐19 patients
| Numbers of sample | RBD positive (%) | NTD positive (%) | |
|---|---|---|---|
| Days 1–7 | 21 | 12 (57.1) | 1 (4.8) |
| Days 8–14 | 27 | 16 (59.3) | 11 (40.7) |
| Days 15–21 | 12 | 12 (100) | 7 (58.3) |
| Days 22–28 | 14 | 14 (100) | 10 (71.4) |
| Days >28 | 153 | 150 (98.0) | 135 (88.2) |
| Total | 227 | 204 (89.9) | 164 (72.2) |
Figure 1.

Patient serological responses to SARS‐CoV‐2 NTD and RBD protein (A and B) Binding of plasma from SARS‐CoV‐2 infected patients to SARS‐CoV‐2 NTD protein (A) and RBD protein (B) were measured during the days symptom after onset by ELISA assay. The mean OD450 ELISA binding values calculated after testing each plasma sample in duplicate are shown. The plamsa sample from healthy donors were used as negative control. The ELISA cutoff value of NTD and RBD protein were 0.3272 and 0.2607, respectively (mean + three standard deviations). (C) Pearson correlation (r) was used to assess the relationship between measured SARS‐CoV‐2 serological binding responses to SARS‐CoV‐2 RBD and NTD protein in the SARS‐CoV‐2 infected patients at consequent time periods. Each sample was tested as duplicates in each assay and the results were confirmed by two independent experiments.
Cross reactivity of the humoral immunity from COVID‐19 patients
The extent of cross‐reactive serological responses to other coronaviruses during SARS‐CoV‐2 infection is not fully understood. Our previous study observed that plasma samples from COVID‐19 patients can cross‐react with the RBD of SARS‐CoV [31]. Here, we further tested the binding of 227 plasma samples of COVID‐19 patients to the NTDs of SARS‐CoV‐2 and SARS‐CoV. Among the 164 samples with positive binding to the NTD of SARS‐CoV‐2, only eight (4.9%) cross‐reacted with the NTD of SARS‐CoV in the ELISA binding assay (Figure 2A). There is no significant correlation in binding between the groups (Pearson correlation = 0.06). In contrast, among 204 samples that showed positive binding to the RBD of SARS‐CoV‐2, 158 (77.5%) cross‐reacted to the RBD of SARS‐CoV. There is a significant correlation in binding between these two RBD antigens (Pearson correlation = 0.43; Figure 2B). This result is consistent with the RBD having a higher sequence conservation compared to NTD. While the RBDs of SARS‐CoV‐2 and SARS‐CoV share 73% amino‐acid sequence identity, their NTDs only share 53% amino‐acid sequence identity (Supporting information Figure 2A and B).
Figure 2.
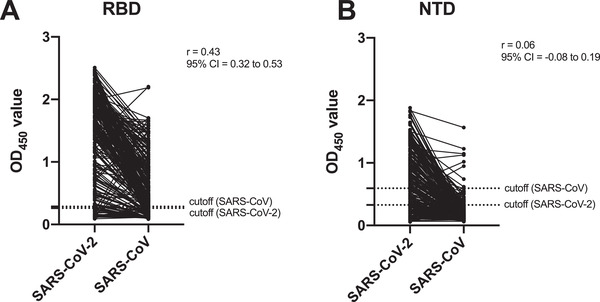
Cross‐reactive serological response to NTD and RBD protein between SARS‐CoV and SARS‐CoV‐2 (A and B) Pearson correlation (r) was used to evaluate the binding capacity of plasma to SARS‐CoV and SARS‐CoV‐2 NTD (A) and RBD (B) protein from 227 SARS‐CoV‐2 infected patients. The ELISA cutoff value of NTD protein to SARS‐CoV and SARS‐CoV‐2 were 0.5939 and 0.3272, and RBD protein to SARS‐CoV and SARS‐CoV‐2 were 0.2867 and 0.2607, respectively (mean + three standard deviations). Each sample was tested as duplicates in each assay and the results were confirmed by two independent experiments.
To explore whether SARS‐CoV‐2 infection can lead to serological responses that cross‐react with other human coronaviruses, we selected 118 plasma samples from the COVID‐19 patients and tested their binding to the spike proteins of all four known human seasonal coronaviruses, namely 229E, NL63, HKU‐1, and OC43. The results were compared to another 118 plasma samples from healthy blood donors that are age‐ and sex‐matched to the COVID‐19 cohort. As our control, the plasma of COVID‐19 patients showed a significantly higher level of binding to the NTD and RBD of SARS‐CoV‐2 compared to that of the healthy controls (Figure 3A and B). Compared to the plasma of healthy controls, the plasma of the COVID‐19 cohort exhibited significantly different of binding to the spike proteins of HKU1, OC43, and NL63 (Figure 3E‐G). However, the difference found in NL63 was much smaller than HKU1 and OC43. In contrast, plasma of healthy controls and COVID‐19 cohort showed no significant difference in binding to the spike proteins of 229E (Figure 3C). We also collected longitudinal plasma samples from six COVID‐19 patients and tested their binding to the NTD and RBD of SARS‐CoV‐2 as well as to the spikes of other human coronaviruses by ELISA (Supporting information Figure 3A‐F). While the increases in binding to the NTD and RBD of SARS‐CoV‐2 were more dramatic, some patients showed modest elevation of serological responses against the spike of different human coronaviruses, especially HKU1 and OC43. Of note, SARS‐CoV, SARS‐CoV‐2, HKU1, and OC43 are beta‐coronavirus, whereas NL63 and 229E are alpha‐coronavirus. Our results suggest that memory B cells with epitopes that are conserved among different beta‐coronaviruses were boosted after SARS‐CoV‐2 infection. Consistently, recent studies have shown that antibodies targeting the S2 domain can acquire broad reactivity among beta‐coronaviruses [32, 33].
Figure 3.
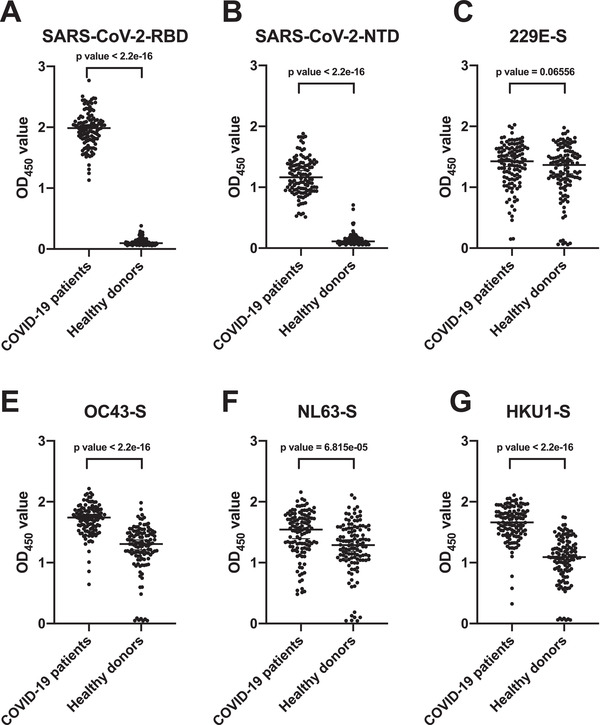
Cross‐reactive serological response to human coronaviruses between COVID‐19 patients and healthy donors (A and B) Binding of plasma samples to SARS‐CoV‐2 NTD (A), SARS‐CoV‐2 RBD (B), 229E‐Spike (C), NL63‐Spike (D), HKU1‐Spike (E), and OC43‐Spike protein (F) were tested by ELISA assay from 118 COVID‐2019 patients and age‐ and sex‐matched healthy donors. The OD450 value from each dot in the figure was taken by means of two replicates in the same experiment. p‐Values were caluated using two‐tailed paired t‐test (***p < 0.001). Error bars repeesent strandard deviation. Each sample was tested as duplicates in each assay and the results were confirmed by two independent experiments.
Immunization of NTD alone in mice does not induce neutralizing antibody
Since NTD neutralizing antibodies have been shown to confer protection to SARS‐CoV‐2 [12, 22, 23, 24, 25, 26], we are interested in evaluating if the NTD protein itself is immunogenic and can potentially be a vaccine candidate. We adopted our previous immunization protocol where BALB/c mice were intraperitoneally (i.p.) immunized twice by SARS‐CoV‐2 or SARS‐CoV NTD protein with Addavax as adjuvant (Wu et al., 2019). Plasma samples were collected 14 days after the second immunization and their bindings to NTD of SARS‐CoV‐2 and SARS‐CoV were measured by ELISA. We found that immunization with SARS‐CoV‐2 NTD could induce homologous and heterologous binding antibodies to the NTD proteins of SARS‐CoV‐2 and SARS‐CoV (Figure 4A). However, no cross‐reactive binding was observed to the SARS‐CoV full spike protein (Figure 4C). Similarly, plasma samples from mice immunized with SARS‐CoV NTD (Figure 4A and C) could cross‐react with SARS‐CoV‐2 NTD protein, but not with the SARS‐CoV‐2 full spike. Although NTD binding antibodies could be induced, no viral neutralizing ability could be found after the immunization of either SARS‐CoV or SARS‐CoV‐2 NTD protein immunization (Figure 4B). As a control, we also tested plasma samples obtained from the mice immunized with live SARS‐CoV‐2 or SARS‐CoV and tested their bindings to NTD proteins [34]. In contrast to NTD immunization, mice immunized with the live SARS‐CoV‐2 or SARS‐CoV can only elicit NTD antibodies to the autologus strain (Figure 4A and C). No cross‐reactivity was found to the spike proteins of NL63, 229E, HKU‐1, and OC43 (Figure 4D). These observations suggest that there is a difference in antigenicity between NTD alone and NTD on the spike protein.
Figure 4.
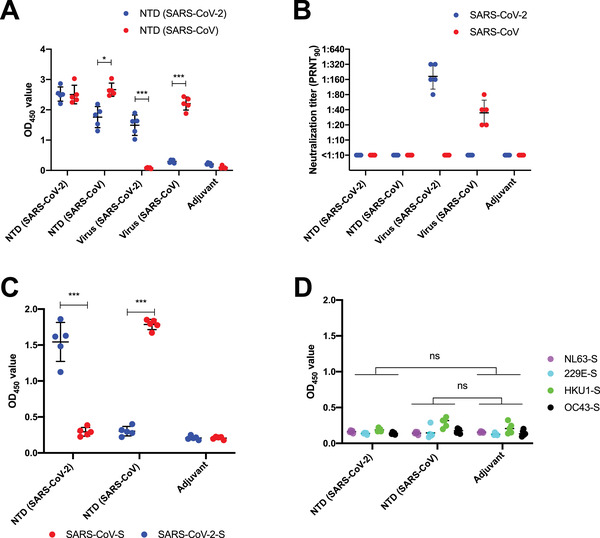
Serological binding and neutralizing capacity against SARS‐CoV and SARS‐CoV‐2 by NTD protein immunization (A) Binding of plasma from SARS‐CoV‐2 NTD protein immunized mice, SARS‐CoV NTD protein immunized mice, live SARS‐CoV‐2 immunized mice and live SARS‐CoV immunized mice against SARS‐CoV and SARS‐CoV‐2 NTD protein were measured by ELISA assay. The mean OD450 values calculated after detecting each plasma sample in duplicate are shown. (B) Neutralization activities of plasma from mice immunized with SARS‐CoV‐2 NTD protein, SARS‐CoV NTD protein, live SARS‐CoV‐2 and live SARS‐CoV were measured. The value from each dot in the figure was tested by the means of two replicates in the same assay. (C) Binding of plasma from SARS‐CoV and SARS‐CoV‐2 NTD protein immunized mice against the full spike of SARS‐CoV‐2 or SARS‐CoV. (D) Binding of plasma from SARS‐CoV and SARS‐CoV‐2 NTD protein immunized mice against NL63‐Spike, 229E‐Spike, HKU1‐Spike and OC43‐Spike protein were tested by ELISA assay. The OD450 value from each dot in the figure was taken by means of two replicates in the same experiment. p‐Values were caluated using two‐tailed t‐test. Error bars represent standard deviation. Each sample was tested as duplicates in each assay and the results were confirmed by two independent experiments.
Putative structural mechanism of altered antigenicity in NTD alone
To further understand the mechanism of differential antibody responses between immunizations with NTD alone and live virus, we performed a structural analysis of the NTD. A cluster of conserved residues on NTD is buried by the RBD on the spike protein (Figure 5A), but is solvent exposed when NTD is presented alone (Figure 5B). In contrast, the solvent exposed surface of NTD on the spike, including the antibody supersite [24] is much less conserved (Figure 5B and C). Together with our observations above, it is possible that, when immunization is performed using NTD, a reasonable percentage of antibodies are elicited to the conserved surface of NTD that is buried when presented on the spike. Besides, NTD is highly N‐glycosylated. It is possible that the N‐glycoforms are different between when NTD is expressed alone and when presented on the spike. Such differences may also contribute to the disparity in antigenicity. Therefore, our structural analysis offer an explanation of (1) why NTD immunization elicits antibodies that cross‐react with heterologous NTD but not heterologous spike protein (Figure 4A and C), and (2) why immunization with NTD but not live virus, which carries the full spike protein, elicit antibodies that cross‐react with heterologous NTD (Figure 2B).
Figure 5.
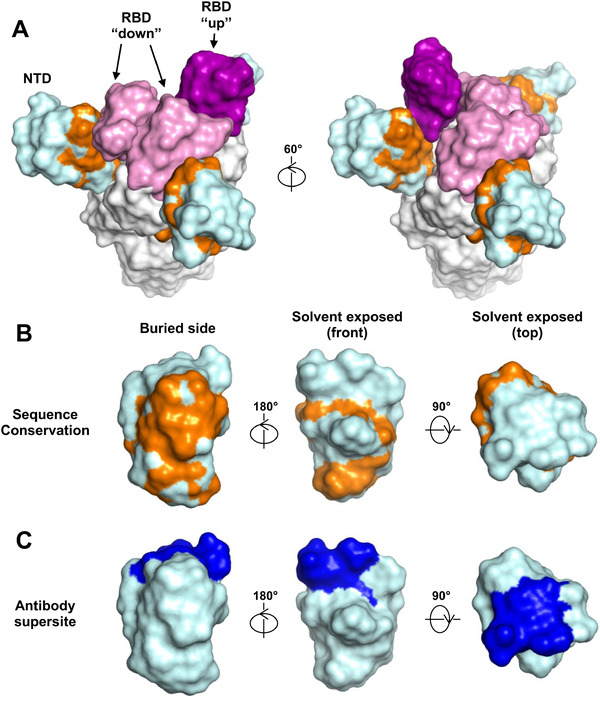
Conservation of NTD protein surface residues between SARS‐CoV‐2 and SARS‐CoV. (A‐B) Surface residues of NTD (cyan) that are conserved between SARS‐CoV‐2 and SARS‐CoV are highlighted in orange on (A) the spike protein where two RBD are in the down conformation (pink) and one RBD is in the up conformation (purple), and on (B) NTD alone. (C) NTD antibody supersites [24] highlighted in blue. Oligomannoses (yellow) were modeled by GlyProt [50].
Discussion
Identification of neutralizing antibodies and their targets on SARS‐CoV‐2 have been a major research area due to the importance for vaccine development. Over the past year, studies have shown that both RBD‐specific and NTD‐specific antibodies can confer potent neutralizing activity [3, 4, 12, 13, 14, 15, 22, 23, 24, 25]. However, while SARS‐CoV‐2 RBD protein can be effective in eliciting neutralizing antibodies [19, 20, 21], our study shows that NTD protein itself is a poor immunogen for eliciting neutralizing antibodies since its antigenicity is altered when expressed alone, where responses may be elicited to epitopes on the NTD that are inaccessible in the full spike protein. Our data substanticate that full spike, which is widely used in the development of mRNA [35, 36] and recombinant viral vectored vaccine [37], is a better immunogen than sudomains for eliciting neutralizing antibodies against SARS‐CoV‐2, especially since spike‐based immunogen is sufficient to elicit high neutralizing titer in human [38, 39]. In addition, the recent emergence of SARS‐CoV‐2 Variants of Concern, such as B.1.1.7 and B.1.351, have acquired mutations in the NTD supersite that can escape many NTD neutralizing antibodies [40, 41], further suggesting that NTD is not a good vaccine target.
Nevertheless, NTD protein can be a useful supplementary tool for serology testing. After SARS‐CoV‐2 infection, both RBD and NTD binding antibodies can be induced in the patient plasma samples after day 14 of symptom onset, suggesting both proteins are suitable for serology testing. In fact, the SARS‐CoV‐2 RBD protein has been using for serological diagnosis [17, 18]. However, RBD‐specific antibodies can be cross‐reactive among SARS‐CoV, SARS‐CoV‐2, and other Sarbecoviruses and may result in false‐positives [34, 42, 43]. Indeed, several cross‐reactive epitopes against RBD also have been identified between SARS‐CoV and SARS‐CoV‐2 [4, 14, 44]. In contrast, our results show that the cross‐reactivity of NTD‐specific antibodies to SARS‐CoV‐2/SARS‐CoV is much lower that RBD‐specific antibodies. Detection of NTD antibodies can be served as a validation assay in population based serology study. A recent study has shown that cross‐reactive antibodies against SARS‐CoV‐2 RBD were found from the blood of individuals collected before the outbreak [45]. It is thus expected that additional assay will be needed to validate the results of population serology screening in countries with prior SARS‐CoV‐1 outbreak or frequent contact with wildlife. In addition, NTD‐based serology can be also useful in animal surveillance of SARS‐like virus.
One interesting finding in our study is that some SARS‐CoV‐2 infected patients showed elevation of serological antibody responses against the spike proteins of another two human coronaviruses, HKU1 and OC43. Immunological imprinting in SARS‐CoV‐2 infected patients due to previous seasonal human coronavirus infection has also been reported [46, 47]. Consistently, two conserved cryptic epitopes located in the S2 domain have recently discovered that enable cross‐neutralization among five human‐infecting beta‐coronaviruses, including SARS‐CoV, SARS‐CoV‐2, MERS, and OC43 [32, 33, 48]. These observations open up the possibility to develop a more universal vaccine for beta‐coronaviruses.
Material and Methods
Virus and cell cultures
Vero and Vero E6 cells were maintained in DMEM medium supplemented with 10% FBS, and 100 U/mL of Penicillin‐Streptomycin. Sf9 cells (Spodoptera frugiperda ovarian cells, female) and High Five cells (Trichoplusia ni ovarian cells, female) were maintained in HyClone insect cell culture medium.
Patient‐derived SARS‐CoV‐2 (BetaCoV/Hong Kong/VM20001061/2020 [KH1]) and SARS‐CoV (strain HK39849, SCoV) were passaged in Vero‐E6 or Vero cells. The virus stock was aliquoted and titrated to determine tissue culture infection dose 50% (TCID50). The neutralization experiments were carried out in a Bio‐safety level 3 (BSL‐3) facility at the School of Public Health, LKS Faculty of Medicine, The University of Hong Kong.
Collection of specimens
Specimens of heparinized blood were collected from the RT‐PCR‐confirmed COVID‐19 patients at the Infectious Disease Centre of the Princess Margaret Hospital, Hong Kong. All study procedures were performed after informed consent. Plasma from healthy blood donors were collected from the Hong Kong Red Cross before the first COVID‐19 case reported on December 1, 2019 (March 2018 to November 2019). The study was approved by the institutional review board of the Hong Kong West Cluster of the Hospital Authority of Hong Kong (approval number: UW20‐169). Day 1 of clinical onset was defined as the first day of the appearance of clinical symptoms. The blood samples were centrifuged at 3000 × g for 10 min at room temperature for plasma collection. All plasma was kept in ‐80°C until used.
Mouse immunization
Six to 10 weeks old BALB/c mice were immunized with two rounds either 15 μg NTD protein or 105 TCID50 live viruses together with 50 μL Addavax, via intraperitoneal (i.p.) route. The boost dose was given to the mice 21 days after the first priming. The plasma samples were collected using heparin tubes on day 14 after the second round of immunization. The experiments were conducted in The University of Hong Kong Biosafety Level 3 (BSL3) facility. The study protocol was carried out in strict accordance with the recommendations and was approved by the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong (CULATR 5422‐20).
Protein expression and purification
The ectodomain (residues 14–1213) with R682G/R683G/R685G/K986P/V987P mutations, receptor‐binding domain (RBD, residues 319–541), and N‐terminal domain (NTD, residues 14 to 305) of the SARS‐CoV‐2 spike protein (GenBank: QHD43416.1), as well as the ectodomain (residues 14–1195) with K968P/V969P mutations, RBD (residues 306–527) and NTD (residues 14–292) of the SARS‐CoV spike protein (GenBank: ABF65836.1) were cloned into a customized pFastBac vector [31, 49]. The RBD and NTD constructs were fused with an N‐terminal gp67 signal peptide and a C‐terminal His6 tag. Recombinant bacmid DNA was generated using the Bac‐to‐Bac system (Life Technologies, Thermo Fisher Scientific). Baculovirus was generated by transfecting purified bacmid DNA into Sf9 cells using FuGENE HD (Promega, Madison, USA) and subsequently used to infect suspension cultures of High Five cells (Life Technologies) at an MOI of 5 to 10. Infected High Five cells were incubated at 28 °C with shaking at 110 rpm for 72 h for protein expression. The supernatant was then concentrated using a Centramate cassette (10 kDa molecular weight cutoff for RBD, Pall Corporation, New York, USA). RBD and NTD proteins were purified by Ni‐NTA Superflow (Qiagen, Hilden, Germany), followed by size exclusion chromatography and buffer exchange to PBS. The spike proteins of 229E, HKU1, NL63, and OC43 were purchased from Sino Biological (China).
Enzyme‐linked immunosorbent assay
A 96‐well ELISA plate (Nunc MaxiSorp, Thermo Fisher Scientific) was first coated overnight with 100 ng per well of purified recombinant protein in PBS buffer. The plates were then blocked with 100 μl of Chonblock blocking/sample dilution ELISA buffer (Chondrex Inc, Redmond, USA) and incubated at room temperature for 1 h. Each human plasma sample was diluted to 1:100 in Chonblock blocking/sample dilution ELISA buffer. Each sample was then added into the ELISA plates for a 2‐h incubation at 37°C. After extensive washing with PBS containing 0.1% Tween 20, each well in the plate was further incubated with the anti‐human IgG secondary antibody (1:5000, Thermo Fisher Scientific) for 1 h at 37°C. The ELISA plates were then washed five times with PBS containing 0.1% Tween 20. Subsequently, 100 μL of HRP substrate (Ncm TMB One; New Cell and Molecular Biotech Co. Ltd, Suzhou, China) was added into each well. After 15 min of incubation, the reaction was stopped by adding 50 μL of 2 M H2SO4 solution and analyzed on a Sunrise (Tecan, Männedorf, Switzerland) absorbance microplate reader at 450 nm wavelength [17].
Plaque reduction neutralization test
Plasma samples were twofold diluted starting from a 1:10 dilution and mixed with equal volumes of around 120 plaque‐forming units (pfu) of SARS‐CoV‐2 or SARS‐CoV as determined by Vero E6 and Vero cells, respectively. After 1‐h incubation at 37°C, the plasma‐virus mixture was added onto cell monolayers seated in a 24‐well cell culture plate and incubated for 1 h at 37°C with 5% CO2. The plasma‐virus mixtures were then discarded and infected cells were immediately covered with 1% agarose gel in DMEM medium. After incubation for 3 days at 37°C with 5% CO2, the plates were formalin fixed and stained by 0.5% crystal violet solution. Neutralization titers were determined by the highest plasma dilution that resulted in >90% reduction in the number of pfus. The test was performed in a BSL3 facility at the University of Hong Kong [17].
Statistical analysis
We defined a sample as ELISA antibody positive if the OD value was 3 standard deviations above the mean of the negative controls. Significance between two groups were determined by Mann‐Whitney test with p‐values lower than 0.05. Correlation between plasma samples were assessed using Pearson correlation coefficients. Two‐tailed paired t‐tests were performed and are shown in Figure 3.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Author contributions
H.L., N.C.W., J.S.M.P., and C.K.P.M. conceived the research idea, planned the study, obtained research funding, analyzed the data, and wrote the manuscript. M.Y., H.L., I.A.W., and N.C.W. expressed and purified the proteins. H.L., R.T.Y.S., Y.W., G.K.Y., Q.T, Y.L., W.L., J.W., and W.W.N performed the experiments. O.T.Y.T organized patient recruitment, data collection, and sampling.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202149234
Abbreviations
- ACE2
angiotensin converting enzyme 2
- COVID‐19
coronavirus disease 2019
- NTD
N‐terminal domain
- RBD
receptor binding domain
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
Supporting information
Supporting information
Acknowledgments
This work was supported by Calmette and Yersin scholarship from the Pasteur International Network Association (H.L.), Bill and Melinda Gates Foundation OPP1170236 and INV‐004923 (I.A.W.), startup funds from the University of Illinois at Urbana‐Champaign (N.C.W.), the US National Institutes of Health (contract no. HHSN272201400006C) (J.S.M.P), National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) Joint Research Scheme (N_HKU737/18) (C.K.P.M. and J.S.M.P), the National Research Foundation of Korea (NRF) grant funded through the Korea government (NRF‐2018M3A9H4055203) (C.K.P.M.), the Research Grants Council of the Hong Kong Special Administrative Region, China (Project no. T11‐712/19‐N) (J.S.M.P) and Guangdong Province International Scientific and Technological Cooperation Projects (grant number 2020A0505100063) (C.K.P.M). We acknowledge the support of the clinicians who facilitated this study, including Drs Wai Shing Leung, Jacky Man Chun Chan, Thomas Shiu Hong Chik, John Yu Hong Chan, Daphne Pui‐Lin Lau, and Ying Man Ho, and the dedicated clinical team at Infectious Diseases Centre, Princess Margaret Hospital, Hospital Authority of Hong Kong and the patients who kindly consented to participate in this investigation.
Contributor Information
Nicholas C. Wu, Email: nicwu@illinois.edu.
Chris K. P. Mok, Email: kapunmok@cuhk.edu.hk.
Data availability statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1. Liu, Q. , Xu, K. , Wang, X. and Wang, W. , From SARS to COVID‐19: What lessons have we learned? J Infect Public Health 2020.13: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu, Y. , Yan, L. M. , Wan, L. , Xiang, T. X. , Le, A. , Liu, J. M. , Peiris, M. et al., Viral dynamics in mild and severe cases of COVID‐19. Lancet Infect Dis 2020. 20: 656–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnes, C. O. , Jette, C. A. , Abernathy, M. E. , Dam, K. A. , Esswein, S. R. , Gristick, H. B. , Malyutin, A. G. et al., SARS‐CoV‐2 neutralizing antibody structures inform therapeutic strategies. Nature 2020.588 7839:682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pinto, D. , Park, Y. J. , Beltramello, M. , Walls, A. C. , Tortorici, M. A. , Bianchi, S. , Jaconi, S. et al., Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature 2020. 583: 290–295. [DOI] [PubMed] [Google Scholar]
- 5. Brouwer, P. J. M. , Caniels, T. G. , van der Straten, K. , Snitselaar, J. L. , Aldon, Y. , Bangaru, S. , Torres, J. L. , Okba, N. M. A. et al., Potent neutralizing antibodies from COVID‐19 patients define multiple targets of vulnerability. Science 2020. 369: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Long, Q. X. , Liu, B. Z. , Deng, H. J. , Wu, G. C. , Deng, K. , Chen, Y. K. , Liao, P. et al., Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med 2020. 26: 845–848. [DOI] [PubMed] [Google Scholar]
- 7. Jiang, H. W. , Li, Y. , Zhang, H. N. , Wang, W. , Yang, X. , Qi, H. , Li, H. et al., SARS‐CoV‐2 proteome microarray for global profiling of COVID‐19 specific IgG and IgM responses. Nat Commun 2020. 11: 3581–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ripperger, T. J. , Uhrlaub, J. L. , Watanabe, M. , Wong, R. , Castaneda, Y. , Pizzato, H. A. , Thompson, M. R. et al., Orthogonal SARS‐CoV‐2 serological assays enable surveillance of low‐prevalence communities and reveal durable humoral immunity. Immunity 2020. 53: 925–933 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dan, J. M. , Mateus, J. , Kato, Y. , Hastie, K. M. , Yu, E. D. , Faliti, C. E. , Grifoni, A. et al., Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science 2021.371 6529:eabf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lau, E. H. Y. , Tsang, O. T. Y. , Hui, D. S. C. , Kwan, M. Y. W. , Chan, W. H. , Chiu, S. S. , Ko, R. L. W. et al., Neutralizing antibody titres in SARS‐CoV‐2 infections. Nat. Commun. 2021. 12: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walls, A. C. , Park, Y. J. , Tortorici, M. A. , Wall, A. , McGuire, A. T. and Veesler, D. , Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 2020. 181: 281–292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu, L. , Wang, P. , Nair, M. S. , Yu, J. , Rapp, M. , Wang, Q. , Luo, Y. et al., Potent neutralizing antibodies against multiple epitopes on SARS‐CoV‐2 spike. Nature 2020. 584: 450–456. [DOI] [PubMed] [Google Scholar]
- 13. Ju, B. , Zhang, Q. , Ge, J. , Wang, R. , Sun, J. , Ge, X. , Yu, J. et al., Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature 2020. 584: 115–119. [DOI] [PubMed] [Google Scholar]
- 14. Liu, H. , Wu, N. C. , Yuan, M. , Bangaru, S. , Torres, J. L. , Caniels, T. G. , van Schooten, J. et al., Cross‐neutralization of a SARS‐CoV‐2 antibody to a functionally conserved site is mediated by avidity. Immunity 2020. 53: 1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seydoux, E. , Homad, L. J. , MacCamy, A. J. , Parks, K. R. , Hurlburt, N. K. , Jennewein, M. F. , Akins, N. R. et al., Analysis of a SARS‐CoV‐2‐infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity 2020. 53: 98–105 e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rogers, T. F. , Zhao, F. , Huang, D. , Beutler, N. , Burns, A. , He, W. T. , Limbo, O. et al., Isolation of potent SARS‐CoV‐2 neutralizing antibodies and protection from disease in a small animal model. Science 2020. 369: 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perera, R. A. , Mok, C. K. , Tsang, O. T. , Lv, H. , Ko, R. L. , Wu, N. C. , Yuan, M. et al., Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), March 2020. Euro Surveill. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Premkumar, L. , Segovia‐Chumbez, B. , Jadi, R. , Martinez, D. R. , Raut, R. , Markmann, A. , Cornaby, C. et al., The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS‐CoV‐2 patients. Sci Immunol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai, L. , Zheng, T. , Xu, K. , Han, Y. , Xu, L. , Huang, E. , An, Y. et al., A universal design of Betacoronavirus vaccines against COVID‐19, MERS, and SARS. Cell 2020. 182: 722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tai, W. , Zhang, X. , Drelich, A. , Shi, J. , Hsu, J. C. , Luchsinger, L. , Hillyer, C. D. et al., A novel receptor‐binding domain (RBD)‐based mRNA vaccine against SARS‐CoV‐2. Cell Res 2020. 30: 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zang, J. , Gu, C. , Zhou, B. , Zhang, C. , Yang, Y. , Xu, S. , Bai, L. et al., Immunization with the receptor‐binding domain of SARS‐CoV‐2 elicits antibodies cross‐neutralizing SARS‐CoV‐2 and SARS‐CoV without antibody‐dependent enhancement. Cell Discov 2020. 6: 61–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chi, X. , Yan, R. , Zhang, J. , Zhang, G. , Zhang, Y. , Hao, M. , Zhang, Z. et al., A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science 2020. 369: 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suryadevara, N. , Shrihari, S. , Gilchuk, P. , Van Blargan, L. A. , Binshtein, E. , Zost, S. L. , Nargi, R. S. et al., Neutralizing and protective human monoclonal antibodies recognizing the N‐terminal domain of the SARS‐CoV‐2 spike protein. Cell 2021. 184 9:2316–2331.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCallum, M. , De Marco, A. , Lempp, F. , Tortorici, M. A. , Pinto, D. , Walls, A. C. , Beltramello, M. et al., N‐terminal domain antigenic mapping reveals a site of vulnerability for SARS‐CoV‐2. Cell 2021. 184 9:2332–2347.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cerutti, G. , Guo, Y. , Zhou, T. , Gorman, J. , Lee, M. , Rapp, M. , Reddem, E. R. et al., Potent SARS‐CoV‐2 neutralizing antibodies directed against spike N‐terminal domain target a single supersite. Cell Host Microbe 2021. 29 5:819–833.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noy‐Porat, T. , Mechaly, A. , Levy, Y. , Makdasi, E. , Alcalay, R. , Gur, D. , Aftalion, M. et al., Therapeutic antibodies, targeting the SARS‐CoV‐2 spike N‐terminal domain, protect lethally infected K18‐hACE2 mice. iScience 2021. 24 5:102479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang, P. , Liu, L. , Iketani, S. , Luo, Y. , Guo, Y. , Wang, M. , Yu, J. et al., Increased resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv 2021. Preprint. 10.1101/2021.01.25.428137. [DOI] [Google Scholar]
- 28. Wang, S. , Qiu, Z. , Hou, Y. , Deng, X. , Xu, W. , Zheng, T. , Wu, P. et al., AXL is a candidate receptor for SARS‐CoV‐2 that promotes infection of pulmonary and bronchial epithelial cells. Cell research 2021: 31: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosa, A. , Pye, V. E. , Graham, C. , Muir, L. , Seow, J. , Ng, K. W. , Cook, N. J. et al., SARS‐CoV‐2 can recruit a heme metabolite to evade antibody immunity. Sci Adv 2021. 7 22:eabg7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen, X. , Pan, Z. , Yue, S. , Yu, F. , Zhang, J. , Yang, Y. , Li, R. et al., Disease severity dictates SARS‐CoV‐2‐specific neutralizing antibody responses in COVID‐19. Signal Transduct Target Ther 2020. 5: 180–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lv, H. , Wu, N. C. , Tsang, O. T. , Yuan, M. , Perera, R. , Leung, W. S. , So, R. T. Y. et al., Cross‐reactive antibody response between SARS‐CoV‐2 and SARS‐CoV infections. Cell Rep. 2020. 31: 107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sauer, M. , Tortorici, M. A. , Park, Y. , Walls, A. C. , Homad, L. , Acton, O. , Bowen, J. E. et al., Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol 2021. doi: 28 6:478–486. 10.1101/2020.12.29.424482. [DOI] [PubMed] [Google Scholar]
- 33. Huang, Y. , Nguyen, A. W. , Hsieh, C.‐L. , Silva, R. , Olaluwoye, O. S. , Wilen, R. , Kaoud, T. S. et al., Identification of a conserved neutralizing epitope present on spike proteins from highly pathogenic coronaviruses. bioRxiv 2021. Preprint. 10.1101/2021.01.31.428824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lv, H. , So, R. T. , Yuan, M. , Liu, H. , Lee, C.‐C. D. , Yip, G. K. , Ng, W. W. et al., Evidence of antigenic imprinting in sequential Sarbecovirus immunization. bioRxiv 2020. Preprint 10.1101/2020.10.14.339465. [DOI] [Google Scholar]
- 35. Jackson, L. A. , Anderson, E. J. , Rouphael, N. G. , Roberts, P. C. , Makhene, M. , Coler, R. N. , McCullough, M. P. et al., An mRNA Vaccine against SARS‐CoV‐2 ‐ Preliminary Report. N Engl J Med 2020. 383: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Polack, F. P. , Thomas, S. J. , Kitchin, N. , Absalon, J. , Gurtman, A. , Lockhart, S. , Perez, J. L. et al., Safety and Efficacy of the BNT162b2 mRNA Covid‐19 Vaccine. N Engl J Med 2020. 383: 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu, S. , Zhong, G. , Zhang, J. , Shuai, L. , Zhang, Z. , Wen, Z. , Wang, B. et al., A single dose of an adenovirus‐vectored vaccine provides protection against SARS‐CoV‐2 challenge. Nat Commun 2020. 11: 4081–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang, Z. , Schmidt, F. , Weisblum, Y. , Muecksch, F. , Barnes, C. O. , Finkin, S. , Schaefer‐Babajew, D. et al., mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature 2021.592 7855:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Widge, A. T. , Rouphael, N. G. , Jackson, L. A. , Anderson, E. J. , Roberts, P. C. , Makhene, M. , Chappell, J. D. et al., Durability of Responses after SARS‐CoV‐2 mRNA‐1273 Vaccination. N Engl J Med 2021. 384: 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang, P. , Nair, M. S. , Liu, L. , Iketani, S. , Luo, Y. , Guo, Y. , Wang, M. et al., Antibody resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7. Nature 2021.593: 130–135. [DOI] [PubMed] [Google Scholar]
- 41. McCallum, M. , De Marco, A. , Lempp, F. A. , Tortorici, M. A. , Pinto, D. , Walls, A. C. , Beltramello, M. et al., N‐terminal domain antigenic mapping reveals a site of vulnerability for SARS‐CoV‐2. Cell 2021.184 9:2332–2347.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rappazzo, C. G. , Longping, V. T. , Kaku, C. I. , Wrapp, D. , Sakharkar, M. , Huang, D. , Deveau, L. M. et al., Broad and potent activity against SARS‐like viruses by an engineered human monoclonal antibody. Science 2021.371: 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cui, J. , Li, F. and Shi, Z.‐L. , Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019. 17: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yuan, M. , Wu, N. C. , Zhu, X. , Lee, C. D. , So, R. T. Y. , Lv, H. , Mok, C. K. P. et al., A highly conserved cryptic epitope in the receptor binding domains of SARS‐CoV‐2 and SARS‐CoV. Science 2020. 368: 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Anderson, E. M. , Goodwin, E. C. , Verma, A. , Arevalo, C. P. , Bolton, M. J. , Weirick, M. E. , Gouma, S. et al., Seasonal human coronavirus antibodies are boosted upon SARS‐CoV‐2 infection but not associated with protection. Cell 2021. 184: 1858–1864 e1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson, E. M. , Goodwin, E. C. , Verma, A. , Arevalo, C. P. , Bolton, M. J. , Weirick, M. E. , Gouma, S. et al., Seasonal human coronavirus antibodies are boosted upon SARS‐CoV‐2 infection but not associated with protection. Cell 2020. 184 7:1858–1864.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aydillo, T. , Rombauts, A. , Stadlbauer, D. , Aslam, S. , Abelenda‐Alonso, G. , Escalera, A. , Amanat, F. et al., Antibody immunological imprinting on COVID‐19 patients. medRxiv 2020. Preprint 10.1101/2020.10.14.20212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Song, G. , He, W.‐t. , Callaghan, S. , Anzanello, F. , Huang, D. , Ricketts, J. , Torres, J. L. et al., Cross‐reactive serum and memory B cell responses to spike protein in SARS‐CoV‐2 and endemic coronavirus infection. bioRxiv 2020. 10.1101/2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wec, A. Z. , Wrapp, D. , Herbert, A. S. , Maurer, D. P. , Haslwanter, D. , Sakharkar, M. , Jangra, R. K. et al., Broad neutralization of SARS‐related viruses by human monoclonal antibodies. Science 2020. 369: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bohne‐Lang, A. and von der Lieth, C. W. , GlyProt: in silico glycosylation of proteins. Nucleic Acids Res 2005. 33: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


