Abstract
Purpose
For this study we aimed to understand if retinal pigment epithelial (RPE) cells express antimicrobial peptide lysozyme as a mechanism to protect the neuroretina from blood-borne pathogens.
Methods
The expression of lysozyme in human and mouse RPE cells was examined by RT-PCR or immune (cyto)histochemistry in cell cultures or retinal sections. RPE cultures were treated with different concentrations of Pam3CSK4, lipopolysaccharides (LPS), staphylococcus aureus-derived peptidoglycan (PGN-SA), Poly(I:C), and Poly(dA:dT). The mRNA expression of lysozyme was examined by qPCR and protein expression by ELISA. Poly(I:C) was injected into the subretinal space of C57BL/6J mice and eyes were collected 24 hours later and processed for the evaluation of lysozyme expression by confocal microscopy. Bactericidal activity was measured in ARPE19 cells following LYZ gene deletion using Crispr/Cas9 technology.
Results
The mRNA and protein of lysozyme were detected in mouse and human RPE cells under normal conditions, although the expression levels were lower than mouse microglia BV2 or human monocytes THP-1 cells, respectively. Immunohistochemistry showed punctate lysozyme expression inside RPE cells. Lysozyme was detected by ELISA in normal RPE lysates, and in live bacteria-treated RPE supernatants. Treatment of RPE cells with Pam3CSK4, LPS, PGN-SA, and Poly(I:C) enhanced lysozyme expression. CRISPR/Cas9 deletion of lysozyme impaired bactericidal activity of ARPE19 cells and reduced their response to LPS and Poly(I:C) stimulation.
Conclusions
RPE cells constitutively express antimicrobial peptide lysozyme and the expression is modulated by pathogenic challenges. RPE cells may protect the neuroretina from blood-borne pathogens by producing antimicrobial peptides, such as lysozyme.
Keywords: innate immunity, antimicrobial peptides, central nervous system (CNS), blood-retina barrier (BRB), blood-borne pathogens, Crispr/Cas9
The central nervous system (CNS), including the neuronal retina is segregated from the systemic immune system by the blood-brain barrier (BBB) or blood-retina barrier (BRB), which protects the CNS and the retina from invading pathogens and immune cell infiltration.1–3 The BBB is formed by tight junctions (TJs) of CNS vascular endothelial cells; whereas the BRB includes the inner BRB (iBRB; i.e. TJs between retinal vascular endothelial cells, and the outer BRB [oBRB]; i.e. the TJs on retinal pigment epithelial [RPE] cells).4,5 In addition to the physical barrier, cells of the BRB and BBB also produce various immune regulatory molecules that can suppress immune cell activation or induce programmed cell death in activated immune cells, a phenomenon known as “immunological barrier.”6–8 Together, the physical and immunological barriers of BBB and BRB ensure that the vital and highly sensitive neurons are not subject to unwanted inflammatory insults.
The integrity and permeability of BRB and BBB, however, are dynamic and can be affected by many factors, including the gut microbiota, chronic inflammation, aging, and even the sleep-wake cycle.9–11 The weakening of the barrier function, even a short period under physiological conditions, may put the CNS and retina under threat to blood-borne pathogens.12 The blood-borne pathogens may penetrate the BBB or BRB by three routes: transcellular entry, paracellular route, and infected leukocytes (so-called “Trojan horse”)12 and the weakening of the barrier function may promote their penetration leading to acute or latent infection. The immunological barrier of BBB or BRB is usually viewed as a mechanism to maintain the immune privilege of the CNS; it is also a major checkpoint for pathogen entry to the CNS parenchyma. Cells of the BBB and BRB, including endothelium, astrocyte, and RPE cell express various pattern recognition receptors, such as Toll-like receptors (TLRs), NOD-like receptors (NLRs), and RIG-like receptors (RLRs).13–15 Activation of the TLRs or NLRs upon engaging with blood-borne pathogens can lead to acute inflammation in the CNS (e.g. meningitis) or the retina (e.g. retinitis), although in many cases the response may be suppressed due to the immunological barrier and pathogens may persist as latent infections.12
The CNS parenchymal cells, such as neurons and astrocytes, can produce amyloid precursor proteins (APPs) that can be enzymatically cleaved into β-amyloid (Aβ),16 and accumulation of Aβ plays an important pathogenic role in Alzheimer's disease.16 Interestingly, Aβ is also an antimicrobial peptide and can protect against microbial infection in mouse and worm models of Alzheimer's disease.17,18 The fact that neurons can produce antimicrobial peptides to fight against invading pathogens led us to speculate that cells of the CNS barrier (i.e. BBB and BRB) may also express antimicrobial enzymes as a mechanism to protect the CNS from blood-borne pathogens.
In this study, we investigated the expression and production of lysozyme, an antimicrobial enzyme widely produced by various cells in particular innate immune cells of animals and humans, in cells of the oBRB (i.e. RPE cells). The oBRB segregates the neuroretina from the highly vascularized choroid. It also critically contributes to the immune privilege (IP) of the subretinal space, for example, by producing various immune checkpoint molecules.8,19 Interestingly, we found that both human and mouse RPE cells constitutively express lysozyme and their production can be further enhanced upon infection.
Materials and Methods
Animals and Subretinal Injection
Adults C57BL/6J mice between 6 and 8 weeks of age were purchased from Silaike Jingda Laboratory Animal Co. Ltd. (Changsha, HN, China) and maintained in a specific pathogen-free (SPF) animal house facility of Central South University on a 12-hour day/night cycle with free access to food and water. All experimental protocols concerning animals in this study were approved by the Animal Welfare and Ethical Review body of Central South University and the procedures were carried out in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. Both male and female mice were used in RPE cell cultures and immunohistochemistry.
The subretinal injection was performed in female C57BL/6J mice using a protocol described previously.20 Briefly, mice were anaesthetized by an intraperitoneal injection of pentobarbital (60 mg/kg; Sigma Aldrich, ShangHai, China), the pupils were dilated with 1% Tropicamide Phenylephrine (Santen Pharmaceutical Co., Ltd., Osaka, Japan). A 1.5 µl of Poly(I:C) (1 μg/μl; InvivoGen, San Diego, CA, USA) was injected into the subretinal space using a 33-gauge beveled needle (Hamilton Bonaduz AG, Switzerland) under a surgical microscope. The same volume of PBS was served as a control.
Cell Culture and Treatment
The human RPE cell line ARPE19 cells and mouse microglia BV2 cells were cultured in Dulbecco's Modified Eagle's Medium and Ham's F12 1:1 (DMEM/F12; Gibco, Shanghai, China) containing 100 U/mL penicillin, 100 μg/mL streptomycin (Thermo Fisher Scientific, Shanghai, China), and 10% fetal bovine serum (FBS; Gibco). The primary mouse RPE (mRPE) cells were isolated from murine eyes and cultured in DMEM with 10% FBS as described previously.21 RPE phenotype was confirmed by RPE65 staining. The human monocyte cell line, THP-1 cells were cultured in RPMI (Thermo Fisher Scientific) with 10% fetal calf serum (FCS).
Treatment with TLR Agonists and dsDNA
ARPE19 or THP-1 cells were seeded in 6-well plates at a density of 1 × 106cells/ well. After reaching confluence, cells were treated with lipopolysaccharide (LPS; 100 ng/mL; Sigma-Aldrich), Pam3CSK4, staphylococcus aureus-derived peptidoglycan (PGN-SA), Poly(I:C), or Poly(dA:dT), all from InvivoGen. Poly(I:C) and Poly(dA:dT) were transfected with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's instructions. Cells were harvested 24 hours later for total RNA or protein extraction. Pam3CSK4 is a synthetic triacylated lipopeptide (LP) that can bind TLR2/1 and is used to mimic the acylated amino terminus of bacterial LPs. PGN-SA is a peptidoglycan preparation from the Gram-positive bacterium S. Aureus and a potent TLR2 agonist. Poly(I:C) is a synthetic double-stranded RNA (dsRNA) and Poly(dA:dT) is a synthetic double-stranded DNA (dsDNA) and they are often used to study immune response to RNA and DNA virus infections.
Knockout Lysozyme in ARPE19 Cells using Clustered Regularly Interspaced Short Palindromic Repeats/Caspase 9
Guide RNA (gRNA)5′-CACCGGAGACAGAAGCACTGATTA-3′ was designed at the MIT Clustered Regularly Interspaced Short Palindromic Repeats/Caspase 9 (CRISPR) web site (http://crispr.mit.edu). The gRNA oligos (Tsingke, BeiJing, China) were phosphorylated, annealed, and cloned into PU6-gRNAcas9puro plasmid (Genepharma, Shanghai, China). All inserts were verified with Sanger sequencing. Then, 2 μg PU6-lysozyme-gRNA cas9puro plasmid was transfected into 2 × 106 ARPE19 cells using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). Forty-eight hours after the transfection, cells were selected with 1 μg/mL puromycin (Thermo Fisher Scientific) for 5 days. The qPCR was used to verify lysozyme expression in CRISPR/Cas9 edited ARPE19 cells. ARPE19 cells transfected with the vector PU6-gRNAcas9puro plasmid were used as controls.
Semi-Quantitative Reverse Transcription PCR and Real-Time qPCR
Total mRNA was extracted from several cultured cells using the Total RNA Kit II (Omega, Norcross, GA, USA) according to the manufacturer's instructions. Then, 1 μg of total RNA was used to synthesize cDNA using the PrimeScript RT Reagent Kit (Vazyme Biotech Co., Nanjing, China). The cDNA of primary human RPE cells was generously gifted from Dr. Gu.22 Semiquantitative reverse transcription PCR and real-time PCR was performed using TaKaRa Taq HS Perfect Mix (Takara Bio, Kusatsu, Japan) and ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech), respectively.
The primers used in this study were designed using the NCBI Primer BLAST system and the primers sequences are detailed in the Table.
Table.
Primer Sequences of Human and Mouse Lysozyme and Beta Actin Genes
| Gene | Sequence | Gene Bank Access No. |
|---|---|---|
| Human | ||
| LYZ | Forward: 5′-ATCAGCCTAGCAAACTGGAT-3′ | NM_000239.3 |
| Reverse:5′-CTCCACAACCTTGAACATAC-3′ | ||
| CCL2 | Forward: 5′-GAGAGGCTGAGACTAACCCA-3′ | NM_002982.4 |
| Reverse: 5′-ATCACAGCTTCTTTGGGACAC-3′ | ||
| VEGFa | Forward: 5′-GTTGTGTGTGTGTGAGTGGTTG-3′ | NM_001025366.3 |
| Reverse: 5′-TTTCTCTTTTCTCTGCCTCCAG-3′ | ||
| TGFB1 | Forward: 5'-CCCAGCATCTGCAAAGCTC-3' | NM_000660.7 |
| Reverse: 5'-GTCAATGTACAGCTGCCGCA-3' | ||
| THBS1 | Forward: 5′-TGCTATCACAACGGAGTTCAGT-3′ | NM_003246.4 |
| Reverse: 5′-GCAGGACACCTTTTTGCAGATG-3′ | ||
| IL6 | Forward: 5′-CCAGCTATGAACTCCTTCTC-3′ | NM_000600.5 |
| Reverse: 5′-GCTTGTTCCTCACATCTCTC-3′ | ||
| IL1B | Forward: 5′-AAGCTGATGGCCCTAAACAG -3′ | NM_000576.3 |
| Reverse: 5′-AGGTGCATCGTGCACATAAG -3′ | ||
| TNF | Forward: 5′-ATGGGCTACAGGCTTGTCACTC-3′ | NM_ 000594 |
| Reverse: 5′-CTCTTCTGCCTGCTGCACTTTG-3′ | ||
| ACTB | Forward: 5′-ACAGAGCCTCGCCTTTGC-3′ | NM_001101.3 |
| Reverse: 5′-ATCACGCCCTGGTGCCT-3′ | ||
| Mouse | ||
| Lyz2 | Forward: 5′-CCTCTGTAGGTCAGTTC-3′ | AH001999.2 |
| Reverse: 5′-ATCAACTGGTCTCCTATAA-3′ | ||
| ACTB | Forward: 5′-CCTTCCTTCTTGGGTATG-3′ | XM_030254057.1 |
| Reverse: 5′-TGTAAAACGCAGCTCAGTAA-3′ | ||
PCR products were confirmed by agarose gel electrophoresis and melting temperature. The density analysis was performed by Image J (National Institutes of Health, Bethesda, MD, USA).
Bacterial Killing Assays
Single colonies of Escherichia coli (E. coli) bacterial strain DH5α (Transgene, Beijing, China) were picked and grown overnight at 37°C in Lysogeny Broth (LB) medium (Sangon, Shanghai, China). The next day, the E. coli cells were diluted at least 1:50 in fresh LB broth and grown to mid-log phase (OD600 between 0.4 and 0.6). To assess the bactericidal activity of ARPE19 cells, 1.8 × 106 of ARPE19 cells were cultured in 6-well plates in DMEM/F12 medium, E. coli were added to the well at multiplicity of infection (MOI) 1:1 (1.8 × 106 CFU/mL). The supernatant was collected 4 hours after the ARPE19 cells/E. coli co-culture. The supernatants were diluted in sterile ddH2O at 1:106 and plated onto LB agar. The ARPE19 cells were harvested and re-suspended in PBS contained 1/1000 Saponin (Sigma-Aldrich) and incubated for 5 minutes at room temperature. The digested cell pellets were plated onto LB agar. All plates were incubated at 37°C for 18 hours and CFU/mL were then calculated.
Enzyme-Linked Immunosorbent Assay
The expression of lysozyme in human RPE (ARPE19) cells and monocytes (THP-1) were measured using the human lysozyme ELISA kit (CUSABIO, Wuhan, China) according to the manufacturer's instructions. In brief, protein lysates were extracted using RIPA Lysis Buffer (Thermo Fisher Scientific) supplemented with cocktails of protease inhibitors and phosphatase inhibitors. The protein concentration was determined by BCA assay (Solarbio, Beijing, China). Equal quantities of protein were loaded into each well of 96 well plates coated with lysozyme-capturing antibody and incubated at room temperature for 2 hours. After washing, 100 μL of anti-lysozyme antibody conjugated with horseradish peroxidase (HRP) was added and incubated for 30 minutes at room temperature, followed by 100 μL of substrate incubation. The plate was read at 450 nm using a microplate reader (Tecan, Männedorf, Switzerland).
Immunofluorescence Staining
Immunofluorescence staining of mouse eye sections was conducted using a protocol described previously.23 In brief, mouse eyes were enucleated and embedded in optimal cutting temperature compound and stored at -80°C until use. Serial cryosections (10 µm thick) were prepared with a cryostat and fixed in 2% paraformaldehyde for 10 minutes. The samples were blocked with 10% goat serum (Servicebio Inc., Wuhan, China) in 2% BSA + 0.1% Triton X-100 in PBS for 1 hour at room temperature. The slides were then incubated overnight with rabbit anti-lysozyme (1:100; Abcam, Cambridge, UK) and rat anti-CD11b (1:100; Novus Biologicals, Abingdon, UK) at 4°C. Rabbit IgG was used as an isotype control to ensure the specificity of lysozyme staining. After thorough washes, samples were incubated with Alexa Fluor 594 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-rat IgG (1:500; Invitrogen, Waltham, MA, USA) for 1 hour at room temperature. Because lysozyme is known to be produced predominately by innate immune cells (e.g. CD11b+ neutrophils and macrophages), we used CD11b antibody to differentiate ocular lysozyme-expressing innate immune cells from nonimmune cells. Samples were imaged using the Zeiss LSM 880 Confocal Microscope (Zeiss, Braunschweig, Germany). The fluorescence intensity of lysozyme in RPE cells from different groups was measured using the ZEN Lite software (Zeiss). The changes in lysozyme expression levels in PSB or Poly(I:C) injected mice were calculated by dividing the fluorescence intensity of each section from treated eyes with the average fluorescence intensity of RPE lysozyme from normal eyes.
RPE cells (including ARPE19 and mRPE) and BV2 cells on coverslips were fixed with pre-cooled methanol for 10 minutes on the ice and blocked with 10% goat serum for 30 minutes at room temperature. The cells were then incubated with rabbit anti-lysozyme (1:100) and rat anti-CD11b (1:100 for BV2 cells) or mouse anti-RPE65 (1:100; Abcam, for RPE cells) overnight at 4°C. After thorough washes, samples were incubated with Alexa Fluor 594 goat anti-rabbit IgG (1:500; Invitrogen), Alexa Fluor 488 goat anti-mouse IgG (1:500; Invitrogen), or Alexa fluor 488 goat anti-rat IgG (1:500; Invitrogen) for 1 hour at room temperature. Nuclei were stained with DAPI (Sigma). All samples were imaged using the Zeiss LSM 880 Confocal Microscope.
Human Tissue
Eyes from human donors were obtained from the Eye Bank of Wuhan Aier Eye Hospital (Wuhan, China). The study protocol was in accordance with the Declaration of Helsinki and the principles set out in the Standard for Eye Bank issued by Ministry of Health of the People's Republic of China for research involving human tissues. The study was approved by the Ethics Review Committee of Aier Eye Hospital Group. Four pairs of eyes from donors aged between 51 and 89 years (3 men and 1 woman) died of accident, old age, colon cancer, or cerebral infarction, respectively, were used in this study. All donors had no history of eye diseases. Eyes were collected within 8 hours after death and fixed in Davidson's fixative solution (Wuhan Servicebio Technology Co., Wuhan, China) for at least 24 hours before processing for histology and immunohistochemistry.
Immunohistochemistry
The fixed human eyeballs were embedded in paraffin and the blocks were cut in 5 µm thickness with a microtome as described previously.24,25 Immunohistochemistry of human eye sections was conducted using the protocol described by us previously.24 In brief, human eye sections were deparaffinized, rehydrated, and immersed in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, and pH 6.0). Antigen retrieval was performed by heating the slides in a pressure cooker for 6 minutes. After unartificial cooling, the sections were processed for immunohistochemistry using rabbit specific HRP/AEC (ABC) Detection IHC Kit (Abcam) following the manufacturer's instructions. In brief, sections were incubated with H2O2 for 15 minutes and followed by blocking buffer for 30 minutes. The samples were then incubated with anti-lysozyme antibody (1:500; Abcam) overnight at 4°C. After thorough washes, samples were incubated with biotinylated goat anti-rabbit IgG(H+L) and streptavidin peroxidase (Abcam) for 10 minutes at room temperature, respectively. The color was developed using the AEC Single Solution substrate (Abcam). Samples were counterstained with hematoxylin (Solarbio Life Sciences, Beijing, China).
Statistical Analysis
All experiments were performed in triplicate. The statistical analysis of the results was performed using Graphpad Prism software (version 8; Graphpad, San Diego, CA). Data were presented as mean ± SD. The difference between the two groups was compared using the unpaired Student t-test. One-way or 2-way ANOVA was used when comparing multiple groups. Any P < 0.05 was considered as statistically significant.
Results
Lysozyme Expression in RPE Cells In Vitro
Semiquantitative reverse transcription PCRs detected mRNA of the lysozyme gene (Lyz in humans) in primary human RPE and ARPE19 line cells (Fig. 1A). Real-time qPCR showed that the expression levels of Lyz in RPE cells were significantly lower than the human macrophage cell line THP-1 cells (Fig. 1B). The mouse form of lysozyme (lyz2) was also detected in primary RPE cells from C57BL/6J mice (see Fig. 1A). The expression levels of Lyz2 mRNA in mRPE was lower than that in mouse microglial cell line BV2 cells by qPCR (Fig. 1C).
Figure 1.
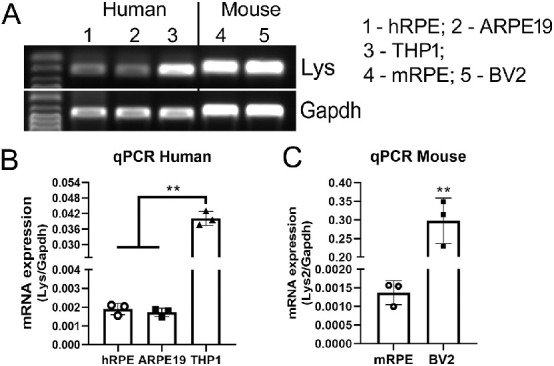
Lysozyme mRNA expression in human and mouse RPE cells. Total RNAs were extracted from human or mouse RPE cells and processed for conventional RT-PCR (A) or qRT-PCR (B, C). Human monocytes THP-1 and mouse microglia BV2 cells were used as positive control cell respectively. (A) Images of agarose gel electrophoresis of PCR products of lysozyme and gapdh genes. (B, C) Real-time RT-PCR showing lysozyme mRNA expression in human RPE and THP-1 cells (B) and mouse RPE and BV2 cells (C). Mean ± SD, **P < 0.001, 1-way ANOVA with Tukey's multiple comparison's tests (B) and unpaired Student t-tests (C).
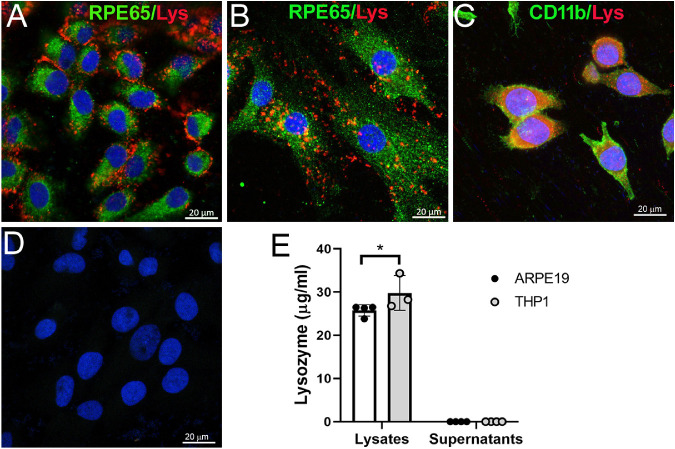
Confocal microscopy of immunolabeled RPE cells showed multiple punctate lysozyme immunoreactivities in the cytoplasm of ARPE19 cells (Fig. 2A) and mouse primary RPE cells (Fig. 2B). Punctate lysozyme staining was also observed in BV2 cells although diffused staining was more frequently observed (Fig. 2C). To understand if lysozyme can be released from RPE cells to the supernatants, we conducted ELISA in the supernatants as well as cell lysates. Interestingly, lysozyme was detected in the lysates of ARPE19 and THP-1 cells but not in their supernatants (Fig. 2E), suggesting that the majority of lysozyme produced by RPE and THP-1 cells remain in the cytosol under normal culture conditions. The level of lysozyme was higher in THP-1 than ARPE19 cells (see Fig. 2E).
Figure 2.
Lysozyme expression in RPE cultures. (A–D) Cultured human ARPE19 cells (A) and mouse primary RPE cells (B) were stained for RPE65 (green) and lysozyme (red) and imaged by confocal microscopy. (C) Mouse BV2 cells were used as a positive control and were stained for CD11b (green) and lysozyme (red). (D) Isotype control staining of ARPE19 cells. (E) Lysozyme levels in the cell lysates and the supernatants of human RPE cells (ARPE19) and monocytes (THP-1) measured by ELISA. Mean ± SD, *P < 0.05. Two-way ANOVA with Sidak's multiple comparison's tests.
Lysozyme Expression in RPE Cells In Vivo
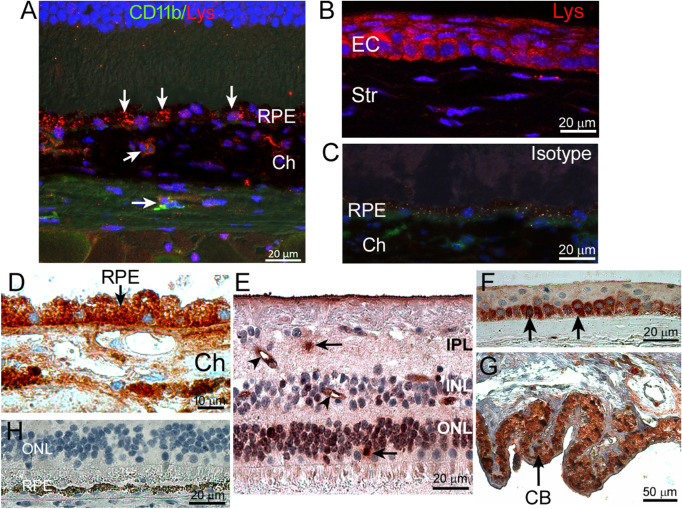
Confocal microscopy of mouse eye sections revealed strong lysozyme expression in the RPE and choroidal cells, including CD11b+ cells (arrows in Fig. 3A). Lysozyme was also detected in mouse corneal epithelial cells (Fig. 3B). Isotype control staining did not show any immunoreactivity (Fig. 3C). Immunohistochemistry of human eyes showed strong immunoreactivity of lysozyme in RPE and choroidal cells (Fig. 3D) and the staining pattern was similar to that in the mouse eyes (see Fig. 3A). Lysozyme was also detected in a small number of retinal cells in the outer nuclear layer (ONL; arrow in Fig. 3A), inner nuclear layer (INL), and inner plexiform layer (IPL; arrow in Fig. 3A), including small blood vessels (arrowheads in Fig. 3E). In addition, corneal epithelial cells, in particular, the basal cells (Fig. 3F) and ciliary body pigment epithelial cells (Fig. 3G) were strongly positive for lysozyme. Isotype control staining did not show any immunoreactivity (Fig. 3H).
Figure 3.
Lysozyme expression in mouse and human eyes. (A, B) Mouse eye sections were stained for CD11b (green), lysozyme (red), and DAPI (blue) and imaged by confocal microscopy. (A) Retinal outer layer showing lysozyme expression (arrows) in RPE and choroid. (B) Lysozyme expression in corneal epithelial cells. (C) Isotype control staining. (D–G) Human eye sections were stained for lysozyme (brown) by immunohistochemistry and imaged by light microscopy. (D) RPE/choroid. Arrow - lysozyme expressing RPE cells. (E) Retina. Arrows – lysozyme positive retinal cells. Arrowheads – lysozyme positive retinal blood vessels. (F) Cornea. Arrows – lysozyme positive basal layer epithelial cells. (G) Ciliary body. Arrow – lysozyme positive ciliary body pigment epithelial cells. (H) Isotype control staining. CB – ciliary body; Ch – choroid; EC – epithelial cells; IPL – inner plexiform layer; INL – inner nuclear layer; OPL – outer nuclear layer; RPE – retinal pigment epithelium; Str – stroma.
Regulation of Lysozyme Expression in RPE Cells
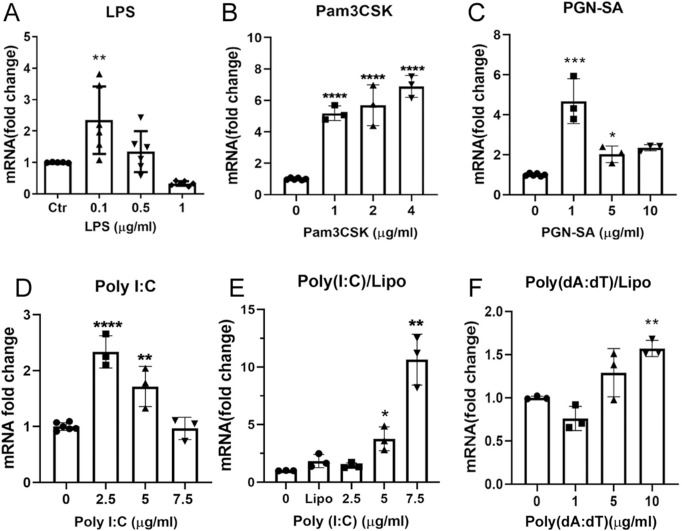
To understand how lysozyme expression in RPE cells is regulated under inflammatory conditions, we treated ARPE19 cells with different pattern recognition receptors (PRRs) agonists, including LPS (TLR4), Pam3CSK (TLR2/TLR1), PGN-SA (TLR2), Poly(I:C), and Poly(dA:dT). Real-time qPCR showed that low concentrations of LPS (0.1 µg/mL) and PNG-SA (1 and 5 µg/mL) significantly increased lysozyme mRNA expression but the effect was not dose-dependent (Figs. 4A, 4C). Pam3CSK (1, 2, and 4 µg/mL) dose-dependently increased lysozyme mRNA expression in RPE cells (Fig. 4B). Naked Poly(I:C), which is recognized by endosome TLR3, induced 1.7 to approximately 2.3-fold increase in lysozyme mRNA expression at the concentrations of 2.5 and 5 µg/mL (Fig. 4D). Poly(I:C) transfected with Lipofectamine 2000, which is sensed by RIG-I/MDA-5, dose-dependently upregulated lysozyme expression and 7.5 µg/mL of transfected Poly(I:C) resulted in a 10-fold increase in lysozyme mRNA (Fig. 4E). When the synthesized dsDNA, Poly(dA:dT) was transfected into RPE cells, a 1.5-fold upregulation of lysozyme mRNA was observed at the dose of 10 µg/mL (Fig. 4F). Our results suggest that the expression of lysozyme in RPE cells is regulated by TLRs and RIG-I/MDA-5 signaling pathways.
Figure 4.
The effect of TLR agonists and synthetic dsRNA, dsDNA on lysozyme mRNA expression in human ARPE19 cells. ARPE19 cells were treated with different concentrations of LPS (A), Pam3CSK (B), PGN-SA (C), naked Poly(I:C), Poly(I:C) transfected with lipofectamine 2000 (E), and Poly(dA:dT) transfected with Lipo2000 (F) for 24 hours. The expression of lysozyme mRNA was examined by real-time qPCR. Mean ± SD, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. One-way ANOVA with Dunnett's multiple comparison's tests.
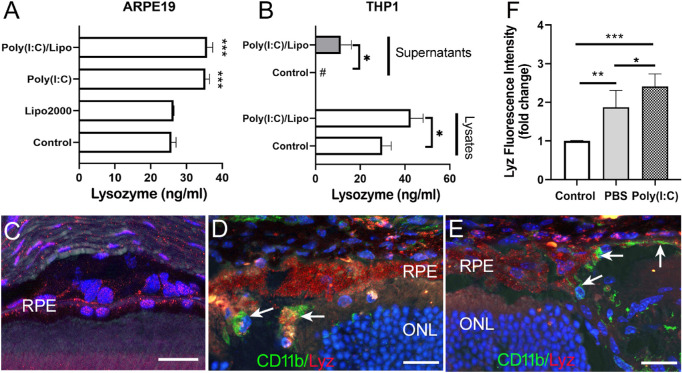
Because Poly(I:C)/Lipo showed the strongest effects on lysozyme mRNA expression in RPE cells, we further examined its effects on lysozyme protein expression. Significantly higher levels of lysozyme were detected in the cell lysate of Poly(I:C) and Poly(I:C)/Lipo treated ARPE19 cells compared to that of control cells by ELISA (Fig. 5A). Lysozyme was not detected in the supernatants of ARPE19 even after Poly(I:C)/Lipo treatment (data not shown). Low levels of lysozyme (4.82 ± 0.68 ng/mL) were detected in the supernatants of ARPE19 cells after challenging with live bacteria for 8 hours. The Poly(I:C)/Lipo treatment significantly increased both the intracellular and secreted form of lysozyme in THP-1 cells (Fig. 5B). Subretinal injection of Poly(I:C) into normal C57BL/6J mice induced CD11b+ cell infiltration (arrows in Fig. 5D). Interestingly, infiltrating CD11b+ cells were also observed in the RPE layer and the outer retina in PBS injected eyes (Fig. 5E), suggesting a nonspecific response to injection-mediated trauma. Both infiltrating CD11b+ cells and RPE cells expressed lysozyme (see Fig. 5D). The fluorescence intensity of lysozyme in RPE cells was significantly increased in subretinal injected eyes compared to naïve eyes (see Figs. 5C–F) and the increment was more pronounced in Poly(I:C) injected eyes (see Fig. 5F).
Figure 5.
The effect of Poly(I:C) on lysozyme protein expression in RPE cells in vitro and in vivo. (A) Human ARPE19 cells were treated with naked Poly(I:C) or Poly(I:C) transfected with lipofectamine 2000 for 24 hours. Cell lysates and supernatants were collected and lysozyme was measured by ELISA. Human monocytes THP-1 were used as a positive control (B). Mean ± SD, #, not detectable. *P < 0.05. ***P < 0.001. One-way ANOVA with Dunnett's multiple comparison's tests (A) and 2-way ANOVA with Sidak's multiple comparison's tests (B). (C–F) Poly(I:C) or PBS was injected into the subretinal space of the mouse eye. Then, 24 hours later, eyes were collected and processed for immunostaining of CD11b (green) and lysozyme (red). (C–E) Representative confocal mages of mouse retina from a control noninjected (C), Poly(I:C) injected (B) and a PBS injected (E) mouse eyes. Arrows in (D) and (E) indicating infiltrating CD11b cells. (F) Fold changes in lysozyme fluorescence intensity of RPE cells in different groups. Mean ± SD, N = 3 to 8 eyes, *P < 0.05; **P < 0.01; ***P < 0.001, 1-way ANOVA with Tukey's multiple comparison test. RPE – retinal pigment epithelial cells; ONL – outer nuclear layer. Scale bar = 20 µm.
Effect of Lysozyme Knockout on ARPE19 Cell Bactericidal Function
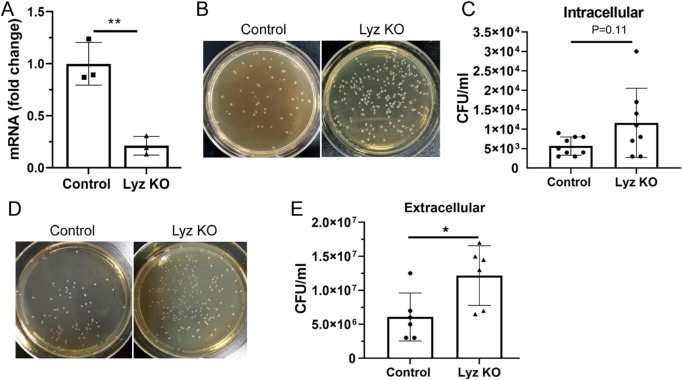
To understand the role of lysozyme in RPE innate defense function, we deleted LYZ gene in ARPE19 cells using CRISPR/Cas9 technology. An 80% reduction in Lyz mRNA expression was observed in RPE cells following CRISPR/Cas9 gene editing (Lyz KO) compared to vector-treated cells (Fig. 6A). When ARPE19 cells were challenged with live pathogenic bacteria, there was a trend of increased intracellular bacterial colonies in Lyz knockout (KO) cells although the difference was not statistically significant (Figs. 6B, 6C). A significantly higher number of extracellular bacterial colonies was observed in Lyz KO cells compared to that in control ARPE19 cells (Figs. 6D, 6E).
Figure 6.
The effect of lysozyme knockout on ARPE19 cell bactericidal function. LYZ gene was deleted in ARPE19 cells using CASPR/Cas9 technique (see Materials and Methods). (A) Lysozyme mRNA expression in vector-treated (Control) and Lysozyme knockout (Lyz KO) cells was examined by qPCR. (B–E) Bactericidal function of ARPE19 cells was conducted using live E. coli. (B) Representative images showing intracellular bacterial colonies in control and Lyz KO cells. (C) Bar/dot figure showing intracellular bacteria in different groups of ARPE19 cells. (D) Representative images showing extracellular bacterial colonies in control and Lyz KO cell cultures. (E) Bar/dot figure showing extracellular bacteria in different groups of ARPE19 cells. Mean ± SD, *P < 0.05; **P < 0.01; unpaired Student's t-tests.
Effect of Lysozyme Knockout on ARPE19 Cell Response to Inflammatory Stimulations
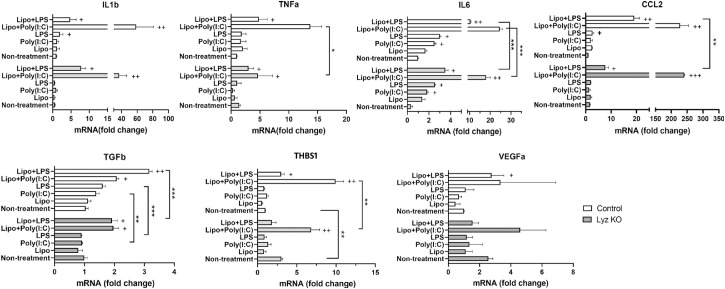
The effect of lysozyme KO on ARPE19 cell response to LPS and Poly(I:C) was examined by qPCR of inflammatory genes (IL1b, TNFa, IL6, and CCL2) and anti-inflammatory/angiogenic genes (TGFb, THBS1, and VEGFa). LPS treatment increased the expression of IL1b, IL6, and CCL2 in control but only IL6 in Lyz KO RPE cells (Fig. 7). A previous study has shown that LPS can be detected by intracellular receptor caspase-4/1126.26 When LPS was incubated together with transfection reagent Lipofectamine 2000 (Lipo + LPS), a more profound response was observed in RPE cells evidenced by significantly increased mRNA expression in all genes tested in our study (see Fig. 7). Deletion of lysozyme blunted Lipo + LPS induced upregulation of IL6, CCL2, and TGFb in ARPE19 cells (see Fig. 7). Naked Poly(I:C) increased IL6 expression in RPE cells (Fig. 7). The highest response was observed in Lipo + Poly(I:C) treated RPE cells and the expression of TNFa, IL6, and THBS1 was significantly lower in Lyz KO cells compared to control RPE cells (Fig. 7B). Our results suggest that deletion of lysozyme reduces the response of ARPE19 cells to inflammatory stimuli.
Figure 7.
The effect of lysozyme knockout on ARPE19 cell immune response. Control (vector-treated) and Lyz KO ARPE19 cells were incubated with LPS or Poly(I:C) in the absence or presence of transfection reagent lipofectamine 2000 for 24 hours. The mRNA expression of IL1b, TNFa, IL6, CCL2, TGFb, Thbs1, and VEGFa was examined by qPCR. Mean ± SD, n = 3, + P < 0.05; ++ P < 0.01 compared to the non-treatment group of the same cell type. *P < 0.05; **P < 0.01; ***P < 0.001 when comparing control with Lyz KO cells. Two-way ANOVA with Sidak's multiple comparison's test.
Discussion
In this study, we show that RPE cells constitutively express antimicrobial peptide lysozyme and the expression can be further enhanced upon pathogen stimulation. We further showed that deletion of lysozyme in ARPE19 cells impaired the bactericidal activity and reduced their response to inflammatory stimuli. RPE cells are believed to safeguard the neuronal retina from potential insults from peripheral circulation by the physical (i.e. TJs of the oBRB) and immunological barriers. Our results suggest that RPE cells may also protect the retina from blood-borne pathogens by producing antimicrobial peptides, such as lysozyme.
Lysozyme is a conserved antimicrobial protein that is critical to host defense. It can catalyze the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in peptidoglycan, thereby compromising the integrity of bacterial cell walls causing lysis of the bacteria.27 Lysozyme is abundant in the blood and liver, in various body fluids, including tears, urine, saliva, and milk, at mucosal surfaces, and in innate immune cells, including macrophages, neutrophils, and dendritic cells.28 Although lysozyme is often released extracellularly, they can also kill intracellular pathogens.28 We found that the majority of lysozyme produced by RPE remain inside the cells. The extracellular lysozyme of RPE was only detectable by ELISA after live bacteria challenging. Deletion of lysozyme using Crispr/Cas9 in ARPE19 cells reduced their bactericidal activity evidenced by significantly higher levels of bacterial load in the supernatants of Lyz KO ARPE19 cells (see Fig. 6). The intracellular bacterial load in Lyz KO ARPE19 cells was slightly but insignificantly higher than that in control ARPE19 cells. Therefore, we are unable to determine whether RPE lysozyme kills the bacteria inside or outside the cells or both. Nevertheless, our results suggest that lysozyme is critically involved in RPE clearance of pathogens in vitro. Further in vivo infection studies will help to understand the role of RPE-derived lysozyme in protecting the retina from blood-borne pathogens.
We found that the expression of lysozyme in RPE cells is modulated by agonists of the cell surface TLRs (e.g. TLR1, TLR2, and TLR4) as well as synthetic dsRNA polymer Poly(I:C). Treatment of RPE cells with Poly(I:C) in vitro or subretinal injection of Poly(I:C) significantly upregulated lysozyme expression in RPE cells. Furthermore, subretinal injection of Poly(I:C) resulted in the infiltration of CD11b+ immune cells, which also express lysozyme. Interestingly, immune cell infiltration, and lysozyme upregulation were also observed in PBS injected eyes (although less severe compared to Poly(I:C) injected eyes), suggesting that injection-induced trauma is sufficient to initiate acute subretinal immune response. The increased expression of lysozyme in RPE cells may be a protective response of the cells against pathogens. The transfected Poly(I:C) showed the strongest effects among all agonists tested in this study, suggesting higher levels of control by RIG-I/MDA5 signaling pathway on lysozyme expression in RPE cells. Apart from the bactericidal function, lysozyme also has antiviral effects, particularly against RNA virus. For example, heat-denatured lysozyme can inactivate murine and human norovirus and hepatitis A virus29; chicken egg lysozyme can suppress the replication of bovine viral diarrhea virus.30 Interestingly, the dsDNA polymer Poly(dA:dT) only induced a mild lysozyme response in cultured RPE cells. Our results suggest that RPE cells may produce lysozyme as a strategy to fight against intracellular pathogens such as RNA virus and bacteria.
In addition to the bacterial/viral killing activity, lysozyme can also modulate immune activation (e.g. enhance or dampen inflammation). The peptidoglycans released from bacteria by lysozyme can further activate other PRRs, including the cytosolic receptors NOD1, NOD2, and TLRs.27 This eventually leads to the production of inflammatory cytokines that provide additional bactericidal activities. In this study, we found that the response of RPE cells to intracellular LPS or Poly(I:C) stimulation was reduced in lysozyme deleted RPE cells (see Fig. 7), suggesting a positive role of lysozyme in modulating RPE immune response to these stimuli. On the other hand, lysozyme can directly bind and neutralize endogenous damage-associated molecular patterns (DAMPs), such as advanced glycation end products and reduce inflammation.31 Furthermore, it has been reported that lysozyme can modulate complement activation32 and reduce the production of oxidative burst in neutrophils.33 The role of lysozyme in modulating RPE immune activation in pathophysiological conditions warrant further investigations.
It is important to note that data presented in this study also have implications in the interpretation of results from LysM-Cre mice. The use of Cre recombinase under the direction of LysM promoter is widely used to edit gene expression in myeloid-derived cells. Because RPE cells also express high levels of lysozyme (lysozyme 2 or LysM in mice), LysM guided gene deletion will also occur in RPE cells. A recent study using LysM-Cre recombination technology reported Lys-Cre expression in retinal microglia and a small population of retinal neurons.34 We also detected a small number of lysozyme-positive cells in the neuroretina, including retinal blood vessels (see Fig. 3D). The expression of lysozyme in retinal and RPE cells should be taken into consideration when interpreting retinal phenotype in LysM-Cre mice.
In summary, our results suggest that apart from the previously recognized physical and immunological barriers, cells of the BRB may also protect the neuronal retina from blood-borne pathogens by producing antimicrobial peptides, such as lysozyme. Our results open up a new avenue of research as further understanding the spectrum of antimicrobial peptides produced by cells of the BRB and BBB as well as their immune regulatory roles will shed light on the pathogenesis of various inflammatory and degenerative neurological diseases.
Acknowledgments
The authors thank Jianing Gu (Aier Eye Institute) for providing human RPE cDNA to this study and Haiping Que (Aier Eye Institute) for technical advices.
Funding from Aier Eye Hospital Group, the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 722717, National Natural Science Foundation of China (81700827), and Natural Science Foundation of Hunan Province, China (2018JJ3002), Hunan Science & Technology Association (2018KX001), and Science & Technology Department of Human Province (2018RS3123).
Disclosure: J. Liu, None; C. Yi, None; W. Ming, None; M. Tang, None; X. Tang, None; C. Luo, None; B. Lei, None; M. Chen, None; H. Xu, None
References
- 1. Hofman P, Hoyng P, vanderWerf F, Vrensen GF, Schlingemann RO.. Lack of blood-brain barrier properties in microvessels of the prelaminar optic nerve head. Invest Ophthalmol Vis Sci . 2001; 42: 895–901. [PubMed] [Google Scholar]
- 2. Wolburg H, Lippoldt A.. Tight junctions of the blood-brain barrier: development, composition and regulation. Vascul Pharmacol . 2002; 38: 323–337. [DOI] [PubMed] [Google Scholar]
- 3. Forrester JV, Xu H.. Good news-bad news: the Yin and Yang of immune privilege in the eye. Front Immunol. 2012; 3: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Naylor A, Hopkins A, Hudson N, Campbell M.. Tight junctions of the outer blood retina barrier. Int J Mol Sci . 2019; 21(1): 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rizzolo LJ, Peng S, Luo Y, Xiao W.. Integration of tight junctions and claudins with the barrier functions of the retinal pigment epithelium. Prog Retin Eye Res . 2011; 30(5): 296–323. [DOI] [PubMed] [Google Scholar]
- 6. Banks WA. The blood-brain barrier as an endocrine tissue. Nat Rev Endocrinol . 2019; 15(8): 444–455. [DOI] [PubMed] [Google Scholar]
- 7. Muoio V, Persson PB, Sendeski MM.. The neurovascular unit - concept review. Acta Physiol (Oxf) . 2014; 210(4): 790–798. [DOI] [PubMed] [Google Scholar]
- 8. Forrester JV, Xu H, Lambe T, Cornall R.. Immune privilege or privileged immunity? Mucosal Immunol . 2008; 1(5): 372–381. [DOI] [PubMed] [Google Scholar]
- 9. Parker A, Fonseca S, Carding SR.. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes . 2020; 11(2): 135–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Logsdon AF, Erickson MA, Rhea EM, Salameh TS, Banks WA.. Gut reactions: How the blood-brain barrier connects the microbiome and the brain. Exp Biol Med (Maywood) . 2018; 243(2): 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keaney J, Campbell M.. The dynamic blood-brain barrier. Febs J . 2015; 282(21): 4067–4079. [DOI] [PubMed] [Google Scholar]
- 12. Forrester JV, McMenamin PG, Dando SJ.. CNS infection and immune privilege. Nat Rev Neurosci . 2018; 19(11): 655–671. [DOI] [PubMed] [Google Scholar]
- 13. Sofroniew MV. Astrocyte reactivity: subtypes, states, and functions in CNS innate immunity. Trends Immunol . 2020; 41(9): 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gorina R, Font-Nieves M, Márquez-Kisinousky L, Santalucia T, Planas AM.. Astrocyte TLR4 activation induces a proinflammatory environment through the interplay between MyD88-dependent NFκB signaling, MAPK, and Jak1/Stat1 pathways. Glia . 2011; 59(2): 242–255. [DOI] [PubMed] [Google Scholar]
- 15. Moses S, Jambulingam M, Madhavan HN.. A pilot study on expression of toll like receptors (TLRs) in response to herpes simplex virus (HSV) infection in acute retinal pigment epithelial cells (ARPE) cells. J Postgrad Med . 2014; 60(3): 243–247. [DOI] [PubMed] [Google Scholar]
- 16. Chen G-f, Xu T-h, Yan Y, et al.. Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacol Sin . 2017; 38(9): 1205–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar DK, Choi SH, Washicosky KJ, et al.. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med . 2016; 8(340): 340ra372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soscia SJ, Kirby JE, Washicosky KJ, et al.. The Alzheimer's disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One . 2010; 5(3): e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen M, Luo C, Zhao J, Devarajan G, Xu H.. Immune regulation in the aging retina. Prog Retin Eye Res . 2019; 69: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Augustine J, Pavlou S, Ali I, et al.. IL-33 deficiency causes persistent inflammation and severe neurodegeneration in retinal detachment. J Neuroinflammation . 2019; 16(1): 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen M, Muckersie E, Robertson M, Fraczek M, Forrester JV, Xu H.. Characterization of a spontaneous mouse retinal pigment epithelial cell line B6-RPE07. Invest Ophthalmol Vis Sci . 2008; 49(8): 3699–3706. [DOI] [PubMed] [Google Scholar]
- 22. Gu J, Wang Y, Cui Z, et al.. The construction of retinal pigment epithelium sheets with enhanced characteristics and cilium assembly using iPS conditioned medium and small incision lenticule extraction derived lenticules. Acta Biomater . 2019; 92: 115–131. [DOI] [PubMed] [Google Scholar]
- 23. Chen M, Copland DA, Zhao J, et al.. Persistent inflammation subverts thrombospondin-1-induced regulation of retinal angiogenesis and is driven by CCR2 ligation. Am J Pathol. 2012; 180(1): 235–245. [DOI] [PubMed] [Google Scholar]
- 24. Liu J, Tang M, Harkin K, et al.. Single-cell RNA sequencing study of retinal immune regulators identified CD47 and CD59a expression in photoreceptors—implications in subretinal immune regulation. J Neurosci Res. 2020; 98(7): 1498–1513. [DOI] [PubMed] [Google Scholar]
- 25. Little K, Llorián-Salvador M, Tang M, et al.. Macrophage to myofibroblast transition contributes to subretinal fibrosis secondary to neovascular age-related macular degeneration. J Neuroinflammation . 2020; 17(1): 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi J, Zhao Y, Wang Y, et al.. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature . 2014; 514(7521): 187–192. [DOI] [PubMed] [Google Scholar]
- 27. Ragland SA, Criss AK.. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog . 2017; 13(9): e1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Callewaert L, Michiels CW.. Lysozymes in the animal kingdom. J Biosci . 2010; 35(1): 127–160. [DOI] [PubMed] [Google Scholar]
- 29. Takahashi M, Okakura Y, Takahashi H, et al.. Heat-denatured lysozyme could be a novel disinfectant for reducing hepatitis A virus and murine norovirus on berry fruit. Int J Food Microbiol . 2018; 266: 104–108. [DOI] [PubMed] [Google Scholar]
- 30. Małaczewska J, Kaczorek-Łukowska E, Wójcik R, Siwicki AK.. Antiviral effects of nisin, lysozyme, lactoferrin and their mixtures against bovine viral diarrhoea virus. BMC Vet Res . 2019; 15(1): 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li YM, Tan AX, Vlassara H.. Antibacterial activity of lysozyme and lactoferrin is inhibited by binding of advanced glycation-modified proteins to a conserved motif. Nat Med . 1995; 1(10): 1057–1061. [DOI] [PubMed] [Google Scholar]
- 32. Ogundele MO. A novel anti-inflammatory activity of lysozyme: modulation of serum complement activation. Mediators Inflamm . 1998; 7: 592871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gordon LI, Douglas SD, Kay NE, Yamada O, Osserman EF, Jacob HS.. Modulation of neutrophil function by lysozyme. Potential negative feedback system of inflammation. J. Clin. Invest . 1979; 64(1): 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fouda AY, Xu Z, Narayanan SP, Caldwell RW, Caldwell RB.. Utility of LysM-cre and Cdh5-cre driver mice in retinal and brain research: an imaging study using tdtomato reporter mouse. Invest Ophthalmol Vis Sci . 2020; 61(3): 51. [DOI] [PMC free article] [PubMed] [Google Scholar]