Abstract
Objectives
To develop and cross-validate a multivariable clinical prediction model to identify invasive bacterial infections (IBI) and to identify patient groups who might benefit from new biomarkers.
Design
Prospective observational study.
Setting
12 emergency departments (EDs) in 8 European countries.
Patients
Febrile children aged 0–18 years.
Main outcome measures
IBI, defined as bacteraemia, meningitis and bone/joint infection. We derived and cross-validated a model for IBI using variables from the Feverkidstool (clinical symptoms, C reactive protein), neurological signs, non-blanching rash and comorbidity. We assessed discrimination (area under the receiver operating curve) and diagnostic performance at different risk thresholds for IBI: sensitivity, specificity, negative and positive likelihood ratios (LRs).
Results
Of 16 268 patients, 135 (0.8%) had an IBI. The discriminative ability of the model was 0.84 (95% CI 0.81 to 0.88) and 0.78 (95% CI 0.74 to 0.82) in pooled cross-validations. The model performed well for the rule-out threshold of 0.1% (sensitivity 0.97 (95% CI 0.93 to 0.99), negative LR 0.1 (95% CI 0.0 to 0.2) and for the rule-in threshold of 2.0% (specificity 0.94 (95% CI 0.94 to 0.95), positive LR 8.4 (95% CI 6.9 to 10.0)). The intermediate thresholds of 0.1%–2.0% performed poorly (ranges: sensitivity 0.59–0.93, negative LR 0.14–0.57, specificity 0.52–0.88, positive LR 1.9–4.8) and comprised 9784 patients (60%).
Conclusions
The rule-out threshold of this model has potential to reduce antibiotic treatment while the rule-in threshold could be used to target treatment in febrile children at the ED. In more than half of patients at intermediate risk, sensitive biomarkers could improve identification of IBI and potentially reduce unnecessary antibiotic prescriptions.
Keywords: epidemiology, therapeutics
What is already known on this topic?
In children, distinction between invasive bacterial and self-limiting infections on only clinical symptoms is unreliable leading to overuse of antibiotics on the one hand, but to missed invasive bacterial infections in others.
Several clinical prediction models including biomarkers have been developed to help decision making by risk prediction of patients at high risk or low risk for bacterial infections, but none predicts the outcome invasive bacterial infections in older children or includes children with chronic conditions.
What this study adds?
We derived and externally validated a clinical prediction model based on clinical predictors from the Feverkidstool (clinical symptoms, C reactive protein) and non-blanching rash, neurological symptoms and comorbidity, to early recognise invasive bacterial infections with data from a large observational European-wide study of febrile children aged 0–18 years.
The rule-out threshold of this model could reduce antibiotic prescription and invasive diagnostics, while the rule-in threshold could be useful to target early treatment for invasive bacterial infections.
In more than half of the patients at intermediate risk, sensitive new biomarkers could reduce diagnostic uncertainty and improve identification of invasive bacterial infections.
Introduction
Children presenting at the emergency department (ED) still die from treatable invasive bacterial infections (IBI) due to delayed or missed diagnosis.1–3 For not missing one child with IBI, antibiotics are prescribed in children with self-limiting viral infections.4 The distinction between bacterial and viral infections based solely on clinical signs and symptoms is unreliable. Although C reactive protein (CRP) and procalcitonin are currently used as markers for bacterial infections, they measure non-specific inflammation and immunological responses. Recent studies focus on proteomic and transcriptomic approaches for finding new discriminators of bacterial and viral infections.5–8 Due to costs and limited resources, it is not feasible to apply new biomarkers to all febrile children. Therefore, prediction models are needed to identify risk groups where biomarkers can improve diagnosis.
Clinical prediction models that include clinical signs and CRP or procalcitonin have been developed to assist decision making in treatment of febrile children,9–15 and have focused on young infants to differentiate between patients at high risk or low risk for IBI (bacteraemia, meningitis, bone/joint infections). No clinical prediction models for IBI exists for older children who are also at risk for IBI.16 17 The Feverkidstool, developed for children aged <16 years, predicts risks for pneumonia and other serious bacterial infections which besides IBIs also includes bacterial infections of the urinary tract, gastrointestinal tract and soft tissue.
Although the Feverkidstool is extensively validated, the original population only included 21 IBI cases and important predictors for IBI such as non-blanching rash or neurological symptoms were not included. Several models yet exist for prediction of bacterial pneumonia and the impact of the original Feverkidstool on antibiotic use in respiratory tract infections is proven.18 Therefore, another model for bacterial pneumonia is not required. Furthermore, prediction of urinary tract infections may be less relevant as sensitive laboratory tests (urinalysis) are readily available for accurate diagnosis at ED visit. In addition, the Feverkidstool is developed in previous healthy children and is therefore not applicable for children with chronic conditions with higher risk of IBI. Hence, a new tool is required for early risk assessment of IBI in febrile children including all age ranges (0–18 years) and chronic conditions.
We aim (1) to derive and cross-validate a clinical prediction model including CRP to identify IBIs in febrile children presenting to different European EDs and (2) to identify patient groups which might benefit from new biomarkers.
Methods
Study design
This study is embedded in MOFICHE (Management and Outcome of Febrile children in Europe), an observational multicentre study, which is part of PERFORM (PErsonalized Risk assessment in Febrile illness to Optimise Real-life Management across the European Union) (www.perform2020.org).
Children aged from 0 to 18 years with temperature ≥38.0°C or fever <72 hours before ED visit were included. Twelve EDs participated in this study: Austria, Germany, Greece, Latvia, the Netherlands (n=3), Spain, Slovenia and the UK (n=3).19 Data were collected for at least 1 year from January 2017 to April 2018. Details of the study design have been described previously.20
For this study, we selected patients with CRP measurement and excluded patients with working diagnosis of urinary tract infections after first assessment at the ED.21 To identify IBI at the earliest opportunity, we included only the first ED visit for patients with IBI who repeatedly visited the ED within the same disease episode. Data were analysed according to a statistical analysis plan (online supplemental appendix 1).
archdischild-2020-319794supp001.pdf (2.7MB, pdf)
Collected data included age, sex, comorbidity (chronic condition expected to last ≥1 year),22 warning signs for identifying risk of serious illness (National Institute for Health and Care Excellence (NICE))23 (consciousness, ill appearance, work of breathing, meningeal signs, focal neurology, non-blanching rash, dehydration) and vital signs (heart rate, respiratory rate, oxygen saturation, temperature, capillary refill time). We collected CRP level (point-of-care or laboratory assay) and microbiologic cultures (blood, cerebrospinal fluid and other) ordered at the ED or at the first day of hospital admission on indication of the physician. Furthermore, we collected data of prescribed antibiotics and admission following ED visit.
Outcome
IBI included bacterial meningitis, bacteraemia and bacterial bone/joint infections, defined as culture or PCR detection of a single pathogenic bacterium in blood, cerebrospinal or synovial fluid. All cultures that were treated as contaminant and cultures growing contaminants were considered non-IBI (online supplemental appendix 2).24 Cultures growing a single contaminant or candida were defined positive in patients with malignancy, immunodeficiency, immunosuppressive drugs or a central catheter, since antimicrobial treatment is needed in these patients.
Model development
Descriptive and univariate logistic regression analyses were performed for children with and without IBI.
Sample size was estimated based on Riley et al.25 Assuming 16 predictors, a prevalence of 0.8% and an expected R2 of 0.0135 (15% of maximum achievable R2), a sample size of 10 587 with 85 cases would be sufficient. For model development,26 27 we considered predefined variables with predictive value for IBI: (1) variables in the Feverkidstool9 (age, sex, temperature, fever duration, tachypnoea and tachycardia defined by Advanced Paediatric Life Support,28 oxygen saturation <94%, capillary refill ≥3 s, work of breathing, ill appearance and CRP value), (2) NICE warnings signs (consciousness, meningeal signs, focal neurology, status epilepticus, non-blanching rash)23 and (3) complex chronic condition (≥2 body systems, malignancy or immunocompromised).22 Consciousness, meningeal signs and focal neurology were combined into a composite variable abnormal neurology. Linearity of continuous variables was assessed using restricted cubic splines. As in the Feverkidstool, age was modelled linear piecewise for children aged <1 year and >1 year and a logarithmic transformation for CRP was used. Outliers were truncated at the 0.01 percentile for temperature (35.7°C) and the 0.99 percentile for CRP (215 mg/L) and fever duration (8 days).
Variable selection was not influenced by the results of the univariate logistic regression analysis, but was performed using least absolute shrinkage and selection operator (LASSO), which reduces the degree of overfitting by shrinking large regression coefficients (detailed methods in online supplemental appendix 3).29 30 The final model was developed on data from all the 12 EDs. For the cross-validation, we created 5 ED groups; 1 group combined the data from the 8 EDs with <10 IBI cases and 4 groups were based on data from EDs with >10 IBI cases per ED: Slovenia, the Netherlands (n=2) and the UK (online supplemental appendix 4). Next, in cross-validation the model was repeatedly derived on four ED groups and validated on the fifth ED group, leading to five different cross-validations.31 The five cross-validations were pooled using a random-effects model. This cross-validation determines model performance most accurately and provides information on the heterogeneity of performance across different settings. This cross-validation is therefore superior to a single external validation.13 31 We assessed the discriminative ability by the area under the receiver operating curve (AUC), and calibration, the agreement between predicted risks and observed cases. We explored the impact of difference in case-mix heterogeneity on the discriminative ability of the model in the internal-external cross-validation. We used decision curve analysis to evaluate the net benefit of the prediction model.32 At different cut-offs for the individual probability of IBI according to the model, we assessed sensitivity, specificity, negative and positive likelihood ratios (LRs). Missing values for the covariates were multiple imputed using the MICE package, resulting in 20 imputation sets (details in online supplemental appendix 3). Sensitivity analysis was performed in the population where missing CRP values were imputed. All analyses were performed in R V.3.6.
Results
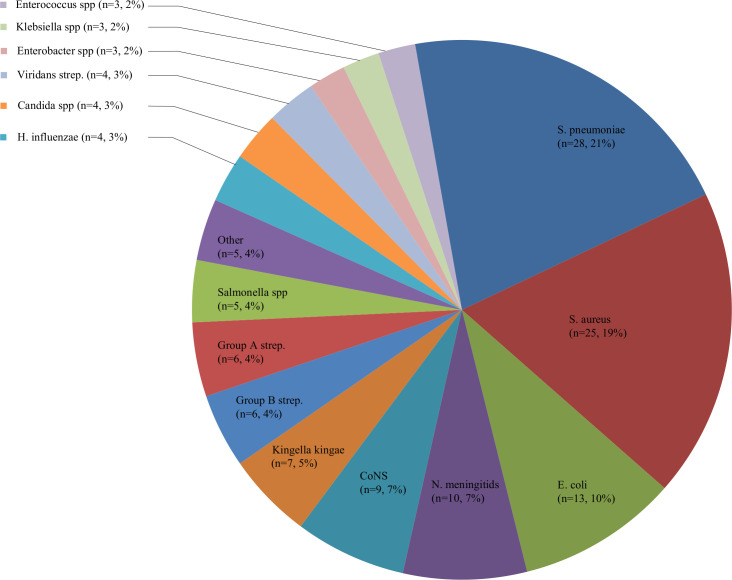
Of 38 480 patients, 17 213 patients had CRP measurements. Patients with CRP measurements were more often ill-appearing and admitted than patients without CRP measurements (online supplemental appendix 5). We excluded 939 urinary tract infections and 6 repeated visits in the same disease period of patients with IBI, resulting in 16 268 patients. Of those, most common infections were the upper respiratory tract (45%), lower respiratory tract (18%), gastrointestinal tract (14%) and undifferentiated fever (9%). IBI was diagnosed in 135 patients (0.8%), and comprised 119 bacteraemias, 15 bacterial meningitis and 9 bone/joint infections (8 patients had concurrent infections). Main pathogens included Streptococcus pneumoniae (21%), Staphylococcus aureus (19%), Escherichia coli (10%), Neisseria meningitidis (7%) and coagulase-negative staphylococcus (7%) (figure 1, online supplemental appendix 6). Complex chronic conditions were present in 37% of patients with IBI vs 6% of patients without IBI. IBI incidence varied from 0.1% to 5.6% of patients per ED (online supplemental appendix 4).
Figure 1.
Identified pathogens for invasive bacterial infections (n=135). CoNS, coagulase-negative staphylococci; spp, species.
Patients with IBI were similar in age and sex compared with patients without IBI. CRP level was higher in the IBI group (median 62 mg/L, IQR 21–144) than in the non-IBI group (median 16 mg/L, IQR 5–45) (p<0.01) (table 1). The majority of IBIs were treated with antibiotics (n=126, 93.3%) at first ED visit and all were treated with antibiotics in the disease course. The associations of the sole predictors with IBI are provided in online supplemental appendix 7.
Table 1.
Characteristics of patients with invasive bacterial infections and patients without invasive bacterial infections
| Invasive bacterial infection (n=135) | No invasive bacterial infection (n=16 133) | |||
| n (%) | Missing | n (%) | Missing | |
| Age in years, median (IQR) | 3.2 (0.8–6.0) | 2.8 (1.4–6.0) | ||
| Female | 76 (56.2) | 8932 (55.4) | ||
| Underlying chronic condition | 2 | 89 | ||
| Any | 68 (50.4) | 3005 (18.6) | ||
| Complex | 50 (37.0) | 1008 (6.2) | ||
| Referred | 96 (71.1) | 3 | 8633 (53.5) | 936 |
| Triage urgency | 5 | 477 | ||
| Low: standard, non-urgent | 41 (30.4) | 9242 (57.3) | ||
| High: immediate, very urgent, intermediate | 89 (65.9) | 6414 (39.8) | ||
| Feverkidstool | ||||
| Temperature in °C, median (IQR) | 38.0 (37.4–38.7) | 3 | 37.8 (37.0–38.5) | 764 |
| Fever duration in days, median (IQR) | 0.5 (0.5–3) | 5 | 1.5 (0.5–3) | 817 |
| Tachypnoea (APLS) | 38 (28.1) | 37 | 3345 (20.7) | 3919 |
| Tachycardia (APLS) | 81 (60.0) | 5 | 5578 (34.6) | 821 |
| Hypoxia <95% | 4 (2.9) | 13 | 749 (4.6) | 2373 |
| Prolonged capillary refill (>3 s) | 8 (5.9) | 29 | 305 (1.9) | 2311 |
| Increased work of breathing | 11 (8.1) | 40 | 887 (5.5) | 2136 |
| Ill appearance | 60 (44.4) | 13 | 4398 (27.3) | 610 |
| CRP in mg/L, median (IQR) | 61 (21–144) | 16 (5–45) | ||
| NICE warning signs | ||||
| Decreased level of consciousness | 6 (4.4) | 137 (0.8) | 141 | |
| Meningeal signs | 8 (5.9) | 24 | 116 (0.7) | 845 |
| Focal neurology | 2 (1.5) | 29 | 95 (0.6) | 1249 |
| Status epilepticus | 0 (0.0) | 8 | 49 (0.3) | 887 |
| Rash: petechiae/non-blanching | 10 (7.4) | 25 | 640 (3.9) | 1183 |
| Blood cultures performed | 134 (99.3) | 3002 (18.6) | ||
| CSF performed | 25 (18.5) | 381 (2.4) | ||
| Admission to the ward >24 hours | 111 (82.2) | 1 | 5879 (36.4) | 159 |
| Admission to the ICU | 10 (7.4) | 125 (0.8) | 17 | |
| Antibiotic treatment following ED visit | 126 (93.3) | 5804 (35.9) | 197 | |
| LSI: airway, breathing or haemodynamic support | 16 (11.9) | 343 (2.1) | ||
APLS, advanced paediatric life support; CRP, C reactive protein; CSF, cerebrospinal fluid; ED, emergency department; ICU, intensive care unit; LSI, life-saving intervention; NICE, National Institute for Health and Care Excellence.
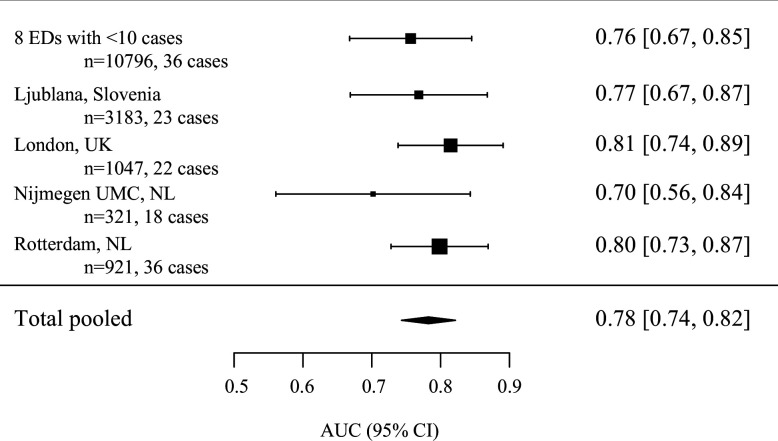
The final model is presented in table 2. This model discriminated well (AUC 0.84 (95% CI 0.81 to 0.88)). In the cross-validation, the model discriminated moderate to well (range AUC 0.76–0.81) yielding a pooled AUC of 0.78 (95% CI 0.74 to 0.82) (figure 2). Calibration was poor to moderate for the different cross-validations (range slope: 0.45–0.81, range intercept −1.2 to 1.0) (online supplemental appendix 8). Apparent calibration was improved by adding an ED-specific variable for high (>2%) versus low (<2%) incidence of IBI (online supplemental appendix 9).
Table 2.
Model specification of multivariate logistic model for IBI
| Coefficients | OR | ||
| (Intercept) | −9.16 | 0.00 | |
| Feverkidstool | Male | −0.19 | 0.83 |
| Age <1 year* | −2.53 | 0.08 | |
| Age ≥1 year* | 0.00 | 1.00 | |
| Temperature | −0.05 | 0.95 | |
| Fever duration in days | −0.15 | 0.86 | |
| Tachypnoea | −0.44 | 0.65 | |
| Tachycardia | 0.69 | 2.00 | |
| Hypoxia | −0.87 | 0.42 | |
| Increased work of breathing | −0.31 | 0.73 | |
| Ill appearance | 0.87 | 2.38 | |
| ln CRP | 0.76 | 2.14 | |
| NICE warning signs | Abnormal neurology | 1.54 | 4.66 |
| Non-blanching rash | 1.38 | 3.96 | |
| Comorbidity | Complex chronic condition | 2.41 | 11.1 |
The risk of children aged <1 year was calculated: β(age <1 year)×age in years.
The risk of children aged ≥1 year was calculated: β(age <1 year)×1+(age in years−1)×β (age ≥ 1 in years).
*Age <1 year and age ≥1 year were calculated linear-piecewise.
CRP, C reactive protein; IBI, invasive bacterial infection; ln, natural log.
Figure 2.
Discriminative value of the prediction model for invasive bacterial infection for five internal-external cross-validations. The model was repeatedly derived on four ED groups, and validated on the fifth ED group which was left out from the derivation. The five cross-validations were pooled using a random-effects model. More details are provided in figure A in online supplemental appendix 3. AUC, area under the receiver operating curve; ED, emergency department; NL, The Netherlands; UK, United Kingdom; UMC, University Medical Centre.
The diagnostic performance was good for the rule-out threshold of 0.1% with sensitivity of 0.97 (95% CI 0.93 to 0.99) and negative LR of 0.09 (95% CI 0.03 to 0.23) (table 3, online supplemental appendix 10). For the rule-in threshold of 2.0%, the model had specificity 0.94 (95% CI 0.94 to 0.95) and positive LR of 8.4 (95% CI 6.9 to 10.0). The intermediate thresholds of 0.1%–2.0% performed poorly (ranges: sensitivity 0.59–0.93, negative LR 0.14–0.47, specificity 0.52–0.88, positive LRs 1.9–4.8) and comprised 9784 (60.1%) patients. The rule-in threshold misclassified four patients with IBI from three different EDs, including two patients with arthritis, and two patients with a sinusitis and pneumonia resulting in bacteraemia. Three of these patients had CRP levels <10 mg/L and symptoms <1 day.
Table 3.
Diagnostic performance of the prediction model for different risk thresholds for invasive bacterial infection
| Risk thresholds (%) |
N below threshold (%) | N above threshold (%) | Sensitivity (95% CI) |
Negative LR (95% CI) |
Specificity (95% CI) |
Positive LR (95% CI) |
| 0.1 | 5495 (33.8) | 10 773 (66.2) | 0.97 (0.93 to 0.99) | 0.09 (0.03 to 0.23) | 0.34 (0.33 to 0.35) | 1.5 (1.4 to 1.5) |
| 0.2 | 8461 (52.0) | 7807 (48.0) | 0.93 (0.87 to 0.96) | 0.14 (0.08 to 0.26) | 0.52 (0.52 to 0.53) | 1.9 (1.9 to 2.1) |
| 0.25 | 9416 (57.9) | 6852 (42.1) | 0.90 (0.84 to 0.95) | 0.17 (0.10 to 0.28) | 0.58 (0.58 to 0.59) | 2.2 (2.0 to 2.3) |
| 0.5 | 12 200 (75.0) | 4068 (25.0) | 0.76 (0.67 to 0.83) | 0.32 (0.24 to 0.44) | 0.75 (0.75 to 0.76) | 3.1 (2.8 to 3.4) |
| 1.0 | 14 224 (87.4) | 2044 (12.6) | 0.59 (0.50 to 0.67) | 0.47 (0.39 to 0.58) | 0.88 (0.87 to 0.88) | 4.8 (4.1 to 5.6) |
| 2.0 | 15 279 (93.9) | 989 (6.1) | 0.48 (0.39 to 0.57) | 0.55 (0.47 to 0.65) | 0.94 (0.94 to 0.95) | 8.4 (6.9 to 10) |
| 5 | 15 831 (97.3) | 437 (2.7) | 0.36 (0.37 to 0.45) | 0.65 (0.57 to 0.74) | 0.98 (0.97 to 0.98) | 15 (12 to 19) |
LR, likelihood ratio.
In sensitivity analysis involving the population with imputed CRP levels (n=37 093, IBI n=135), model development yielded similar coefficients (online supplemental appendix 11).
Discussion
Based on the Feverkidstool and important predictors for early recognition of IBI, we derived and cross-validated a clinical prediction tool, in febrile children at different European EDs. The prediction model discriminated well between patients with and without IBI. The risk threshold of 0.1% has good rule-out value for IBI and thus decreases the risk of missing an IBI. The higher risk thresholds of >2.0% have good rule-in value and these thresholds can be used to identify patients at high risk of IBI to target treatment. The large number of patients with intermediate risk of 0.1%–2.0% for IBI is expected to benefit most from sensitive biomarkers.
Strengths of this study include the participation of 12 European EDs based in 8 countries with a broad population of febrile children of all ages and chronic conditions. Furthermore, we performed five cross-validations which provided us insight in heterogeneity between EDs, and improves the generalisability of our results. Second, we included a large number of IBI cases, while previous studies did not have sufficient cases to define a prediction model exclusively for IBI.9–11 Furthermore, our model involves accessible predictors as clinical symptoms and CRP level, which will facilitate implementation in practice. We provide clinical case examples of the model (online supplemental appendix 12) and, to help physicians to use this model in practice, a web-based digital calculator will be developed.
Our study has some limitations. First, we focused our study on patients who had CRP measurement on indication. This involved more severe illness than patients without CRP measurement. However, the CRP group reflect patients with diagnostic uncertainty and is more likely to benefit from a clinical prediction model. All patients with IBI had CRP measurement, leading to inclusion of all eligible IBIs in the main analysis. In our sensitivity analysis, predictors were similar in the model developed on imputed CRP levels. Therefore, model performance was not influenced by selection of patients with CRP measurement. Second, diagnostic tests were ordered according to usual care. If patients with an IBI did not have cultures taken >24 hours after hospital admission, this was not included in the data and these patients could have been misclassified as non-IBI. Since diagnostic workup is in general performed at the ED or <24 hours after presentation, this misclassification is minimised. Third, due to the low incidence of IBI, model performance was evaluated in cross-validation with a lower number of cases than is optimal for validation (100 cases).33 34Although discrimination of the model was good in the cross-validations, calibration was poor to moderate. The low incidence of IBI and other case-mix differences not taken into account by our model may have influenced model performance in the cross-validation. Our range of IBI incidence (range EDs 0.1%–5.6%) was comparable with IBI incidence in other studies including febrile population of all age ranges (range 0.4%–4.5%).9 11 35 Fourth, due to limited measurements of systolic blood pressure (14.7%) and procalcitonin in our cohort (1.6%), we were not able to include these as predictor. Lastly, data on individual immunisation status were not available and were not included in the model. In the clinical assessment of febrile patients, immunisation status should be taken into account.
Patients with and without IBI were discriminated well in the cross-validations. Calibration was poor to moderate indicating discrepancy between model predictions and the observed risk of IBI. Addition of the ED covariate of low/high incident IBI improved calibration, indicating that model performance is influenced by the likelihood of IBI in the ED. Therefore, ED incidence should be included in the model.
Clinical prediction models involving older children are the Feverkidstool and Irwin’s model, and predict pneumonia and other serious bacterial infections separately, whereas our model focuses on IBI. Discrimination of our model in cross-validation (pooled AUC: 0.78 (95% CI 0.74 to 0.82) was better compared with one external validation and similar to another external validation of the Feverkidstool for other serious bacterial infection.9 11 Unlike our study, these models were not based on an European-wide ED population. We recommend to use the Feverkidstool to guide antibiotic prescription in suspected lower respiratory tract infections18 and to use our model in febrile children to predict IBI. These two models, the original Feverkidstool and our model will be integrated in one electronic decision tool. For both implementation of the Feverkidstool and our model, measurement of (point-of-care) CRP is necessary. We do not recommend CRP measurement in all febrile children, but since CRP level is an important discriminator in bacterial and viral illness, measurement should be easily accessible to aid in the decision-making process at the ED.
Missing and undertreatment of IBI in children can lead to morbidity and mortality. Current practice is to start antibiotic treatment in patients at risk for bacterial infection awaiting culture results which take >48 hours. Since the low incidence of IBI, this leads to overuse of antibiotics and resources. The balance of not missing IBIs and overtreating self-limiting infections is delicate. Therefore, clinical prediction models can help in decision making at the ED. Our study showed that the low-risk threshold can be helpful to rule-out IBI and to reduce invasive diagnostics and antibiotic use.
Starting early treatment is key to prevent adverse outcomes due to IBI. The high risk threshold of >2.0% can be used for targeted treatment with intravenous antibiotics. Although our model was able to identify 38% of the study population as low or high risk, diagnostic uncertainty exist for the intermediate group (60%). In our study, this intermediate group with diagnostic uncertainty was estimated as 25% of the population of febrile children presenting to the ED, including patients without CRP measurement. Additional diagnostics including procalcitonin, repeated CRP measurement36 or novel sensitive biomarkers may be helpful in the decision making for this intermediate risk group. The potential benefit of additional diagnostics using these risk thresholds will need to be evaluated in future studies.
Conclusion
Based on the Feverkidstool and important clinical predictors, we derived and cross-validated a clinical prediction model for early detection of IBI in febrile children in an European-wide cohort. Where the rule-in threshold of this model could target early treatment to reduce adverse outcomes from IBI, the rule-out threshold has the potential to reduce unnecessary use of invasive diagnostics and antibiotics. However, more than half of the population was at intermediate risk. In this group, sensitive, new biomarkers could improve identification of IBI and could potentially reduce unnecessary antibiotic use.
Acknowledgments
The authors would like to thank the members of the PERFORM consortium as listed in online supplemental appendix 13.
Footnotes
Twitter: @rgnijman, @CarrolEnitan
Correction notice: This paper has been amended since it was published online. The data sharing statement has been updated.
Contributors: Conceptualisation: NNH, DB, RGN, DN, JAH, AB, UvB, EC, IE, ME, MvdF, RdG, BK, EL, IM, FM-T, MP, FS, MT, DZ, WZ, ML, CV, HAM. Data curation: NNH, DB, RGN, JAH, AB, UvB, EC, IE, ME, MvdF, RdG, JAH, BK, EL, IM, FM-T, MP, FS, MT, DZ, WZ, ML, CV, HAM. Formal analysis: NNH, DN. Methodology: NNH, DB, RGN, DN, JAH, AB, UvB, EC, IE, ME, MvdF, RdG, BK, EL, FM-T, DN, MP, MT, DZ, CV, WZ, ML, HAM. Supervision: CV, HAM. Writing—original draft: NNH. Writing—review and editing: NNH, DB, RN, DN, JAH, AB, UvB, EC, IE, ME, MvdF, RdG, BK, EL, IM, FM-T, MP, FS, MT, DZ, WZ, ML, CV, HAM.
Funding: This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 668303. The research was supported by the National Institute for Health Research Biomedical Research Centres at Imperial College London, Newcastle Hospitals NHS Foundation Trust and Newcastle University.
Disclaimer: The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Competing interests: None declared.
Patient and public involvement statement: The design of the study did not involve patients. The study results will be disseminated to members of the public, patients and experts through social media and networks.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
An anonymized data set containing individual participant data is available in a public data repository: https://data.hpc.imperial.ac.uk/resolve/?doi=7549. DOI: 10.14469/hpc/7549. For inquiries to obtain the full dataset, please contact the data manager of the PERFORM consortium (Tisham.de08@imperial.ac. uk).
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The study was approved by all the participating hospitals. No informed consent was needed for this study. Austria (Ethikkommission Medizinische Universitat Graz, ID: 28-518 ex 15/16), Germany (Ethikkommission Bei Der LMU München, ID: 699-16), Greece (Ethics committee, ID: 9683/18.07.2016), Latvia (Centrala medicinas etikas komiteja, ID: 14.07.201 6. No. Il 16-07-14), Slovenia (Republic of Slovenia National Medical Ethics Committee, ID: ID: 0120-483/2016-3), Spain (Comité Autonómico de Ética de la Investigación de Galicia, ID: 2016/331), The Netherlands (Commissie Mensgebonden onderzoek, ID: NL58103.091.16), UK (Ethics Committee, ID: 16/LO/1684, IRAS application no. 209035, confidentiality advisory group reference: 16/CAG/0136).
References
- 1. Wolfe I, Cass H, Thompson MJ, et al. Improving child health services in the UK: insights from Europe and their implications for the NHS reforms. BMJ 2011;342:d1277. 10.1136/bmj.d1277 [DOI] [PubMed] [Google Scholar]
- 2. Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 2015;385:430–40. 10.1016/S0140-6736(14)61698-6 [DOI] [PubMed] [Google Scholar]
- 3. Pruitt CM, Neuman MI, Shah SS, et al. Factors associated with adverse outcomes among febrile young infants with invasive bacterial infections. J Pediatr 2019;204:177–82. 10.1016/j.jpeds.2018.08.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van de Maat J, van de Voort E, Mintegi S, et al. Antibiotic prescription for febrile children in European emergency departments: a cross-sectional, observational study. Lancet Infect Dis 2019;19:382–91. 10.1016/S1473-3099(18)30672-8 [DOI] [PubMed] [Google Scholar]
- 5. Herberg JA, Kaforou M, Wright VJ, et al. Diagnostic test accuracy of a 2-Transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA 2016;316:835–45. 10.1001/jama.2016.11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oved K, Cohen A, Boico O, et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS One 2015;10:e0120012. 10.1371/journal.pone.0120012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Houten CB, de Groot JAH, Klein A, et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (opportunity): a double-blind, multicentre, validation study. Lancet Infect Dis 2017;17:431–40. 10.1016/S1473-3099(16)30519-9 [DOI] [PubMed] [Google Scholar]
- 8. Gómez-Carballa A, Cebey-López M, Pardo-Seco J, et al. A qPCR expression assay of IFI44L gene differentiates viral from bacterial infections in febrile children. Sci Rep 2019;9:11780. 10.1038/s41598-019-48162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nijman RG, Vergouwe Y, Thompson M, et al. Clinical prediction model to aid emergency doctors managing febrile children at risk of serious bacterial infections: diagnostic study. BMJ 2013;346:f1706. 10.1136/bmj.f1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuppermann N, Dayan PS, Levine DA, et al. A clinical prediction rule to identify febrile infants 60 days and younger at low risk for serious bacterial infections. JAMA Pediatr 2019;173:342–51. 10.1001/jamapediatrics.2018.5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Irwin AD, Grant A, Williams R, et al. Predicting risk of serious bacterial infections in febrile children in the emergency department. Pediatrics 2017;140. 10.1542/peds.2016-2853. [Epub ahead of print: 05 Jul 2017]. [DOI] [PubMed] [Google Scholar]
- 12. Gomez B, Mintegi S, Bressan S, et al. Validation of the "Step-by-Step" Approach in the Management of Young Febrile Infants. Pediatrics 2016;138. 10.1542/peds.2015-4381. [Epub ahead of print: 05 Jul 2016]. [DOI] [PubMed] [Google Scholar]
- 13. Vos-Kerkhof Ede, Gomez B, Milcent K, et al. Clinical prediction models for young febrile infants at the emergency department: an international validation study. Arch Dis Child 2018;103:1033–41. 10.1136/archdischild-2017-314011 [DOI] [PubMed] [Google Scholar]
- 14. Leroy S, Bressan S, Lacroix L, et al. Refined Lab-score, a risk score predicting serious bacterial infection in febrile children less than 3 years of age. Pediatr Infect Dis J 2018;37:387–93. 10.1097/INF.0000000000001915 [DOI] [PubMed] [Google Scholar]
- 15. Aronson PL, Shabanova V, Shapiro ED, et al. A prediction model to identify febrile infants ≤60 days at low risk of invasive bacterial infection. Pediatrics 2019;144. 10.1542/peds.2018-3604. [Epub ahead of print: 05 Jun 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burman C, Serra L, Nuttens C, et al. Meningococcal disease in adolescents and young adults: a review of the rationale for prevention through vaccination. Hum Vaccin Immunother 2019;15:459–69. 10.1080/21645515.2018.1528831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watson RS, Carcillo JA, Linde-Zwirble WT, et al. The epidemiology of severe sepsis in children in the United States. Am J Respir Crit Care Med 2003;167:695–701. 10.1164/rccm.200207-682OC [DOI] [PubMed] [Google Scholar]
- 18. van de Maat JS, Peeters D, Nieboer D, et al. Evaluation of a clinical decision rule to guide antibiotic prescription in children with suspected lower respiratory tract infection in the Netherlands: a stepped-wedge cluster randomised trial. PLoS Med 2020;17:e1003034. 10.1371/journal.pmed.1003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borensztajn D, Yeung S, Hagedoorn NN, et al. Diversity in the emergency care for febrile children in Europe: a questionnaire study. BMJ Paediatr Open 2019;3:e000456. 10.1136/bmjpo-2019-000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagedoorn NN, Borensztajn DM, Nijman R, et al. Variation in antibiotic prescription rates in febrile children presenting to emergency departments across Europe (MOFICHE): A multicentre observational study. PLoS Med 2020;17:e1003208. 10.1371/journal.pmed.1003208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21., Roberts KB, Subcommittee on Urinary Tract Infection, Steering Committee on Quality Improvement and Management . Urinary tract infection: clinical practice guideline for the diagnosis and management of the initial UTI in febrile infants and children 2 to 24 months. Pediatrics 2011;128:595–610. 10.1542/peds.2011-1330 [DOI] [PubMed] [Google Scholar]
- 22. Simon TD, Cawthon ML, Stanford S, et al. Pediatric medical complexity algorithm: a new method to stratify children by medical complexity. Pediatrics 2014;133:e1647–54. 10.1542/peds.2013-3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The National Institute for Health and Care Excellence (NICE) . Fever in under 5S: assessment and initial managment CG160 may 2013, 2013. Available: https://www.nice.org.uk/guidance/cg160 [Accessed Aug 2017]. [PubMed]
- 24. Pneumonia Etiology Research for Child Health (PERCH) Study Group . Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet 2019;394:757–79. 10.1016/S0140-6736(19)30721-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Riley RD, Snell KI, Ensor J, et al. Minimum sample size for developing a multivariable prediction model: PART II - binary and time-to-event outcomes. Stat Med 2019;38:1276–96. 10.1002/sim.7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su T-L, Jaki T, Hickey GL, et al. A review of statistical updating methods for clinical prediction models. Stat Methods Med Res 2018;27:185–97. 10.1177/0962280215626466 [DOI] [PubMed] [Google Scholar]
- 27. Steyerberg EW, Borsboom GJJM, van Houwelingen HC, et al. Validation and updating of predictive logistic regression models: a study on sample size and shrinkage. Stat Med 2004;23:2567–86. 10.1002/sim.1844 [DOI] [PubMed] [Google Scholar]
- 28. Advanced Life Support Group . Advanced paediatric life support: the practical approach. 5th edn. Wiley, 2011. [Google Scholar]
- 29. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc Series B 1996;58:267–88. [Google Scholar]
- 31. Steyerberg EW, Harrell FE. Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol 2016;69:245–7. 10.1016/j.jclinepi.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565–74. 10.1177/0272989X06295361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Calster B, Nieboer D, Vergouwe Y, et al. A calibration hierarchy for risk models was defined: from utopia to empirical data. J Clin Epidemiol 2016;74:167–76. 10.1016/j.jclinepi.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 34. Collins GS, Ogundimu EO, Altman DG. Sample size considerations for the external validation of a multivariable prognostic model: a resampling study. Stat Med 2016;35:214–26. 10.1002/sim.6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Craig JC, Williams GJ, Jones M, et al. The accuracy of clinical symptoms and signs for the diagnosis of serious bacterial infection in young febrile children: prospective cohort study of 15 781 febrile illnesses. BMJ 2010;340:c1594. 10.1136/bmj.c1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wasserman A, Karov R, Shenhar-Tsarfaty S, et al. Septic patients presenting with apparently normal C-reactive protein: a point of caution for the ER physician. Medicine 2019;98:e13989. 10.1097/MD.0000000000013989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
archdischild-2020-319794supp001.pdf (2.7MB, pdf)
Data Availability Statement
An anonymized data set containing individual participant data is available in a public data repository: https://data.hpc.imperial.ac.uk/resolve/?doi=7549. DOI: 10.14469/hpc/7549. For inquiries to obtain the full dataset, please contact the data manager of the PERFORM consortium (Tisham.de08@imperial.ac. uk).