Abstract
Clear cell renal cell carcinoma (ccRCC) is one of the most prevalent renal malignant cancer, whose survival rate and quality of life of patients are still not satisfactory. Nevertheless, the TNM staging system currently used in clinical cannot make accurate survival predictions and precise treatment decisions for ccRCC patients. Therefore, there is an urgent need for more reliable biomarkers to identify high-risk subgroups of ccRCC patients to guide timely intervention and treatment. Recently, MiRNAs have been shown to be closely related to the procession of a variety of tumors, and they have high stability in various tissues, which makes them suggested to have the potential as a prognostic biomarker of ccRCC. In this study, by analyzing and processing the miRNAs expression profile of ccRCC patients from the TCGA database, we finally constructed an excellent miRNAs signature and verified it through a variety of methods. In order to build a more accurate and reliable clinical predictive model, we integrated the miRNAs signature with other prognostic-related clinical parameters to construct a nomogram. Functional enrichment analysis showed that miRNAs in the signature may regulate the genes involved in the Hippo signaling pathway, Tight junction, and Wnt signaling pathway to cause different prognoses of ccRCC patients, which may provide a reference for subsequent basic research and targeted therapy. To conclude, our study constructed a useful miRNAs signature, which allows the prognosis stratification for ccRCC patients and thereby guides the timely and effective interventions on high-risk patients. At the same time, this study also found the potential biological pathways involved in the procession of ccRCC.
Keywords: clear cell renal cell carcinoma, differentially expressed genes, prognosis, bioinformatics
Introduction
Renal carcinoma is one of the most common malignant tumors of the urinary system. In 2020, there were more than 73000 new cases and 14000 new deaths of renal carcinoma in the United States. 1 According to statistics, around one-third of patients with renal carcinoma present local or distant metastasis. Besides, for local renal cell carcinoma (RCC) patients who have undergone nephrectomy, approximately one quarter of them have distant relapses. 2 RCC is the most common type of renal cancer, accounting for more than 90%, and the morbidity of RCC is steadily increasing every year. 3 The pathological type of RCC consists of clear cell renal cell carcinoma (ccRCC), papillary adenocarcinoma, chromophobe cell carcinoma, Xp11.2 translocation RCC, and unclassified RCC, etc. 4 They all originate from renal tubular epithelial cells, but there are highly heterogeneous in biological characteristics and response to treatment between different histological subtypes. 5 The most common histological subtype of RCC is ccRCC (about 70%-80%), which is highly aggressive and has a poor prognosis. 5 Identification of the patients with poor prognosis early is of great help for timely and accurate clinical intervention and treatment. Unfortunately, there is currently a lack of specific biomarkers for screening such patients. Thus, it is very imperative to explore the prognosis-related specific biomarkers of ccRCC patients and develop novel treatments for this fatal disease to improve its prognosis.
MiRNAs are a class of short, single-stranded non-coding RNAs, composed of about 22 nucleotides, which negatively regulate gene expression by mediating mRNAs degradation or inhibiting their translation. 6 Recently, a large number of studies have confirmed that miRNAs are involved in the growth, migration, invasion, angiogenesis, and the regulation of tumor microenvironment in various tumors. 7,8 Besides, some miRNAs have been shown to hold the potential value for diagnosis and prognosis of a variety of tumors. 9 A study that a 17-miRNA signature could effectively classify the ccRCC tissues and the adjacent normal tissues, and further research has found that the decreased expression of miR-204-5p and miR-139-5p was associated with an increased risk of recurrence in ccRCC patients. 10 Meng et al found that the up-regulation of miR-125b-2-3p is closely related to lymphatic invasion, distant metastasis and poor survival of ccRCC patients. 11 Additionally, as for ccRCC patients, the high expression of miR-210-3p, miR-210, miR-146a-5p, miR-141 and the low expression of miR-194, miR-124-3p, miR-30a-5p are also found to be associated with the poor prognosis. 12,13 However, studies on miRNAs as biomarkers to predict the prognosis of ccRCC are still not enough. The diagnostic ability of most miRNAs signatures are not ideal and have not been applied in clinical practice. Therefore, finding and constructing a more accurate and effective miRNAs signature with prognostic value of ccRCC is of great clinical significance.
Herein, we first downloaded the miRNAs expression data and corresponding clinical information of ccRCC patients from The Cancer Genome Atlas database (TCGA) database. After differential expression analysis, univariate Cox regression, Least Absolute Shrinkage and Selection Operator (LASSO) regression analysis, and multivariate Cox regression analysis, 5 miRNAs with optimal prognostic value were screened out. Then, we constructed a 5-miRNA signature and validated the prognostic ability of our model by multiple methods. To build a more accurate and reliable clinical predictive model, we integrated the miRNAs signature and other prognosis-related clinical parameters to construct a nomogram. Finally, in order to explore the potential biological processes related to prognosis, we performed functional enrichment analysis on the downstream target genes of the 5 miRNAs. The result indicated that Hippo signaling pathway, Tight junction, and Wnt signaling pathway may be related to the differential prognosis of ccRCC patients.
Methods
Data Sources and Screening of Differentially Expressed miRNAs
Firstly, the expression data of miRNAs and the corresponding clinical information of ccRCC patients in the Kidney Clear Cell Carcinoma (KIRC) subset of TCGA (http://tcga.cancer.gov) database were downloaded for analysis. These expression data were obtained from the sequencing results of 545 tumor tissues and 71 adjacent non-tumor tissues. Then, we excluded the patients without complete clinical information and the characteristics of the rest ccRCC patients were shown in Table S1. Finally, the differentially expressed miRNAs (DE-miRNAs) between tumor tissues and adjacent non-tumor tissues were identified using the “edgeR” package in R software with |log2FC|>1 and adjusted P < 0.05. 14
Identify a Potential Prognosis-Related miRNAs Signature
We randomly divided patients into 2 groups according to the ratio of 7:3. The expression data of 1 group (Training set) was used to construct a signature of miRNAs with prognostic value, while the expression data of another group (Testing set) was used to validate the signature we constructed. Firstly, a univariate Cox regression analysis (Hazard Ratio ≠ 1, P < 0.05) was performed on the expression data of the training set (n = 364) to identify the prognosis-related miRNAs (Pr-miRNAs). Then, LASSO regression analysis was conducted to sub-select the Pr-miRNAs with the “glmnet” package in R software, which can scale down the regression coefficient forcedly. 15 1000-times cross-validations were performed and the optimal penalty parameter λ was determined to increase the reliability and objectivity. 16 Next, a multivariate Cox regression analysis was performed on the sub-selected Pr-miRNAs to get the optimal prognostic miRNAs. Finally, we combined the expression levels of optimal prognostic miRNAs and their regression coefficients to establish a formula of risk score:
The Exp is the expression level of miRNAs if patient i, while the coef means its regression coefficient.
Evaluation and Validation of the miRNAs Model
We stratified all patients in the training set into high- and low-risk groups according to the median risk score. Kaplan-Meier (K-M) survival analysis was used to compare the survival differences between the above 2 groups and the time-dependent receiver operating characteristic (ROC) curve was used to evaluate the prognostic accuracy of miRNAs signature. 17 Besides, we also compared the survival differences between the high- and low-risk groups in different clinical subgroups (including ages, genders, histologic grades, and pathologic stages) to measure the prognostic performance of our miRNAs signature.
To validate the miRNAs signature’s prognostic power and applicability, we performed the above formula on the patients in the testing set (n = 152) and the entire set (n = 516) similarly. The KM survival analysis and ROC curve were also used to evaluate our miRNAs signature.
Identification of Independent Prognostic Factors and Nomogram Construction
To determine whether the miRNAs signature is an independent prognostic factor of ccRCC. We set the survival of the patients as the dependent variable, while the risk score ages of patients, gender, histologic grades, and pathologic stages as independent variables to perform univariate Cox regression analysis. As for variables with statistical differences (P < 0.05), we further conducted multivariate Cox regression analysis on these variables to determine if they are independent prognostic factors of ccRCC. In order to build an accurate and reliable clinical predictive model, we constructed a nomogram by integrating all independent prognostic factors to predict the survival of ccRCC patients at 1-, 3-, and 5-year by using the “rms” package of the R software. 18 The nomogram can integrate various prognostic factors to predict the outcome of individual clinical events, it was often used to measure the prognosis of patients.
Evaluate and Validate the Nomogram
For our nomogram, calibration plots were used to evaluate its performance. In the calibration curve, X-axis represents the predicted survival rate of our nomogram while Y-axis represents the actual survival rate respectively and the 45° dotted line represents the best outcome. Then, KM survival analysis and time-related ROC analysis were also conducted to measure the predictive power of the nomogram.
Identification of Downstream Target Genes
The possible target genes of the 5 optimal prognostic miRNAs were predicted by the miRDB online database (http://www.mirdb.org/), which can predict and annotate mammalian miRNA target genes. At the same time, we downloaded mRNAs expression data of ccRCC and adjacent nontumor renal specimens from the TCGA database and screened out the differentially expressed mRNAs between ccRCC and adjacent non-tumor specimens. Integrating the predicted target genes with the differentially expressed mRNAs, we finally obtained the possible downstream target genes of 5 optimal prognostic miRNAs.
Functional Enrichment Analysis
To find out the potential mechanisms involved in the different prognosis of the ccRCC, we used the “clusterProfiler” package in R software to perform the gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on those possible downstream target genes of 5 optimal prognostic miRNAs. 19 P < 0.05 was considered as statistically significant.
Statistical Analysis
R software (version 3.6.3) was used to perform the statistical analyses in this study. All statistical tests were 2 sides and P < 0.05 were statistically significant.
Results
DE-miRNAs in ccRCC
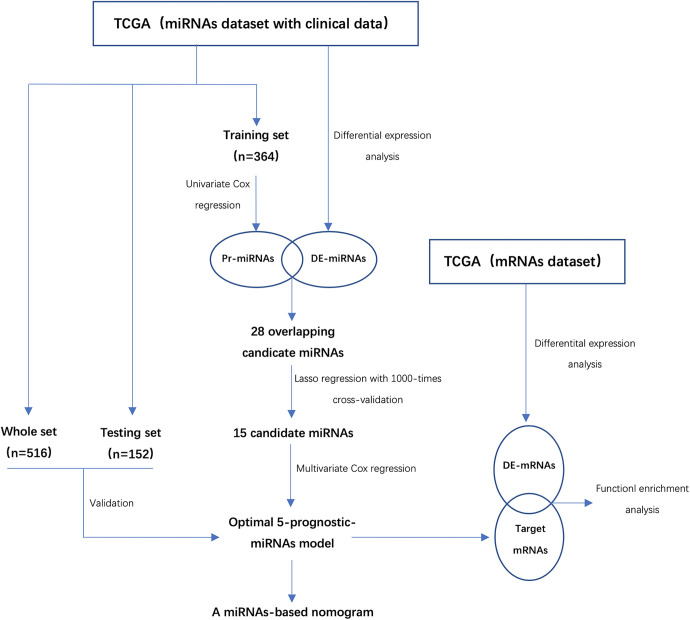
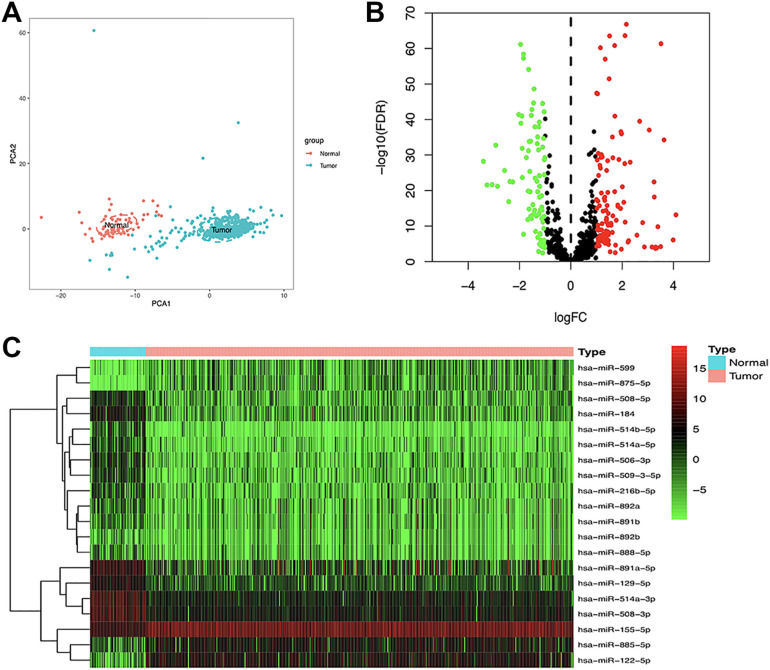
Figure 1 showed the flowchart of our study. Firstly, a total of 2089 miRNAs were obtained from the raw sequencing data of ccRCC patients in the TCGA dataset. Principal component analysis (PCA) showed that there was a significant difference in the expression profile of miRNAs between tumor and adjacent non-tumor tissues (Figure 2A). Subsequently, we identified a total of 221 DE-miRNAs between the 2 tissues, including 115 up-regulated DE-miRNAs and 96 down-regulated DE-miRNAs (Figure 2B). Figure 2C showed the heatmap of the top 20 DE-miRNAs in tumor and adjacent nontumor tissues.
Figure 1.
The flowchart of this study. DE-miRNAs indicates differentially expressed miRNAs; Pr-miRNAs, prognosis-related miRNAs; DE-mRNAs, differentially expressed mRNAs; TCGA, the cancer genome atlas.
Figure 2.
Screening of the DE-miRNAs. A, PCA plot of the expression profile of miRNAs between tumor and normal tissue. B, Volcano plot of DE-miRNAs between tumor and normal tissue. C, Heatmap of the DEmiRNAs. DE-miRNAs indicates differentially expressed miRNAs; PCA, principal component analysis.
Identify a Prognosis-Related miRNAs Signature
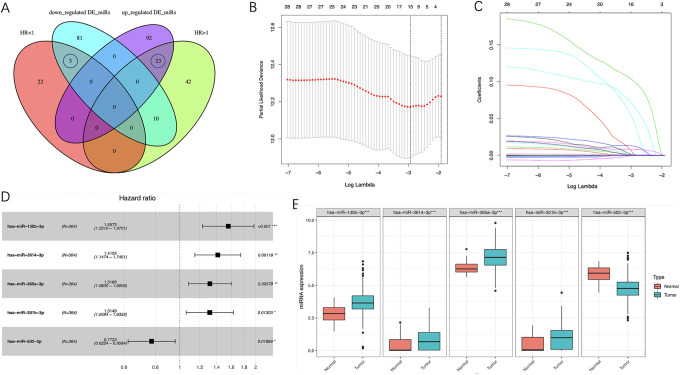
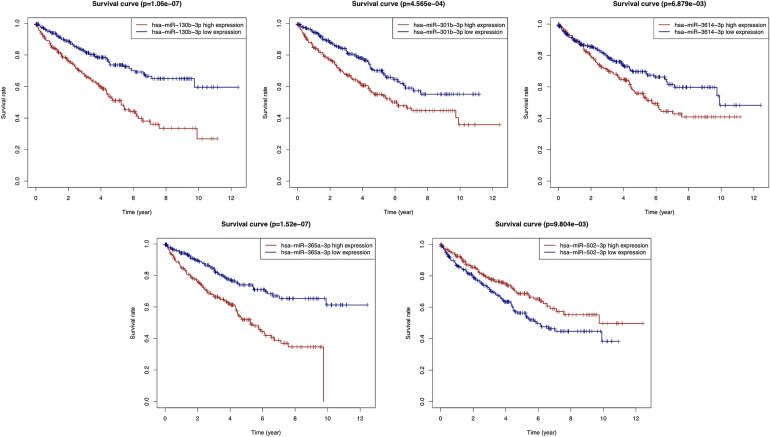
According to our screening criteria, a total of 102 Pr-miRNAs were screened in the training set, of which 75 with HR >1 and 27 with HR <1. By intersecting the 75 prognostic miRNAs (HR>1) with the 115 up-regulated miRNAs and the 27 prognostic miRNAs (HR<1) with the 96 down-regulated miRNAs, we obtained 28 miRNAs (Figure 3A). To screen out the miRNAs with better prognostic value, we performed LASSO regression with 1000-times cross-validations on these miRNAs and obtained 15 candidate miRNAs (Figure 3B-C). Then, these candidate miRNAs were further performed on multivariate Cox regression analysis. Finally, 5 miRNAs with the optimal prognostic value were identified (Figure 3D). Figure 3E shows the expression levels of the optimal prognosis-related miRNAs in tumor and adjacent non-tumor tissues. The prognostic power of each optimal prognosis-related miRNAs was shown in Figure 4. As Figure 4 showed, higher expressions of the hsa-miR-130b-3p, hsa-miR-301b-3p, hsa-miR-3614-3p and hsa-miR-365a-3p were associated with worse prognosis. While the higher expression of the hsa-miR-502-3p was associated with a better prognosis. Furthermore, we established a risk score formula by combining the expression levels of these 5 optimal prognosis-related miRNAs and their corresponding regression coefficients (Risk score = Exp(miR-130b-3p)*0.44297908 + Exp(miR-301b-3p)*0.27378521 + Exp(miR-3614-3p)*0.34747177 + Exp(miR-365a-3p)*0.275049 − Exp(miR-502-3p)*0.25833766), and calculated the risk score of all 525 patients according to the formula.
Figure 3.
Identification of 5 optimal Pr-miRNAs and their expression levels in ccRCC. A, Venn diagram of 28 Pr-miRNAs. B, 15 candidate miRNAs obtained by LASSO regression with 1000-times cross-validation using minimum λ value. C, LASSO coefficients profiles of 28 Pr-miRNAs. D, 5 optimal Pr-miRNAs got by multivariate Cox regression analysis. E, Expression pattern of the 5 optimal Pr-miRNAs between tumor and normal tissues. Pr-miRNAs indicates prognosis-related miRNAs; ccRCC: clear cell renal cell carcinoma; LASSO, least absolute shrinkage and selection operator.
Figure 4.
Kaplan-Meier survival analysis of miR-130b-3p, miR-301b-3p, miR-3614-3p, miR-365a-3p, and miR-502-3p.
Evaluation and Validation of the 5-miRNAs Signature
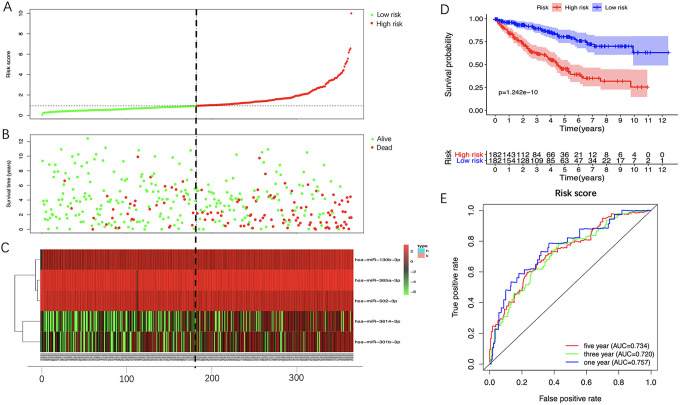
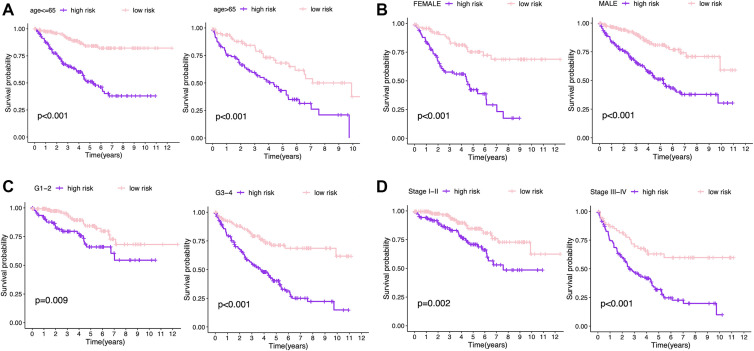
We stratified patients into 2 groups (high- and low- risk) based on the median risk score (0.9354) in training set (Figure 5A). The survival status of all patients was shown in Figure 5B while the heatmap of the expression levels of the 5 optimal prognosis-related miRNAs was shown in Figure 5C. The KM survival curve showed that high-risk patients hold worse overall survival (OS) than low-risk patients (Figure 5D). The ROC curve indicated that the 5-miRNAs signature had a reliable prognostic accuracy with the area under the curve (AUC) was 0.757 at 1 year, 0.720 at 3 years, and 0.734 at 5 years (Figure 5E). Besides, we measured the prognostic performance of the signature between different clinical subgroups and the results also showed that the OS of high-risk patients was significantly worse than that of low-risk patients (Figure 6).
Figure 5.
Prognostic analysis of 5-miRNA signature in the training set. The dotted line represented the median risk score and stratified the ccRCC patients into low- and high-risk group. A, The curve of risk score. B, Survival status of the ccRCC patients. C, Heatmap of the expression levels of the 5 optimal Pr-miRNAs in low- and high-risk group. D, K-M survival analysis of the 5-miRNA signature. E, Time-dependent ROC analysis of the 5-miRNA signature. CcRCC indicates clear cell renal cell carcinoma; Pr-miRNAs, prognosis-related miRNAs; K-M, Kaplan-Meier; ROC, receiver operating characteristic.
Figure 6.
Kaplan-Meier survival analysis of the 5-miRNA signature in different subgroups including (A) younger than 65 years old, older than 65 years old; (B) male, female; (C) grade1/2, grade3/4; (D) stage III, stage III-IV. K-M indicates Kaplan-Meier.
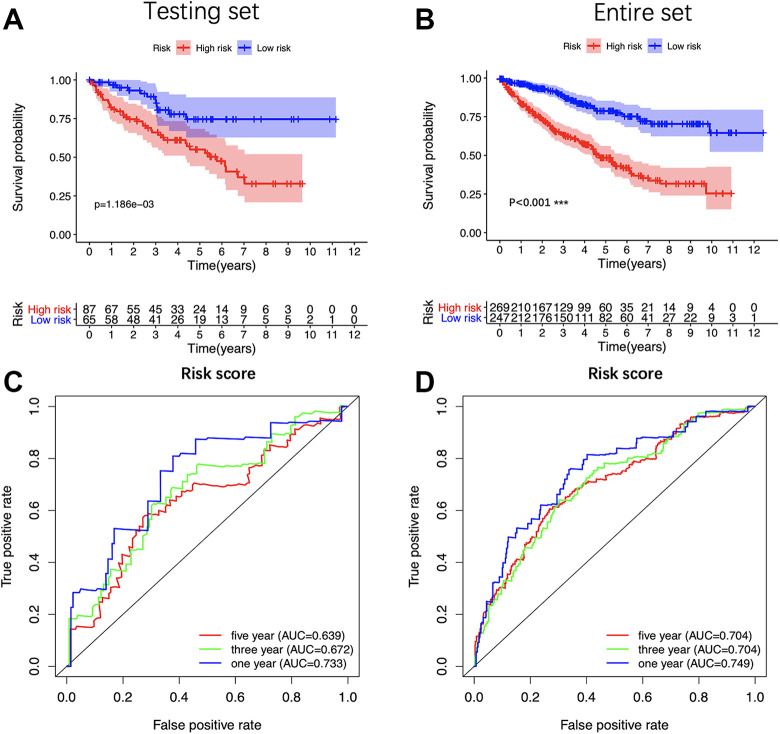
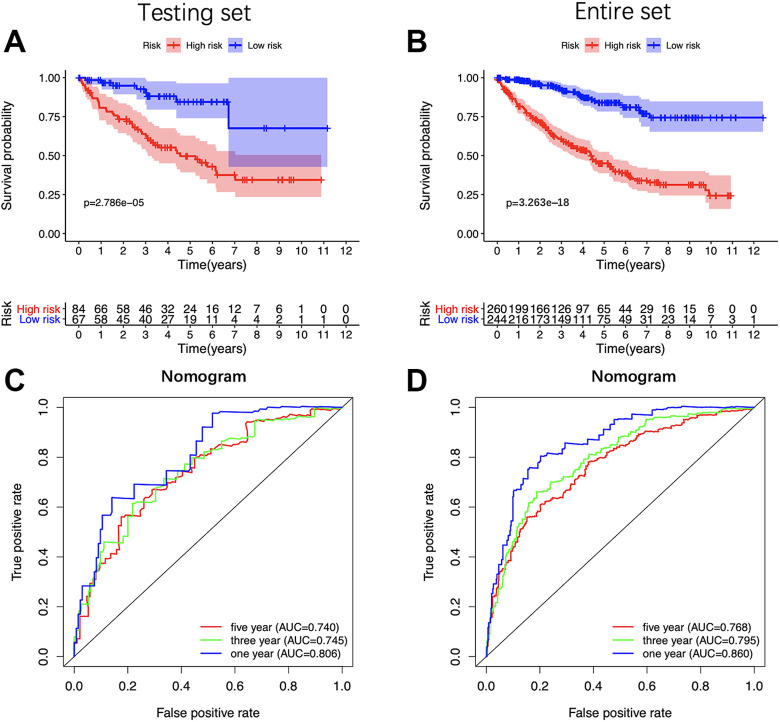
In order to validate the constructed 5-miRNAs signature, we further applied the risk score formula to the testing set and the entire set. Consistent with the training set, both sets presented that high-risk patients hold worse OS than low-risk patients (Figure 7A and B). The ROC curve showed that the AUC of the testing set was 0.733 at 1 year, 0.672 at 3 years, 0.639 at 5 years; and the AUC of the entire set was 0.749 at 1 year, 0.704 at 3 years, 0.704 at 5 years. (Figure 7C and D). In summary, the signature constructed by us had good prognostic power for ccRCC patients.
Figure 7.
Validation of the 5-miRNA signature. Kaplan-Meier survival analysis of the 5-miRNA signature in internal testing set (A) and the entire set (C). Time-dependent ROC analysis of the 5-miRNA signature in internal testing set (B) and the entire set (D). K-M indicates Kaplan-Meier; ROC, receiver operating characteristic.
Construction, Calibration and Validation of the Nomogram
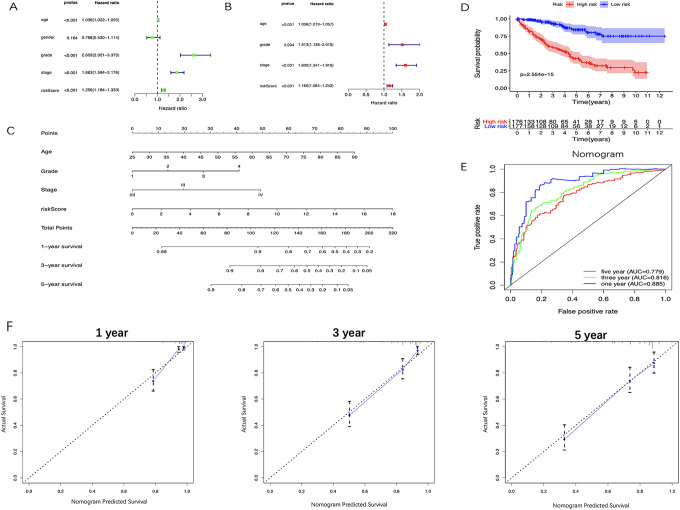
The results of the univariate analysis showed that the age of patients, the histologic grades, the pathologic stages and the risk score based on the 5-miRNA signature were all negatively correlated with the prognosis of ccRCC patients. The prognosis of older patients with the higher histological grade, the later pathological stage and the higher risk score seem to be worse. In contrast, the gender of the patients was not associated with the prognosis (Figure 8A). The 5-miRNA signature and other clinical parameters (ages of patients, histologic grades, and pathologic stages) were identified as the independent prognostic factors for ccRCC through multivariate analysis (Figure 8B). Then, a nomogram consists of these independent prognostic factors was established to predict the survival at 1-, 3- and 5-year of ccRCC patients (Figure 8C). Besides, the KM survival analysis and the ROC curve indicated that the nomogram has a better prognostic performance than the 5-miRNAs signature (Figure 8E and F) and consistent results were got in testing set and the entire set (Figure 9A-D).
Figure 8.
Identifying the independent prognostic factors and construction of miRNA-based nomogram. A, Forrest plot of Univariate Cox regression analysis in ccRCC. B, Forrest plot of multivariate Cox regression analysis in ccRCC. C, Nomogram integrated 5 miRNA-based risk score, age, grade, and stage. D, The calibration plot of the nomogram for agreement test between 1-, 3- and 5-year OS prediction and the actual outcome. E, OS of the high-risk group was significantly worse than that of the low-risk group. F, Time-dependent ROC curves of the nomogram. CcRCC indicates clear cell renal cell carcinoma; OS, overall survival; ROC, receiver operating characteristic.
Figure 9.
Validation of the miRNA-based nomogram. Kaplan-Meier survival analysis of the nomogram in the testing set (A) and the entire set (C). Time-dependent ROC analysis of the nomogram in the testing set (B) and the entire set (D). K-M indicates Kaplan-Meier; ROC, receiver operating characteristic.
Biological Pathways Related to the Prognosis
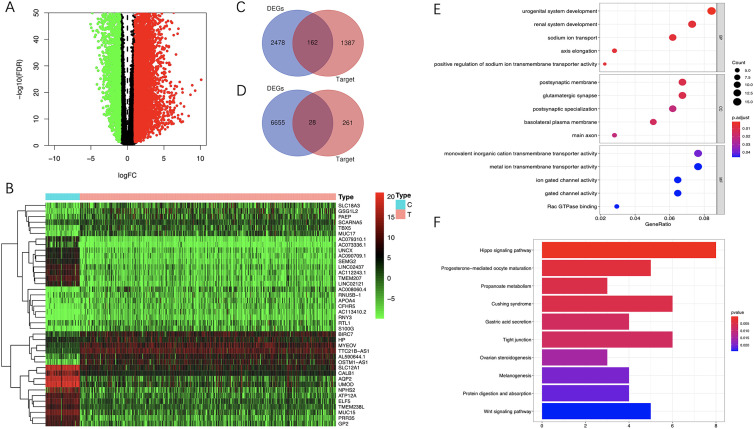
To explore the biological functions and potential mechanisms related to prognosis, we predicted the target genes of 5 miRNAs in the miRDB online database. 1549 target genes were predicted for the 4 up-regulated miRNAs, while 289 target genes were predicted for the 1 down-regulated miRNAs. By analyzing the mRNAs expression data of ccRCC in the TCGA database, A total of 6683 up-regulated mRNAs and 2640 down-regulated mRNAs were screened out between ccRCC and adjacent non-tumor tissues. The target genes of up-regulated miRNAs were intersected with the down-regulated mRNAs, and the target genes of down-regulated miRNAs were intersected with the up-regulated mRNAs. Finally, 190 possible downstream target genes were obtained (Figure 10A-D).
Figure 10.
Functional enrichment analysis. A, Volcano plot of DE-mRNAs between tumor and normal tissue. B, Heatmap of the DE-mRNAs. C, Intersecting the target genes of up-regulated miRNAs with the downregulated mRNAs. D, Intersecting the target genes of down-regulated miRNAs with the up-regulated mRNAs. E, GO enrichment analysis of the possible downstream target genes. F, KEGG enrichment analysis of the possible downstream target genes. GO indicates gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DE-mRNAs, differentially expressed mRNAs.
The GO and KEGG enrichment analysis were performed to identify the potential biological functions of these target genes. The results of GO showed that these target genes were mainly enriched in biological processes such as urogenital system development, renal system development, sodium ion transport, axis elongation, positive regulation of sodium ion transmembrane transporter activity, etc (Figure 10E). KEGG pathway enrichment analysis results showed that these target genes were mainly enriched in Hippo signaling pathway, Propanoate metabolism, Tight junction and Wnt signaling pathway (Figure 10B).
Discussion
Nowadays, although the treatment of ccRCC has been standardized, the mortality of patients has not been significantly improved. 2 In order to reduce the mortality of ccRCC patients and improve their prognosis, it is sensible to develop effective biomarkers or construct accurate models with prognostic value for ccRCC patients. Studies have found that a variety of miRNAs were involved in the occurrence and progression of ccRCC, and some of them have also been proved to be closely related to the prognosis of ccRCC. Combining their stability in multiple organizations, they were supported to be effectively prognostic biomarkers for ccRCC. Thus, the objective of this study is to construct a miRNAs-based model with a strong prognostic ability to help identify the high-risk ccRCC patients with poor prognosis, and then guide timely and effective intervention and treatment, which finally improve the prognosis and quality of life of these patients.
We obtained DE-miRNAs between tumor tissues and adjacent non-tumor tissues by analyzing the sequencing data in the TCGA database. After a series of regression analyses, we constructed a 5-miRNA signature, including miR-130b-3p, miR-3614-3p, miR-365a-3p, miR-301b-3p, and miR-502-3p, which is validated as an independent prognostic factor of ccRCC. Patients were stratified into 2 groups according to the risk score formula, and KM survival analysis and ROC curve have confirmed the performance of our signature. We further validated the miRNAs signature in testing set, entire set and different clinical types of patients. To establish a more reliable and individualized clinical prediction method, we incorporated the 5-miRNA signature with other independent prognostic factors to construct a nomogram. The predicted outcome of the nomogram showed a good agreement with the actual outcome in the calibration plots, and KM survival analysis and ROC curve analysis also showed that the constructed nomogram had higher prognostic efficiency. Finally, by integrating the predicted target genes of 5 miRNAs in miRDB online database and differentially expressed genes of ccRCC patients in the TCGA database, we obtained the potential downstream target genes of these 5 miRNAs, and further performed the functional enrichment analysis on these genes to explore the potential biological pathways involved in the different prognosis of the ccRCC.
It is worth noting that the 5 optimal prognostic miRNAs screened in our study have also been reported to be related to the ontogeny, progression and prognosis of tumors in other literature. Peng et al found that the high expression of miR-130 family was closely related to the poor survival of cancer patients, especially patients with gastric cancer or hepatocellular carcinoma. 20 A study also found that the signaling cascade involved in miR-130b-3p can mediate an interaction between cancer cells and M2 macrophages in the tumor microenvironment to accelerate the progression of gastric cancer. 21 Besides, the up-regulated miR-130b-3p in hepatocellular carcinoma can promote the progression and angiogenesis of hepatocellular carcinoma, which is significantly associated with the poor prognosis of patient. 22 Interestingly, miR-130b-3p also holds the ability to inhibit the progression of tumors. Shui et al proved that miR-130b-3p can directly target Notch ligand Delta-like 1 to inhibit the invasion and metastasis of breast cancer cells. 23 Another study found that exosomal miR-130b-3p targeted serine/threonine-protein kinase 1 through the p53 signal pathway could inhibit medulloblastoma tumorigenesis. 24 As for miR-3614-3p, a study has reported that the overexpression of it can down-regulate the expression of TRIM25 to inhibit the growth of breast cancer cells. 25 MiR-365a-3p was proved to inhibit the expansion and metastasis of colorectal cancer cells partly via inhibiting the expression of ADAM10 and JAK/STAT signal. 26,27 In addition, miR-365a-3p could also prevent the progression of pancreatic cancer through inhibiting NF-κB pathway. 27 MiR-365a-3p is also regarded as an oncogene, which play an important role in the proliferation, migration and invasion of lung cancer, gastric cancer and laryngeal squamous cell carcinoma lung cancer cells. 28 -30 As an oncogene, miR-301b-3p is involved in the progression of hepatocellular carcinoma, gastric cancer and prostate cancer through a variety of different mechanisms. 31 -33 As for miR-502-3p, it can target SET to inhibit the proliferation, migration and invasion of hepatocellular carcinoma. 34
To explore the biological functions and potential mechanisms related to prognosis, we performed the functional enrichment analysis on the possible downstream target genes of the 5 optimal prognostic miRNAs. The results of GO showed that these target genes were mainly associated with the urogenital system development, renal system development and sodium ion transport. KEGG analysis results showed that these target genes were mainly enriched in the Hippo signal pathway, tight junction and Wnt signal pathway. Hippo signal pathway is actually a kind of tumor-suppressive pathway, which is involved in cell growth, proliferation, differentiation, apoptosis and regeneration, and its imbalance is closely related to the occurrence and progression of a variety of tumors. 35,36 YAP, a downstream target of the Hippo signal pathway, whose overexpression was associated with the poor prognosis of hepatocellular carcinoma. 37 Besides, the Hippo pathway and its upstream and downstream effectors were also involved in regulating the development and tumorigenesis of the liver and pancreas. 38 Wnt signal pathway, like Hippo pathway, was first found in drosophila, which is highly conserved in evolution and is closely related to cancer. 39,40 Wnt pathway was also involved in the occurrence and progression of a variety of tumors such as colorectal cancer, melanoma, bladder cancer, prostate cancer and leukemia, which was highly related to cancer cell metastasis and immune response in the tumor microenvironment. 39,40 Tight junctions were involved in maintaining cell polarity, and tight junction proteins were related to the regulation of cell proliferation, transformation and metastasis by recruiting signal proteins. 41 Bhat et al summarized the interaction mechanism and related molecular signals between tight junction protein claudin 1-20 and a variety of cancers, and emphasized the important role of tight junction proteins in tumors. 41 Our results suggested that the 5 miRNAs might affect the prognosis of ccRCC patients mainly by regulating the expression of genes enriched in the above 3 signal pathways. This provides a reference for improving the prognosis of high-risk patients by targeted regulation of the above 3 signaling pathways.
In conclusion, our study constructed a 5-miRNAs signature with prognostic value for ccRCC patients, which is an independent prognostic factor for patients with ccRCC. Furthermore, we integrated our miRNAs signature and other prognosis-related clinical parameters to construct a nomogram, which can predict the prognosis of ccRCC patients more accurate and reliable. Besides, we also found that the potential mechanism of different prognosis of ccRCC patients caused by potential downstream target genes of the 5 miRNAs may be related to Hippo, tight junction and Wnt signal pathways. However, there are still some limitations. This study is based on bioinformatics data analysis, and the results are needed to further validate in multicenter and prospective studies in the future. It is pretty necessary to explore the potential mechanisms related to our signature in the ontogeny, development and prognosis of ccRCC, which may provide a new field of vision for risk stratification, early intervention and targeted therapy in patients with ccRCC.
Supplemental Material
Supplemental Material, sj-pdf-1-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-2-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-3-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-4-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-5-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Abbreviations
- RCC
renal cell carcinoma
- ccRCC
clear cell renal cell carcinoma
- TCGA
The Cancer Genome Atlas
- LASSO
Least Absolute Shrinkage and Selection Operator
- DE-miRNAs
differentially expressed miRNAs
- Pr-miRNAs
prognosis-related miRNAs
- K-M
Kaplan-Meier
- ROC
receiver operating characteristic
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PCA
Principal component analysis; OS, overall survival.
Footnotes
Authors’ Note: Jiyue Wu and Feilong Zhang contributed equally to this work and should be listed as first co-authors. Our study did not require an ethical board approval because it did not contain human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Feilong Zhang, MM  https://orcid.org/0000-0001-6781-199X
https://orcid.org/0000-0001-6781-199X
Wei Wang, MD  https://orcid.org/0000-0003-2642-3338
https://orcid.org/0000-0003-2642-3338
Supplemental Material: Supplemental material for this article is available online
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2. Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med. 2017;376(4):354–366. doi:10.1056/NEJMra1601333 [DOI] [PubMed] [Google Scholar]
- 3. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers.2017;3:17009. doi:10.1038/nrdp.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diaz-Montero CM, Rini BI, Finke JH. The immunology of renal cell carcinoma. Nat Rev Nephrol. 2020;16(12):721–735. doi:10.1038/s41581-020-0316-3 [DOI] [PubMed] [Google Scholar]
- 5. Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22(11):2089–2100. doi:10.1101/gr.131110.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi:10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 7. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. doi:10.1158/2159-8290.CD-15-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J, Yao F, Xiao Z, Sun Y, Ma L. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev. 2018;37(1):5–15. doi:10.1007/s10555-017-9712-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H, Wang Z, Ma R, Wu J, Feng J. MicroRNAs as biomarkers for the progression and prognosis of colon carcinoma. Int J Mol Med. 2018;42(4):2080–2088. doi:10.3892/ijmm.2018.3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shu X, Hildebrandt MA, Gu J, et al. MicroRNA profiling in clear cell renal cell carcinoma tissues potentially links tumorigenesis and recurrence with obesity. Br J Cancer. 2017;116(1):77–84. doi:10.1038/bjc.2016.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meng X, Liu K, Xiang Z, Yu X, Wang P, Ma Q. MiR-125b-2-3p associates with prognosis of ccRCC through promoting tumor metastasis via targeting EGR1. Am J Transl Res. 2020;12(9):5575–5585. [PMC free article] [PubMed] [Google Scholar]
- 12. He YH, Chen C, Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233(6):4458–4465. doi:10.1002/jcp.26347 [DOI] [PubMed] [Google Scholar]
- 13. Schaefer A, Stephan C, Busch J, Yousef GM, Jung K. Diagnostic, prognostic and therapeutic implications of microRNAs in urologic tumors. Nat Rev Urol. 2010;7(5):286–297. doi:10.1038/nrurol.2010.45 [DOI] [PubMed] [Google Scholar]
- 14. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi:10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 16. Goeman JJ. L1 penalized estimation in the cox proportional hazards model. Biom J. 2010;52(1):70–84. doi:10.1002/bimj.200900028 [DOI] [PubMed] [Google Scholar]
- 17. Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi:10.1111/j.0006-341x.2000.00337.x [DOI] [PubMed] [Google Scholar]
- 18. Eng KH, Schiller E, Morrell K. On representing the prognostic value of continuous gene expression biomarkers with the restricted mean survival curve. Oncotarget. 2015;6(34):36308–36318. doi:10.18632/oncotarget.6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng Z, Duan F, Yin J, Feng Y, Yang Z, Shang J. Prognostic values of microRNA-130 family expression in patients with cancer: a meta-analysis and database test. J Transl Med. 2019;17(1):347. doi:10.1186/s12967-019-2093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Meng W, Yue P, Li X. M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J Exp Clin Cancer Res. 2020;39(1):134. doi:10.1186/s13046-020-01626-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Liao Y, Wang C, Yang Z, et al. Dysregulated Sp1/miR-130b-3p/HOXA5 axis contributes to tumor angiogenesis and progression of hepatocellular carcinoma. Theranostics. 2020;10(12):5209–5224. doi:10.7150/thno.43640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shui Y, Yu X, Duan R, et al. miR-130b-3p inhibits cell invasion and migration by targeting the notch ligand delta-like 1 in breast carcinoma. Gene. 2017;609:80–87. doi:10.1016/j.gene.2017.01.036 [DOI] [PubMed] [Google Scholar]
- 24. Huang S, Xue P, Han X, et al. Exosomal miR-130b-3p targets SIK1 to inhibit medulloblastoma tumorigenesis. Cell Death Dis. 2020;11(6):408. doi:10.1038/s41419-020-2621-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Z, Tong D, Han C, et al. Blockade of miR-3614 maturation by IGF2BP3 increases TRIM25 expression and promotes breast cancer cell proliferation. EBioMedicine. 2019;41:357–369. doi:10.1016/j.ebiom.2018.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hong YG, Xin C, Zheng H, et al. miR-365a-3p regulates ADAM10-JAK-STAT signaling to suppress the growth and metastasis of colorectal cancer cells. J Cancer. 2020;11(12):3634–3644. doi:10.7150/jca.42731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yin L, Xiao X, Georgikou C, et al. MicroRNA-365a-3p inhibits c-Rel-mediated NF-κB signaling and the progression of pancreatic cancer. Cancer Lett. 2019;452:203–212. doi:10.1016/j.canlet.2019.03.025 [DOI] [PubMed] [Google Scholar]
- 28. Wang Y, Zhang S, Bao H, et al. MicroRNA-365 promotes lung carcinogenesis by downregulating the USP33/SLIT2/ROBO1 signalling pathway. Cancer Cell Int. 2018;18:64. doi:10.1186/s12935-018-0563-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geng J, Liu Y, Jin Y, et al. MicroRNA-365a-3p promotes tumor growth and metastasis in laryngeal squamous cell carcinoma. Oncol Rep. 2016;35(4):2017–2026. doi:10.3892/or.2016.4617 [DOI] [PubMed] [Google Scholar]
- 30. Gao M, Liu L, Zhang D, Yang Y, Chang Z. Long non-coding RNA NEAT1 serves as sponge for miR-365a-3p to promote gastric cancer progression via regulating ABCC4. Onco Targets Ther. 2020;13:3977–3985. doi:10.2147/OTT.S245557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo Y, Yao B, Zhu Q, et al. MicroRNA-301b-3p contributes to tumour growth of human hepatocellular carcinoma by repressing vestigial like family member 4. J Cell Mol Med. 2019;23(8):5037–5047. doi:10.1111/jcmm.14361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fan H, Jin X, Liao C, Qiao L, Zhao W. MicroRNA-301b-3p accelerates the growth of gastric cancer cells by targeting zinc finger and BTB domain containing 4. Pathol Res Pract. 2019;215(11):152667. doi:10.1016/j.prp.2019.152667 [DOI] [PubMed] [Google Scholar]
- 33. Zheng H, Bai L. Hypoxia induced microRNA-301b-3p overexpression promotes proliferation, migration and invasion of prostate cancer cells by targeting LRP1B. Exp Mol Pathol. 2019;111:104301. doi:10.1016/j.yexmp.2019.104301 [DOI] [PubMed] [Google Scholar]
- 34. Jin H, Yu M, Lin Y, et al. MiR-502-3P suppresses cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting SET. Onco Targets Ther. 2016;9:3281–3289. doi:10.2147/OTT.S87183 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Wang Y, Xu X, Maglic D, et al. Comprehensive molecular characterization of the hippo signaling pathway in cancer. Cell Rep. 2018;25(5):1304–1317.e5. doi:10.1016/j.celrep.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moon S, Yeon Park S, Woo Park H. Regulation of the hippo pathway in cancer biology. Cell Mol Life Sci. 2018;75(13):2303–2319. doi:10.1007/s00018-018-2804-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu MZ, Yao TJ, Lee NP, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115(19):4576–4585. doi:10.1002/cncr.24495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kong D, Zhao Y, Men T, Teng CB. Hippo signaling pathway in liver and pancreas: the potential drug target for tumor therapy. J Drug Target. 2015;23(2):125–133. doi:10.3109/1061186X.2014.983522 [DOI] [PubMed] [Google Scholar]
- 39. Murillo-Garzon V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14(11):683–696. doi:10.1038/nrurol.2017.144 [DOI] [PubMed] [Google Scholar]
- 40. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473. doi:10.1038/onc.2016.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhat AA, Uppada S, Achkar IW, et al. Tight junction proteins and signaling pathways in cancer and inflammation: a functional crosstalk. Front Physiol. 2018;9:1942. doi:10.3389/fphys.2018.01942 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-2-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-3-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-4-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-5-tct-10.1177_15330338211027923 for A Novel miRNA-Based Model Can Predict the Prognosis of Clear Cell Renal Cell Carcinoma by Jiyue Wu, Feilong Zhang, Jiandong Zhang, Zejia Sun, Changzhen Hao, Huawei Cao and Wei Wang in Technology in Cancer Research & Treatment