Abstract
The fall of 2020 brought several new variants of SARS-CoV-2 circulating across the globe, and the steadily increasing COVID-19 cases are responsible for the emergence of these variants. All the SARS-CoV-2 variants reported to date have multiple mutations in the spike (S) protein, specifically in the receptor-binding domain (RBD). Here, we employed an integrated computational approach involving structure and sequence based predictions to study the effect of naturally occurring variations in the S-RBD on its stability and ACE2 binding affinity. The hotspot stabilizing residue mutations N501I, N501Y, Q493L, Q493H and K417R, strengthen the RBD-ACE2 complex by modulating the interaction statistics at the interface. Thus, we report here some critical mutations that could increase the binding affinity of the SARS-CoV-2 RBD with ACE2, increasing the viral infectivity and pathogenicity. Understanding the effect of these mutations will help in developing potential vaccines and therapeutics.
Keywords: SARS-CoV-2, Mutation, Spike protein, Receptor binding domain, RBD-ACE2 interactions, COVID-19, Hotspot residues, SARS-CoV-2 variants
Graphical abstract
1. Introduction
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the global pandemic causing the coronavirus disease 2019 (COVID-19) is a massive threat to public health and the economy. It has already infected a large number of people globally, causing common cold-like symptoms to severe complications such as shortness of breath, chest pain and loss of speech or movement, eventually leading to acute respiratory distress syndrome and acute respiratory failure (Clinical characteristics, 2021; Mokhtari et al., 2020). Despite the approval of various vaccines for emergency use there is a constant rise in the covid-19 cases worldwide.
Since the start of the COVID-19 pandemic, genomic variations are observed in SARS-CoV-2 across different geographical regions (Mercatelli and Giorgi, 2020). Förster et al. in their study with genomic data from the Global Initiative on Sharing All Influenza Data (GISAID) database, identified three main variants, which they named, A, B and C. The A and C type was common in Europe and America, whereas type B was specific to East Asia (Forster et al., 2020). Later, six major clades (basal, D614G, L84S, L3606F, D448del and G392D) and 14 subclades were identified by analyzing genome variants of SARS-CoV-2 from all over the world (Koyama et al., 2020). Among all, the most common clade identified was the D614G variant in the spike (S) protein. A group has described 11 major mutation events which define five major clades, namely, G614, S84, V251, I378 and D392 from SARS-CoV-2 clinical samples. They also report several non-synonymous mutations in the S protein, which may have functional consequences (Guan et al., 2020). The first SARS-CoV-2 variant of concern (VOC) was reported in December 2020 from the United Kingdom (UK) (O |-CoV-2 Variants, 2021). This SARS-CoV-2 B.1.1.7 lineage (a.k.a. 20I/501Y.V1 VOC, 202012/01) with multiple S protein mutations (deletion 69–70, deletion 144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) spread rapidly across South East England and London (Ecdc, 2020). This lineage was associated with higher transmission rates and increased mortality (Davies et al., 2021a, 2021b). Soon after, the B.1.351 lineage (a.k.a. 20H/501Y.V2) was identified in Nelson Mandela Bay, South Africa, with multiple S protein mutations, including K417N, E484K and N501Y (Science Brief). Another variant, P.1 (a.k.a. 20 J/501Y.V3) emerged in Brazil, contains mutations K417T, E484K and N501Y in the RBD with evidence to affect transmissibility and antigenic profile (Science Brief; Spike E484K mutation in t, 2020). All these SARS-CoV-2 VOC carry a common mutation N501Y in the receptor binding domain (RBD) of the S protein. More recently, another variant reported from India has been added in the list of VOC by the WHO. The lineage is B.1.617.2 (Delta) with mutations, L452R, T478K, D614G and P681R in the S protein (Tracking-CoV-2 varia, 2021). A variant named Delta plus or AY.1 has been identified in India with the point mutation K417N. This variant is reported to be the mutant of the Delta strain (B.1.617.2) of SARS-CoV-2 (New “Delta Plus” variant). Mutations occurring in the S protein are being continuously reported; however, their impact on the virus's virulence, transmission, and antigenicity remains elusive.
The SARS-CoV-2 S protein is the major antigenic determinant responsible for the virulence and infectivity of the virus (Walls et al., 2020). This homotrimeric protein mediates the entry of the virus particles into the host cell via the angiotensin converting enzyme 2 (ACE2) receptor (Hoffmann et al., 2020; Verma et al., 2020). The RBD in the S1 subunit of S protein has a receptor binding motif (RBM), which specifically interacts with the ACE2 receptor and initiates the viral entry. Structural analysis has revealed the atomic details of interactions between the RBM and ACE2 (Shang et al., 2020). The interface involves interaction between 17 and 20 amino acids from SARS-CoV-2 RBM and ACE2, respectively (Lan et al., 2020a). The two virus binding hotspots (hotspot-31 and hotspot-353) previously identified in the SARS-CoV-ACE2 interface are also characterized for the SARS-CoV-2-ACE2 interface. These two hotspots were first determined while understanding the structural basis of the major species barrier between civets and human SARS-CoV infections (Li, 2008; Wu et al., 2012). The hotspot-31 is stabilized by Q493 at SARS-CoV-2 RBM, whereas the residues Y505, G502, G496 and N501 supports the hotspot-353 at the SARS-CoV-2-ACE2 interface (Shang et al., 2020; Verma and Subbarao, 2021). The only salt bridge between the SARS-CoV-2 and ACE2 is formed by K417 and D30 of ACE2. These hotspot-stabilizing residues contribute to the high ACE2 binding affinity of SARS-CoV-2 (Shah et al., 2020). Various naturally occurring viral variants have been reported with the mutations at the amino acids directly involved in the ACE2 interaction (Li et al., 2020). However, the effect of such mutations on the protein function and structure remain largely unclear. In this study, we have employed a computational approach to investigate the effects of naturally occurring viral variations in the RBD, specifically in the RBM, on its stability and binding affinity with the ACE2 receptor. We further identified the stabilizing effects of RBM variants with mutations in the residues supporting the hotspot region at the SARS-CoV-2-ACE2 interface.
2. Methodology
2.1. Data collection and structure preparation
We collected the information of the viral variations from 2019 Novel Coronavirus Resource (2019nCoVR) (nCo- 2019 Novel Corona, 2019). This database integrates the information of SARS-CoV-2 strains found worldwide (Zhao et al., 2020; Song et al., 2020). We collected the data of all the S protein viral variants for which the mutation sites were located in the RBD. As of December 30, 2020, we gathered 289 viral variations in the RBD, of which 25 were from the 17 key amino acids critical for ACE2 interaction.
The X-ray crystal structure of SARS-CoV-2 RBD in complex with ACE2 was downloaded from the RCSB Protein data bank with PDB ID 6M0J (B - 6M0J: Crystal, 2021; Lan et al., 2020b). The structure was further prepared by the addition of missing amino acid side chains, hydrogen atoms and missing loop regions. Finally, the structure was minimized with the OPLS3e force field. All these steps were carried out using the protein preparation wizard of the Schrodinger suit (v2019.1) (Protein Preparation Wizar, 2020). The coordinates of the RBD were saved in a separate file for the stability predictions. The RBD-ACE2 complex was used to determine their binding affinity upon mutations. All the mutations were introduced in the RBD structure and RBD-ACE2 complex individually with the maestro interface of the Schrodinger suit (v2019.1) and the structures were saved for further calculations (Maestro | Schrödinger).
2.2. Stability and binding affinity predictions
We predicted the effect of each variation (mutation) on the stability of RBD using the site directed mutator (SDM2) server (SDM, 2021). SDM is a knowledge based approach that uses the environment-specific substitution tables (ESSTs) to calculate the stability difference score (pseudo ΔΔG) between the wild type (WT) and the mutant protein structures (Pandurangan et al., 2017). The minimized RBD PDB structure and the list of mutations (289 point mutations) were given as the input in the webserver. A negative ΔΔG value corresponds to mutations predicted to be destabilizing, and a positive value suggests that the mutation can stabilize the protein.
For further binding affinity predictions, we selected only key 25 mutations corresponding to the amino acids directly involved in ACE2 interaction. We calculated the binding free energy of all these mutant RBD-ACE2 complexes and the WT complex using the Molecular Mechanics/Generalized Born Surface Area (MMGBSA) approach. MMGBSA calculations were carried out using the HawkDock server (HawkDock Server, 2021). It is a webserver that employs MMGBSA to predict the binding free energy and decompose the free energy contribution to the binding free energy of the protein-protein complex per residue (Hou et al., 2011; Sun et al., 2014; Chen et al., 2016). Hence, we also analyzed the contribution of mutant residues to the total binding energy of the complex.
2.3. Sequence based approach to analyze mutation effect on protein function
After carrying out the structural analysis to understand protein stability and binding affinity, we performed sequence based analysis to predict the functional effect of these mutations. Single amino acid substitutions are the most frequent type of mutations observed in the SARS-CoV and SARS-CoV-2 S protein affecting the protein function. We used the PredictSNP server to predict the effect of RBD single amino acid variants on its function (PredictSNP, 2021). PredictSNP is a consensus classifier combining the datasets from six different tools, MAPP, PhD-SNP, PolyPhen-1, PolyPhen-2, SIFT and SNAP (Bendl et al., 2014). This consensus classifier gives significantly improved and accurate predictions over the individual tools. The amino acid sequence of the SARS-CoV-2 S protein (P0DTC2) was downloaded from UniProt (S - Spike glycoprotein pr, 2019). The S RBD protein sequence was submitted to the PredictSNP server in FASTA format. The analysis was carried out for variations that increase the binding affinity between the RBD and ACE2, as explained in the previous section.
2.4. Interaction analysis and molecular dynamics simulation
We analyzed the RBD-ACE2 complex to understand the change in interactions between the RBM and ACE2 upon mutations stabilizing the complex. The EMBL-EBI PDBsum was used to generate the interaction plot of the complexes (Bsum home page. http://, 2021). We used PyMol to visualize the complexes and generate figures (PyL | pymol. org). Further, we carried out molecular dynamics (MD) simulation to confirm the structural impact of the predicted stabilizing mutants. MD simulation of the selected RBD mutants was performed using GROMACS v2019.4 (Berendsen et al., 1995). Total 12 different simulation systems, including the RBD mutants in apo-form (receptor unbound) and the receptor bound form were subjected to 100 nanoseconds (ns) simulation. We parameterized the protein with AMBER99SB force field and defined a cubic box around it with 1.0 nm distance from the box edges (Hornak et al., 2006). The TIP3P water model was used to solvate the system, and a neutral system was achieved by replacing the required water molecules with Na+/Cl− ions (Mahoney and Jorgensen, 2000). Subsequently, 50,000 steps of energy minimization were carried out for each system using steepest descent minimization algorithm. The minimized systems were equilibrated in two steps, NVT (constant number of particles, volume and temperature) and NPT (constant number of particles, pressure, and temperature) for 100 ps. After the successful equilibration of the systems, the final MD run was carried out for 100 ns with a time step of 2 fs (fs). The MD trajectories were analyzed using Gromacs analysis tools, and Xmgrace was used to create the 2D plots (PJ, 2005).
3. Results and discussion
3.1. Effects of RBD viral variations on its stability and ACE2 binding affinity
Based on the SARS-CoV-2 genome sequences deposited in 2019nCoVR, 2961 mutation sites are identified in S protein as of December 30, 2020. We downloaded the information of all the S protein mutations occurring specifically in the RBD region. A total of 289 mutations were examined to predict their effect on the structural stability of S-RBD. The SDM2 server predicted the ΔΔG values mapped to each mutation of the RBD. The ΔΔG <0 and ΔΔG >0 corresponds to the reduced and increased protein stability, respectively (Fig. 1 b). Among the 289 individual mutations only 37% were predicted to increase the stability of the RBD, whereas 63% decreased the protein stability (Fig. 1a). Further, we carried out a detailed analysis of the mutations occurring at the residue positions directly involved in interaction with ACE2. Point mutations occurring in the critical interacting residues in the RBM could affect the binding affinity between ACE2 and RBD. Therefore, we predicted the binding free energy of these 25 RBD mutants and ACE2 complex through the MMGBSA approach (Fig. 2 ). Table 1 summarizes the effects of these mutations on RBD stability and ACE2 binding affinity.
Fig. 1.
Effect of viral variations on RBD stability. (a) Contribution of 289 RBD variations on protein stability. (b) The ΔΔG values of selected RBD variations and their stability effect.
Fig. 2.
Binding free energy of the RBD-ACE2 complex. (a) The binding free energy of the RBD-ACE2 complexes with variations in the RBD. (b) Effect of RBD variation on ACE2 binding affinity.
Table 1.
Effect of selected viral variations on RBD stability and ACE2 binding.
| Viral variation | ΔΔG | RBD Stability | ACE2-RBD Binding energy (kcal/mol) | ACE2 binding affinity |
|---|---|---|---|---|
| N501Y | 0.53 | Increased | −67.86 | High |
| Y453F | −0.5 | Reduced | −60.9 | High |
| G446D | −4.12 | Reduced | −55.95 | Low |
| G446V | −2.64 | Reduced | −60.16 | High |
| L455F | −0.09 | Reduced | −61.61 | High |
| A475V | −0.12 | Reduced | −61.19 | High |
| K417N | −1.34 | Reduced | −51.25 | High |
| F486I | 0.31 | Increased | −58.55 | Low |
| F486L | 0.38 | Increased | −58.09 | Low |
| N501S | 0.17 | Increased | −61.04 | High |
| N501I | 0.8 | Increased | −61.69 | High |
| N501T | −0.01 | Reduced | −62.15 | High |
| Y505C | −0.06 | Reduced | −52.76 | Low |
| Q493L | 0.66 | Increased | −60.06 | High |
| Q493K | −0.04 | Reduced | −56.58 | Low |
| G446S | −4.11 | Reduced | −60.21 | High |
| T500S | −0.15 | Reduced | −55.72 | Low |
| Y505H | 0.09 | Increased | −57.21 | Low |
| K417R | 0.3 | Increased | −62.04 | High |
| Y495F | −0.12 | Reduced | −59.86 | High |
| Y449F | 0.02 | Increased | −55.3 | Low |
| F456L | −0.2 | Reduced | −57.33 | Low |
| F456Y | 0.26 | Increased | −59.55 | Low |
| Y489H | −0.78 | Reduced | −57.04 | Low |
| Q493H | 0.29 | Increased | −60.28 | high |
The mutation at position G446 to G446D, G446S and G446V shows the maximum destabilizing effect on the RBD with ΔΔG values, −4.12, −4.11 and −2.64 kcal/mol, respectively (Fig. 1b). This observation signifies the importance of G446 at the loop region to provide flexibility in the RBM. Additionally, the mutation G446D compared to G446S and G446V shows a noteworthy effect on the binding affinity between RBD and ACE2. The charged amino acid D at position 446 contributes negatively with the residue binding free energy of 2.13 kcal/mol to the total binding free energy of RBD-ACE2 complex (Fig. S1). The RBD-ACE2 WT complex has a total binding free energy of −59.66 kcal/mol. Hence, the binding free energy of the WT RBD-ACE2 complex increases from −59.66 kcal/mol to −55.95 kcal/mol for the mutant G446D RBD-ACE2 complex (Fig. 2). Since, the amino acids S and V are neutral as G; therefore, this could be a possible reason for no significant change in the binding free energy of the RBD-ACE2 complex for these mutants. The mutant K417N has the next minimum ΔΔG value, i.e. −1.34 kcal/mol, corresponding to the reduced structural stability. A single salt bridge is formed at the ACE2-RBD interface by residues D30 and K417 of ACE2 and RBD, respectively. Asparagine being a polar residue fails to interact with D30 of ACE2, resulting in the disruption of the salt bridge and also a decrease in the non-bonded contacts. Consistently, the variant K417N has the highest binding free energy (−51.25 kcal/mol) with a difference of −8.41 kcal/mol from the WT (−59.66 kcal/mol), resulting in the lowest binding affinity between the ACE2 and RBD complex (Fig. 2). In contrast, R at this position has a positive ΔΔG value of 0.3 kcal/mol, signifying stabilizing effect on the protein structure. The K417R mutant of RBD has a much lower binding free energy for ACE2 interaction (−62.04 kcal/mol), and an increased number of contacts between the complex result in high binding affinity between the two proteins.
The point mutations L455F, A475V and T500S have negligible destabilizing effect on the RBD structure with the ΔΔG values −0.09, −0.17 and −0.15 kcal/mol, respectively (Fig. 1b). However, T when replaced with S at position 500 reduces the binding affinity between RBD and ACE2, whereas the variants L455F and A475V increase their binding affinity (Fig. 2). Similarly, when Y, a polar amino acid at positions 453 and 495 is replaced with F a non-polar amino acid may slightly destabilize the RBD but increases the binding affinity of RBD towards ACE2. The mutation at position 486 from F to I or L have ΔΔG values 0.31 and 0.38 kcal/mol, respectively; corresponding to increased RBD stability, while decreasing the binding affinity. The residue F at position 456 when replaced with less non-polar L or polar Y results in decreased RBD-ACE2 affinity (Fig. 2). Hence, these observations suggest that increase in the non-polar content of the RBD structure at specific residue positions may increase its affinity for ACE2. The mutation of the aromatic residue Y at the position 489 to H and 505 to H or C leads to reduced RBD-ACE2 binding affinity. The naturally occurring variants with change at residue position N501 to N501Y, N501S, N501I and N501T show neutral to stabilizing effect on the RBD structure. Notably, all these mutants also have lower ACE2-RBD binding free energy than the WT complex. The mutation N501Y has been found in several VOC strains of SARS-CoV-2 such as B.1.1.7, B.1.351 and P.1. The point mutation N501Y has a stabilizing effect on the RBD with ΔΔG value 0.53 kcal/mol and result in the highest binding affinity between RBD and ACE2 (−67.86 kcal/mol). However, the other mutations N501T, N501I and N501S have the binding free energy −62.15, −61.69 and −61.04 kcal/mol, respectively. The variation N501I also has the maximum ΔΔG value 0.8 kcal/mol with highest stabilizing effect on the RBD. Similarly, the point mutations Q493L has ΔΔG 0.66 kcal/mol and Q493H has the value 0.29 kcal/mol resulting in an increase in structural stability of the RBD. Contrarily, the variants N501T and Q493K do not have significant effect on the protein structure but change the binding free energy of the complex, with N501T increasing the binding affinity and Q493K decreasing the affinity of the complex. As a result, point mutations in the S protein RBM can have a major impact on ACE2 binding. We also predicted the binding free energy for the two VOC with double mutation in their RBD. The variant B.1.617 with RBD mutations E484Q and L452R in complex with ACE2 has the total binding free energy of −64.89 kcal/mol. Whereas, the variant B.1.1.7 (N501Y and E484K) RBD-ACE2 complex has the binding free energy of −67.76 kcal/mol. Hence, both these variants have a lower binding free energy than the WT complex suggesting that these mutations in the RBD increase the binding affinity between the RBD and ACE2.
3.2. Consensus prediction of RBD mutation effect on protein function
Among the 25 point mutations in the RBM, 13 were reported to increase the binding affinity between RBD and ACE2. The variants with more negative binding free energy (high binding affinity) than the WT complex are N501Y, N501T, K417R, N501I, L455F, A475V, N501S, Y453F, Q493H, G446S, G446V, Q493L and Y495F. Hence, we further analyzed if these mutants alter the S protein function by performing sequence based analysis. The consensus classifier PredictSNP was used to predict the functional effect of these point mutations. The web server PredictSNP integrates other six tools which classify the effect of mutations as neutral or deleterious. In accordance with the consensus prediction, a mutation was classified as deleterious only when it is predicted as deleterious by more than three tools. We generated these predictions for the 13 mutations upon which the binding free energy of the RBD-ACE2 complex is decreased, ultimately increasing the binding affinity. The effect of mutations predicted by all the six tools as well as the consensus classifier are shown in Table 2 . Only two point mutations, Y495F and G446V, were predicted to be deleterious by the consensus classifier. The mutation Y495F and G446V also reduced the RBD stability according to their ΔΔG values predicted in the previous section. All other 11 mutations (N501I, N501S, N501T, N501Y, Q493L, Q493H, A475V, L455F, G446S and K417R) were predicted to have a neutral effect on the protein function. Therefore, the function of the RBD will remain unaffected upon these point mutations. Finally, we identified a set of mutations with increased RBD stability, higher RBD-ACE2 binding affinity than WT, and a neutral effect on protein function. Interestingly, all these characteristics were observed for mutants at the residue positions involved in the hotspot stabilization at the RBD-ACE2 interface. The S protein with RBD variants N501I, N501S, N501Y, Q493L, Q493H and K417R are predicted to increase RBD stability, ACE2 binding affinity and have no effect on its function. The residue N501 supports the hotspot-353 and Q493 stabilizes the hotspot-31 at the SARS-CoV-2 RBD-ACE2 interface. The residue K at position 417 makes a salt bridge essential for both proteins to make a stable complex. Hence, the SARS-CoV-2 viral variants with these mutations may have increased affinity for ACE2, affecting the viral infectivity and transmission.
Table 2.
Effect of SARS-CoV-2 RBD variants (with lower RBD-ACE2 binding free energy than WT) on protein function.
| Viral variant | PredictSNP | MAPP | PhD-SNP | PolyPhen-1 | PolyPhen-2 | SIFT | SNAP |
|---|---|---|---|---|---|---|---|
| N501I | Neutral | Deleterious | Neutral | Neutral | Neutral | Neutral | Deleterious |
| N501S | Neutral | Neutral | Neutral | Deleterious | Neutral | Neutral | Neutral |
| N501T | Neutral | Neutral | Neutral | Neutral | Neutral | Neutral | Neutral |
| N501Y | Neutral | Neutral | Neutral | Neutral | Neutral | Deleterious | Deleterious |
| Y495F | Deleterious | Deleterious | Neutral | Neutral | Deleterious | Deleterious | Deleterious |
| Q493L | Neutral | Neutral | Neutral | Neutral | Neutral | Neutral | Deleterious |
| Q493H | Neutral | Neutral | Neutral | Neutral | Neutral | Neutral | Deleterious |
| A475V | Neutral | Deleterious | Neutral | Neutral | Neutral | Deleterious | Deleterious |
| L455F | Neutral | Neutral | Neutral | Neutral | Neutral | Neutral | Deleterious |
| Y453F | Neutral | Deleterious | Neutral | Neutral | Neutral | Neutral | Deleterious |
| G446S | Neutral | Neutral | Neutral | Neutral | Deleterious | Neutral | Deleterious |
| G446V | Deleterious | Deleterious | Deleterious | Neutral | Deleterious | Neutral | Deleterious |
| K417R | Neutral | Deleterious | Neutral | Neutral | Neutral | Deleterious | Neutral |
3.3. Comparative analysis of RBD-ACE2 interface of WT and mutants
The overall statistics of interface between the RBD of SARS-CoV-2 and ACE2 is similar to that of the SARS-CoV RBD and ACE2. However, RBM sequence variations have resulted in enhanced receptor binding in SARS-CoV-2. Specific hotspot regions stabilize molecular interactions between the S-RBD and the receptor ACE2 at the interface. Variations occurring at these interface regions may alter the binding affinity of RBD for the ACE2 receptor. Our findings report naturally occurring mutations in the hotspot region, increasing the protein stability and ACE2 binding affinity. To understand the change in interface statistics between the WT RBD-ACE2 complex and the reported mutants (N501I, N501Y, Q493L, Q493H and K417R) we analyzed detailed molecular interactions of these complexes (Fig. 3 a and Fig. 3b). The number of interface residues is 20 and 17 for ACE2 and RBD, respectively, which remains unchanged upon individually occurring point mutations N501I, N501Y, Q493L, Q493H, K417R in the RBM (Table S1). In the WT complex, the hotspot-31 (K31 of ACE2) is stabilized by non-bonded interactions of Q493 of the RBD. On the other hand, hotspot-353 (K353 of ACE2) is supported by G502 and G496 forming hydrogen bonds as well as Y505 and N501 making non-bonded contacts. Another critical electrostatic interaction occurs between the oppositely charged residues D30 of ACE2 and K417 of RBD, making a salt bridge along with a hydrogen bond at the interface. The single amino acid variation N501I and N501Y leads to an increased number of contacts at the hotspot-353. However, the total interface area remains the same, keeping all other interactions identical. Isoleucine at position 501 increases the number of contacts with K353 and Y41, supporting the hotspot-353. Similarly, Y at this position makes a large number of atom to atom contacts with K353 and Y41. The amino acid N, when replaced by Y at 501, creates a side chain-side chain hydrogen bond with K353. Hence, increasing the intermolecular interactions between the RBD and ACE2.
Fig. 3.
RBD-ACE2 interface interactions. (a) Dimplot showing the interactions between the RBD (WT and mutant N501I, N501Y) and ACE2. (b) Dimplot showing the interactions between the RBD (mutant Q493L, Q493H and K417R) and ACE2.
Glutamine at position 493 is the only residue interacting with K31 and E35 giving stability to the central region of the RBD-ACE2 interface and hotspot-31. The mutant Q493L does not make any considerable change at the interface; rather, it interacts with H34 of ACE2 instead of E35. Although, if Q is replaced by H, at position 493, the interaction network near hotspot-31 becomes much stronger with increased molecular contacts. The ionizable side chain of the H493 makes a salt bridge of distance 2.94 Å with E35 and numerous non-bonded contacts with K31 and E35 of ACE2. Thus, this mutation favours stable ACE2 and RBD interactions. The ACE2 and RBD are supported by a single salt bridge between D30 and K417 at the interface. Amino acid variation at this RBD position may disrupt the salt bridge leading to decreased ACE2 binding affinity. However, the distance of the salt bridge between D30 and K417 is reduced from 2.90 Å to 1.25 Å, when K at 417 is replaced with R. There is also an increase in the number of atom to atom contacts between D30 and R417 (11) in comparison to D30 and K417 (3).
To further support our results, we determined the contribution of each residue to the total binding free energy of the ACE2 and RBD complex. The residue Y at position 501 contributes seven times more to the absolute binding free energy than N at this position (Fig. 4 a). An increase in both the electrostatic and Van der Waals energy results in the total residue energy of −8.13 kcal/mol compared to −1.4 kcal/mol for the WT. Similarly, I at 501 also contributes positively to the total binding free energy of the ACE2-RBD complex. The residue K417, when replaced with R, do not show any significant change in the residue energy contribution (Fig. 4b). However, there was a slight increase in the Van der Waals energy term to the total residue energy contribution. At position 493, the WT residue Q contributes −2.36 kcal/mol to the total binding free energy. In contrast, L at the same position has a total contribution of −2.71 kcal/mol due to less positive polar solvation energy. Moreover, H493 has a positive contribution of Van der Waals and electrostatic energy terms due to increased polar contacts at the interface. The interactions at the hotspot regions of the ACE2 and RBD complex in WT and mutants are shown in Fig. 4. Clearly, these mutations have a positive effect on the RBD and ACE2 interaction.
Fig. 4.
Effect of residue mutations on RBD-ACE2 interactions. Per residue energy decomposition for (a) N501, N501Y and N501I, (b) K417 and K417R, (c) Q493, Q493L and Q493H. Structural representation of interactions at RBD-ACE2 interface upon mutation. (d) N501Y, (e) WT, (f) Q493L, (g) N501I, (h) K417R and (i) Q493H.
3.4. Stability analysis and essential dynamics
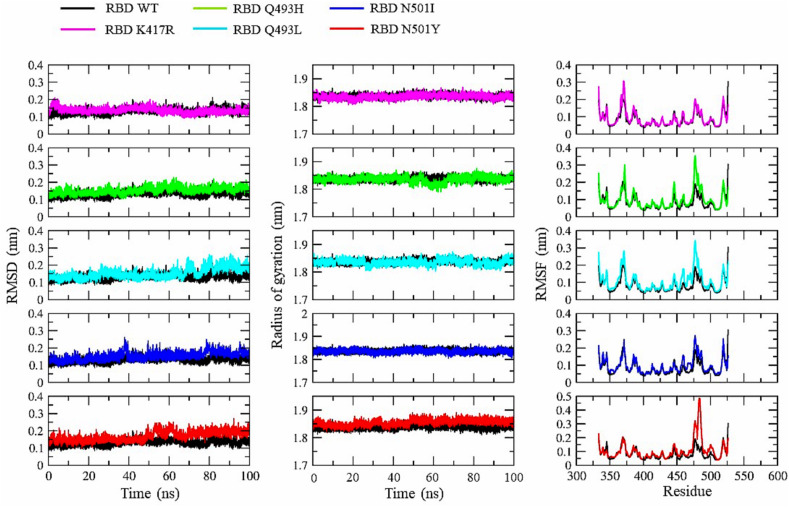
We carried out MD simulation of the RBD and the RBD-ACE2 complexes of WT and mutants to understand the time evolution of these molecular structures. All the 12 systems, RBD WT, RBD with single amino acid substitutions viz, K417R, Q493L, Q493H, N501I, and N501Y, RBD + ACE2 WT, RBD K417R + ACE2, RBD Q493L + ACE2, RBD Q493H + ACE2, RBD N501I + ACE2, RBD N501Y + ACE2, were simulated separately for 100 ns. We evaluated the effect of all these single amino acid substitutions on RBD structure with and without ACE2 to support our findings from the previous section. We determined the root mean square deviation (RMSD) throughout the simulation to investigate the RBD stability (Fig. 5 ). The RMSD of WT RBD and the mutants K417R, Q493H, N501I, N501Y lies relatively in the same range and converges at the end with no significant deviations during the simulation. However, a slight fluctuation was observed in the RBD Q493L during the last 30ns of the simulation period.
Fig. 5.
The Root Mean Square Deviation (RMSD), Radius of gyration (Rg) and Root Mean Square Fluctuation (RMSF) for the RBD WT and mutant structures.
Further, to observe the structural compactness of the RBD upon point mutations, we calculated the radius of gyration (Rg) of the protein structure. The Rg values for all the structures was between 1.8 and 1.9 nm suggesting that mutations have no effect on protein folding and compactness (Fig. 5). Finally, we calculated root mean square fluctuation (RMSF) of the Cα atoms of all the RBD residues to explore the flexibility of WT and mutant structures. Two major fluctuation peaks were observed in the WT RBD, one between the residues 475–487 corresponding to the loop region in the RBM and another in the core domain spanning the residues 365–374. Similar peaks were observed for all the RBD mutants with higher fluctuations for residues 475–487 at the loop region in the RBM. The overall flexibility of the RBD in both WT and mutants were comparable, and hence, no major fluctuation was observed for the mutated residues (Fig. 5).
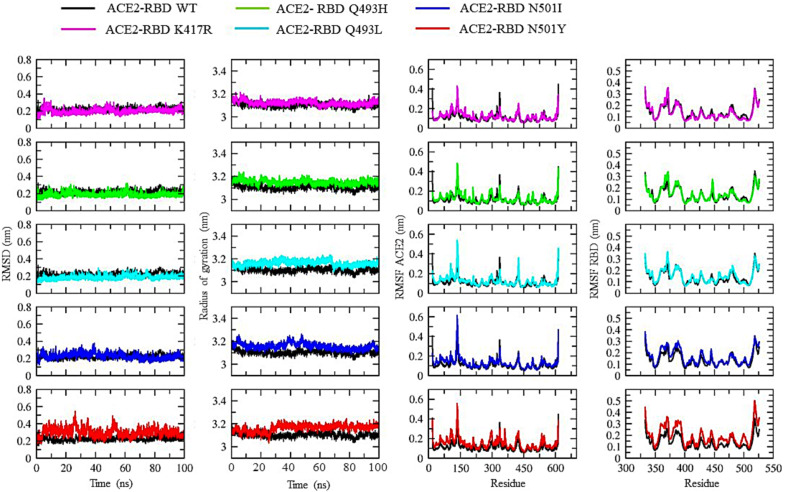
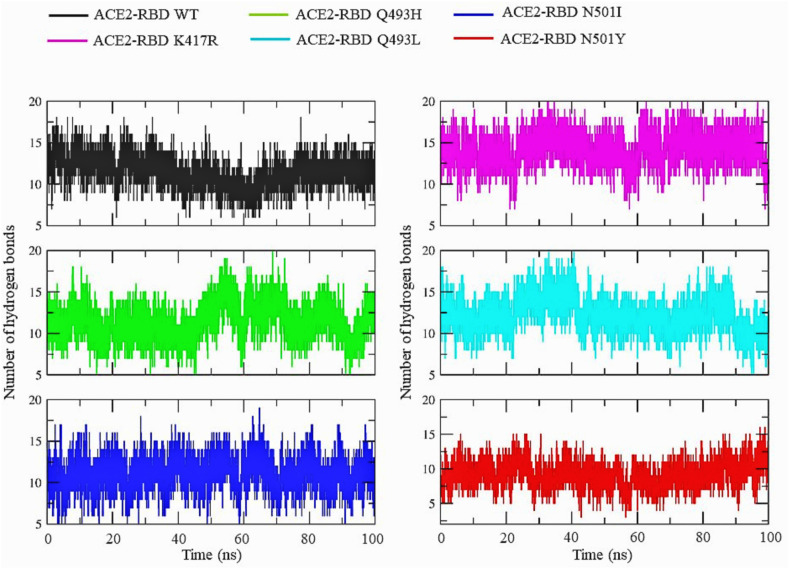
We further evaluated all the parameters for the RBD-ACE2 WT and mutant complexes (Fig. 6 ). The RMSD for the WT complex and mutant complexes K417R, Q493H, Q493L and N501I was stable throughout the 100 ns simulation. For N501Y, the backbone RMSD fluctuated within a small range between 20 and 60 ns. The Rg for all the complexes was between 3.10 and 3.25 nm, indicating a similar degree of compactness irrespective of the RBD mutation. The residue fluctuations in the RBD and ACE2 when simulated together as a complex were evaluated by plotting their RMSF values. Various small fluctuation peaks were observed in the ACE2 corresponding to the flexible loop regions located between the residues 134–140, 334–339 and 414–435. There was no major fluctuation peak observed in the RBM in both WT and mutant complexes; instead, two consecutive fluctuations were observed in the flexible regions of the core domain. We also evaluated the average number of hydrogen bonds between ACE2 and the RBD throughout the simulation period (Fig. 7 ). The average number of stable hydrogen bonds in all the complexes were 10–11. However, the maximum number of hydrogen bonds were observed in the case of the K417R mutant complex. Our simulation results confirmed the stability of the RBD structure as well as the ACE2 and RBD complexes for the mutations K417R, Q493H, Q493L, N501I and N501Y occurring in the SARS-CoV-2 S protein variants.
Fig. 6.
The Root Mean Square Deviation (RMSD), Radius of gyration (Rg) and Root Mean Square Fluctuation (RMSF) for the WT and mutant structure of RBD-ACE2 complexes.
Fig. 7.
The average number of hydrogen bonds between RBD and ACE2 throughout the molecular dynamics simulation.
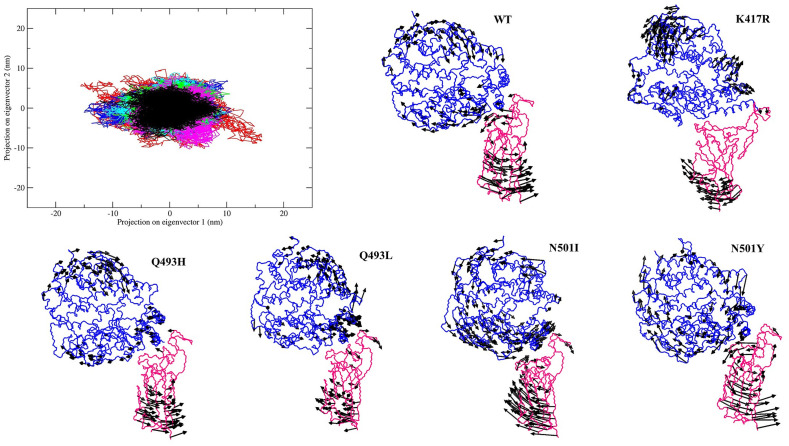
Lastly, we used the atomic trajectories of WT and mutant systems to understand the biologically relevant principal motions of these protein molecules. We performed essential dynamics analysis to get information about the collective motion of the protein complexes during the simulation. The first two eigenvectors or principal components (PCs), PC1 and PC2 were evaluated by diagonalizing the covariance matrix of the eigenvectors. The projection of eigenvector 1 and 2 (PC1 and PC2) is shown in Fig. 8 , defining the essential subspace of the protein dynamics. The motion of WT ACE2-RBD complex and mutant complexes K417R, Q493H and Q493L shows a small conformational space in comparison to N501I and N501Y mutant complexes. The confined phase space of these ACE2-RBD complexes indicate stability of the complexes. However, there is a slight increase in the conformational space of the N501I and N501Y mutant complexes along PC1 suggesting that these mutations in the RBD may increase some flexibility in the ACE2-RBD complex. Although, the protein complexes reach the equilibrated state only after few more navigations along the PC1. Hence, to understand the motion of both the proteins along the first principal movement we generated the porcupine plots for the WT and all the mutant complexes (Fig. 8). The length of the arrows shows the magnitude of the motion, and the arrow tip shows the direction. In the WT system, the motion of ACE2 was circular in a clockwise direction, whereas the RBD projection shows an anticlockwise trend. For the K417R mutant system, the motion was observed in the core domain of RBD and the upper helical region of the ACE2 not involved in the RBD interaction. This indicates restricted movements at the interface and may lead to higher ACE2-RBD stability. The magnitude of motion in both Q493H and Q493L complexes was observed to be lower than the WT. In the Q493H complex, the direction of RBM motion was towards ACE2, suggesting a higher binding affinity between them. On the other hand, N501I and N501Y complexes show higher magnitude of motion in both RBD and ACE2. However, for N501Y, the directions were identical to the WT complex, but the RBM possessed large movements towards ACE2, indicating increased binding affinity. Therefore, our simulations confirmed the stability of the mutant complexes, and the results were in agreement with our protein stability and binding affinity analysis. Thus, the mutations K417R, Q493H, Q493L, N501I and N501Y in the RBM increase the RBD stability and the ACE2 binding affinity.
Fig. 8.
Essential dynamics of the WT and mutant RBD-ACE2 complexes. The conformational landscape of WT and mutant complexes showing the projection on eigenvector 1 and eigenvector 2 (projection along principal components, PC1 and PC2). WT (Black); magenta (K417R); green (Q493H); cyan (Q493L); blue (N501I); red (N501Y). The porcupine plots of the first eigenvector for all the complexes are shown. The arrows indicate the direction and magnitude of motion of the proteins. ACE2 and RBD are shown in blue and magenta color, respectively.
4. Conclusion
The SARS-CoV-2 has flourished into numerous different variants giving rise to substantive changes in the virus behaviour. The position of the genomic variation plays a vital role as most of the mutations may have little or no effect on the virus infectivity or transmissibility, while some may develop as more contagious strains (The effects of virus vari, 2021). The maximum number of variations in the SARS-CoV-2 genome has been observed in the gene encoding S protein. Spike protein is the major antigenic determinant to elicit an immune response by producing antibodies against the virus. Hence, the likelihood of the virus mutating with variations in the S protein increases. Initially a SARS-CoV-2 variant with mutation D614G in the S protein circulated across the globe and became the predominant form of the virus with increased infectivity and transmissibility (Zhang et al., 2020a, 2020b). Another critical mutation, N501Y has been identified in several SARS-CoV-2 VOC, i.e. the B.1.1.7 (UK), B.1.351 (South African) and P.1 (Brazil). The variation N501Y occurs in the S-RBD, more specifically at the hotspot stabilizing residue of the RBM. Thus, we investigated the effect of such variations occurring in the RBD and specifically at the residue positions involved in the ACE2 binding. Out of 25 mutations in the key amino acids critical for protein interaction, 13 were predicted to increase the affinity for ACE2. We predicted that the mutation N501Y might increase the RBD stability and its binding affinity for ACE2. The aromatic amino acid Y at position 501 contributes by increasing the electrostatic energy at the interface and stabilizing hotspot-353 of ACE2. Moreover, we observed that point mutations in the RBM could modulate the stability and binding affinity between the RBD and ACE2. However, increased RBD stability, high ACE2 binding affinity and no deleterious effect on protein function were observed for mutations N501I, N501S, N501Y, Q493L, Q493H and K417R. Since the residues N501, Q493 and K417 are critical for ACE2 interaction, any change at these positions will directly affect the ACE2 binding. Hence, these hotspot stabilizing residues at the RBM are an attractive target for therapeutic development against the novel coronavirus. Moreover, there are two major limitations in our study that could be addressed in future research. First, the study and results only focuses on the single point mutations occurring in the RBD. It will be interesting to understand the effect of double and triple mutations occurring in the RBD, which maybe helpful in determining the importance of each residue contributing in the receptor binding. The second limitation concerns the lack of experimental validation of our computational results. However, our study provides valuable preliminary data to carry out experiments concerning the effect of point mutations occurring in the S-RBD. Therefore, to address the COVID-19 pandemic with continuous emergence of new variants of SARS-CoV-2, it is important to study the effect of these variations to tackle this deadly virus.
CRediT authorship contribution statement
Jyoti Verma: Formal analysis, Writing – original draft. Naidu Subbarao: Formal analysis, Writing – original draft.
Acknowledgment
We acknowledge the financial assistance provided by University Grant Commission in the form of Junior Research Fellowship (JRF) to Jyoti Verma.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2021.06.009.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Energy contribution of residues to the total binding free energy of RBD-ACE2 complex. Contribution of WT residues and variants at the same positions are shown in single graph.
References
- Maestro | Schrödinger.
- PyMOL | pymol.Org.
- RCSB PDB - 6M0J Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2. https://www.rcsb.org/structure/6M0J [DOI] [PubMed]
- Bendl J., Stourac J., Salanda O., et al. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Van Der Spoel D., Van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- PDBsum home page http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html
- Chen F., Liu H., Sun H., et al. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Phys. Chem. Chem. Phys. 2016;18:22129–22139. doi: 10.1039/c6cp03670h. [DOI] [PubMed] [Google Scholar]
- Clinical characteristics of COVID-19 https://www.ecdc.europa.eu/en/covid-19/latest-evidence/clinical
- Davies N.G., Abbott S., Barnard R.C., et al. 2021. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group . 2021. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7; pp. 1–5. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecdc . 2020. Suggested Citation: European Centre for Disease Prevention and Control. Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Sadykov M., Mfarrej S., et al. A genetic barcode of SARS-CoV-2 for monitoring global distribution of different clades during the COVID-19 pandemic. Int. J. Infect. Dis. 2020;100:216–223. doi: 10.1016/j.ijid.2020.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HawkDock Server http://cadd.zju.edu.cn/hawkdock/
- Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., et al. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Genet. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-cov-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82:6984–6991. doi: 10.1128/jvi.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney M.W., Jorgensen W.L. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000;112:8910–8922. doi: 10.1063/1.481505. [DOI] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhtari T., Hassani F., Ghaffari N., et al. COVID-19 and multiorgan failure: a narrative review on potential mechanisms. J. Mol. Histol. 2020;51:613–628. doi: 10.1007/s10735-020-09915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2019nCoVR . 2019. Novel Coronavirus Resource.https://bigd.big.ac.cn/ncov/ [Google Scholar]
- New “Delta Plus” Variant of SARS-CoV-2 Identified. Is it a Concern for India? - Coronavirus Outbreak News.
- WHO | SARS-CoV-2 variants 2021. http://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/ WHO.
- Pandurangan A.P., Ochoa-Montaño B., Ascher D.B., Blundell T.L. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017;45:W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pj . Oregon Graduate Institute of Science and Technology B; 2005. Turner Center for Coastal and Land-Margin Research. XMGRACE. [Google Scholar]
- PredictSNP Predict SNP effect! https://loschmidt.chemi.muni.cz/predictsnp/
- Protein Preparation Wizard | Schrödinger https://www.schrodinger.com/protein-preparation-wizard
- S - spike glycoprotein precursor - severe acute respiratory syndrome coronavirus 2 (2019-nCoV) - S gene & protein. https://www.uniprot.org/uniprot/P0DTC2
- Science Brief: Emerging SARS-CoV-2 Variants | CDC.
- SDM Predict effects of mutation on protein stability. http://marid.bioc.cam.ac.uk/sdm2/prediction
- Shah M., Ahmad B., Choi S., Woo H.G. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput. Struct. Biotechnol. J. 2020;18:3402–3414. doi: 10.1016/j.csbj.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020:1–4. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Ma L., Zou D., et al. The global landscape of SARS-CoV-2 genomes, variants, and haplotypes in 2019nCoVR. Genom. Proteomics Bioinf. 2020 doi: 10.1016/j.gpb.2020.09.001. S1672-0229(20)30131-5. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil SARS-CoV-2 coronavirus/nCoV-2019 genomic epidemiology - virological. 2020. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584
- Sun H., Li Y., Tian S., et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014;16:16719–16729. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- The effects of virus variants on COVID-19 vaccines. https://www.who.int/news-room/feature-stories/detail/the-effects-of-virus-variants-on-covid-19-vaccines?gclid=CjwKCAjwxuuCBhATEiwAIIIz0YdYlN6K2AybPJ8ye-3TI_Psj-a8ZCTNRO67RubKmnabS0_3OOwvYxoC6igQAvD_BwE
- Tracking SARS-CoV-2 variants https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- Verma J., Subbarao N. A comparative study of human betacoronavirus spike proteins: structure, function and therapeutics. Arch. Virol. 2021;166:697–714. doi: 10.1007/s00705-021-04961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma J., Subbarao N., Rajala M.S. Envelope proteins as antiviral drug target. J. Drug Target. 2020:1–7. doi: 10.1080/1061186X.2020.1792916. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Peng G., Wilken M., et al. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 [Google Scholar]
- Zhao W.M., Song S.H., Chen M.L., et al. The 2019 novel coronavirus resource. Yi chuan = Hered. 2020;42:212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Energy contribution of residues to the total binding free energy of RBD-ACE2 complex. Contribution of WT residues and variants at the same positions are shown in single graph.