Abstract
Background The Coronavirus Disease 2019 (COVID-19) had become a Public Health Emergency of International Concern with more than 90 million confirmed cases worldwide. Therefore, this study aims to establish a predictive score model of progression to severe type in patients with COVID-19. Methods This is a retrospective cohort study of 151 patients with COVID-19 diagnosed by nucleic acid test or specific serum antibodies from February 13, 2020, to March 14, 2020, hospitalized in a COVID-19-designed hospital in Wuhan, China. Results Of the 151 patients with average age of 63 years, 64 patients were male (42.4%), and 29 patients (19.2%) were classified as severe group. Multivariate analysis showed that age > 65 years (odds ratio [OR] = 9.72, 95%CI: 2.92–32.31, P < 0.001), lymphocyte count ≤ 1.1 × 109/L (OR = 3.42, 95%CI: 1.24–9.41, P = 0.017) and AST > 35 U/L (OR = 3.19, 95%CI: 1.11–9.19, P = 0.032) were independent risk factors for the disease severity. The area under curve (AUC) of receiver operating characteristic curve of the probabilities of the composite continuous variable (age + lymphocyte + AST) is 0.796. Finally, a predictive score model called ALA was established, and its AUC was 0.83 (95%CI: 0.75–0.92). Using a cutoff value of 9.5 points, the positive and negative predictive values were 54.1% (38–70.1%) and 92.1% (87.2–97.1%), respectively. Conclusion The ALA score model can quickly identify severe patients with COVID-19, so as to help clinicians to better choose accurate management strategy.
Keywords: COVID-19, Wuhan, ALA score, Severe pneumonia, Risk factors
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a globally important infectious disease [1]. As of January 21, 2021, the whole world has been involved in the coronavirus disease 2019 (COVID-19) pandemic, there were 95,612,831 confirmed cases of COVID-19 reported globally, and 2,066,176 patients died [2].
Some studies had shown that the common symptoms of COVID-19 were fever, cough, myalgia, fatigue and so on [3–5]. Severe respiratory infection symptoms could be observed in some severe cases in the early stage of COVID-19, then rapidly progressed to acute respiratory distress syndrome (ARDS), acute respiratory failure, and other serious complications resulting in the more than 20% mortality [4, 6]. However, it is difficult to identify the severe patients accurately in the early stage of admission. Therefore, in this study, we evaluated and compared the demographic characteristics and initial clinical laboratory data between severe type and non-severe type in patients with COVID-19 diagnosed by real-time reverse transcription-polymerase chain reaction (RT-PCR) or serological assays on initial admission in order to establish a predictive score model for severe type, which could help medical staffs quickly identify the severe patients at the early stage of admission, so as to give accurate management for them.
Methods
Study Population
This is a retrospective cohort study of 151 patients with COVID-19 pneumonia treated or managed by Harbin Medical Team to aid Hubei province from February 13, 2020 to March 14, 2020, hospitalized in Wuhan NO.1 hospital, China. Of the 151 patients, 138 patients discharged from this hospital. All patients lived in Wuhan and had a history of exposure to confirmed case of COVID-19. RT-PCR was tested in all patients, and serological assays including specific IgM antibody and IgG antibody of SARS-COV-2 were tested in 103 patients. All patients were diagnosed with COVID-19 pneumonia according to National Health Commission of the People's Republic of China interim guidance [7]. Patients with COVID-19 pneumonia were divided into two subtypes according to the degree of disease severity. The severe type was defined according to any of the following criteria: (1) respiratory distress, respiratory rate (RR) ≥ 30 breaths/min; (2) percutaneous oxygen saturation ≤ 93% at rest; (3) artery partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤ 300 mmHg (1 mmHg = 0.133 kPa); (4) death. The non-severe type was defined as those who met none of these criteria. This study was reviewed and approved by the Medical Ethical Committee of the Second Affiliated Hospital of Harbin Medical University (Ethical review approval number: KY2020-011).
Data Collection
The clinical symptoms, comorbidities and clinical laboratory data from electronic medical records were extracted. Routine blood tests (white blood cell count, lymphocyte count, platelets count, neutrophils count) were performed on the blood samples. Clinical chemical tests include AST (aspartate aminotransferase), ALT (alanine aminotransferase), albumin, glucose, LDH (lactate dehydrogenase), creatine kinase, serum creatinine, BUN (blood urea nitrogen). The initial laboratory indices of some patients were not perfect because our team went to support another hospital. Noneffective antibiotic treatment was defined according to the following: no declining temperature or no improvement of symptoms even worse after 48 to 72 h antibiotic treatment. To identify SARS-CoV-2 infection, RT-PCR was used for the testing of nasopharyngeal swab samples obtained from all patients at admission according to the recommendation by the National Institute for Viral Disease Control and Prevention (China) [8]. In addition, serum from 103 patients was collected to detect SARS-CoV-2 specific IgG antibody and IgM antibody.
Statistical Analysis
SPSS Statistics 25 (IBM, SPSS) was used for statistical analysis. Normal distributed continuous data were described by mean ± standard deviation (SD) and compared using student’s t-test. Abnormal distributed continuous data were described by median (interquartile range, IQR) and compared using Mann–Whitney test. For categoric data, Chi-square test or the Fisher exact test was used for the comparison. Logistic regression analysis was used to estimate the odds ratio (OR) for statistically significant correlation between risk factors and severity. Receiver operating characteristic (ROC) analysis was used to estimate the diagnostic accuracy of risk factors for disease severity. Bilateral test (the test level α = 0.05) was used, and P < 0.05 was considered statistically significant.
The establishment of predictive score model was performed using R software, version 3.6.2 (R Foundation for Statistical Computing, Vienna, Austria). In order to establish the predictive model better, the missing data are replaced by “unknown”. All the variables at a statistically significant level (p < 0.05) after multivariate logistic regression analysis were candidates for formulation of a nomogram based on proportionally converting each multivariate regression coefficient to a 0- to 100-point scale by using the “rms” package of R (https://CRAN.R-project.org/package=rms). The predictive performance of the nomogram was measured by concordance index (C-index) and calibration with 1000 bootstrap samples to decrease the overfit bias [9].
For convenience of clinical use, a novel scoring model including age, lymphocyte and AST was established, and their relevant points were determined by above multivariate logistic regression to reflect their weights of impact on the severe type. Lymphocyte count and AST were categorized based on their normal ranges. The performance of the scoring model was assessed by using ROC curves. The area under curve (AUC) and optimal cutoff values were determined and assessed according to the sensitivity, specificity, predictive values and likelihood ratios.
Results
Demographic Characteristics
The study was made up of 151 patients(64 men and 87 women) with an average age of 63 years (Table 1). Of the 151 patients, 103 patients with COVID-19 were diagnosed by RT-PCR testing; 48 patients with COVID-19 were diagnosed by serological specific antibody testing. A total of 122 patients (80.8%) were non-severe, defined as the non-severe group, and 29 patients (19.2%) were severe, defined as severe group. The common self-reported symptoms at onset of illness were fatigue (n = 106 [70.2%]), cough (n = 103 [68.2%]), fever (n = 101 [66.9%]), productive cough (n = 42 [27.8%]), and dyspnea (n = 29 [19.2%]). Eighty-five patients (56.3%) have at least one underlying disease. Common comorbidities include hypertension (n = 60 [39.7%]), diabetes (n = 32 [21.2%]), cardiovascular disease (n = 18 [11.9%]), chronic obstructive pulmonary disease (COPD, n = 6 [4.0%]) and cancer (n = 5 [3.3%]).
Table 1.
Comparison of the demographic characteristics between severe and non-severe patients with COVID-19
| Demographic characteristics | All patients (n = 151) | Severe Group (n = 29, 19.2%) | Non-severe Group (n = 122, 80.8%) | P-value |
|---|---|---|---|---|
| Age (y), mean | 63 ± 14 | 72 ± 14 | 61 ± 13 | < 0.001 |
| < 65 | 80 (53.0) | 5 (17.2) | 75 (61.5) | < 0.001 |
| ≥ 65 | 71 (47.0) | 24 (82.8) | 47 (38.5) | |
| Gender | 0.475 | |||
| Male | 64 (42.4) | 14 (48.3) | 50 (41.0) | |
| Female | 87 (57.6) | 15 (51.7) | 72 (59.0) | |
| Clinical symptoms | ||||
| Cough | 103 (68.2) | 17 (58.6) | 86 (70.5) | 0.217 |
| Productive cough | 42 (27.8) | 6 (20.7) | 36 (29.5) | 0.341 |
| Fever | 101 (66.9) | 18 (62.1) | 83 (68.0) | 0.540 |
| Dyspnea | 29 (19.2) | 20 (69.0) | 9 (7.4) | < 0.001 |
| Fatigue | 106 (70.2) | 24 (82.8) | 82 (67.2) | 0.100 |
| Comorbidities | ||||
| Any | 85 (56.3) | 17 (58.6) | 68 (55.7) | 0.778 |
| COPD | 6 (4.0) | 1 (3.4) | 5 (4.1) | 1.000 |
| Hypertension | 60 (39.7) | 13 (44.8) | 47 (38.5) | 0.533 |
| Diabetes | 32 (21.2) | 9 (31.0) | 23 (18.9) | 0.149 |
| Cardiovascular disease | 18 (11.9) | 2 (6.9) | 16 (13.1) | 0.528 |
| Cancer | 5 (3.3) | 1 (3.4) | 4 (3.3) | 1.000 |
| Other chronic history | 6 (4.0) | 5 (17.2) | 1 (0.8) | 0.001 |
| Noneffective antibiotic treatment | 25 (16.6) | 10 (34.5) | 15 (12.3) | 0.010 |
Values are presented as mean ± standard deviation or number (%). COVID-19, coronavirus disease 2019; COPD chronic obstructive pulmonary disease
The average ages of severe group and non-severe group were 72 and 61, respectively (P < 0.001). Compared with the non-severe group, more patients presented with dyspnea (9 [7.4%] vs. 20 [69.0%], P < 0.001) in severe group. Noneffective antibiotic treatment was more likely to be observed in the severe patients compared with non-severe patients (10 [34.5%] vs. 15 [12.3%], P = 0.010). There were no statistical significances for other clinical features between the two groups.
Clinical Laboratory Data
Clinical laboratory data of patients with COVID-19 are summarized in Table 2. The levels of lymphocytes (0.83 [0.62, 1.37] × 109 /L vs 1.55 [1.13, 1.95] × 109/L; P < 0.001), eosinophils (0.01 [0.00, 0.07] × 109/L vs 0.1 [0.06, 0.19] × 109 /L; P = 0.001), albumin/globulin ration (0.96 ± 6.84 vs 1.32 ± 0.29; P < 0.001), albumin (29.2 [26.48, 32.1] g/L vs 34 [30.5, 38.5] g/L; P < 0.001), serum potassium (3.7 [3.0, 4.2] mmol/L vs 4.2 [4.0, 4.3] mmol/L; P = 0.022), serum sodium (136.1 [133.8, 142.1] mmol/L vs 140.7 [140.1, 141.25] mmol/L; P = 0.044) and serum calcium (2.03 [1.94, 2.09] mmol/L vs 2.16 [2.01, 2.22] mmol/L; P = 0.005) were significantly lower in severe patients compared with non-severe patients. Significantly higher levels of white blood count (7.69 [6.07, 11.16] × 109/L vs 5.87 [4.87, 7.29] × 109/L; P = 0.001), neutrophils count (7.60 [4.13, 12.98] × 109/L vs 3.4 [2.53, 4.32] × 109/L; P = 0.001), AST (37 [25, 59] U/L vs 23 [18, 31] U/L; P < 0.001), total bilirubin (16.82 ± 6.63 umol/L vs 11.84 ± 3.64 umol/L; P = 0.029), LDH (342 [195, 721] U/L vs 201 [174, 248] U/L; P < 0.001) and BUN (8.85 [6.75, 12.85] mmol/L vs 4.25 [3.38, 5.25] mmol/L; P = 0.001) were observed in patients with severe type compared with those with non-severe type. There were no significant differences for other laboratory indices between the two groups.
Table 2.
Comparison of clinical laboratory data between severe and non-severe patients with COVID-19
| Tests in study population | Severe group n = 29 | Non-severe group n = 122 | P-value |
|---|---|---|---|
| Hematologic | |||
| White blood cells, × 109/L | 28 (7.69; 6.07, 11.16) | 107 (5.87; 4.87, 7.29) | 0.001 |
| Lymphocytes, × 109/L | 28 (0.83; 0.62, 1.37) | 107 (1.55; 1.13, 1.95) | < 0.001 |
| ≤ 1.1 × 109/L | 17 (60.7) | 26 (24.3) | < 0.001 |
| Neutrophils, × 109/L | 16 (7.60; 4.13, 12.98) | 23 (3.4; 2.53, 4.32) | 0.001 |
| Red blood cells, × 1012/L | 14 (3.89; 3.46, 5.03) | 23 (4.02; 3.78, 4.47) | 0.914 |
| Mononuclear cells, × 109 /L | 14 (0.58; 0.40, 0.73) | 23 (0.53; 0.5, 0.7) | 0.817 |
| Eosinophils, × 109 /L | 14 (0.01; 0.00, 0.07) | 23 (0.1; 0.06, 0.19) | 0.001 |
| ≤ 0.02 × 109 /L | 10 (71.4) | 0 (0.0) | < 0.001 |
| Platelets, × 109 /L | 27 (235; 109, 301) | 107 (239; 193, 287) | 0.393 |
| Haemoglobin, g/L | 14 (126.9 ± 25.56) | 23 (123.35 ± 13.46) | 0.582 |
| Basophil percentage, % | 13 (0.2; 0.1, 0.2) | 23 (0.3; 0.2, 0.4) | 0.055 |
| Biochemical | |||
| AST, U/L | 27 (37; 25, 59) | 107 (23; 18, 31) | < 0.001 |
| > 35U/L | 14 (51.9) | 19 (17.8) | < 0.001 |
| ALT, U/L | 26 (35.5; 15, 46.75) | 107 (23; 16, 42) | 0.145 |
| Total protein, g/L | 14 (60.21 ± 6.09) | 22 (69.92 ± 7.63) | 0.272 |
| Albumin, g/L | 28 (29.2; 26.48, 32.1) | 107 (34; 30.5, 38.5) | < 0.001 |
| ≤ 40 g/L | 26 (92.9) | 92 (86.0) | 0.524 |
| Globulin, g/L | 13 (31.20; 26.70, 33.70) | 23 (28.1; 24.4, 30.6) | 0.060 |
| A/G | 14 (0.96 ± 6.84) | 23 (1.32 ± 0.29) | < 0.001 |
| Total bilirubin, umol/L | 12 (16.82 ± 6.63) | 23 (11.84 ± 3.64) | 0.029 |
| Glucose, mmol/L | 9 (5.7; 4.65, 12.25) | 48 (5.25; 4.9, 6.5) | 0.443 |
| LDH, U/L | 19 (342; 195, 721) | 83 (201; 174, 248) | < 0.001 |
| > 250U/L | 7 (36.8) | 20 (24.1) | 0.001 |
| Creatine kinase, U/L | 18 (58; 39.75, 125) | 82 (51.5; 42, 100.5) | 0.680 |
| Serum creatinine, umol/L | 16 (88; 55.5, 140.25) | 22 (61.5; 54, 73.5) | 0.052 |
| BUN, mmol/L | 16 (8.85; 6.75, 12.85) | 22 (4.25; 3.38, 5.25) | 0.001 |
| Uric acid, umol/L | 15 (335; 181, 475) | 20 (303; 233.75, 372.75) | 0.730 |
| Total carbon dioxide, mmol/L | 15 (25.29 ± 5.62) | 18 (27.71 ± 3.27) | 0.157 |
| Ion | |||
| K+, mmol/L | 15 (3.7; 3.0, 4.2) | 17 (4.2; 4.0, 4.3) | 0.022 |
| Na+, mmol/L | 15 (136.1; 133.8, 142.1) | 17 (140.7; 140.1, 141.25) | 0.044 |
| Cl−, mmol/L | 14 (103.26 ± 10.80) | 17 (105.94 ± 4.42) | 0.397 |
| Ca2+, mmol/L | 14 (2.03; 1.94, 2.09) | 18 (2.16; 2.01, 2.22) | 0.005 |
| ≤ 2 mmol/L | 6 (42.9) | 3 (16.7) | 0.132 |
| Mg2+, mmol/L | 13 (0.9; 0.83, 0.98) | 18 (0.92; 0.86, 0.95) | 0.540 |
Data are presented as n (mean ± SD) or n (median; IQR) or number (%). COVID-19, coronavirus disease 2019; ALT alanine aminotransferase; AST aspartate aminotransferase; A/G albumin/globulin ration; LDH lactate dehydrogenase; BUN blood urea nitrogen
Risk factors of the Severe COVID-19
As shown in Table 3, univariate analysis found that age > 65 years (OR = 7.66, 95%CI: 2.73–21.46, P < 0.001), lymphocytes ≤ 1.1 × 109/L (OR = 4.81, 95%CI: 2–11.58, P < 0.001) and AST > 35 U/L (OR = 4.99, 95%CI: 2.02–12.31, P < 0.001) were significantly associated with the severe type. Multivariate analysis showed that age > 65 years (OR = 9.72, 95%CI: 2.92–32.31, P < 0.001), lymphocyte count ≤ 1.1 × 109/L (OR = 3.42, 95%CI: 1.24–9.41, P = 0.017) and AST > 35 U/L (OR = 3.19, 95%CI: 1.11–9.19, P = 0.032) were independent factors for the severe type.
Table 3.
Univariate and multivariate analysis for risk factors of severe type in patients with COVID-19
| Clinical characteristics | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| OR (95%CI) | P-value a | OR (95%CI) | P-value a | |
| Age(years) | ||||
| ≤ 65 | Reference | Reference | ||
| > 65 | 7.66 (2.73,21.46) | < 0.001 | 9.72 (2.92,32.31) | < 0.001 |
| Comorbidity | ||||
| Without | Reference | |||
| Any | 1.13 (0.5,2.56) | 0.779 | ||
| Lymphocyte (× 109/L) | ||||
| > 1.1 | Reference | Reference | ||
| ≤ 1.1 | 4.81 (2,11.58) | < 0.001 | 3.42 (1.24,9.41) | 0.017 |
| Unknown | 0.49 (0.06,4.09) | 0.511 | 0.28 (0.01,8.38) | 0.465 |
| Eosinophils, × 109 /L | ||||
| > 0.02 | Reference | |||
| ≤ 0.02 | 244,632,670.97 (0, Inf) | 0.988 | ||
| Unknown | 0.87 (0.26,2.87) | 0.821 | ||
| AST, U/L | ||||
| ≤ 35 | Reference | Reference | ||
| > 35 | 4.99 (2.02,12.31) | < 0.001 | 3.19 (1.11,9.19) | 0.032 |
| Unknown | 0.9 (0.18,4.41) | 0.899 | 2.17 (0.14,34.37) | 0.584 |
| Albumin, g/L | ||||
| > 40 | Reference | |||
| ≤ 40 | 2.12 (0.46,9.87) | 0.339 | ||
| Unknown | 0.5 (0.04,6.12) | 0.588 | ||
| Total bilirubin, umol/L | ||||
| ≤ 24 | Reference | |||
| > 24 | 13,242,969.58 (0, Inf) | 0.987 | ||
| Unknown | 0.39 (0.16,0.97) | 0.044 | ||
| Ca2+, mmol/L | ||||
| > 2 | Reference | |||
| ≤ 2 | 3.75 (0.73,19.14) | 0.112 | ||
| Unknown | 0.27 (0.1,0.75) | 0.012 | ||
AST aspartate aminotransferase
aLogistic regression analysis
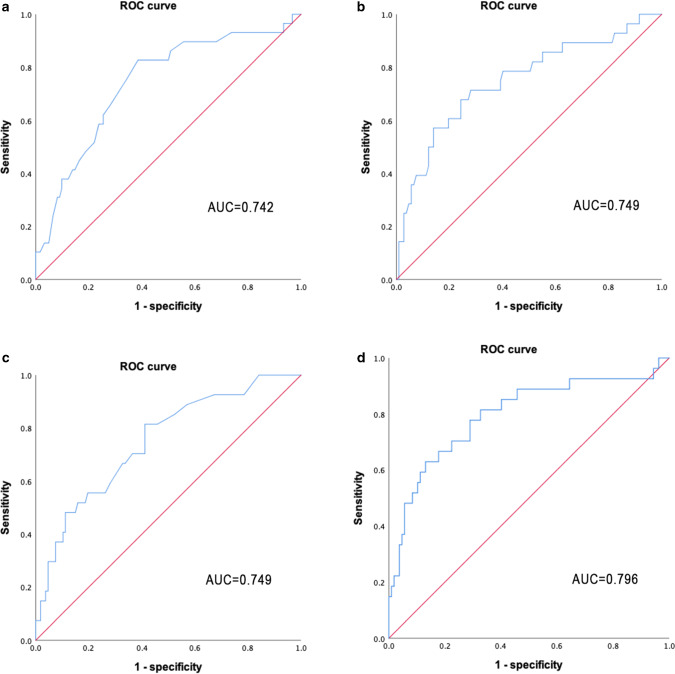
Based on the ROC curve analysis (Fig. 1a–c), the AUC of age, lymphocyte, and AST were 0.742 (95%CI: 0.639–0.844; P < 0.001), 0.749 (95%CI: 0.640–0.859; P < 0.001), and 0.749 (95%CI: 0.647–0.850; P < 0.001), respectively. When the three continuous variables were composited, the AUC of the probabilities of the composite variable is 0.796 (95%CI: 0.690–0.903; P < 0.001, Fig. 1d).
Fig. 1.
Receiver operator characteristic analysis of age a, lymphocyte b, AST c and composite variable (age + lymphocyte + AST, d) for predicting severe pneumonia. NOTE: AST, aspartate aminotransferase; AUC, area under curve
Formulation of Nomogram and Establishment of a Prediction Scoring Model
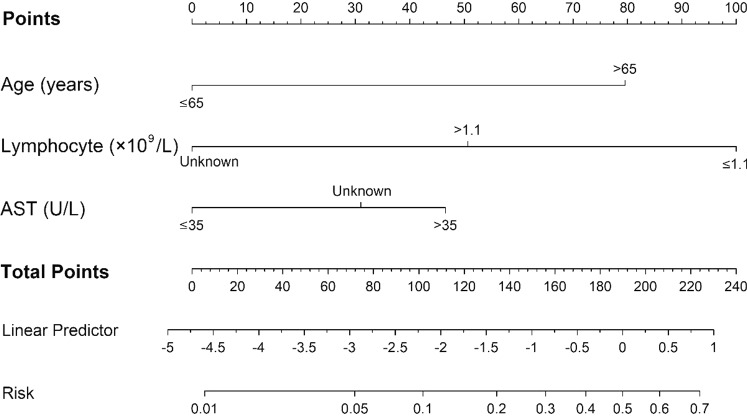
A predictive nomogram was formulated based on above independent risk factors associated with severe group and validated using the bootstrap method internally (Fig. 2). The nomogram demonstrated good accuracy in estimating the risk of progression of illness, with a C-index of 0.83 (Fig. 3).
Fig. 2.
Nomogram to estimate the risk of severity in patients with COVID-19
Fig. 3.
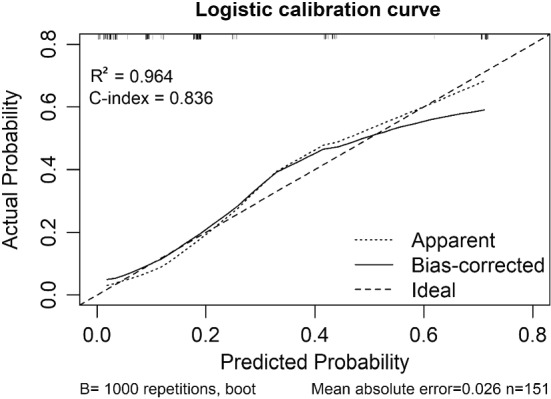
The calibration curves for predictions in patients with COVID-19
In order to identify severe patients quickly, a new scoring model called ALA (age, lymphocyte and AST) was established according to the results of nomogram, which scores from 5 to 13 points. For age scores, there were two levels: ≤ 65 years and > 65 years. For lymphocyte scores, there were three levels: ≤ 1.1 × 109/L, > 1.1 × 109/L, and unknown. For AST scores, there were three levels: ≤ 35 U/L, > 35 U/L, and unknown (Table 4). ROC analysis was used to assess the performance of the ALA model, the AUC was 0.83 (95%CI: 0.75–0.92). Using a cutoff value of 9.5 points, the sensitivity and specificity were 69.0% (52.1–85.8%) and 86.1% (79.9–92.2%), the positive and negative predictive values were 54.1% (38–70.1%) and 92.1% (87.2–97.1%), respectively (Table 5).
Table 4.
The calculators of ALA points
| Characteristics | Points |
|---|---|
| Age (years) | |
| ≤ 65 years | 2 |
| > 65 years | 5 |
| Lymphocyte (× 109/L) | |
| > 1.1 × 109 /L | 2 |
| ≤ 1.1 × 109 /L | 4 |
| Unknown | 1 |
| AST (U/L) | |
| ≤ 35 U/L | 2 |
| > 35 U/L | 4 |
| Unknown | 3 |
Table 5.
Predictive accuracy of the ALA model for the severity of COVID-19
| Variable | Enrolled patients (n = 151) |
|---|---|
| AUROC | 0.83 (0.75–0.92) |
| Cutoff value (95% CI) | 9.5 |
| Sensitivity, % | 69.0 (52.1–85.8) |
| Specificity, % | 86.1 (79.9–92.2) |
| Accuracy, % | 82.8 (82.6–83.0) |
| Positive predictive value, % | 54.1 (38.0–70.1) |
| Negative predictive value, % | 92.1 (87.2–97.1) |
| Positive likelihood ratio | 4.95 (2.99–8.19) |
| Negative likelihood ratio | 0.36 (0.21–0.62) |
Area under receiver operating characteristic, AUROC
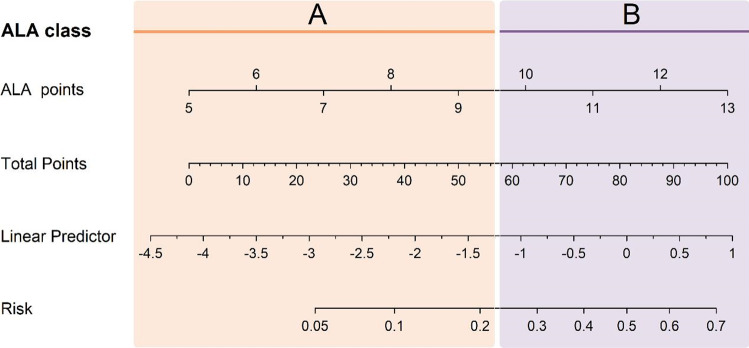
Furthermore, ALA scores are divided into two risk levels based on the probability of severity. Low risk (Class A) was defined as follow: patients with 5–9 points had less than 22% probabilities to develop severe type; and high risk (Class B) was defined as follow: patients with 10–13 points had 22–70% probabilities to develop severe type (Fig. 4).
Fig. 4.
Nomogram for ALA estimation of severity risk
Discussion
At present, COVID-19 has become a major threat to global health [10]. Most patients with COVID-19 have mild symptoms and good prognosis, whereas some cases rapidly progressed to severe pneumonia even died. In this study, of the 151 patients with COVID-19, 29 patients (19.2%) were included in the severe group. Fever, cough, and fatigue are the common symptoms in patients with COVID-19. Our data differentiated from the recent report showed SARS-CoV-2 was more likely to affect females, and the average age of patients with COVID-19 appeared older [6]. Whether the differences are due to regional differences, which requires further multi-center research.
In this study, severe patients were older and less likely to be treated with antibiotics than non-severe patients. And severe patients had lower levels of albumin, lymphocytes, eosinophils, and higher levels of neutrophils compared with non-severe patients. These differences of demographic characteristics were consistent with results noted in some studies [4, 11], whereas this study suggested that antibiotic treatment was more noneffective in severe patients. In addition, this study also found that organ dysfunction (e.g. elevated level of AST, LDH, BUN) was significantly associated with severe type in patients infected by SARS-CoV-2, which is consistent with results of some published studies [12–15].
Furthermore, multivariate analysis showed that age > 65 years, lymphocyte counts ≤ 1.1 × 109/L and AST > 35 U/L were independent risk factors for severe type. Internationally, health authorities and governments are warning older people that they are at higher risk for more severe and potentially fatal diseases linked to COVID-19. The mortality data from the Oxford COVID-19 Evidence Service showed that the mortality was 3.6% for patients aged 60–69 years, 8.0% for those aged 70–79 years and 14.8% for those aged over 80 years [16]. In addition, many studies had verified that viral hyperinflammation and dysregulation of immune responses (e.g. decreased level of lymphocytes) were highly involved in the pathological process of COVID-19 [12–15, 17, 18]. Besides, SARS-CoV-2 might directly bind to ACE2 positive cholangiocytes and cause liver damage [19], and the pathological results of autopsy revealed that degeneration of hepatocytes, focal necrosis with neutrophil infiltration, lymphocytic and monocyte infiltration in the hepatic manifold area, and micro-thrombosis were common in the liver of dead patients with COVID-19 [7]. Therefore, the three risk factors (age, lymphocyte and AST) will be good predictors for severe type. In this study, the composite variable (age + lymphocyte + AST) had a good diagnostic value for severe patients according to ROC curve (area under curve = 0.796).
At present, more than 2 million patients died due to COVID-19 [2]; thereby, it is important to early identify patients with high risk for severity. Here, we constructed a risk scoring system (ALA) according to age, lymphocyte counts and AST levels, which could rapidly identify patients with high risk for severe type. Of the 151 patients, 116 (76.8%) patients had 5–9 points (class A), which was less likely to develop the severe type. And 35 (23.2%) patients had 10–13 points (class B), which indicated that these patients were more likely to develop the severe type. Therefore, patients with 10–13 points should be considered to be early transferred to tertiary centers and be played more attention. In addition, we should continuously monitor the blood routine, coagulation and biochemical indicators of the class B patients to prevent the possibility of disease progression. This score will be helpful for clinicians to make appropriate decisions and optimize the use of hospital resources.
This study has several limitations. First of all, the relatively small sample size and the lack of laboratory data of some patients may have a certain impact on the statistical results. Second, it is a retrospective, single-center study; therefore, a prospective study or a multi-center study is needed to verify the reliability of the ALA score model in future. Third, medical records collected from the first batch of hospital tests are not representative of the entire hospitalization process. Fourth, the sensitivity and specificity of the scoring model may be further improved by adding other specific variables.
Conclusion
Our findings suggested that age, lymphocytes and AST were independent risk factors for the severity of COVID-19. The ALA scoring model could help medical staffs to identify the severe COVID-19 patients quickly at the early stage of admission, so as to give accurate management for these patients.
Acknowledgements
We thank Wuhan NO.1 Hospital for providing the medical records.
Author Contributions
SD, ZS, LL, WQ and ZX performed data collection. YY and RJ performed data analysis and edited manuscripts. YD, WM and SY prepared the first manuscript draft, validated data collection, refined the research idea, performed data analysis and edited manuscripts. CH and LY developed the research idea, refined the research idea, validated data collection and edited manuscripts. CH and LY are the guarantors of the manuscript.
Funding
None.
Declarations
Conflict of interest
The author declares that they have no conflict of interest.
Ethical approval
This study was reviewed and approved by the Medical Ethical Committee of the Second Affiliated Hospital of Harbin Medical University (KY2020-011).
Footnotes
Menghan Wang, Dongping Yu and Yu Shang contribute equally to this article.
Contributor Information
Yupeng Li, Email: 954203558@qq.com.
Hong Chen, Email: chenhong744563@aliyun.com.
References
- 1.W.H. Organization: Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (2020). Feb 11
- 2.W.H. Organization: Coronavirus (COVID-19) data. https://www.who.int/data#reports (2021). Accessed 21 January
- 3.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. Risk Factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Shang Y, Yang Y, Wang M, Yu D, Su D, Liu L, Wang Q, Zhang X, Ren J, Chen H. Factors associated with a positive severe acute respiratory syndrome coronavirus 2 testing in suspected cases presenting with pneumonia: a retrospective cohort study in a single medical center. Respir. Intern. Rev. Thorac. Dis. 2020;99(9):739–747. doi: 10.1159/000508398. [DOI] [PubMed] [Google Scholar]
- 6.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Health Commission of the People's Republic of China: Update on novel coronavirus-infected pneumonia situation on March 3, 2020. . http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml (2020). Accessed 3,Mar
- 8.National Institute for Viral Disease Control and Prevention (China): Specific primers and probes for detection 2019 novel coronavirus. http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html (2020)
- 9.Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur. Heart J. 2014;35(29):1925–1931. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biscayart C, Angeleri P, Lloveras S, Chaves T, Schlagenhauf P, Rodriguez-Morales AJ. The next big threat to global health? 2019 novel coronavirus (2019-nCoV): What advice can we give to travellers?—Interim recommendations January 2020, from the Latin-American society for Travel Medicine (SLAMVI) Travel Med. Infect. Dis. 2020;33:101567. doi: 10.1016/j.tmaid.2020.101567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, Cuomo-Dannenburg G, Thompson H, Walker PGT, Fu H, Dighe A, Griffin JT, Baguelin M, Bhatia S, Boonyasiri A, Cori A, Cucunuba Z, FitzJohn R, Gaythorpe K, Green W, Hamlet A, Hinsley W, Laydon D, Nedjati-Gilani G, Riley S, van Elsland S, Volz E, Wang H, Wang Y, Xi X, Donnelly CA, Ghani AC, Ferguson NM. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian J, Jin X, Hao S, Cai H, Zhang S, Zheng L, Jia H, Hu J, Gao J, Zhang Y, Zhang X, Yu G, Wang X, Gu J, Ye C, Jin C, Lu Y, Yu X, Yu X, Ren Y, Qiu Y, Li L, Sheng J, Yang Y. Analysis of epidemiological and clinical features in older patients with corona virus disease 2019 (COVID-19) out of Wuhan. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, He W, Yu X, Hu D, Bao M, Liu H, Zhou J, Jiang H. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oxford COVID-19 Evidence Service: Global COVID-19 Case Fatality Rates. . https://www.phc.ox.ac.uk/covid-19/evidence-service/reviews/global-covid-19-case-fatality-rates) (2020). Accessed 7,Oct
- 17.Lagunas-Rangel FA. Neutrophil-to-Lymphocyte ratio and Lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. J. Med. Virol. 2020 doi: 10.1002/jmv.25819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W, Tian DS. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai, X.Q.; L, H.; Zhang, Y.; Han, W.Y.; Lu Z.; Ke, A.W.; Zhou, J.; Shi, G.M.; Fang, N.; Fan, J.; Cai, J.B.; Fan, J.; Lan, F.: Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. 10.1101/2020.02.03.931766 (2020). Accessed 04,Feb