Abstract
Background
Hereditary hearing loss (HHL) is the most common sensory deficit, which highly afflicts humans. With gene sequencing technology development, more variants will be identified and support genetic diagnoses, which is difficult for human experts to diagnose. This study aims to develop a machine learning-based genetic diagnosis model of HHL-related variants of GJB2, SLC26A4 and MT-RNR1.
Methods
This case-control study included 1898 subjects, among which 1354 were HHL patients and 544 were carriers. Risk assessment models were established based on variants at 144 sites in three genes related to HHL by building six machine learning (ML) models. We compared the ML models with the genetic risk score (GRS) and expert interpretation (EI) to verify the clinical performance.
Findings
Among the six ML models, the support vector machine (SVM) showed the best performance. For the prediction of HHL-related gene sites in subjects with variants, the area under the receiver operating characteristic (AUC) of the SVM model was 0.803 (0.680–0.814) in the 10-fold stratified cross-validation and 0.751 (0.635–0.779) in external validation. The predicted results were better than both EI and GRS. Furthermore, 11 sites were identified as the smallest feature set that can be accurately predicted.
Interpretation
The developed SVM model has great potential to be an efficient and effective tool for HHL prediction when high throughput sequencing data are available.
Keywords: Hereditary hearing loss, Machine learning, Genetic risk score, Genetic diagnosis
Research in context.
Evidence before this study
Hereditary hearing loss (HHL) is the most common sensory defect in the world. At present, 224 genes related to HHL have been reported, but only GJB2, SLC26A4 and MT-RNR1 have been clinically tested in most countries. The development of sequencing technology will bring more genetic data, and it is urgent to build more accurate automatic diagnosis models to help doctors solve the difficulties caused by the explosion of clinical genetic diagnosis data in the future.
Added value of this study
We established the HHL gene diagnosis model based on machine learning, and compared with GRS model and human expert genetic counseling model, the model performed significantly better. We also found correlations between several unreported variants.
Implications of all the available evidence
The proposed method can help improve the efficiency of clinical workflow and accelerate the process of incorporating more genes into HHL clinical gene diagnosis.
Alt-text: Unlabelled box
1. Introduction
Hearing loss is the most common sensory defect in the world. It affects more than 500 million people worldwide, accounting for approximately 6.8% of the world's population [1]. Hereditary hearing loss (HHL) refers to hearing loss caused by genetic and chromosomal abnormalities [2]. HHL accounts for 50% of the hearing loss cases. Non-syndromic hearing loss accounts for 70% of the hereditary hearing loss cases, among which 75 to 80% are inherited as autosomal recessive traits, 20% as autosomal dominant traits, less than 2% are X-linked and less than 1% are mitochondrial inherited [3]. Autosomal dominant-associated hearing loss is mostly delayed onset, progressive, while that caused by autosomal recessive inheritance is characterized by the early onset and severe disease [2]. The existence of complex interactions between genetic and environmental factors, pathogenic variants at multiple sites, incomplete explicit variants and benign variants at HHL-associated variants and a large number of disease susceptibility sites, makes the accurate genetic evaluation of patients carrying multiple HHL susceptibility variants difficult.
To date, more than 224 genes related to syndromic and non-syndromic hearing loss have been discovered (https://deafnessvariationdatabase.org/). With the advancement in next-generation sequencing (NGS) technology and the cost reduction, it has become technically feasible to sequence a large number of genes in a diagnostic test. This is particularly relevant for diseases with high genetic and phenotypic heterogeneity, such as hearing loss [5]. The resulting data are available in a structured genetic database and have been used to influence the diagnosis of inherited diseases, including HHL [4]. With the number of HHL-involved genes continually increasing, we attempt to address the difficulties facing human experts in the clinical diagnosis of deafness that result from the comprehensive genetic testing using large-scale parallel sequencing.
Establishing the genetic diagnosis of HHL is an essential part of the clinical evaluation for HHL patients and their families. Identifying the genetic cause of hearing loss could potentially provide families with prognostic information and guide future medical management [6]. However, it is very difficult for human experts to observe all the 224 HHL-associated genes at the same time. In order to cope with the increasing size of sequencing data and number of detected gene variants, it is necessary to establish a predictive model based on genetic data that can perform automatic diagnosis. Building evaluation models, like the genetic risk score or other machine learning (ML) methods-based models, can help to precisely guide the disease predictive diagnostics and evaluate the disease risk [7]. The GRS model is a method that incorporates genetic susceptibility factors into the risk score to evaluate the effects of these factors in the model. It is one of the important methods to evaluate the ability of risk prediction in epidemiological research. ML methods have been widely used in different medical situations to improve the performance of the diagnostic tests and identify risk factors associated with complex diseases [8,9]. These methods have a more powerful role when dealing with large, complex and disparate data, which allows data-informed decision-making to provide faster next-generation diagnosis and treatment to the patients, reducing the costs and scale [10].
Only few studies have been so far conducted on the application of ML methods for personalized HHL gene predictive diagnosis or to compare the predictive accuracy and reliability of ML methods with traditional genetic risk assessment models and expert interpretation. The genetic diagnosis of HHL still includes only a few common genes. For example, Nele Hilgert's selection criteria in DNA diagnostics is GJB2, SLC26A4 and WFS1 [11]. However, the three common genes for the clinical genetic diagnosis of HHL in China include GJB2, SLC26A4 and MT-RNR1 genes [12], which also represent the most common cause of HHL. Variants in the GJB2 gene accounted for 18.31% of the patients with HHL, while those in the SLC26A4 gene accounted for 13.73% [13]. The use of ototoxic drugs is believed to be the main cause of hearing loss in China. Several variants in MT-RNR1 are related to the increased sensitivity to ototoxicity, which is induced by aminoglycosides [14]. The establishment of a practical and efficient identification model of the HHL high-risk population is of important clinical significance in the screening process.
In this study, we present an ML model for the automatic diagnosis of HHL using genetic variant data, then we compare its performance with the traditional genetic risk assessment models and expert advice. We collected the diagnostic advice based on the genetic sequencing results from experts in the Department of Otolaryngology, Head and Neck Surgery at the Shanghai Ninth People's Hospital, Shanghai Jiaotong University Medical College, and we compared the results with the etiology diagnosis and clinical evaluation of hearing loss from the American College of Medical Genetics and Genomics to verify the reliability of our model [15].
2. Methods
Due to the retrospective nature of our study, the need for a signed informed consent was waived. Our research is divided into four steps, and the process flowchart of the whole work is shown in Fig. 1.
Fig. 1.
Study flowchart. In Step 1, we preprocess the data and divide the resulting data into a training set and a test set. In Step 2, we train six machine learning models with 10-fold cross-validation on the training set and test them on the test set. In Step 3, we compare the SVM model with the GRS and EI models among the six machine learning models. In Step 4, we find the model that can accurately predict the HHL minimal site set. DT, decision tree; SVM, support vector machine; RF, random forest; KNN, k-nearest neighbor; ADA, adaptive Boosting; MLP, multilayer perceptron; GRS, genetic risk score; EI, expert interpretation.
2.1. Sample collection
The HHL genetic sequencing data were obtained from the electronic medical records of 2660 patients and their immediate family members from October 2009 to April 2017 in the Department of Otolaryngology, Head and Neck Surgery at the Shanghai Ninth People's Hospital, Shanghai Jiaotong University Medical College. In addition, there were 895 patients and their immediate family members from the Disabled Persons' Federation. All the samples had been screened using the conventional Sanger sequencing, which has been the preferred screening method for genetic research and clinical genetic diagnosis since the chain termination method for DNA sequencing was first proposed in 1975 [16].
Due to the way the data were collected, most samples were HHL. In order to alleviate this imbalance in the data, the result of a community's NGS hotspot screening was obtained in batches from the same center. Until August 11, 2020, 1257 community members had been genetically tested at the Shanghai Ninth People's Hospital. There was an overlap ratio of 96% between the NGS hotspots and Sanger sequencing sites. The details of all the sites are shown in Supplementary Tables S1 and S2. All the individuals had undergone bilaterally hearing screening. Hearing loss was classified as mild ( > 25–40 dB), moderate (41–55 dB), moderate-severe (56–70 dB), severe (71–90 dB) or profound ( > 90 dB) [17]. According to the degree of hearing loss that was recorded, the individuals were divided into patients and carriers. Patients are individuals with hearing loss for which variants were detected in any of the three genes. Carriers are healthy individuals for which variants were detected in any of the three genes.
This study was performed according to a protocol approved by the ethics committee of the Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences. Reference number is 2017-806.The researchers who conducted the analysis were blinded to the data collection and did not participate in it. All the collected data and identity information remain strictly confidential.
2.2. Preprocessing
The purpose of the diagnostic model is to predict whether the variants in the HHL sites are pathogenic. The model construction started by filtering the sample data from the three centers as follows. First, all wild-type genotypes were excluded. Then, we deleted the values of the missing gene test results. Finally, the sample data were divided into carriers and HHL patients according to whether they passed the hearing screening test or not. Overall, we obtained a total of 144 nucleotide variants in three genes, including the 47 variants of GJB2, 90 variants of SLC26A4 and 7 variants of MT-RNR1; the details are shown in the Supplementary Material. In the end, 1898 samples were organized in two sets of data: (1) 1354 patients with GJB2, SLC26A4 and MT-RNR1 hot spot variants who failed the hearing screening test and were diagnosed as HHL; (2) 544 people with GJB2, SLC26A4 and MT-RNR1 hot spot variants who passed the hearing screening test and were diagnosed as HHL gene carriers. An internal 10-fold stratified cross-validation dataset included 918 HHL patients and 411 carrier controls collected from the Shanghai Ninth People's Hospital, while the external validation dataset included a total of 436 HHL cases and 133 carrier controls collected from The Disabled Persons' Federation and Community screening. Table 1 summarizes the clinical characteristics of each dataset.
Table 1.
Demographic in internal 10-fold cross validation and external validation.
| Internal 10-fold cross validation(n = 1329) | External validation(n = 569) | ||
|---|---|---|---|
| Data source | Hospital electronic medical records | Disabled persons' federation | Community screening |
| Years | 2009.10–2017.4 | 2011.8–2017.4 | 2020.5–2020.8 |
| Sequencing methods | Sanger sequencing | Sanger sequencing | NGS hotspot screening |
| Age in years | 18 ± 15 | 34 ± 12 | 27 ± 4 |
| Gender | |||
| Male | 682 | 243 | / |
| Female | 647 | 223 | 103 |
To develop the ML models, we used integer coding to first encode the classified data into numbers, i.e., no mutation, heterozygous and homozygous were all mapped to different integers. The feature set was composed of the 144 variants in the three genes. Some ML algorithms are sensitive to data scaling, and the MLP (multilayer perceptron) requires all the input features to vary within a similar range, so we used the StandardScaler method to ensure that each feature had an average value of less than 0 and a variance of 1. The SVM requires all feature to be roughly in the same range, and the commonly used scaling method was MinMaxScaler, which scales all the features to the range between 0 and 1. We used logistic regression analysis to establish the GRS model. It is important to note that having several correlated variables in the model results in collinearity, which is a possible explanation for the non-significant association [18]. Therefore, we conducted linkage disequilibrium analysis to find out the gene sites that are not completely independent of inheritance. We also performed a spearman correlation analysis on 144 sites.
2.3. Model development and validation
Having more accurate genetic diagnosis and prediction models enables further development of the personalized medical care. The genetic risk score and ML are the two main methods to predict the disease risk.
In the ML method, predictions are made using a complex set of statistical and computational algorithms, by mathematically mapping the complex associations between a set of risk gene variation sites and complex disease phenotypes. Based on the eigenvalues of the 144 gene sites, we built six commonly used ML models using the Python Scikit-learn package and compared their performance on the HHL genetic diagnosis task. These models included the decision tree (DT), support vector machine (SVM), random forest (RF), k-nearest neighbor (KNN), adaptive Boosting (ADA) and multilayer perceptron (MLP) algorithms. These six ML algorithms are supervised learning algorithms. KNN represented the simplest classifier. The algorithm finds the nearest data point in the training dataset to make predictions about new gene variant data. Regarding SVM, after the kernel function transformation, the optimal hyperplane of SVM maximizes the separation between different categories, which is suitable for the classification of small and medium-sized complex datasets. DT is a widely used model for classification and regression tasks, which mainly suffers from overfitting the training data. RF is a collection of many decision trees, which generally yields lower variance and better model generalization than a single DT. AdaBoost is also an ensemble method, but it pays more attention to the training examples with insufficient pre-fitting and increases the relative weight of misclassified training examples. MLP is a relatively simple feed forward neural network used for classification and categorization. It is often precisely adjusted and only suitable for specific usage scenarios [[19], [20], [21], [22]].
To achieve better generalization ability of the model, we used 10-fold stratified cross-validation in the training set. The stratified cross-validation was performed as follows: we divided the data so that the ratio between the categories of the observations in each part is the same as the entire dataset ratio. The grid search method was then to try all possible parameter combinations we are concerned with to optimize the hyperparameters.
The genetic risk score (GRS) analysis is a common method to study the genetic structure and relationship of complex diseases. GRS is a summation of genotypic scores of disease-associated variants combining the genetic effects of multiple causal variants [23]. It is currently widely used in various diseases, and single nucleotide polymorphism (SNP) is often used as a genetic susceptibility factor for risk assessment [24,25]. In order to correspond to the machine learning model, in addition to SNPs, we also included the pathogenic sites as genetic susceptibility factors in our study. Two methods are usually used to create the GRS: a simple count method and a weighted method [26]. The second method was chosen because the risk contributions of the variants to HHL were not equal.
The related disease model is shown in Formula (2). A logistic regression model was fitted to obtain the ROC curve and AUC parameters. GRS may be utilized in the evaluation of the genetic risk for HHL.
| (1) |
| (2) |
where D = 1 means that the sample is a case, D = 0 means that the sample is a healthy control, G represents a set vector of the number of risk alleles at a genetic susceptibility site, and denotes the risk allele for the ith genetic susceptibility site.
2.4. Expert interpretation
In order to further evaluate the performance of the developed ML models, we used the joint advice and suggestions on the deafness genetic screening results from three experienced experts from the Department of Otorhinolaryngology, Head and Neck Surgery at the Ninth People's Hospital, Shanghai Jiaotong University School of Medicine. The three experts were attending doctors with 20 years, 22 years, and 35 years of experience, respectively. The experts reviewed the variants consulted in the DVD and ClinVar databases. The expert consultation points on the results of deafness genetic screening were typical, and compared with the etiology diagnosis and clinical evaluation of hearing loss from the American College of Medical Genetics and Genomics, the latter only tests GJB2 and GJB6 when suspecting HHL [15].
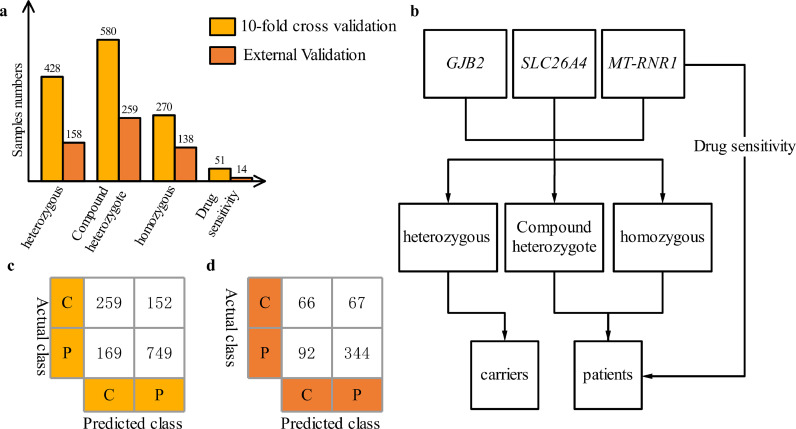
As shown in Fig. 2, the genotypes were divided into three types, and then specific incomplete explicit genetic susceptibility variants were discussed. The p.V37I variant is an incomplete explicit variant. In Table S3, p.V37I homozygote or combination with at any pathogenic variants of GJB2 (V37I/V37I or V37I/XX) are considered to have a high probability of hereditary deafness. When building the EI model, the program predicted the case to belong to a patient when the probability was greater than 50%. The GJB2 and SLC26A4 homozygous and compound heterozygous genotypes are deafness-causing genotypes, and MT-RNR1 variants are drug-sensitive genotypes, these two were diagnosed as HHL, while GJB2 and SLC26A4 heterozygotes were diagnosed as carriers. Detailed advice on the results of complete deafness genetic screening can be found in Supplemental Table S3. The diagnostic report was based on the keywords recommended in the consultation. By comparing the result of the sample's genotype based on the genetic counseling suggestion with the result of the actual disease, a confusion matrix was obtained. The sensitivity and specificity obtained by calculating the confusion matrix were plotted as the expert's performance in the ROC curve under the same dataset.
Fig. 2.
Diagnosing HHL through expert interpretation on internal 10-fold cross-validation and external validation samples. (a) Genotype distribution of internal 10-fold cross-validation and external validation data; (b) process diagram of the expert interpretation; (c) and (d) are the confusion matrix of internal and external validation data, respectively. C, carriers; P, patients.
2.5. Model evaluation criteria
To evaluate the performance of different algorithms and then select the most suitable methods for a specific task with the classification objective, we evaluated the results of different models according to the classic indices.
Accuracy is the proportion of properly classified samples, defined as:
| (3) |
Sensitivity is the rate of correctly detected HHL samples, defined as:
| (4) |
Precision is the accuracy of HHL prediction, defined as:
| (5) |
is the harmonic average of accuracy and sensitivity, since both precision and sensitivity are considered, defined as:
| (6) |
The predictive performance of all model types was evaluated by two important classifier performance evaluation indicators in machine learning: the receiver operating characteristic curve (ROC) and area under the curve (AUC) [27].
2.6. Statistical analysis
Statistical analysis was performed by using Python (version 3.3.6) and IBM SPSS software (version 21.0).
2.7. Role of funding source
The funders had no role in the study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. Machine learning model performance
The accuracy, sensitivity, precision and f1-score of the HHL diagnostic models established by the six ML models of DT, SVM, RF, KNN, ADA and MLP in 10-fold cross-validation and external validation are shown in Table 2. A comparison of the ROC curve and AUC value of the model under different algorithms is shown in Fig. 3. High sensitivity means a low missed diagnosis rate; high specificity means a low misdiagnosis rate. In clinical screening, we tend to sacrifice specificity for sensitivity. The AUC value of the SVM model in Fig. 3 is similar to that of the RF model, but the SVM model has a better sensitivity. Overall, the SVM model had the best detection performance, with an AUC of 0.803 (0.680–0.814) and 0.751 (0.635–0.779) in the 10-fold cross-validation and external validation, respectively.
Table 2.
The performance of the six ML models for NSHL risk assessment in 10-fold cross validation and external validation.
| Method | Accuracy (95%CI) | Sensitivity (95%CI) | Precision (95%CI) | F1-score (95%CI) | |
|---|---|---|---|---|---|
| 10-fold cross-validation | DT | 78.1%(71.9–84.3%) | 83.0%(77.4–87.5%) | 86.0%(80.6–90.2%) | 84.5%(79.0–88.8%) |
| SVM | 81.4% (74.6–88.2%) | 92.1%(89.3–94.9%) | 84.3%(78.9–89.7%) | 88.0%(83.8–92.2) | |
| RF | 78.6%(71.8–85.4%) | 85.7%(80.3–89.8%) | 83.5%(78.0–87.9%) | 84.6%(79.1–88.8%) | |
| KNN | 76.1%(68.1–84.1%) | 84.3%(78.9–88.7%) | 83.3%(77.7–87.7%) | 83.8%(78.3–88.2%) | |
| ADA | 79.2%(72.4–86.0%) | 85.2%(79.8–89.4%) | 84.1%(78.6–88.4%) | 84.6%(79.2–88.9%) | |
| MLP | 70.9%(62.3–79.5%) | 86.4%(80.8–91.9%) | 84.7%(79.1–88.6%) | 85.5%(79.9–90.2%) | |
| External validation | DT | 77.0%(67.7–86.3%) | 97.3%(91.6–99.3%) | 78.7%(70.7–85.0%) | 87.0%(79.8–91.6%) |
| SVM | 81.2%(73.3–89.1%) | 92.5%(85.4–96.5%) | 80.5%(72.2–86.9%) | 86.1%(78.2–91.4%) | |
| RF | 79.8%(70.8–88.8%) | 86.0%(77.6–91.7%) | 82.1%(73.5–88.5%) | 84.0%(75.5–91.4%) | |
| KNN | 77.9%(67.9–87.9%) | 86.9%(78.7–92.4%) | 80.9%(72.3–87.4%) | 83.8%(75.4–89.8%) | |
| ADA | 79.6%(68.5–90.7%) | 88.8%(80.9–93.8%) | 81.9%(73.4–88.2%) | 85.2%(77.0–90.9%) | |
| MLP | 79.8%(68.4–91.2%) | 86.3%(77.8–91.9%) | 82.0%(73.3–88.1%) | 84.1%(75.5–90.0%) |
Fig. 3.
Comparison of the receiver operating characteristic curve in six ML models in (a) internal 10-fold cross validation and (b) external validation. The AUC values obtained using the respective method are included in the parentheses, and the dotted line indicates an AUC of 0.5.
3.2. Comparing the GRS models and machine learning models with expert interpretations
In order to evaluate the performance of the ML models compared with the actual clinical discrimination situation and the traditional genetic risk assessment model, we compared the GRS model and machine learning model with the expert interpretation in the same ROC curve evaluation system. Fig. 4 shows the ROC curves and AUC values of the HHL identification models established by the SVM model, GRS model and expert interpretation (EI) in the 10-fold cross validation and external validation.
Fig. 4.
Comparison of the receiver operating characteristic curve by the SVM model, GRS model and EI in (a) internal 10-fold cross validation and (b) external validation.
The HHL-associated sites were screened using a binary logistic model, and a genetic risk model was constructed based on the GRS. By building haplotypes, the analysis of pairwise linkage disequilibrium showed the two most frequent SNPs, p.V27I and p.E114G, to be tightly linked, and p.I203T to be in complete linkage with p.V27I [28]. In the Spearman correlation analysis on 144 sites, the parts with significant correlations are shown in Fig. 5. The results showed that p.V27I and p.E114G have a significant positive correlation (two-tailed test, p < 0.01), which is consistent with the linkage disequilibrium analysis and previously published reports [29], but c.235delC was negatively correlated with p.V27I and p.E114G. This correlation was not published before, and we report it here for the first time. Finally, to calculate the score, only the sites with the most significant main effect were included; thus, the c.235delC, p.E114G and p.I203T variants were deleted when establishing the GRS model. By the above-mentioned preconditioning, all the remaining sites were considered to be independent predictors of the HHL status. We used the forward stepwise selection method based on the likelihood ratio test of the maximum partial likelihood estimation as the criterion to eliminate the variables.
Fig. 5.
Heat map between the significantly related sites in a total of 144 sites.
Since clinicians cannot make a subjective estimate of the probability of a sample suffering from HHL, we cannot compare the performance of the statistical models with that of the best human experts, and there is currently no publicly established procedural discriminant standard for this purpose. In this work, we collected the consultation points for the genetic screening results of the clinical experts for the discriminative diagnosis. According to these results, the HHL and carrier samples were classified, and the confusion matrix was then constructed. In the 10-fold cross-validation, the EI sensitivity was 0.816 and the 1-specificity was 0.371. In the external verification, the EI sensitivity was 0.789 and the 1-specificity was 0.530.
The results show that the SVM has a better HHL prediction effect than that of the GRS model and EI when operating with the same 1-specificity value. In the 10-fold cross-validation with a 1-specificity of 0.371, the SVM model showed a sensitivity improvement of 6.2 and 21.1% compared with the result of EI and GRS, respectively. In the external verification, the SVM showed a sensitivity improvement of 9.5% compared with the result of EI and 32.4% compared with the result of GRS.
3.4. Minimum sites set based on the model evaluation
In order to identify the variants that have a significant impact on machine learning models and assess the importance of these variants in HHL genes, it is necessary to determine the minimum sites set. The importance of the features among all variants was scored using DT and RF. Feature importance here refers to the degree to which each feature (variant) is important in the classification decisions. Based on the different ranking results of the two above-mentioned models, the sites with a feature importance greater than 0.001 were screened out. The sites with feature importance below 0.001 had little effect on the classification decisions and AUC value. As a result, the DT model screened out 77 sites, while the RF model screened out 62 sites.
We adopted the strategy of sequentially adding one site and using the SVM model for prediction. Through the process of one-by-one addition, we obtained the corresponding AUC value to evaluate the performance of different numbers of sites on the SVM model. The results are shown in Fig. 6. In the sorted DT model, the highest AUC of 0.784 was obtained using 40 sites (Fig. 6a), while in the sorted RF model, the highest AUC of 0.737 was obtained using 31 sites (Fig. 6b). Then, the curve plateaued regardless of further sites’ addition. Through careful scrutiny, we selected the DT models, since an AUC value of 0.769 was obtained using 11 sites. The minimum sites number of the accurate prediction results and their importance plot are also shown in Table 3 [12,30,31]. Polymorphic sites and incomplete penetrance sites accounted for the first, second, fifth and eleventh sites, i.e., four of the eleven sites. The highest allele frequencies of the three polymorphism sites of p.V27I, p.E114G and p.I203T are observed in East Asian populations (http://cdgc.eargene.org/). These sites are benign genotypes in the DVD database and variant of undetermined significance in the ClinVar database. The p.I203T site is a benign genotype in both DVD and ClinVar databases. Therefore, in the EI model, all samples with benign variations were classified as carriers. However, in the predicted results of the SVM model, some samples carrying p.V27I, p.E114G and p.I203T were classified as patients.
Fig. 6.
The evaluation of the SVM prediction effect based on the one-by-one site addition method. (a) Feature importance ordered by DT; (b) feature importance ordered by RF.
Table 3.
The minimum number of variants required to accurately predict the results and their importance plot.
 |
4. Discussion
The 2014 ACMG guidelines for the diagnosis of hearing loss recommend incorporating genetic testing early in the diagnostic protocols for the evaluation of HHL [15]. Due to Hearing Loss's high Genetic heterogeneity, Oza et al. provide a comprehensive illustration of the newly specified ACMG hearing loss rules [32]. Many studies continue to identify new HHL variants associated with hearing loss using massively parallel sequencing [6,33]. With the discovery of variants, there is an urgent need to adjust the clinical methods of gene diagnosis. The application of machine learning in gene-based diagnosis can obviously reduce the artificial part and expand the screening range, which is not inferior to the expert diagnosis. Some researchers have recently made some progress in the application of ML methods to build hearing loss predictive models. Hildebrand et al. developed a tool called AudioGene, a supervised support vector ML algorithm that uses audiometric data to predict the autosomal dominant non-syndromic hearing loss genotypes, and achieved an average accuracy of 88% using the data of 360 patients [34,35]. Weininger et al. used the AudioGene tool to stratify 141 patients with progressive hearing loss [36]. However, this tool, which was developed in 2008, is a product of the low development and high cost of gene sequencing instruments. The third-generation sequencing technology began to be widely used around 2014. Chang et al. developed three ML models, based on the algorithms of classification tree, logistic regression (LR) and random forest (RF), to predict the cochlear dead areas of 380 hearing loss patients with different causes [37]. By screening noise-induced hearing loss (NIHL)-associated single nucleotide polymorphisms (SNPs), Zang et al. constructed genetic risk prediction models for NIHL [38]. However, there is no risk assessment or prediction of HHL based on a large amount of gene sequencing data, and the amount of research data is relatively small, which calls for further studies to improve the achieved stage.
Our study may be the first to establish a genetic risk assessment model for HHL combined with the state-of-the-art ML techniques. In this study, we began by determining three HHL genes and the corresponding specific sites that are commonly used for the clinical diagnosis in the region. In our correlation analysis, we found that c.235delC was significantly negatively correlated with p.V27I and p.E114G, which had not been reported before. Then, we conducted HHL prediction models using ML, GRS and EI in internal 10-fold cross-validation and external validation. The SVM model showed the best predictive performance in classifying carriers and HHL cases. Additionally, we also found the minimum sites set that could effectively identify the high risk for HHL based on the features represented in the 11 most important sites. The main reason of why the performance of the EI model was not as good as that of the SVM model is the presence of polymorphic sites and incomplete penetrance sites. Among the top eleven most important sites in the SVM model, three sites are polymorphic, which are p.V27I, p.E114G and p.I203T. These three polymorphic sites were recommended as benign variants in the EI model, while the ML classification model tends to classify them as pathogenic. Although they are widely regarded as benign variants, there is still some controversy. Chen et al. observed the correlation between variants and hearing phenotypes of 300 HHL infants and 484 normal infants to indicate that homozygote p.V27I/p.E114G is associated with mild and moderate hereditary hearing loss [29]. Choi et al. used in vitro approaches to suggest that p.E114G homozygotes or compound heterozygotes carrying p.E114G with other variants may cause HHL. Besides, p.V27I may compensate for the p.E114G variant's deleterious effect in hemichannel activities [39]. Ambrosi et al. proved that the p.I203T variant formed instable hemichannels [40]. Therefore, further studies on the genotyping of p.V27I, p.E114G and p.I203T are needed.
The DNA predictive accuracy of risk assessment is limited by the broad heritability. However, we do not fully understand the impact of genetic risk sites, and the role of DNA sequences in the genetic risk [41]. Compared with the scoring-based methods, one of the advantages of using ML methods is that it can be directly applied to pre-calculations without a complete understanding of which sites affect the genetic risk and odds ratios. Due to the ability of ML algorithms to process multidimensional data compared with GRS, it has an improved prediction ability of complex diseases [7]. The field of Big Data has revolutionized the diagnostic field of HHL, and modern sequencing technologies have made a significant impact on the patient care and support for personalized medicine [5]. More than 200 genes are known to be associated with HHL in the form of autosomal dominant, autosomal recessive, X-linked and MT-RNR1 genes. The expert interpretation we collected came from only three experienced experts. A much larger number of experts with a broader range of experience are likely to have given a more representative view. Although in this work, we have built a machine learning diagnostic model based on three HHL genes only, our study proves that the diagnosis of hereditary hearing loss using ML models is very promising. More variants will be considered for the clinical diagnosis in the future.
In conclusion, we built predictive models with the clinical HHL gene sites using different ML algorithms and traditional risk assessment algorithms. Then, they were evaluated against the predictive performance of human experts. The results showed that SVM had the best performance in the genetic diagnosis according to the comprehensive evaluation of ML algorithms and had a relatively high performance compared with the EI and GRS models. In addition, based on the feature importance, we obtained the minimum sites set that can be accurately predicted. The p.V27I, p.E114G and p.I203T sites in the minimum sites set may be likely pathogenic. Hence, we believe that this SVM model could help to rapidly diagnose HHL, improve the clinical efficiency and reduce the cost; it might even be widely applied in the clinics. With the rapid development of gene sequencing techniques, ML predictive models are very important for the future clinical diagnosis of HHL.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgments
Contributors
XL conceived the study, designed the model, and analyzed the data. FL and KH contributed to data acquisition and collection. WX contributed to the manuscript. TY provided additional pathological insights. JC provided guidance for data analysis. XC supervised all aspects of the project implementation. HW provided important guidance for the development of data analysis, as well as the writing, revision and finalization of the paper. XL wrote the manuscript. XL and WX verified the underlying data. All authors read, edited, and approved the final manuscript.
Acknowledgments
We would like to acknowledge all participants in this work. This work was supported by the National Key Research and Development Program (2017YFC1001800).
Data sharing statement
All individual participant data on which the results reported in this article were based will be made available free of charge on the Mendeley Data (http://dx.doi.org/10.17632/6mh8mpnbgv.1) after de-identification. There are no restrictions to obtaining the data.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103322.
Contributor Information
Xiaohe Chen, Email: chenxh@sibet.ac.cn.
Hao Wu, Email: haowu@sh-jei.org.
Appendix. Supplementary materials
References
- 1.Wilson B.S., Tucci D.L., Merson M.H., O'Donoghue G.M. Global hearing health care: new findings and perspectives. Lancet. 2017;390(10111):2503–2515. doi: 10.1016/S0140-6736(17)31073-5. [DOI] [PubMed] [Google Scholar]
- 2.Shearer A.E., Hildebrand M.S., Smith R.J. University of Washington, Seattle; 2017. Hereditary hearing loss and deafness overview. GeneReviews®[Internet] [Google Scholar]
- 3.Smith R.J.H., Bale J.F., White K.R. Sensorineural hearing loss in children. Lancet. 2005;365(9462):879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 4.Abou Tayoun A.N., Al Turki S.H., Oza A.M. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med. 2016;18(6):545–553. doi: 10.1038/gim.2015.141. [DOI] [PubMed] [Google Scholar]
- 5.Vona B., Muller M., Dofek S., Holderried M., Lowenheim H., Tropitzsch A. A big data perspective on the genomics of hearing loss. Laryngo Rhino Otol. 2019;98(S 01):S32–S81. doi: 10.1055/a-0803-6149. [DOI] [PubMed] [Google Scholar]
- 6.Shearer A.E., DeLuca A.P., Hildebrand M.S. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho D.S.W., Schierding W., Wake M., Saffery R., O'Sullivan J. Machine learning SNP based prediction for precision medicine. Front Genet. 2019;10:267. doi: 10.3389/fgene.2019.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang H., Tsui B.Y., Ni H. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat Med. 2019;25(3):433–438. doi: 10.1038/s41591-018-0335-9. [DOI] [PubMed] [Google Scholar]
- 9.Lin D., Chen J., Lin Z. A practical model for the identification of congenital cataracts using machine learning. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.102621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toh T.S., Dondelinger F., Wang D. Looking beyond the hype: applied AI and machine learning in translational medicine. EBioMedicine. 2019;47:607–615. doi: 10.1016/j.ebiom.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilgert N., Smith R.J., Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681(2-3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y., Huang S., Deng T. Mutation spectrum of common deafness-causing genes in patients with non-syndromic deafness in the xiamen area. China PLOS One. 2015;10(8) doi: 10.1371/journal.pone.0135088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y., You Y., Huang D. Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in China. J Transl Med. 2009;7:79. doi: 10.1186/1479-5876-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheffield A.M., Smith R.J.H. The epidemiology of deafness. Cold Spring Harb Perspect Med. 2019;9(9) doi: 10.1101/cshperspect.a033258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alford R.L., Arnos K.S., Fox M. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med. 2014;16(4):347–355. doi: 10.1038/gim.2014.2. [DOI] [PubMed] [Google Scholar]
- 16.Sanger F. The croonian lecture, 1975. Nucleotide sequences in DNA. Proc R Soc Lond B Biol Sci. 1975;191(1104):317–333. doi: 10.1098/rspb.1975.0131. [DOI] [PubMed] [Google Scholar]
- 17.Raymond M., Walker E., Dave I., Dedhia K. Genetic testing for congenital non-syndromic sensorineural hearing loss. Int J Pediatr Otorhinolaryngol. 2019;124:68–75. doi: 10.1016/j.ijporl.2019.05.038. [DOI] [PubMed] [Google Scholar]
- 18.Stoltzfus J.C. Logistic regression: a brief primer. Acad Emerg Med. 2011;18(10):1099–1104. doi: 10.1111/j.1553-2712.2011.01185.x. [DOI] [PubMed] [Google Scholar]
- 19.Bi Q., Goodman K.E., Kaminsky J., Lessler J. What is machine learning? A primer for the epidemiologist. Am J Epidemiol. 2019;188(12):2222–2239. doi: 10.1093/aje/kwz189. [DOI] [PubMed] [Google Scholar]
- 20.Deo R.C. Machine learning in medicine. Circulation. 2015;132(20):1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handelman G.S., Kok H.K., Chandra R.V., Razavi A.H., Lee M.J., Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284(6):603–619. doi: 10.1111/joim.12822. [DOI] [PubMed] [Google Scholar]
- 22.Boulesteix A.L., Wright M.N., Hoffmann S., Konig I.R. Statistical learning approaches in the genetic epidemiology of complex diseases. Hum Genet. 2020;139(1):73–84. doi: 10.1007/s00439-019-01996-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Ning Y., Zhang F. PCA-based GRS analysis enhances the effectiveness for genetic correlation detection. Brief Bioinform. 2019;20(6):2291–2298. doi: 10.1093/bib/bby075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabana N.A., Ashiq S., Ijaz A. Genetic risk score (GRS) constructed from polymorphisms in the PON1, IL-6, ITGB3, and ALDH2 genes is associated with the risk of coronary artery disease in Pakistani subjects. Lipids Health Dis. 2018;17(1):224. doi: 10.1186/s12944-018-0874-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bossini-Castillo L., Villanueva-Martin G., Kerick M. Genomic risk score impact on susceptibility to systemic sclerosis. Ann Rheum Dis. 2021;80(1):118–127. doi: 10.1136/annrheumdis-2020-218558. [DOI] [PubMed] [Google Scholar]
- 26.Cornelis M.C., Qi L., Zhang C. Joint effects of common genetic variants on the risk for type 2 diabetes in US men and women of European ancestry. Ann Intern Med. 2009;150(8):541–550. doi: 10.7326/0003-4819-150-8-200904210-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fawcett T. An introduction to ROC analysis. Recognit Lett. 2006;27(8):861–874. [Google Scholar]
- 28.Kim H.J., Park C.H., Kim H.J. Sequence variations and haplotypes of the GJB2 gene revealed by resequencing of 192 chromosomes from the general population in Korea. Clin Exp Otorhinolaryngol. 2010;3(2):65–69. doi: 10.3342/ceo.2010.3.2.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W.X., Huang Y., Yang X.L. The homozygote p.V27I/p.E114G variant of GJB2 is a putative indicator of nonsyndromic hearing loss in Chinese infants. Int J Pediatr Otorhinolaryngol. 2016;84:48–51. doi: 10.1016/j.ijporl.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 30.Wu C.C., Tsai C.H., Hung C.C. Newborn genetic screening for hearing impairment: a population-based longitudinal study. Genet Med. 2017;19(1):6–12. doi: 10.1038/gim.2016.66. [DOI] [PubMed] [Google Scholar]
- 31.Dai P., Yu F., Han B. GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med. 2009;7:26. doi: 10.1186/1479-5876-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oza A.M., DiStefano M.T., Hemphill S.E. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat. 2018;39(11):1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azaiez H., Booth K.T., Ephraim S.S. Genomic landscape and mutational signatures of deafness-associated genes. Am J Hum Genet. 2018;103(4):484–497. doi: 10.1016/j.ajhg.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildebrand M.S., DeLuca A.P., Taylor K.R. A contemporary review of AudioGene audioprofiling: a machine-based candidate gene prediction tool for autosomal dominant nonsyndromic hearing loss. Laryngoscope. 2009;119(11):2211–2215. doi: 10.1002/lary.20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor K.R., Deluca A.P., Shearer A.E. AudioGene: predicting hearing loss genotypes from phenotypes to guide genetic screening. Hum Mutat. 2013;34(4):539–545. doi: 10.1002/humu.22268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weininger O., Warnecke A., Lesinski-Schiedat A., Lenarz T., Stolle S. Computational analysis based on audioprofiles: a new possibility for patient stratification in office-based otology. Audiol Res. 2019;9(2):230. doi: 10.4081/audiores.2019.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y.S., Park H., Hong S.H., Chung W.H., Cho Y.S., Moon I.J. Predicting cochlear dead regions in patients with hearing loss through a machine learning-based approach: a preliminary study. PLoS One. 2019;14(6) doi: 10.1371/journal.pone.0217790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Ni Y., Liu Y. Screening of noise-induced hearing loss (NIHL)-associated SNPs and the assessment of its genetic susceptibility. Environ Health. 2019;18(1):30. doi: 10.1186/s12940-019-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi S.Y., Lee K.Y., Kim H.J. Functional evaluation of GJB2 variants in nonsyndromic hearing loss. Mol Med. 2011;17(5-6):550–556. doi: 10.2119/molmed.2010.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ambrosi C., Walker A.E., Depriest A.D. Analysis of trafficking, stability and function of human connexin 26 gap junction channels with deafness-causing mutations in the fourth transmembrane helix. PLoS One. 2013;8(8):e70916. doi: 10.1371/journal.pone.0070916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Los Campos G., Vazquez A.I., Hsu S., Lello L. Complex-trait prediction in the era of big data. Trends Genet. 2018;34(10):746–754. doi: 10.1016/j.tig.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.