Abstract
Background
Accurate predictions of distant metastasis (DM) in locally advanced rectal cancer (LARC) patients receiving neoadjuvant chemoradiotherapy (nCRT) are helpful in developing appropriate treatment plans. This study aimed to perform DM prediction through deep learning radiomics.
Methods
We retrospectively sampled 235 patients receiving nCRT with the minimum 36 months’ postoperative follow-up from three hospitals. Through transfer learning, a deep learning radiomic signature (DLRS) based on multiparametric magnetic resonance imaging (MRI) was constructed. A nomogram was established integrating deep MRI information and clinicopathologic factors for better prediction. Harrell's concordance index (C-index) and time-dependent receiver operating characteristic (ROC) were used as performance metrics. Furthermore, the risk of DM in patients with different response to nCRT was evaluated with the nomogram.
Findings
DLRS performed well in DM prediction, with a C-index of 0·747 and an area under curve (AUC) at three years of 0·894 in the validation cohort. The performance of nomogram was better, with a C-index of 0·775. In addition, the nomogram could stratify patients with different responses to nCRT into high- and low-risk groups of DM (P < 0·05).
Interpretation
MRI-based deep learning radiomics had potential in predicting the DM of LARC patients receiving nCRT and could help evaluate the risk of DM in patients who have different responses to nCRT.
Funding
The funding bodies that contributed to this study are listed in the Acknowledgements section.
Keywords: Locally advanced rectal cancer, Distant metastasis, Neoadjuvant chemoradiotherapy, Deep learning radiomics, Magnetic resonance imaging
Research in Context.
Evidence before this study
Neoadjuvant chemoradiotherapy before total mesorectal excision has no effect on the distant metastasis free survival of locally advanced rectal cancer patients. In view of the different therapeutic effects of neoadjuvant chemoradiotherapy, it is still controversial which patients should receive adjuvant chemotherapy to reduce the risk of distant metastasis. Previous studies have pointed out that clinicopathological factors and imaging information were related to the prognosis of locally advanced rectal cancer patients. In addition, deep learning radiomics shows good prospects in prognostic prediction. Thus, this study aimed to develop a deep learning radiomic model to accurately predict distant metastasis of locally advanced rectal cancer patients receiving neoadjuvant chemoradiotherapy, which can help clinicians formulate appropriate treatment plans.
Added value of this study
In this multicentre study, we first established a deep learning radiomic signature based on multiparametric magnetic resonance imaging. Tested by an external validation cohort, deep learning radiomic signature is a prognostic factor independent of other clinical characteristics. Subsequently, a nomogram combining deep MRI information with clinicopathologic factors was constructed. While achieving a better predictive performance, the nomogram can divide patients into high- and low-risk groups of distant metastasis, regardless of their responses to neoadjuvant chemoradiotherapy.
Implications of all the available evidence
By well predicting the distant metastasis of locally advanced rectal cancer patients receiving neoadjuvant chemoradiotherapy, the deep learning radiomic signature and the nomogram proposed in this study can be used as clinical diagnostic aids to achieve personalized treatment options.
Alt-text: Unlabelled box
1. Introduction
Most patients with locally advanced rectal cancer (LARC) receive a standard treatment including neoadjuvant chemoradiotherapy (nCRT) followed by total mesorectal excision (TME) [1]. Although nCRT before TME shows high effectiveness on local control, there is no effect on overall survival [2,3]. Since the main factor affecting survival is distant metastases (DM), the guidelines recommend adjuvant chemotherapy after TME to reduce the risk of DM [4,5]. However, there is still controversy on which patients should receive adjuvant chemotherapy. A pooled analysis [6] indicated that applying adjuvant chemotherapy for all LARC patients receiving nCRT and TME is a simple and pragmatic strategy, but it carries the risk of overtreatment. Patients with pathologic complete response (pCR) after nCRT may not benefit from subsequent treatment. A recent study has shown that patients with ypT0-2N0, the downstaging subgroup that have favorable oncological prognosis, will not benefit from postoperative adjuvant chemotherapy, either [7]. Contrarily, some studies suggested that adjuvant chemotherapy can improve overall survival of pCR patients by potentially eradicating residual micrometastatic disease [8,9].
Many researchers have conducted research on the prognosis of LARC and found that clinicopathological factors and medical imaging are both valuable for prognosis prediction [10], [11], [12], [13], [14], [15], [16]. Using clinicopathological factors, the severity of neural invasion has been proven to be a crucial prognostic factor in rectal cancer [10]. Furthermore, combining different clinicopathological factors to construct clinical nomograms could predict DM in patients with LARC [11,12]. On the other hand, among medical imaging techniques, non-invasive magnetic resonance imaging (MRI) is routinely used, which can provide a wealth of prognosis information for LARC. Tumor morphology information has been shown to be strongly associated with LARC prognosis [13], [14], [15]. Radiomics based on manually defined features could further extract high throughput information in MRI to predict DM in LARC [16]. However, the prognostic prediction models constructed by the above studies were for all LARC patients, regardless of whether the patients received nCRT. Given that nCRT is currently a common strategy for treating LARC patients, it is necessary to establish a model that focuses on the prediction of DM in patients receiving nCRT.
In previous studies, different methods have been used to mine prognostic information in medical images [17,18], among which deep learning radiomics has rapidly become a methodology of choice for analyzing medical images due to its unique advantages. Deep learning uses backpropagation algorithms to train machines in obtaining intricate structures in raw data [19]. The intricate structures extracted from medical images by deep learning are usually related to specific tasks, while handcrafted features lack this specificity. Early use of deep learning radiomics focused on lesions classification and detection and had at times achieved human expert-level performance [20,21]. Recent studies show that deep learning radiomics can also extract prognostic-related information from medical images and have good prospects in survival prediction tasks [22], [23], [24], involving multiple imaging modalities and application areas. For these reasons, deep learning radiomics may have potential in predicting DM in patients with LARC.
In this study, we developed a multiparametric-MRI signature based on deep learning radiomics for DM prediction in patients with LARC receiving nCRT and TME. For better prediction performance, an integrating nomogram combined deep MRI information and clinicopathologic factors was further constructed. The predictive performance of the signature and nomogram were tested on an external validation cohort. With the nomogram, we evaluated the risk of DM in patients with different responses to nCRT.
2. Methods
2.1. Ethics statement
The study was conducted in accordance with the guidelines of the Declaration of Helsinki, and was approved by the Ethics Committee of Yunnan Cancer Hospital (KY201824), The Sixth Affiliated Hospital of Sun Yat-sen University (E2019076) and Fudan University Shanghai Cancer Center (050432-4-1212B). Patient consent was waived due to the retrospective design of this study.
2.2. Patients
The imaging and clinical information of the patients in this multicentre study was collected from three different hospitals in China. A total of 235 consecutive patients with biopsy-proven rectal cancer between August 2012 and March 2015 were included. Locally advanced disease was determined based on pre-treatment CT of the chest and abdomen and pelvis MRI. All patients received complete treatment for nCRT before TME. Radiation therapy was administered over a period of five weeks, with a total dose of 45-50 Gy in 25-28 fractions. An Oral Capecitabine treatment was performed simultaneously with radiation therapy (dose 820 mg/m2, twice per day). The average time between nCRT and TME was 6-8 weeks, all patients achieved complete (R0) resection. In order to train the deep learning radiomic model and verify its robustness, we grouped 170 patients from two hospitals into a primary cohort and 65 patients from another hospital into an external validation cohort. Other details of patient recruitment and exclusion are shown in Fig. S1.
2.3. Imaging data acquisition and Processing
All MRI examinations were performed with endorectal coils, within one week before the patients’ colonoscopy. The imaging protocols included T2-weighted imaging (T2WI) and diffusion weighted imaging (DWI). From DWI images with b-values of 0 and 1000 s/mm2, apparent diffusion coefficient (ADC) maps were generated. Details regarding the MRI acquisition parameters are shown in Table S1.
In order to avoid interference between redundant information on MRI sequences, we used the ITK-SNAP software [25] to manually delineate regions of interest (ROI) on each lesion of T2W slices and DW slices (ROIs on ADC map slices were copied from the corresponding ROIs on DW slices with a b-value of 1,000 s/mm2). Other details of the ROI delineation are summarized in Supplementary Methods. The minimum bounding rectangle containing ROI information was extracted from each slice and resized to 112*112. This operation could make the ROIs fit the Convolutional Neural Network (CNN) structure and reduce the impact of different voxel spacing between different centers. In addition, all ROIs were normalized with z-score so that get a standard normal distribution of image intensities. Based on the resized and normalized ROIs, we generated T2W/ADC bounding boxes of size 112*112*20 for each patient. The bounding boxes were the final input to the deep learning radiomic model. If the bounding box contained less than 20 ROI patches, all-zero matrices of size 112*112 were added to it.
2.4. Deep learning radiomic signature construction and validation
While pre-trained deep learning models have been successful in computer vision and natural language processing tasks, transfer learning is an effective method for applying these pre-trained models to medical image analysis [26], In this study, we used the primary cohort to fine-tune ResNet18 [27] pre-trained on ImageNet. The channels of the T2W/ADC bounding boxes were reduced to three by 1*1 convolutions, so the boxes could fit to the input shape of the pre-trained model. Since the pre-trained model is more widely used in classification tasks, we used a two-step approach to predict DM. First, patients were divided into two groups according to whether DM occurred within three years after surgery (patients who were lost to follow-up within three years were excluded). Based on the binary classification problem, the deep learning radiomic models were trained on the T2W bounding boxes and ADC bounding boxes of 162 patients respectively, then tested on the external validation cohort of 62 patients. Through the trained models, the probability of each patient (including the censored patients) developing DM within three years after surgery was obtained. The probabilities predicted by T2W slices and ADC slices were called T2W-prob and ADC-prob, respectively. The sum of these two probabilities was defined as DL-prob (range from [0, 2]). Subsequently, we used DL-prob to build multiparametric MR radiomic signature (also called deep learning radiomic signature (DLRS) in this study) through the Cox proportional hazard model.
The optimal cut-off value of DLRS was calculated by the Youden Index of the primary cohort. Through time-dependent receiver operating characteristic (ROC) analysis, Kaplan-Meier (K-M) survival analysis and Harrell's concordance index (C-index), we evaluated the prognostic value of DLRS.
2.5. Individualized DM prediction nomogram construction and model performance evaluation
The combination of clinicopathologic factors with deep information of multiparametric MRI may further improve the model's predictive performance. We firstly used prognostic clinicopathologic factors to construct a clinical model; we then combined DL-prob and clinicopathologic factors to construct a combination prediction model in comparison. The models were constructed in the primary cohort based on a Cox regression analysis, and the combination model was converted into an individualized DM prediction nomogram [28].
Model performance evaluation methods included C-index, net reclassification improvement (NRI) and integrated discrimination improvement (IDI) [29]. Decision curve analysis (DCA) [30], clinical impact analysis and the ROC component analysis [31] were performed to further quantify the benefits of the nomogram in clinical applications.
2.6. Prognostic evaluation of patients with different responses to nCRT based on nomogram
In this study, the responses to nCRT were divided into good response and poor response. Specifically, patients with a good response to nCRT were defined as the pCR cohort and downstaging (ypT0-2N0) cohort. Patients with a poor response to nCRT were defined as the non-pCR cohort and non-downstaging cohort. Through subgroup analysis, we investigated whether patients with different DM risks could be stratified in these cohorts using the nomogram. More specifically, the Youden Index-based cut-off of the nomogram was used to divide the patients into high- and low- risk groups for DM. Then, the postoperative DMFS of patients in different risk groups were evaluated based on a K-M survival analysis.
2.7. Availability of data and materials
Due to the privacy of patients, the MRI data and clinical information related to patients cannot be available for public access but can be obtained from the corresponding author on reasonable request. The experiment and implementation details are described in detail in the Methods, which can be replicated with public libraries.
2.8. Statistical analysis
We used R software (version 3.4.0) and SPSS software (version 21) to perform all statistical analyses in this study. Expression of continuous variables was mean ± SD. The Mann-Whitney U test was used to compare differences between two groups of continuous variables. The Chi-square test or Fisher exact test was used to compare differences between two groups of categorical variables. The relationship between clinicopathologic factors or MRI information and DMFS was evaluated by univariate and multivariate Cox analyses. The difference between K-M curves was compared using the Log-Rank test. For all results of statistical analysis, P < 0·05 (two-sided tests) was considered significant.
2.9. Role of funding source
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
3. Results
3.1. Patient clinical characteristics
Clinical characteristics of patients in different cohorts are summarized in Table 1. Distant metastasis positivity within three years after surgery were 21·2% (36/170) and 29·2% (19/65) in the primary and validation cohorts. Among all 235 patients, there were 165 men (70·21%) and 70 women (29·79%), and the average age of all patients was 54·93 (SD, 11·16). The histologic variants of rectal adenocarcinoma included mucinous adenocarcinoma and signet ring cell carcinoma. Among them, there were three cases of mucinous adenocarcinoma and two cases of signet ring cell carcinoma in the primary cohort; four cases of signet ring cell carcinoma in the external validation cohort.
Table 1.
Clinical characteristics of patients in the primary and validation cohorts.
| Characteristic | Primary cohort (n=170) | Validation cohort (n=65) | P-value | ||
| Age(years, mean ± SD) | 54.82 ± 10.85 | 55.23 ± 12.01 | 0.602 | ||
| Gender (%) | 0.634 | ||||
| Male | 121 | 71.18% | 44 | 67.69% | |
| Female | 49 | 28.82% | 21 | 32.31% | |
| Tumor location (%) | <0.001 | ||||
| High | 4 | 2.35% | 2 | 3.08% | |
| Mid | 100 | 58.82% | 17 | 26.15% | |
| Low | 66 | 38.82% | 46 | 70.77% | |
| cT stage (%) | <0.001 | ||||
| cT3 | 130 | 76.47% | 29 | 44.62% | |
| cT4 | 40 | 23.53% | 36 | 55.38% | |
| Lymph node status (%) | <0.001 | ||||
| LN negative | 25 | 14.71% | 28 | 43.08% | |
| LN positive | 145 | 85.29% | 37 | 56.92% | |
| CEA (%) | 0.612 | ||||
| Normal | 130 | 76.47% | 47 | 72.31% | |
| Elevated | 40 | 23.53% | 18 | 27.69% | |
| Surgical approach (%) | |||||
| Laparotomy | 79 | 46.47% | 57 | 87.69% | <0.001 |
| Laparoscopy | 91 | 53.53% | 8 | 12.31% | |
| ypT stage (%) | 0.842 | ||||
| ypT0 | 24 | 14.12% | 7 | 10.77% | |
| ypT1 | 14 | 8.24% | 3 | 4.62% | |
| ypT2 | 26 | 15.29% | 11 | 16.92% | |
| ypT3 | 94 | 55.29% | 39 | 60.00% | |
| ypT4 | 12 | 7.06% | 5 | 7.69% | |
| ypN stage (%) | 0.021 | ||||
| ypN0 | 113 | 66.47% | 34 | 52.31% | |
| ypN1 | 35 | 20.59% | 25 | 38.46% | |
| ypN2 | 22 | 12.94% | 6 | 9.23% | |
| Adjuvant chemotherapy (%) | 0.060 | ||||
| Yes | 160 | 94.12% | 56 | 86.15% | |
| No | 10 | 5.88% | 9 | 13.85% | |
| Adjuvant radiotherapy (%) | 0.298 | ||||
| Yes | 6 | 3.53% | 5 | 7.69% | |
| No | 164 | 96.47% | 60 | 92.31% | |
| Distant metastasis (%) | 0.031 | ||||
| Yes | 42 | 24.71% | 26 | 40.00% | |
| No | 128 | 75.29% | 39 | 60.00% | |
| Locoregional recurrence (%) | 0.502 | ||||
| Yes | 7 | 4.12% | 4 | 6.15% | |
| No | 163 | 95.88% | 61 | 93.85% | |
Note: P-values were calculated by Mann-Whitney U test, Chi-square test or Fisher exact test.
3.2. Prognostic performance of DLRS
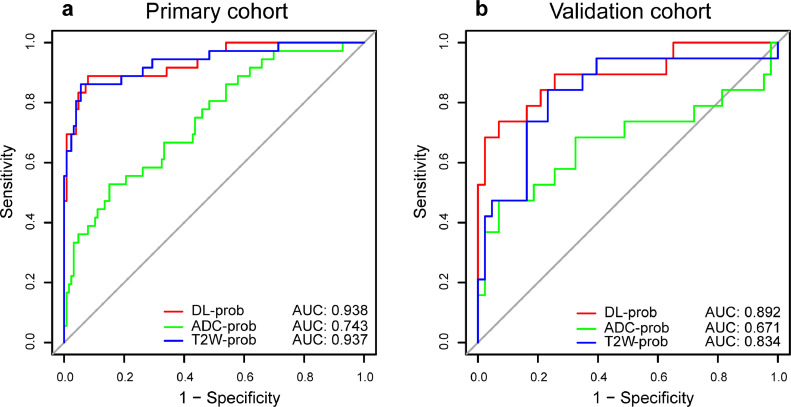
The trained deep learning radiomic models performed well on the classification task of whether DM occurred in LARC patients within three years after surgery (Fig. 1a-b). Among them, DL-prob had the best discrimination performance, with an AUC of 0·892 in the validation cohort. This indicated that the DL-prob was closely related to the prognosis of LARC patient and proved the feasibility of using it to construct a reliable prognostic signature. The effect of different sizes of bounding boxes and fine-tuned ROIs on the classification performance of the models is shown in Fig. S2.
Fig. 1.
ROC curves of prognostic performance with different deep learning models of the (a) primary cohort (n = 162) and (b) external validation cohort (n = 62). ROC receiver operating characteristic; AUC area under receiver operating characteristic curve; T2W T2-weighted; ADC apparent diffusion coefficient.
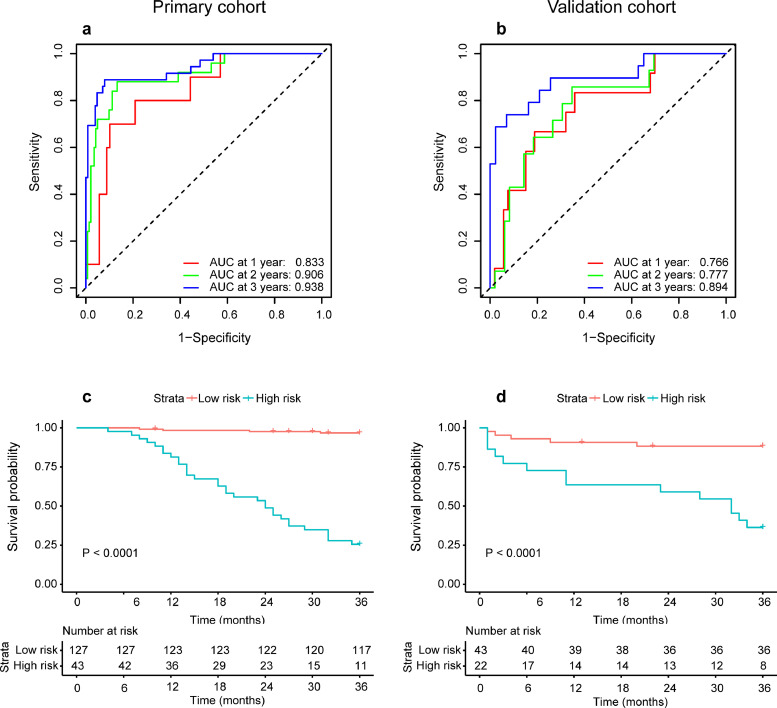
According to C-index and time-dependent ROC analysis in the validation cohort, signatures constructed based on different binary classification MR-prob had good prognostic predictive performance. Meanwhile, DLRS established by DL-prob had better performance compared to T2W/ADC radiomic signature (established by T2W-prob/ADC-prob). In the validation cohort, DLRS AUC of three-year DMFS was 0·894 (Fig. 2b), and the DLRS C-index of DMFS was 0·747 (95% CI, 0·665-0·830) (Table 2). The time-dependent ROC curves of T2W radiomic signature and ADC radiomic signature are shown in Fig. S3. According to DLRS, the patients were further divided into high- and low-risk groups for DM. The optimal cut-off score for DLRS was set to 1·784 based on the primary cohort. The distribution of clinicopathologic characteristics according to high and low DLRS are shown in Table S2. Through the K-M survival analysis, the stratification effect of DLRS was significant in both the primary and the validation cohort (all P < 0·001, Log-Rank test) (Fig. 2c-d).
Fig. 2.
Predictive performance of DLRS for DMFS. (a) and (b) are time-dependent ROC curves for one year, two years and three years of the primary cohort (n = 170) and external validation cohort (n = 65). (c) and (d) are K-M curves for stratifying high- and low-risk patients of DM of the primary cohort (P < 0·0001, log-rank test) and external validation cohort (P < 0·0001, log-rank test). The numbers of patients at risk for each time step are shown in the bottom. DLRS deep learning risk signature; DMFS distant metastasis free survival; ROC receiver operating characteristic; AUC area under receiver operating characteristic curve.
Table 2.
Model performances in the primary and validation cohorts.
|
Primary cohort |
Validation cohort |
|||||
| Model | C-index | 95%CI | C-index | 95%CI | ||
| Nomogram | 0.865 | 0.814-0.916 | 0.775 | 0.695-0.856 | ||
| Clinical | 0.714 | 0.632-0.797 | 0.601 | 0.487-0.732 | ||
| DLRS | 0.851 | 0.795-0.906 | 0.747 | 0.665-0.830 | ||
| T2W | 0.854 | 0.802-0.905 | 0.729 | 0.640-0.819 | ||
| ADC | 0.688 | 0.615-0.761 | 0.599 | 0.473-0.724 | ||
| NRI | 95%CI | P-value | NRI | 95%CI | P-value | |
| Nomogram vs. Clinical | 0.599 | 0.501-0.697 | <0.001* | 0.395 | 0.195-0.595 | <0.001* |
| DLRS vs. Clinical | 0.529 | 0.406-0.651 | <0.001* | 0.286 | 0.089-0.483 | 0.004* |
| IDI | 95%CI | P-value | IDI | 95%CI | P-value | |
| Nomogram vs. Clinical | 0.409 | 0.310-0.508 | <0.001* | 0.260 | 0.117-0.402 | < 0.001* |
| DLRS vs. Clinical | 0.384 | 0.275-0.493 | <0.001* | 0.187 | 0.023-0.352 | 0.026* |
Note: *P-value < 0.05, P-values were calculated by NRI test and IDI test.
Abbreviations: DLRS, deep learning radiomic signature; T2W, T2-weighted; ADC, apparent diffusion coefficient.
3.3. Combination model construction and individualized nomogram prognostic performance evaluation
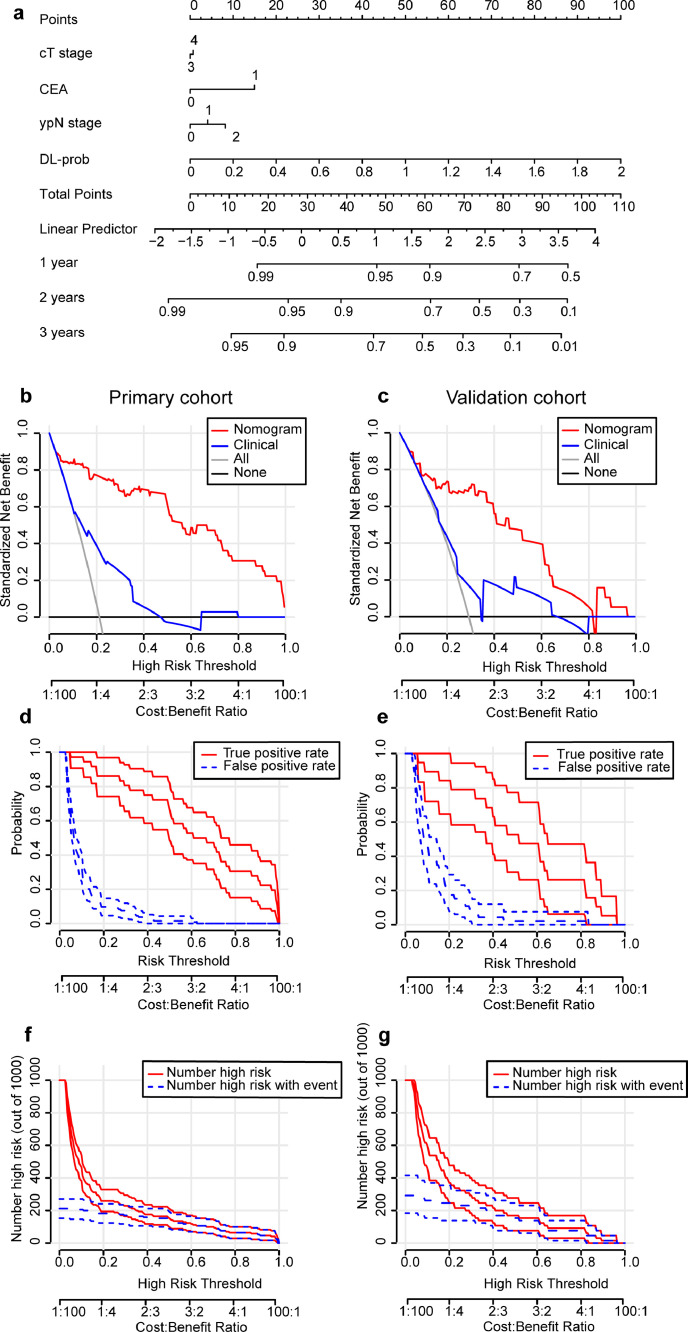
Based on univariate and multivariable Cox analyses, the clinical model was established using cT stage, CEA and ypN stage (Table S3). These three clinicopathological factors had a significant prognostic value for DMFS (P < 0·05, Likelihood ratio test). Then we combined DL-prob with the above clinicopathological factors to establish a combination model. Through a multivariate Cox analysis, DL-prob showed its great prognostic value (HR 14·387, 95% CI, 7·428-27·864; P < 0·001, Likelihood ratio test). The nomogram converted from the combination model is shown in Fig. 3a.
Fig. 3.
Integrated nomogram and evaluation of the nomogram in multi centers. (a) is a nomogram for individual prediction of DMFS combined with deep MRI information and clinicopathological factors. (b) and (c) are the decision curves of integrated nomogram/clinical model of the primary cohort (n = 170) and external validation cohort (n = 65). (d) and (e) are the plots of true- and false-positive rates of the primary cohort and external validation cohort, as functions of the risk threshold for integrated nomogram. (f) and (g) are clinical impact curves for 1000 random patients based on the integrated nomogram of the primary cohort and external validation cohort. 95% confidence intervals constructed via bootstrapping is displayed on both sides of the ROC components plot or clinical impact plot. ROC receiver operating characteristic; DMFS distant metastasis free survival; MRI magnetic resonance imaging.
Compared with other models, the model based on nomogram discrimination showed significantly improved performance. The C-index of the nomogram was the highest in the validation cohort (Table 2), which was 0·775 (95% CI, 0·695-0·856) compared with the DLRS C-index of 0·747 (95% CI, 0·665-0·830 P = 0·026, Student t test) and the clinical model C-index of 0·601 (95% CI, 0·487-0·732 P = 0·008, Student t test). The calculated NRI and IDI further proved that the prognostic predictive performance of nomogram and DLRS was better than the clinical model. According to the quantitative results, the nomogram improved the predictive performance more significantly (Table 2). The standardized net benefit (sNB) was used as a performance metric in the decision curve, and the application of the nomogram showed excellent benefits in the relevant threshold range (Fig. 3b-c). In order to evaluate the predictive value of nomogram in further clinical applications, we provided ROC components plot and clinical impact plot as supplements (Fig. 3d-g).
3.4. Prognostic evaluation of patients with different responses to nCRT based on nomogram
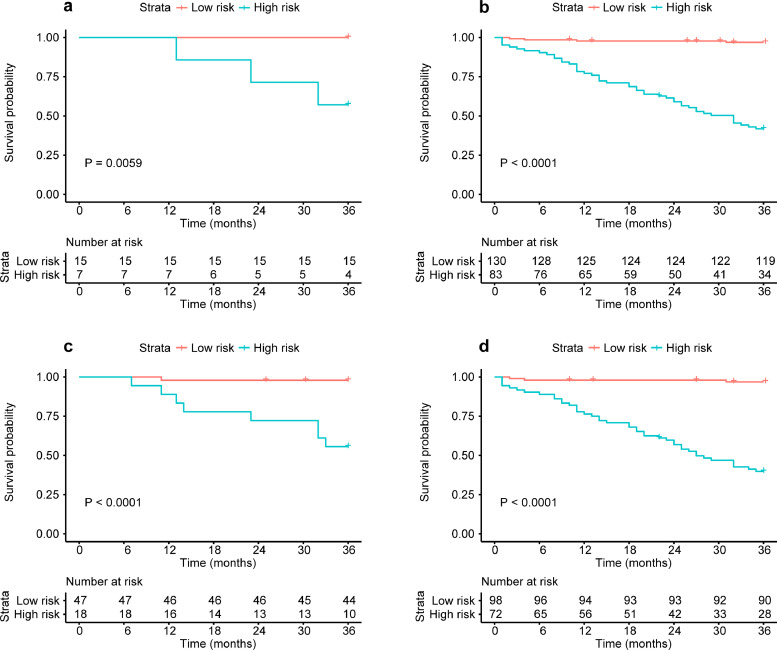
Through further subgroup analysis based on the nomogram, high- and low-risk patients of DM in the pCR cohort (P = 0·006, Log-Rank test), non-pCR cohort (P < 0·001, Log-Rank test), downstaging cohort (P < 0·001, Log-Rank test) and non-downstaging cohort (P < 0·001, Log-Rank test) could all be significantly stratified (Fig. 4a-d). In this study, 22/65 patients achieved a pCR/downstaging after nCRT, of which 7/18 were defined as high-risk for DM through nomogram. Meanwhile, there were 213/170 patients with non-pCR/non-downstaging, of which 130/98 were defined as low-risk for DM through nomogram. The results of subgroup analysis of other clinicopathologic characteristics (including cT stage, lymph node status, ypT stage and ypN stage) are shown in Fig. S4.
Fig. 4.
Nomogram-based K-M curves of patients with different responses to nCRT. (a) and (b) are the K-M DMFS curves for pCR (P = 0•0059, log-rank test) and non-pCR (P < 0•0001, log-rank test) patient subgroup. (c) and (d) are the K-M DMFS curves for downstaging (ypT0-2N0) (P < 0•0001, log-rank test) and non-downstaging (P < 0•0001, log-rank test) patient subgroup. The numbers of patients at risk for each time step are shown in the bottom. nCRT neoadjuvant chemoradiotherapy; DMFS distant metastasis free survival; pCR pathologic complete response.
4. Discussion
Accurate prognosis prediction is very beneficial for the choice of treatment and risk stratification for cancer patients. In this multicentre study, we developed and validated a deep learning radiomics-based multiparametric-MRI signature to predict DMFS in patients with LARC receiving nCRT and TME. As a risk factor independent of clinicopathological factors (such as TNM stage), DLRS performed well in DM prediction and high/low risk stratification of patients, which demonstrated the value of MRI-based deep learning radiomics in prognosis prediction. Furthermore, combining DL-prob extracted from multiparametric MRI with clinicopathological factors to construct a nomogram could achieve better prognostic prediction effect compared to only using image information or clinical factors.
For LARC patients with different response to nCRT, the choice of subsequent treatment options has always been controversial [[6], [7], [8], [9],11]. If all patients are treated with postoperative adjuvant chemotherapy, the risk of overtreatment will increase; if patients with good response are considered to be unable to benefit from adjuvant chemotherapy and receive no follow-up treatment, the DMFS of patients with poor prognosis cannot be guaranteed. In this study, our nomogram can serve as a prognostic prediction tool to well stratify the high- and low-risk patients of DM regardless of their response to nCRT, thereby promoting more precise treatment options. In the good response cohort, the proportions of patients classified as high-risk for DM by the nomogram were 31·8% (7/22, pCR patients) and 27·7% (18/65, downstaging patients). In the poor response cohort, the proportions of patients classified as low-risk for DM by the nomogram were 61·0% (130/213, non-pCR patients) and 57·6% (98/170, non-downstaging patients). The above findings suggested that the risk of DM may not be judged simply based on the patient's response to nCRT. In the good response cohort, the high-risk patients in the pCR/downstaging cohorts have a relatively high probability of 42·9% (3/7) and 44·4% (8/18) of DM within three years. It is inappropriate to give up postoperative adjuvant therapy for these high-risk patients. It is worth noting that in the cohort of patients with pCR, all patients classified as low-risk according to nomogram did not develop distant metastases within three years after surgery. The above findings indicate that in future clinical applications, if patients with good response are defined as low-risk for DM by the nomogram, then close observation can be used to replace adjuvant therapy for their treatment strategy. If they are defined as high-risk for DM, the decision not to offer adjuvant therapy should be made with caution.
In this study, the information of multiparametric-MR scans was extracted by deep learning radiomics. Previous studies have shown that according to machine learning methods, corresponding features can be extracted from MR scans and used to predict the prognosis of patients with LARC receiving neoadjuvant therapy (One example is the prediction of therapeutic responses [32], [33], [34]). Tumors are heterogeneous both on genetic and histopathological levels, which is reflected in intratumoral spatial variation [35]. Due to intrinsic aggressive biology or treatment resistance, tumors with high heterogeneity show poor prognosis [36]. Radiomics noninvasively captures risk-related intratumoral and intertumoral heterogeneity in voxels, identifies radiographic phenotypes, and provides additional prognostic information. A previous study pointed out that survival-related imaging features were also associated with cellular events such as angiogenesis and peritumoral infiltration [37]. In addition, the identification of tumor subtypes of distinct survival through radiomics analysis [38] and the investigation of the correlation between radiomics features and dysregulated signaling pathways [39] have provided a potential biological basis for the difference in prognostic outcomes. However, these methods were limited by the predefined features designed by domain experts and may ignore image features related to specific tasks. For the survival prediction task of this article, we defined patients as high- and low-risk groups according to the time point of distant metastasis after surgery and trained our CNN accordingly. Our CNN was based on the residual structure, which can solve the problem of gradient disappearance [27]. Meanwhile, making the data set contain complete tumor information, the use of early stopping and proper dropout can effectively improve the robustness of the model. Then we used the trained CNN to obtain binary classification MR-prob from T2W and ADC slices, and these probabilities were considered to be highly correlated with prognosis. In a previous study, the radiomic features of tissue were particularly sensitive to different scan parameters, and the features related to tumor showed lower sensitive to the scanner manufacturer and magnetic field [40]. In this study, we found that after proper preprocessing, MR images performed with different scanners will not have a significant influence on the performance of the deep learning radiomic model. (Fig. S5; all P > 0·05, Delong test). Grad-CAM based visualization of network prediction and Spearman correlation heat map between MR-prob and clinicopathological factors are shown in Fig. S6. Through T2W-prob, ADC-prob and DL-prob, we established prognostic models and obtained radiomic signatures for single-modality and multiparametric MR scans. We further found through different evaluation methods that the multiparametric MR radiomic signature had the best prognostic performance among all MR signatures. This indicated that different types of medical images can reflect tumor information from different views and can complement each other. The combination of medical images can provide better temporal and spatial matching of tumor volume in different modalities. Combining the radiomic features extracted from PET-CT and MRI for efficacy evaluation and prognosis prediction has been proposed previously [41], [42], [43]. From a methodological perspective, the DL model has great potential to combine different imaging information for prognostic prediction, because the captured information is usually related to a specific task. However, due to the design of this study was retrospective and the imaging data was from different centers, it is difficult to construct a model by combining MRI and other types of imaging. We would like to explore it in the future prospective study.
It has become a consensus that clinicopathological characteristics are closely related to the prognosis of cancer patients. For example, TNM staging and the severity of neural invasion, etc. have all been confirmed to be related to the prognosis of LARC patients [10,11]. Although the N staging in MRI may sometimes be underestimated. Some studies have demonstrated that combining imaging features and clinicopathological risk factors can improve the predictive performance of a prognostic model [44,45]. In this study, we drew the same conclusion by combining several prognosis-related clinicopathological factors and multiparametric-MR scan information to establish a prognostic model. When comparing it with the clinical model through NRI and IDI, the improvement in prediction accuracy was significant (all P < 0·001, NRI test, IDI test).
There are some limitations to this study. First, since our study involved cohorts from multiple institutions, there were differences in some clinicopathologic factors among patients in different cohorts. Secondly, the number of cases included in the study was limited, and the patient population in this study was from China. Therefore, although tests have been conducted in different centers, further large-scale international validation is needed for the application of the model to patients of other ethnicities. Finally, information contained in pathological images and genomes may be helpful in prognostic prediction tasks, which suggests that the larger well-designed prospective studies with multiple information should be done in the future.
In conclusion, we designed a multiparametric MR signature based on deep learning radiomics to predict the DM of LARC patients receiving nCRT and TME. In addition, we combined deep multiparametric MR information and clinicopathological factors to construct a nomogram for further clinical applications and better predictive performance. Both DLRS and nomogram showed good DM predictive performance. With the nomogram, we can evaluate the risk of DM in patients who have different responses to nCRT.
Contributors
Conception and design: Zhenyu Liu, Tong Tong, Xiaochun Meng and Jie Tian;
Collection and assembly of data: Dafu Zhang, Zhenhui Li, Peiyi Xie, Yingying Ding, Weixing Dai and Guoxiang Cai;
Verification of the underlying data: Xiangyu Liu, Zhenyu Liu and Kai Sun;
Development of methodology: Xiangyu Liu, Dafu Zhang, Wei Wei, Kai Sun and Zhenchao Tang;
Data analysis and interpretation: Xiangyu Liu, Zhenhui Li, Dafu Zhang, Zhenyu Liu and Kai Sun;
Writing – original draft: Xiangyu Liu, Zhenyu Liu, Zhenhui Li and Jie Tian;
Final approval of manuscript: All authors.
Declaration of Competing Interest
The authors have declared that no competing interest exists.
Acknowledgments
Acknowledgements
This paper is supported by the National Key R&D Program of China under Grant Nos. 2017YFA0205200, 2017YFA0700401, the National Natural Science Foundation of China under Grant Nos. 81922040, 92059103, 81930053, and 82001914, Chinese Academy of Sciences under Grant No. QYZDJ-SSW-JSC005, KFJ-STS-ZDTP-059. The Youth Innovation Promotion Association CAS (grant number 2019136). The Project of High-Level Talents Team Introduction in Zhuhai City (Zhuhai HLHPTP201703). Guangdong Natural Science Foundation (No.2021A1515011795). The authors would like to acknowledge the instrumental and technical support of Multi-modal biomedical imaging experimental platform, Institute of Automation, Chinese Academy of Sciences.
Data sharing statement
Due to the privacy of patients, the related data cannot be available for public access but can be obtained from Tong Tong (t983352@126.com), Xiaochun Meng (mengxch3@mail.sysu.edu.cn) and Jie Tian (jie.tian@ia.ac.cn) upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103442.
Contributor Information
Tong Tong, Email: t983352@126.com.
Xiaochun Meng, Email: mengxch3@mail.sysu.edu.cn.
Jie Tian, Email: jie.tian@ia.ac.cn.
Appendix. Supplementary materials
References
- 1.Li Y, Wang J, Ma X. A review of neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Int J Biol Sci. 2016;12:1022. doi: 10.7150/ijbs.15438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer R, Liersch T, Merkel S. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol. 2012;30:1926–1933. doi: 10.1200/JCO.2011.40.1836. [DOI] [PubMed] [Google Scholar]
- 3.Peeters KCMJ, Marijnen CAM, Nagtegaal ID. The TME trial after a median follow-up of 6 years: increased local control but no survival benefit in irradiated patients with resectable rectal carcinoma. Ann Surg. 2007;246:693–701. doi: 10.1097/01.sla.0000257358.56863.ce. [DOI] [PubMed] [Google Scholar]
- 4.Glynne-Jones R, Wyrwicz L, Tiret E. Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28:22–40. doi: 10.1093/annonc/mdx224. iv. [DOI] [PubMed] [Google Scholar]
- 5.Benson AB, Venook AP, Al-Hawary MM. Rectal cancer, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2018;16:874–901. doi: 10.6004/jnccn.2018.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maas M, Nelemans PJ, Valentini V. Adjuvant chemotherapy in rectal cancer: defining subgroups who may benefit after neoadjuvant chemoradiation and resection: a pooled analysis of 3,313 patients. Int J Cancer. 2015;137:212–220. doi: 10.1002/ijc.29355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Huang Y, Sun G. Rectal cancer patients with downstaging after neoadjuvant chemoradiotherapy and radical resection do not benefit from adjuvant chemotherapy. Ann Transl Med. 2020;8 doi: 10.21037/atm-20-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dossa F, Acuna SA, Rickles AS. Association between adjuvant chemotherapy and overall survival in patients with rectal cancer and pathological complete response after neoadjuvant chemotherapy and resection. JAMA Oncol. 2018;4:930–937. doi: 10.1001/jamaoncol.2017.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polanco PM, Mokdad AA, Zhu H, Choti MA, Huerta S. Association of adjuvant chemotherapy with overall survival in patients with rectal cancer and pathologic complete response following neoadjuvant chemotherapy and resection. JAMA Oncol. 2018;4:938–943. doi: 10.1001/jamaoncol.2018.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceyhan GO, Liebl F, Maak M. The severity of neural invasion is a crucial prognostic factor in rectal cancer independent of neoadjuvant radiochemotherapy. Ann Surg. 2010;252:797–804. doi: 10.1097/SLA.0b013e3181fcab8d. [DOI] [PubMed] [Google Scholar]
- 11.Valentini V, Van Stiphout RGPM, Lammering G. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol. 2011;29:3163–3172. doi: 10.1200/JCO.2010.33.1595. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Lin H, Lu X. A nomogram to predict distant metastasis after neoadjuvant chemoradiotherapy and radical surgery in patients with locally advanced rectal cancer. J Surg Oncol. 2017;115:462–469. doi: 10.1002/jso.24522. [DOI] [PubMed] [Google Scholar]
- 13.Dresen RC, Beets GL, Rutten HJT. Locally advanced rectal cancer: MR imaging for restaging after neoadjuvant radiation therapy with concomitant chemotherapy part I. Are we able to predict tumor confined to the rectal wall? Radiology. 2009;252:71–80. doi: 10.1148/radiol.2521081200. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Lee JM, Hong SH. Locally advanced rectal cancer: added value of diffusion-weighted MR imaging in the evaluation of tumor response to neoadjuvant chemo-and radiation therapy. Radiology. 2009;253:116–125. doi: 10.1148/radiol.2532090027. [DOI] [PubMed] [Google Scholar]
- 15.Patel UB, Taylor F, Blomqvist L. Magnetic resonance imaging–detected tumor response for locally advanced rectal cancer predicts survival outcomes: mercury experience. J Clin Oncol. 2011;29:3753–3760. doi: 10.1200/JCO.2011.34.9068. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Meng X, Zhang H. Predicting distant metastasis and chemotherapy benefit in locally advanced rectal cancer. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambin P, Rios-Velazquez E, Leijenaar R. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litjens G, Kooi T, Bejnordi BE. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 19.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 20.Esteva A, Kuprel B, Novoa RA. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulshan V, Peng L, Coram M. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Jin C, Yu H. Development and validation of a deep learning CT signature to predict survival and chemotherapy benefit in gastric cancer: a multicenter, retrospective study. Ann Surg. 2020 doi: 10.1097/SLA.0000000000003778. [DOI] [PubMed] [Google Scholar]
- 23.Kather JN, Krisam J, Charoentong P. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 2019;16:1–22. doi: 10.1371/journal.pmed.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosny A, Parmar C, Coroller TP. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15 doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yushkevich PA, Piven J, Hazlett HC. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Shin H-C, Roth HR, Gao M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He K, Zhang X, Ren S, Sun J. Proc IEEE Conf Comput Vis pattern Recognit. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 28.Grimes DA. The nomogram epidemic: resurgence of a medical relic. Ann Intern Med. 2008;149:273–275. doi: 10.7326/0003-4819-149-4-200808190-00010. [DOI] [PubMed] [Google Scholar]
- 29.Pencina MJ, D'Agostino Sr RB, D'Agostino Jr RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 30.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerr KF, Brown MD, Zhu K, Janes H. Assessing the clinical impact of risk prediction models with decision curves: guidance for correct interpretation and appropriate use. J Clin Oncol. 2016;34:2534. doi: 10.1200/JCO.2015.65.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Zhang XY, Shi YJ. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res. 2017;23:7253–7262. doi: 10.1158/1078-0432.CCR-17-1038. [DOI] [PubMed] [Google Scholar]
- 33.Tang Z, Zhang X-Y, Liu Z. Quantitative analysis of diffusion weighted imaging to predict pathological good response to neoadjuvant chemoradiation for locally advanced rectal cancer. Radiother Oncol. 2019;132:100–108. doi: 10.1016/j.radonc.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Yi Y, Liu Z. Radiomics-based pretherapeutic prediction of non-response to neoadjuvant therapy in locally advanced rectal cancer. Ann Surg Oncol. 2019;26:1676–1684. doi: 10.1245/s10434-019-07300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davnall F, Yip CSP, Ljungqvist G. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Höckel M, Knoop C, Schlenger K. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiother Oncol. 1993;26:45–50. doi: 10.1016/0167-8140(93)90025-4. [DOI] [PubMed] [Google Scholar]
- 37.Macyszyn L, Akbari H, Pisapia JM. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol. 2015;18:417–425. doi: 10.1093/neuonc/nov127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Cui Y, Sun X. Unsupervised clustering of quantitative image phenotypes reveals breast cancer subtypes with distinct prognoses and molecular pathways. Clin Cancer Res. 2017;23:3334–3342. doi: 10.1158/1078-0432.CCR-16-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J, Zhang S, Li KK-W. Incremental prognostic value and underlying biological pathways of radiomics patterns in medulloblastoma. EBioMed. 2020;61 doi: 10.1016/j.ebiom.2020.103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha A, Yu X, Sahoo D, Mazurowski MA. Effects of MRI scanner parameters on breast cancer radiomics. Expert Syst Appl. 2017;87:384–391. doi: 10.1016/j.eswa.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giannini V, Mazzetti S, Bertotto I. Predicting locally advanced rectal cancer response to neoadjuvant therapy with 18 F-FDG PET and MRI radiomics features. Eur J Nucl Med Mol Imaging. 2019;46:878–888. doi: 10.1007/s00259-018-4250-6. [DOI] [PubMed] [Google Scholar]
- 42.Lucia F, Visvikis D, Desseroit M-C. Prediction of outcome using pretreatment 18 F-FDG PET/CT and MRI radiomics in locally advanced cervical cancer treated with chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2018;45:768–786. doi: 10.1007/s00259-017-3898-7. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Franc BL, Harnish RJ. Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer. 2018;4:1–13. doi: 10.1038/s41523-018-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Liang C, He L. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34:2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 45.Dinapoli N, Barbaro B, Gatta R. Magnetic resonance, vendor-independent, intensity histogram analysis predicting pathologic complete response after radiochemotherapy of rectal cancer. Int J Radiat Oncol Biol Phys. 2018;102:765–774. doi: 10.1016/j.ijrobp.2018.04.065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to the privacy of patients, the MRI data and clinical information related to patients cannot be available for public access but can be obtained from the corresponding author on reasonable request. The experiment and implementation details are described in detail in the Methods, which can be replicated with public libraries.