Abstract
Background
Metabolic syndrome (MetS), a cluster of factors associated with risks of developing cardiovascular diseases, is a public health concern because of its growing prevalence. Considering the combination of concomitant components, their development and severity, MetS phenotypes are largely heterogeneous, inducing disparity in diagnosis.
Methods
A case/control study was designed within the NuAge longitudinal cohort on aging. From a 3-year follow-up of 123 stable individuals, we present a deep phenotyping approach based on a multiplatform metabolomics and lipidomics untargeted strategy to better characterize metabolic perturbations in MetS and define a comprehensive MetS signature stable over time in older men.
Findings
We characterize significant changes associated with MetS, involving modulations of 476 metabolites and lipids, and representing 16% of the detected serum metabolome/lipidome. These results revealed a systemic alteration of metabolism, involving various metabolic pathways (urea cycle, amino-acid, sphingo- and glycerophospholipid, and sugar metabolisms…) not only intrinsically interrelated, but also reflecting environmental factors (nutrition, microbiota, physical activity…).
Interpretation
These findings allowed identifying a comprehensive MetS signature, reduced to 26 metabolites for future translation into clinical applications for better diagnosing MetS.
Keywords: Metabolic syndrome, Metabolomics, Deep phenotyping, Lipidomics, Metabolic signature
Research in context.
Evidence before this study
Prior association studies linking metabolites and lipids with MetS (i) have been limited in terms of molecular species profiled, (ii) lacked of considering the interaction between metabolisms as well as with extrinsic factors, and (iii) were very rarely issued from longitudinal studies.
Added value of this study
Our deep phenotyping approach, along with a 3-year follow-up design, provides robust and integrated insights into MetS mechanisms and proposes new candidate biomarkers within an optimized statistically, analytically and biologically refined associated molecular signature.
Implications of all the available evidence
These findings highlight the interest of a comprehensive molecular signature as marker of MetS, that should be validated for future translation into clinical applications for better diagnosing MetS.
Alt-text: Unlabelled box
1. Introduction
The metabolic syndrome (MetS), defined as a cluster of risk factors for cardiovascular disease (CVD), has been recognized for decades with a rising prevalence worldwide [1]. The main culprits of this rise are the aging of the population and the complex interactions between lifestyle factors such as unhealthy dietary habits and sedentarity, leading to overweight and obesity [[2], [3], [4]]. Because several clinical definitions co-exist [5] among health organizations (e.g. National Cholesterol Education Program (NCEP), International Diabetes Federation (IDF), World Health Organization (WHO)), the true prevalence of MetS is difficult to reliably establish. However, MetS comprises elevated blood pressure, dyslipidemia, including hypertriglyceridemia and reduced blood levels of high-density lipoprotein cholesterol (HDL-C), fasting hyperglycemia, and central adiposity. It is now accepted that it represents a global public health concern with a worldwide prevalence reaching one third of US adults having MetS and over 45% by the age of 60 [1,6]. There is also a consensus regarding the presence of multiple metabolic risk factors for CVD and type 2 diabetes (T2D) [7,8]. Moreover, considering the combination of concomitant components, and their development and severity profile, patients identified with MetS are largely heterogeneous, inducing a disparity in the diagnosis and therapeutic approach [9]. A better characterization of pathophysiological alterations associated with MetS could therefore contribute to improve diagnosis and better syndrome delineation.
In this context, metabolomics and lipidomics have emerged over the last decade as powerful tools for the analysis of phenotypes, providing key insights into modified metabolic pathways and better understanding of pathophysiological processes [10,11]. Indeed, metabolic profiles allow getting an integrated view of metabolism because of a sensitive detection of molecular changes over time, resulting from the interaction between intrinsic and extrinsic factors. Metabolites, used as single targets or in combination within a comprehensive signature, are thus promising biomarkers to reveal metabolic dysfunctions. Metabolomics has therefore been widely applied for metabolic disease phenotyping and candidate biomarker discovery as well as pathophysiological exploration of underlying mechanisms [12,13]. However, even if studies on T2D have been among the main drivers in this chronic metabolic disease research field using these global approaches for biomarker research, few studies focussed on MetS and often consisted in targeted approaches with a restricted number of detected metabolites [14]. Consequently, an integrated vision of metabolic derangements is lacking along with a limited capacity of study comparisons [15].
In the present study, a 3-year follow-up design of stable subjects within an observational longitudinal cohort, as well as a deep phenotyping approach based on a multiplatform strategy involving metabolomics and lipidomics untargeted methods, were set up, with the objective to better characterize metabolic perturbations in MetS and define a comprehensive MetS signature stable over time in older men.
2. Materials
2.1. The NuAge cohort and subject selection
The present study was designed within the 5-year observational Quebec Longitudinal Study on Nutrition and Successful Aging (NuAge). The cohort was constituted of 1793 men and women in good general health, selected from three age groups (68–72, 73–77, 78–82) at recruitment. French or English-speaking community-dwelling participants were committed to give fasting blood, undergo several direct measures annually, and to answer questionnaires related to food and health biannually. The NuAge database comprises large qualitative and quantitative data related to anthropometry/body composition, nutrition/dietary intakes, numerous markers of physical, clinical and cognitive status, physical activity, functional autonomy and social functioning. Methodological description of measures, questionnaires and blood test, processing and storing have been described in Gaudreau et al. [16].
2.1.1. Ethical approval
All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the NuAge study. The NuAge Study has been approved by the Research Ethics Board (REB) of both the Geriatric University Institutes of Montreal and Sherbrooke Research Centers. The management framework of the actual NuAge Database and Biobank has been approved by the REB of the CIUSSS-de-l'Estrie-CHUS (protocol #2019-2832).
2.1.2. Subject selection
A case/control study on MetS was designed within the NuAge cohort, with serum samples collected at two time points (recruitment 2003–2005 (T1) and 3 years later (T4)), with the objective to identify a metabolic signature of MetS, stable over time using a multiplatform lipidomic/metabolomics approach. In this context, an optimized subject selection strategy was developed. Briefly, the selection was based on the presence and number of MetS criteria, and their stability over the three years. It was performed among the 853 males as it has been recognized that in the province of Quebec, men have more risk factors of MetS than women [[17], [18], [19], [20]]. MetS was defined using the following criteria - thresholds defined for men [5,21]: elevated waist circumference (≥ 102 cm, WC); high blood pressure (systolic > 130 mmHg and/or diastolic > 85 mmHg) or antihypertensive drug treatment with history of hypertension, elevated fasting blood glucose (≥ 5.7 mM) or drug treatment for hyperglycemia (oral hypoglycemic, insulin); high circulating triglyceride levels (≥ 1.7 mM) or drug treatment (fibrates, nicotinic acid); and reduced-HDL-cholesterol (< 1.0 mM) or drug treatment (fibrates, nicotinic acid). Regarding the study objectives, only stable subjects over time were included. Using these five criteria, subjects with unstable (changing status) or uncertain MetS status (due to missing values) over time were then excluded. Cases were defined as having three or more of the MetS criteria, while controls were defined as having less than three MetS criteria at each time point. It resulted in identifying 61 incident cases and 88 controls. Concerning control individuals, it was important to exclude extreme subjects that could generate false negative results. Therefore, in agreement with clinicians, controls with seven or more drug treatments were excluded [22]. Moreover, value outliers were analyzed. Because no time effect was observed for the quantitative variables defining MetS, individuals with mean extreme values for MetS biological variables over time, outside the range defined by the mean (T1 to T4) ± 1.5 interquartile range (IQR) were excluded. Finally, this strategy ended up selecting 61 cases and 62 controls. Because it is known that metabolomic profiles are modified by age, it was checked that there was no significant age difference between cases and controls to avoid a potential bias. To do so, three experimental classes were defined according to the age distribution (67–72 years old (n = 25 vs 22), 73–77 years old (n = 22 vs 24), 78–84 years old (n = 15 vs 15)), and the size balance between age class in both groups was checked using Fisher's Exact Test.
2.1.3. Epidemiological data
Fifty-eight quantitative variables evaluated at T1 and T4 were considered to precisely describe the selected population: 23 biochemical parameters, 8 clinical variables, 25 nutritional data and finally 2 scores related to physical activity (Physical Activity Scale for the Elderly (PASE) questionnaire; [23]) and health-related quality of life (using physical (PCS) component summary score derived from the Medical Outcome Study 36-item Short Form Health Survey [SF-36] questionnaire; [24,25]). In particular, nutritional data consisted in intake data obtained from the mean of two to three non-consecutive 24 h dietary recalls (24-HR) [26], as well as in a validated Canadian global dietary quality index, the Canadian Healthy Eating Index (C-HEI) [27]. This index is based on intake of four food groups: grain products, fruits and vegetables, milk products, meat and alternatives, and five other items: % of energy as total fat intake and saturated fat intake, cholesterol, salt and diet variety. The total score ranges from 0 to 100, with higher scores indicating whether the nutritional quality of the diet is closer to the Canadian guidelines for healthy eating.
2.2. Randomization of biological samples
Following sample selection and in perspective of multiplatform analyses, sample preparation and analytical sequence had to be carefully built. In metabolomics, analytical sequences are usually randomized using a Williams-Latin-Square strategy defined according to the main factors of the study, as well as potential confounding factors linked to sampling conditions. In the present work, samples were randomized using this strategy, defined first according to the main factor of the study (MetS), considering the sum of the annual number of MetS criteria between the two time points (T1 to T4), (divided in 4 groups: 0–3; 3–7; 11–14; 15–20; 0 being no positive criteria over the 3 years and 20 for 5 positive criteria over this period). This randomization was used both for sample preparations and analyses.
3. Methods
Seven complementary untargeted metabolomics methods based on 3 different analytical platforms, Ultra High-Performance Liquid Chromatography coupled to High-Resolution Mass Spectrometry (LC-MS), Gas Chromatography coupled to High-Resolution Mass Spectrometry (GC-MS), and Nuclear Magnetic Resonance spectroscopy (NMR), were used to characterize the MetS phenotypic spectrum.
Quality control samples were designed and prepared to control for potential bias due to sample preparation or analytical drifts. Since in untargeted metabolomics hundreds to thousands of metabolites are detected, the use of internal standards for each metabolite is almost impossible and pooled quality control (QC) samples are recognized to be the most appropriate approach [28]. In the present study, these QC samples consisted in a pool of human serum samples extracted independently and subsequently diluted 1/2, 1/4 and 1/8. All analytical sequences were standardized: at least three blank (solvent) samples and five pooled QC samples were injected for column conditioning. Then, the stability of the analytical system was monitored using these QC, injected one time at the beginning of each analytical sequence and thereafter every 10 samples.
3.1. Data production
3.1.1. Ultra high-performance liquid chromatography coupled to high-resolution mass spectrometry (LC-MS)
Three methods were performed to maximize the serum metabolome coverage: reversed phase LC-MS (C18) analysis complemented by hydrophilic interaction chromatography (HILIC) to allow the detection of polar metabolites and an untargeted lipidomics approach using a reverse phase LC-MS (C8) to profile a large set of lipid species.
3.2. C18-based system (C18Pos and C18Neg)
Serum samples (100 µL) were slowly thawed on ice at room temperature. Proteins precipitation was performed by addition of 200 µL of ice-cold methanol (MeOH). This mixture was vortexed and placed at −20 °C for 30 min. After a 10 min centrifugation (4 °C, 15,493 g, Sigma 3-16PK, Fischer Bioblock Scientific), the supernatant was divided into three 45 µL aliquots, dried completely (EZ2.3 Genevac, Biopharma Technologies France) and stored at −80 °C until further analysis. Just before analysis, 150 µL of injection solvents (water and acetonitrile 50/50 + 0.1% Formic Acid) was added to the dry fraction. A pooled QC sample was prepared by mixing 5 µL from each extracted sample. This sample preparation was automated on a Freedom EVO200 TECAN robot (Tecan Trading AG, Switzerland,), enabling liquid handling with a high repeatability (CV≤0.75%).
Metabolic profiles were determined using an U3000 liquid chromatography system (Thermo Fisher Scientific, San Jose, CA, USA) coupled to a high-resolution Bruker Impact HDII UHR-QTOF (Bruker Daltonics, Wissembourg, France) equipped with an electrospray source (ESI). Chromatographic separation was performed on a Waters HSS T3 column (150 × 2.1 mm, 1.8 µm) at 0.4 mL/min, 30 °C and using an injection volume of 5 µL. Mobile phases A and B were water and acetonitrile with 0.1% formic acid, respectively. The gradient elution was 0% B (2 min), 0–100% B (13 min), 100% B (7 min), 100–0% B (0.1 min) and 0% B (3.9 min for re-equilibration). The mass resolving of the mass spectrometer was 50,000 full width at half maximum (FWHM) at m/z 1222. Samples were analyzed in the positive and negative ionization modes (C18Pos, C18Neg). Capillary and end plate offset voltages were set at 2500 V and 500 V for the ESI source. The drying gas temperature was 200 °C and nebulization gas flow was 10 L/min. Mass spectrum data was acquired in full-scan mode over mass range 50–1000 mass-to-charge ratio (m/z).
3.3. HILIC-based system (HILICneg)
Metabolite extraction was performed from 50 µL of serum following methanol-assisted protein precipitation as previously described [29]. Briefly, 200 μL of methanol containing internal standards at 3.75 µg/mL (Dimetridazole, 2-amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl) propanoic acid (AMPA), 2-methyl-4-chlorophenoxyacetic acid (MCPA), Dinoseb (Sigma-Aldrich, Saint-Quentin Fallavier, France)) were added to 50 µL of serum. The resulting samples were then left on ice for 90 min until complete protein precipitation. After centrifugation (20,000 g, 15 min, 4 °C), supernatants were collected and dried under a nitrogen stream using a TurboVap instrument (Thermo Fisher Scientific, Courtaboeuf, France) and stored at −80 °C until analysis. Dried extracts were resuspended in 150 µL of ammonium carbonate 10 mM pH10.5/acetonitrile (40:60). After reconstitution, the tubes were vortexed, incubated in an ultrasonic bath for 5 min on ice, and centrifuged (20,000 g, 15 min, 4 °C). A volume of 95 µL of the supernatant was transferred into 0.2 mL vials. External standard solution (5 µL; mixture of 9 authentic chemical standards covering the mass range of interest: 13C-glucose, 15N-aspartate, ethylmalonic acid, amiloride, prednisone, metformin, atropine sulfate, colchicine, imipramine) was added to all samples in order to check for consistency of analytical results in terms of signal and retention time stability throughout the experiments. The QC samples were prepared by mixing 20 µL of each extracted sample. QC samples were injected every 5 samples.
Metabolic profiling experiments were performed using an U3000 liquid chromatography system coupled to an Exactive mass spectrometer from Thermo Fisher Scientific (Courtaboeuf, France) fitted with an electrospray source operating in the negative ion mode. Chromatographic separation was performed on a Sequant ZICpHILIC column (5 µm, 2.1 × 150 mm, Merck, Darmstadt, Germany) maintained at 15 °C for improved peak shape and chromatographic separation of nucleotidic metabolites [30,31], and also equipped with an on-line prefilter (Thermo Fisher Scientific, Courtaboeuf, France). Mobile phases A and B were an aqueous buffer of 10 mM ammonium carbonate in water adjusted to pH 10.5 with ammonium hydroxide, and 100% acetonitrile, respectively. The flow rate was 200 µL/min. Chromatographic elution was achieved under the following gradient conditions: isocratic step of 2 min at 80% B, followed by a linear gradient from 80 to 40% of phase B from 2 to 12 min. The chromatographic system was then rinsed for 5 min at 0% B, and the run ended with an equilibration step of 15 min (80% B). The Exactive mass spectrometer was operated with a capillary voltage set at −3 kV and a capillary temperature set at 280 °C. The sheath gas pressure and the auxiliary gas pressure (nitrogen) were at 60 and 10 arbitrary units, respectively. The mass resolving power of the analyzer was 50,000 (FWHM) at m/z 200, for singly charged ions. The detection was achieved from m/z 75 to 1000.
3.4. Lipidomic untargeted approach (LIPIDO)
Serum samples were extracted using an adapted method to that previously described [32]. Briefly, 100 μL of serum was added to 490 μL of CHCl3/MeOH 1:1 (v/v) and 10 μL of internal standard mixture. Samples were vortexed for 60 s, sonicated for 30 s using an ultrasonic probe (Bioblock Scientific Vibra Cell VC 75,185, Thermo Fisher Scientific Inc., Waltham, MA, USA) and incubated for 2 h at 4 °C with mixing. Seventy-five μL of H2O was then added and samples were vortexed for 60 s before centrifugation at 15,000 g for 15 min at 4 °C. The upper phase (aqueous phase), containing gangliosides, lysoglycerophospholipids, and short chain glycerophospholipids, was transferred into a glass tube and dried under a stream of nitrogen. The protein disk interphase was discarded and the lower lipid-rich phase (organic phase) was pooled with the dried upper phase and the mixture dried under nitrogen. Samples were resuspended with 100 μL of a solution CHCl3/MeOH 1:1 (v/v). Ten μL were 100-fold diluted in a solution of MeOH/isopropanol/H2O 65:35:5 (v/v/v) before injection.
Lipidomic profiles were determined using an Ultimate 3000 liquid chromatography system (Thermo Fisher Scientific, San Jose, CA, USA) coupled to a high resolution Thermo Orbitrap Fusion (Thermo Fisher Scientific, San Jose, CA, USA) equipped with an electrospray source (ESI). Chromatographic separation was performed on a Phenomenex Kinetex C8 column (150 × 2.1 mm, 2.6 µm) at 0.4 mL/min, 60 °C and using an injection volume of 10 µL. Mobile phases A and B were H2O/MeOH 60:40 (v/v), 0.1% formic acid and isopropanol/MeOH 90:10 (v/v), 0.1% formic acid in negative ionization mode (LIPIDOneg), respectively. Ammonium formate (10 mM) was added to both mobile phases in the positive ionization mode (LIPIDOpos) in order to detect glycerolipids and cholesteryl-esters under [M+NH4]+ form. The gradient elution was solvent B was maintained for 2.5 min at 32%, from 2.5 to 3.5 min it was increased to 45% B, from 3.5 to 5 min to 52% B, from 5 to 7 min to 58% B, from 7 to 10 min to 66% B, from 10 to 12 min to 70% B, from 12 to 15 min to 75% B, from 15 to 19 min to 80% B, from 19 to 22 min to 85% B, and from 22 to 23 min to 95% B; from 23 to 25 min, 95% B was maintained; from 25 to 26 min solvent B was decreased to 32% and then maintained for 4 min for column re-equilibration. The mass resolving power of the mass spectrometer was 240,000 (FWHM) for MS experiments. Samples were analyzed in both positive and negative ionization modes. The ESI source parameters were as follows: the spray voltage was set to 3.7 kV and −3.2 kV in positive and negative ionization mode, respectively. The heated capillary was kept at 360 °C and the sheath and auxiliary gas flow were set to 50 and 15 (arbitrary units), respectively. Mass spectra were recorded in full-scan MS mode from m/z 50 to m/z 2000.
3.4.1. Gas chromatography coupled to high-resolution mass spectrometry (GCMS)
Serum samples were slowly thawed at 4 °C overnight. Four hundred µL of ice-cold methanol (−20 °C) were added to 100 µL serum sample and the mixture was vortexed. After protein precipitation, samples were kept at −20 °C for 30 min and then centrifuged (Sigma 3–16PK, Fischer Bioblock Scientific) at 20,627 g for 10 min at 4 °C. Two hundred and fifty µL of supernatant were transferred into a 2 mL amber glass vial. After the addition of 10 µL of [13C1]-l-valine (200 µg/mL), samples were evaporated under EZ2.3 Genevac (Biopharma Technologies France). At the same time and in parallel, a control derivatization sample (serum substituted by milliQ water) was prepared in order to remove the background noise produced during sample pre-processing, derivatization, and GC/MS analysis. The dry residues were dissolved with addition of 80 µL of methoxylamine solution (15 mg/mL in pyridine) to each vial, vortexed vigorously for 1 min and incubated for 24 h at 37 °C (in order to inhibit the cyclization of reducing sugars and the decarboxylation of α-keto acids). Then, 80 µL of N,O-bis(Trimethylsilyl)trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) as catalyst were added into the mixture for derivatization (60 min, 70 °C). Before injection, 50 µL of derivatized mixture were transferred in a glass vial containing 100 µL heptane. QC pool samples were prepared using 10 µL of each extracted and derivatized samples.
Metabolic profiles were obtained using an Agilent 7890B Gas Chromatograph coupled to an Agilent Accurate Mass QTOF 7200 equipped with a 7693A Injector (SSL) Auto-Sampler (Agilent Technologies, Inc). Separation was achieved on a fused silica column HP-5MS UI 30 m x 0.25 mm i.d. chemically bonded with a 5% phenyl-95% methylpolysiloxane cross-linked stationary phase (0.25 µm film thickness) (Agilent J & W Scientific, Folsom, CA, USA). Helium was used as a carrier gas at a flow rate of 1 mL/min. Two µl of derivatized sample was injected using 1:20 split. Temperatures of injector, transfer line, and electron impact (EI) ion source were set to 250 °C, 280 °C and 230 °C, respectively. The initial oven temperature was 60 °C for 2 min, ramped to 140 °C at a rate of 10 °C/min, to 240 °C at a rate of 4 °C/min, to 300 °C at a rate of 10 °C/min and finally held at 300 °C for 8 min. Agilent ‘‘retention time locking” (RTL) was applied to control the reproducibility of retention times. [13C1]-l-valine was used to lock the GC method [33]. The electron energy was 70 eV and mass data were collected in a full scan mode (m/z 55-700) using a resolving power of 7000 (FWHM) to m/z 464 (perfluorotributylamine, PFTBA). Acquisition rate was 5 spectra/sec with acquisition time of 200 msec/spectrum. Four heptane blanks were injected at the beginning of each sequence, followed by four pool samples, and then one pool sample and one derivatization control sample after each set of 10 samples. Initially tune and calibrate the system were performed using PFTBA with acquisition conditions 2 GHz EDR with N2 (1.5 mL/min) and the limits for average PPM error were 3.0 and maximum error: 8.0. Also, a calibration was made between each sample.
3.5. Nuclear magnetic resonance spectroscopy (NMR)
Serum aliquots (50 µL) were slowly thawed at room temperature on ice. One hundred µL of phosphate buffer (0.2 M, pH 7.0) prepared in deuterium oxide (D2O) were added to the aliquots, and each sample was vortexed and centrifuged for 15 min at 4500 g and 150 µL of the supernatants were transferred into the 3 mm NMR tubes.
All 1H NMR spectra of serum samples were obtained on a Bruker Avance III HD spectrometer (Bruker, Karlsruhe, Germany) operating at 600.13 MHz for 1H resonance frequency and equipped with an inverse detection 5 mm CQPCI 1H-31P-13C-15N cryoprobe connected to a cryoplatform and a cooled SampleJet sample changer. Spectra were acquired at 300 K using the Carr-Purcell-Meiboom-Gill (CPMG) spin-echo pulse sequence with a total spin-echo delay of 240 msec to attenuate broad signals from proteins and lipoprotein and a 2 s relaxation delay. A water suppression signal was achieved by pre-saturation during the relaxation delay. The spectral width was set to 20 ppm for each spectrum, and 256 scans were collected with 32 K points. Free induction decays were multiplied by an exponential window function before Fourier Transform. The spectra were manually phased and calibrated to the lactate signal (δ 1.33 ppm), and the baseline was corrected using TopSpin 3.2 software (Bruker, Karlsruhe, Germany).
3.6. Data treatment
Following metabolomic/lipidomic analyses, some samples were identified as missing, because of problem in sample preparation or missing data (1 for C18Pos, 6 for HILIC, 4 for Lipidomic and 1 for GCMS). All the obtained raw data from metabolic profiles were processed to yield a data matrix containing variables and peak intensities. All the data treatments were performed separately for each analytical method as individual datasets, under the Galaxy web-based platform Worflow4Metabolomics (W4M) [34] to ensure the standardization and reproducibility of the data treatment workflows.
3.6.1. Data extraction and pre-processing for MS
First, raw data were extracted using XCMS [35], followed by quality checks and signal drift correction according to the strategy described by van der Kloet et al. [36] based on the use of pooled QC samples, to yield a data matrix containing retention times, masses, and peak intensities that have been corrected for batch effects. These steps include noise filtering, automatic peak detection, and chromatographic alignment. In particular, all XCMS extractions used a “minfrac” parameter of 0.2 to keep variables if present in at least 20% of the samples, since a huge variability of profiles in the selected individuals was expected. Due to a high degree of correlations between the two lipidomic extracted datasets, they were merged for further data processing. After signal drift and batch effect correction within the six datasets, metabolite MS signals were then filtered using the following criteria: ratio of chromatographic peak areas of samples over blanks (above 3), correlation between QC pool dilution factors and areas of chromatographic peaks (over 0.7), repeatability of QC pool samples (CVs under 30%) and ratio of QC pool sample CVs over biological sample CVs (below 1).
3.6.2. NMR data pre-processing
The NMR spectra were imported in the Amix software (version 3.9.15, Bruker, Rheinstetten, Germany) for data integration. A variable size bucketing was performed based on graphical pattern (74 buckets) and each bucket was then integrated.
3.6.3. Filtration
During the analysis, metabolites produce several analytical features corresponding to signals derived from different adduct ions generated in the ESI process, signals from the presence of isotopes in the molecule, signals from in-source fragmentation processes, and to different peaks from the same molecule in NMR. The data extraction step results in thousands of features present in the final datasets with a high degree of correlation, which is a constraint for the use of various data mining and statistical methods. For example, analytical redundancy highly affects multiple testing correction. Indeed, having non-independent variables (coming from the same metabolite) lead to an over-correction of data that can hide potentially relevant information. Therefore, the analytical redundancy inside each of the 6 datasets was reduced in the present study. In metabolomics, filtering was technique-specific but with a common characteristic to reduce correlation above 0.90 and to select one single representative per group, as being the most intense signal for MS data and the purest one for NMR. This procedure was conducted using the Analytic Correlation Filtration (ACorF) tool [37] within W4M, with a manual selection of the representative feature only for NMR. In lipidomics, this step was performed according to the workflow previously described [32]. Briefly, a first automatic feature annotation was achieved through using an in silico database containing the exact masses corresponding to pseudo-molecular ions ([M + H]+, [M-H]− and [M-2H]2−), adducts([M+NH4]+, [M+Na]+,[M-H+CHO2]−), and in source fragments ([M + H−H2O]+) ions along with their corresponding 13C and double 13C isotopes. Furthermore, specific retention time windows for each lipid class were also added by examining retention times of species containing the longest and the shortest fatty acyl chains. Then, annotated lipid species were thus kept if (i) their 13C isotope was detected and aligned in time (± 5 s), and (ii) all related ions (i.e. pseudo-molecular ions, adduct ions and/or in source fragments, either as monoisotopic or 13C and 2×13C isotopes) had the same retention time as a reference ion specific of a lipid class/subclass (± 5 s, and ±10 s between the two ionization modes after merging the corresponding peaktables). In addition, the relative isotopic abundance (RIA) between the monoisotopic ion and its corresponding 13C isotope, were automatically calculated and compared to theoretical ones. Annotated lipid species with an RIA error higher than 30% were filtered out. This threshold of 30% was selected since RIA errors of all internal standards were below this value. The two lipidomic peaktables obtained in both positive and negative ionization modes were merged, because of their high degree of correlations, due to the detection of specific lipid classes in both modes (i.e.: lysophosphatidylcholines, phosphatidylcholines and sphingomyelins) under [M + H]+ and [M-H+CHO2]− forms, respectively. The two peaktables were aligned according the retention time at ± 10 s.
3.7. Statistical analyses
All statistical analyses were performed after data pre-processing and filtration of the individual 6 datasets.
3.7.1. Measurement of serum metabolomes in the NuAge MetS sub-cohort
Correlation analyses were performed to give an overview of links between detected metabolites/lipids in serum, both at the level of method datasets and individual variables. First, the RV coefficients [38] were used to provide insight into the global association between datasets using the R software (version 3.4.1) [39], with the R-package FactoMineR [40]. This coefficient [38] is a multivariate generalization of the squared Pearson correlation coefficient, defining a scale of similarity between two matrices and measure to what degree the different datasets give the same view on the samples [41]. Second, to investigate individual correlations between detected features, pair-wise Spearman correlation coefficients between variables were calculated using the Between Table Correlation tool available in the W4M and a network analysis was done. The significant correlation coefficients >0.7 (after Benjamini-Hochberg correction) were filtered and a graphical representation of Spearman correlation network was made with Cytoscape [42].
3.7.2. Metabolite and lipid levels modulated with MetS
Individuals in this study were selected stable regarding their MetS status. Nonetheless we could expect that part of their metabolism was affected by time. Thus, the impact of time on the metabolomic/lipidomic datasets was also evaluated. As no interaction effect was observed between status and time, linear mixed models (LMM) were performed to analyze repeated measures, considering fixed effect factors (time, status (case/control), and their interactions) and subject as random effect, using the module available in W4M. In order to verify that the LMM assumptions were met, we considered the different residuals of LMM. The assumption of homogeneity of variance of the residuals was checked for each fixed factor using a Levene test. Then, the normality of the conditional residuals and random effect residuals were verified using quantile-quantile plot. The linearity of fixed effects was checked as proposed by Singer et al. [43] using plot of the marginal predictions vs standardized marginal residues. A p-value threshold of 0.05 after Benjamini-Hochberg (BH) correction was considered to detect variables strongly affected by status and time. Similar statistical analyses were performed on epidemiological data.
3.7.3. Identification of a comprehensive molecular MetS signature
The objective of the present study was to identify a limited number of metabolites that could together reflect the MetS status. In this context, a variable selection was first performed based on the methodology developed by Rinaudo et al. [44], using the biosigner module available in the W4M Galaxy instance [34] on each individual dataset. The aim was to focus on the variables, which significantly contribute to the performance of the discrimination. As feature selection may be affected by correlations between variables, a Pearson correlation filter on each dataset (over 0.8) was applied beforehand. All variables selected by biosigner with at least one of three classifiers (Partial Least Squares Discriminant Analysis (PLS-DA), Support Vector Machine (SVM), and Random Forest (RF)) were first considered. Then, this process was repeated five times to cope for the selection variability induced by the bootstrap effect of the methodology. The unions over the five repetitions were included in individual predictive subsets. In a second step the selected variables of each subsets were integrated into a common PLS-DA model to characterize the discriminant power of the comprehensive signature by combining the 6 individual predictive subsets. For all PLS analyses, unit variance scaling (UV) was applied to variable intensities. All PLS models were defined using the 7-fold cross validation method. The prediction power of the model was assessed using the Q2 parameter. To check that PLS components could not lead to a correct classification by chance, a permutation evaluation was carried out (n = 200). For each test, samples are randomly assigned to each experimental group, a PLS model is carried out and R2Y and Q2 are computed. The result of the tests is displayed on a validation plot, which shows the correlation coefficient with the original non-permuted sample, having a value 1 on the horizontal axis and R2Y and Q2 values on the vertical axis. Logically, permuted samples must lead to poor predictive models with lower Q2 values compared to the true model.
In a perspective of future clinical application, an optimized reduced signature was then proposed. To fulfill this objective, the redundancy between methods was eliminated (correlation coefficient > 0.8), keeping the most robust variable (highest intensity, best peak purity). In a second step, this signature was restricted to the strictly formally identified compounds. The prediction model performance was evaluated using a confusion matrix, cross-validated error rates (using 200 repetitions of random training/test splits), and areas under ROC curves (AUC) [45] using the R software (version 3.6.2) [R package “pROC” [46]] with a CI estimated with the DeLong's method [47].
3.8. Metabolite annotation
The metabolite annotation was first conducted computationally using W4M and then, all annotations involved manual curation and interpretation of spectra.
Metabolites contributing to the discrimination of the MetS phenotype were first identified using in-house databases, containing the reference spectra of more than 2000 authentic standard compounds analyzed in the same analytical conditions, and providing a comprehensive spectral information (i.e. protonated or deprotonated molecules, adducts and in-source fragment ions for LC-HRMS, or molecular ions as well as major fragments for GCMS). Metabolite annotation was first performed by using these spectral databases according to accurately measured masses within MS spectra and chromatographic retention times. Confirmation of metabolite annotation in LC-HRMS was then accomplished by running additional LC-MS/MS experiments using a Dionex Ultimate chromatographic system combined with a Q-Exactive mass spectrometer (Thermo Fisher Scientific) under non-resonant collision-induced dissociation conditions using higher-energy C-trap dissociation (HCD) in both positive and negative ion modes, conducted on the same QC samples, and with the instrument set in the targeted acquisition mode, using inclusion lists. Resulting MS/MS spectra were then manually matched to those included in the in-house spectral database and acquired using different collision energies. Confirmation of metabolite annotation in GC-MS was done by matching electron impact spectra, as well as using reports from the literature.
Then, the remaining unknown compounds were identified on the basis of their exact masses which were compared to those registered in Metlin (https://metlin.scripps.edu; [48]), in the Human Metabolome Database (HMDB; www.hmdb.ca; [49]), in Massbank (https://massbank.eu/MassBank/; [50]), in Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/; [51]), or in the National Institute of Standard and Technology (NIST; https://www.nist.gov/srd/nist-special-database-14; [52]). Database queries were performed with a mass error of 0.005 Da, and a retention time difference of 0.1 min for the in-house databases. Database results were confirmed using appropriate standards when available, isotopic patterns, and mass fragmentation analyses. For unidentified ions, the number of plausible elemental compositions were restricted to a small number (or uniquely identified) with the support of additional chemical information, i.e. the molecular formula of the precursor ions, reports from the literature [53], and knowledge of possible metabolic pathways. Metabolites were classified accordingly to Sumner et al. [54] concerning the levels of confidence in the identification process: identified (confirmed by an authentic chemical standard analyzed under the same conditions, with the match at least two orthogonal criteria among accurate measured mass, retention time and MS/MS or EI(MS) spectrum), putatively annotated (spectral similarity with public/commercial spectral libraries), putatively characterized compound classes or unknown.
It is important to note that only very few standards of lipid species are commercially available compared to the large diversity of endogenous lipid species present in complex biological matrices. Therefore, results of in-house database queries were filtered, according to the workflow described in Seyer et al. [32], taking into account retention time ranges of each lipid class, as well as isotope patterns, for selection of relevant lipid species, as previously described in the data filtration section. Finally, all HCD mass spectra resulting from the additional MS/MS experiments, were manually inspected to identify specific diagnostic ions and to confirm the structure of lipid species [55] (see Supplemental Fig. 2), that were named following the LipidMaps nomenclature [56].
Spectral assignments were based on matching 1D and 2D NMR data to reference spectra in a homemade reference database, as well as with other databases (http://www.bmrb.wisc.edu/metabolomics/; http://www.hmdb.ca/), and reports from the literature [57].
3.9. Extraction of modulated metabolic network
To link metabolites identified in untargeted metabolomics/lipidomics experiments within the context of genome-scale reconstructed metabolic networks, the metabolites described as modulated after LMM, stable over time, and identified or annotated, were mapped into the human genome-scale metabolic network Recon2.2 [58]. This network contains 7785 reactions and 6047 metabolites. In order to map the modulated metabolites on this network, we first retrieved their ChEBI identifier and then search for their matching identifier in the Recon2.2 network using the "identifier matcher tool" in MetExplore. This tool allows performing both an exact matching (to find the exact metabolite in the network corresponding to the modulated metabolite from the experimental dataset) and an ontology-based matching (to make the link with a corresponding more generic class metabolite, when the exact same metabolite cannot be retrieved in the network) [59]. In the metabolic network, each metabolite is assigned to several different cellular compartments. However, because current global and untargeted metabolomics approaches do not provide information on cellular localization of metabolites, we chose to consider only cytosolic metabolites. In order to focus on the most likely modulated part of the network, we first selected all the metabolic pathways in which at least one modulated metabolite was found, while excluding pathways involving only transport reactions. Forty-one pathways, including 2753 reactions, were selected. Then, a metabolic sub-network extraction was performed from the modulated metabolites. It consists in computationally identifying among the previously selected reactions, the ones that are more likely to be related to the modulated metabolites. The algorithm computes the lightest path between each pair of metabolites in the dataset. The lightest path is a sequence of reactions and metabolites connecting two metabolites and minimizing a topological criterion in the network [60,61]. For one dataset, the related sub-network is thus the union of all the lightest paths between metabolites present in this dataset. Pathway enrichment analyses were performed to assess whether the modulated metabolites were significantly over-represented in a metabolic pathway. Pathway enrichment statistics were performed using the one-tailed exact Fisher test, with a BH correction for multiple tests, using the metabolic pathways defined in Recon2.2. All computational and visualization tasks were performed within MetExplore web server based on the Recon2.2 metabolic network (biosource id #4311) [62,63].
Role of funding sources: All metabolomics and lipidomics analyses (data collection) were funded by the MetaboHUB French infrastructure (ANR-INBS-0010). Funders had no role in study design, data analysis, interpretation or writing of report.
4. Results
4.1. Overview of study population
Fifty-eight quantitative variables in total were considered to precisely describe the selected population: 23 biochemical parameters, 8 clinical variables, 25 nutritional data (essentially related to macronutrient intake and selected nutrients described as being related with MetS), and finally 2 scores related to physical activity and global health (see Materials and Methods). As defined in the subject selection process (see Materials and Methods), MetS status of individuals was stable over the three years follow-up (for the 4 time points considered). Behind the stability of MetS status, the clinical parameters associated with MetS, analyzed at T1 and T4, were found stable over time, with a slight improvement for some of them (i.e. significant reduction of systolic blood pressure, fasting glucose and triglycerides (TG)). Differences of most of the MetS criteria quantitative variables were highly significant between cases and controls (BH corrected p-values from 10−5 to 10−21, Table 1; Supplementary Fig. 1). The main descriptive data, as well as results from linear mixed models, are presented in Table 1 and Supplementary Tables 1a, 1b. They showed that all the subjects were globally stable over time, not only for clinical values of MetS criteria, as already emphasized, but also for the main parameters related to physical activity, nutrition and health-related quality of life (physical (PCS) component summary score). Regarding MetS status, results showed that MetS subjects were less healthy and active than controls, with all global scores related to physical activity (PASE) and health-related quality of life (PCS) found significant (corrected p-values = 0.02 and 3.6 × 10−3, respectively). Moreover, cases showed also significant lower total energy and carbohydrate intakes (corrected p-value = 0.025 and 6.8 × 10−3, respectively). In addition, despite the fact that total dietary fiber intake was at the limit of significance, the evaluation of the consumption of cereal products, based on the Canadian Food Guide recommended intakes for grain products (Canadian-Healthy Eating Index, C-HEI) was significantly lower in cases in comparison to controls (corrected p-value = 0.052 and 0.013, respectively).
Table 1.
Overview of the study population.
| Controls |
Cases |
Corrected p-value time (BH) | Corrected p-value MetS status (BH) | |||
|---|---|---|---|---|---|---|
| T1 | T4 | T1 | T4 | |||
| n | 62 | 62 | 61 | 61 | – | – |
| Age (yrs) | 73.5 ± 4.1 (62) | – | 74.1 ± 3.6 (61) | – | 1.00 | 0.34 |
| Body weight (kg) | 71.0 ± 8.0 (62) | 69.9 ± 7.8 (62) | 87.7 ± 12.5 (61) | 87.4 ± 13.3 (61) | 0.04 | 6.2 × 10−14 |
| BMI (kg/m2) | 25.1 ± 2.3 (62) | 24.8 ± 2.4 (62) | 30.5 ± 3.7 (61) | 30.6 ± 3.7 (61) | 0.37 | 1.5 × 10−16 |
| Waist circumference (cm) | 93.3 ± 6.9 (62) | 92.8 ± 6.9 (62) | 109.9 ± 8.9 (61) | 110.8 ± 9.5 (61) | 0.67 | 6.2 × 10−21 |
| Fasting serum glucose (mM) | 5.08 ± 0.44 (62) | 4.86 ± 0.58 (62) | 6.66 ± 1.45 (61) | 6.54 ± 1.21 (61) | 0.04 | 2.1 × 10−15 |
| Fasting TG (mM) | 1.23 ± 0.47 (50) | 1.18 ± 0.40 (53) | 2.23 ± 1.01 (51) | 1.94 ± 0.86 (51) | 0.04 | 1.6 × 10−8 |
| Fasting HDL-C (mM) | 1.43 ± 0.45 (50) | 1.50 ± 0.34 (53) | 1.13 ± 0.29 (56) | 1.16 ± 0.26 (56) | 0.74 | 1.1 × 10−5 |
| SBP (mmHg) | 126.2 ± 16.6 (62) | 120.9 ± 18.4 (62) | 138.4 ± 15.8 (61) | 133.7 ± 19.3 (61) | 0.02 | 4.4 × 10−5 |
| DBP (mmHg) | 71.8 ± 9.9 (62) | 73.9 ± 8.1 (62) | 74.7 ± 8.9 (61) | 73.6 ± 9.4 (61) | 0.69 | 0.47 |
| Leucoc (109/L) | 5.61 ± 1.29 (62) | 5.97 ± 1.57 (62) | 6.32 ± 1.22 (61) | 6.55 ± 1.34 (61) | 0.02 | 2.0 × 10−2 |
| Lympho (109/L) | 1.49 ± 0.45 (62) | 1.54 ± 0.46 (62) | 1.75 ± 0.43 (61) | 1.76 ± 0.63 (61) | 0.50 | 2.0 × 10−2 |
| SF-36-Physical Component Summary Score* (PCS) | 52.8 ± 5.8 (62) | 52.3 ± 6.0 (61) | 49.7 ± 8.0 (61) | 46.7 ± 9.2 (61) | 0.01 | 3.6 × 10−3 |
| Physical activity (PASE score) | 125.4 ± 51.9 (62) | 124.7 ± 53.5 (59) | 107.1 ± 55.2 (61) | 94.6 ± 50.5 (57) | 0.41 | 2.3 × 10−2 |
| Energy (kCal/day) | 2179 ± 524 (62) | 2251 ± 576 (62) | 1935 ± 462 (60) | 2026 ± 506 (59) | 0.08 | 2.5 × 10−2 |
| Carbohydrate (g/day) | 269.6 ± 68.8 (62) | 272.3 ± 73.6 (62) | 231.7 ± 64.0 (60) | 238.4 ± 62.5 (59) | 0.45 | 6.8e-3 |
| Protein (g/day) | 87.5 ± 22.8 (62) | 89.3 ± 28.2 (62) | 83.1 ± 20.4 (60) | 82.9 ± 22.6 (59) | 0.75 | 0.31 |
| Lipid (g/day) | 78.5 ± 23.9 (62) | 83.8 ± 26.7 (62) | 71.9 ± 23.4 (60) | 80.3 ± 27.0 (59) | 0.02 | 0.35 |
| C-HEI-Cereals (score: 0–10) | 7.99 ± 2.25 (62) | 7.72 ± 2.26 (62) | 6.66 ± 2.05 (60) | 6.99 ± 2.20 (60) | 0.88 | 1.3 × 10−2 |
| Total dietary fiber (g/day) | 23.6 ± 9.4 (62) | 24.9 ± 10.7 (62) | 19.8 ± 7.5 (60) | 21.4 ± 7.2 (59) | 0.10 | 5.2 × 10−2 |
Mean ± SD; linear mixed model p-value (after Benjamini-Hochberg (BH) correction for 58 parameters). Bold: corrected p-value < 0.05.
T-scores based on a mean of 50 and a SD of 10.
4.2. Serum metabolomes in the NuAge sub-cohort
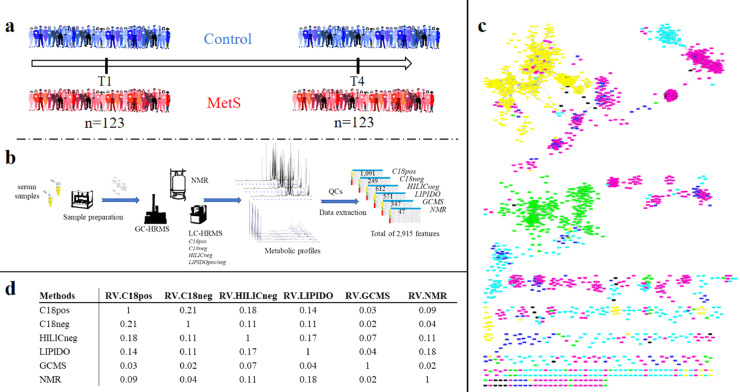
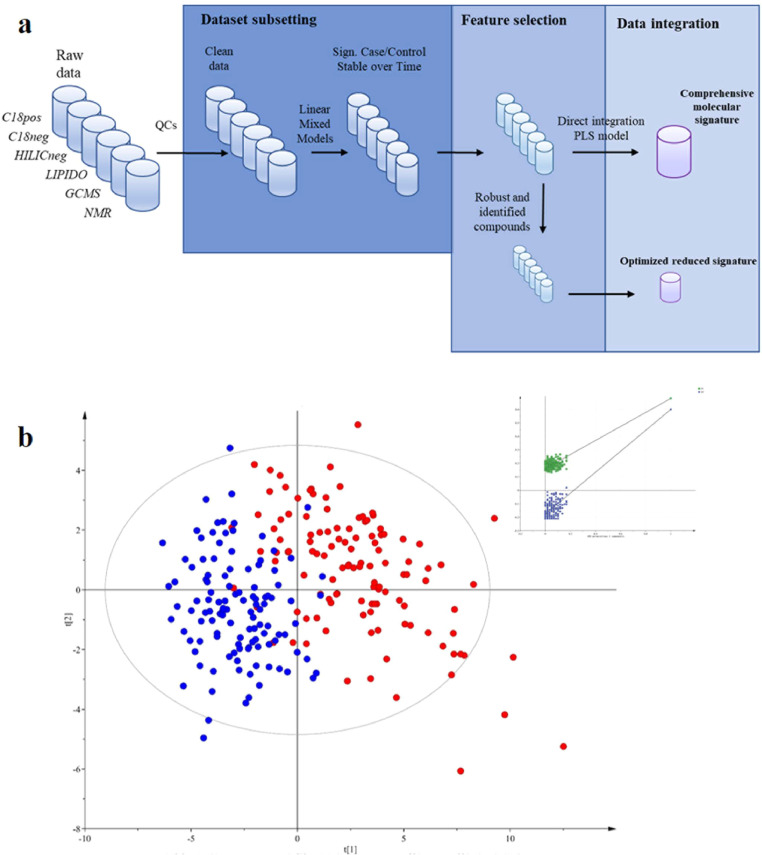
Given the high diversity of metabolites present over a wide concentration range in biological samples, proper MetS evaluation requires a broad metabolome coverage. The use of complementary technologies, combining both Nuclear Magnetic Resonance (NMR) and high-resolution mass spectrometry (HRMS), as well as different chromatographic systems, including gas, reverse-phase and hydrophilic interaction chromatography with detection in both positive and negative electrospray ionization modes, allowed covering both polar and apolar compounds for relevant and comprehensive metabolome and lipidome analysis (see Materials and Methods). On this basis, a full workflow was developed for serum analysis, allowing the sample preparation and the measurement of a wide diversity of metabolites from the more polar ones to lipids (Fig. 1a and b).
Fig. 1.
Study design and multiplatform metabolomics data generation.
a: Experimental design of the MetS case/control study, involving the follow-up of 123 stable subjects over 3 years.
b: Analytical workflow based on seven complementary untargeted metabolomics methods using 3 different analytical platforms (LC- and GC-HRMS, NMR) for the serum analysis of the 123 subjects collected at two time points. Data analysis resulted in 2915 metabolite and lipid related features, detected across at least 20% of the subjects over time.
c: The similarity of method blocks evaluated using RV coefficients (a matrix correlation coefficient (see method section)) after Multiple Factorial Analysis. d: Spearman correlation network between the 2915 metabolite and lipid features from the six metabolomic/lipidomic datasets (significant correlation >0.7). Nodes correspond to features obtained using the different analytical platform (Pink: C18pos; Dark Blue: C18neg; Green: GCMS; Blue: HILICneg; Yellow: Lipidomics; Black: NMR). Two nodes are connected by an edge if correlation is significant between the corresponding features (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
After data processing using a reproducible online Galaxy workflow [34], consisting in automatic peak detection, alignment and integration, quality control steps and removal of redundant spectral features, a total of 2915 metabolite and lipid related robust features (mean CV in QCs = 13%), were detected across at least 20% of the subjects over time (Fig. 1b). Correlation analyses of still unannotated species were performed to give an overview of links between detected features. The similarity of the various retrieved datasets evaluated using RV coefficients [38] between matrices was found to be low (< 0.18), in accordance with the complementary serum metabolome coverage of the 6 chosen methods (Fig. 1c). This result was also confirmed by the low level of strong correlations between individual metabolite and lipid features detected in the different platforms, with only 7% of Spearman correlation coefficients found significant (after Benjamini-Hochberg (BH) correction) and > 0.7, as shown in Fig. 1d (and supplementary data). This network highlights both analytical correlations (shared features between methods) and biological ones (that could come from different metabolites involved in the same metabolic pathways).
4.3. Metabolite and lipid levels modulated with MetS
This 3-year follow-up and the analyses performed at two time points (T1 and T4) enabled us to study the serum metabolome modulations by MetS and robustly delineate the associated metabolic perturbations, in relation with clinical parameters and some potentially interacting extrinsic factors, such as diet and physical activity.
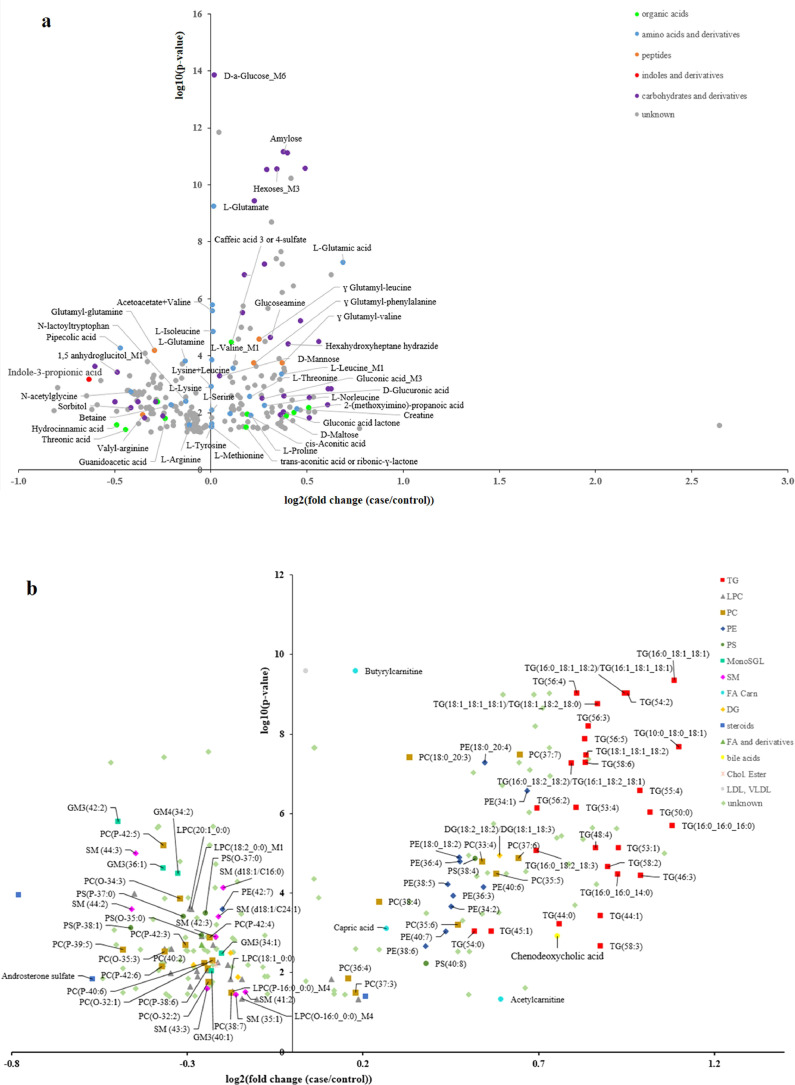
In regards to the characteristics of the study population, consistent results were observed with a wide range of metabolites modulated by MetS and stable over time. More precisely, 476 metabolite and lipid features were found significant between cases and controls, and independent of time, which represents around 16% of the detected metabolome/lipidome. Fig. 2 (a and b, Datasets EV2 and EV3) highlights the magnitude of these changes as well as their respective significance (248 metabolites and 228 lipids). Among them, 158 were successfully identified or annotated, and 26 were attributed to putatively characterized classes (e.g. by spectral similarity to known compounds of a chemical class), as exemplified in Supplementary Fig. 2. Only nine compounds are shared metabolites detected by several methods (reported as metabolite_Mx in Datasets EV2 and EV3). Moreover, six metabolites are detected still with redundant spectral features (mainly carbohydrates), due to interference of co-eluting compounds (reported as metabolite_Fx in Datasets EV2 and EV3). Globally 50% of these molecular features are positively associated with MetS, with a high confidence (LMM corrected p-value < 10−5) for the identified carbohydrates, amino acids and some lipids (mainly TGs, corrected p-value < 10−7). Among the negatively modulated molecules, metabolites from food (e.g. 1,5-anhydroglucitol), microbial metabolism (e.g. indole-3-propionic acid, pipecolic acid) and several amino acid derivatives and peptides (corrected p-values ranging from 10−3 to 10−5) were highlighted. When grouping lipids into common classes, only TGs and gangliosides were found significantly different (fold change (log2 (case /control), FC) = 0.94, corrected p-value = 7.4 × 10−10; FC = −0.32, corrected p-value = 9 × 10−6, respectively). Some other lipid classes, such as phosphatidylcholines (PC), phosphatidylethanolamines (PE) or fatty acids (FA) were found very heterogeneous, with still individual compounds highly significantly modulated (PC(18:0_20:3), FC = 0.33, corrected p-value = 3.8 × 10−8; PE(18:0_20:4), FC = 0.5, corrected p-value = 5.2 × 10−8).
Fig. 2.
The effect of MetS on metabolome/lipidome.
Volcano-plots representing metabolites (a) and lipids (b) found significantly different (linear mixed model p-values adjusted with BH correction) between cases and controls, and independent of time: x-axis represents the magnitude of changes (log2 mean fold change (case/control)), y-axis the confidence (-log10 corrected p-value) for the 248 metabolites and 228 lipids related features. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
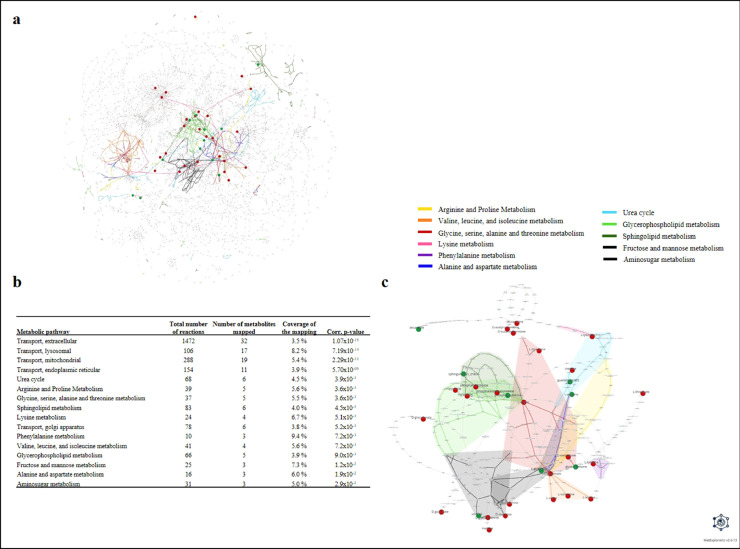
To explore and visualize the underlying mechanisms, a network analysis was performed. First, the ChEBI identifiers of the 132 modulated unique metabolites stable over time (after removal of remaining redundancy from spectral features and shared metabolites), identified or annotated, were retrieved and then, their matching identifiers in the Recon2.2 network [58], were searched. Among these metabolites, 21 were "exactly" matched with network metabolites, whereas 68 were matched with a more generic compound and the remaining without any match. This is especially the case for lipid compounds such as the different triglyceride species (e.g. TG(46:0), TG(48:0)) which are all represented by the "M_tag_hs" metabolite in the network corresponding to the generic triglyceride class. Therefore, several of the modulated metabolites matched to the same network metabolite. The complete list of the 89 modulated metabolites along with their ChEBI and matching network identifiers is provided in Supplementary Table 2. Those 89 metabolites were mapped into the human genome-scale metabolic network Recon2.2 corresponding to 43 unique metabolites in the network. Fig. 3a displays the localization of these metabolites within the global metabolic network, highlighting the pathways in which they are preferentially located. It provides a global view of the modulated pathways, as well as their inter-connections. The predicted modulated pathways obtained after the over-representation analysis are highlighted on Fig. 3b. To identify more specifically the parts of the networks that are modulated by MetS, and based on the hypothesis that the reactions connecting the modulated metabolites within the network are the most likely to be associated with the disease perturbation, we extracted the metabolic sub-network corresponding to the union of all the lightest paths between metabolites of interest (Fig. 3c, Supplementary Tables 3 and 4). This sub-network suggests that the metabolome/lipidome changes rely essentially on the modulation of pathways mainly related to amino acid pathways (arginine/proline metabolism, glycine/serine/alanine/threonine metabolism, lysine metabolism, phenylalanine metabolism, valine/leucine/isoleucine metabolism, and alanine/aspartate metabolism) and urea cycle. Other key pathways including metabolites and reactions related to sphingolipid metabolism, glycerophospholipid metabolism, fructose and mannose, and aminosugar metabolism were also found to be implicated (Fig. 3).
Fig. 3.
Visualization of predicted modulated reactions network analysis.
a: Visualization of the modulated metabolites within the Recon2.2 metabolic network. Only reactions belonging to metabolic pathways with at least one discriminant metabolite (except for transport reactions) were included in the visualization (2753 reactions). Reactions are represented by squares and metabolites by circles. Metabolites identified as significantly modulated by MetS (mapped metabolites) are colored in red. Metabolic pathways significantly enriched in modulated metabolites are highlighted in colors.
b: Metabolic pathways significantly enriched in modulated metabolites. Pathway enrichment analysis was performed on the set of metabolites identified as significantly modulated by MetS. The p-values were obtained by performing a hypergeometric test followed by a Benjamini-Hochberg correction.
c: Modulated metabolic sub-network. The metabolic sub-network was extracted by gathering the union of all lightest paths between each pair of mapped metabolites (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
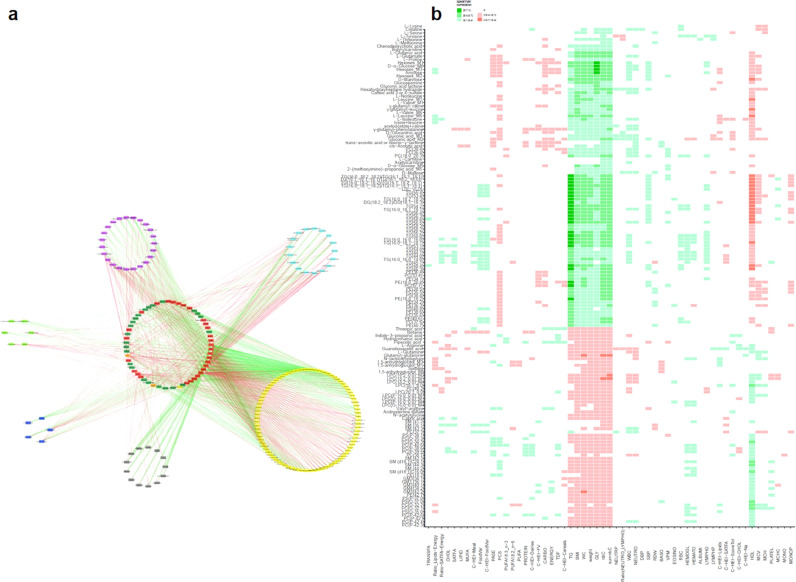
4.4. Relationships between modulated metabolites/lipids and clinical parameters
Correlation analyses contributed to explore the links between the molecular signature and biochemical/clinical/nutritional and general health parameters. Fig. 4a (and Supplementary data) presents the correlation network of significant correlations between the modulated metabolites and lipids by MetS (after removal of redundant spectral features) and all the other parameters considered. It shows that when using this multiplatform approach, metabolomics gives a global metabolic view of MetS considering its intrinsic and lifestyle factors (related in this case to nutrition and physical activity). More precisely, Fig. 4b (and Supplementary data) highlights the links between almost all significantly modulated metabolites and the five individual clinical criteria defining MetS, without that much specificity (i.e. a metabolite chemical family being related to several MetS criteria), revealing the interconnection of underlying metabolic processes and MetS components. Moreover, it shows some associations between around 20% of the metabolites and physical activity evaluation. Regarding nutrition, global scores or food intake data do not reach to capture the complexity of changes at the individual metabolite level. However, some specific associations can be observed between some food groups and particular metabolites, as interestingly between fiber/cereal consumptions and microbiome metabolites (e.g. indole-3-propionic acid, pipecolic acid).
Fig. 4.
Exploration of the links between metabolic profiles and phenotypic parameters.
Spearman correlations (coefficients significantly different from 0 after BH correction) between the stable modulated metabolites/lipids by MetS and the 58 phenotypic quantitative variables. Green: positive correlations; Red: negative correlations.
a: Global view of the resulting network organized by methods: Pink: C18pos; Dark Blue: C18neg; Green: GCMS; Blue: HILICneg; Yellow: Lipidomics; Black: NMR. Dark Green: nutritional data; Orange: scores related to physical activity and health-related quality of life; Red: clinical data.
b: Detailed view showing individual correlation (variables re-organized by hierarchical clustering analysis). Metabolite_M1: detected in C18pos; Metabolite_M2: detected in C18neg; Metabolite_M3: HILICneg; Metabolite_M4: detected in LIPIDO; Metabolite_M5: detected in GCMS; Metabolite_M6 detected in NMR (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
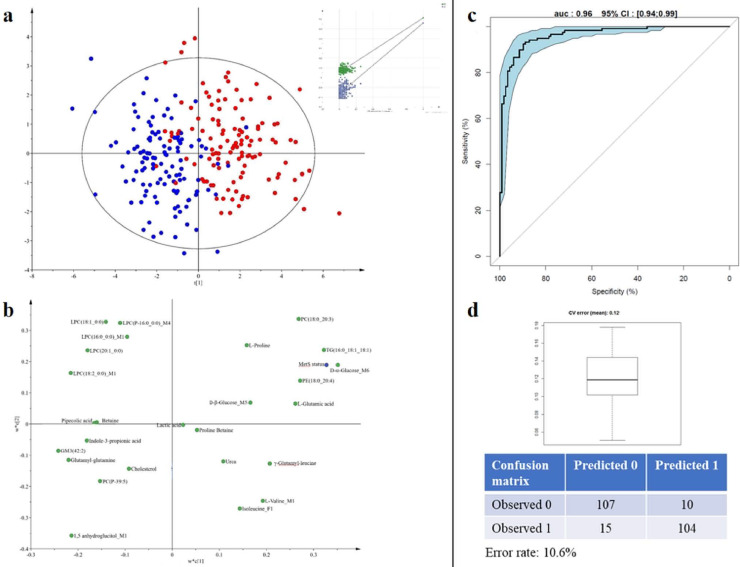
4.5. Identification of a comprehensive molecular MetS signature
A full feature selection strategy was developed to build a comprehensive molecular MetS signature, stable over time (Fig. 5a). After LMM, all the variables stable over time regardless of their link with MetS (i.e. 2176 variables in total for the six datasets) were submitted to a feature selection step. The biosigner recursive feature selection method [44] was applied to each of the six datasets (after filtration of intrinsic correlations) to select the smallest feature subsets which significantly contribute to the MetS prediction using 3 binary classifiers with distinct mathematical backgrounds (Partial Least Square-Discriminant Analysis (PLS-DA), Random Forest, and Support Vector Machine), and repeated five times (see Materials and Methods). For each dataset, the metabolites most contributing to prediction, selected by at least one of the three methods, and being part of the union over the five repetition, were retained. This approach allows selecting the necessary and sufficient variables for MetS prediction, mainly due to its ability to combine large number of data permutations with three different multivariate methods. It resulted in six subsets, including a selection from 2 to 39 variables. These six predictive subsets were then integrated into a PLS-DA model to obtain a comprehensive molecular MetS signature (that includes 102 variables, see Supplementary data) and characterize its discriminant power. The resulting validated model allowed a good discrimination between cases and controls (R2Y = 0.68 and Q2Cum = 0.60). Fig. 5b shows the score plot and the permutation test.
Fig. 5.
Identification of a comprehensive molecular MetS signature.
a: Feature selection strategy developed to build a comprehensive molecular MetS signature, stable over time. b: Score plot of the Partial Least Squares Discriminant Analysis performed on all subjects at both times, on the 102 variables issued from feature selection, showing the MetS phenotype effect (R2Y = 0.68 and Q2 = 0.60) with the graphical result of the 200-permutation test. Graphical representation using the SIMCA software – Umetrics AB, 2015.
In a perspective of future clinical application where a short list of robust biomarkers is needed, an optimized reduced signature was then proposed. Of the 102 statistically significant variables of interest, 41 were successfully identified or annotated, and 6 attributed to a characterized compound class. Then, the spectral redundancy (6 variables for 2 metabolites) and the redundancy between methods were eliminated (9 variables for 4 shared metabolites), keeping the most robust variable (highest intensity, best peak purity) as representative. Restricting this signature to the most robust unique annotated compounds, the final model (R2Y = 0.61 and Q2Cum = 0.55; Fig. 6a) included 26 metabolites, namely, D-α- and D-β-glucose, lactic acid, 1,5-anhydroglucitol, L-glutamic acid, L-valine, L-proline, isoleucine, betaine, proline betaine, glutamyl glutamine, γ-glutamyl leucine, urea, pipecolic acid, indole-3-propionic acid, LPC(16:0_0:0), LPC (P-16:0_0:0), LPC(18:1_0:0), LPC(18:2_0:0), LPC(20:1_0:0), PE(18:0_20:4), PC(18:0_20:3), PC(P-39:5), TG(16:0_18:1_18:1), GM3(42:2), and total cholesterol (Fig. 6b, and Supplementary data). The loading plot highlighted the importance of these metabolites in the discrimination. This model still allowed a very good prediction (AUC = 0.96, CI [0.94–0.99]) with 11% of error rate (12% using cross validated estimation) (Fig. 6c and d). To illustrate the complementarity of the present signature to clinical variables, correlation analyses between the predicted Y and the individual clinical criteria related to MetS were performed. Results (Supplemental Table 5) showed correlation coefficients ranging from −0.48 to 0.77, which is consistent with the additional metabolic information brought by the present signature.
Fig. 6.
Reduced MetS signature.
a: Score plot of the Partial Least squares Discriminant Analysis performed on the most robust unique annotated compounds for all subjects at both times (R2Y=0.58 and Q2=0.56) with the graphical result of the 200-permutation evaluation.
b: Loading plot associated with the score plot of the PLS-DA. This plot corresponds to the projection of each variable on the latent components of the PLS-DA model. The farthest a variable is from the plot origin, the highest importance it has to the discrimination. As the twin of the score plot, the loading plot enables to grasp similarity between variables in the contribution to discrimination (proximity on the plot) as well as to assess which group of individuals is more likely to have a high intensity value of the metabolite. Status: MetS status; Metabolite_M1: detected in C18pos; Metabolite_M2: detected in C18neg; Metabolite_M3: HILICneg; Metabolite_M4: detected in LIPIDO; Metabolite_M5: detected in GCMS; Metabolite_M6 detected in NMR; Metabolite_F: variable related to a metabolite fragment.
c: ROC curve of the PLS-DA model (11% adjusted misclassification rate, AUC=0.96, CI:[0.94–0.99]). d: box-plot of the error rate corresponding to the 200 cross-validated models (12% error rate, with adjusted confusion matrix). Graphical representation using the SIMCA software – Umetrics AB, 2015.
5. Discussion
Metabolic syndrome is a complex health condition responsible for the concurrence of several metabolic abnormalities and cardiovascular disturbances. Even if several clinical definitions co-exist, its increasing worldwide prevalence, with now above 1 billion people affected worldwide, particularly in the aging population with chronic metabolic diseases, makes it a global public health concern [[64], [65], [66]]. In this context, the present study objective was to deeply investigate and characterize the MetS phenotype. Our strategy was based on analysing MetS subjects stable over a 3-year follow-up and on performing a large phenotyping using complementary untargeted metabolomics/lipidomics platforms. The type of design selected (i.e., comparing each individual with himself, and with a control group) is of major interest in metabolic disease research as it increases the ability to identify potential biomarkers by reducing the impact of confounding factors. Using this multiplatform approach allowed then generating novel integrated insights into MetS mechanisms and proposing new candidate biomarkers within an optimized statistically, analytically and biologically refined associated molecular signature.
The multiplatform untargeted strategy allowed a broad metabolome coverage, delivering a comprehensive picture of the MetS with no a priori knowledge the identity of analytes. Indeed, advances in analytical techniques, and particularly the increase of their resolving and separation powers, significantly impacted metabolomics research allowing higher sensitivity and deeper metabolic coverage [66]. In the present study, we successfully measured 2915 metabolite and lipid features in serum samples from 123 older men at two time points using 6 distinct analytical platforms. Results showed, as expected, good complementarity of the 6 methods, allowing to detect a large diversity of metabolites, from very polar molecules such as carbohydrates, or amino acids to very apolar compounds such as TG. This complementarity was also evidenced at each data treatment steps, first by the low correlation level between method datasets, and between individual metabolite and lipid related features, and second by the small number of shared metabolites and lipids detected by several methods. Among them, it is interesting to note that carbohydrates and derivatives, often poorly detected in LC-HRMS methods, are more precisely characterized (because of enhanced separation performance) by GC-HRMS and more quantitatively assessed by NMR.
The significantly altered metabolites captured by the multi-metabolomic platforms were integrated to reveal the modulated metabolic pathways and get potential information about the involved mechanisms. The contextualization of metabolites into pathways and diseases is a difficult task that stems from several of the following aspects. Firstly, despite the use of the most recent databases and tools, a large part of the metabolome remains unknown and identifiers of annotated species have to be harmonized, as well as ontologies between different research fields to fully map identified metabolites and nodes in metabolic networks [59]. This partial mapping, together with some annotation uncertainties (especially for lipids), can lead to limitation of biological interpretation [59]. Secondly, the complexity of interpretation lies in the fact that metabolites are involved in multiple overlapping pathways. Thirdly, the challenge in metabolic network analysis resides in the quality of the reconstructed networks, as well as the choice of the algorithm to extract sub-networks [60]. Eventually, although pathway enrichment analysis is useful in identifying the metabolic pathways in which modulated metabolites are overrepresented and which are therefore more likely to be modulated, the results of this analysis must be interpreted with caution as the different state-of-the-art metabolomics methods and databases allow covering only a limited part of the metabolic network, resulting in some uncovered or partially covered metabolic pathways that would never be highlighted because of analytical limits [67]. In the present study, the mapping of modulated metabolites into the highly curated human metabolic network Recon2.2, performed with the MetExplore tool [62], revealed that among the specific identified modulated metabolic pathways, changes in the global metabolism of amino acids is of key importance in MetS. In fact, the 6 metabolic pathways linked to amino acids, showed that glutamate, glutamine, and serine play central roles in energy and nitrogen metabolisms. These observations are in accordance with several publications that have linked their metabolisms to insulin resistance and impaired glucose tolerance [68,69]. More precisely, glutamate, as part of 5/11 modulated pathways, was found as one of the most modulated metabolites (corrected p-value = 5.8 × 10−10). The role of glutamine and glutamate (as well as their ratio) on insulin and glucagon activity have been widely reported [70,71]. Moreover, it has been shown that serine and glycine nitrogen groups contribute in multiple transamination reactions via glutamate formation [72]. Secondly, branched-chain amino acid (BCAA) metabolism was identified as highly modulated (with corrected LMM p-values ranging from 2 × 10−6 to 2 × 10−4) as well as some gamma-glutamyl-derivatives (corrected p-value < 2 × 10−4) formed in the glutathione dependent transport of some amino acids. Some previous work indicated that these modulations could also reflect an effect on the transport of these amino acids [73]. These results are in accordance with a large body of literature associating an increase of BCAA pathway to obesity and related metabolic dysfunction, as well as to insulin resistance and diabetes [69,[74], [75], [76], [77]]. Multi-organ complex metabolic interplaying processes seem to be linked to these changes and the full mechanisms still remain to be elucidated [[77], [78], [79]]. In addition, the gut microbiota may play a role in the supply of BCAA to the body [[79], [80], [81]].
Moreover, serine is a central amino acid linked not only to other amino acids but also to lipids with the phosphatidylserine family. In particular, it is the precursor for the biosynthesis of the large family of sphingolipids, which includes sphingomyelins and gangliosides. In fact, among this last subclass, the ganglioside GM3 circulating levels were reported as affected by both glucose and lipid abnormalities, even if the mechanisms are still unclear [82,83]. In the present study, some GM3 species were found highly negatively modulated (GM3 (42:2) corrected LMM p-value = 2 × 10−6; GM3 (36:1) corrected p-value = 2 × 10−5), with significant positive correlation with HDL-Chol and negative with glucose levels. Such findings may confer a protection role to the GM3 species against cardiometabolic risk. In particular, GM3 (42:2), often reported in the literature as GM3(d18:1/24:1), has been identified as the most attractive target for metabolic screening of MetS risk factors [84]. Analysis of the different lipid families and subclasses have been shown to provide insights into the pathophysiology of MetS and all of its clinical components [13]. In lipidomics dataset, TG were found to be the most modulated family, as expected in the context of MetS and dyslipidemia. Lysophosphatidylcholine species (LPCs) as major bioactive lipid components of oxidized LDL-cholesterol, are known to play a key role in inflammation processes [85,86]. In particular, LPC levels were described as decreased with insulin resistance, independently of obesity [87]. More specifically, two main modulated species in our study (namely LPC(18:2_0:0) and LPC(16:0_0:0), corrected LMM p-value = 2 × 10−4 and 3 × 10−3, respectively) were found as being associated with metabolic health in obesity [88]. Another study deeply investigated the positive associations between LPC levels and whole-body insulin sensitivity and concluded that they were related to muscle insulin sensitivity, rather than hepatic insulin resistance [89]. Moreover, van der Kolk et al. [90] recently highlighted that lower fasting plasma levels of several LPCs, are associated with muscle insulin resistance rather than obesity per se, with a key importance of LPC(18:1_0:0), LPC(18:2_0:0), LPC(16:0_0:0), as found in the present study. Furthermore, the modulation of the glycerophospholipid pathway also reflects the contribution of PCs from two major subclasses that were found inversely modulated, namely diacyl-phosphatidylcholines (PCaa) and the ether-bond PCs. In fact, PCaa were described as essential for the VLDL particles and HDL hepatic formation and secretion [89,91]. Ether-bound PCs were already reported to be negatively correlated with MetS [89] with a decrease in oxidative stress level, as they can act as antioxidants to prevent lipoprotein oxidation.
Additionally to LPCs involved in glucose metabolism/uptake, circulating hexoses were found highly modulated (corrected LMM p-values between 10−6 and 10−14) as well as the galactose pathway. Associations between plasma hexose levels and dysglycemia/risk for T2D have been already reported and it was suggested that excess glucose could increase fluxes towards secondary conversion pathways with for example, enlarged fructose concentrations [89,92]. Fructose, glucose, and galactose are all precursors of glycogen and the importance of galactose as a substrate for hepatic glycogen synthesis has been described [93]. In consequence, these increased sugar concentrations may again, reflect some insulin resistance and/or β-cell dysfunction in our older adult cases. While metabolic syndrome and insulin resistance have been associated with increased gluconeogenesis [70,94], the importance of glutamine as a renal gluconeogenic substrate is in agreement with the observed decreased in circulating level of glutamine in association with the modulation of the glucose pathway. Furthermore, the observed decreased level of 1,5-anhydroglucitol, previously described in prediabetes and T2D, was related to its renal loss stimulated under hyperglycemic conditions [73,95,96].
Finally, in contrast to Ostojic et al. [97], the present case/control comparison showed that plasma levels of the amino acid derivative, guanidinoacetic acid, were significantly down-regulated in cases with a significant increased level of creatine. This might suggest an increase of creatine synthesis from guanidinoacetic acid. However, it is important to keep in mind that creatine levels can also be modulated by numerous factors, in addition to the guanidinoacetic acid-creatine axis.
As metabolome does also reflect some modulation by extrinsic factors, nutritional data must be considered. Indeed, subject characteristics revealed that MetS subjects appear to be under some nutritional ‘control’ with less energy and carbohydrate intakes as usually recommended. The negative modulations of exogenous sugars evaluated from metabolomics data, are in accordance with these results. Moreover, data from dietary intakes suggested that cases were consuming less fibres/cereals, in agreement with beneficial effects of fibres on glycemic control/insulin resistance [98]. In serum metabolome, the indole-3-propionic acid level was found positively correlated with dietary fiber ingestion as previously described [99]. This metabolite, known to be a microbial product of tryptophan, was described as part of the link between diet, intestinal microbiota, insulin, and glucose metabolism through its role in modulating secretion of glucagon-like peptide (GLP)−1 [100]. As reported by De Mello et al. [99], it was directly associated with fiber intake mainly originated from whole grains. Indeed, the type of carbohydrate ingested and pH can modulate the production of indole metabolites by the intestinal microflora. Our results, together with the lower consumption of grain products in cases, suggest similar mechanisms. In addition, pipecolic acid which originates mainly from the catabolism of dietary lysine by intestinal bacteria [101], was found positively correlated with cereals and fiber intakes, consistently with previous studies associating pipecolic acid with a whole grain-enriched diet [102] as well as with total and soluble fiber intakes [103].
Finally, activation/prevalence of gut microbial species, bile acid reabsorption, and dietary factors have been shown to change the composition of the bile acid pool: among their numerous functions, bile acids are involved in the regulation of energy expenditure, glucose and lipid metabolism. Chenodeoxycholic acid (corrected LMM p-value=10−3) in particular, has been shown to be increased in insulin resistance and obesity [104,105].
In addition to this biological contextualization, different alternative integration methods have been developed in systems biology to reduce the gap between the high amount of generated data and the knowledge of complex biological systems. Performing large-scale metabolomics requires special attention, in particular because of the high dimensionality of data [106]. Special attention must therefore be paid to standard operating procedures for data production and data processing [107] to be able to extract meaningful information [108]. In the present strategy, a mid-level data fusion (i.e. selection of variables from each metabolomic/lipidomic dataset before integration) was performed to build the MetS comprehensive signature. First, removing analytical redundancies both facilitates annotation of metabolites, which is a complex step in the metabolomics workflow, and avoids over correction during multiple testing. Then, as described in different studies, variable selection is of major interest, both to limit the over fitting and increase robustness of the models, maximize potentiality of each complementary method, and summarize the biological information of importance [109]. Finally, contextualization of signatures using enrichment and network analyses are of great value for a functional integration and the identification of molecular mechanisms involved in the pathophysiology. Consequently, in the present study, the refinement of the comprehensive signature, performed both in terms of measurement reliability, but also by showing the consistent association between the modulated metabolites/lipids and the underlying biological mechanisms, is increasing the value of the proposed biomarker combination within the reduced signature for further investigation and possible clinical application [110].
This study has limitation in term of sample size and male subject inclusion only. Nevertheless, the NuAge cohort quality in terms of follow-up and phenotypic characteristics as well as the present study being performed at two-time points, contributes to the robustness of the results. Additionally, the use of such a large serum phenotyping approach involving complementary analytical platforms is time and expertise consuming and the associated high-costs limit such discovery studies. Evidently, the proposed biomarker combination within the reduced signature will need validation for its qualification in various populations. The present study will contribute to fulfill this objective and accelerate the translation into clinics, with the possibility of implementing such a reduced signature using a targeted strategy based on a single available and robust analytical device, with manageable analytical issues and reasonable costs.
From a clinical perspective, the interest of a large phenotyping strategy, as the one reported here, to better characterize disease phenotypes and discover potential candidate biomarkers and signatures that can become after validation, new tools for diagnosis, is now a recognized approach [111]. Then, in a subsequent step, the combination of the assessment of an individual health status from metabolomics/lipidomics together with clinical measurements [112] will contribute to improve clinical diagnosis and refine the different phenotypes associated with MetS components. Indeed, disentangling the relative contribution of different factors (e.g. diet, medical co-morbidities, fasting versus fed state, age, sex) to circulating metabolite levels is required to understand the functional role of novel metabolites, and assess the potential value of selected markers in association with clinical outcomes [113]. Finally, this will allow developing systems medicine by combining high-throughput large scale measurements at multi-scale levels over time with different types of clinical information [114].
6. Contributors
BC, JAM, ET, CJu, PG and EP-G designed the study. BC, NL, ET, MP, PG, and EP-G performed subject selection. CC, FC, BeC, CJo, BL, CM, and EC-V generated raw data, pre-processed and annotate them (for NMR, HILIC-HRMS, lipidomics, C18-HMRS and GC-HRMS, respectively). SM processed the data and performed statistical analyses. MB, JFM, MP and ET supervised statistical analyses. FJ, NP and FV performed network analyses. FF and EP-G supervised metabolomics analyses. BC and EP-G draft the manuscript. PG and JAM were part of the initial co-principal investigators of the NuAge study and are part of the NuAge Database and Biobank Steering Committee. PG is the scientific director of the NuAge Biobank. All authors contributed to writing, critically revisiting and approved the submitted version of the manuscript.
7. Data sharing statement
All de-identified data supporting this manuscript are available in Supplementary data.
Declaration of Competing Interest
All authors declare they have no conflict of interest.
Acknowledgments
The NuAge Study was supported by a research grant from the Canadian Institutes of Health Research (CIHR; MOP-62842). The actual NuAge Database and Biobank, containing data and biologic samples of 1753 NuAge participants (from the initial 1793 participants), are supported by the Fonds de recherche du Québec (FRQ; 2020-VICO-279753), the Quebec Network for Research on Aging, a thematic network funded by the Fonds de Recherche du Québec - Santé (FRQS) and by the Merck-Frost Chair funded by La Fondation de l'Université de Sherbrooke. All metabolomics and lipidomics analyses were funded and performed within the MetaboHUB French infrastructure (ANR-INBS-0010).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103440.
Appendix. Supplementary materials
References
- 1.Saklayen M.G. The global epidemic of the metabolic syndrome. Curr Hypertens Rep. 2018;20(2):12. doi: 10.1007/s11906-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godos J., Zappala G., Bernardini S., Giambini I., Bes-Rastrollo M., Martinez-Gonzalez M. Adherence to the Mediterranean diet is inversely associated with metabolic syndrome occurrence: a meta-analysis of observational studies. Int J Food Sci Nutr. 2017;68(2):138–148. doi: 10.1080/09637486.2016.1221900. [DOI] [PubMed] [Google Scholar]
- 3.Edwardson C.L., Gorely T., Davies M.J. Association of sedentary behaviour with metabolic syndrome: a meta-analysis. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0034916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie H., Roser M. Obesity. https://ourworldindata.org/obesity; 2017.
- 5.Alberti K.G., Eckel R.H., Grundy S.M. Harmonizing the metabolic syndrome: a joint interim statement of the international diabetes federation task force on epidemiology and prevention; national heart, lung, and blood institute; american heart association; world heart federation; international atherosclerosis society; and international association for the study of obesity. Circulation. 2009;120(16):1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 6.Aguilar M., Bhuket T., Torres S., Liu B., Wong R.J. Prevalence of the metabolic syndrome in the United States, 2003-2012. JAMA. 2015;313(19):1973–1974. doi: 10.1001/jama.2015.4260. [DOI] [PubMed] [Google Scholar]
- 7.Mottillo S., Filion K.B., Genest J. The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. J Am Coll Cardiol. 2010;56(14):1113–1132. doi: 10.1016/j.jacc.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill S., O'Driscoll L. Metabolic syndrome: a closer look at the growing epidemic and its associated pathologies. Obes Rev. 2015;16(1):1–12. doi: 10.1111/obr.12229. [DOI] [PubMed] [Google Scholar]
- 9.Day C. Metabolic syndrome, or what you will: definitions and epidemiology. Diabetes Vasc Dis Res. 2007;4(1):32–38. doi: 10.3132/dvdr.2007.003. [DOI] [PubMed] [Google Scholar]
- 10.Ramautar R., Berger R., van der Greef J., Hankemeier T. Human metabolomics: strategies to understand biology. Curr Opin Chem Biol. 2013;17(5):841–846. doi: 10.1016/j.cbpa.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 11.Lindon J.C., Nicholson J.K. The emergent role of metabolic phenotyping in dynamic patient stratification. Expert Opin Drug Metab Toxicol. 2014;10(7):915–919. doi: 10.1517/17425255.2014.922954. [DOI] [PubMed] [Google Scholar]
- 12.Dumas M.E., Kinross J., Nicholson J.K. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology. 2014;146(1):46–62. doi: 10.1053/j.gastro.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Meikle P.J., Christopher M.J. Lipidomics is providing new insight into the metabolic syndrome and its sequelae. Curr Opin Lipidol. 2011;22(3):210–215. doi: 10.1097/MOL.0b013e3283453dbe. [DOI] [PubMed] [Google Scholar]
- 14.Guasch-Ferre M., Hruby A., Toledo E. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39(5):833–846. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monnerie S., Comte B., Ziegler D., Morais J.A., Pujos-Guillot E., Gaudreau P. Metabolomic and lipidomic signatures of metabolic syndrome and its physiological components in adults: a systematic review. Sci Rep. 2020;10(1) doi: 10.1038/s41598-019-56909-7. Uk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudreau P., Morais J.A., Shatenstein B. Nutrition as a determinant of successful aging: description of the Quebec longitudinal study nuage and results from cross-sectional pilot studies. Rejuvenation Res. 2007;10(3):377–386. doi: 10.1089/rej.2007.0596. [DOI] [PubMed] [Google Scholar]
- 17.Statistique C. Proportion des personnes de 12 ans et plus ayant reçu un diagnostic d'hypertension, selon le groupe d’âge et selon le sexe, Québec, 2013–4.
- 18.Statistique Canada c. Proportion des personnes de 12 ans et plus ayant reçu un diagnostic d'hypertension, selon le groupe d’âge et selon le sexe, Québec, 2013–4.
- 19.Statistique C. Statistiques de santé et de bien-être selon le sexe. principaux problèmes de santé chroniques. 2012.
- 20.Statistique C. Tableau 13-10-0096-20 Indice de masse corporelle, embonpoint ou obèse, autodéclaré corrigé, adulte, selon le groupe d'âge (18 ans et plus).
- 21.Tan M.C., Ng O.C., Wong T.W., Joseph A., Chan Y.M., Heja A.R. Prevalence of metabolic syndrome in type 2 diabetic patients: a comparative study using WHO, NCEP ATP III, IDF and harmonized definitions. Health. 2013;5(10):1689–1696. [Google Scholar]
- 22.Masnoon N., Shakib S., Kalisch-Ellett L., Caughey G.E. What is polypharmacy? A systematic review of definitions. BMC Geriatr. 2017;17(1):230. doi: 10.1186/s12877-017-0621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washburn R.A., Smith K.W., Jette A.M., Janney C.A. The physical activity scale for the elderly (PASE): development and evaluation. J Clin Epidemiol. 1993;46(2):153–162. doi: 10.1016/0895-4356(93)90053-4. [DOI] [PubMed] [Google Scholar]
- 24.Farivar S.S., Cunningham W.E., Hays R.D. Correlated physical and mental health summary scores for the SF-36 and SF-12 Health Survey, V.I. Health Qual Life Outcomes. 2007;5:54. doi: 10.1186/1477-7525-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ware J.E., Kosinski M., Keller S.D. 2nd Ed. QualityMetric, Inc; 2001. SF-36® Physical and Mental Health Summary Scales: A Manual for Users. Boston, MA: Lincoln, RI. [Google Scholar]
- 26.Shatenstein B., Gauvin L., Keller H. Individual and collective factors predicting change in diet quality over 3 years in a subset of older men and women from the NuAge cohort. Eur J Nutr. 2016;55(4):1671–1681. doi: 10.1007/s00394-015-0986-y. [DOI] [PubMed] [Google Scholar]
- 27.Shatenstein B., Nadon S., Godin C., Ferland G. Diet quality of Montreal-area adults needs improvement: estimates from a self-administered food frequency questionnaire furnishing a dietary indicator score. J Am Diet Assoc. 2005;105(8):1251–1260. doi: 10.1016/j.jada.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Broadhurst D., Goodacre R., Reinke S.N. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics. 2018;14(6):72. doi: 10.1007/s11306-018-1367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boudah S., Olivier M.F., Aros-Calt S. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2014;966:34–47. doi: 10.1016/j.jchromb.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 30.Aros-Calt S., Muller B.H., Boudah S. Annotation of the staphylococcus aureus metabolome using liquid chromatography coupled to high-resolution mass spectrometry and application to the study of methicillin resistance. J Proteome Res. 2015;14(11):4863–4875. doi: 10.1021/acs.jproteome.5b00697. [DOI] [PubMed] [Google Scholar]
- 31.Johnsen E., Wilson S.R., Odsbu I. Hydrophilic interaction chromatography of nucleoside triphosphates with temperature as a separation parameter. J Chromatogr A. 2011;1218(35):5981–5986. doi: 10.1016/j.chroma.2011.01.066. [DOI] [PubMed] [Google Scholar]
- 32.Seyer A., Boudah S., Broudin S., Junot C., Colsch B. Annotation of the human cerebrospinal fluid lipidome using high resolution mass spectrometry and a dedicated data processing workflow. Metabolomics. 2016;12:91. doi: 10.1007/s11306-016-1023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao X.F., Pujos-Guillot E., Sebedio J.L. Development of a quantitative metabolomic approach to study clinical human fecal water metabolome based on trimethylsilylation derivatization and GC/MS analysis. Anal Chem. 2010;82(15):6447–6456. doi: 10.1021/ac1006552. [DOI] [PubMed] [Google Scholar]
- 34.Giacomoni F., Le Corguille G., Monsoor M. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics. 2015;31(9):1493–1495. doi: 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tautenhahn R., Bottcher C., Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Kloet F.M., Bobeldijk I., Verheij E.R., Jellema R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J Proteome Res. 2009;8(11):5132–5141. doi: 10.1021/pr900499r. [DOI] [PubMed] [Google Scholar]
- 37.Monnerie S., Petera M., Lyan B., Gaudreau P., Comte B., Pujos-Guillot E. Analytic correlation filtration: a new tool to reduce analytical complexity of metabolomic datasets. Metabolites. 2019;9(11) doi: 10.3390/metabo9110250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert P., Escoufier Y. Unifying tool for linear multivariate statistical-methods - Rv-coefficient. J R Stat Soc. 1976;25(3):257–265. C-App. [Google Scholar]
- 39.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: a language and environment for statistical computing. [Google Scholar]
- 40.Le S., Josse J., Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25(1):1–18. [Google Scholar]
- 41.Smilde A.K., van der Werf M.J., Bijlsma S., van der Werff-van der Vat B.J., Jellema R.H. Fusion of mass spectrometry-based metabolomics data. Anal Chem. 2005;77(20):6729–6736. doi: 10.1021/ac051080y. [DOI] [PubMed] [Google Scholar]
- 42.Cline M.S., Smoot M., Cerami E. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer J.M., Rocha F.M.M., Nobre J.S. Graphical tools for detecting departures from linear mixed model assumptions and some remedial measures. Int Stat Rev. 2017;85(2):290–324. [Google Scholar]
- 44.Rinaudo P., Boudah S., Junot C., Thevenot E.A. biosigner: a new method for the discovery of significant molecular signatures from omics data. Front Mol Biosci. 2016;3:26. doi: 10.3389/fmolb.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia J.G., Broadhurst D.I., Wilson M., Wishart D.S. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9(2):280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robin X., Turck N., Hainard A. pROC: an open-source package for R and S plus to analyze and compare ROC curves. BMC Bioinform. 2011;12 doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 48.Guijas C., Montenegro-Burke J.R., Domingo-Almenara X. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 2018;90(5):3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wishart D.S., Feunang Y.D., Marcu A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–DD17. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horai H., Arita M., Kanaya S. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 51.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Babushok V.I., Linstrom P.J., Reed J.J. Development of a database of gas chromatographic retention properties of organic compounds. J Chromatogr A. 2007;1157(1–2):414–421. doi: 10.1016/j.chroma.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 53.Harvey D.J., Vouros P. Mass Spectrometric Fragmentation of trimethylsilyl and related alkylsilyl derivatives. Mass Spectrom Rev. 2020;39(1–2):105–211. doi: 10.1002/mas.21590. [DOI] [PubMed] [Google Scholar]
- 54.Sumner L.W., Amberg A., Barrett D. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colsch B., Fenaille F., Warnet A., Junot C., Tabet J.C. Mechanisms governing the fragmentation of glycerophospholipids containing choline and ethanolamine polar head groups. Eur J Mass Spectrom. 2017;23(6):427–444. doi: 10.1177/1469066717731668. [DOI] [PubMed] [Google Scholar]
- 56.Liebisch G., Fahy E., Aoki J. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J Lipid Res. 2020;61(12):1539–1555. doi: 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicholson J.K., Foxall P.J., Spraul M., Farrant R.D., Lindon J.C. 750MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67(5):793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 58.Swainston N., Smallbone K., Hefzi H. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12(7) doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poupin N., Vinson F., Moreau A. Improving lipid mapping in genome scale metabolic networks using ontologies. Metabolomics. 2020;16(4) doi: 10.1007/s11306-020-01663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frainay C., Jourdan F. Computational methods to identify metabolic sub-networks based on metabolomic profiles. Brief Bioinform. 2017;18(1):43–56. doi: 10.1093/bib/bbv115. [DOI] [PubMed] [Google Scholar]
- 61.Faust K., Dupont P., Callut J., van Helden J. Pathway discovery in metabolic networks by subgraph extraction. Bioinformatics. 2010;26(9):1211–1218. doi: 10.1093/bioinformatics/btq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cottret L., Frainay C., Chazalviel M. MetExplore: collaborative edition and exploration of metabolic networks. Nucleic Acids Res. 2018;46(W1):W495–W502. doi: 10.1093/nar/gky301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chazalviel M., Frainay C., Poupin N. MetExploreViz: web component for interactive metabolic network visualization. Bioinformatics. 2018;34(2):312–313. doi: 10.1093/bioinformatics/btx588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ervin R.B. Prevalence of metabolic syndrome among adults 20 years of age and over, by sex, age, race and ethnicity, and body mass index: united States, 2003-2006. Natl Health Stat Rep. 2009;(13):1–7. [PubMed] [Google Scholar]
- 65.Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Pract. 2014;2014 doi: 10.1155/2014/943162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Ranasinghe P., Mathangasinghe Y., Jayawardena R., Hills A.P., Misra A. Prevalence and trends of metabolic syndrome among adults in the asia-pacific region: a systematic review. BMC Public Health. 2017;17 doi: 10.1186/s12889-017-4041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frainay C., Schymanski E.L., Neumann S. Mind the gap: mapping mass spectral databases in genome-scale metabolic networks reveals poorly covered areas. Metabolites. 2018;8(3) doi: 10.3390/metabo8030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sookoian S., Pirola C.J. Alanine and aspartate aminotransferase and glutamine-cycling pathway: their roles in pathogenesis of metabolic syndrome. World J Gastroentero. 2012;18(29):3775–3781. doi: 10.3748/wjg.v18.i29.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.'t Hart L.M., Vogelzangs N., Mook-Kanamori D.O. Blood metabolomic measures associate with present and future glycemic control in type 2 diabetes. J Clin Endocr Metab. 2018;103(12):4569–4579. doi: 10.1210/jc.2018-01165. [DOI] [PubMed] [Google Scholar]
- 70.Cheng S.S., Rhee E.P., Larson M.G. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–U132. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cabrera O., MC Jacques-Silva, Speier S. Glutamate is a positive autocrine signal for glucagon release. Cell Metab. 2008;7(6):545–554. doi: 10.1016/j.cmet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alves A., Bassot A., Bulteau A.L., Pirola L., Morio B. Glycine metabolism and its alterations in obesity and metabolic diseases. Nutrients. 2019;11(6) doi: 10.3390/nu11061356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suhre K., Meisinger C., Doring A. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS ONE. 2010;5(11) doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Newgard C.B., An J., Bain J.R. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang T.J., Larson M.G., Vasan R.S. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–U83. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferrannini E., Natali A., Camastra S. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62(5):1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lynch C.J., Adams S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newgard C.B. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15(5):606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.White P.J., Newgard C.B. Branched-chain amino acids in disease. Science. 2019;363(6427):582–583. doi: 10.1126/science.aav0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saad M.J.A., Santos A., Prada P.O. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiology. 2016;31(4):283–293. doi: 10.1152/physiol.00041.2015. [DOI] [PubMed] [Google Scholar]
- 81.Bloomgarden Z. Diabetes and branched-chain amino acids: what is the link? J Diabetes. 2018;10(5):350–352. doi: 10.1111/1753-0407.12645. [DOI] [PubMed] [Google Scholar]
- 82.Sato T., Nihei Y., Nagafuku M. Circulating levels of ganglioside GM3 in metabolic syndrome: a pilot study. Obes Res Clin Pract. 2008;2(4):231–238. doi: 10.1016/j.orcp.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 83.Huynh K., Barlow C.K., Jayawardana K.S. High-throughput plasma lipidomics: detailed mapping of the associations with cardiometabolic risk factors. Cell Chem Biol. 2019;26(1) doi: 10.1016/j.chembiol.2018.10.008. 71-+ [DOI] [PubMed] [Google Scholar]
- 84.Veillon L., Go S., Matsuyama W. Identification of ganglioside GM3 molecular species in human serum associated with risk factors of metabolic syndrome. PLoS ONE. 2015;10(6) doi: 10.1371/journal.pone.0129645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aiyar N., Disa J., Ao Z.H. Lysophosphatidylcholine induces inflammatory activation of human coronary artery smooth muscle cells. Mol Cell Biochem. 2007;295(1–2):113–120. doi: 10.1007/s11010-006-9280-x. [DOI] [PubMed] [Google Scholar]
- 86.Lusis AJ. Atherosclerosis. Nature. 2000;407(6801):233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Surowiec I., Noordam R., Bennett K. Metabolomic and lipidomic assessment of the metabolic syndrome in Dutch middle-aged individuals reveals novel biological signatures separating health and disease. Metabolomics. 2019;15(2):23. doi: 10.1007/s11306-019-1484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tonks K.T., Coster A.C.F., Christopher M.J. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity. 2016;24(4):908–916. doi: 10.1002/oby.21448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Floegel A., Stefan N., Yu Z.H. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van der Kolk B.W., Vogelzangs N., Jocken J.W.E. Plasma lipid profiling of tissue-specific insulin resistance in human obesity. Int J Obesity. 2019;43(5):989–998. doi: 10.1038/s41366-018-0189-8. [DOI] [PubMed] [Google Scholar]
- 91.Cole L.K., Vance J.E., Vance D.E. Phosphatidylcholine biosynthesis and lipoprotein metabolism. BBA Mol Cell Biol. 2012;1821(5):754–761. doi: 10.1016/j.bbalip.2011.09.009. L. [DOI] [PubMed] [Google Scholar]
- 92.Fiehn O., Garvey W.T., Newman J.W., Lok K.H., Hoppel C.L., Adams S.H. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS ONE. 2010;5(12) doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nuttall F.Q., Gannon M.C. Dietary management of type 2 diabetes: a personal odyssey. J Am Coll Nutr. 2007;26(2):83–94. doi: 10.1080/07315724.2007.10719589. [DOI] [PubMed] [Google Scholar]
- 94.Stumvoll M., Perriello G., Meyer C., Gerich J. Role of glutamine in human carbohydrate metabolism in kidney and other tissues. Kidney Int. 1999;55(3):778–792. doi: 10.1046/j.1523-1755.1999.055003778.x. [DOI] [PubMed] [Google Scholar]
- 95.Drogan D., Dunn W.B., Lin W.C. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case control study. Clin Chem. 2015;61(3):487–497. doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- 96.Pujos-Guillot E., Brandolini M., Petera M. Systems metabolomics for prediction of metabolic syndrome. J Proteome Res. 2017;16(6):2262–2272. doi: 10.1021/acs.jproteome.7b00116. [DOI] [PubMed] [Google Scholar]
- 97.Ostojic S.M., Vranes M., Loncar D., Zenic N., Sekulic D. Guanidinoacetic acid and creatine are associated with cardiometabolic risk factors in healthy men and women: a cross-sectional study. Nutrients. 2018;10(1) doi: 10.3390/nu10010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Consortium I. Dietary fibre and incidence of type 2 diabetes in eight European countries: the EPIC-InterAct Study and a meta-analysis of prospective studies. Diabetologia. 2015;58(7):1394–1408. doi: 10.1007/s00125-015-3585-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Mello V.D., Paananen J., Lindstrom J. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci Rep. 2017;7 doi: 10.1038/srep46337. Uk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chimerel C., Emery E., Summers D.K., Keyser U., Gribble F.M., Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine l cells. Cell Rep. 2014;9(4):1202–1208. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fujita T., Hada T., Higashino K. Origin of d- and l-pipecolic acid in human physiological fluids: a study of the catabolic mechanism to pipecolic acid using the lysine loading test. Clin Chim Acta. 1999;287(1–2):145–156. doi: 10.1016/s0009-8981(99)00129-1. [DOI] [PubMed] [Google Scholar]
- 102.Hanhineva K., Lankinen M.A., Pedret A. Nontargeted metabolite profiling discriminates diet-specific biomarkers for consumption of whole grains, fatty fish, and bilberries in a randomized controlled trial. J Nutr. 2015;145(1):7–17. doi: 10.3945/jn.114.196840. [DOI] [PubMed] [Google Scholar]
- 103.Lecuyer L., Dalle C., Micheau P. Untargeted plasma metabolomic profiles associated with overall diet in women from the SU.VI.MAX cohort. Eur J Nutr. 2020;59(8):3425–3439. doi: 10.1007/s00394-020-02177-5. [DOI] [PubMed] [Google Scholar]
- 104.Cariou B., Chetiveaux M., Zair Y. Fasting plasma chenodeoxycholic acid and cholic acid concentrations are inversely correlated with insulin sensitivity in adults. Nutr Metab. 2011;8 doi: 10.1186/1743-7075-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palau-Rodriguez M., Tulipani S., Isabel Queipo-Ortuno M., Urpi-Sarda M., Tinahones F.J., Andres-Lacueva C. Metabolomic insights into the intricate gut microbial-host interaction in the development of obesity and type 2 diabetes. Front Microbiol. 2015;6:1151. doi: 10.3389/fmicb.2015.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brown M., Dunn W.B., Ellis D.I. A metabolome pipeline: from concept to data to knowledge. Metabolomics. 2005;1(1):39–51. [Google Scholar]
- 107.Dunn W.B., Broadhurst D., Begley P. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 108.Alyass A., Turcotte M., Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genomics. 2015;8 doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grissa D., Petera M., Brandolini M., Napoli A., Comte B., Pujos-Guillot E. Feature selection methods for early predictive biomarker discovery using untargeted metabolomic data. Front Mol Biosci. 2016;3:30. doi: 10.3389/fmolb.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morrow D.A., de Lemos J.A. Benchmarks for the assessment of novel cardiovascular biomarkers. Circulation. 2007;115(8):949–952. doi: 10.1161/CIRCULATIONAHA.106.683110. [DOI] [PubMed] [Google Scholar]
- 111.Yurkovich J.T., Tian Q., Price N.D., Hood L. A systems approach to clinical oncology uses deep phenotyping to deliver personalized care. Nat Rev Clin Oncol. 2020;17(3):183–194. doi: 10.1038/s41571-019-0273-6. [DOI] [PubMed] [Google Scholar]
- 112.Dammann O., Gray P., Gressens P., Wolkenhauer O., Leviton A. Systems epidemiology: what's in a name? Online J Public Health Inform. 2014;6(3):e198. doi: 10.5210/ojphi.v6i3.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cui L., Lu H.T., Lee Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom Rev. 2018;37(6):772–792. doi: 10.1002/mas.21562. [DOI] [PubMed] [Google Scholar]
- 114.Hansen J., Iyengar R. Computation as the mechanistic bridge between precision medicine and systems therapeutics. Clin Pharmacol Ther. 2013;93(1):117–128. doi: 10.1038/clpt.2012.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.