SUMMARY
The fusion of digits or toes, syndactyly, can be part of complex syndromes, including Van der Woude syndrome. A subset of Van der Woude cases is caused by dominant negative mutations in the epithelial transcription factor Grainyhead like-3 (GRHL3) and Grhl3−/− mice have soft tissue syndactyly. Although impaired interdigital cell death of mesenchymal cells causes syndactyly in multiple genetic mutants, Grhl3−/− embryos had normal interdigital cell death, suggesting alternative mechanisms for syndactyly. We found that in digit separation, the overlying epidermis forms a migrating interdigital epithelial tongue (IET) as the epithelium invaginates to separate the digits. Normally, the non-adhesive surface periderm allows the IET to bifurcate as the digits separate. In contrast, in Grhl3−/− embryos, the IET moves normally between the digits but fails to bifurcate because of abnormal adhesion of the periderm. Our study identifies epidermal developmental processes required for digit separation.
Keywords: Limb Development, Epithelial Mechanism, Digit Separation, Periderm, Syndactyly, interdigital cell death, Embryonic Epidermis, Grainyhead like-3, Van der Woude syndrome
Graphical Abstract
eTOC Blurb
Kashgari et al. identify epidermal developmental process required for digit separation. During this process, epidermal cells actively migrate downward into the underlying mesenchyme forming an interdigital epithelial tongue (IET). Fusion of the digits is prevented by the non-adhesive periderm cells on the epithelial surface.
INTRODUCTION
Classical studies in model organisms found that digit separation is mediated by interdigital mesenchymal cell death (Hurle & Colvee, 1982; Lorda-Diez et al., 2015; Montero & Hurlé, 2010). Such tissue elimination occurs via multiple cell death mechanisms including caspase-dependent apoptosis, lysosomal-mediated cell death, cell senescence, and necrosis (Chautan et al., 1999; Montero & Hurlé, 2010; Stewart et al., 2000; Zuzarte-Luis et al., 2006). The cell death begins in the distal interdigital mesenchyme, underneath the developing epidermis, where a signaling loop between epidermally expressed Fibroblast growth factor 8 (FGF-8) and Bone morphogenic proteins (Bmps) controls cell survival in the underlying mesenchyme.
Work on mouse mutants with soft tissue syndactyly has attributed the digit fusion to impaired interdigital cell death. Even developmental regulators that are selectively expressed in the digit epithelium are thought to signal to the underlying mesenchyme to effect digit separation. Yet, early ultrastructural studies of digit separation in chickens and humans suggested the possibility of an active role for the epidermis (Hurle & Colvee, 1982; Kelley, 1973). Such mechanisms, however, remain to be elucidated.
Van der Woude syndrome is characterized by cleft lip and/or cleft palate. In addition, 7-8% of Van der Woude patients have limb defects, most commonly soft tissue syndactyly (Peyrard-Janvid et al., 2014)., suggesting shared developmental mechanisms in oral-clefting and digit separation. Whereas mutations in the transcription factor gene Interferon Regulatory Factor 6 (Irf6) cause most Van der Woude cases, dominant-negative mutations in the transcription factor gene Grainyhead-like 3 (Grhl3) cause a subset of the remaining cases (Kondo et al., 2002; Peyrard-Janvid et al., 2014; Richardson, Dixon, Jiang, & Dixon, 2009; Wang et al., 2016).
The embryonic epidermis is covered by periderm, a single layer of epithelial cells that are shed prior to birth. Lacking cell adhesion molecules on their apical surfaces, periderm cells prevent adhesions of embryonic body parts in utero. In the oral epithelium, IRF6 is required for periderm formation whereas GRHL3 seems to be required for periderm function (de la Garza et al., 2013; Hammond, Dixon, & Dixon, 2017; Kousa et al., 2017; Richardson et al., 2009; Richardson et al., 2014). It is well established that oral clefting in Van der Woude syndrome is caused by abnormal adhesion of periderm cells, but the cause of syndactyly in Van der Woude patients is unknown.
Similar to Van der Woude patients, mice deleted for the epidermally-expressed Grhl3 have syndactyly, leading us to investigate epithelial mechanisms in digit separation. We found that despite normal interdigital cell death, Grhl3−/− embryos exhibit syndactyly, suggesting that mechanisms in addition to interdigital cell death are required for digit separation. We show that a specialized epithelial structure, the interdigital epithelial tongue (IET), forms at the leading edge of the epithelium invaginating into the mesenchyme between the digits dorsally and ventrally. The IET formation supports the notion that active epithelial movement contributes to digit separation. At the same time, periderm covering the developing digits is required to prevent epithelial fusion of adjacent digits, a mechanism regulated by GRHL3.
RESULTS
GRHL3 is required for normal digit separation
Although mice germline-deleted for Grhl3 (Grhl3−/−) die at birth with an open neural tube and epidermal barrier defects (Kousa et al., 2019;Ting et al., 2003; Yu et al., 2006), mice deleted for Grhl3 in the epidermis (Krt14-Cre;Grhl3fl/fl) survive into adulthood (Gordon et al., 2014). Up to a quarter of these mice have soft tissue syndactyly (Figure 1A) similar to that found in 7% of VWS patients with GRHL3 mutations (Peyrard-Janvid et al., 2014), indicating that epithelial expression of Grhl3 is required for normal digit separation and that Grhl3 deletion can model syndactyly in Van der Woude syndrome.
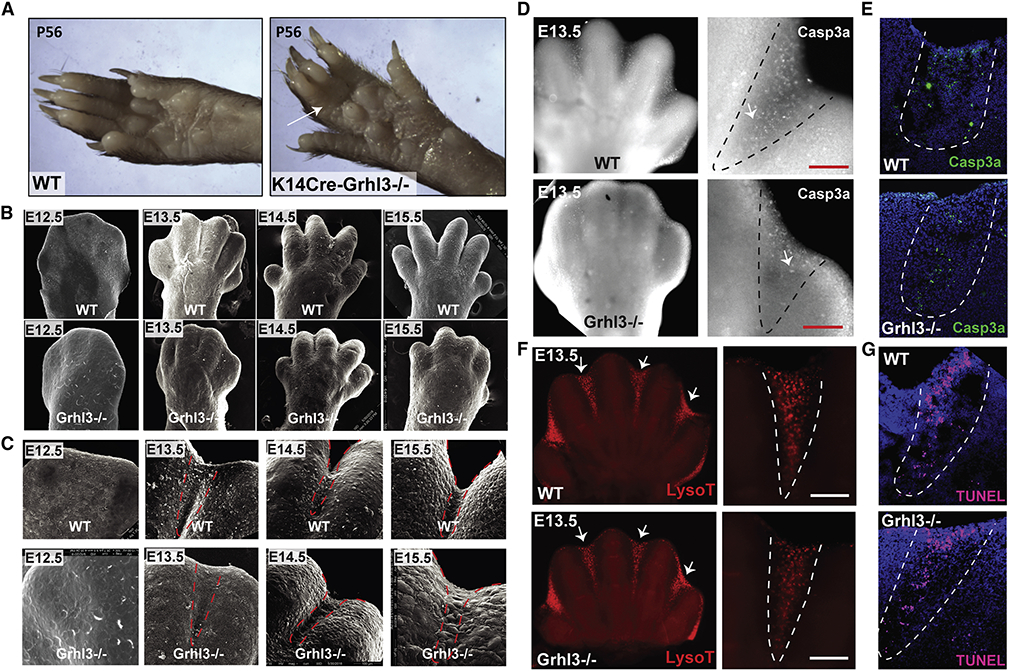
Figure 1. Syndactyly in Grhl3−/− embryos is not caused by impaired interdigital cell death.
(A) Forelimbs of WT mice (left) and mice with epithelial deletion of Grhl3 (right) at postnatal day 56. Arrow indicates soft-tissue syndactyly between the third and fourth digit in Krt14-Cre;Grhl3fl/fl mice. (B) Scanning EM of WT and Grhl3−/− forelimbs at E12.5, E13.4, E14.5 and E15.5, showing defective digit separation in Grhl3−/− embryos. (C) Higher-magnification scanning EM images of the third and fourth digit in WT and Grhl3−/− forelimbs indicate fusion between the digits at E14.5 and E15.5 in Grhl3−/− embryos. Dashed-lines indicate the interdigital zone. (D) Wholemount staining of E13.5 forelimbs with Cleaved caspase-3 (gray) in WT and Grhl3−/− embryos. (right panel) Higher-magnification of the interdigital zone indicated by dashed-lines. (E) Immunofluorescence staining of Cleaved caspase-3 (green) in E13.5 limb sections of WT embryos (top) and Grhl3−/− embryos (bottom). Dashed-lines indicate interdigital zone. (F) Lysotracker staining of WT and Grhl3−/− forelimbs at E13.5. Arrows point to the interdigital zone. The right panels are magnified views of the left images. (G) TUNEL staining (magenta) of E13.5 limb sections from WT and Grhl3−/− embryos. Dashed-lines indicate the interdigital zone.
To understand the role of Grhl3 in digit separation, we studied digit development in Grhl3−/− mouse embryos at key developmental stages: E12.5, E13.5, E14.5, and E15.5 (Figure 1B, C, S1A and Movie 1). We limited our analyses to the developmental stages before the re-fusion of the digits that starts around E16 in mice (Maconnachie, 1979).
As a consequence of cell death in the interdigital zone, wild-type (WT) forepaws form an interdigital indentation between E12.5 and E13.5 (Figure 1B, C; upper rows). A similar interdigital indentation was detected in Grhl3−/− embryos (Figure 1B, C; lower rows), suggesting that Grhl3 loss does not affect interdigital cell death. In WT mice, digit separation proceeds from E13.5 to E15.5 (Figure 1B,C; upper panels) as previously described (Hiscock et al., 2017; Maconnachie, 1979). By contrast, in Grhl3−/− embryos, digit separation proceeds more slowly between E13.5 and E14.5, and there is beginning re-fusion of the digits at E15.5 (Figure 1B, C; lower panels, Figure S1b, c and Movie 1). We found a similar digit separation delay in the foot pads of Grhl3−/− embryos (Figure S1B). These phenotypes are highly consistent; syndactyly was found in 100% of fore- and hind limbs of Grhl3−/− embryos. Together, these data indicate that GRHL3 is required for digit separation during limb development.
Interdigital cell death progresses normally in Grhl3−/− embryos
Research with mouse mutants indicates that defective programmed cell death in the interdigital mesenchyme underlies most cases of soft tissue syndactyly (Hernández-Martínez & Covarrubias, 2011) . This is even true for regulators restricted to the overlying epidermis; those are proposed to signal to the interdigital mesenchyme to control cell death (Bečić et al., 2016; Bosch et al., 2018; Kaltcheva et al., 2016; Pajni-Underwood et al., 2007). Hence, we initially hypothesized that GRHL3, which is expressed in the epidermis, regulates cell death signals sent from the overlying epidermis to the underlying interdigital mesenchyme.
Immunofluorescence staining of E13.5 forelimbs with an antibody against cleaved caspase-3, a common marker for caspase-dependent apoptosis, shows a mild decrease in caspase 3-positive cells in the interdigital zone of Grhl3−/− embryo (Figure 1D, E). In contrast, overall interdigital cell death as indicated by lysosomal activation (Figure 1F) and DNA fragmentation (Figure 1G) is unaffected in Grhl3−/− mice. These findings are consistent with the normal interdigital indentation in Grhl3−/− forepaws at E13.5 (Figure 1B, C and Figure S1A, B). Together, these results indicate that death of the interdigital mesenchyme is normal in the absence of GRHL3, despite the mild decrease in caspase-3 cleavage; the mechanisms that induce death of the interdigital mesenchyme are highly redundant and partially caspase-independent (Chautan et al., 1999; Zuzarte-Luis et al., 2006).
Digits in Grhl3−/− embryos are joined by an epithelial sheet
As our data suggest that mechanisms independent of interdigital cell death cause defective digit separation in Grhl3−/− embryos, we next investigated the nature of the interdigital fusion in E15.5 Grhl3−/− embryos. To obtain a complete picture of the syndactyly, we embedded WT and Grhl3−/− E15.5 forelimbs in plastic, followed by serial thin sectioning (2 μm) and staining with the epithelial marker keratin 5 (Parfitt et al., 2012). Serial sections were then tiled, and mosaic images digitally assembled followed by 3D reconstruction—this allowed large scale imaging of the entire forepaw (Figure 2A and Movie 2). Using imaging software, we could then slice the 3D reconstructions along two different planes: YZ axis, dorsal-ventral; and XY axis, proximal-distal (Figure 2B).
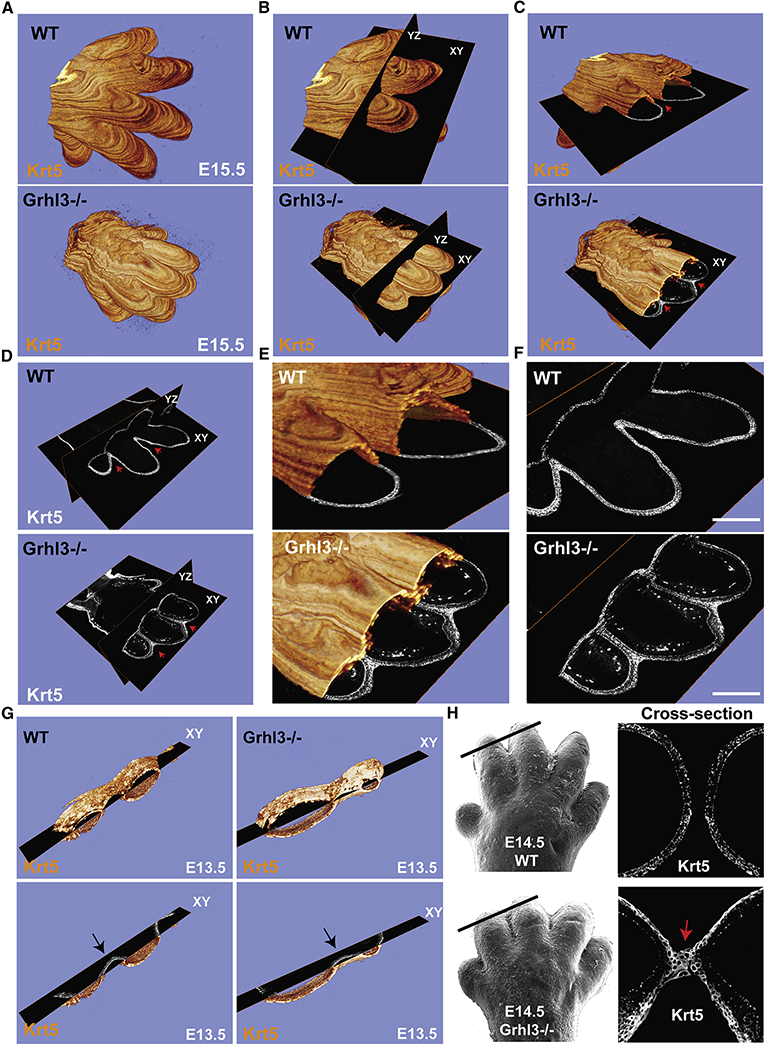
Figure 2. Interdigital epithelial fusion causes syndactyly in Grhl3−/− embryos.
(A) Three-dimensional (3D) reconstructions of E15.5 whole forelimbs from WT (top panel) and Grhl3−/− (bottom panel) embryos stained with the epithelial marker Keratin-5 (orange). (B) Representation of ortho-slices orientation of the 3D forelimb (YZ: Dorsal to ventral, XY: proximal to distal). (C) Cross-section views of the 3D reconstructions (XY: proximal to distal). Red arrows indicate complete separation of the epithelia and the digits in WT (top panel), and epithelial fusion between the unseparated digits in Grhl3/− (bottom panel). (D-F) Ortho-slices of the 3D reconstruction showing the epithelia in WT (top panel) and Grhl3−/− (bottom panel) embryos in two different planes (YZ: Dorsal to ventral, XY: proximal to distal). Red arrows indicate the interdigital epithelia in two ortho-slices planes. (G) Snapshots of 3D reconstructions stained with Keratin-5 (orange) of WT and Grhl3−/− forelimbs at E13.5. Bottom images show a single plane of the XY axis. Arrows point to the interdigital epithelia, indicated by keratin-5 staining (gray), in WT and Grhl3−/− embryos. (H) Top left, scanning EM image of WT E14.5 forelimb (reused from figure 1). Black line indicates the location of the slice. Top right, the indicated slice show Keratin-5 staining (gray). Bottom left, scanning EM image of Grhl3−/− E14.5 forelimb (reused from figure 1). Black line indicates the location of the slice. Bottom right, the indicated slice shows keratin-5 staining (gray). Red arrow indicates the interdigital epithelial fusion.
Using this method, we discovered that the digits in Grhl3−/− embryos are primarily joined by an epithelial fusion, not by interdigital mesenchyme that failed to undergo cell death (Figure 2C and Movie 3). The two ortho-slice views indicate that the epithelial fusion of the digits in Grhl3−/− embryos is observed in both directions, dorsal-ventral and proximal-distal (Figure 2D-F). No interdigital epithelial fusion was detected at E13.5 in Grhl3−/− embryos (Figure 2G), consistent with normal digit appearance at this time-point (Figure 1B, C). Beginning interdigital epithelial fusion was detected at E14.5 in Grhl3−/− mice (Figure 2H), consistent with the onset of defective digit separation as observed by scanning electron microscopy (EM) (Figure 1B, C). Based on these data, we hypothesize that in addition to interdigital cell death, mechanisms intrinsic to the epidermis are required for digit separation.
Formation of an interdigital epithelial tongue in normal digit separation
Investigating potential epidermal mechanisms in digit separation, we noticed the formation of a multi-layered interdigital epithelial tongue (IET) in the WT E15.5 forelimb 3D reconstructions (Figure 3A and Movie 2). The IET forms between the digits at the leading edge of the epidermis as it invaginates from the ventral and the dorsal side (Figure 3A). IET formation is first detected at E14.5 (Figure 3B, S2A). As the dorsal and ventral IETs invaginate, they meet, fuse, and then bifurcate, allowing separation of the digits; these events progress in a distal-to-proximal direction (Figure 3A, B).
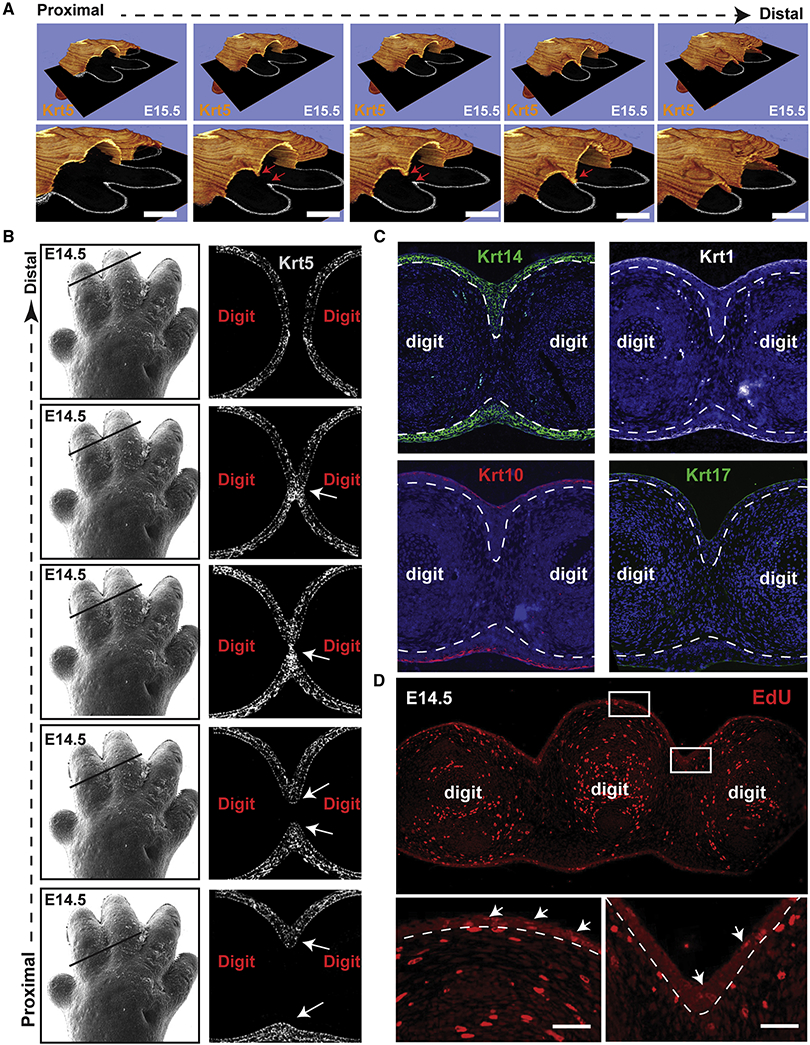
Figure 3. Formation of an interdigital epithelial tongue (IET) in normal digit separation.
(A) Series of images showing the interdigital epithelia stained with keratin-5 (orange) in continuous ortho-slices of 3D reconstructions in WT forelimbs at E15.5. Lower panels show higher magnification images. Arrows indicate the IET moving from the dorsal and ventral side. (B) left panel, scanning EM images of WT E14.5 forelimb (from figure 1). Right panel, continuous series of (proximal-to-distal) E14.5 mouse forelimb sections stained with Keratin-5 (gray). Arrows point to the IET. (C) Immunofluorescence staining showing the expression of several epithelial markers, KRT14, KRT1, KRT10 and KRT17, in the forming interdigital epithelial tongue in the E14.5 limb. (D) Proliferating cells indicated by EdU positive staining in WT forelimbs at E14.5. Arrows point to EdU positive cells in the dorsal epidermis and in the interdigital zone.
Despite being multilayered, the IET is composed of basal-like keratinocytes only as indicated by the expression of KRT14 and the lack of expression of the differentiated keratinocyte markers KRT1 and KRT10 and the periderm marker KRT17 (Figure 3C). The differentiated keratinocyte layer and the periderm cover the surface of the skin, including the IET, but do not extend into the IET. EdU staining of forelimb sections shows active cell proliferation along the basal epidermis, including in (although not preferentially) the IET (Figure 3D). The IET is reminiscent of other developmental structures involved in collective migration of epithelia (Kashgari et al., 2018; Scarpa & Mayor, 2016), suggesting that it may play an active role in the separation of the digits.
Modeling suggests that directed cell migration plays a role in the formation of the IET
Next, we modeled IET formation, testing the influence of cell proliferation and active migration—processes that are important in the early formation of epithelial appendages such as hair follicles and the mammary gland (Hens & Wysolmerski, 2005; Mikkola & Millar, 2006). We considered two possible cellular models driving IET formation (Figure 4A). Model 1 posits that epidermal cell proliferation along with cell death in the underlying interdigital mesenchyme are sufficient for forming a multi-layered epithelial tongue. Model 2 posits that in addition to cell proliferation and interdigital cell death, the overlying epidermal cells need to actively migrate into the interdigital space to form a multi-layered epithelial tongue.
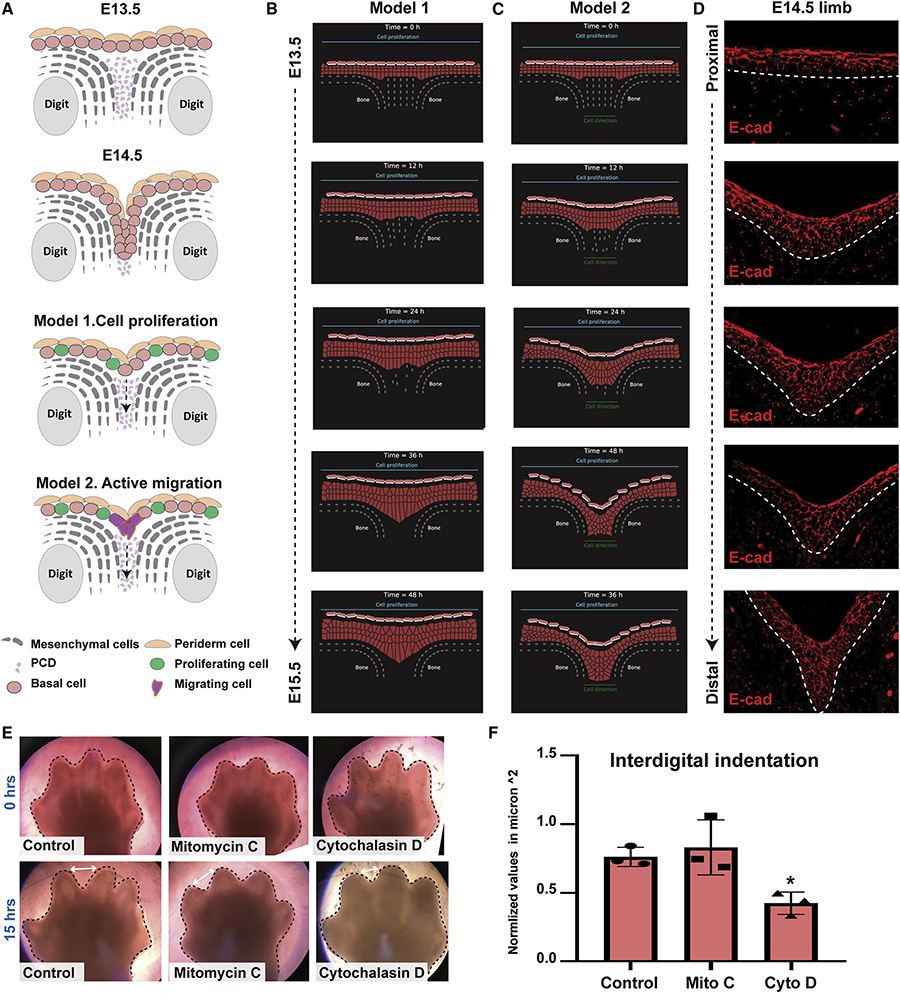
Figure 4. Mathematical modeling suggests that directed cell migration plays a role in the formation of the IET.
(A) Schematic representations of two proposed models for interdigital epithelial tongue formation during digit separation. Model 1: interdigital mesenchymal regression by cell death and active cell proliferation of epithelial cells in the overlying epidermis. Model 2: in addition to the factors in model 1, active cell migration of epithelial cells at the leading-edge is present. (B-C) The computational model simulating embryonic hand development during the span of 48 hours. Cells are represented by points connected by generalized Morse potentials to model biomechanical forces. Epithelial cells (pink), Periderm cells (light pink), and Mesenchymal cells (grey). (B) Model 1: Epithelial cell proliferation alone is insufficient to induce IET formation. (C) Model 2: Active cell migration (indicated by the green line at the bottom), in addition to cell proliferation, is sufficient for the epithelial to invade the cavity between the two digits and form the interdigital epithelial tongue. (D) E-cadherin staining of interdigital epithelial in WT forelimb sections at E14.5 suggests that active cell migration is indeed present in the developing limb. (E) Bright-field images of ex-vivo cultured forelimbs from E13.5 embryos at 0hrs (top) and 15hrs (bottom) of Mitomycin-C (Mito C) and Cytochalasin-D (Cyto D) treatment (N=3/group). Control group was incubated with media alone. (F) Measurements of the interdigital indentation at 15hrs post-incubation. Values were normalized to the interdigital indentation at 0hrs. Statistical significance was determined using one-way ANOVA (*p=0.01).
To test these predictions, we constructed a two-dimensional multiscale model of the epithelial invagination between two digits along the YZ axis (dorsal-ventral) to simulate digit separation over time (E13.5-E14.5) (Figure 4B, C, and Movies 4,5). We combined a cell-centered Voronoi model with the sub-cellular element method into a hybrid. Epithelial and mesenchymal cells are modeled with the Voronoi model, where each cell is represented by a single point (Barton et al., 2017; Bi et al., 2016; Kennedy et al., 2016; Mosaffa et al., 2018). The periderm cells are modeled by a hybrid of the subcellular element method (Christley et al., 2010; Gord et al., 2014; Newman, 2005; Sandersius & Newman, 2008), where additional nodes represent the surface molecules. To model death of the underlying mesenchymal cells, we randomly removed mesenchymal nodes at constant time intervals. In addition, at constant time intervals one epithelial cell divides randomly. We found that proliferation of epidermal keratinocytes concurrent with mesenchymal interdigital cell death (Model 1) is insufficient to induce IET formation (Figure 4B and Movie 4). In contrast, when an attractive force was added in the interdigital zone (Model 2), epidermal cells moved downwards collectively, forming a multilayered epithelial structure between the digits (Figure 4C and Movie 5) reminiscent of the IET (Figure 4D). Thus, these computational models suggest that active migration of epidermal cells, concomitantly with cell proliferation and interdigital cell death, is required for complete digit separation.
To test predictions of the computational model, we asked if chemical inhibitors blocking proliferation or migration altered the separation of the digits in limb explants. We cultured embryonic forelimbs from E13.5 WT embryos in media containing either Mitomycin-C, an inhibitor of cell proliferation, or Cytochalasin-D, an inhibitor of cell migration (Figure 4E). Whereas Mitomycin-C did not have an effect, Cytochalasin-D significantly delayed the progress of interdigital indentation (Figure 4 E,F), supporting the importance of cell migration for digit separation. Together, the computational modeling and the data suggest that active migration of interdigital epidermal cells is necessary for complete digit separation during embryonic development.
IET formation is mediated by collective cell migration of surface epidermal cells during digit separation
Based on the previous results, we hypothesized that digit separation requires collective cell migration where a cohesive group of epithelial cells moves directionally as observed in the development of many organs, including blood vessels, mammary gland, and Drosophila trachea (Ilina & Friedl, 2009; Scarpa & Mayor, 2016).
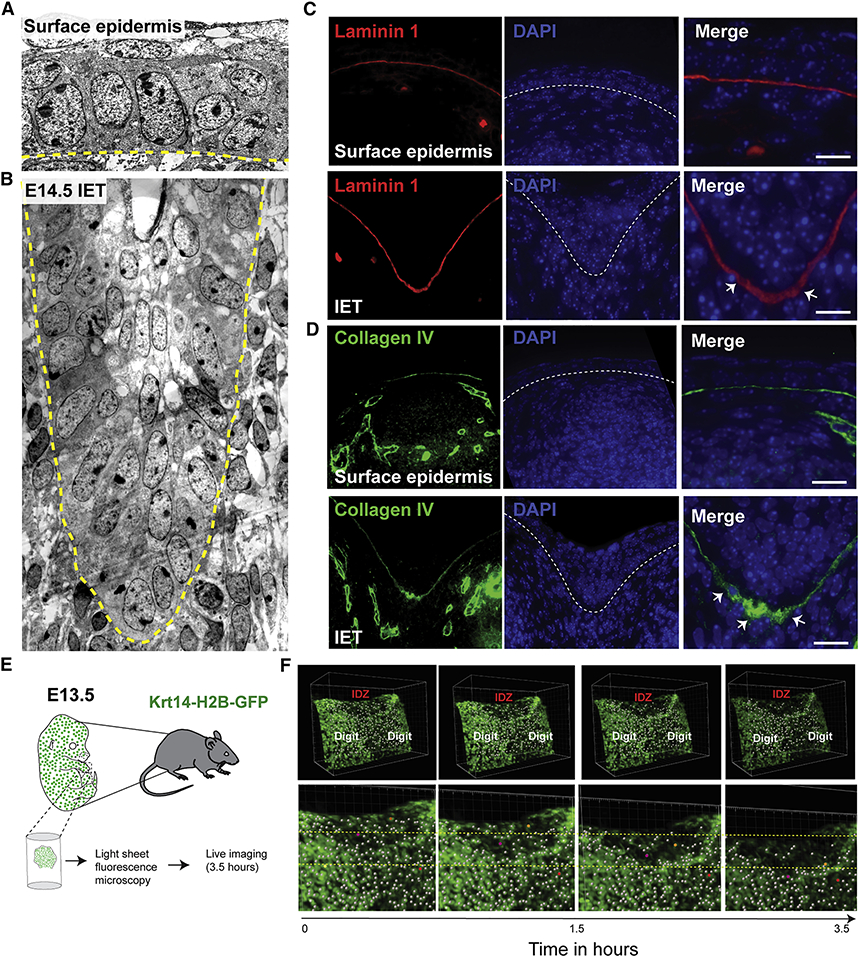
Changes in cell shape, including front-rear cell polarity, and changes in the surrounding extracellular matrix are prototypical features of active cell migration. Thus, we next investigated whether cells at the IET leading edge exhibit migratory characteristics by comparing transmission EM images of the IET with the surface epidermis (Figure 5A, B). In the surface epidermis, basal keratinocytes exhibit a cuboidal shape with nuclei oriented mostly perpendicular to the basal lamina (Figure 5A and S3A, B). In contrast, basal keratinocytes in the IET show elongated cell shape with nuclei oriented toward the leading edge, slanting horizontally relative to the basal lamina (Figure 5B and S3A, B). These data suggest that the IET represents a collectively migrating, multilayered epithelial structure.
Figure 5. Active cell migration of surface epidermal cells during digit separation.
(A) EM showing the surface epidermis and (B) the IET in E14.5 forelimbs from WT embryos. Yellow dashed line indicates the basement membrane. (C-D) Immunofluorescence staining with the basement membrane marker Laminin-1 (red) and Collagen IV (green). (E) Schematic representation of ex-vivo live imaging of E13.5 embryonic forelimb using light-sheet fluorescence microscopy. (F) Snapshots of tracked cells at the interdigital zone (IDZ) over time (top panel). Higher magnification views of the IDZ with labeled migrating cells (orange and purple) and non-migrating cell (red) (bottom panel).
Changes in the expression of adhesion molecules is also a feature of collectively migrating epithelia (Ilina & Friedl, 2009; Theveneau & Mayor, 2012). Typically, leader cells exhibit low E-cadherin expression, whereas cells distal from the IET maintain their E-cadherin expression. Unlike E-cadherin, P-cadherin is highly expressed in migrating leader cells (Plutoni et al., 2016), where it activates the small Rho GTPase Cdc42, a promoter of actin polymerization and microtubule re-organization required for front-rear cell polarity. Immunofluorescence staining shows basolateral expression of P-cadherin in basal keratinocytes located distally from the leading edge of the IET, whereas keratinocytes at the leading edge show increased expression of P-cadherin in the apical cell surface adjacent to the basement membrane (Figure S3C). The increased expression of P-cadherin at the front of IET leader cells suggests that signaling pathways involved in cell polarity and active migration are activated in IET leading edge.
We also observed signs of IET-extracellular matrix interactions. The Laminin 1-containing basement membrane is thicker and punctate at the leading edge of the IET (Figure 5C), whereas it is thinner and continuous more distally, away from the leading edge. Furthermore, the expression of Collagen IV, a wound-enriched basement membrane component, was increased at the IET leading edge (Figure 5D). These findings suggest active basement membrane remodeling at the IET’s leading edge, similar to that found in association with actively migrating epidermal keratinocytes in the migrating wound front (Bonnans et al., 2014; Kelley, 1973).
Proteolytic enzymes, including matrix metalloproteinases, degrade the basement membrane and the underlying extracellular matrix to facilitate collective cell migration (Martins et al., 2013). Consistent with this notion, the expression of the matrix metalloproteinase Mmp3 is specifically upregulated underneath the IET leading edge in the interdigital mesenchyme (Figure S3D).
To visualize migration of epidermal cells during digit separation, we used light-sheet fluorescence microscopy to monitor movement of Krt14-H2B-GFP-labeled keratinocytes in forelimb explant cultures isolated from embryos at E13.5 (Figure 5E, S3E and Movie 6). We observed migration of epidermal cells at the interdigital zones between the developing digits whereas cells located distally, above the digit, are stationary (Figure 5f and Movie 7). In addition, ortho-slices of the developing explants show thickening of the epidermis at the interdigital zone, not detected in ortho-slices away from the interdigital zone (Figure S3F, G and Movies 8,9). Together, these data show that in response to the underlying interdigital mesenchymal cell death, the epidermis invaginate from the dorsal and ventral sides, forming a multilayered interdigital epithelial tongue, the IET, that actively migrates into the underlying mesenchyme to promote digit separation.
GRHL3 is required for normal periderm development
Having gained understanding of epithelial mechanisms in normal digit separation, we turned our attention back to the Grhl3−/− embryos—to understand how GRHL3 regulates digit separation. At E15.5, the fused digits in Grhl3−/− embryos contain an epidermal sheet between them rather than mesenchymal tissue we would have expected had defective interdigital cell death caused the syndactyly in Grhl3−/− embryos (Figure 2A and Movie 3). In the light of these findings and our data on normal digit separation, we hypothesized that in Grhl3−/− embryos the IET invaginates normally between the digits, but as the IET dives into the mesenchyme, the epidermis covering the adjacent digits adheres together, creating an epithelial sheet fusing the digits. Going back to E14.5 Grhl3−/− limbs, when impaired digit separation is first evident, we discovered that the IETs form and migrate normally from the dorsal and ventral side, meeting in the middle (Figure S4A).
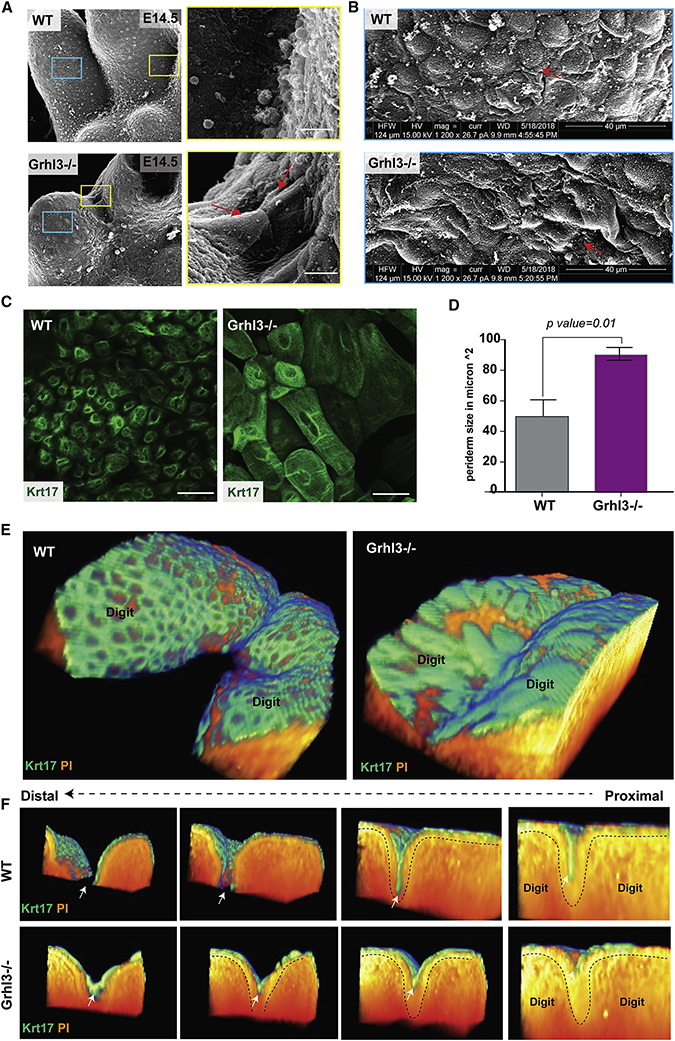
The adhesion of embryonic surfaces is prevented by the periderm, the outermost layer of the embryonic epidermis (Hammond et al., 2017; Richardson et al., 2014). Consistent with the notion of the adjacent digits adhering to each other in Grhl3−/− embryos, scanning EM views of the ventral E14.5 forepaws show a surface cell on a forming digit tightly adhering to a surface cell on the adjacent digit; this phenomenon is not observed in WT embryos (Figure 6A). These same images also indicate morphological difference of the surface skin in Grhl3−/− embryos compared to WT; the Grhl3−/− epidermal surface is smoother with larger cells than found on the WT surface (Figure 6B).
Figure 6. GRHL3 is required for normal periderm development.
(A) Scanning EM (ventral views) of WT and Grhl3−/− forelimbs at E14.5. The right panel shows high-magnification images of the yellow-box area indicating abnormal adhesion between surface cells covering adjacent digits. (B) High-magnifications (5020X) images of the blue-box area showing abnormal morphology and size of surface cells in Grhl3−/− epidermis compared to WT embryos. (C) Whole mount immunofluorescence staining of periderm cells (Keratin-17-green) on the dorsal surface of forelimbs in WT and Grhl3−/− embryos at E15.5. (D) Average periderm cell size in WT and Grhl3−/− epidermis at E14.5 (N=4 embryos/genotype). (E) 3D-views of two adjacent digits in WT and Grhl3−/− embryonic forelimbs at E15.5 stained with periderm marker KRT17 (green) and phalloidin (orange). (F) Series of images showing transverse views of the 3D reconstruction, moving from proximal-to-distal direction. Dashed lines indicate the basal lamina. White arrows indicate the periderm layer.
Next, we used immunostaining with periderm markers to test whether the periderm forms in Grhl3−/− embryos. Based on KRT17 expression, the periderm layer is formed at E13.5 and persists at least to E15.5 (Figure S4B), indicating that GRHL3 is not required for specification of periderm at the indicated developmental stages. Similar findings were obtained with the late periderm marker KRT6 at several developmental stages, although it appears to be delayed in the Grhl3−/− periderm (Figure S4C). The appearance of Grhl3−/− periderm cells, however, is abnormal, showing a spikey cell surface (Figure S4B). Transmission EM of the forepaw epidermis at E14.5 indicates that in WT mice, the periderm cells have an endothelial-like shape and are larger than basal and intermediate keratinocytes (Figure S4D, left panel). By contrast, periderm cells in the Ghrl3−/− epidermis exhibit irregular cell shape, characterized by expansion of the cytoplasm and membrane protrusions (Figure S4D, right panel). In addition, wholemount KRT17 staining showed that the periderm cells in Grhl3−/− embryos are significantly larger than WT periderm cells; this abnormality is found across the entire forepaw surface, including in the interdigital areas (Figure 6C, D). These data indicate that GRHL3 is dispensable for the formation of periderm cells but required for their normal appearance. These findings could be relevant to the syndactyly in Grhl3−/− embryos because periderm has non-adhesive properties, preventing pathological epithelial adhesions in the developing embryo (Hammond et al., 2017; Richardson et al., 2014).
To better understand the location of the periderm in the developing digits and in relation to the IET, we stained WT and Grhl3−/− E15.5 forelimbs with the periderm marker KRT17 and generated 3D-reconstructions (Figure 6E). Transverse views, moving from proximal to distal of these reconstructions, show that in the WT embryos, the periderm cells cover the digits and the surface of the IET without entering into the IET, keeping the digits separated due to the periderm’s non-adhesive property (Figure 6F, arrow). In Grhl3−/− embryos, the IET forms normally, but the periderm cells on the surface abnormally adhere to each other, resulting in epithelial fusion in the interdigital area (Figure 6F). We sometimes note a few periderm cells intermingling in the epithelial sheet fusing the digits (Figure S4A).
Together, these data indicate that periderm cells play a key role in mediating the separation of the interdigital epithelia during digit separation and that Grhl3 is required for normal periderm development preventing epithelial fusion.
GRHL3 is required for asymmetric expression of adhesion complex proteins in periderm cells; role in the non-adhesive function of the digit periderm
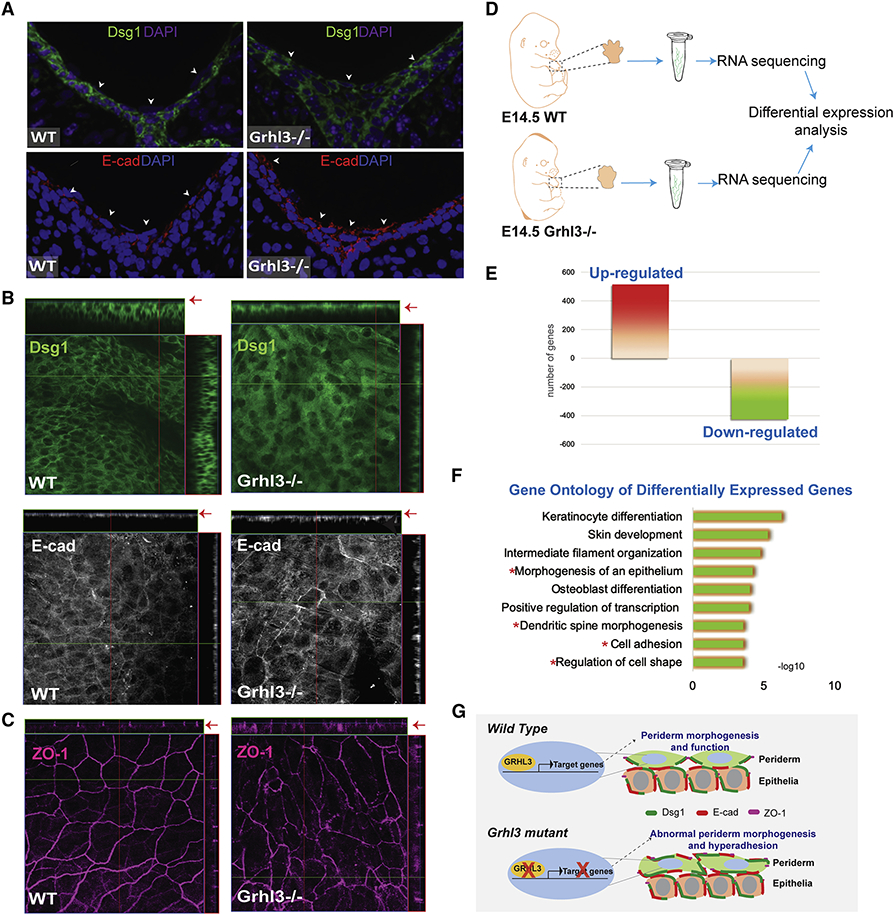
The non-adhesive function of periderm depends on the exclusion of adhesion complexes from the periderm’s apical surface (Figure S4E). Given the abnormal adhesion observed in the interdigital periderm in Grhl3−/− embryos, we tested whether GRHL3 is required for normal expression and localization of adhesion complex proteins in the periderm. Immunofluorescence staining of transverse sections in WT E14.5 forelimbs clearly show Dsg1 and E-cadherin expression around intermediate epidermal cells, but no expression on the apical surfaces of periderm cells in the interdigital area (Figure 7A, left panel). In contrast, in Ghrl3−/− embryos, Dsg1 and E-cadherin are abnormally expressed on the apical surfaces of interdigital periderm cells (Figure 7A, right panel), mirroring observations from the oral periderm in Grhl3−/− mutant mice (Peyrard-Janvid et al., 2014). The abnormal expression of Dsg1 and E-cadherin in the periderm apical surface is not exclusive to the interdigital area, but found throughout the Ghrl3−/− forepaw epidermis (Figure 7B). These data indicate that GRHL3 restricts the localization of adhesion complexes away from the apical surface of the developing periderm, including the limb epidermis.
Figure 7. GRHL3 is required for the non-adhesive function of periderm.
(A) Immunofluorescence staining of desmoglein-1 (green) and E-cadherin (Red) in E14.5 forelimb cross-sections from WT and Grhl3−/− embryos. Arrows indicate surface periderm cells and point to the expression of Dsg1 (Green) and E-cad (Red) on the apical side of periderm cells in Grhl3−/− embryos. Magenta and Blue indicate nuclear staining. (B) Whole-mount staining of adhesion complex proteins desmoglein-1 (green) and E-cadherin (gray) at the dorsal epidermis of E14.5 forelimb in WT and Grhl3−/− embryos. Arrows point to the surface cells in the Ortho-slice. (C) Whole-mount staining of the tight junction protein, Zonula occludens-1 (ZO-1; magenta) at the dorsal epidermis of E14.5 forelimb in WT and Grhl3−/− embryos. (D) Schematic representation of the experimental design and RNA isolation preformed for RNA-seq analysis (N=3/genotype). (E) Chart showing the number of significantly upregulated and down-regulated genes in Grhl3−/− compared to WT embryos. (F) Gene Ontology of the differentially expressed genes in Grhl3−/− forepaws at E14.5. Red asterisks indicate GO categories associated with abnormal epithelial morphology. (G) Schematic model depicting GRHL3-transcriptional regulation during periderm development. During embryonic development of the epidermis, GRHL3 in periderm cells directly or/and indirectly regulates the expression of genes that are required to restrict the expression of adhesion molecules from the apical surface, promoting the anti-adhesive function of the periderm. In the absence of GRHL3, periderm cells development is impaired, with abnormal expression of adhesion molecules on the apical surface, resulting in abnormal adhesion of periderm cells.
Periderm cell polarity is controlled by the apicolateral expression of tight junctions which restrict adhesion complex expression from the apical surface (Figure S5E). In WT forepaw epidermis, the tight junction protein ZO-1 is precisely located at the apicolateral junctions, leading to the formation of the highly polarized, endothelial cell-like periderm cells (Figure 7C and S4D). In contrast, Ghrl3−/− periderm cells have a punctate expression of ZO-1 that is not restricted to the apicolateral membrane; it is also found in the basolateral side of the cell, where ZO-1 is not normally expressed (Figure 7C). The data suggest that GRHL3 in the periderm regulates the expression of genes that are necessary for periderm differentiation and development.
Transcriptomic profiling of WT and Ghrl3−/− forepaws at E14.5 (Figure 7D) identified 942 genes that were differentially expressed in Ghrl3−/− embryos; of these differentially expressed genes, 515 genes were upregulated and 427 genes were down-regulated (Figure 7E). Differentially expressed genes were enriched in functional categories such as keratinocytes differentiation and skin development, as well as morphogenesis of an epithelium, cell adhesion and regulation of cell shape (Figure 7F). Genes with a role in cell adhesion included Dsg3, nectin2 and Cdh1, and with a role in periderm development included Irf6 and Stratifin (Table 1). Although the transcriptomic analysis was not exclusive to periderm cells, these data are consistent with GRHL3 regulating periderm genes with roles in adhesion and cell shape (Figure 7G).
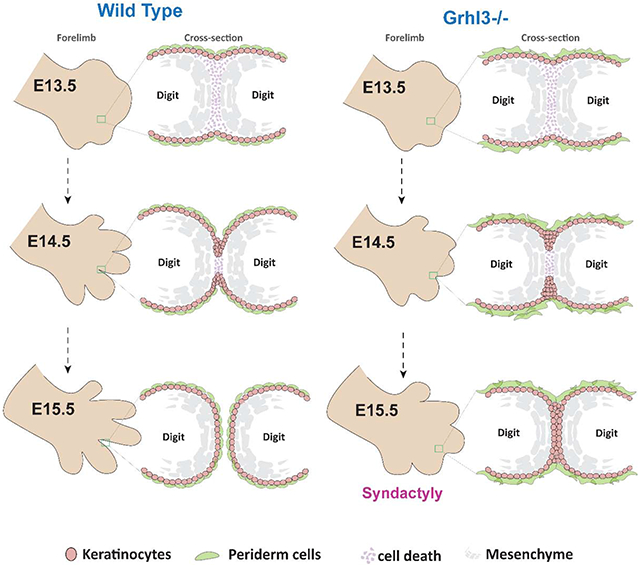
Altogether, our findings highlight the temporal phases of epithelial mechanisms in digit separation. These mechanisms are initiated by active cell migration of surface epidermal cells into the interdigital mesenchyme. These basal-like epidermal cells form a multilayered epithelial tongue, the IET, between the developing digits. As the IET invaginates the interdigital zone, periderm cells on one digit that lack adhesion complexes on their apical surfaces repel periderm cells on the adjacent digit. These non-adhesive properties lead to IET bifurcation and ultimately digit separation. Thus, normal periderm development is essential during digit separation, which explains why and how abnormal periderm formation in Ghrl3−/− mice causes syndactyly.
DISCUSSION
Here we uncover epithelial mechanisms required for digit separation in mammals. Our studies indicate that in addition to interdigital mesenchymal cell death, collective migration of an epidermal structure, the IET, mediates digit separation. Additionally, we show that the epidermal transcription factor GRHL3 controls periderm differentiation, which is required to prevent fusion of the migrating interdigital epithelium and syndactyly.
While previous studies have noticed thickening of the invaginating epithelium (Wong et al., 2012) and others have commented on possible epithelial mechanisms (Hurle & Colvee, 1982; Kelley, 1970; Kelley, 1973; Pautou, 1975), researchers have focused on programmed cell death of the interdigital mesenchyme as a key event in digit separation. In fact, most mouse mutants with soft tissue syndactyly have been ascribed to decreased or altered interdigital cell death (Hernández-Martínez & Covarrubias, 2011; Hurle & Colvee, 1982; Kelley, 1970; Kelley, 1973).
The work on interdigital cell death has identified three main signaling pathways: Bmps, Fgfs, and retinoic acid (RA). During limb growth, Fgf8 released from the epithelium promotes mesenchymal cell proliferation and inhibits mesenchymal cell death by decreasing the levels of the cell death-inducing retinoic acid (RA). Interdigital cell death is initiated by the activation of the Bmp signaling pathway through Bmpr1a in the epithelium, which downregulates Fgf8 levels, leading to increased RA levels in the interdigital mesenchyme and cell death.
Our findings indicate that mechanisms distinct from interdigital cell death are also required for digit separation. Three-dimensional reconstructions of the developing distal limbs at three key stages of digit separation—E13.5, E14.5, and E15.5—show epithelial mechanisms important for digit separation. As the interdigital mesenchyme is removed through cell death, the epidermis invaginates from the ventral and the dorsal side. The epithelial leading edge forms a multi-layered epithelial structure, the IET. The IETs from the ventral and dorsal sides eventually meet and separate. As the interdigital cell death progresses in a distal to proximal direction, the progress of the dorsal and ventral epithelial invaginations also progresses in a distal to proximal direction.
The epidermal invagination between the digits and the formation of the IET shares some similarities with the development of skin appendages, including mammary gland and hair follicles (Mikkola & Millar, 2006). Modeling the morphogenetic mechanisms underlying the formation of such epithelial outgrowths, we found that in addition to epithelial cell proliferation and interdigital mesenchymal cell death, a migration force was required for the formation and movement of the IET. Which led us to study the IET as a collectively migrating epithelial front, a developmental phenomenon involved in the formation of many organs during embryogenesis such as epithelial tube branching required for blood vessels, lungs and nephric ducts (Scarpa & Mayor, 2016). In collectively migrating epithelia, cell-cell adhesions are essential for coordinating the collective movement and maintaining the mechanical integrity of the epithelial cohort. And chemotactic cues in the surrounding extracellular matrix are required to guide the directionality of the migration. In addition, changes in actin-cytoskeleton and cell polarity of leader cells are required for active migration. Interestingly, mice lacking the actin-based motor molecule Myosin 10 (Myo10), known to promote leading edge cell migration, exhibit soft tissue syndactyly, similar to what we have observed in Grhl3−/− mice (Tokuo et al., 2018). In addition to our data, these recent findings support a role for active cell migration in the separation of the digits.
Consistent with this model, keratinocytes at the IET leading edge exhibit morphological features of leader cells with cell elongation and nuclei orientated toward the direction of migration rather than being perpendicular to the basal lamina. Downregulation of E-cadherin and up-regulation of P-cadherin at the leading edge, which we observed in the IET, parallels the expression pattern of adhesion molecules during active cell migration in epithelial morphogenesis and invasive cancers (Friedl & Gilmour, 2009; Lee et al., 2006). Additionally, we observed modifications of the basement membrane and the mesenchymal extracellular matrix at the leading edge of the IET consistent with active migration. In addition, we could observe movement of epidermal cells into the interdigital space in forepaw explant cultures and inhibition of digit separation of explant cultures treated with a chemical migration inhibitor. Although the data cited above suggest the importance of active epithelial migration, we cannot rule out the possibility that the epithelial indentation is also driven by the contracting underlying mesenchyme.
The formation of the IET brings the epidermis that covers adjacent digits into close contact. But because the outermost surface of the epidermis is covered by periderm cells, a non-adhesive layer, adherence of the digits in prevented. Periderm achieves this non-adhesive function by restricting the expression of adhesion complexes to the basolateral surface (Figure 7G). In Grhl3−/− embryos, the apical surface of periderm cells aberrantly expresses adhesion complexes making these cells susceptible to adhesion. Consequently, as the IET moves between the digits the epidermis from adjacent digits adhere to each other, creating elongated epithelial sheets between fused digits.
Abnormal epithelial fusion of the oral epithelia is caused by disrupted oral periderm development, whereas soft-tissue fusion of the digits has been linked solely to impaired interdigital cell death. In mouse embryos, Grhl3 expression is limited to the epithelia, including the periderm, and to some cartilage cells in the developing digits, but absent in the mesenchyme (Auden et al., 2006; Kudryavtseva et al., 2003). The occurrence of syndactyly in Van der Woude patients suggests a common mechanism in epithelial fusion regulated by GRHL3. Interestingly, mutations in other genes expressed in the periderm, including Stratifin and Jagged 2, also cause syndactyly (Herron et al., 2005; Jiang et al., 1998), suggesting that defects in other signaling pathways in the epidermis may cause syndactyly through similar mechanisms as GRHL3.
In conclusion, we propose new epithelial mechanisms in digit separation. Our data show how GRHL3 regulates the function of periderm cells at the cellular level and also explain why patients with GRHL3 mutations exhibit syndactyly. It will be of interest in future studies to determine whether syndactyly in mouse mutants for other epidermally-expressed developmental regulators also involve the epithelial mechanisms we have described here.
STAR★ METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bogi Andersen (Bogi@uci.edu). All unique/stable reagents generated in this study are available from the Lead Contact. There are restrictions to the availability of (Krt14-H2B-GFP mouse line) due to a material transfer agreement with the Rockefeller University.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice were housed and maintained in accordance with protocols approved by the University Laboratory Animal Resources (ULAR). Transgenic mouse strains used in these studies were on the C57BL/6 background: Grhl3−/− (Yu et al., 2006), Grhl3fl/fl, Krt14-Cre;Grhl3fl/fl, and Grhl3-Cre;LacZ (Gordon et al., 2014). Skin-specific deletion of Grhl3 in C57BL/6J/ mice was generated by crossing Grhl3 floxed mice with Tg (KRT14-cre)1Amc/J (stock# 004782). Offsprings were bred to produce WT, heterozygous and homozygous-Grhl3-flox Cre-positive mice (Krt14-Cre;Grhl3−/−, Krt14-Cre;Grhl3fl/− and Krt14-Cre;Grhl3fl/fl), respectively. Grhl3-Cre;LacZ reporter mice were purchased from MMRRC. K14-H2B-GFP mice were obtained from Elaine Fuchs laboratory at Rockefeller University (Adam et al., 2018).
MATERIALS AND METHODS
Scanning and Transmission Electron Microscopy
Timed pregnancies were used to collect mouse embryos at several developmental stages. Embryo stages were confirmed with morphological criteria. Forelimbs were harvested and fixed with 0.25% Glutaraldehyde for 24 hrs in 4°C, followed by post-fixation with 2% Osmium Tetroxide.
To prepare for Scanning EM imaging, the specimen was dehydrated through series of ethanol and then substituted with pure acetone before drying at CO2 critical point using Leica CPD300. After coating with 4nm of Platinum/Palladium with Leica sputter coater ACE200, the specimen was examined using Thermafisher Quanta3D and the collected images were then evaluated using NIH ImageJ software.
To prepare sample for Transmission Electron Microscopy imaging, the specimen was dehydrated through series of ethanol after post-fixation and followed by resin infiltration. Forelimbs were then embedded in Spurr low viscosity resin and cured for 72 hours in 65°C oven. The EM blocks were trimmed and sliced into 70nm sections using Leica UC7 Ultramicrotome. The sections were collected with slot copper grids and stained with both lead citrate and uranyl acetate before imaging with a JEOL 2100F microscope. The montage images were collected with SerialEM software and further processed using IMOD software package.
Plastic-embedded limbs for 3D reconstructions
Embryonic limbs were collected at the indicated time points and fixed in 4% PFA at 4°C overnight. The next day, limbs were embedded in low melting point agarose (3%). All tissues were dehydrated serially in 50%, 70%, 95%, and 100% ethanol for 30 mins at each concentration at room temperature. After dehydration, tissues were infiltrated with BMMA resin (Polyscience, Warrington PA) at 4°C, over three days, displacing the ethanol in an ascending BMMA concentration gradient from 25% to 75% BMMA in ethanol. The tissue was then submerged and rotated in a 100% BMMA for 24 hrs at 4°C before the polymerization stage (Parfitt et al., 2012). Each limb was then placed in a BEEM gelatin capsule with fresh BMMA and sealed tightly. For polymerization, the capsules were placed under the UV light for 20 hrs in Pelco UCV2 cryo chamber (Ted Pella, Redding CA) set at 4C. Sections (2 um) were serially cut using a diamond knife (DiAtome, Hatfield, PA) on a Leica EM UC7 ultramicrotome.
Immunofluorescence staining of plastic sections and 3D reconstructions
Slides of thin sections embedded in plastic were permeabilized with permeabilization buffer (0.3% Triton-X in PBS) for 10-15 mins and blocked with blocking buffer (0.1% Triton-X + 0.4% Bovine Serum Albumin in 1XPBS). Sections were incubated with the primary antibody (listed in the Key source table) overnight at 4C. The next day, slides were washed with 1XPBS three times, followed by incubation with anti-goat-Alexa Fluor 488, donkey-anti-rabbit- Alexa Fluor 555, donkey-anti-Armenian for 1 hour in room temperature. For nuclear staining, slides were incubated with DAPI solution for 1 min, then mounted with 50% glycerol. For re-staining plastic slides, mounting solution was removed by 1XPBS, and slides were incubated in stripping buffer (glycerine) for 1-2 hrs at 60°C, followed by several 1XPBS washes. For 3D reconstructions, a continuous series of stained slides were imaged using Leica DMI6000B fully automated inverted fluorescent microscope (Leica Microsystems Inc., Buffalo Grove, IL). Images were aligned and 3D constructs were generated using AMIRA software.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti Cleaved Caspase-3 (Asp 175) | Cell Signaling Technology | Cat# 9661 |
| Rabbit monoclonal anti E-cadherin (24E10) | Cell Signaling Technology | Cat# 3195 |
| Rabbit monoclonal anti P- cadherin (C13F9) | Cell Signaling Technology | Cat# 2189 |
| Rat monoclonal anti Laminin 1 A&B chains (Clone AL-2) | Millipore Sigma | Cat# MAB1904 |
| Mouse monoclonal anti Mmp3 (Clone 4F10) | Thermofisher | Cat# MA5-17123; RRID: AB2538594 |
| Rabbit monoclonal anti Desmoglein (Clone EPR6766) | Abcam | Cat# ab124798; RRID: AB124798 |
| Red DND-99 LysoTracker® | Thermofisher | Cat# L7528 CAS: 67-68-5 |
| Rabbit polyclonal anti Keratin 1 (Clone Poly19056) | Biolegend | Cat# 905601; RRID: AB2565051 |
| Rabbit monoclonal anti Keratin 5 (Clone EP1601Y) | Abcam | Cat# ab52635; RRID: AB52635 |
| Rabbit polyclonal anti Keratin 6A (Clone Poly19057) | Biolegend | Cat# 905701; RRID: AB2565052 |
| Mouse monoclonal anti Cytokeratin 10 (Clone DE-K10) | Thermofisher | Cat# MA5-13705; RRID: AB10979022 |
| Rabbit polyclonal anti Keratin 14 (Clone Poly19053) | Biolegend | Cat# 905301; RRID: AB2565048 |
| Rabbit monoclonal anti Keratin 17 (Clone D12E5) XP® | Cell Signaling Technology | Cat# 12509 |
| Rabbit polyclonal Collagen IV antibody (ab6586) | Abcam | Cat#Ab6586 |
| Rhodamine Phalloidin | Thermofisher | Cat#R415 |
| Chemicals, Peptides and Recombinant Proteins | ||
| EdU (5-ethynyl-2'-deoxyuridine) | Thermofisher | Cat# C10337 |
| TdT (terminal deoxynucleotidyl Transferase) | Thermofisher | Cat# C10617 |
| Mitomycin C | Sigma-Aldrich | Cat# M4287 |
| Cytochalasin D | Sigma-Aldrich | Cat# C2618 |
| BGJb Medium (Gibco) | Thermofisher | Cat# 12591038 |
| Critical Commercial Assays | ||
| Click-iT™ Plus TUNEL Assay for In Situ Apoptosis Detection, Alexa Fluor™ 488 dye | Thermofisher | Cat# C10617 |
| Click-iT™ EdU Alexa Fluor™ 488 Imaging Kit | Thermofisher | Cat# C10337 |
| Experimental Models: Organisms/ Stains | ||
| Mouse: Krt14Cre:Grhl3 fl/fl | W Gordon et.al (2014) | N/A |
| Mouse: Grhl3−/− mice | Yu et.al (2006) | N/A |
| Mouse: Krt14-Cre | The Jackson Laboratory | 004782 |
| Mouse Mouse: Krt14-H2B-GFP | Fuchs et al., 2018 | N/A |
| Oligonucleotides | ||
| Primer: Get-1 Forward: GAA TTA CAA GTC TGT GCC ACC A Reverse: ATT TGC TGA CCT TTT TCT GAG C |
Eurofins Genomics | N/A |
| Primer: Grhl3-flox Forward: CCC CTG AGC AGT TGG AAT A Reverse: TGG CCA CAC TGA CAA GAG AG |
Eurofins Genomics | N/A |
| Primer: Cre Forward: GCA CTG ATT TCG ACC AGG TT Reverse: CCC GGC AAA ACA GGT AGT TA |
Eurofins Genomics | N/A |
| Software and Algorithms | ||
| Amira-Avizo Software | Thermofisher | Version 2019.1 |
| OriginLab | https://www.originlab.com | Version 2019B |
| GraphPad Prism 7.01 | GraphPad software | scientific-software/prism/ |
| Fiju (ImageJ) | https://fiji.sc/ | N/A |
| Kallisto | Pachter lab | kallisto 0.43.0 |
| EdgeR | Robinson et al., 2010 | Bioconductor |
| Imaris 3/4D Image Visualization and Analysis | Imaris | Version 9.5 |
| Deposited Data | ||
| Bulk RNAseq data | This study | GSE141009 |
| Sequence-Based Reagents | ||
| Illumina TruSea® Stranded mRNA | Illumina | Cat#RS-122-2101 |
Whole-mount staining and immunohistochemistry of embryonic mouse limbs
Whole limbs were collected at the indicated time points and fixed with 4% PFA overnight at 4°C. Post fixation, limbs were washed with 1XPBS several times, 20 mins each wash, permeabilized with permeabilization buffer (triton-X in PBS) for 30-45 mins, and blocked with blocking buffer for 2-3 hrs. Limbs were then incubated with primary Antibody 1-2 days. On the third day, limbs were washed with 1XPBS for 20-30 mins each and then incubated with secondary antibody for 3-4 hrs at RT or for 1 day at 4C in the dark. Limbs were mounted in glycerol. Mounted whole-limbs were imaged using Zeiss LSM 510 (Carl Zeiss, Jena, Germany) and femtosecond laser (Chameleon, Coherent Inc., Santa Clara, CA).
For immunofluorescence staining of sections, cryosections (8 um thick) of limbs and dorsal skin were collected on Fisherbrand™ Superfrost™ Plus Microscope Slides. Slides were fixed with cold acetone for 13 mins, followed by 3 washed with 1XPBS, then fixed again with 4% PFA for 10 mins at RT. After fixation, slides were permeabilized with permeabilization buffer (triton-X in PBS) for 10-15 mins and blocked with blocking buffer for 1 hr at RT. Slides were incubated with primary antibody for 2 hrs at RT or overnight at 4°C. The next day, slides were washed three times with 1XPBS and then incubated with a secondary antibody for 1 hr at RT in the dark. Slides were mounted with DAPI mount and imaged using Leica DMI6000B fully automated inverted fluorescent microscope (Leica Microsystems Inc., Buffalo Grove, IL)
For EdU staining, pregnant female mice were injected with 0.5 mg/ml of EdU and sacrificed 3 hrs post injection. Embryos were collected and limbs were dissected and embedded in plastic. EdU staining was performed on plastic section using Click-iT EdU Alexa Fluor 488 Imaging Kit - Thermo Fisher Scientific.
Cell death detection
For LysoTracker staining, embryonic limbs at E13.5 were dissected and Lysotracker staining was performed using LysoTracker Red (LT:Molecular Probes). Briefly, dissected forelimbs were incubated with LysoTracker Red at 37°C for 30 min; protected from light (Eshkar-Oren et al., 2015). Confocal imaging was performed using Zeiss LSM 510 (Carl Zeiss, Jena, Germany) and femtosecond laser (Chameleon, Coherent Inc., Santa Clara, CA). For the TUNEL assay, embryonic limbs at E13.5 were dissected and embedded in OCT. Frozen sections (8 um) were stained using Click-iT™ Plus TUNEL Assay for In Situ Apoptosis Detection, Alexa Fluor™ 488 dye, (Thermofisher, Cat#C10617) as described by the manufacturer.
Mathematical Modeling
We construct a two-dimensional multiscale model of the developing limb along the dorsal-ventral axis to simulate the limb separation over time. In the cell-centered and Voronoi models each cell is represented by a single point. To model the biomechanical forces inside a tissue these nodes are connected by generalized Morse potentials that exercise forces on nearby nodes such that the cells move (Figure S3). We model the mesenchymal cells by a single point and visualize them as ellipsoids (gray in Figure S3) Epithelial cells are also represented by a single point, but visualized by the Voronoi tessellation (red in Figure S3) of the point cloud. Periderm cells were modeled with a modified subcellular element method. Each periderm cell is represented by a central node surrounded by 11 surface molecules along the sides and basal surface of an elongated hexagon. The modification from the traditional subcellular element method is that the nodes representing a cell have constant positions relative to each other, modeling a rigid body. The central node and surface molecules are connected to all other nodes in the model by different Morse potentials. The sum of all the forces leads to a lateral translation and rotation of each periderm cell. The mesenchymal cells surrounding the bones are inactive and immobile but pose reflective boundary conditions on the moving cells. One mesenchymal cell is removed every 500 sec and one cell division every 250 sec. To model a directed cell migration, an attractive potential for all cells is placed in the middle between the two bones. At constant time intervals one epithelial cell divides randomly. To model the apoptosis of the mesenchymal cells we randomly remove a mesenchymal node at constant time intervals. The digit development is then simulated by solving the differential equations resulting from the force equations numerically using Euler’s method.
Ex-vivo limb culture
Forelimbs were isolated from WT embryos at E13.5. The limbs were cultured in 96-well plate and incubated in BGJb media (Gibco BRL) supplemented with 10%FBS and (penicillin/streptomycin) antibiotic mix. Inhibitor-treated Limbs were incubated with 10 uM of Mitomycin-C and 10 uM of Cytochalasin-D with media for 15 hours. Images of the cultured limbs were collected at 0hr and 15hrs post inhibitor treatment using standard bright-field microscopy.
Live imaging with Light-Sheet Fluorescence Microscopy
Forelimbs were isolated from Krt14-H2B-GFP embryos at E13.5. The limb was placed in a transparent capillary tube and embedded between a sterile low melt agarose (0.08%). The capillary was next placed inside the light-sheet chamber (Z-Ziess), incubated in Phenol free media with 10% FBS. Z-stacks of the limb were collected every 5 mins for 3.5 hours. All the captured images that were collected over time were processed using IMARS software.
RNA sequencing
Forepaws were isolated from E14.5 WT and Grhl3−/− embryos (N=3/genotype) and immediately snap-frozen in liquid nitrogen. Frozen limbs where grinded using mortars and pastels followed by resuspension in Trizol. RNA extraction was performed according to the manufacturer’s instructions (Direct-zol RNA Purification Kits). Total RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip and Nanodrop absorbance ratios for 260/280nm and 260/230nm. Library construction was performed according to the Illumina TruSeq® Stranded mRNA Sample Preparation Guide. The input quantity for total RNA was 500ng and mRNA was enriched using oligo dT magnetic beads. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. The multiplexed libraries were sequenced on four lanes using single end 100 cycles chemistry on the HiSeq 4000. The version of HiSeq control software was HCS 3.4.0.38 with real time analysis software, RTA 2.7.7. Sequencing quality of FastQ files was determined using FASTQC. Transcript assembly was processed using Kallisto and list of differentially expressed genes was generated using EdgeR. Gene ontology analysis was performed using DAVID Functional Annotation Bioinformatics Microarray Analysis. GEO accession number GSE141009.
QUANTIFICATION AND STATISITCAL ANALYSIS
Measurements of interdigital indentation in limb explants
Interdigital indentation between digit 2 and digit 3 was measured by drawing a circle that fits the tip of digits and measuring the distance from the lowest point in the indentation between digits 2 and 3 along the radius from the center to the circle (Yang et al., 2013) at 0 hour and 15 hours post incubation with chemical inhibitors (N=3 forelimbs/per group). The value of interdigital indentation at 15 hours was normalized to the value interdigital indentation at 0 hours in each limb explant. Statistical analysis was performed using one-way ANOVA (Prism).
Measurements of nucleus orientation
The orientation of basal epidermal cells located in the IET and in the surface epidermis was determined by measuring the angular plane of the nucleus relative to the basement membrane. Cells (n=90) were categorized as either perpendicular or non-perpendicular, wherein perpendicular cells possessed an angular measurement of 90±30 degrees relative to the basement membrane and non-perpendicular cells possessed angles between 0±60 degrees and above 120 degrees relative to the basement membrane (Lechler & Fuchs, 2005).. Angular measurements were processed using IMAGEJ 1.51r. Epidermal cell angles were then plotted as radial histograms using Origin 2019b (OriginLab) from data binned into 15° increments.
Measurements of Periderm cell size
Images (40X) of whole-mount immunofluorescence staining of the limb skin with keratin-17 were used to measure the size of periderm cells. Images were calibrated with a micrometer scale bar taken at the same magnification. The cells were processed through IMAGEJ 1.51r with a conversion factor of 5.26 pixel/μM. Cell area measurements (μM2) were collected by circumscribing each cell membrane using the segmented line tool (n=118). The collected measurements were then processed in Microsoft Excel. For each genotype, 3-4 embryos were analyzed. Statistical analysis was performed using student’s T-test.
DATA AND CODE AVAILABILITY
RNA sequencing data generated in this study are available at Gene Expression Omnibus (GEO) accession number GSE141009.
Supplementary Material
Movie 1. Related to Figure 1. A morphed time-lapse of forelimb digit separation during development.
Movie 2. Related to Figure 2. 3D reconstruction of WT E15.5 limb epithelia.
Movie 3. Related to Figure 2. 3D reconstruction of Grhl3−/− E15.5 limb epithelia.
Movie 4. Related to Figure 4. A time-course (48h) representation of Voronoi models featuring cell proliferation.
Movie 5. Related to Figure 4. A time-course (48h) representation of Voronoi models featuring cell migration.
Movie 6. Related to Figure 5. Live imaging of Krt14-H2B-GFP cells in a cultured embryonic forelimb explant (E13.5) over time (3.5 hours).
Movie 7. Related to Figure 5. Cell-tracking in a cultured embryonic forelimb explant (E13.5) over time (3.5 hours).
Movie 8. Related to Figure 5. Ortho-slice view of a 3D stack at the interdigital zone.
Movie 9. Related to Figure 5. Ortho-slice view of a 3D stack at the digit zone.
Highlights.
Epidermal cells actively migrate into the interdigital mesenchyme
Active migration of epidermal cells is required for digit separation
Non-adhesive property of periderm cells prevents epithelial fusion between the digits
GRHL3 is required for normal periderm development and function
Acknowledgments:
Many thanks to Thor Andersen for creating time-course morph movies of WT and Grhl3−/− embryonic limbs. We thank the personnel at UC Irvine Material and Research Institute (Dr. Ich Tran and Dr. Xing Li) for their services in Scanning and transmission electron microscopy. We thank Dr. Adeela Syed in UCI Optical Biology Core for Light-sheet Florescence Microscopy. This work was supported by NIH grants R01AR44882 and R21AR069962 and the Irving Weinstein Foundation (to B. Andersen); and NSF grant DMS1763272, a grant from the Simons Foundation (594598, QN), and NIH grant U01AR073159 (to Q. Nie) and a grant from Research to Prevent Blindness, Inc. RPB 203478 (to J. Jester).
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam RC, Yang H, Ge Y, Lien W-H, Wang P, Zhao Y, Polak L, Levorse J, Baksh SC, Zheng D, & Fuchs E (2018). Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression. Cell Stem Cell, 22(3), 398–413.e7. 10.1016/j.stem.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, & Jane SM (2006). Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expression Patterns, 6(8), 964–970. 10.1016/j.modgep.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Barton DL, Henkes S, Weijer CJ, & Sknepnek R (2017). Active Vertex Model for cell-resolution description of epithelial tissue mechanics. PLOS Computational Biology, 13(6), e1005569. 10.1371/journal.pcbi.1005569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bečić T, Bilan K, Mardešić S, Vukojević K, & Saraga-Babić M (2016). Spatiotemporal distribution of proliferation, proapoptotic and antiapoptotic factors in the early human limb development. Acta Histochemica, 118(5), 527–536. 10.1016/j.acthis.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Bi D, Yang X, Marchetti MC, & Manning ML (2016). Motility-driven glass and jamming transitions in biological tissues. Physical Review. X, 6(2). 10.1103/PhysRevX.6.021011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnans C, Chou J, & Werb Z (2014). Remodelling the extracellular matrix in development and disease. Nature Reviews. Molecular Cell Biology, 15(12), 786–801. 10.1038/nrm3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch PJ, Fuller LC, & Weiner JA (2018). An essential role for the nuclear protein Akirin2 in mouse limb interdigital tissue regression. Scientific Reports, 8(1), 12240. 10.1038/s41598-018-30801-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chautan M, Chazal G, Cecconi F, Gruss P, & Golstein P (1999). Interdigital cell death can occur through a necrotic and caspase-independent pathway. Current Biology: CB, 9(17), 967–970. [DOI] [PubMed] [Google Scholar]
- Christley S, Lee B, Dai X, & Nie Q (2010). Integrative multicellular biological modeling: A case study of 3D epidermal development using GPU algorithms. BMC Systems Biology, 4, 107. 10.1186/1752-0509-4-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshkar-Oren I, Krief S, Ferrara N, Elliott AM, & Zelzer E (2015). Vascular patterning regulates interdigital cell death by a ROS-mediated mechanism. Development (Cambridge, England), 142(4), 672–680. 10.1242/dev.120279 [DOI] [PubMed] [Google Scholar]
- Friedl P, & Gilmour D (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nature Reviews. Molecular Cell Biology, 10(7), 445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Gord A, Holmes WR, Dai X, & Nie Q (2014). Computational modelling of epidermal stratification highlights the importance of asymmetric cell division for predictable and robust layer formation. Journal of the Royal Society, Interface, 11(99). 10.1098/rsif.2014.0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WM, Zeller MD, Klein RH, Swindell WR, Ho H, Espetia F, Gudjonsson JE, Baldi PF, & Andersen B (2014). A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. The Journal of Clinical Investigation, 124(12), 5205–5218. 10.1172/JCI77138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond NL, Dixon J, & Dixon MJ (2017). Periderm: Life-cycle and function during orofacial and epidermal development. Seminars in Cell & Developmental Biology. 10.1016/j.semcdb.2017.08.021 [DOI] [PubMed] [Google Scholar]
- Hens JR, & Wysolmerski JJ (2005). Key stages of mammary gland development: Molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Research, 7(5), 220. 10.1186/bcr1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Martínez R, & Covarrubias L (2011). Interdigital cell death function and regulation: New insights on an old programmed cell death model. Development, Growth & Differentiation, 53(2), 245–258. 10.1111/j.1440-169X.2010.01246.x [DOI] [PubMed] [Google Scholar]
- Herron BJ, Liddell RA, Parker A, Grant S, Kinne J, Fisher JK, & Siracusa LD (2005). A mutation in stratifin is responsible for the repeated epilation ( Er ) phenotype in mice. Nature Genetics, 37(11), 1210. 10.1038/ng1652 [DOI] [PubMed] [Google Scholar]
- Hiscock TW, Tschopp P, & Tabin CJ (2017). On the Formation of Digits And Joints During Limb Development. Developmental Cell, 41(5), 459–465. 10.1016/j.devcel.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurle JM, & Colvee E (1982). Surface changes in the embryonic interdigital epithelium during the formation of the free digits: A comparative study in the chick and duck foot. Journal of Embryology and Experimental Morphology, 69, 251–263. [PubMed] [Google Scholar]
- Ilina O, & Friedl P (2009). Mechanisms of collective cell migration at a glance. Journal of Cell Science, 122(18), 3203–3208. 10.1242/jcs.036525 [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, & Gridley T (1998). Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes & Development, 12(7), 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltcheva MM, Anderson MJ, Harfe BD, & Lewandoski M (2016). BMPs are direct triggers of interdigital programmed cell death. Developmental Biology, 411(2), 266–276. 10.1016/j.ydbio.2015.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashgari G, Huang Y, & Andersen B (2018). Embryonic Development of the Epidermis. In Reference Module in Biomedical Sciences. Elsevier. 10.1016/B978-0-12-801238-3.65811-7 [DOI] [Google Scholar]
- Kelley RO (1970). An electron microscopic study of mesenchyme during development of interdigital spaces in man. The Anatomical Record, 168(1), 43–53. 10.1002/ar.1091680104 [DOI] [PubMed] [Google Scholar]
- Kelley RO (1973). Fine structure of the apical rim—Mesenchyme complex during limb morphogenesis in man. Development, 29(1), 117–131. [PubMed] [Google Scholar]
- Kennedy RC, Ropella GE, & Hunt CA (2016). A cell-centered, agent-based framework that enables flexible environment granularities. Theoretical Biology & Medical Modelling, 13, 4. 10.1186/s12976-016-0030-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RLLF, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, … Murray JC (2002). Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nature Genetics, 32(2), 285–289. 10.1038/ng985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousa YA, Zhu H, Fakhouri WD, Lei Y, Kinoshita A, Roushangar RR, Patel NK, Agopian AJ, Yang W, Leslie EJ, Busch TD, Mansour TA, Li X, Smith AL, Li EB, Sharma DB, Williams TJ, Chai Y, Amendt BA, … Schutte BC (2019). The TFAP2A-IRF6-GRHL3 genetic pathway is conserved in neurulation. Human Molecular Genetics, 28(10), 1726–1737. 10.1093/hmg/ddz010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryavtseva EI, Sugihara TM, Wang N, Lasso RJ, Gudnason JF, Lipkin SM, & Andersen B (2003). Identification and characterization of Grainyhead-like epithelial transactivator (GET-1), a novel mammalian Grainyhead-like factor. Developmental Dynamics, 226(4), 604–617. 10.1002/dvdy.10255 [DOI] [PubMed] [Google Scholar]
- Lechler T, & Fuchs E (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature, 437(7056), 275–280. 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, & Thompson EW (2006). The epithelial–mesenchymal transition: New insights in signaling, development, and disease. The Journal of Cell Biology, 172(7), 973–981. 10.1083/jcb.200601018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorda-Diez CI, Montero JA, Garcia-Porrero JA, & Hurle JM (2015). Interdigital tissue regression in the developing limb of vertebrates. International Journal of Developmental Biology, 59(1-2–3), 55–62. 10.1387/ijdb.150065jh [DOI] [PubMed] [Google Scholar]
- Maconnachie E (1979). A study of digit fusion in the mouse embryo. Journal of Embryology and Experimental Morphology, 49, 259–276. [PubMed] [Google Scholar]
- Martins VL, Caley M, & O’Toole EA (2013). Matrix metalloproteinases and epidermal wound repair. Cell and Tissue Research, 351(2), 255–268. 10.1007/s00441-012-1410-z [DOI] [PubMed] [Google Scholar]
- Mikkola ML, & Millar SE (2006). The Mammary Bud as a Skin Appendage: Unique and Shared Aspects of Development. Journal of Mammary Gland Biology and Neoplasia, 11, 187–203. 10.1007/s10911-006-9029-x [DOI] [PubMed] [Google Scholar]
- Montero JA, & Hurlé JM (2010). Sculpturing digit shape by cell death. Apoptosis, 15(3), 365–375. 10.1007/s10495-009-0444-5 [DOI] [PubMed] [Google Scholar]
- Mosaffa P, Rodríguez-Ferran A, & Muñoz JJ (2018). Hybrid cell-centred/vertex model for multicellular systems with equilibrium-preserving remodelling. International Journal for Numerical Methods in Biomedical Engineering, 34(3). 10.1002/cnm.2928 [DOI] [PubMed] [Google Scholar]
- Newman TJ (2005). Modeling multicellular systems using subcellular elements. Mathematical Biosciences and Engineering: MBE, 2(3), 613–624. [DOI] [PubMed] [Google Scholar]
- Pajni-Underwood S, Wilson CP, Elder C, Mishina Y, & Lewandoski M (2007). BMP signals control limb bud interdigital programmed cell death by regulating FGF signaling. Development (Cambridge, England), 134(12), 2359–2368. 10.1242/dev.001677 [DOI] [PubMed] [Google Scholar]
- Parfitt GJ, Xie Y, Reid KM, Dervillez X, Brown DJ, & Jester JV (2012). A Novel Immunofluorescent Computed Tomography (ICT) Method to Localise and Quantify Multiple Antigens in Large Tissue Volumes at High Resolution. PLOS ONE, 7(12), e53245. 10.1371/journal.pone.0053245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautou PM-P (1975). Morphogenèse de l’autopode chez l’embryon de poulet. Development, 34(2), 511–529. [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, Rautio J, Pegelow M, Karsten A, Basel-Vanagaite L, Gordon W, Andersen B, Svensson T, Murray JC, Cornell RA, … Schutte BC (2014). Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. American Journal of Human Genetics, 94(1), 23–32. 10.1016/j.ajhg.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutoni C, Bazellieres E, Borgne-Rochet ML, Comunale F, Brugues A, Séveno M, Planchon D, Thuault S, Morin N, Bodin S, Trepat X, & Gauthier-Rouvière C (2016). P-cadherin promotes collective cell migration via a Cdc42-mediated increase in mechanical forces. The Journal of Cell Biology, 212(2), 199–217. 10.1083/jcb.201505105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, & Dixon MJ (2009). Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Human Molecular Genetics, 18(14), 2632–2642. 10.1093/hmg/ddp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Hammond NL, Coulombe PA, Saloranta C, Nousiainen HO, Salonen R, Berry A, Hanley N, Headon D, Karikoski R, & Dixon MJ (2014). Periderm prevents pathological epithelial adhesions during embryogenesis. The Journal of Clinical Investigation, 124(9), 3891–3900. 10.1172/JCI71946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandersius SA, & Newman TJ (2008). Modeling cell rheology with the Subcellular Element Model. Physical Biology, 5(1), 015002. 10.1088/1478-3975/5/1/015002 [DOI] [PubMed] [Google Scholar]
- Scarpa E, & Mayor R (2016). Collective cell migration in development. The Journal of Cell Biology, 212(2), 143–155. 10.1083/jcb.201508047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Yi S, Kassabian G, Mayo M, Sank A, & Shuler C (2000). Changes in expression of the lysosomal membrane glycoprotein, LAMP-1 in interdigital regions during embryonic mouse limb development, in vivo and in vitro. Anatomy and Embryology, 201(6), 483–490. 10.1007/s004290050335 [DOI] [PubMed] [Google Scholar]
- Theveneau E, & Mayor R (2012). Cadherins in collective cell migration of mesenchymal cells. Current Opinion in Cell Biology, 24(5), 677–684. 10.1016/j.ceb.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SB, Wilanowski T, Auden A, Hall M, Voss AK, Thomas T, Parekh V, Cunningham JM, & Jane SM (2003). Inositol- and folate-resistant neural tube defects in mice lacking the epithelial-specific factor Grhl-3. Nature Medicine, 9(12), 1513–1519. 10.1038/nm961 [DOI] [PubMed] [Google Scholar]
- Tokuo H, Bhawan J, & Coluccio LM (2018). Myosin X is required for efficient melanoblast migration and melanoma initiation and metastasis. Scientific Reports, 8(1), 10449. 10.1038/s41598-018-28717-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun Y, Huang Y, Pan Y, Jia Z, Ma L, Ma L, Lan F, Zhou Y, Shi J, Yang X, Zhang L, Jiang H, Jiang M, Yin A, Cheng J, Wang L, Yang Y, & Shi B (2016). Association study between Van der Woude Syndrome causative gene GRHL3 and nonsyndromic cleft lip with or without cleft palate in a Chinese cohort. Gene, 588(1), 69–73. 10.1016/j.gene.2016.04.045 [DOI] [PubMed] [Google Scholar]
- Wong YL, Behringer RR, & Kwan KM (2012). Smad1/Smad5 signaling in limb ectoderm functions redundantly and is required for interdigital programmed cell death. Developmental Biology, 363(1), 247–257. 10.1016/j.ydbio.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Bassuk AG, & Fritzsch B (2013). Pricklel stunts limb growth through alteration of cell polarity and gene expression: Prickle1 Stunts Limb Growth. Developmental Dynamics, 242(11), 1293–1306. 10.1002/dvdy.24025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, & Andersen B (2006). The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Developmental Biology, 299(1), 122–136. 10.1016/j.ydbio.2006.07.015 [DOI] [PubMed] [Google Scholar]
- Zuzarte-Luis V, Berciano MT, Lafarga M, & Hurlé JM (2006). Caspase redundancy and release of mitochondrial apoptotic factors characterize interdigital apoptosis. Apoptosis: An International Journal on Programmed Cell Death, 11(5), 701–715. 10.1007/s10495-006-5481-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Related to Figure 1. A morphed time-lapse of forelimb digit separation during development.
Movie 2. Related to Figure 2. 3D reconstruction of WT E15.5 limb epithelia.
Movie 3. Related to Figure 2. 3D reconstruction of Grhl3−/− E15.5 limb epithelia.
Movie 4. Related to Figure 4. A time-course (48h) representation of Voronoi models featuring cell proliferation.
Movie 5. Related to Figure 4. A time-course (48h) representation of Voronoi models featuring cell migration.
Movie 6. Related to Figure 5. Live imaging of Krt14-H2B-GFP cells in a cultured embryonic forelimb explant (E13.5) over time (3.5 hours).
Movie 7. Related to Figure 5. Cell-tracking in a cultured embryonic forelimb explant (E13.5) over time (3.5 hours).
Movie 8. Related to Figure 5. Ortho-slice view of a 3D stack at the interdigital zone.
Movie 9. Related to Figure 5. Ortho-slice view of a 3D stack at the digit zone.
Data Availability Statement
RNA sequencing data generated in this study are available at Gene Expression Omnibus (GEO) accession number GSE141009.