Abstract
Background
Aberrant methylation is a known cause of cancer initiation and/or progression. There is scant data on the genome-wide methylation pattern of non-functioning pancreatic neuroendocrine tumors (NFPanNETs) and in sporadic and hereditary NFPanNETs.
Methods
Thirty-three tissue samples were analyzed, including sporadic (n=9), von Hippel-Lindau [VHL]-related and multiple endocrine neoplasia type 1 [MEN1]-related NFPanNETs (n=10 each), which were compared to normal islet cells (NI, n=4). Genome-wide CpG methylation profiling was performed using the Infinium MethylationEPIC BeadChips Assay and analyzed by R-based tools.
Results
On unsupervised hierarchical clustering, sporadic and MEN1-related NFPanNETs clustered together, and the VHL group in a separate cluster. MEN1-related NFPanNET had a higher rate of hypermethylated CpG sites comparing to sporadic and VHL-related tumor groups. Differentially methylated region (DMR) analysis confirmed the higher rate of hypermethylation in MEN1-related tumors. Moreover, in integrated analysis of gene expression data in the same tumor samples, we found downregulated gene expression in most genes that were hypermethylated. In a CpG Island Methylator Phenotype (CIMP) analysis, three genes were identified and confirmed to have downregulated gene expression, SFRP5 in sporadic NFPanNET, and CDCA7L and RBM47 in MEN1-related NFPanNET.
Conclusions
MEN1 NFPanNETs have a higher rate of genome-wide hypermethylation compared with other NFPanNET subtypes. The similarity between pathways enriched on methylation analysis of known genes involved in NFPanNET tumorigenesis suggest a key role for aberrant methylation in the pathogenesis of NFPanNETs.
Keywords: pancreatic neuroendocrine tumor, DNA methylation, MEN-1, VHL, nonfunctioning
Precis
There is scant data on the genome-wide methylation pattern of sporadic and hereditary non-functioning pancreatic neuroendocrine tumors (NFPanNETs).
In the current analysis we compared genome-wide methylome of NFPanNETs of patients with and without hereditary syndromes, and found a higher rate of hypermethylation in those related to multiple endocrine neoplasia type 1, and hypomethylation in von Hippel-Lindau-related tumors.
Introduction
The incidence of pancreatic neuroendocrine tumors (PanNETs) has been increasing in the last decades, currently with an incidence of 0.48 cases per 100,000 inhabitants annually in the United States (1–3). PanNETs are categorized as functioning (secreting compounds that lead to a clinical syndrome) and nonfunctioning tumors (NFPanNET), the latter being more prevalent. Functional PanNETs, even when localized and small, require treatment to control the clinical manifestations, and morbidity and mortality caused by the excess hormone production. In contrast, small, low-grade NFPanNETs do not always require surgical or medical intervention, as they may have an indolent clinical course, but the natural history of such tumors is heterogenous and unpredictable. The current recommendations for treatment of syndromic NFPanNETs are mostly based on the risk of metastatic disease, using tumor growth and size as a surrogate marker for aggressiveness 1.
PanNETs can occur sporadically or in the context of an inherited cancer syndrome. These include Multiple Endocrine Neoplasia Type 1 (MEN1), von Hippel-Lindau (VHL), Neurofibromatosis type 1, Tuberous Sclerosis Complex and Cowden syndrome 1–4. About 40% of patients with MEN1 5, and about 15% of patients with VHL 1 develop NFPanNETs during their lifetime. Syndromic PanNETs may have a less aggressive natural history than their sporadic counterparts 6–8. Hence the criteria used for sporadic NFPanNETs risk stratification do not accurately predict the metastatic potential and/or aggressive behavior of NFPanNETs in patients with VHL and MEN1 9–13. The biologic mechanism underlying this difference is not well understood 1, 14.
The genes responsible for the major inherited cancer syndromes associated with NFPanNETs (VHL, MEN1) have been well characterized. In VHL, a broad genotype-phenotype association was demonstrated for PanNET, with missense and/or exon 3 pathogenic variants associated with high risk of disease progression and metastasis 6, 15. In contrast, no clear genotype-phenotype association has been found in MEN1. Also, a growing number of studies have characterized the main genes and molecular pathways associated with the tumorigenesis of sporadic PanNETs 16, 17, and reported a high rate of somatic mutations in MEN1, DAXX, ATRX, and PTEN.
The best characterized epigenetic event involved in cancer initiation and/or progression is DNA methylation of cytosines, by DNA methyltransferases. The dinucleotide sequence of cytosine followed by guanine is termed CpG, and a high-density cluster of CpGs are referred to as CpG island. Approximately 60% of CpG islands are in the 5’ regulatory (promoter) regions of genes, and have key role in transcription regulation, although CpG islands that are not in promoter regions may also be targets for de novo methylation in cancer. Furthermore, differential CpG methylation in DNA enhancer regions have also been implicated in cancer and altered gene expression 18. DNA methylation affects a wide range of molecular and cellular processes involved in cancer initiation and progression, such as apoptosis, cell cycle, DNA damage repair, growth factor response, signal transduction, and more 19.
Gene-specific CpG promoter hypermethylation has been reported in a variety of NETs including the pancreas, pituitary, and adrenal glands. The genes with dysregulated methylation included CDKN2A, CDKN2B, APC, CTNNB1, HIC1, RIZ1, MEG3, MLH1 and RASSF1A 20–23. Furthermore, specific methylation analysis has shown a high rate of promoter hypermethylation in MEN1-related tumors in an analysis of 56 genes 24. To our knowledge, the genome-wide methylation profiling of NFPanNETs has not been reported, both sporadic and syndromic, as well as, an integrated analysis of methylation and gene expression. Furthermore, the epigenetically regulated pathway(s) involved in NFPanNETs are unknown. Such data could be useful for better understanding the pathobiology of NFPanNETs and for identifying new markers of disease behavior, and markers for diagnosis of NFPanNETs, and can help identify new targets for treatment of both syndromic and sporadic NFPanNETs with epigenetics modulating drugs 25, 26.
Given the limited data on the importance of methylation in NET and the lack of genome-wide methylome data of inherited and sporadic NFPanNETs, we investigated the genome-wide methylome of sporadic and syndromic (MEN1 and VHL) NFPanNETs with integrated gene expression data in the same samples.
Materials and Methods
Patients
The study protocol was approved by the institutional review board, and written informed consent was obtained from all patients. Patients were diagnosed with VHL or MEN1 based on either germline DNA analysis revealing a pathogenic variant, or according to established clinical criteria for VHL 15 and MEN1 27. Patients clinical data were retrieved from their medical records, and the tumor (non) functional status was determined by clinical features, and by biochemical testing as clinically indicated. Tissue samples included 9 sporadic NFPanNET, 10 MEN1 NFPanNET, 10 VHL NFPanNET and 4 NI samples. Detailed clinical and histopathologic data of the study cohort are summarized in Table 1.
Table 1.
Clinical and histopathologic data of patients with NFPanNET in the study cohort
| Sporadic | VHL | MEN-1 | |
|---|---|---|---|
| Tumor WHO grade (Grade 1/Grade 2) n[%] |
5/4 [56/44] | 6/4 [60/40] | 5/5 [50/50] |
| Metastatic disease (Yes/No/Unknown) n[%] |
|||
| Lymph nodes | 0/9/0 [0/100/0] | 1/6/3 [10/60/30] | 2/6/2 [20/60/20] |
| Distant metastases | 0/9/0 [0/100/0] | 1/9/0 [10/90/0] | 2/8/0 [20/80/0] |
| Tumor diameter (cm) median[range] |
3.5 [3.0–7.0] | 2.0 [0.9–5.0] | 2.5 [0.7–3.2] |
| Tumor site in the pancreas (Head/Uncinate/Body/Tail) n[%] |
6/0/1/2 [60/0/10/20] | 5/1/1/3 [50/10/10/30] | 3/1/1/5 [30/10/10/50] |
NFPanNET, non-functioning pancreatic neuroendocrine tumor; VHL, von Hippel-Lindau; MEN-1, multiple endocrine neoplasia type 1; WHO, World Health Organization
Tissue procurement
Tissue samples were obtained at the time of surgical resection. Samples were snap frozen and immediately stored at −80°C. Tissue sections adjacent to those used for RNA and DNA isolation were stained using hematoxylin-eosin to confirm the diagnosis and a tumor cell content >80%. All tissues included in the study underwent secondary histologic review by an endocrine pathologist (MMQ).
Normal islet cell samples
Normal islet cell (NI) samples were obtained from the Division of Transplantation, Department of Surgery at the University of Alabama at Birmingham. Pancreas were recovered, with informed consent, from cadaveric donors after in situ vascular perfusion with University of Wisconsin solution at 4°C, as part of a multiorgan procurement. The pancreatic islets were isolated by a semiautomated method and purified using the Cobe 2991 cell processor (Gambro BCT, Lakewood, CO). The number of islets within each size class was converted to the standard number of islets of 150-μm diameter, equal in volume to the sample. Purity was assessed by comparing the relative quantity of dithizone-stained endocrine tissue with unstained exocrine tissue. Only islets with greater than 90% viability and greater than 60% purity were used.
DNA extraction and processing
Frozen NFPanNET tissue samples were sectioned and DNA was extracted using the DNeasy blood and tissue kit (QIAGEN, Valencia, CA) according to the manufacturer protocol. DNA quality was determined using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE) and quantified using the Quant-iT PicoGreen dsDNA Assay (Life Technologies, Grand Island NY).
Gene expression analysis
Gene expression analysis was performed as described previously 28. Briefly, sample labeling, and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies). Total RNA from each sample was linearly amplified and labeled. The labeled complementary RNAs (cRNAs) were purified using the RNeasy Mini Kit (Qiagen), and the labeled cRNAs were measured using the NanoDrop ND-1000 spectrophotometer. One microgram of each labeled cRNA was fragmented, hybridized and scanned using the Agilent DNA Microarray Scanner (Agilent Technologies).
Methylation data processing and analysis
Methylation analysis and data quality control were performed on R version 3.4.4 and R Studio Version 1.0.153, using the minfi package 29, which was modified for the analysis of Illumina EPIC arrays 30. All comparisons were based on the M values calculated by the minfi package from the corresponding beta values. The minfi package was also used for functional normalization 31, and for differentially methylated probe (DMP, at each CpG site) detection.
Regions with higher and lower M values compared to NI values were considered hyper- and hypomethylated, respectively. DMPs with at least 2-fold change with a false discovery rate adjusted p-value < 0.05 were considered significantly different and were included for further analysis. Pathway analysis using DMPs was performed using the ReacromePA package 32.
Differentially methylated regions (DMRs) were detected using the bumphunter package 33, based on a maximal gap of 300, M value ratio of 0.55 and 500 permutations. Significantly different regions were considered based on an area family-wise error rate (FWER) <0.05. CpG island methylator phenotype (CIMP) analysis was done to detect genes with a statistically significant hypermethylation in gene promoter regions compared to NI samples based on the DMR analysis results. Promoter regions were annotated using the matchGenes function of bumphunter package, with default parameters.
To confirm that our results were not biased by NI impurity, we analyzed the methylation status of different NFPanNETs based the raw M values. We compared the rate of CpG hypomethylation and hypermethylation (mean M-value < −2 or > 2, respectively). Methylation levels were compared using the analysis of variances (ANOVA) with the Tukey adjustment method (Supplementary Figure 1).
To assess the full extent of the implications of altered methylation on gene expression, we performed an ENCODE based analysis for differential methylation in gene enhancers. All DMRs were compared with the RegulomeDB (http://www.regulomedb.org). We considered coordinates as enhancers if they were verified by expression quantitative trait loci (RegulomoeDB score of 1f or above). The enhancers detected were further verified by up- and down-regulation of gene expression in hypo- and hypermethylated gene enhancers, respectively.
The heatmap and unsupervised hierarchical clustering analysis presented in Figure 1 were analyzed using the Partek Genomic suite 7.17 software (Partek Inc., St. Louis, MO).
Figure 1. (A).
Heatmap of unsupervised cluster analysis based on methylation M values for the top 5000 most variable methylated probes. The sporadic and MEN1-related tumors clustered together and VHL-related NFPanNET separately. (B) DMPs by genomic regions of CpG sites. For each group upper/lower panels represent hyper/hypomethylation, respectively, the y-axis represents delta-M value (dM) vs. normal pancreatic islet-cells, the blue line (grey area) represents mean (standard deviation) of dM.
DMP, differentially methylated probes; NFPanNETs, non-functioning pancreatic neuroendocrine tumor; VHL, von Hippel-Lindau; MEN1, multiple endocrine neoplasia type 1; dM, delta-M value
Results
Different methylation pattern between subgroups of NFPanNETs
A total of 15,844 CpG sites were found to be differentially methylated between NFPanNETs and NIs. Unsupervised clustering analysis, using the top 5000 DMPs, showed clustering of the VHL NFPanNETs together and a separate cluster of sporadic and MEN1 NFPanNETs (Figure 1A). A total of 4666, 2780 and 25633 DMPs in the sporadic, VHL and MEN1 groups, respectively, were found to be significantly different with a delta M value > 2 (hypermethylated) or < −2 (hypomethylated) compared to NI samples. The rate of hypomethylated CpG sites was significantly higher among VHL-related NFPanNET (66.3%) compared with both sporadic (33.7%) and MEN-1-related NFPanNET (37.8%, p<0.001 for both comparisons) (Figure 1B). We further analyzed the methylation patterns by overall cancer stage including tumor size and lymph node metastasis, and tumor grade, and found no statistically significant DMPs.
Pathway analysis
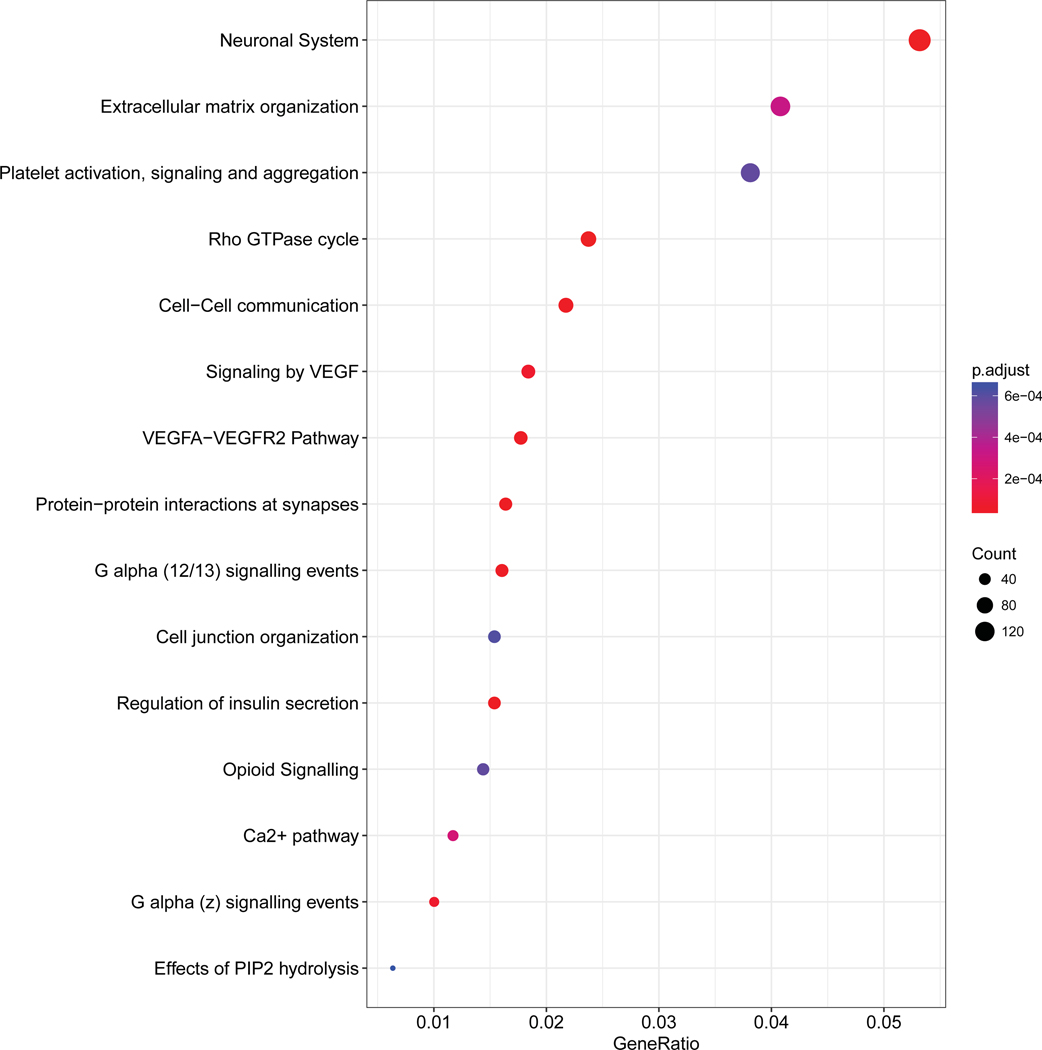
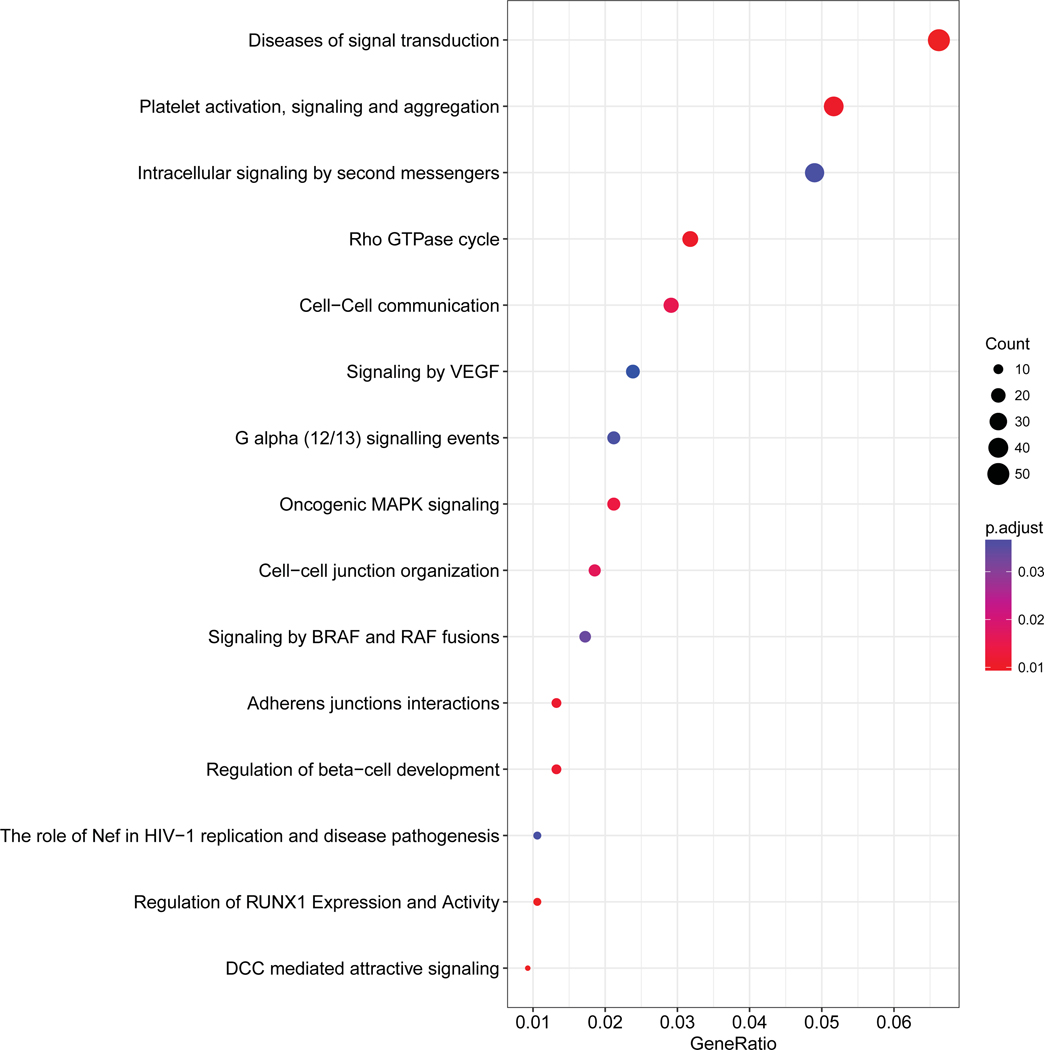
We performed pathway analysis based on the genes annotated in the DMP positions. In sporadic NFPanNETs, the main pathways enriched included the transcription regulator RUNX3, PLC-related pathway, DAG/IP3 signaling, regulation of insulin secretion, and calcium-related pathways. In VHL-related tumors, among the most enriched pathways were intracellular signal transduction pathways, VEGF-related pathway, regulation of beta−cell development and RUNX1 expression and activity. Whereas in MEN1-related tumors, neuronal pathways, VEGF signaling, insulin secretion regulation, calcium dependent pathway, G alpha and pophatidylinositol-4,5-bisphosphate (PIP2)-related pathway were enriched according to the aberrantly methylated genes (Figure 2).
Figure 2.
Pathway analysis based on the differentially methylated probe analysis. For each group, the fifteen most enriched pathways are shown.
NFPanNETs, non-functioning pancreatic neuroendocrine tumor; VHL, von Hippel-Lindau; MEN-1, multiple endocrine neoplasia type 1
Differentially methylated regions (DMRs)
We performed a DMR analysis to define key genomic regions more robustly. DMR analysis detected 34 DMRs in sporadic (18/16 hyper-/hypomethylated), 32 DMRs in VHL-related (9/23 hyper-/hypomethylated) and 66 DMRs in MEN1-related NFPanNETs (56/10 hyper-/hypomethylated), as compared to NI samples (Figure 3A). The total number of genes annotated for these regions were 34 in the sporadic group, 26 in the VHL group and 66 in the MEN1 group. Of them, 16 (8/8), 13 (4/9) and 29 (26/3) were associated with significantly differential gene expression compared with NI samples (Figure 3B). We found 5, 1 and 19 in 8, 4 and 26 hypermethylated genes in the sporadic, VHL and MEN1 groups, respectively, associated with downregulated gene expression, and 7, 7 and none of the 8, 9 and 3 hypomethylated genes were associated with upregulated gene expression, respectively (Figure 3B).
Figure 3.
Number of overlapping genes with hyper- and hypomethylation between sporadic, VHL and MEN1 NFPanNETs (A), and gene expression difference in NFPanNETs vs. normal islet cells, in the different groups (B), in significantly hyper and hypomethylated genes based in DMR analysis (left and right in each group, respectively). The filled color indicates up- (blue) or down-regulation (red) of gene expression.
NFPanNETs, non-functioning pancreatic neuroendocrine tumors; VHL, von Hippel-Lindau; MEN-1, multiple endocrine neoplasia type 1; DMR, differentially methylated regions
Candidate genes for CpG island methylator phenotype (CIMP)
To characterize a possible CIMP in NFPanNET, we compared methylation levels between each group and NI samples, and identified several genes with significantly hypermethylated promoter regions 34. The analysis was based on the DMR analysis results and detected eight genes with promoter hypermethylation in the MEN1 group, two in the sporadic group and none in the VHL group. To validate these findings, we evaluated gene expression data in the same samples as used for the methylation analysis. We found downregulated gene expression (based on a fold change <1.5 compared with NI samples, with a p-value < 0.05 of an unpaired t-test) in one of the two genes with promoter hypermethylation in the sporadic group (SFRP5) and 2 of the 8 genes in the MEN1 group (CDCA7L and RBM47) (Figure 4).
Figure 4.
Gene promoter hypermethylation in sporadic NFPanNET (SFRP5) and MEN-1 NFPanNET (CDCA7L and RBM47), compared with normal islet cells. The methylation status of the genes was inversely associated with differential gene expression.
NFPanNETs, non-functioning pancreatic neuroendocrine tumors; VHL, von Hippel-Lindau; MEN-1, multiple endocrine neoplasia type 1; DMR, differentially methylated regions
Aberrant methylation of enhancer regions
Significantly different methylation levels between NFPanNET and NI samples were detected in four gene enhancer regions (Table 2). The gene enhancer for PTPRN2 was found to be significantly hypomethylated based on the DMR analysis in all three groups (sporadic, MEN1 and VHL), and was associated with upregulated PTPRN2 expression in both sporadic (fold change of 12.7, p-value < 0.001) and MEN1 (fold change of 3.7, p-value = 0.003) NFPanNETs. In addition, hypomethylation of the enhancer for TRPV2 was found in the VHL group, with upregulated gene expression (fold change of 4.7, p-value < 0.001). Enhancer hypermethylation was found in two genes (SLC1A5, PQLC2) in MEN1-related NFPanNETs, SLC1A5, was associated with downregulated gene expression (fold change of 3.0, p-value = 0.004).
Table 2.
Gene enhancers with significantly different methylation status in non-functioning pancreatic neuroendocrine tumors.
| Group | Methylation status | Coordinate | dbSNP ID | RegulomeDB score | Gene regulated |
|---|---|---|---|---|---|
| VHL | Hypo | chr7 157361733 |
rs221294 | 1b | PTPRN2 |
| VHL | Hypo | chr17 16318931 |
rs3813769 | 1f | TRPV2 |
| Sporadic | Hypo | chr7 157361733 |
rs221294 | 1b | PTPRN2 |
| MEN-1 | Hyper | chr19 47288149 |
rs8105903 | 1a | SLC1A5 |
| MEN-1 | Hyper | chr1 18807136 |
rs2992757 | 1d | PQLC2 |
| MEN-1 | Hypo | chr7 157361733 |
rs221294 | 1b | PTPRN2 |
VHL, von Hippel-Lindau; MEN-1 multiple endocrine neoplasia type 1 syndrome
Discussion
In this study, we analyzed the genome-wide methylation pattern of NFPanNETs, aiming to characterize aberrant methylation in these tumors, and possible differences between sporadic vs. hereditary NFPanNETs. We have shown that in MEN1 NFPanNETs, there are significantly more hypermethylated genomic positions as compared to sporadic and VHL NFPanNETs. In pathway analysis based on genes with an aberrant methylation patterns, pathways associated with the specific signaling pathways, such as VEGF in VHL, were identified. Finally, we also found three genes in which significant CpG promoter hypermethylation was detected, with downregulated gene expression. We identified altered methylation in several gene enhancers that was associated with altered gene expression in NFPanNETs.
The gene responsible for the MEN1 syndrome, MEN1, is located on chromosome 11q13 35. Germline MEN1 mutations are detected in 70–90% of patients with a familial presentation of MEN1 36. Menin, the protein encoded by MEN1, participates in epigenetic control through direct interaction with protein-complexes and through regulation of non-coding RNAs. Altered menin function leads to epigenetic aberrations and consequently to tumorigenesis, possibly explaining the weak genotype-phenotype association based on MEN1 mutation analysis 20. Our findings of a high rate of methylation aberrations in MEN-1-related tumors compared with sporadic and VHL-related tumors further support the role of menin in the regulation of DNA methylation.
The results of the pathway analysis emphasize the possible key role of methylation in the pathogenesis of NFPanNETs. In all three groups, pathways found to be enriched are associated with NET pathogenesis and/or with islet cell function. These include insulin secretion regulation in sporadic and MEN1-related groups, and beta cell differentiation in the VHL group. The two transcription factors RUNX1 37 and RUNX3 38, enriched in the VHL and sporadic groups, respectively, were previously reported to be altered in NET. Both PIP2 39 and DAG/IP3 40, are involved in the PI3K/AKT/mTOR pathway, which was reported by Scarpa and associates as one of the four main pathways of NFPanNET pathogenesis 17. While Scarpa et al. showed that mutations in genes involved in these pathways are common, we now show that epigenetic alterations of genes involved in these pathways may be an alternative mechanism for dysregulation of these same pathways in NFPanNETs.
We identified three candidate genes for CIMP, based on promoter hypermethylation and downregulated gene expression. SFRP5 (secreted frizzle-related protein 5), that was detected in sporadic NFPanNET in our analysis is associated with Wnt/ß-catenin signaling. Altered methylation of SFRP5 was reported in esophageal squamous-cell carcinoma 41 and in treatment-resistance leukemia 42. SFRP5 has also been reported to be associated with beta cell function 43. Promoter hypermethylation was detected in MEN1-related NFPanNETs in two genes: CDCA7L and RBM47. The former encodes the cell division cycle-associated 7-like protein, which interacts with c-Myc, a pathway altered regulated by menin 44. CDCA7L was associated with hepatocellular carcinoma, and was found to have altered methylation and gene expression in pediatric pineal tumors 45. The tumor-suppressive role of RBM47 (RNA binding motif 47) was reported in colorectal and breast cancer 46, 47, and in lung adenocarcinomas 48. Validation of the aberrant methylation of these genes’ promoter region in an independent cohort in the future will be important to define CIMP in NFPanNET subtypes.
We have identified altered methylation in several gene enhancers. Among them the enhancer for the PTPRN2 gene was found hypomethylated in all three groups. The gene PTPRN2 (protein tyrosine phosphatase, receptor type N2) encodes the transmembrane protein PTP IA-2beta, which is highly expressed in the pancreatic islets, and known as a target antigen in autoimmune diabetes mellitus 49. Altered methylation of PTPRN2 has been reported in hepatocellular carcinoma 50 and lung cancer 51, but PTPRN2 enhancer hypomethylation has not been reported in NETs. Enhancer methylation status has been implicated in cancer initiation 18, in defining subtypes of breast cancer 52, and in cancer plasticity and cancer survival 53. However, to the best of our knowledge this is the first report evaluating enhancer methylation status and gene expression in NETs. The common hypomethylation of PTPRN2 enhancer in all three groups of NFPanNET suggests this as a possible common pathogenic pathway in these tumors.
We have shown that aberrant methylation was associated with altered gene expression and thus likely to play an important role in NFPanNET initiation and or progression even in the presence of driver mutations. Moreover, the overlap in pathways between the different groups in our study suggests a common epigenetic pathogenic mechanism.
The main limitation of our study is the relatively small number of samples, stemming from the low incidence of sporadic NFPanNET and especially the hereditary types. The comparison between methylation status of the NFPanNET to NI samples may be affected by sample purity. Impurity of NI samples may lead to false positive methylation difference in genes with differential expression in the pancreatic islet vs. exocrine pancreas (the probable contaminant). To control for such bias, we performed independent comparison of methylation levels excluding the islet cell samples, and found hypermethylation of sporadic and MEN1-related NFPanNET and hypomethylation of the VHL-related tumors, which supports the results of the comparison between NFPanNETs and NI samples.
In conclusion, our study supports the key role of altered DNA methylation in NFPanNET tumorigenesis, which was further supported by the associated gene expression changes in the differentially methylated genes. The methylome of NFPanNETs is distinct from NI and between VHL and sporadic and MEN1-related NFPanNETs. The involvement of common pathways in genes with altered methylation status suggest that epigenetic cancer therapy may be an effective treatment strategy for NFPanNETs that fail standard therapy.
Supplementary Material
Acknowledgments
This research was supported by the intramural research program of the Center for Cancer Research, National Cancer Institute.
Footnotes
The authors declare that they have nothing to disclose.
References
- 1.Keutgen XM, Hammel P, Choyke PL, Libutti SK, Jonasch E, Kebebew E. Evaluation and management of pancreatic lesions in patients with von Hippel-Lindau disease. Nat Rev Clin Oncol. 2016;13: 537–549. [DOI] [PubMed] [Google Scholar]
- 2.Bilimoria KY, Tomlinson JS, Merkow RP, et al. Clinicopathologic features and treatment trends of pancreatic neuroendocrine tumors: analysis of 9,821 patients. J Gastrointest Surg. 2007;11: 1460–1467; discussion 1467–1469. [DOI] [PubMed] [Google Scholar]
- 3.Horton WA, Wong V, Eldridge R. Von Hippel-Lindau disease: clinical and pathological manifestations in nine families with 50 affected members. Arch Intern Med. 1976;136: 769–777. [DOI] [PubMed] [Google Scholar]
- 4.Kloppel G, Perren A, Heitz PU. The gastroenteropancreatic neuroendocrine cell system and its tumors: the WHO classification. Ann N Y Acad Sci. 2004;1014: 13–27. [DOI] [PubMed] [Google Scholar]
- 5.Thakker RV. Multiple endocrine neoplasia--syndromes of the twentieth century. J Clin Endocrinol Metab. 1998;83: 2617–2620. [DOI] [PubMed] [Google Scholar]
- 6.Tirosh A, Sadowski SM, Linehan WM, et al. Association of VHL Genotype With Pancreatic Neuroendocrine Tumor Phenotype in Patients With von Hippel-Lindau Disease. JAMA Oncol. 2018;4: 124–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sallinen V, Le Large TY, Galeev S, et al. Surveillance strategy for small asymptomatic non-functional pancreatic neuroendocrine tumors - a systematic review and meta-analysis. HPB (Oxford). 2017;19: 310–320. [DOI] [PubMed] [Google Scholar]
- 8.Yao JC, Hassan M, Phan A, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26: 3063–3072. [DOI] [PubMed] [Google Scholar]
- 9.Blansfield JA, Choyke L, Morita SY, et al. Clinical, genetic and radiographic analysis of 108 patients with von Hippel-Lindau disease (VHL) manifested by pancreatic neuroendocrine neoplasms (PNETs). Surgery. 2007;142: 814–818; discussion 818 e811–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlesworth M, Verbeke CS, Falk GA, Walsh M, Smith AM, Morris-Stiff G. Pancreatic lesions in von Hippel-Lindau disease? A systematic review and meta-synthesis of the literature. J Gastrointest Surg. 2012;16: 1422–1428. [DOI] [PubMed] [Google Scholar]
- 11.Hammel PR, Vilgrain V, Terris B, et al. Pancreatic involvement in von Hippel-Lindau disease. The Groupe Francophone d’Etude de la Maladie de von Hippel-Lindau. Gastroenterology. 2000;119: 1087–1095. [DOI] [PubMed] [Google Scholar]
- 12.Libutti SK, Choyke PL, Bartlett DL, et al. Pancreatic neuroendocrine tumors associated with von Hippel Lindau disease: diagnostic and management recommendations. Surgery. 1998;124: 1153–1159. [DOI] [PubMed] [Google Scholar]
- 13.Sadowski SM, Triponez F. Management of pancreatic neuroendocrine tumors in patients with MEN 1. Gland Surg. 2015;4: 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Mestier L, Gaujoux S, Cros J, et al. Long-term Prognosis of Resected Pancreatic Neuroendocrine Tumors in von Hippel-Lindau Disease Is Favorable and Not Influenced by Small Tumors Left in Place. Ann Surg. 2015;262: 384–388. [DOI] [PubMed] [Google Scholar]
- 15.Tirosh A, Lakis ME, Green P, et al. In-silico VHL Gene Mutation Analysis and Prognosis of Pancreatic Neuroendocrine Tumors in von Hippel-Lindau Disease. J Clin Endocrinol Metab. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scarpa A, Chang DK, Nones K, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543: 65–71. [DOI] [PubMed] [Google Scholar]
- 18.Aran D, Hellman A. DNA methylation of transcriptional enhancers and cancer predisposition. Cell. 2013;154: 11–13. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal SK, Debelenko LV, Kester MB, et al. Analysis of recurrent germline mutations in the MEN1 gene encountered in apparently unrelated families. Hum Mutat. 1998;12: 75–82. [DOI] [PubMed] [Google Scholar]
- 20.Iyer S, Agarwal SK. Epigenetic regulation in the tumorigenesis of MEN1-associated endocrine cell types. J Mol Endocrinol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg D, Akerstrom G, Westin G. Evaluation of CDKN2C/p18, CDKN1B/p27 and CDKN2B/p15 mRNA expression, and CpG methylation status in sporadic and MEN1-associated pancreatic endocrine tumours. Clin Endocrinol (Oxf). 2008;68: 271–277. [DOI] [PubMed] [Google Scholar]
- 22.Modali SD, Parekh VI, Kebebew E, Agarwal SK. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol Endocrinol. 2015;29: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J Clin Endocrinol Metab. 2005;90: 2179–2186. [DOI] [PubMed] [Google Scholar]
- 24.Conemans EB, Lodewijk L, Moelans CM, et al. DNA methylation profiling in MEN1-related pancreatic neuroendocrine tumors reveals a potential epigenetic target for treatment. Eur J Endocrinol. 2018. [DOI] [PubMed] [Google Scholar]
- 25.de Lera AR, Ganesan A. Epigenetic polypharmacology: from combination therapy to multitargeted drugs. Clin Epigenetics. 2016;8: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lines KE, Stevenson M, Filippakopoulos P, et al. Epigenetic pathway inhibitors represent potential drugs for treating pancreatic and bronchial neuroendocrine tumors. Oncogenesis. 2017;6: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thakker RV, Newey PJ, Walls GV, et al. Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1). J Clin Endocrinol Metab. 2012;97: 2990–3011. [DOI] [PubMed] [Google Scholar]
- 28.Keutgen XM, Kumar S, Gara S, et al. Transcriptional alterations in hereditary and sporadic nonfunctioning pancreatic neuroendocrine tumors according to genotype. Cancer. 2018;124: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortin JP, Triche TJ, Jr., Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33: 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortin JP, Labbe A, Lemire M, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12: 477–479. [DOI] [PubMed] [Google Scholar]
- 33.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41: 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jover R, Nguyen TP, Perez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandrasekharappa SC, Guru SC, Manickam P, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276: 404–407. [DOI] [PubMed] [Google Scholar]
- 36.Agarwal SK. The future: genetics advances in MEN1 therapeutic approaches and management strategies. Endocr Relat Cancer. 2017;24: T119–T134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofsli E, Wheeler TE, Langaas M, Laegreid A, Thommesen L. Identification of novel neuroendocrine-specific tumour genes. Br J Cancer. 2008;99: 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold CN, Sosnowski A, Schmitt-Graff A, Arnold R, Blum HE. Analysis of molecular pathways in sporadic neuroendocrine tumors of the gastro-entero-pancreatic system. Int J Cancer. 2007;120: 2157–2164. [DOI] [PubMed] [Google Scholar]
- 39.Zarebczan B, Chen H. Signaling mechanisms in neuroendocrine tumors as targets for therapy. Endocrinol Metab Clin North Am. 2010;39: 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shawl AI, Park KH, Kim UH. Insulin receptor signaling for the proliferation of pancreatic beta-cells: involvement of Ca2+ second messengers, IP3, NAADP and cADPR. Islets. 2009;1: 216–223. [DOI] [PubMed] [Google Scholar]
- 41.Kishino T, Niwa T, Yamashita S, et al. Integrated analysis of DNA methylation and mutations in esophageal squamous cell carcinoma. Mol Carcinog. 2016;55: 2077–2088. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Wang X, Hu R, et al. Methylation of SFRP5 is related to multidrug resistance in leukemia cells. Cancer Gene Ther. 2014;21: 83–89. [DOI] [PubMed] [Google Scholar]
- 43.Carstensen-Kirberg M, Hatziagelaki E, Tsiavou A, et al. Sfrp5 associates with beta-cell function in humans. Eur J Clin Invest. 2016;46: 535–543. [DOI] [PubMed] [Google Scholar]
- 44.Wu G, Yuan M, Shen S, et al. Menin enhances c-Myc-mediated transcription to promote cancer progression. Nat Commun. 2017;8: 15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Ramirez M, Hernandez-Jimenez AJ, Guerrero-Guerrero A, et al. Pediatric pineal germinomas: Epigenetic and genomic approach. Clin Neurol Neurosurg. 2017;152: 45–51. [DOI] [PubMed] [Google Scholar]
- 46.Rokavec M, Kaller M, Horst D, Hermeking H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci Rep. 2017;7: 4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanharanta S, Marney CB, Shu W, et al. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakurai T, Isogaya K, Sakai S, et al. RNA-binding motif protein 47 inhibits Nrf2 activity to suppress tumor growth in lung adenocarcinoma. Oncogene. 2016;35: 5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J, Li Q, Xie H, et al. Identification of a second transmembrane protein tyrosine phosphatase, IA-2beta, as an autoantigen in insulin-dependent diabetes mellitus: precursor of the 37-kDa tryptic fragment. Proc Natl Acad Sci U S A. 1996;93: 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gentilini D, Scala S, Gaudenzi G, et al. Epigenome-wide association study in hepatocellular carcinoma: Identification of stochastic epigenetic mutations through an innovative statistical approach. Oncotarget. 2017;8: 41890–41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wielscher M, Vierlinger K, Kegler U, Ziesche R, Gsur A, Weinhausel A. Diagnostic Performance of Plasma DNA Methylation Profiles in Lung Cancer, Pulmonary Fibrosis and COPD. EBioMedicine. 2015;2: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleischer T, Tekpli X, Mathelier A, et al. DNA methylation at enhancers identifies distinct breast cancer lineages. Nat Commun. 2017;8: 1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bell RE, Golan T, Sheinboim D, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26: 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.